microRNA319-regulated TCP transcription factors influence leaf development in distinct ways in central and marginal parts of the organ.
Abstract
The characteristic leaf shapes we see in all plants are in good part the outcome of the combined action of several transcription factor networks that translate into cell division activity during the early development of the organ. We show here that wild-type leaves have distinct transcriptomic profiles in center and marginal regions. Certain transcripts are enriched in margins, including those of CINCINNATA-like TCPs (TEOSINTE BRANCHED, CYCLOIDEA and PCF1/2) and members of the NGATHA and STYLISH gene families. We study in detail the contribution of microRNA319 (miR319)-regulated TCP transcription factors to the development of the center and marginal regions of Arabidopsis (Arabidopsis thaliana) leaves. We compare in molecular analyses the wild type, the tcp2 tcp4 mutant that has enlarged flat leaves, and the tcp2 tcp3 tcp4 tcp10 mutant with strongly crinkled leaves. The different leaf domains of the tcp mutants show changed expression patterns for many photosynthesis-related genes, indicating delayed differentiation, especially in the marginal parts of the organ. At the same time, we found an up-regulation of cyclin genes and other genes that are known to participate in cell division, specifically in the marginal regions of tcp2 tcp3 tcp4 tcp10. Using GUS reporter constructs, we confirmed extended mitotic activity in the tcp2 tcp3 tcp4 tcp10 leaf, which persisted in small defined foci in the margins when the mitotic activity had already ceased in wild-type leaves. Our results describe the role of miR319-regulated TCP transcription factors in the coordination of activities in different leaf domains during organ development.
Leaves are impressive examples for the plasticity found in plant development. The size and shape of leaves not only vary between different species but also depend considerably on plant age and the environment. Leaves are generated at the flanks of the shoot apical meristem. They first appear as rod-shaped primordia, which then expand and grow to form flat laminas through the activity of a marginal meristem (Donnelly et al., 1999). Cell divisions in the growing leaf lamina are maintained mainly by a plate meristem and occur first throughout the organ and then become restricted to a region in the proximal part of the organ, until ceasing rather abruptly (Donnelly et al., 1999; Beemster et al., 2005; Kazama et al., 2010; Andriankaja et al., 2012; Powell and Lenhard, 2012; Rodriguez et al., 2014). Dispersed meristematic cells producing stomata and vascular cells continue to proliferate for a longer period of time (White, 2006; Andriankaja et al., 2012). After cell proliferation stops, cell enlargement becomes the driving force for organ growth. The coordination of cell division and cell expansion throughout the organ is a key to establish leaf size and shape. Around this time of transition from division to expansion, chloroplasts already are developing and genes involved in photosynthesis are being activated and, in turn, influence cell proliferation (Andriankaja et al., 2012).
The TCP transcription factors are a plant-specific family of transcription factors, named after the first identified members, TEOSINTE BRANCHED, CYCLOIDEA, and PCF1/PCF2 (Kosugi and Ohashi, 1997; Cubas et al., 1999). In Arabidopsis (Arabidopsis thaliana), 24 family members have been identified, which can be further classified according to similarities in the TCP domain and biological functions (Nicolas and Cubas, 2016). Five TCPs, TCP2, TCP3, TCP4, TCP10, and TCP24, and their homologs in different species are regulated by the evolutionarily conserved microRNA (miRNA) miR319 (Palatnik et al., 2003). High levels of miR319 down-regulate these TCPs and cause important changes in Arabidopsis leaf morphogenesis and the generation of crinkled leaves (Palatnik et al., 2003; Koyama et al., 2007; Efroni et al., 2008). A triple mutant in miR319-regulated TCPs also affects leaf development severely as well as mutations in the snapdragon (Antirrhinum majus) gene CINCINNATA (CIN), a TCP4 homolog (Nath et al., 2003; Schommer et al., 2008). The effects of miR319-regulated TCPs in the generation of crinkles are enhanced by three other TCPs: TCP5, TCP13, and TCP17 (usually referred to as CIN-like TCPs). These three TCPs lack the regulation by miR319 but are closely related to the miRNA-regulated family members, based on similarities in their amino acid sequences (Efroni et al., 2008; Li, 2015). Transcriptome analyses of leaves with modified TCP levels have shown that their differentiation program is modified (Efroni et al., 2008). However, TCPs also have been shown to directly activate MIR396b and CYCLIN DEPENDENT KINASE INHIBITOR1 (ICK1; Schommer et al., 2014), which are known to regulate cell proliferation (Wang et al., 2000; Rodriguez et al., 2010). Furthermore, miR319-regulated TCPs are involved in hormone biosynthesis and response (for review, see Nicolas and Cubas, 2016).
Leaf margins can be smooth, lobed, or serrated. The typical shape that is acquired varies between species and depends on plant age. How the margin is defined and which genes are involved in constructing the typical shape of a leaf is not well understood. Various studies describe genes and mutants or transgenic lines that affect margin development, such as ICK1 overexpressors, yucca mutants or plants with changes in the CUP SHAPED COTYLEDON (CUC)/miR164 balance, and pin formed1, which affect the degree of serration of the Arabidopsis leaf (Nikovics et al., 2006; Kawamura et al., 2010; Engelhorn et al., 2012; Steiner et al., 2012). NGATHA (NGA) and STYLISH (STY) genes, initially described by their function in gynoecium development, also have been found to be important in the generation of leaf margins. Both gene families promote auxin biosynthesis (Sohlberg et al., 2006; Martínez-Fernández et al., 2014), and multiple knockouts have leaves with more serrations than the wild type (Kuusk et al., 2002; Beemster et al., 2005; Alvarez et al., 2009, 2016; Trigueros et al., 2009; Ballester et al., 2015). Recent studies show that simultaneous down-regulation of miR319-regulated TCPs and NGAs results in dramatic leaf phenotypes with indefinite growth at the margins (Alvarez et al., 2016).
Transcriptome studies have been carried out at different stages of leaf development, and their analyses have led to the identification of genes involved in cell proliferation and differentiation in different leaves (Beemster et al., 2005; Schmid et al., 2005; Efroni et al., 2008) as well as genes involved in the transition from primary to secondary morphogenesis (Andriankaja et al., 2012). Detailed studies during the progression of leaf development using whole organs have identified markers of leaf differentiation, implicating functions for the TCPs in the regulation of developmental timing, especially in the control of early differentiation events during leaf development (Efroni et al., 2008).
Interestingly, previous transcriptomic studies have been performed in whole leaves, which might underscore the events occurring in specific leaf domains. Here, we isolate a quadruple knockout, tcp2 tcp3 tcp4 tcp10, which has crinkled margins similar to or stronger than the jaw-D mutant that overexpresses miR319. In contrast, tcp double mutants have larger leaves with no obvious appearance of crinkles. We performed transcriptomic studies on the margins and central parts of leaves from the wild type and tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 mutants. We found that the reduction of TCP activity had general effects on the transcriptome of the organ, and we detected domain-specific differences. We also found a strong activation of cell proliferation markers in leaf margins that had not been detected in previous transcriptomic studies using whole leaves. We found that most of the genes enriched in leaf margins are affected by the down-regulation of miR319-regulated TCPs. Still, a group of margin-specific genes with an overrepresentation of transcription factors acts independently of TCP control. Our data provide new insights into leaf development and the role of miRNA-regulated TCPs in the coordination of organ growth.
RESULTS
Differential Effects of tcp Double and Quadruple Mutants on Leaf Development
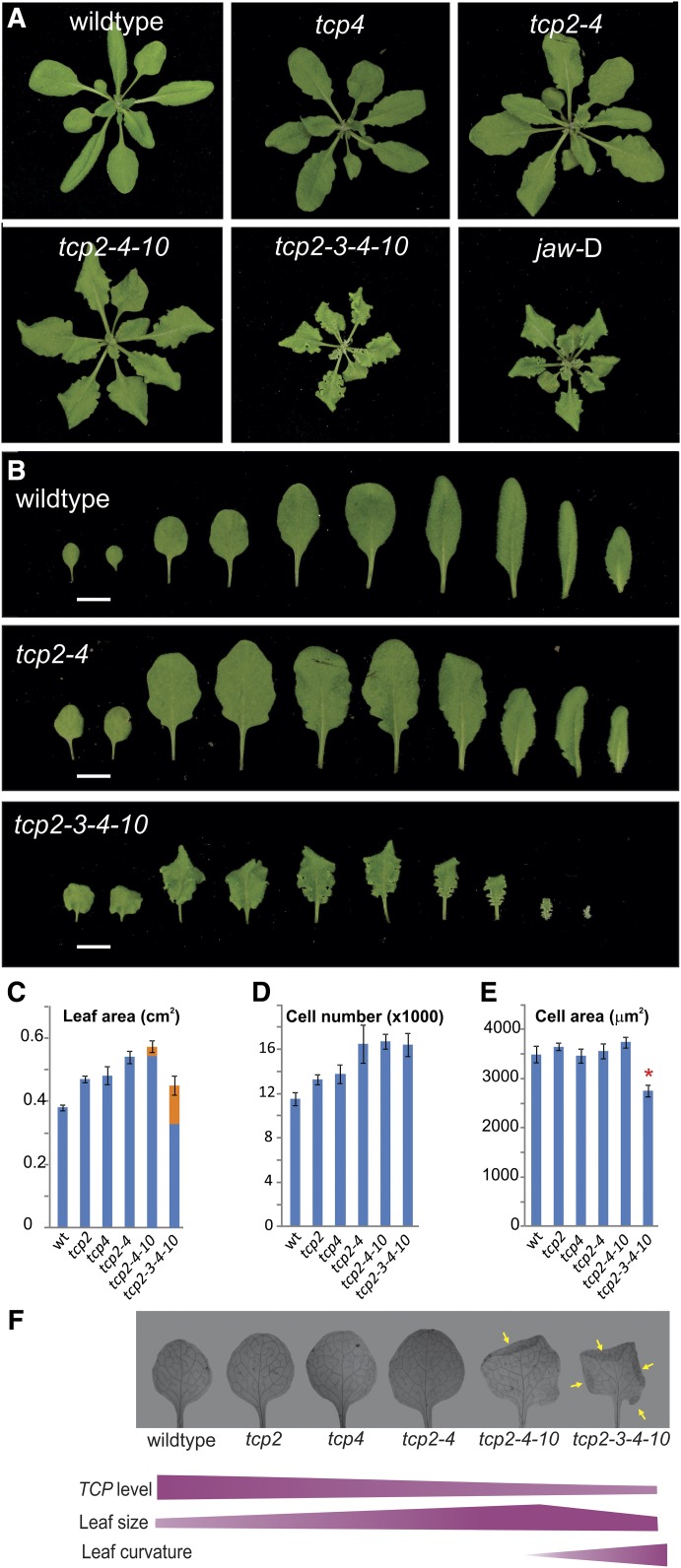
To study the functions of miR319-regulated TCPs, we characterized a series of loss-of-function tcp insertional knockout mutants, including single, double, triple, and quadruple mutants (Fig. 1). The leaves and rosettes of tcp single mutants were slightly larger than those of the wild type (Fig. 1). In contrast to the small effects on the size of the single tcp knockouts, we observed a significant increase of leaf size in tcp2 tcp4 double knockouts (Fig. 1). The tcp quadruple mutant had a different phenotype, with strongly crinkled leaves (Fig. 1). While changes in the leaf curvature are seen in all quadruple knockout leaves, we observed that crinkles and small folds at the margins become more predominant starting with leaf 5 and onward. We determined the area of the crinkles by flattening the leaves and considering the areas that were folded one or more times on top of each other and entered them as multiples into the calculations of the total leaf area (Fig. 1). Analysis of the first leaf revealed that the complete area was smaller than that in the wild type, if not considering folded areas (Fig. 1). Yet, the tcp quadruple mutant had smaller leaves than the double mutant (Fig. 1).
Figure 1.
Effects of decreasing TCP levels on leaf size and shape. A, Four-week-old rosettes of Arabidopsis plants with decreased levels of TCP activity. B, Disassembled rosettes of the indicated genotypes. C, Leaf area of and two of the indicated genotypes. Marked in orange are the contributions of folded regions to the total leaf area (see “Materials and Methods”). D, Number of cells in the same leaves as in C. E, Cell area of the same leaves as in C. The asterisk indicates statistical significance of the difference according to P < 0.01 by Student’s t test. wt, Wild type. F, Cleared and flattened leaves of the different genotypes that have reduced TCP activity. Yellow arrows indicate folded regions. Violet bars illustrate how leaf size and curvature develop with respect to the gradual reduction of TCP activity. The mutants tcp2 tcp4, tcp2 tcp4 tcp10, and tcp2 tcp3 tcp4 tcp10 are labeled tcp2-4, tcp2-4-10, and tcp2-3-4-10, respectively.
The triple mutant tcp2 tcp4 tcp10 had an intermediate phenotype between the double and quadruple mutants. The leaves already had some crinkles (Fig. 1), although to a much lower extent than in the quadruple tcp mutant. Leaves of tcp2 tcp4 tcp10 were larger than those of the wild type but similar to those of tcp2 tcp4, even after considering folded regions (Fig. 1).
We analyzed the first leaf at the cellular level. The number of palisade parenchyma mesophyll cells increased in single and double mutants with respect to wild-type leaves (Fig. 1). As the cell size remained fairly constant in these leaves (Fig. 1), the increase of leaf size in tcp single and double mutants correlated directly with an increase in cell number. A different scenario was observed in plants with a strong decrease in TCP activity. First, the cell number did not increase steadily with the combination of tcp mutants; rather, the total number of cells in double, triple, and quadruple tcp knockouts was similar (Fig. 1). We also noticed that the cell size in the tcp2 tcp3 tcp4 tcp10 quadruple knockout was reduced (Fig. 1), which explains its reduced leaf area (Fig. 1). Therefore, a mild decrease in TCP activity such as seen in single and double mutants caused an increase in cell number, which, in turn, produced larger leaves. Further loss of TCP activity triggered additional changes in leaf morphogenesis, namely a modification of organ curvature and a reduction of cell expansion.
Transcriptome Analysis of Leaf Margins and Inner Lamina
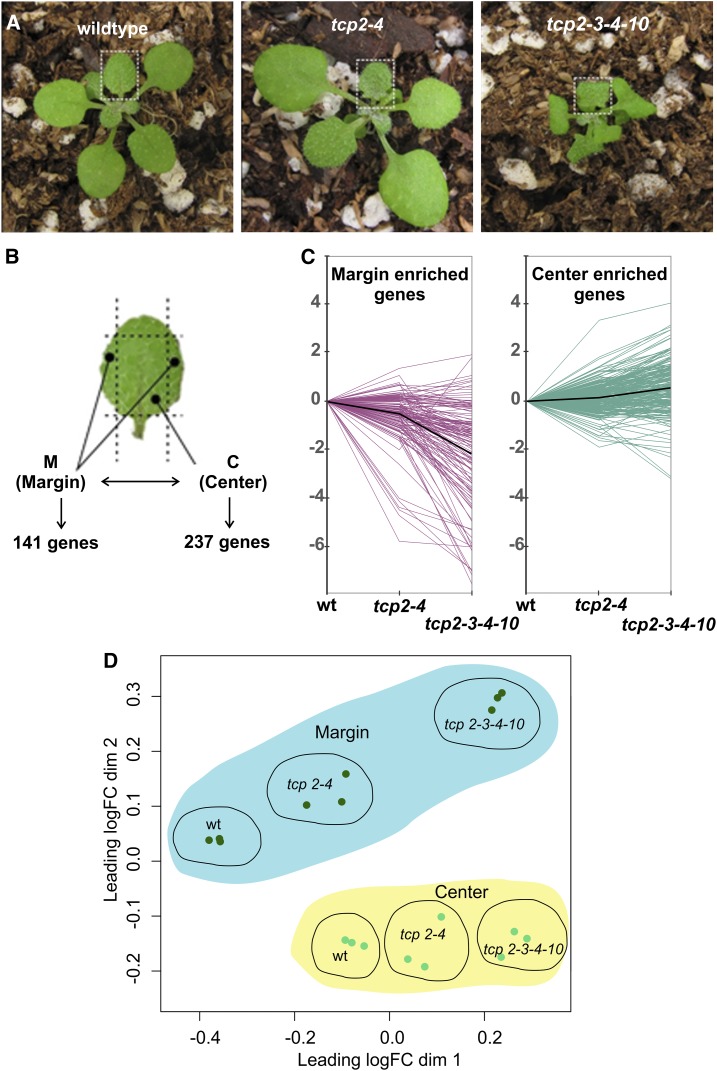
Previous transcriptomic analyses have focused on whole developing leaves or leaves dissected into proximal and distal parts (Efroni et al., 2008; Li et al., 2010; Pettkó-Szandtner et al., 2015; Nicolas and Cubas, 2016). Here, we decided to analyze the plant transcriptome in the margins and the center of the leaf. To do this, we collected developing fifth leaves of the wild type and tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 mutants (Fig. 2; for details, see “Materials and Methods”). The leaves were dissected with the help of a stereomicroscope, as shown in Figure 2, and the marginal or central areas were collected. Biological triplicates representing the margins and central regions of more than 50 individual plants were subjected to transcriptome analysis by RNA sequencing (RNAseq) (Anders et al., 2013).
Figure 2.
Transcriptome analysis of leaf domains of the wild type and tcp mutants. A, Rosettes with emerging leaf 5 as used for transcriptome analysis by RNAseq experiments. B, Schematic display of regions that were used as marginal and center samples to obtain RNA for the RNAseq experiments. In the wild type, 237 center- and 141 margin-enriched genes were identified. C, Graphical display showing fold change of the behavior of genes that were determined in B to be margin or center enriched in wild-type samples (wt) or in tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 mutants. The black lines represent the average expression levels of all margin- and center-enriched genes. D, Multidimensional scaling plot for count data (all genes). Distances correspond to leading log fold change (logFC) between each pair of RNA samples.
In a first step, we focused on the analysis of the wild-type samples. We identified 141 transcripts enriched in the margins and 237 enriched in the center samples (at least 2-fold change in expression levels and false discovery rate < 0.01; Fig. 2; Supplemental Table S1), indicating that the expression profile of the leaf is not homogenous but that margins and center parts have their specific signature.
Among the 141 genes enriched in wild-type margins, we detected 13 transcription factors of the WRKY, BHLH-like, NGA, STY, and TCP gene families (Supplemental Table S1). Interestingly, several of those have been shown to affect leaf shape and size when misregulated (Kuusk et al., 2002, 2006; Alvarez et al., 2006, 2009; Efroni et al., 2008; Trigueros et al., 2009; Ballester et al., 2015; Lee et al., 2015).
We inspected the 141 genes that are enriched in wild-type margins also in the margins of the tcp mutants. We found that the margin-specific genes decreased their expression in the double and quadruple tcp mutant margins (Fig. 2C). In contrast, we observed that the expression of 237 center-enriched genes from wild-type leaves was less affected in the centers of the tcp mutant leaves (Fig. 2C; Supplemental Table S1). These results are consistent with the morphological changes observed in the margins of the plants with low TCP activity.
An analysis of the six different samples in the RNAseq experiment displayed in multidimensional scaling, and smear plots further showed that the margins of the quadruple tcp mutant were strikingly different in their expression profiles from the wild type and the double tcp knockout (Fig. 2; Supplemental Fig. S1). However, we also detected that the tcp2 tcp4 knockout, which does not appear to have morphologically changed margins compared with the wild type, on the molecular level displayed more changes in the marginal than in the central part of the leaf (Fig. 2; Supplemental Fig. S1). Therefore, our analysis detected approximately 140 genes enriched in the borders of developing leaves and showed that miR319-regulated TCPs are necessary to ensure a correct regulation of many of those.
Differential Control of Gene Expression by TCPs in Leaf Domains: Down-Regulated Genes in tcp Mutants
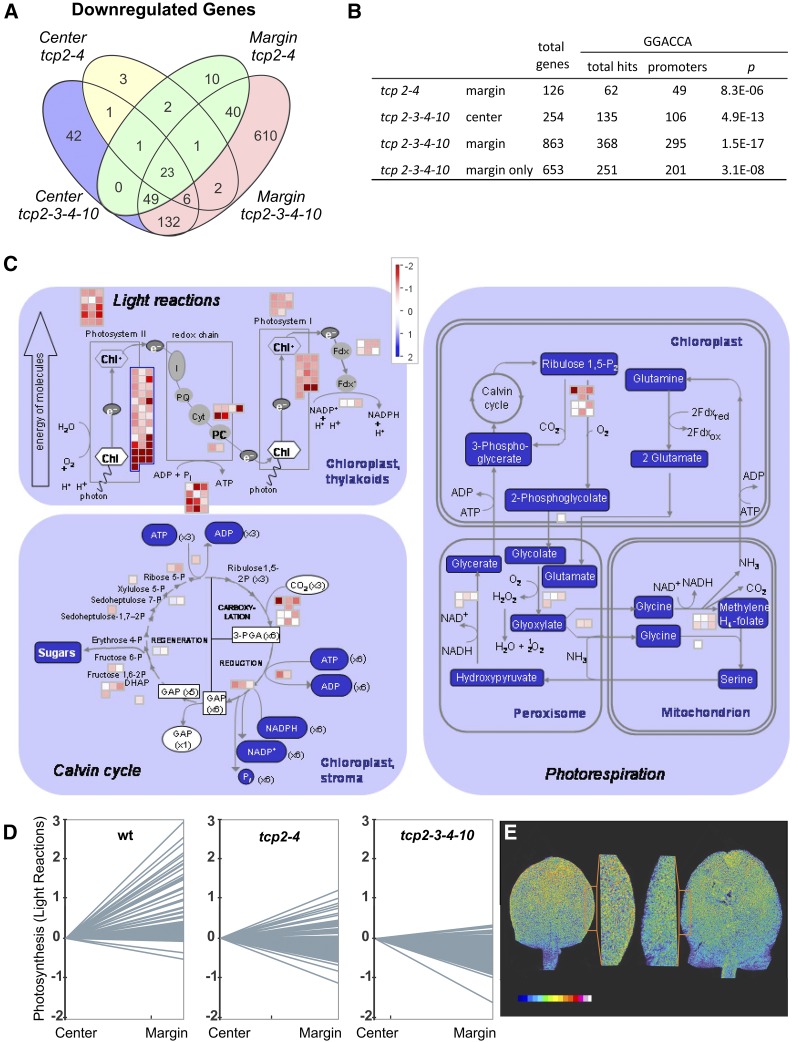
Next, we inspected the genes that were down-regulated in the margins and the center of the tcp mutant leaves compared with the wild type (Supplemental Table S2). We found that 863 genes were down-regulated in the margins of the quadruple tcp mutants and 254 in the center of the organ (Fig. 3; Supplemental Table S2). That tcp2 tcp3 tcp4 tcp10 had more changes in the margin than in the center is consistent with mutant leaves having crinkles and changes in leaf curvature, especially along the border regions. A total of 210 genes were down-regulated in both margin and center of the tcp2 tcp3 tcp4 tcp10 mutants (Fig. 3), and most of the genes affected in tcp2 tcp4 were a subgroup of those affected in the quadruple knockout (Supplemental Table S2), demonstrating that the double mutant reflects to a certain extent a partial state of TCP activity between the wild type and the tcp2 tcp3 tcp4 tcp10 quadruple mutant.
Figure 3.
Coordination of gene expression related to photosynthesis by miR319-regulated TCPs. A, Venn diagram showing the intersections of down-regulated genes in margins and centers of tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 compared with the wild type. B, Promoter analysis of down-regulated genes in tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 center and margin regions for overrepresentation of the TCP binding motif GGACCA. C, Graphical display of MapMan analysis of gene expression for photosynthesis-related genes in tcp2 tcp3 tcp4 tcp10 margins compared with the wild type. D, Graphical display of relative expression changes of photosynthesis-related genes between center and marginal regions in the wild type (wt), tcp2 tcp4, and tcp2 tcp3 tcp4 tcp10 as determined by RNAseq experiments. The mutants tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 are labeled tcp2-4 and tcp2-3-4-10, respectively. E, Chlorophyll contents in developing young leaves as estimated by their fluorescence using a light scanning confocal microscope. Left, Wild type with closeup; right, tcp2 tcp3 tcp4 tcp10 with closeup. The colored bar indicates the fluorescence intensity profile used from blue (low fluorescence) to white (high fluorescence).
MiR319-regulated TCPs bind a core sequence, GGACCA (Schommer et al., 2008). We observed that the presence of this sequence was enriched in the promoters of the down-regulated genes (defined as 1 kb upstream of the transcription start) in tcp2 tcp4 margins and in the promoters of tcp2 tcp3 tcp4 tcp10 center and margin samples (Fig. 3; Supplemental Table S3). Among the down-regulated genes were validated TCP targets such as LIPOXYGENASE2, ICK1, and the ARABIDOPSIS RESPONSE REGULATOR16 (Supplemental Table S2; Schommer et al., 2008, 2014; Efroni et al., 2013); noteworthy also was the NGA2 gene, whose activity had been shown to be reduced in the miR319a-overexpressing jaw-D background (Ballester et al., 2015).
Analysis of Gene Ontology (GO) term enrichment in the down-regulated genes in the center and the margins of tcp2 tcp3 tcp4 tcp10 leaves revealed that response to stimuli was the most frequent category (Supplemental Table S4). Most interesting, we saw that among down-regulated genes in the quadruple tcp mutant margins were many genes related to photosynthesis (Fig. 3; Supplemental Table S2). It was found recently that many photosynthesis-related genes start being expressed close to the time point when cell proliferation stops during leaf development (Andriankaja et al., 2012).
We then looked at the expression of photosynthesis-related genes (light reactions) in the center and margin of wild-type plants and found that many of them were more strongly expressed in the margins than in the center (Fig. 3). However, when we looked at the relative activity levels of these genes in the tcp knockouts, we found an opposite pattern of expression, with a tendency of photosynthetic genes to be expressed at higher levels in the center compared with the margins (Fig. 3). As expected, the effect was stronger in the quadruple than in the double tcp mutant, but it was already clear in tcp2 tcp4 (Fig. 3). Overall, these results strongly suggest that TCPs are necessary to coordinate the maturation program of the photosynthetic machinery in the margin and center of the leaf. We estimated the chlorophyll levels in chloroplasts of developing leaves of the wild type and the tcp quadruple knockout by analyzing the fluorescence intensity with a laser scanning confocal microscope. We visualized the distribution of chloroplasts and displayed the state of maturation of the leaf in terms of photosynthesis in its different areas. In agreement with other studies (Andriankaja et al., 2012) in the wild type, more chlorophyll had accumulated in the distal parts of the leaf and along the margins when compared with the center of the leaf. In the quadruple knockout mutant, we detected a lower level of chlorophyll fluorescence in the distal part of the leaf, indicating a delayed maturation of chloroplasts (Fig. 3). Furthermore, and in contrast to the wild type, less chlorophyll fluorescence was emitted from the marginal regions compared with the central part of the leaf (Fig. 3). This confirmed our results toward photosynthesis-related genes that we had obtained from the RNAseq experiments and showed the delayed maturation in general and, especially, in the marginal regions of the quadruple knockout leaves. However, we did not find an enrichment of the GGACCA motif in the promoters of the photosynthetic genes, suggesting that the effect of the TCPs is indirect or at least not caused by their direct binding.
TCPs Repress Cell Proliferation and Early Leaf Developmental Programs in the Margins
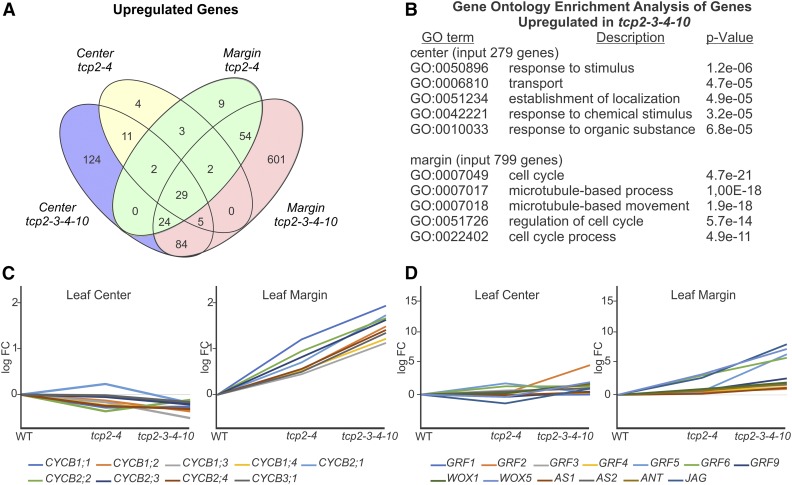
Next, we analyzed up-regulated genes in the tcp knockouts. We found that, in the center region of the tcp2 tcp4 mutant, only 56 genes increased their expression levels (Fig. 4), while at the same time, 279 genes were up-regulated in the tcp2 tcp3 tcp4 tcp10 blade (Fig. 4). Forty-seven of the 56 genes that were enriched in tcp2 tcp4 also were up-regulated in tcp2 tcp3 tcp4 tcp10 (Fig. 4; Supplemental Table S5), confirming that the genes that change their expression levels in the double mutant are largely a subgroup of those affected in the quadruple mutant.
Figure 4.
Up-regulation of the leaf developmental program and mitotic genes in tcp mutant margins. A, Venn diagram showing the intersections of up-regulated genes in margins and centers of tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 compared with the wild type. B, Output of GO enrichment analysis for up-regulated genes in tcp2 tcp3 tcp4 tcp10 margin and center regions. C, Display of expression changes of B-type cyclin genes in tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 center and marginal leaf samples in RNAseq analysis. The y axis shows log fold change (logFC) compared with the wild type (WT). D, Display of expression changes of selected genes that are active in early stages of leaf development in tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 center and marginal leaf samples. The y axis shows log fold change of mutant samples compared with the wild type. The mutants tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 are labeled tcp2-4 and tcp2-3-4-10, respectively.
Drastic were the changes in the margins of the tcp2 tcp3 tcp4 tcp10 quadruple mutant compared with the wild type, as 799 genes were up-regulated (Fig. 4; Supplemental Table S5). Again, the difference between tcp2 tcp4 and the wild type was much less pronounced, with only 123 genes being enriched (Fig. 4; Supplemental Table S5). An analysis for GO term enrichment within the genes up-regulated in the center region of the tcp2 tcp3 tcp4 tcp10 mutants revealed GO term enrichment related to stimulus (Fig. 4; Supplemental Table S4). We obtained a different view once we analyzed genes up-regulated in the margins. The most up-regulated categories were related to cell cycle- and microtubule-based processes, as required during mitosis (Fig. 4; Supplemental Table S6). All Arabidopsis CYCLINB genes were up-regulated in the margins of tcp2 tcp3 tcp4 tcp10 leaves, while they were unchanged or diminished in the center (Fig. 4; Supplemental Table S6). This is interesting, as B-type cyclins control the G2-to-M transition of the cell cycle (Polyn et al., 2015). We extended the analysis to a list of mitosis-specific genes (Menges et al., 2005) and other genes that have already been described to be involved in early leaf development and obtained similar results, finding them to be expressed at higher levels in the tcp2 tcp3 tcp4 tcp10 compared with the wild-type margin (Fig. 4; Supplemental Table S6).
JAGGED was the most up-regulated gene in tcp quadruple knockout margins, and GROWTH REGULATING FACTOR5 and WUSCHEL-RELATED HOMEOBOX5 were among the top 10 (Fig. 4; Supplemental Table S5). An analysis of genes known to participate in leaf development revealed the up-regulation of ASYMMETRIC LEAVES1, ASYMMETRIC LEAVES2, AINTETGUMENTA, and WUSCHEL-RELATED HOMEOBOX1 (Fig. 4; Supplemental Tables S5 and S6). Most interestingly, these genes changed in the margins of the quadruple knockouts but were largely unaffected in the central leaf regions (Fig. 4; Supplemental Table S6). Overall, these results suggest that processes related to early leaf development and cell division remain active specifically in the margins of the tcp2 tcp3 tcp4 tcp10 leaves when they are already shut down in the wild-type leaf or the central zones of the tcp mutant leaves.
Interaction between Cell Proliferation Programs and miR319-Regulated TCPs in Different Leaves
Previous studies in snapdragon cin mutants and young leaves of miR319 overexpressors have shown a delay in the repression of B-type cyclin activity mostly in the organ margins, which has been associated with a corresponding change in leaf curvature, although microarray studies of whole leaves did not reveal an increase in mitotic activity (Nath et al., 2003; Palatnik et al., 2003; Efroni et al., 2008). That crinkles at the leaf margins of jaw-D or quadruple tcp mutants increase in leaves that are generated later during the plant’s life cycle and that cell proliferation markers represent the most up-regulated group of genes in our transcriptomic experiments in leaf 5 margins prompted us to look in more detail into the interaction between cell proliferation and the miR319-TCP network during the development of different leaves.
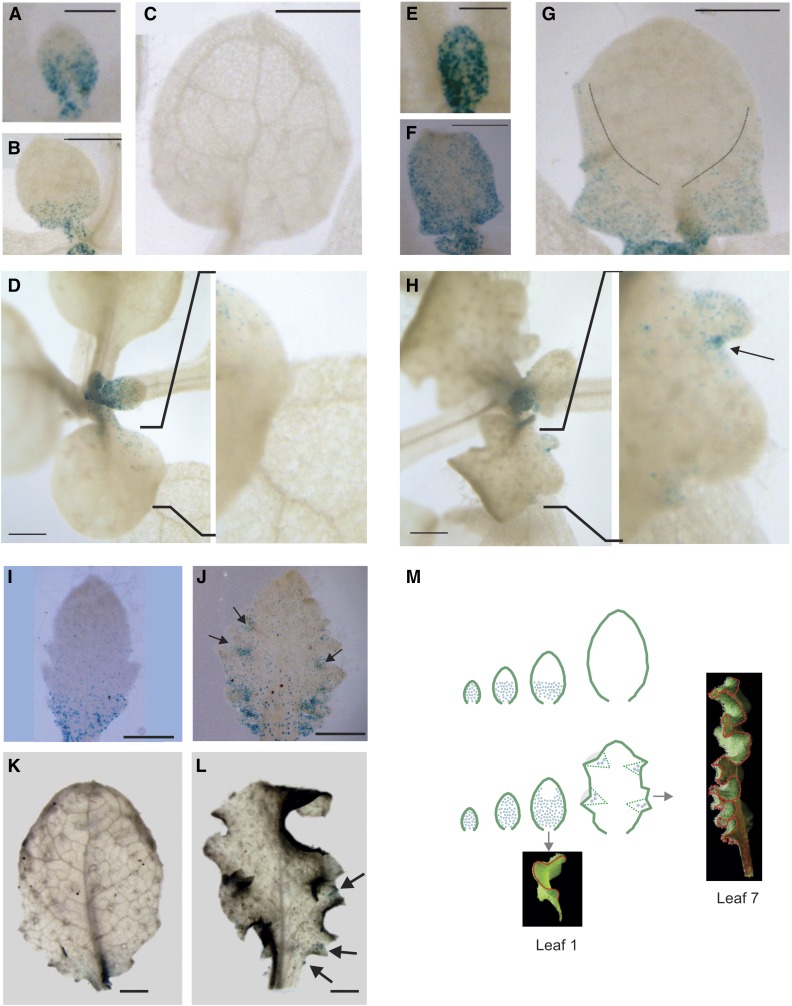
To study this, we overexpressed miR319a, which causes the same phenotypic effects as the tcp2 tcp3 tcp4 tcp10 mutant (Fig. 1), in the context of a CYCLINB1;1:GUS reporter line, which labels cells undergoing mitosis (Donnelly et al., 1999). In wild-type plants, the reporter is expressed initially in the whole leaf primordia but then becomes restricted to the base of young developing leaves, until cell proliferation stops and leaves continue to grow by cell expansion (Fig. 5; Donnelly et al., 1999). In the first true leaves, we observed that, upon overexpression of miR319, cell proliferation was extended to the margins (Fig. 5) and lasted longer than in the wild type, in agreement with previous observations (Nath et al., 2003; Efroni et al., 2008).
Figure 5.
Activation of discrete foci expressing CYCLINB1;1:GUS upon TCP down-regulation. A to L, GUS assays. A to D, I, and K, Young developing leaves of a CYCLINB1;1:GUS reporter line. E to H, J, and L, Young leaves of the same CYCLINB1;1:GUS reporter overexpressing miR319a, resulting in crinkly leaves. A, Leaf 1 at 7 d after sowing (DAS). B, Leaf 1 at 9 DAS. C, Leaf 1 at 11 DAS. D, Leaf 4. E, Leaf 1 of miR319 overexpressor at 8 DAS. F, Leaf 1 of miR319 overexpressor at 10 DAS. G, Leaf 1 of miR319 overexpressor at 12 DAS. The dashed lines delimit the domain containing cycling cells. H, Leaf 4 of miR319 overexpressor. I, Leaf 7. J, Leaf 7 of miR319 overexpressor. K and L, More mature leaves of the wild type (K) and miR319a overexpressor (L). M, Models showing the effect of miR319-regulated TCPs on cell division in the developing leaf of the wild type (top) and tcp quadruple knockouts or miR319-overexpressing plants (bottom). Blue dots indicate proliferating cells. Bars = 0.2 mm (A, E, I, and J), 0.5 mm (B, C, F, and G), and 1 mm (D, H, K, and L).
However, a closer inspection of the B-type cyclin CYCLINB1;1 behavior in the fourth leaf of Arabidopsis plants overexpressing miR319 revealed additional changes in the pattern of proliferating cells in these leaves. We found that discrete regions at the margins of the leaf harbored cells undergoing mitosis, in contrast to wild-type leaves (Fig. 5, see insets). This effect was even more pronounced in young seventh leaves (Fig. 5). Therefore, Arabidopsis leaves with low TCP activity do no simply have a delay in the mitotic arrest front, but cell proliferation continues to be activated in specific subdomains at the leaf margins. The foci of dividing cells persisted for extended periods, even after proliferation in wild-type leaves had ceased completely (Fig. 5). Leaves produced by older plants normally have more serrations than the younger ones (Poethig, 2003). In the case of miR319 overexpressors, we also noticed that more discrete foci harboring proliferating cells were detected in later rather than in earlier leaves, which is consistent with a larger number of crinkles in these leaves. We propose that probably an extension of the cell proliferation, such as seen in the first leaves of Arabidopsis or snapdragon, would cause a change in leaf curvature, but the formation of discrete foci would result in crinkles along the leaf margins, as seen in leaves produced later during the development of miR319 overexpressors or tcp mutants (Fig. 5).
Identification of Margin-Specific Genes Independent of TCPs
In our analysis, we had identified 237 genes enriched in the center and 141 genes enriched in the margins of wild-type leaves (Fig. 2; Supplemental Table S1). We performed the same analysis with the margin and center samples of the tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 leaves and searched for center- and margin-enriched genes in the mutants. We found that, of the 237 genes enriched in the center of the wild-type leaves, 154 (approximately 65%) were still enriched in the center of the quadruple tcp mutant leaves (Supplemental Table S1; Supplemental Fig. S2). In contrast, when we looked at the expression of the 141 margin-specific genes that we identified in wild-type leaves, we observed that only 31 continued to be enriched in the margins of quadruple tcp mutants (Supplemental Table S1; Supplemental Fig. S2).
We focused then on the genes that were enriched in the margins of both wild-type and tcp quadruple mutant leaves, suggesting that the expression of these genes will be at least partially independent of the miR319-regulated TCPs. Interestingly, among the 31 genes (Table I), we found eight transcription factors belonging to the groups of NGA, STY, and CIN-like TCPs. More specifically, we identified NGA1, NGA3, STY1, STY2, and SHI-RELATED SEQUENCE4, as well as the three CIN-like TCPs TCP5, TCP13, and TCP17, to be enriched in both wild-type and mutant margins (Table I). Also interesting was the identification of the auxin biosynthesis gene YUCCA2 as margin enriched, as it is thought to be regulated by NGAs (Trigueros et al., 2009). The STY gene family consists of eight members, three of which we determined to be margin enriched. Noteworthy, some of these genes already have been described to play a role in margin development either of the gynoecium or the leaf. So have multiple sty mutants and plants with reduced NGA activity more strongly serrated leaves than the wild type (Kuusk et al., 2002, 2006; Alvarez et al., 2009).
Table I. Expression levels of margin-specific genes independent of TCP activity.
Expression values of double (2KO) and quadruple (4KO) knockout centers and margins are expressed as normalized counts per million and fold change of margin samples compared with center samples. n/d, Not detected; n/s, nonsignificant; WT, wild type.
| Code | Gene | Normalized Counts per Million |
Normalized Counts per Million |
Fold Change |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Center |
Margin |
Margin/Center |
||||||||
| WT |
2KO |
4KO |
WT |
2KO |
4KO |
WT |
2KO |
4KO |
||
| Transcription factors | ||||||||||
| AT1G01030 | NGA3 | 2.80 ± 0.38 | 1.31 ± 0.86 | 0.51 ± 0.09 | 6.79 ± 0.82 | 5.01 ± 0.64 | 3.43 ± 0.91 | 2.41 | 4.16 | 6.49 |
| AT2G18120 | SRS4 | 0.81 ± 0.07 | 0.92 ± 0.39 | 1.26 ± 0.32 | 2.25 ± 0.52 | 2.26 ± 0.67 | 4.04 ± 0.73 | 2.78 | n/s | 3.15 |
| AT2G46870 | NGA1 | 8.77 ± 1.10 | 4.43 ± 1.33 | 2.47 ± 0.90 | 25.54 ± 2.62 | 18.93 ± 2.09 | 13.2 ± 0.46 | 2.92 | 4.45 | 5.33 |
| AT3G02150 | TCP13 | 5.79 ± 1.52 | 5.97 ± 1.53 | 5.04 ± 1.44 | 12.19 ± 2.24 | 13.34 ± 1.10 | 10.75 ± 2.04 | 2.11 | 2.19 | 2.09 |
| AT3G51060 | STY1 | 0.12 ± 0.00 | 0.09 ± 0.09 | n/d | 1.18 ± 0.39 | 1.24 ± 0.52 | 0.47 ± 0.10 | 8.45 | 11.60 | 39.15 |
| AT4G36260 | STY2 | 0.16 ± 0.31 | 0.04 ± 0.07 | 0.31 ± 0.31 | 2.23 ± 0.88 | 1.85 ± 0.20 | 3.44 ± 1.13 | 12.25 | 30.58 | 10.81 |
| AT5G08070 | TCP17 | 1.46 ± 0.24 | 1.01 ± 0.52 | 1.52 ± 0.69 | 4.34 ± 0.68 | 4.59 ± 0.69 | 4.45 ± 0.60 | 2.95 | 4.30 | 2.94 |
| AT5G60970 | TCP5 | 4.99 ± 0.50 | 4.93 ± 0.53 | 5.53 ± 1.56 | 11.73 ± 2.31 | 13.31 ± 1.93 | 14.05 ± 1.35 | 2.32 | 2.62 | 2.58 |
| Hormones and metabolites | ||||||||||
| AT1G56710 | Pectin lyase superfamily | 0.08 ± 0.16 | 0.04 ± 0.07 | 0.13 ± 0.19 | 2.69 ± 0.75 | 3.01 ± 1.99 | 0.92 ± 0.33 | 27.44 | 50.85 | 6.17 |
| AT1G70560 | CKRC1 | 3.09 ± 0.20 | 2.82 ± 0.45 | 1.51 ± 0.38 | 13.77 ± 0.19 | 9.17 ± 1.60 | 5.77 ± 1.03 | 4.45 | 3.35 | 3.78 |
| AT2G04160 | AIR3 | 10.50 ± 1.78 | 5.93 ± 0.79 | 0.67 ± 0.31 | 53.78 ± 8.03 | 26.16 ± 2.61 | 3.35 ± 0.53 | 5.09 | 4.44 | 5.03 |
| AT2G23170 | GH3.3 | 0.20 ± 0.16 | 0.20 ± 0.29 | 1.36 ± 0.32 | 1.23 ± 0.79 | 0.27 ± 0.26 | 4.37 ± 0.86 | 5.72 | n/s | 3.12 |
| AT2G43840 | UGT74F1 | 1.42 ± 0.41 | 0.97 ± 0.35 | 0.54 ± 0.03 | 3.56 ± 0.22 | 1.54 ± 0.43 | 1.59 ± 0.44 | 2.50 | n/s | 2.90 |
| AT3G13380 | BRL3 | 4.74 ± 1.56 | 4.48 ± 1.40 | 2.58 ± 0.82 | 10.09 ± 2.46 | 7.36 ± 0.97 | 5.26 ± 0.15 | 2.13 | n/s | 2.02 |
| AT4G13260 | YUC2 | 2.35 ± 0.44 | 2.21 ± 1.23 | 1.28 ± 0.44 | 8.06 ± 1.22 | 6.71 ± 0.96 | 4.81 ± 0.31 | 3.38 | 3.20 | 3.79 |
| AT5G26220 | GCT2;1 | 1.02 ± 0.66 | 3.49 ± 2.98 | 1.46 ± 0.65 | 4.55 ± 1.91 | 7.08 ± 4.55 | 6.83 ± 3.32 | 4.43 | n/s | 4.67 |
| AT5G28030 | DES1 | 2.76 ± 0.51 | 1.84 ± 0.21 | 0.58 ± 0.27 | 15.33 ± 4.13 | 11.13 ± 0.41 | 5.63 ± 1.33 | 5.49 | 5.98 | 9.43 |
| AT5G59130 | Subtilase family | 15.83 ± 0.21 | 18.76 ± 4.4 | 12.99 ± 3.19 | 34.49 ± 3.43 | 39.81 ± 3.05 | 30.67 ± 3.51 | 2.18 | 2.14 | 2.37 |
| Transport related | ||||||||||
| AT1G74810 | BOR5 | 5.28 ± 1.11 | 7.87 ± 0.34 | 9.96 ± 2.35 | 11.43 ± 0.83 | 13.87 ± 1.1 | 20.41 ± 0.66 | 2.15 | 1.75 | 2.10 |
| AT2G36590 | PROT3 | 3.97 ± 0.30 | 3.24 ± 0.49 | 2.35 ± 0.70 | 10.30 ± 0.56 | 6.95 ± 2.13 | 5.28 ± 1.23 | 2.59 | 2.11 | 2.21 |
| AT4G24120 | YSL1 | 7.14 ± 1.58 | 6.17 ± 0.93 | 4.98 ± 1.94 | 14.61 ± 6.32 | 18.42 ± 1.55 | 10.61 ± 1.42 | 2.03 | 2.96 | 2.08 |
| AT5G17700 | MATE efflux family | 16.59 ± 3.74 | 16.27 ± 2.94 | 6.13 ± 1.85 | 43.7 ± 1.04 | 50.25 ± 7.07 | 19.62 ± 3.36 | 2.64 | 3.14 | 3.25 |
| AT5G50790 | SWEET10 | 0.61 ± 0.23 | 0.43 ± 0.27 | 0.14 ± 0.07 | 2.00 ± 0.55 | 3.42 ± 0.83 | 1.85 ± 0.57 | 3.29 | 7.08 | 12.71 |
| AT5G55930 | OPT1 | 9.00 ± 3.88 | 7.36 ± 1.61 | 4.18 ± 0.75 | 23.03 ± 5.30 | 19.51 ± 1.28 | 9.59 ± 1.31 | 2.57 | 2.61 | 2.28 |
| Others | ||||||||||
| AT1G22900 | Disease-resistant responsive | 0.12 ± 0.24 | 0.15 ± 0.04 | 0.14 ± 0.28 | 6.15 ± 0.12 | 5.00 ± 0.48 | 1.16 ± 0.79 | 44.65 | 32.28 | 7.97 |
| AT2G47880 | Glutaredoxin family | 2.68 ± 1.56 | 3.13 ± 1.51 | 0.77 ± 0.11 | 30.60 ± 5.42 | 26.33 ± 6.45 | 2.16 ± 0.12 | 11.39 | 8.63 | 2.84 |
| AT3G05730 | Unknown | 15.44 ± 3.12 | 4.55 ± 2.44 | 4.39 ± 3.06 | 652.26 ± 60.06 | 341.85 ± 26.96 | 11.08 ± 1.97 | 42.17 | 76.86 | 2.52 |
| AT3G09520 | EXO70H4 | 3.21 ± 0.94 | 3.61 ± 1.14 | 3.11 ± 0.57 | 7.41 ± 1.86 | 6.13 ± 1.39 | 7.72 ± 1.50 | 2.33 | n/s | 2.44 |
| AT3G16120 | Dynamin light chain t1 family | 0.97 ± 0.12 | 1.18 ± 0.44 | 1.64 ± 0.50 | 2.66 ± 0.76 | 3.79 ± 0.76 | 5.71 ± 0.75 | 2.76 | 3.26 | 3.40 |
| AT3G16660 | Pollen Ole e 1 | 1.58 ± 0.15 | 0.36 ± 0.19 | 0.65 ± 0.53 | 69.08 ± 5.14 | 36.25 ± 3.71 | 3.27 ± 0.41 | 43.25 | 93.08 | 5.20 |
| AT5G37540 | Eukaryotic Asp protease family | 2.68 ± 0.41 | 2.57 ± 0.19 | 2.35 ± 0.24 | 5.86 ± 1.80 | 5.67 ± 1.97 | 5.84 ± 1.13 | 2.21 | 2.10 | 2.48 |
We suggest that at least part of the identified group of 31 genes that continued to be expressed in the margins of the tcp mutants likely represent pathways that are involved in margin development in a manner independent of miR319-regulated TCPs. This would also be in agreement with the additive or synergistic effects observed in margin development when overexpressing miR319 and simultaneously decreasing other CIN-like TCPs (Efroni et al., 2008) or NGA genes (Alvarez et al., 2016). Still, several of these genes quantitatively decrease in tcp mutants (Table I), suggesting that complex regulatory networks might exist that relate these transcription factors. In good agreement, NGA2 was enriched in the margins of the quadruple knockout compared with its blade region, albeit its level of expression was decreased (Supplemental Table S7).
Vasculature Development Is Impaired in the tcp2 tcp3 tcp4 tcp10 Mutant
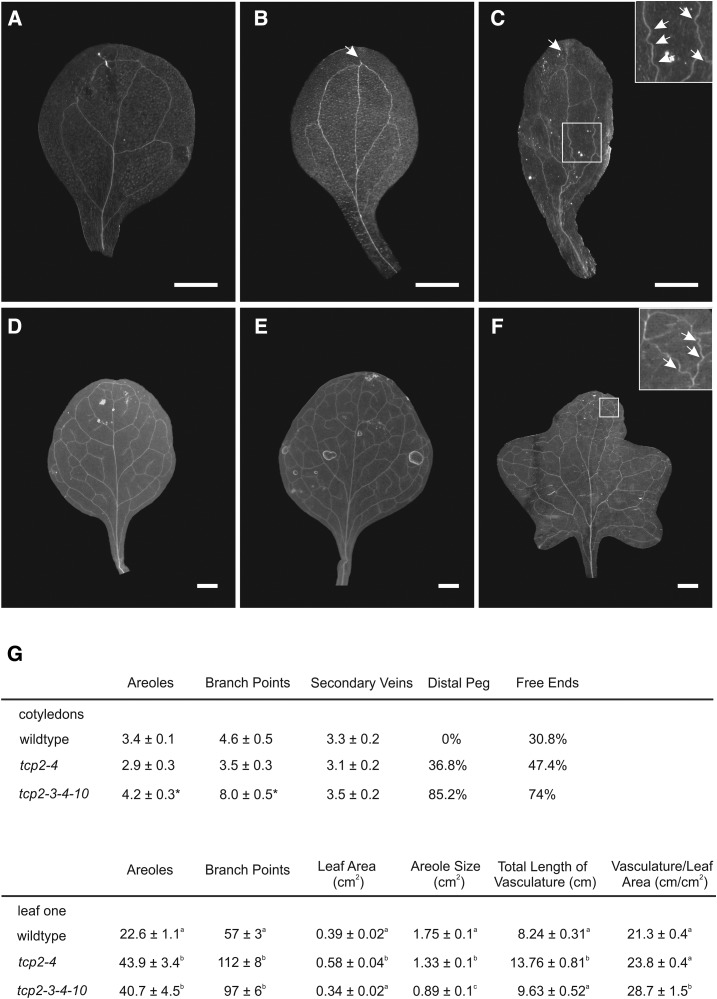
In the analysis of the RNAseq experiments, we also observed that several genes related to auxin biosynthesis or signaling were down-regulated in the margins of the tcp2 tcp3 tcp4 tcp10 mutant, including 19 SMALL AUXIN UPREGULATED RNA genes and eight INDOLE-3-ACETIC ACID INDUCIBLE genes (Supplemental Table S2). Auxin is known to play an important role in the establishment of the vasculature (Scarpella et al., 2006), and observations of alterations in the vasculature organization also have been reported in studies with TCP3-SRDX constructs (Li and Zachgo, 2013). We analyzed the venation patterns in the tcp2 tcp4 double and tcp2 tcp3 tcp4 tcp10 quadruple mutants compared with the wild type. In the Columbia wild-type cotyledon, we usually observed one midvein and secondary vein loops parting from it, generating four areoles (Fig. 6). In the tcp2 tcp4 and tcp2 tcp3 tcp4 tcp10 mutants, we observed a significant occurrence of distal pegs, ectopic vein pieces distal from the closing loops that end unconnected and close to the distal margin of the cotyledon (Fig. 6). We further observed an increased tendency for secondary veins ending unconnected and, therefore, not generating proximal areoles. However, the total average number of areoles was slightly higher in the quadruple mutant, due to an increased number of branch points (Fig. 6). Furthermore, the quadruple mutant veins were often not straight but had a wiggly appearance (Fig. 6, insets).
Figure 6.
Modification of the venation pattern in tcp mutants. A to C, Cleared cotyledons of the wild type (A), tcp2 tcp4 (B), and tcp2 tcp3 tcp4 tcp10 (C). D to F, Cleared first leaves of the wild type (D), tcp2 tcp4 (E), and tcp2 tcp3 tcp4 tcp10 (F). G, Table summarizing the scored parameters describing the venation pattern. Bars = 1 mm.
We also analyzed the vasculature of the first true leaf, which is more complex than that of cotyledons. We found that the numbers of branch points and areoles increased significantly in the double and quadruple mutants with respect to the wild type (Fig. 6). Moreover, the vasculature per leaf area increased and, in turn, decreased the average size of the areoles in double and quadruple knockouts (Fig. 6). Taken together, these data demonstrated that the tcp2 tcp3 tcp4 tcp10 leaf had a more extended vascular system than the wild-type leaf and that these differences are already seen between the wild type and tcp2 tcp4 double mutants.
DISCUSSION
In this study, we analyzed the effects of reduced TCP activity on different domains of the growing leaf. We employed a set of cumulative single and multiple mutants with decreasing TCP levels, focusing on tcp2 tcp4 double mutants that have large flat leaves and tcp2 tcp3 tcp4 tcp10 quadruple mutants with strongly crinkled organs. We dissected the leaf center and marginal regions. In a first step, our analyses revealed many genes that were enriched in either margins or central areas of developing wild-type Arabidopsis leaves. In subsequent analyses of the tcp double and quadruple mutant samples, we identified both organ-wide and margin-specific effects of the TCPs. Most conspicuously, cell proliferation genes were significantly up-regulated only in the margins of the tcp mutants.
Spatial Control of Gene Expression by TCP Transcription Factors
Leaf size initially increased with loss of TCP activity. However, it did not continue to increase steadily in triple and quadruple knockouts, but rather stagnated. Leaves from quadruple tcp mutants were smaller than those from double mutants and had mesophyll cells that were significantly smaller than those of their wild-type and double mutant counterparts. The reduced cell size is likely the consequence of a disturbed or delayed cell differentiation in the quadruple knockout leaves, which is in agreement with other reports describing a role for TCPs in the control of differentiation (Efroni et al., 2008; Sarvepalli and Nath, 2011a, 2011b).
Previous work has focused on the analysis of whole leaves or transverse organ sections (Efroni et al., 2008; Li et al., 2010; Pettkó-Szandtner et al., 2015), reflecting the developmental program along the proximodistal axis of the leaf. Here, we focus on the mediolateral axis of the organ by performing a comparative analysis of leaf margins and center regions. Using this approach, we detected large numbers of genes differentially expressed in the center or margin of the leaf. Mutations in the TCPs affected gene activity in both the center and margin of the leaves; however, margins were much more affected even in tcp2 tcp4 double mutants that have flat leaves, confirming a primary role of these transcription factors in the developmental control of the leaf margins. Even though tcp mutants have largely affected leaf margins, we detected a small group of genes that continued to be enriched in wild-type and tcp mutant leaf margins. Several of the margin-enriched genes detected here have already been described with biological functions related to margins. They include, for example, eight transcription factor genes of the CIN-like TCPs, NGA and STY gene families, and hormone biosynthetic genes such as YUCCA2. Down-regulation of the CIN-like TCPs TCP5, TCP13, and TCP17 has been shown to synergistically enhance the effects of miR319-regulated TCPs in the control of leaf growth (Efroni et al., 2008). NGA and STY genes have been known to be important in the generation of margins, initially of the gynoecium but also in leaf development. They influence organ shape, as seen in sty1 sty2 double knockouts as well as quadruple nga1 nga2 nga3 nga4 knockouts, which also have leaves that are larger and more serrated than the wild type. In contrast, overexpressors of STY1 or NGA genes have smooth leaf margins (Kuusk et al., 2002; Alvarez et al., 2009; Trigueros et al., 2009; Lee et al., 2015). NGA genes also seem to regulate YUCCA2 (Trigueros et al., 2009), which we identified to be margin enriched. Recently, a combined knockdown of miR319-regulated TCPs and NGA genes has been shown to have a dramatic additive effect on leaf development with undetermined growth of the margins (Alvarez et al., 2016). In this elegant study, it was concluded that miR319-regulated TCPs and NGAs work redundantly in the inhibition of meristematic activity in the leaf margins and its proximal part. It may be plausible to think that simultaneous down-regulation of the margin-enriched genes identified here together with the miR319-regulated TCPs also would cause synergistic effects in margin development. Furthermore, it is worthwhile to note that all three groups of transcription factors have not only been implicated in leaf development, affecting shape and vasculature, but also in gynoecium or fruit development, suggesting that, indeed, they may be able to entertain close interactions (Palatnik et al., 2003; Kuusk et al., 2006; Martínez-Fernández et al., 2014).
The origination of the vascular pattern in the leaf is closely related to the establishment of leaf shape and to the presence of auxin convergence points in the leaf margins (Scarpella et al., 2006, 2010). Therefore, disturbed margin development may go hand in hand with defects in auxin signaling or biosynthesis and may be the reason for the observed alterations in the tcp mutant vasculature.
A careful analysis of the group of down-regulated genes in the tcp mutant margins compared with the center samples revealed that it contained many genes related to photosynthesis. This is in contrast to what is seen in the wild type, where, in general, photosynthesis-related genes were expressed at higher levels in the margins than in the center region of the leaf. We visualized by confocal microscopy that the distribution of chlorophyll in the leaf was as predicted by the transcriptome analysis and confirmed a delayed maturation of the photosynthetic apparatus in the tcp2 tcp3 tcp4 tcp10 leaf, especially in the marginal regions.
Simultaneous down-regulation of miR319-regulated TCPs and NGA genes results in strongly crinkled leaves with white margins, indicating the lack of chlorophyll (Alvarez et al., 2016). Interestingly, we observed a change in the relative expression domain of photosynthetic genes also in tcp2 tcp4 double mutants, suggesting that TCP activity quantitatively regulates the expression of photosynthesis-related genes. The activation of photosynthesis happens upon the decision of differentiation very early in leaf development, and chloroplast differentiation is linked to the control of cell proliferation (Andriankaja et al., 2012). Therefore, a delay in the activation of photosynthesis-related genes upon the reduction or lack of TCP activity aids further in the role of the TCPs as coordinators of cell proliferation and differentiation with specific roles in the mediolateral axis of the leaf blade. Our results also are in agreement with earlier observations that indicated that TCPs trigger the differentiation of leaf cells (Efroni et al., 2008).
Although the NGA genes were identified as margin-enriched genes in wild-type and in tcp2 tcp3 tcp4 tcp10 margins, the expression levels of all detected NGA genes were reduced in the double and quadruple tcp mutant samples compared with the wild type in both center and marginal regions. These results are consistent with previous data showing that TCP2 and TCP3 can bind to a conserved promoter element of all NGA genes in yeast experiments and also to the NGA3 promoter in transient Nicotiana benthamiana assays and activate it (Ballester et al., 2015). Overall, these results suggest that the miRNA-regulated TCPs partially control NGA expression, both in the center and margins of the leaf (Ballester et al., 2015; this study), and that the two groups of transcription factors synergistically control margin development (Alvarez et al., 2016). The margin-specific STY genes did not show a uniform down-regulation in the tcp knockout samples, which suggests that their functions in margin development might be less connected to the TCPs than those of the NGAs.
Margin-Specific Control of Cell Proliferation by miR319-Regulated TCPs
While previous transcriptomic analyses did not detect changes in the expression of cell cycle genes upon overexpression of miR319 (Efroni et al., 2008; Schommer et al., 2008), our specific analysis of young leaf margins identified a major up-regulation of genes known to participate in cell proliferation in the tcp mutants. An analysis for GO term enrichment in the tcp2 tcp3 tcp4 tcp10 margin samples resulted in rankings with GO terms related to cell cycle- and microtubule-based processes being the top overrepresented groups.
An interaction between the TCP network and the cell cycle machinery with other factors involved in the control of plant age has been suggested. Recent results point toward the TCPs interacting with SQUAMOSA PROMOTER BINDING PROTEIN-LIKE transcription factors (Rubio-Somoza et al., 2014), which might provide further explanations for different effects of the TCPs in different leaves. Analysis of a CYCLINB1;1 reporter revealed changes in its expression pattern that were dependent on the point of appearance of the leaf during the plant’s ontogeny. Leaves of plants overexpressing miR319 and of tcp2 tcp3 tcp4 tcp10 knockouts that are produced later in the plant’s life history have strong crinkles at their margins in addition to a general change in leaf curvature. In the first pair of true leaves, we found that cell proliferation was extended toward the margins and detectable for a longer period of time, similar to what has been described in previous studies (Nath et al., 2003; Efroni et al., 2008). However, in higher order leaves, we observed that cell proliferation was maintained in discrete foci along the leaf margins until later stages of leaf development. We propose that theses foci are responsible for the localized generation of extra cells that cause the conspicuous phenotype of crinkled leaves, as observed in adult plants with low TCP activity, due to the overexpression of miR319 or loss of function of multiple TCP genes.
The changes in cell proliferation observed in the margins of plants with very low TCP activity are consistent with a role of the miR319-regulated TCPs in the control of the marginal meristem of the leaf (Donnelly et al., 1999; Alvarez et al., 2016). However, despite their prominent effects on margin development, the TCPs do have functions in the control of cell proliferation in the leaf blade. Leaves of the single and double knockouts that do not obviously change their morphological appearance are larger than wild-type leaves. Furthermore, the snapdragon cin mutant has been described to show bulky growth of the intervein sections (Nath et al., 2003), which might suggest that, also in the leaf blade, some cells remain with mitotic activity that should have ceased to proliferate to achieve the neutral curvature of the blade.
That TCP2, TCP3, TCP4, and TCP10 were not identified as margin-enriched genes hints that their tissue-specific effects may not be achieved directly through their transcriptional regulation. Recent studies show that TCPs can interact and form protein complexes with other transcription factors. In a global analysis where dimerization abilities of all Arabidopsis TCP family members were tested by yeast two-hybrid assays, it turned out that miR319-regulated TCPs have a strong tendency to heterodimerize with TCP5, TCP13, and TCP17 (Danisman et al., 2013). Curiously, the transcripts of these exact three members of the TCP family were identified in our analyses to be margin enriched. It might be that heterodimers between the miR319-regulated TCPs and the closely related TCP5, TCP13, and TCP17 are necessary to guarantee normal leaf development in the margins. In addition, it has been suggested that changes in leaf morphogenesis during the plant life cycle can be achieved by the competition of transcription factor complexes between TCPs and members of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE and CUC transcription factor families. The function of TCPs may be to sequester CUC2 and CUC3 proteins in order to prevent functional CUC dimers from building (Rubio-Somoza et al., 2014), with CUC2 especially being expressed in the marginal leaf regions (Nikovics et al., 2006). Furthermore, TCP4 interacts with other proteins like TCP INTERACTOR CONTAINING EAR MOTIF PROTEIN1, which then leads to the establishment of functional complexes that regulate transcription (Tao et al., 2013). TCP4 also has been shown to interact with chromatin-remodeling complexes harboring BRHAMA ATPases (Efroni et al., 2013). Taken together, it seems possible that the formation of different protein complexes may be one of the ways in which the TCPs achieve their various ways of action throughout the plant’s life cycle and in different regions of the leaf.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) accession Columbia wild-type and mutant plants were grown at 23°C in long-day conditions (16 h of light/8 h of dark). tcp2, tcp4, and tcp10 insertional mutants and the cyclinB reporter have been described in earlier work (Schommer et al., 2008; Rodriguez et al., 2010). The tcp3 insertional mutant was obtained from the Arabidopsis Biological Resource Center (stock no. CS855978).
Measurement of Leaf Size
Leaves were detached and mounted between two glass plates before photographing with illumination from below. Leaf circumferences were calculated with ImageJ software, and the folded areas that were identified by darker color in the image were calculated correspondingly.
Measurement of Cell Size and Number and Observation of Vasculature
To obtain paradermal views of palisade cells, leaves were fixed with FAA (5 v/v formaldehyde, 10 v/v acetic acid, 50 v/v ethanol) and cleared with chloral hydrate solution as described (Horiguchi et al., 2005). Palisade leaf cells were observed using differential interference contrast microscopy.
GUS Assays
To visualize the activity of the reporters, transgenic plants were subjected to GUS staining, according to Donnelly et al. (1999). Stained tissue was transferred to 70 (v/v) ethanol.
RNAseq Experiments
For this experiment, seeds were surface sterilized, sown on plates with Murashige and Skoog agar medium, and stratified for at least 2 d. To synchronize the developmental stages of the three genotypes at tissue collection, an offset of 2 d was generated before placing in the greenhouse the plates of the next plant line, starting with tcp2 tcp3 tcp4 tcp10 and ending with Columbia. Once in the greenhouse, plates were grown at 23°C in long-day conditions (16 h of light/8 h of dark) for 5 days, at which point seedlings were transplanted to individual soil pots. At emergence of the seventh leaf, the fifth leaf of each plant was harvested at Zeitgeber time 6, dissected on a cold plate into center and margin samples as shown in Figure 2, and transferred to separate tubes containing 200 μL of RNALater solution (Ambion; no. AM7020). Each center and margin sample consisted of the center and margin portions, respectively, of the fifth leaf of 15 to 20 different individual plants. This was performed by triplicates for each line. After tissue harvest, RNALater was removed by placing the contents of each tube on a piece of sterile absorbent paper, and the sample was immediately flash frozen in liquid nitrogen. Total RNA was extracted using TRIzol (Invitrogen; no. 15596-018), treated with Turbo DNase (Ambion; no. AM2238), and repurified by phenol:chloroform:isoamyl alcohol (25:24:1, v/v) extraction. Total RNA integrity was confirmed on the BioAnalyzer (Agilent). Barcoded libraries were constructed using the Illumina TruSeq RNA kit with an average of 1 μg of total RNA as starting material. Library quality was monitored on the BioAnalyzer and then sequenced as 100-bp single-end reads using an Illumina HiSeq sequencer. Resulting RNAseq raw data were processed as described elsewhere (Anders et al., 2013), and differential expression was assessed with edgeR version 3.8.6 (Robinson et al., 2010). Differentially expressed genes were selected based on a minimum fold change of ±2 and a false discovery rate < 0.01 (i.e. P value adjusted for multiple testing with the Benjamini-Hochberg procedure). GO analysis was performed with agriGO (Du et al., 2010; Tian et al., 2017). Functional enrichments were visualized with MapMan (Thimm et al., 2004).
Accession Numbers
The RNAseq data discussed in this article have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE99854 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE99854).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. M versus A plots for RNAseq data.
Supplemental Figure S2. Center- and margin-enriched transcripts.
Supplemental Table S1. Center- and margin-enriched genes.
Supplemental Table S2. Down-regulated genes in tcp mutants.
Supplemental Table S3. Identification of GGACCA boxes in down-regulated genes.
Supplemental Table S4. GO term enrichment analysis in center and marginal parts of tcp mutants.
Supplemental Table S5. Up-regulated genes in tcp mutants.
Supplemental Table S6. Mitosis- and early leaf development-associated genes in tcp mutants.
Supplemental Table S7. Expression levels of NGA and STY genes.
Acknowledgments
RNAseq experiments were carried out in Detlef Weigel’s laboratory under the supervision of Ignacio Rubio-Somoza and with the help of Claude Becker. We thank members of the RNA Biology laboratory for critical discussions.
Footnotes
The work was mainly supported by grants from the Argentine Ministry of Science to C.S. (PICT2013-2763) and J.F.P. (PICT2016-0761). It was supported by an EMBO Short-Term Fellowship to E.G.B. and CONICET fellowships to E.G.B. and U.C. J.F.P. and R.E.R. are members of CONICET. C.S. is contracted by CONICET. Work performed at the Max Planck Institute was supported by SFB1101 to Detlef Weigel.
References
- Alvarez JP, Furumizu C, Efroni I, Eshed Y, Bowman JL (2016) Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5: e15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y (2009) The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21: 1373–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786 [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Ballester P, Navarrete-Gómez M, Carbonero P, Oñate-Sánchez L, Ferrándiz C (2015) Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol Plant 155: 21–32 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Danisman S, van Dijk AD, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent GC, Immink RG (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot 64: 5673–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D (2013) Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J, Reimer JJ, Leuz I, Göbel U, Huettel B, Farrona S, Turck F (2012) Development-related PcG target in the apex 4 controls leaf margin architecture in Arabidopsis thaliana. Development 139: 2566–2575 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Kawamura E, Horiguchi G, Tsukaya H (2010) Mechanisms of leaf tooth formation in Arabidopsis. Plant J 62: 429–441 [DOI] [PubMed] [Google Scholar]
- Kazama T, Ichihashi Y, Murata S, Tsukaya H (2010) The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol 51: 1046–1054 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusk S, Sohlberg JJ, Long JA, Fridborg I, Sundberg E (2002) STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129: 4707–4717 [DOI] [PubMed] [Google Scholar]
- Kuusk S, Sohlberg JJ, Magnus Eklund D, Sundberg E (2006) Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J 47: 99–111 [DOI] [PubMed] [Google Scholar]
- Lee BH, Kwon SH, Lee SJ, Park SK, Song JT, Lee S, Lee MM, Hwang YS, Kim JH (2015) The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol Biol 89: 529–538 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Li S. (2015) The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal Behav 10: e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S (2013) TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76: 901–913 [DOI] [PubMed] [Google Scholar]
- Martínez-Fernández I, Sanchís S, Marini N, Balanzá V, Ballester P, Navarrete-Gómez M, Oliveira AC, Colombo L, Ferrándiz C (2014) The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front Plant Sci 5: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Nicolas M, Cubas P (2016) TCP factors: new kids on the signaling block. Curr Opin Plant Biol 33: 33–41 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pettkó-Szandtner A, Cserháti M, Barrôco RM, Hariharan S, Dudits D, Beemster GT (2015) Core cell cycle regulatory genes in rice and their expression profiles across the growth zone of the leaf. J Plant Res 128: 953–974 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2003) Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Polyn S, Willems A, De Veylder L (2015) Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M (2012) Control of organ size in plants. Curr Biol 22: R360–R367 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Debernardi JM, Palatnik JF (2014) Morphogenesis of simple leaves: regulation of leaf size and shape. Wiley Interdiscip Rev Dev Biol 3: 41–57 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D (2014) Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24: 2714–2719 [DOI] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011a) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011b) Interaction of TCP4-mediated growth module with phytohormones. Plant Signal Behav 6: 1440–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Barkoulas M, Tsiantis M (2010) Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E (2006) STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J 47: 112–123 [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45: W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros M, Navarrete-Gómez M, Sato S, Christensen SK, Pelaz S, Weigel D, Yanofsky MF, Ferrándiz C (2009) The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21: 1394–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- White DW. (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]