A transmembrane protein conserved broadly in plants and animals promotes antiviral silencing by enhancing the amplification of virus-derived small interfering RNAs.
Abstract
Small interfering RNAs (siRNAs) are processed from virus-specific dsRNA to direct antiviral RNA interference (RNAi) in diverse eukaryotic hosts. We have recently performed a sensitized genetic screen in Arabidopsis (Arabidopsis thaliana) and identified two related phospholipid flippases required for antiviral RNAi and the amplification of virus-derived siRNAs by plant RNA-dependent RNA polymerase1 (RDR1) and RDR6. Here we report the identification and cloning of ANTIVIRAL RNAI-DEFECTIVE2 (AVI2) from the same genetic screen. AVI2 encodes a multispan transmembrane protein broadly conserved in plants and animals with two homologous human proteins known as magnesium transporters. We show that avi2 mutant plants display no developmental defects and develop severe disease symptoms after infection with a mutant Cucumber mosaic virus (CMV) defective in RNAi suppression. AVI2 is induced by CMV infection, particularly in veins, and is required for antiviral RNAi and RDR6-dependent biogenesis of viral siRNAs. AVI2 is also necessary for Dicer-like2-mediated amplification of 22-nucleotide viral siRNAs induced in dcl4 mutant plants by infection, but dispensable for RDR6-dependent biogenesis of endogenous transacting siRNAs. Further genetic studies illustrate that AVI2 plays a partially redundant role with AVI2H, the most closely related member in the AVI2 gene family, in RDR1-dependent biogenesis of viral siRNAs and the endogenous virus-activated siRNAs (vasi-RNAs). Interestingly, we discovered a specific genetic interaction of AVI2 with AVI1 flippase that is critical for plant development. We propose that AVI1 and AVI2 participate in the virus-induced formation of the RDR1/RDR6-specific, membrane-bound RNA synthesis compartment, essential for the biogenesis of highly abundant viral siRNAs and vasi-RNAs.
Posttranscriptional gene silencing (PTGS; Napoli et al., 1990; Lindbo et al., 1993) directs a potent antiviral defense mechanism in plants against both RNA and DNA viruses (Baulcombe, 2004; Ding, 2010; Zhang et al., 2015; Wang, 2015). In Arabidopsis (Arabidopsis thaliana), antiviral silencing against RNA viruses is initiated after dsRNA replicative intermediates are processed by Dicer-like4 (DCL4) and DCL2 into 21- and 22-nt primary small interfering (siRNAs), respectively (Xie et al., 2004; Bouché et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Diaz-Pendon et al., 2007). These virus-derived siRNAs (vsiRNAs) subsequently act as specificity determinants to guide specific antiviral silencing mainly by Argonaute1 (AGO1) and AGO2 although AGO4, AGO5, AGO7, and AGO10 also display antiviral activities in some cases (Morel et al., 2002; Qu et al., 2008; Wang et al., 2011; Brosseau and Moffett, 2015; Carbonell and Carrington, 2015; Garcia-Ruiz et al., 2015; Brosseau et al., 2016; Fang and Qi, 2016). A shared feature of distinct gene silencing mechanisms mediated by siRNAs in plants, including antiviral silencing, is the production of secondary siRNAs, which are processed from dsRNA precursors synthesized de novo by cellular RNA-dependent RNA polymerases (RDRs; Diaz-Pendon et al., 2007; Donaire et al., 2008; Borges and Martienssen, 2015). In Arabidopsis, antiviral silencing requires the secondary vsiRNAs amplified by either RDR1 or RDR6, although primary vsiRNAs produced in rdr1 rdr6 double mutant plants exhibit antiviral activity (Garcia-Ruiz et al., 2010; Wang et al., 2010; Guo et al., 2013; Zhang et al., 2015).
Much is known about the antiviral function of the genes shared among the small RNA silencing pathways (Baulcombe, 2004; Ding, 2010; Zhang et al., 2015). However, less is known about the factors that are essential for antiviral silencing, but are dispensable for endogenous RNA silencing critical for plant development. Recently, we developed a sensitized genetic screen for the identification of, to our knowledge, new host genes in the antiviral silencing pathway in Arabidopsis (Guo et al., 2017) after extensive characterization of two Cucumber mosaic virus (CMV) mutants not expressing the viral suppressor of RNA silencing (VSR), the 2b protein (Diaz-Pendon et al., 2007; Wang et al., 2010, 2011). CMV-Δ2b contains three nucleotide substitutions that prevent the translation of VSR-2b without altering the carboxyl-terminal sequence of the viral RNA-dependent RNA polymerase 2a protein (RdRP-2a) encoded by the genomic RNA 2 in the −1 overlapping reading frame (Ding et al., 1994). In contrast, CMV-2aTΔ2b contains a 295-nt deletion in the VSR-2b coding region that also results in the carboxyl-terminal truncation of RdRP-2a (Soards et al., 2002). Both CMV-Δ2b and CMV-2aTΔ2b cause symptomless systemic infection in wild-type Col-0 plants and become as virulent as the wild-type virus (Fny strain) in dcl2 dcl4 double mutants (Ziebell and Carr, 2009; Wang et al., 2010). Both mutant viruses also cause virulent systemic infection in rdr1 rdr6 double mutant plants; However, although rdr6 single mutant plants are as susceptible as rdr1 rdr6 double mutant plants to CMV-Δ2b, they are as resistant as wild-type plants to CMV-2aTΔ2b (Wang et al., 2010, 2011). Thus, whereas the C-terminal truncation of RdRP-2a does not cause obvious defects in virus replication, it enhances the susceptibility of CMV-2aTΔ2b to RDR1-dependent antiviral silencing, which fails to suppress CMV-Δ2b infection in rdr6 single mutant plants. A recent study has demonstrated that the Tomato bushy stunt virus (TBSV) RNA replication compartment is protective against antiviral silencing (Kovalev et al., 2017), suggesting that the C-terminal truncation of RdRP-2a may decrease such protection of CMV-2aTΔ2b from RDR1-dependent antiviral silencing.
Several antiviral RNAi defective (avi) mutants have been identified by screening T-DNA insertion mutants of Arabidopsis that develop disease symptoms after CMV-Δ2b infection (Guo et al., 2017). The causal mutation of avi1 was not linked to T-DNA insertion and thus was cloned by a mapping-by-sequencing approach (Guo et al., 2017). AVI1 was renamed as ALA2 because it encodes one of the 12 Arabidopsis aminophospholipid transporting ATPases (ALAs) with its flippase activity previously characterized in yeast (López-Marqués et al., 2010). Our studies have shown that ALA2 acts together with ALA1 to enhance the amplification of vsiRNAs by either RDR1 or RDR6 (Guo et al., 2017). However, both ALA1 and ALA2 are dispensable for the biogenesis of the endogenous, RDR6-dependent transacting siRNAs (tasi-RNAs) or microRNAs (miRNAs; Guo et al., 2017). Notably, ALA1 and ALA2 are required for the biogenesis of the virus-activated siRNAs (vasi-RNAs), a recently described class of RDR1-dependent endogenous siRNAs that are induced by CMV and Turnip mosaic virus (TuMV) to target diverse host genes for silencing by a DCL4-AGO2 pathway (Cao et al., 2014). Interestingly, ALA1 has also been identified independently by screening for enhancers of rdr6 via infection with CMV-2aTΔ2b (Zhu et al., 2017).
Here we report the identification and cloning of AVI2 from the same sensitized genetic screen. Encoding a multispan transmembrane protein conserved broadly in plants and animals, AVI2 was not previously characterized in plants and was disrupted by a T-DNA insertion in the avi2 mutant plants. CMV-Δ2b infection induced severe disease symptoms in the avi2 mutant plants that were associated with an increased virus accumulation compared to wild-type plants. Further genetic characterization demonstrated that AVI2 and its homolog AVI2H are required for RDR6- and/or RDR1-dependent production of vsiRNAs and vasi-RNAs in the antiviral silencing response. However, both avi2 and avi2h mutant plants exhibited no defects in either development or the biogenesis of miRNAs and tasi-RNAs, indicating a specific function in antiviral silencing. Interestingly, combining the avi2 allele with either ala1 or ala2 in double mutants revealed a genetic interaction between AVI2 and ALA1 to control plant development. We propose that plants have evolved specific genes to enhance antiviral silencing by up-regulating the biogenesis of viral siRNAs and vasi-RNAs.
RESULTS
Identification and Cloning of AVI2 from Genetic Screen
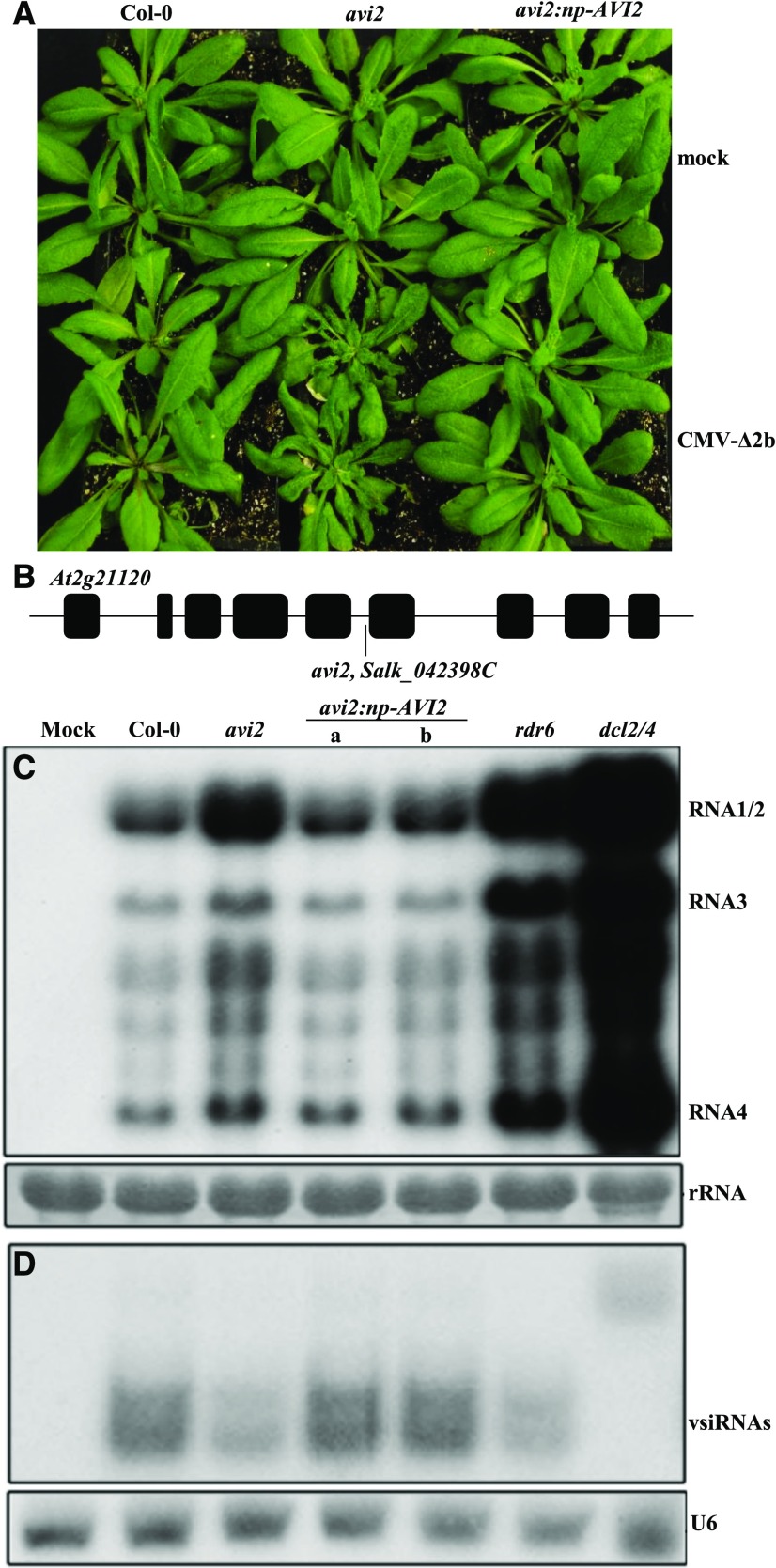
Similar to avi1 mutant plants, we found that avi2 mutant plants exhibited no visible developmental defects and developed severe disease symptoms after infection with CMV-Δ2b (Fig. 1A), which is derived from Fny-CMV, a subgroup I strain of CMV (Wang et al., 2011). Characterization of the enhanced disease susceptibility to CMV-Δ2b in F2 plants from backcrosses with wild-type Col-0 plants indicated that avi2 corresponded to a single recessive mutation and that the enhanced virus susceptibility phenotype of avi2, originally identified from Salk_042398C, cosegregated with the T-DNA inserted into the fifth intron of an uncharacterized gene, AT2g21120 (Fig. 1B). Northern-blot hybridization showed that the enhanced susceptibility phenotype of avi2 mutant plants was correlated with an increased accumulation of CMV-Δ2b compared to that in Col-0 plants (Fig. 1C). We have shown that CMV-Δ2b replicates to approximately 18- and 37-fold higher levels in rdr1 rdr6 and dcl2 dcl4 double mutant plants than in wild-type Col-0 plants, respectively (Wang et al., 2011). By comparison, CMV-Δ2b replicated in the infected avi2 plants to levels comparable to that in rdr6 plants, but lower than that in dcl2 dcl4 mutant plants (Fig. 1C). To confirm the T-DNA insertion as the causal mutation of avi2, AT2g21120 under the control of its native promoter (np) was transformed into the homozygous avi2 mutant plants and the progeny plants from two independent lines were assayed for the susceptibility to CMV-Δ2b. Northern-blot hybridization revealed that both lines of the transgenic avi2 plants accumulated low levels of the viral genomic and subgenomic RNAs similar to that found in Col-0 plants infected with CMV-Δ2b (Fig. 1C). Consistently, the avi2 plants expressing wild-type AT2g21120 did not develop disease symptoms after CMV-Δ2b infection (Fig. 1A). These results together showed that the enhanced virus susceptibility of avi2 mutant plants was caused by a loss-of-function mutation in AT2g21120, which was named as AVI2.
Figure 1.
avi2 mutant plants are defective in antiviral silencing. A, Wild-type Col-0 (left) and avi2 mutant plants (middle) were photographed three weeks after mock inoculation (top two rows) or infection with CMV-∆2b (under two rows). Also examined were plants from two independent lines of avi2 mutant plants carrying a transgene for the genomic AVI2 gene driven by its native promoter (right). B, avi2 mutant plants (Salk_042398C) contained a T-DNA insertion in the 5th intron in the At2g21120 gene designated as AVI2. C, Northern-blotting detection of CMV-∆2b genomic and subgenomic RNAs in the upper systemically infected leaves three weeks after inoculation. 25S rRNA in the same membrane was stained to show equal loading. D, Northern-blot analysis to compare the accumulation of the negative-strand vsiRNAs in the same set of RNA samples examined in (C). The same membrane was probed for U6 RNA to show equal loading.
AVI2 Encodes a Multispan Transmembrane Protein and Is Induced by Virus Infection
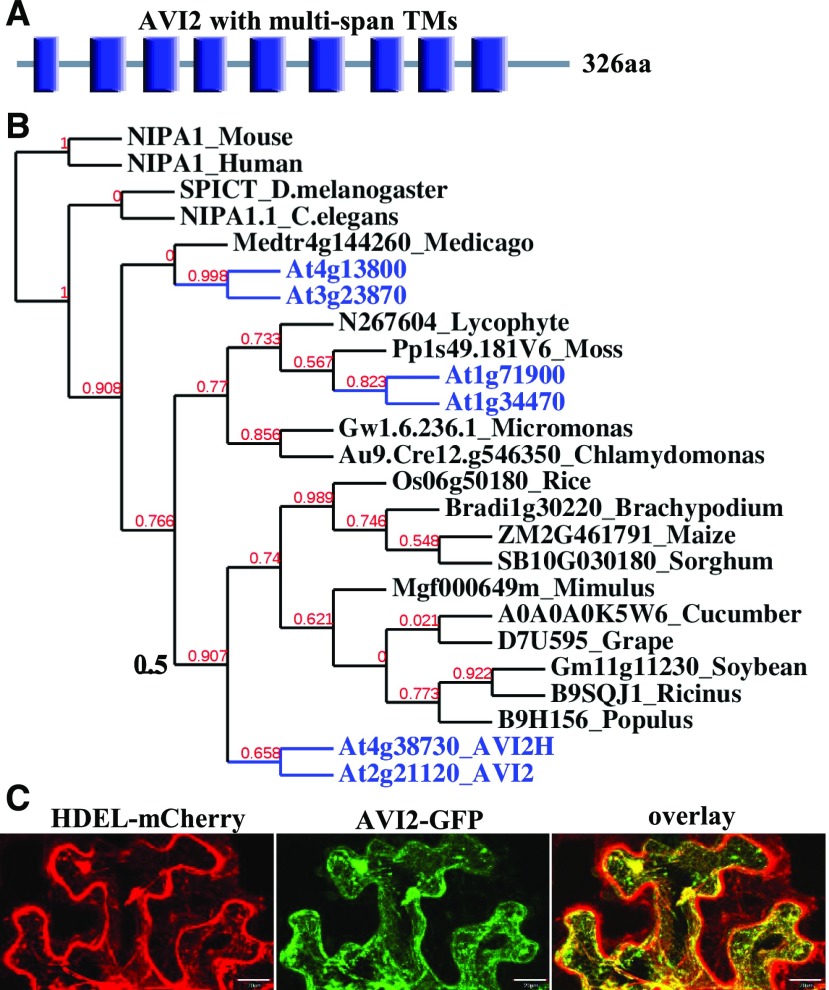
AVI2 protein contains nine transmembrane domains plus a domain of unknown function (DUF803; Fig. 2A). Little is known about the expression and function of AVI2, although genes closely related to AVI2 are conserved broadly in monocot and dicot plants (Fig. 2B). After coinfiltration with plasmids expressing GFP-tagged AVI2 and an ER-targeted HDEL-mCherry into the leaves of Nicotiana benthamiana, we detected colocalization of GFP-AVI2 with the ER marker HDEL-mCherry in the infiltrated leaves (Fig. 2C), suggesting an association of AVI2 with the ER membrane.
Figure 2.
AVI2 encodes a multispan transmembrane protein widely distributed in plants and animals. A, Nine transmembrane domains (blue bar) in AVI2 protein. B, AVI2 homologs in plants and animals with the six proteins from Arabidopsis shown in blue. C, Confocal microscopy of N. benthamiana leaf epidermal cells agroinfiltrated with constructs expressing HDEL-mCherry (an ER marker, left) and AVI2-GFP (middle).
Northern-blot hybridization showed that the accumulation of AVI2 mRNA was detectable in Col-0 plants after CMV-Δ2b infection, but not after mock infection (Fig. 3A). Low or undetectable expression of AVI2 under normal growth conditions without viral infection was consistent with the observation that avi2 plants showed no developmental defects (Fig. 1A). The accumulation of AVI2 mRNA was not detectable by northern-blot hybridization in avi2 mutant plants after either mock inoculation or CMV-Δ2b infection (Fig. 3A), suggesting that the avi2 was a null allele. AVI2 was also induced by infection with CMV-2aTΔ2b (Fig. 3A), which contains a 295-nt deletion in the VSR-2b coding region that also results in the C-terminal truncation of the viral RdRP-2a (Soards et al., 2002; Wang et al., 2011). Moreover, we found that wild-type Fny-CMV infection similarly induced the accumulation of AVI2 mRNA (Fig. 3A), suggesting that AVI2 is also active during the virulent infection and strong silencing suppression by Fny-CMV.
Figure 3.
AVI2 expression is induced by CMV infection. A, Northern-blotting detection of AVI2 mRNA in Col-0 or avi2 plants after mock inoculation or two weeks after infection with CMV-∆2b, CMV-2aT∆2b, or wild-type Fny strain of CMV. B, Col-0 plants carrying a GUS transgene driven by the promoter of AVI2 were stained to reveal GUS enzymatic activity after mock inoculation (left) or CMV-∆2b infection (right) at two weeks postinoculation.
We further generated transgenic Col-0 plants expressing β-glucuronidase (GUS) under the control of the AVI2 np. We found that the AVI2np-GUS transgene expressed at low or undetectable levels in Arabidopsis plants without virus infection (Fig. 3B). However, GUS expression was strongly induced by infection with either CMV-Δ2b (Fig. 3B) or Fny-CMV (Supplemental Fig. S1) in the upper systemically infected leaves, especially in the veins. Our findings together suggest that AVI2 is transcriptionally induced by CMV infection.
Functional Analysis of the AVI2 Gene Family in Antiviral Immunity
The genome of Arabidopsis encodes five additional genes highly homologous to AVI2 with the most closely related member (AT4g38730, designated as AVI2H) exhibiting 74% amino acid sequence identity with AVI2 (Fig. 2B). We obtained T-DNA insertion mutants for all of the five members of the AVI2 family, including two independent mutants for AVI2H (Fig. 4A). We found that avi2h-1 and avi2h-2 mutants (Fig. 4B) as well as the remaining four mutants, Salk_043567 (AT1g34470), Salk_122824 (AT1g71900), Salk_051233C (AT3g23870), and cs853195 (AT4g13800), exhibited no visible developmental defects and did not develop disease symptoms after infection with either CMV-Δ2b or CMV-2aTΔ2b. These results indicate that none of the five AVI2 homologs plays an essential role in plant development or antiviral response.
Figure 4.
Susceptibility of avi2 - avi2h single and double mutants to CMV-∆2b and CMV-2aT∆2b, which are silenced by the RDR6 pathway predominantly and by both RDR1 and RDR6 pathways, respectively. A, The positions of T-DNA insertion in AVI2H in avi2h-1 and avi2h-2, two allelic mutants of AVI2H. B, Plants of the indicated genotypes were photographed two weeks after mock inoculation (top row) or infection with CMV-2aT∆2b (middle row) or CMV-∆2b (bottom row). C, Northern-blotting detection of the viral genomic and subgenomic RNAs in the upper systemically infected leaves two weeks after inoculation. 25S rRNA in the same membrane was stained to show equal loading. D, Northern-blot analysis of small RNAs to compare the accumulation of the negative-strand vsiRNAs, the vasiRNAs targeting LHCB1.3 mRNA or 25S rRNA, TasiR255, or TasiR1511. The same membrane was further probed for U6 RNA to show equal loading.
We next generated two double mutants by combining avi2 with either avi2h-1 (SALK_056627) or avi2h-2 (SALK_126137) and examined their susceptibility to CMV-Δ2b. We found no obvious differences in either the symptom severity (Fig. 4B) or virus accumulation levels (Fig. 4C) among avi2h single mutant plants and avi2 avi2h-1 and avi2 avi2h-2 double mutants after infection with CMV-Δ2b. These results suggest that AVI2H did not contribute to the AVI2-mediated host defense against CMV-Δ2b, known to be suppressed predominantly by RDR6-dependent antiviral silencing (Wang et al., 2011).
We further compared the responses of the single and double mutant plants to CMV-2aTΔ2b, known to be effectively suppressed by either RDR1- or RDR6-dependent antiviral silencing (Wang et al., 2010, 2011; Guo et al., 2017). We found that CMV-2aTΔ2b was less virulent and replicated to lower levels in avi2 mutant plants than CMV-Δ2b did (Fig. 4, B and C). However, CMV-2aTΔ2b replicated to higher levels and caused more severe dwarfing in both avi2 avi2h-1 and avi2 avi2h-2 double mutants than in avi2 single mutant plants (Fig. 4, B and C). By comparison, both of the double mutants were as susceptible to CMV-2aTΔ2b as the single avi2 mutant to CMV-Δ2b. These findings showed that whereas AVI2 was indispensable for the host defense against CMV-Δ2b, it acted in a partially redundant manner with AVI2H to suppress CMV-2aTΔ2b infection.
AVI2 and AVI2H Act to Enhance the Biogenesis of vsiRNAs and vasiRNAs
The above results showed that avi2/avi2h single and double mutant plants displayed differential susceptibility to CMV-Δ2b and CMV-2aTΔ2b, known to trigger distinct pathways of antiviral silencing. These observations suggest that AVI2 and AVI2H act in the antiviral silencing response. To examine this hypothesis, we compared the accumulation of vsiRNAs in these mutant plants after infection with the mutant viruses by northern-blot hybridization because vsiRNAs are a central component of antiviral silencing. We used the same DNA oligomer probes selected previously to detect the negative-strand vsiRNAs (Guo et al., 2017; Wang et al., 2010, 2011). We have shown previously that CMV-Δ2b is repressed in Arabidopsis plants by 21- and 22-nt vsiRNAs that are produced, respectively, by DCL4 and DCL2 and amplified predominantly by RDR6 (Diaz-Pendon et al., 2007; Wang et al., 2011). As expected, 21- and 22-nt vsiRNAs were undetectable in dcl2 dcl4 double mutants and in the absence of RDR6-dependent amplification of vsiRNAs, accumulated to lower levels in rdr6 plants than in Col-0 plants even though CMV-Δ2b replicated to higher levels in rdr6 plants (Fig. 1, C and D). Similar to rdr6 plants, we also detected an inverse relationship between the accumulation levels of the virus and its vsiRNAs in avi2 mutant plants, which accumulated a lower level of 21- and 22-nt vsiRNAs, but supported a higher level of CMV-Δ2b replication than Col-0 plants did (Fig. 1, C and D). Importantly, the repression of CMV-Δ2b in avi2 mutant plants by the wild-type AVI2 transgene was correlated with an enhanced accumulation of 21- and 22-nt vsiRNAs (Fig. 1, C and D). Moreover, CMV-Δ2b infection induced no major difference in the accumulation of vsiRNAs in either avi2 avi2h-1 or avi2 avi2h-2 double mutant plants compared to avi2 mutant plants (Fig. 4D). These findings indicated that whereas AVI2H was dispensable, AVI2 played an essential role in antiviral silencing against CMV-Δ2b by enhancing RDR6-dependent amplification of vsiRNAs.
We found that the accumulation of vsiRNAs was also markedly reduced in avi2 single mutant plants compared to either Col-0 plants (Fig. 4D) or rdr1 and rdr6 single mutant plants after infection with CMV-2aTΔ2b. The accumulation level of vsiRNAs remained low in avi2 avi2h-1 and avi2 avi2h-2 double mutant plants even through CMV-2aTΔ2b replicated to markedly higher levels than in avi2 single mutant plants (Fig. 4D). These results indicated that the amplification of vsiRNAs by both RDR1 and RDR6 to target CMV-2aTΔ2b was defective in these double mutant plants. Our findings suggest that in addition to an essential role in RDR6-dependent amplification of vsiRNAs, AVI2 also played a partially redundant role with AVI2H in antiviral silencing against CMV-2aTΔ2b by enhancing RDR1-dependent biogenesis of vsiRNAs.
In addition to the amplification of vsiRNAs, RDR1 and RDR6 are also required for the biogenesis of the endogenous vasi-RNAs and tasi-RNAs, respectively (Cao et al., 2014; Peragine et al., 2004; Vazquez et al., 2004). We detected no obvious changes in the accumulation of two different tasi-RNAs in Col-0, and avi2, avi2 avi2h-1, and avi2 avi2h-2 plants after the infection with either CMV-Δ2b or CMV-2aTΔ2b (Fig. 4D). However, we found that vasi-RNAs accumulated to lower levels in avi2 plants than in Col-0, avi2h-1, and avi2h-2 plants after infection with CMV-2aTΔ2b (Fig. 4D). Notably, both vasi-RNAs became undetectable in avi2 avi2h-1 and avi2 avi2h-2 double mutant plants infected with CMV-2aTΔ2b (Fig. 4D). In Arabidopsis, RDR1-dependent antiviral silencing and RDR1-dependent vasi-RNAs are not induced by CMV-Δ2b infection (Wang et al., 2010, 2011; Guo et al., 2017). Consistently, we found that vasi-RNAs remained undetectable in avi2/avi2h single and double mutant plants after infection with CMV-Δ2b (Fig. 4D). These results suggest that although neither played a role in the biogenesis of tasi-RNAs, AVI2 acted in a partially redundant manner with AVI2H in the RDR1-dependent biogenesis of vasi-RNAs.
Distinct Roles of AVI2 and AVI2H in the Biogenesis of vsiRNAs and vasiRNAs by RDR6 and/or RDR1
We next conducted additional genetic studies to verify the distinct functional roles of AVI2 and AVI2H in RDR1- and RDR6-dependent antiviral silencing. To this end, we generated avi2 rdr1, avi2 rdr6, and avi2 dcl4 double mutants and compared the infection with CMV-2aTΔ2b in these mutants with the respective single mutants as well as the rdr1 rdr6 and dcl2 dcl4 plants characterized previously. Consistent with previous studies (Diaz-Pendon et al., 2007; Wang et al., 2010), CMV-2aTΔ2b replicated to high levels in rdr1 rdr6 double mutant plants whereas rdr1 and rdr6 single mutants exhibited no disease symptoms, accumulated high levels of vsiRNAs, and supported low levels of virus replication (Fig. 5). Similar to rdr1 rdr6 plants, avi2 rdr1 double mutant plants also developed severe disease symptoms, supported high levels of virus replication, and produced extremely low levels of vsiRNAs after CMV-2aTΔ2b infection (Fig. 5, A to C). These results indicated that RDR6-dependent biogenesis of vsiRNAs was suppressed in avi2 rdr1 plants by the avi2 allele as effectively as in rdr1 rdr6 plants by the rdr6 allele, and thus further supported an essential role of AVI2 in RDR6-dependent antiviral silencing concluded from the characterization of CMV-Δ2b infection in Col-0, rdr6, and avi2 plants (Fig. 1).
Figure 5.
AVI2 and AVI2H function in the biogenesis of vsiRNAs and vasiRNAs mediated by RDR6 and/or RDR1. A, Wild-type (Col-0), single, and double mutant plants were photographed two weeks after mock inoculation or infection with CMV-2aT∆2b. B, Northern-blotting detection of the viral genomic and subgenomic RNAs in the upper systemically infected leaves two weeks after inoculation. 25S rRNA in the same membrane was stained to show equal loading. C and D, Northern-blot analysis of small RNAs to compare the accumulation of the negative-strand vsiRNAs (C) and the vasiRNAs to target four different endogenous genes (D). The same membrane was further probed for U6 RNA to show equal loading.
Unlike the rdr1 allele, introducing the rdr6 allele into avi2 mutant plants only weakly enhanced the accumulation and symptom severity of CMV-2aTΔ2b compared to avi2 single mutant plants (Fig. 5, A and B). We noted that vsiRNAs remained abundant in avi2 rdr6 plants (Fig. 5C). These findings suggested weak suppression of RDR1-dependent antiviral silencing against CMV-2aTΔ2b in avi2 rdr6 plants by the avi2 allele. This result was consistent with a partially redundant role of AVI2 with AVI2H in RDR1-dependent biogenesis of vsiRNAs and vasi-RNAs, concluded above from the characterization of CMV-2aTΔ2b infection in avi2-avi2h single and double mutant plants (Fig. 4).
We found that CMV-2aTΔ2b replicated to higher levels and was more virulent in avi2 dcl4 double mutants than in avi2 and dcl4 single mutant plants (Fig. 5, A and B). We reported previously that RDR6-dependent 22-nt secondary vsiRNAs made by DCL2 accumulated to very high levels in dcl4 mutant plants infected with CMV-Δ2b (Wang et al., 2011). Infection with CMV-2aTΔ2b also induced production of highly abundant 22-nt vsiRNAs in dcl4 mutant plants, but not in dcl2 dcl4 double mutant plants (Fig. 5C). Remarkably, these DCL2-dependent 22-nt vsiRNAs accumulated to markedly reduced levels in avi2 dcl4 double mutant plants than in dcl4 single mutant plants, even though CMV-2aTΔ2b replicated to higher levels in the double mutant plants (Fig. 5, B and C). These findings suggest that AVI2 is also necessary for the virus-induced amplification of DCL2-dependent 22-nt vsiRNAs in the absence of DCL4.
As expected from the previous work (Cao et al., 2014), all of the four vasiRNAs induced by CMV-2aTΔ2b became undetectable in RDR1-defective single and double mutants (Fig. 5D). We found that expression of the AVI2 transgene in avi2 mutant plants restored the accumulation of the vasiRNAs induced by CMV-2aTΔ2b (Fig. 5D). However, all of the four vasiRNAs remained detectable in avi2 rdr6 double mutant plants as found in avi2 mutant plants (Fig. 5D), consistent with a redundant role of AVI2H in the biogenesis of vasi-RNAs (Fig. 4D). Compared to the vasi-RNAs targeting the protein-coding genes, however, the 22-nt vasi-RNAs targeting 25S rRNA were induced to extremely high levels in dcl4 mutant plants by CMV-2aTΔ2b and the unusually strong induction was suppressed by the avi2 allele in avi2 dcl4 double mutant plants (Fig. 5D). These findings further indicate an independent function for AVI2 in the biogenesis of vasi-RNAs.
Genetic Interaction between AVI1 and AVI2
None of the single and double avi2/avi2h mutants displayed developmental defects (Figs. 1 and 2; Supplemental Fig. S2A). In contrast, whereas ala1 and ala2 single mutants isolated previously from the same genetic screen develop normally as Col-0 plants, ala1 ala2 double mutant plants exhibit severe developmental defects (Guo et al., 2017). Notably, we detected similar developmental defects in avi2 ala2 double mutant plants, but not in avi2 ala1 double mutant plants (Fig. 6A), revealing a specific genetic interaction between AVI2 and ALA2 (AVI1) that plays a role in plant development. After infection with CMV-Δ2b, however, both avi2 ala2 and avi2 ala1 double mutant plants developed more severe disease symptoms compared to the single mutant plants (Fig. 6B), although only avi2 ala1 plants supported higher levels of virus accumulation (Supplemental Fig. S2B), suggesting that symptom severity of the double mutants was not determined solely by either the genetic interaction or virus accumulation levels. We further compared infection of the same panel of mutants with the wild-type Q strain of CMV, a subgroup II strain of CMV much less virulent than Fny-CMV in Col-0 plants (Diaz-Pendon et al., 2007; Paulukatis and Garcia-Arenal, 2003). As found previously with Fny-CMV infection (Guo et al., 2017), Q-CMV did not replicate to higher levels in any of the silencing-defective mutant plants compared with Col-0 plants due to effective silencing suppression by VSR-2b (Supplemental Fig. S2C). Nevertheless, we observed a similar pattern of enhanced disease susceptibility of the single and double mutants to Q-CMV and CMV-Δ2b (Fig. 6, B and C), indicating a role for the identified genes in disease tolerance against virus infection.
Figure 6.
Genetic interaction between AVI2 and AVI1. Wild-type (Col-0), avi2 ala2(avi1), and ala1 ala2 mutant plants were photographed approximately one month after germination (A), or three weeks after the infection with Fny-CMV-∆2b (B) or wild-type Q-CMV (C) with avi2, ala2(avi1), ala1, and rdr1 rdr6 mutant plants as controls.
DISCUSSION
The induction of antiviral silencing and active silencing suppression by VSRs represent an evolutionary arms race between hosts and pathogens. Whereas VSR expression is essential for virus infection in wild-type plants and becomes dispensable in mutant plants defective in antiviral silencing, strong silencing suppression by wild-type viruses often prevents detection of the antiviral activity of the host antiviral silencing pathway. For example, Fny-CMV and TuMV do not replicate to higher levels in plants after the elimination of the secondary vsiRNAs either alone or together with the primary vsiRNAs by multiple genetic mutations (Garcia-Ruiz et al., 2010; Guo et al., 2017). In this study, we characterized the avi2 mutant plants identified from a genetic screen sensitized by using a mutant virus unable to suppress antiviral silencing (Guo et al., 2017). The systemic infection of wild-type Col-0 plants with CMV-Δ2b is symptomless and is associated with low levels of virus accumulation because of the enhanced susceptibility to antiviral silencing directed by vsiRNAs amplified predominantly by RDR6 pathway (Wang et al., 2011). We found that in contrast to Col-0 plants, avi2 plants developed severe disease symptoms after CMV-Δ2b infection, which was correlated with an increased level of virus accumulation. We demonstrate that the defect of avi2 plants in antiviral defense was caused by a T-DNA insertion in AVI2 and restored by the expression of a wild-type AVI2 transgene.
Our detailed genetic analyses strongly indicate that AVI2 acts in the antiviral silencing defense mechanism by enhancing the production of secondary vsiRNAs. We showed that the biogenesis of secondary vsiRNAs to target CMV-Δ2b predominantly by RDR6-dependent pathway was defective in avi2 mutant plants, similar to rdr6 single mutant plants (Fig. 1). We also demonstrated strong suppression of RDR6-dependent biogenesis of secondary vsiRNAs by the avi2 allele in avi2 rdr1 double mutant plants infected with CMV-2aTΔ2b, similar to rdr1 rdr6 double mutant plants (Fig. 5). Moreover, infection of dcl4 mutant plants with CMV-Δ2b induces RDR6-dependent amplification of 22-nt vsiRNAs made by DCL2 (Wang et al., 2011), which may be similar to the secondary siRNAs found in dcl4 plants to target transgenes undergoing RDR6-dependent PTGS (Parent et al., 2015). We found that the amplification of these 22-nt vsiRNAs induced in dcl4 plants by CMV-2aTΔ2b infection was strongly suppressed by the avi2 allele in avi2 dcl4 double mutant plants (Fig. 5). These findings together support an essential role for AVI2 in RDR6-dependent biogenesis of vsiRNAs (Supplemental Fig. S2).
Functional analysis of the small AVI2 gene family in Arabidopsis showed that AVI2 also played a partially redundant role in RDR1-dependent biogenesis of both vsiRNAs and vasi-RNAs with AVI2H (Supplemental Fig. S2), which shared the highest sequence similarity with AVI2 (Fig. 2). We found that avi2 single mutant plants produced markedly reduced levels of vsiRNAs and were more susceptible than Col-0, rdr1, and rdr6 plants to CMV-2aTΔ2b (Figs. 4 and 5), which unlike CMV-Δ2b, is repressed equally efficiently by RDR1 and RDR6 pathways (Wang et al., 2011). However, we observed robust replication of CMV-2aTΔ2b and strong suppression of vsiRNA production only in avi2 avih double mutants (Fig. 4). Weak suppression of RDR1-dependent antiviral silencing against CMV-2aTΔ2b by the avi2 allele was also observed in avi2 rdr6 double mutant plants (Fig. 5). Moreover, in response to infection with either CMV-2aTΔ2b or VSR-expressing TuMV, Arabidopsis plants produce highly abundant RDR1-dependent vasi-RNAs (Cao et al., 2014). These vasi-RNAs target diverse endogenous genes for silencing in a pathway dependent on DCL4 and AGO2, but not RDR6, DCL2, or AGO1, which also participate in the silencing of RNA viruses (Baulcombe, 2004; Cao et al., 2014; Carbonell and Carrington, 2015; Ding, 2010; Zhang et al., 2015). The presence of the avi2 allele, but neither of the avi2h alleles, strongly suppressed the induction of RDR1-dependent vasi-RNAs by CMV-2aTΔ2b infection (Figs. 4 and 5). However, vasi-RNAs became hardly detectable in avi2 avi2h double mutants (Fig. 4). Thus, AVI2 also played a partially redundant role with AVI2H in the biogenesis of RDR1-dependent vasi-RNAs, similar to its role in RDR1-dependent amplification of vsiRNAs (Supplemental Fig. S3).
These activities of AVI2 and AVI2H exhibit shared features with those described recently for ALA1 and ALA2 (Supplemental Fig. S3) identified from the same genetic screen, both of which are required for antiviral silencing and RDR6- and/or RDR1-dependent biogenesis of vsiRNAs and vasi-RNAs (Guo et al., 2017). For example, both avi2 avi2h and ala1 ala2 double mutants are highly susceptible to CMV-2aTΔ2b and severely defective in the biogenesis of both vsiRNAs and vasi-RNAs. The expression of both ALA1 and AVI2 in Arabidopsis plants is induced by CMV infection and the induction of AVI2 seems especially strong in the veins (Fig. 3), suggesting a possible role in the host inhibition of virus movement observed previously in the infection with VSR-deficient mutants of CMV (Ding et al., 1995; Lewsey et al., 2009). Although none of the identified AVI genes has an essential endogenous function, construction of double mutants revealed a unique role for the specific genetic interaction between AVI2 and ALA1 in plant development (Fig. 6). Interestingly, our analyses also indicate a role of AVI1 and AVI2 in disease tolerance against virus infection that is not associated with reduced virus load.
AVI2 encodes a multispan transmembrane protein that is broadly conserved in plants and animals. AVI2 is highly homologous to human NIPA1 and NIPA2 proteins thought to function as magnesium transporters. Mutations in NIPA1/2 are associated with a neurodegenerative disorder characterized by a slow, gradual, progressive weakness and spasticity of the lower limbs (Goytain et al., 2007, 2008). It is currently unknown whether AVI2 and/or AVI2H act as magnesium transporters in plants. We have proposed recently that the phospholipid transporter activity of ALA1/ALA2 plays a role in the formation of a membrane-bound, RDR1/RDR6-specific RNA synthesis compartment similar to that assembled from viral RdRP (den Boon et al., 2010; Guo et al., 2017). The putative magnesium transporter activity of AVI2 and AVI2H may have an impact on the synthesis of vsiRNA and vasi-RNA precursors by RDR1 and RDR6 because RNA synthesis by TBSV RdRP is influenced by the cytosolic concentration of Mn2+ and Mg2+ ions (Jaag et al., 2010). The available data suggest that AVI2, ALA1, and ALA2 are all localized to ER when they are expressed alone in N. benthamiana cells (López-Marqués et al., 2010, 2012). Thus, it is possible that AVI1 (ALA2) and AVI2 identified from our genetic screens may facilitate the virus-induced formation of the RDR1/RDR6-specific RNA synthesis compartment on distinct host membrane domains, essential for the synthesis of the dsRNA precursors of only the highly abundant vsiRNAs and vasi-RNAs.
MATERIALS AND METHODS
Plant Materials, Viruses, and Infection Assays
Salk homozygous T-DNA insertion mutant pools CS27942 and CS27952, as well as Salk_056627 (avi2h-1), Salk_126137 (avi2h-2), Salk_043567 (At1g34470), Salk_122824 (At1g71900), Salk_051233C (At3g23870), and cs853195 (At4g13800) were obtained from the Arabidopsis Information Resource Center. The mutant lines for Arabidopsis rdr1-1, rdr6-15, dcl2-1, and dcl4-2 were described in Wang et al. (2011), and ala1 and ala2-1 as well as ala1 ala2-1 were described in Guo et al. (2017). Additional double and triple mutants were generated by genetic crosses and genotyped. Plants were grown at 23°C with 9-h light. Plants with approximately 10 to 11 leaves were infected with virus by the mechanic rubbing method. Three expanded leaves of each plant were infected. Primers for mutant genotyping were listed in Supplemental Table S1. CMV-Δ2b was described previously (Wang et al., 2011), in which three AUG codons at the first (start codon), 8th, and 18th positions of 2b ORF encoded by wild-type RNA 2 of Fny strain of CMV were mutated to ACG so that the amino acids encoded in the −1 overlapping 2a ORF were not altered. CMV-2aTΔ2b contained a 295-nt deletion in the 2b coding sequence, which also results in the C-terminal truncation of the viral RNA-dependent RNA polymerase (Wang et al., 2011). Wild-type Fny-CMV and Q-CMV were as described (Soards et al., 2002; Diaz-Pendon et al., 2007). The conditions for plant growth, virus infection by mechanical inoculation with purified virions, RNA extraction from the upper noninoculated leaves for northern-blot hybridization, and the probes used to detect the viral genomic and subgenomic RNAs, the viral siRNAs, vasiRNAs, tasiRNAs, and miRNAs were as described previously (Guo et al., 2017).
Complementation Analysis of avi2 Mutant Plants
Genomic AVI2 (AT2G21120) gene from 1500 bp upstream of the ATG to the end of the ORF was amplified from Col-0 genomic DNA with Phusion DNA polymerase (M0530S; New England Biolabs) and cloned into pent/D-TOPO (Invitrogen). The final plasmids np-AVI2 was obtained through the LR reactions between the pENTR-AVI2 and pEARLEYGATE301 (Earley et al., 2006) and transformed into avi2 mutant plants by the Agrobacterium-mediated floral-dipping method. Two independent lines selected for phosphinothricin resistance were compared to Col-0 and avi2 mutant plants for susceptibility to CMV-Δ2b and CMV-2aTΔ2b. The primers used to amplify genomic AVI2 DNA were listed in Supplemental Table S1.
Transient AVI2 Expression in Nicotiana benthamiana Leaves and Confocal Microscopy
AVI2 cDNA was amplified from Col-0 plants for the construction of entry vectors pENTR/d-Topo, which were then transferred to Gateway destination expression vector pGWB506 for N-GFP tagged fusion proteins (Nakagawa, et al., 2007) using Gateway LR Clonase enzyme mix (Invitrogen Cat. no. 11791019). EHA105 agrobacteria expressing AVI2-GFP and HDEL-mCherry (an ER marker) were coinfiltrated into the leaves of N. benthamiana. Two days after infiltration, epidermal cells of the coinfiltrated leaves were examined for GFP fusion protein expression under a TCS SP5 confocal microscope (Leica) through a 20× water-immersion objective lens. GFP was imaged with 488-nm excitation and a detection window of 495 nm to 535 nm whereas HDEL-mCherry was imaged with 584-nm excitation and a detection window of 600 nm to 620 nm using the Sequential Scanning Module under the software LAS AF (Leica).
In Vivo Expression Analysis
The promoter region of AVI2 (from the start codon to 1500 bp upstream) was cloned into entry vectors pENTR/d-Topo for the construction of the final plasmid AVI2np-GUS after LR reaction between p-ENTR/d-topo-npAVI2 and pGWB433. The primers used to amplify the np of AVI2 were listed in Supplemental Table S1. AVI2np-GUS was transformed into Col-0 by the Agrobacterium-mediated floral-dipping method. In vivo analysis of GUS expression in AVI2np-GUS transgenic plants was carried out according to the published protocol (Stangeland and Salehian, 2002). AVI2np-GUS transgenic plants were inoculated with buffer or CMV-Δ2b and examined for GUS expression two weeks after inoculation. The accumulation of AVI2 transcripts in the upper noninoculated leaves of Col-0 and avi2 mutant plants two weeks after inoculation with either buffer or CMV-Δ2b as well as in Col-0 plants after infection with CMV-Δ2b, CMV-2aTΔ2b, or wild-type CMV (Fny-CMV) was determined by northern-blot analysis using a probe specific to AVI2 mRNA. The primers used to amplify the probe were listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ▪▪▪.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used in this work.
Supplemental Figure S1. proAVI2-GUS was induced after the infection of wild-type CMV.
Supplemental Figure S2. Comparing the development of the single and double mutant plants and their susceptibility to CMV-∆2b or Q-CMV.
Supplemental Figure S3. A model for AVI2/AVI2H function.
Footnotes
This project was supported by grants from the US Department of Agriculture Research Service (6659-22000-025), the US-Israel Binational Agricultural Research and Development Fund, and by the Agricultural Experimental Station of the University of California, Riverside (to S.-W.D.).
References
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau C, El Oirdi M, Adurogbangba A, Ma X, Moffett P (2016) Antiviral defense involves AGO4 in an Arabidopsis-Potexvirus interaction. Mol Plant Microbe Interact 29: 878–888 [DOI] [PubMed] [Google Scholar]
- Brosseau C, Moffett P (2015) Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 27: 1742–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW (2014) Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci USA 111: 14613–14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Carrington JC (2015) Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol 27: 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- den Boon JA, Diaz A, Ahlquist P (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW (2007) Suppression of antiviral silencing by Cucumber Mosaic Virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. (2010) RNA-based antiviral immunity. Nat Rev Immunol 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Ding SW, Anderson BJ, Haase HR, Symons RH (1994) New overlapping gene encoded by the Cucumber Mosaic Virus genome. Virology 198: 593–601 [DOI] [PubMed] [Google Scholar]
- Ding SW, Li WX, Symons RH (1995) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J 14: 5762–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martínez-García B, Martínez-Priego L, Pagán I, Llave C (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol 82: 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fang X, Qi Y (2016) RNAi in plants: an Argonaute-centered view. Plant Cell 28: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Carbonell A, Hoyer JS, Fahlgren N, Gilbert KB, Takeda A, Giampetruzzi A, Garcia Ruiz MT, McGinn MG, Lowery N, Martinez Baladejo MT, Carrington JC (2015) Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip Mosaic Virus infection. PLoS Pathog 11: e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and Dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytain A, Hines RM, El-Husseini A, Quamme GA (2007) NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem 282: 8060–8068 [DOI] [PubMed] [Google Scholar]
- Goytain A, Hines RM, Quamme GA (2008) Functional characterization of NIPA2, a selective Mg2+ transporter. Am J Physiol Cell Physiol 295: C944–C953 [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang R, Wang J, Ding SW, Lu R (2013) Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci USA 110: 16085–16090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Lu J, Wang X, Zhan B, Li W, Ding SW (2017) Lipid flippases promote antiviral silencing and the biogenesis of viral and host siRNAs in Arabidopsis. Proc Natl Acad Sci USA 114: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag HM, Pogany J, Nagy PD (2010) A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe 7: 74–81 [DOI] [PubMed] [Google Scholar]
- Kovalev N, Inaba JI, Li Z, Nagy PD (2017) The role of co-opted ESCRT proteins and lipid factors in protection of tombusviral double-stranded RNA replication intermediate against reconstituted RNAi in yeast. PLoS Pathog 13: e1006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey M, Surette M, Robertson FC, Ziebell H, Choi SH, Ryu KH, Canto T, Palukaitis P, Payne T, Walsh JA, Carr JP (2009) The role of the Cucumber Mosaic Virus 2b protein in viral movement and symptom induction. Mol Plant Microbe Interact 22: 642–654 [DOI] [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Hanisch S, Meffert K, Buch-Pedersen MJ, Jakobsen MK, Pomorski TG, Palmgren MG (2010) Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA α-subunit. Mol Biol Cell 21: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués RL, Poulsen LR, Palmgren MG (2012) A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a β-subunit. PLoS One 7: e33042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palukaitis P, García-Arenal F (2003) Cucumoviruses. Adv Virus Res 62: 241–323 [DOI] [PubMed] [Google Scholar]
- Parent JS, Bouteiller N, Elmayan T, Vaucheret H (2015) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81: 223–232 [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soards AJ, Murphy AM, Palukaitis P, Carr JP (2002) Virulence and differential local and systemic spread of Cucumber Mosaic Virus in tobacco are affected by the CMV 2b protein. Mol Plant Microbe Interact 15: 647–653 [DOI] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Biol Rep 20: 107–114 [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Wang A. (2015) Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW (2010) RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wu Z, Li Y, Wu J (2015) Biogenesis, function, and applications of virus-derived small RNAs in plants. Front Microbiol 6: 1237 10.3389/fmicb.2015.01237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gao H, Xu G, Wu D, Song S, Jiang H, Zhu S, Qi T, Xie D (2017) Arabidopsis ALA1 and ALA2 mediate RNAi-based antiviral immunity. Front Plant Sci 8: 422 10.3389/fpls.2017.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell H, Carr JP (2009) Effects of Dicer-like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by Cucumber Mosaic Virus and a mutant virus lacking the 2b counter-defence protein gene. J Gen Virol 90: 2288–2292 [DOI] [PubMed] [Google Scholar]