Tomato CALCINEURIN B-LIKE PROTEIN 10 (SlCBL10) ensures plant growth by regulating proper distribution of Na+ and Ca2+ in the shoot apical meristem and developing organs under salt stress.
Abstract
Characterization of a new tomato (Solanum lycopersicum) T-DNA mutant allowed for the isolation of the CALCINEURIN B-LIKE PROTEIN 10 (SlCBL10) gene whose lack of function was responsible for the severe alterations observed in the shoot apex and reproductive organs under salinity conditions. Physiological studies proved that SlCBL10 gene is required to maintain a proper low Na+/Ca2+ ratio in growing tissues allowing tomato growth under salt stress. Expression analysis of the main responsible genes for Na+ compartmentalization (i.e. Na+/H+ EXCHANGERs, SALT OVERLY SENSITIVE, HIGH-AFFINITY K+ TRANSPORTER 1;2, H+-pyrophosphatase AVP1 [SlAVP1] and V-ATPase [SlVHA-A1]) supported a reduced capacity to accumulate Na+ in Slcbl10 mutant leaves, which resulted in a lower uploading of Na+ from xylem, allowing the toxic ion to reach apex and flowers. Likewise, the tomato CATION EXCHANGER 1 and TWO-PORE CHANNEL 1 (SlTPC1), key genes for Ca2+ fluxes to the vacuole, showed abnormal expression in Slcbl10 plants indicating an impaired Ca2+ release from vacuole. Additionally, complementation assay revealed that SlCBL10 is a true ortholog of the Arabidopsis (Arabidopsis thaliana) CBL10 gene, supporting that the essential function of CBL10 is conserved in Arabidopsis and tomato. Together, the findings obtained in this study provide new insights into the function of SlCBL10 in salt stress tolerance. Thus, it is proposed that SlCBL10 mediates salt tolerance by regulating Na+ and Ca2+ fluxes in the vacuole, cooperating with the vacuolar cation channel SlTPC1 and the two vacuolar H+-pumps, SlAVP1 and SlVHA-A1, which in turn are revealed as potential targets of SlCBL10.
The development of crop plants tolerant to abiotic stress is crucial to meet the growing food demand through sustainable agriculture. Along their life cycle, plants need to balance development and adaptive responses to unfavorable conditions, salinity being one of the most severe factors limiting the productivity of agricultural crops (Flowers et al., 2010; Shabala, 2013). Significant advances have been made in the study of genes involved in salt stress tolerance and ion homeostasis (Maathuis, 2014), especially in the model species Arabidopsis (Arabidopsis thaliana), whereas knowledge about species of agronomic interest such as tomato (Solanum lycopersicum) remains scarce. Salt tolerance is determined by the ability of the plant to regulate Na+ transport rate from the root to the shoot through the xylem and by the capacity to accumulate Na+ ion into the vacuoles of the adult leaves and stem, which allows the plants to protect young developing tissues from Na+ toxicity (Shabala, 2013; Maathuis, 2014). Na+ efflux is mediated by the plasma membrane Na+/H+ antiporter SALT OVERLY SENSITIVE 1 (SOS1; Hasegawa, 2013), whereas HIGH-AFFINITY K+ TRANSPORTER (HKT) proteins, particularly those belonging to class I (Platten et al., 2006), are critical determinants of Na+ unloading from xylem vessels to other cells in the stele (Hasegawa, 2013). In tomato, two HKT1-like isoforms have been identified, SlHKT1;1 and SlHKT1;2, which underlie a major tomato quantitative trait locus for Na+/K+ homeostasis (Asins et al., 2013). Mainly, SlHKT1;2 has been involved in the regulation of Na+ movement from root to shoot through xylem and therefore in the Na+ concentration in leaves under saline conditions (Almeida et al., 2014; Asins et al., 2015). Compartmentalization in the vacuole of Na+ ions is an effective mechanism to avoid the toxic effects of Na+ in the cytoplasm (Maathuis, 2014). The transport of Na+ from the cytoplasm into the vacuole occurs via tonoplast Na+/H+ EXCHANGERs (NHXs). Four NHX isoforms have been identified in tomato; among them, LeNHX3 and LeNHX4 show the strongest induction upon salinity (Gálvez et al., 2012). In addition, LeNHX3 has been associated with a quantitative trait locus for Na+ concentration in leaves (Villalta et al., 2008). In Arabidopsis, the Na+ compartmentalization process mediated by vacuolar Na+/H+ antiporters is driven by the electrochemical gradient of protons across the tonoplast generated by the vacuolar H+ pumps, H+- pyrophosphatase AVP1 and V-ATPase (Maeshima, 2000; Hasegawa, 2013). Two full-length cDNA clones (SlVHA-A1 and SlVHA-A2) coding for two isoforms of the V-ATPase catalytic subunit (V-ATPases A1 and A2) have been isolated in tomato. In response to salinity, the abundance of the SlVHA-A1 transcript in leaves was nearly doubled with respect to control conditions, while SlVHA-A2 did not change and was mostly expressed in roots (Bageshwar et al., 2005).
Salt tolerance in plants also required a proper balance of Ca2+ and Na+ ions (Manaa et al., 2013). Thus, it has been well documented that Ca2+ has a direct inhibitory effect on Na+ entry into the cell by decreasing Na+ influx through nonselective cation channels and acting as a counter-cation inside storage organelles (Shabala et al., 2005). However, a Ca2+ deficit situation can occur in plants growing under salinity, since the elevated concentration of Na+ hinders Ca2+ uptake by roots (Zhai et al., 2015). The large central vacuole of a typical mature cell is by far the largest intracellular Ca2+ storage in plants; therefore, the mobilization of Ca2+ vacuolar reservoirs by the plant in this unfavorable situation is crucial to maintain the growth of young tissues (Bonales-Alatorre et al., 2013). A steady-state level of vacuolar Ca2+ depends on the balance between active Ca2+ import to vacuoles and vacuolar channels mediating Ca2+-induced Ca2+ release (Conn et al., 2011). CATION EXCHANGER (CAX) are ion transporters located on the tonoplast membrane (Hirschi, 1999; Manohar et al., 2011), and several studies in Arabidopsis have suggested they play a critical role in plant adaptation to certain stresses such as salinity (Cheng et al., 2003; Park et al., 2005). These antiporters also use the driving force of the proton gradient generated by the vacuolar pumps (V-ATPase and AVP1) to accumulate Na+ into the vacuole against its electrochemical gradient. In addition, the TWO-PORE CHANNEL 1 (TPC1) gene encodes for most prominent cation Slow Vacuolar channel, which represents the major cation conductance of the largest organelle in most plant cells (Kintzer and Stroud, 2016), mainly Ca2+, but also other ions such as K+ and Na+ (Hedrich and Marten, 2011). In Arabidopsis, it has been proven that TPC1 mediates a voltage-activated Ca2+ influx in leaf cells (Furuichi et al., 2001), contributing to the cytosolic calcium elevation and therefore to stress signaling (Hedrich and Marten, 2011; Choi et al., 2014).
CALCINEURIN B-LIKE PROTEIN 10 (CBL10), the last CBL family member to be identified so far, has also been involved in the regulation of salt stress response in Arabidopsis (Kim et al., 2007; Quan et al., 2007). CBLs are EF-hand Ca2+ protein sensors and upon Ca2+ binding, they undergo conformational changes to associate with a group of CBL-INTERACTING PROTEIN KINASES (CIPKs; for review, see Kolukisaoglu et al., 2004; Luan, 2009; Kim, 2013). Different combinations of CBLs and CIPKs complexes may generate temporal and spatial specificity in Ca2+ signaling, integrating various stimuli to determine cellular responses (Batistic et al., 2010). Previous studies have determined that CBL10 interacts and recruits CIPK24 (SOS2) toward the tonoplast, speculating that the CBL10-CIPK24 complex might phosphorylate and activate a tonoplast Na+ channel or transporter yet unknown to transport cytosolic Na+ into the vacuole (Kim et al., 2007; Quan et al., 2007; Waadt et al., 2008; Lin et al., 2009). Moreover, a recent study has also demonstrated that Arabidopsis CBL10 is critical for reproductive development under salt stress, although this function occurs independently from SOS2 interaction (Monihan et al., 2016). Likewise, Kang and Nam (2016) have provided an additional explanation for the positive role of CBL10 in salt tolerance by regulating sensitivity to brassinosteroids. Following the initial discovery of CBL10 in Arabidopsis, a homolog has been reported in Populus and attributed similar functions (Tang et al., 2014). A CBL10 homolog has also been reported in tomato, and its function in pathogen response within the reactive oxygen species signaling pathway has been demonstrated (de la Torre et al., 2013). However, the role for SlCBL10 in the regulation of abiotic stress responses in tomato remains unexplored. Furthermore, the relationship of the CBL10 gene with other genes involved in the regulation of ion homeostasis (SOS1, HKT1s, AVP1, VHA-A1, and TPC1) has not been established so far in any species. This study reports the identification of the tomato Slcbl10 knock-out mutant, which exhibited very high salt sensitivity. The functional characterization of this mutant revealed a new role of the SlCBL10 gene in the salt tolerance of tomato by balancing Na+ and Ca2+ homeostasis.
RESULTS
Isolation of the pms916 Salt Hypersensitive T-DNA Mutant and Molecular Cloning of the Tagged Gene
An in vitro phenotypic screening to identify mutants with altered salt stress responses was performed in a tomato T-DNA mutant collection by growing T2 segregating families in a basal culture medium (SCM) supplemented with 100 mm NaCl (for details, see “Materials and Methods”). As a result, the pms916 (protecting meristem from salt stress 916) mutant was isolated, which exhibited abnormal thickening of the hypocotyl, severe inhibition of vegetative development, and a collapse of the apical meristem after 20 d of salt treatment (20 DST; Fig. 1A). A similar mutant phenotype was observed in three additional repeated assays (Experiments E1, E2, and E3 in Supplemental Table S1) performed using identical salt stress conditions. Subsequently, the salt sensitivity of the pms916 mutant was corroborated in vivo by growing T2 plants under greenhouse conditions (Experiments E4 and E5 in Supplemental Table S1). In all the in vitro and in vivo experiments, genetic analysis indicated that pms916 mutation was inherited as monogenic and recessive (Supplemental Table S1).
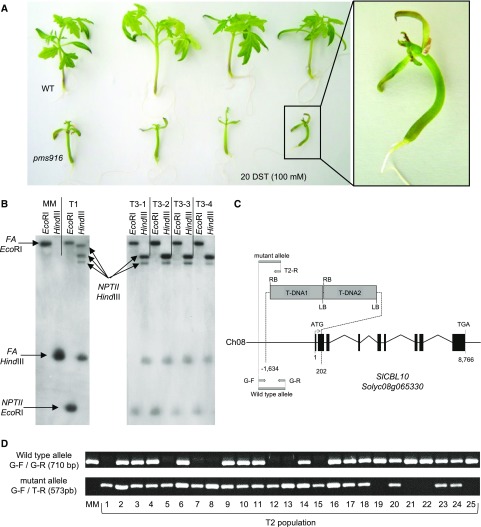
Figure 1.
Molecular cloning of pms916 salt hypersensitive T-DNA mutant. A, Phenotype of pms916 mutant plants grown in vitro after 20 DST of 100 mm NaCl. B, Southern-blot analysis using a chimeric probe that includes the complete coding sequence of the NPTII gene fused to 811 pb of the coding sequence of FA gene (used as hybridization positive control). C, Genomic organization of the SlCBL10 gene and the two T-DNA copies inserted in a head-to-tail tandem orientation in the pms916 mutant. The tandem T-DNA insertion resulted in a 1,836-bp deletion between −1,634 and 202 bp in the SlCBL10 gene. Number 1 indicates the translation start site, and 8,766 indicates the last nucleotide of the coding region. Exons are depicted as black boxes; the lines between boxes are introns. The dotted lines indicate the position where the insertion is located. The gray arrows indicate the primers used for genotyping the T2 population. G-F and G-R: specific genomic forward and reverse primers, respectively, used to amplify the wild-type allele (without T-DNA insertion). G-F and T2-R: specific genomic forward and specific T-DNA border primers, respectively, used to amplify the mutant allele (carrying the T-DNA insertion). D, Genotyping of T2 individuals. All T2 plants homozygous for the mutant allele (1, 5, 7, 8, 12, 13, and 15) displayed pms916 mutant phenotype, while T2 plants heterozygous (2, 3, 4, 6, 9, 10, 11, 14, 16, 17, 18, 20, 23, and 24) and homozygous for the wild-type allele (19, 21, 22, and 25) showed wild-type phenotype.
Southern-blot analysis showed that the original T1 plant carrying the pms916 mutation harbored three T-DNA copies (Fig. 1B). To identify lines with a single T-DNA insertion, T3 progenies were used for the kanamycin sensitivity test. The results showed a 3:1 (tolerant:sensitive) segregation for kanamycin response in 4 of the 10 progenies evaluated, indicating a single-locus insertion of the T-DNA. These four T3 progenies were grown in vitro under identical salt conditions as described above (SCM + 100 mm NaCl). From each T3 progeny, a single plant showing a pms916 mutant phenotype was analyzed by Southern blotting. The results indicated that two T-DNA insertions were present in all mutant plants (Fig. 1B). Genomic regions flanking T-DNA insertions were cloned by anchor-PCR, and the sequencing analysis revealed that the two T-DNA copies were inserted in a head-to-tail tandem orientation (Fig. 1C). Hence, both T-DNA copies were inherited as a single locus, which agreed with the results of the kanamycin sensitivity test. The T-DNA tandem insertion was located on chromosome 8 of the tomato genome, within the CALCINEURIN B-LIKE PROTEIN 10 (SlCBL10) gene (Solyc08g065330). The inserted fragment caused a deletion of 1,836 bp between 1,634 bp upstream and 202 bp downstream of the translation start codon of the SlCBL10 gene (Fig. 1C), preventing, in all likelihood, the translation of a functional protein.
A PCR cosegregation analysis was then carried out to determine whether the T-DNA insertion correlated with the mutant phenotype. For this purpose, the SlCBL10 genotype of 25 T2 individuals was determined using G-F, G-R, and T2-R primer combinations (Fig. 1, C and D; Supplemental Table S2). Among the 18 wild-type individuals, 14 plants were heterozygous and 4 homozygous for the wild-type allele, while the remaining 7 mutant plants carried the mutant allele in the homozygous state (Fig. 1D). These results strongly supported that the disruption of the SlCBL10 gene by the T-DNA insertion was responsible for the pms916 mutant phenotype and therefore, the tomato pms916 insertional mutant was renamed as Slcbl10.
Phenotypic Characterization of Slcbl10 Mutant Plants
To more deeply characterize the Slcbl10 mutant phenotype, T3 homozygous and azygous lines for the Slcbl10 mutation, both belonging to the same T2 family, were grown in a hydroponic system as described in the “Materials and Methods” section. In the absence of salt stress, Slcbl10 mutant plants grown normally with the only exception of a slight chlorosis at the margins of some young leaflets at the moment when plants have developed a few number of leaves. The sensitive phenotype was accentuated as mutant plants grew, showing bulging and thickening of the leaflets forming the shoot apex (Supplemental Fig. S1).
Short-term hydroponic salt treatment (HST) assays showed that salinity caused severe damages in the aerial part of Slcbl10 mutant plants in both young and adult plants (for further details, see “Materials and Methods”). Vegetative growth of Slcbl10 homozygous plants was arrested at young stages (HSTy assay), and they showed swelling and curved appearance of leaves, chlorosis at edge of leaflet, and apical collapse (Fig. 2A). Salt treatment also induced growing abnormalities in Slcbl10 adult plants (HSTa assay). Thus, after 2 DST, mainly young apical tissues became burnt and wilted, and subsequently mutant plants stopped growing as a consequence of apical collapse after just 6 DST (Fig. 2B). However, it was surprising that leaves and stems at basal positions on Slcbl10 mutant plants did not show evident symptoms of salt sensitivity. Instead, their external appearances were similar to that of wild-type plants after 6 DTS (Fig. 2B), indicating that in a first instance, loss of SlCBL10 function mainly affects shoot apex and growing tissues although finally the whole plant is affected and dies from apical collapse.
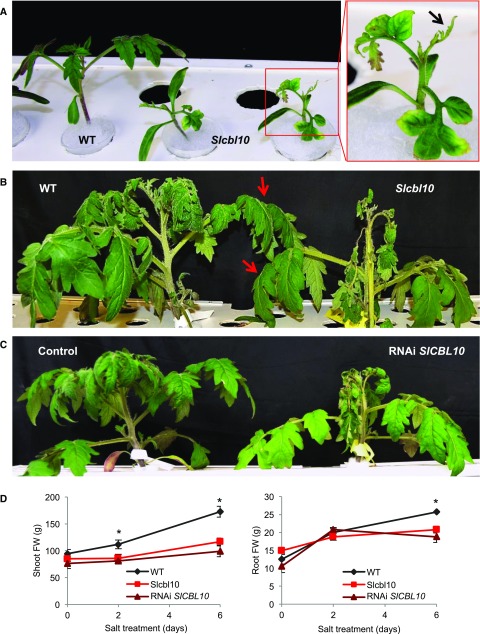
Figure 2.
SlCBL10 protects the tomato shoot apex and young tissues from salt stress conditions. A, Wild-type and Slcbl10 mutant plants grown in a hydroponic system and salt-treated at cotyledon development stage (HSTy assay). Framed in red a Slcbl10 mutant plant severely damaged by salinity, particularly in the shot apex (arrow). B, Phenotype of adult wild-type and Slcbl10 mutant plants grown in a hydroponic system and salt treated at the fifth fully developed leaf stage (HSTa assay). Note that shoot apex and young tissues of mutant plants are severely affected by salinity while adult leaves display a similar appearance to wild-type (pointed with red arrows). C, Phenotype of RNAi SlCBL10 and control plants subjected to hydroponic salt treatment (HSTa assay conditions). Note that RNAi SlCBL10 plant phenocopies the mutant phenotype under saline conditions. D, Changes in shoot and root fresh weights of wild-type, Slcbl10 mutant, and RNAi SlCBL10 plants during salt treatment. Values are the mean ± se of two independent assays, each with three biological replicates. Asterisks indicate significant differences (Student’s t test, P < 0.05).
To corroborate that the salt hypersensitivity phenotype of the Slcbl10 mutant was due to the loss of SlCBL10 gene function, two clonal replicates of 14 independent RNA intereference (RNAi) SlCBL10 lines and 13 independent regenerants (these latter obtained under the same conditions except for the use of the RNAi gene construct [control plants]) were grown and characterized under hydroponic salt treatment (HSTa assay conditions). All RNAi SlCBL10 lines displayed salt stress sensitivity, showing an altered phenotype almost equal to that of mutant plants (Fig. 2C), including decreased fresh weight of shoots and roots (Fig. 2D). These results confirmed that the T-DNA mutation affecting the SlCBL10 gene was responsible for the salt-hypersensitivity initially observed in the pms916 mutant.
Functional Complementation of the Arabidopsis cbl10 Mutant Line
To test whether the SlCBL10 gene is an ortholog of the Arabidopsis CBl10, the entire open reading frame corresponding to the putative SlCBL10 was cloned under the control of a 35S promoter. This construct was used to transform and test a functional complementation of the Arabidopsis cbl10 knock-out T-DNA line (Quan et al., 2007). Salt tolerance of three independent transgenic lines overexpressing SlCBL10 in a cbl10 genetic background was compared to that of wild-type (Col-0) and the cbl10 mutant plants (Fig. 3A). While growth in 100 mm NaCl is more severely compromised in the cbl10 mutant than in wild-type plants, heterologous expression of SlCBL10 in this mutant genetic background restores growth to wild-type levels (Fig. 3B). Previous studies have shown that despite being hypersensitive to salinity, the Arabidopsis cbl10 mutant accumulated less Na+ after high salt exposure (Kim et al., 2007). Accordingly, results here obtained demonstrated that the expression of SlCBL10 restored Na+ accumulation to wild-type levels in cbl10 mutant plants (Fig. 3C). In conclusion, SlCBL10 was able to reiterate CBL10 function in Arabidopsis, indicating that SlCBL10 is a true ortholog of the CBL10 gene.
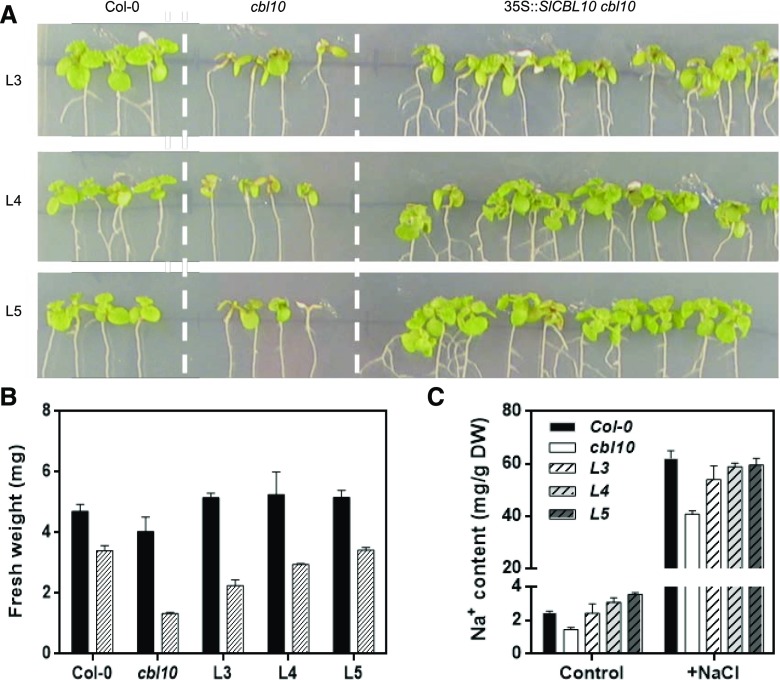
Figure 3.
Ectopic SlCBL10 expression restores salt tolerance in the Arabidopsis cbl10 mutant. Five-day-old seedlings of wild-type (Col-0), cbl10 mutant, and three transgenic lines (L3-5) overexpressing SlCBL10 in an Arabidopsis cbl10 mutant genetic background were transferred to MS supplemented with 100 mm NaCl. A, Representative plants 10 d after transfer. B, Fresh weight per seedling. Black bars, without NaCl treatment; hatched bars 100 mm NaCl treatment. C, Sodium accumulation in plants of the indicated genotypes after 10 d.
The SlCBL10 Gene Is Differentially Expressed in Tomato Tissues and Is Induced by Salt Stress
Changes of SlCBL10 expression induced by salt stress were further analyzed in adult wild-type plants grown in a hydroponic system (HSTa assay conditions). Salinity induced an increase of SlCBL10 expression in all tissues analyzed (Fig. 4A). In shoot, the highest expression levels were detected in upper adult leaves (3-fold increase), followed by young leaves and stems (2-fold increase). The lowest SlCBL10 expression was found in roots, although a significant increase was also detected in salt-treated plants.
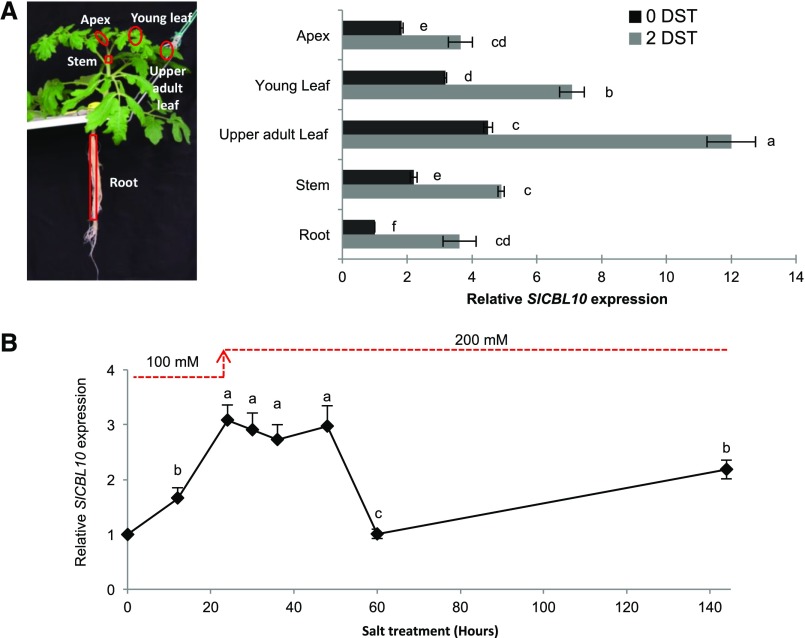
Figure 4.
Expression pattern of SlCBL10 gene in wild-type plants under salt stress conditions. Wild-type plants grown in a hydroponic system and salt-treated at the fifth fully developed leaf stage (HSTa assay). A, Levels of SlCBL10 transcripts were quantified by RT-qPCR in apex, young leaf, upper adult leaf (first fully developed leaf), stem (first internode), and root of wild-type plants developed in absence of salt (0 DST) and after 2 DST. B, Time-course analysis of SlCBL10 gene expression during 6 d of salt treatment (144 h) was analyzed in upper adult leaf. Values are the mean ± se of two independent assays, each with three biological replicates. Different lowercase letters indicate significant differences determined by ANOVA (P < 0.05).
The temporal effect of salt stress on SlCBL10 expression was analyzed in the first developed leaf of wild-type plants from the same experiment mentioned above (Fig. 4B). Transcript levels of SlCBL10 were significantly induced by salt treatment after 12 h, reaching the maximum levels between 24 and 48 h. Later, SlCBL10 expression decreased to the basal levels found in the absence of salt stress after 60 h of treatment. At 6 DST, a further increase of SlCBL10 expression was observed, therefore suggesting that two phases of induction may occur during exposure to salt stress.
Na+ Distribution Pattern Is Altered in Slcbl10 Mutant Plants
To ascertain whether the salt sensitivity phenotype was associated with changes in Na+ distribution patterns induced by salt stress, ion contents were analyzed in several adult plant tissues from the HSTa assay (Fig. 5A). Results showed that although Na+ uptake at whole level was lower in Slcbl10 mutant than in wild-type plants (25% and 20% lower in mutant after 2 DST and 6 DST, respectively), the Na+ distribution pattern along the mutant plant was completely altered. In wild-type plants, Na+ is preferentially accumulated in roots and later in adult leaves and stems to prevent Na+ from reaching toxic levels in young developing tissues. In fact, the lowest Na+ accumulation was found in the shoot apex of wild-type plants in salt conditions. Contrarily, the Slcbl10 mutant did not retain Na+ properly in roots or in adult leaves or stems as 30% less Na+ is detected in these tissues, and therefore similar Na+ contents were found along the mutant plants. The incapacity of the Slcbl10 mutant to retain Na+ in adult vegetative tissues allows the ion to reach the apex in higher concentration, 70% higher than in wild type, after 6 DST.
Figure 5.
The salt-hypersensitivity phenotype of Slcbl10 mutant is associated with an altered Na+ long-distance distribution promoted by an impaired capacity to compartmentalize Na+ into leaf vacuole. Wild-type and Slcbl10 mutant plants were cultivated in a hydroponic system under salt conditions (HSTa assay). A, Na+ content and (B) Na+/K+ ratio were analyzed in apex, upper adult leaves (first fully developed leaf), stem (first internode), and root after 2 and 6 DST. C, Relative expression of key genes involved in long-distance Na+ distribution (SlSOS2, SlSOS1, and SlHKT1;2) and in Na+ compartmentalization into the vacuole (SlNHX3, SlNHX4, SlAV1, and SlVHA-A1) was analyzed in upper adult leaves at 0 DST and after 2 DST. Values are the mean ± se of two independent assays, each with three biological replicates. Asterisks indicate significant differences (Student’s t test, P < 0.05).
An opposite tendency was observed for K+ distribution. Thus, during salt treatment, the K+ content was higher in roots, upper adult leaves, and stems of mutant plants than in wild-type plants, while a similar K+ content was detected in the apex (Supplemental Fig. S2). This fact promoted that the Na+/K+ ratio was lower in roots and upper adult leaves of the Slcbl10 mutant but higher in the apex due to a higher Na+ accumulation in these tissues (Fig. 5B). Together, these results strongly suggest that salt stress induces significant alterations in Na+ and K+ homeostasis of Slcbl10 mutant plants.
Transcript levels of main genes involved in Na+ homeostasis were analyzed in the first developed leaf and roots (Fig. 5C). With this purpose, expression of genes responsible for the uptake and long-distance Na+ transport in tomato, SlSOS1, and SlHKT1s (Olías et al., 2009; Asins et al., 2013), as well as genes involved in Na+ accumulation into vacuole (LeNHX3, LeNHX4 [Gálvez et al., 2012], SlAVP1 and SlVHA-A1 genes [Gaxiola et al., 2007]) were determined. In addition, expression of SlSOS2 gene, which has been related to both long-distance Na+ transport and Na+ accumulation process (Huertas et al., 2013; Olías et al., 2009), was also analyzed. Although no differences in the expression levels of these genes were detected between Slcbl10 mutant and wild-type plants when they were grown under control conditions, some significant changes were found when plants grew under salt stress, mainly affecting leaf tissue (Fig. 5C). Salinity induced up-regulation of all analyzed genes involved in Na+ compartmentalization, that is LeNHX3, LeNHX4, SlSOS2, SlAVP1, and SlVHA-A1, in wild type but not in mutant leaves where only LeNHX3 and SlVHA-A1 were up-regulated. Furthermore, the SlHTK1;2 gene was significantly down-regulated in leaves of mutant plants. In roots, the only remarkable difference was the induction by salinity of SlSOS1 expression in mutant but not in wild-type plants, which indicated that Na+ extrusion from root to external medium was increased when SlCBL10 expression is knocked-out (Supplemental Fig. S3).
Disruption of SlCBL10 Gene Affects Ca2+ Homeostasis under Salt Stress
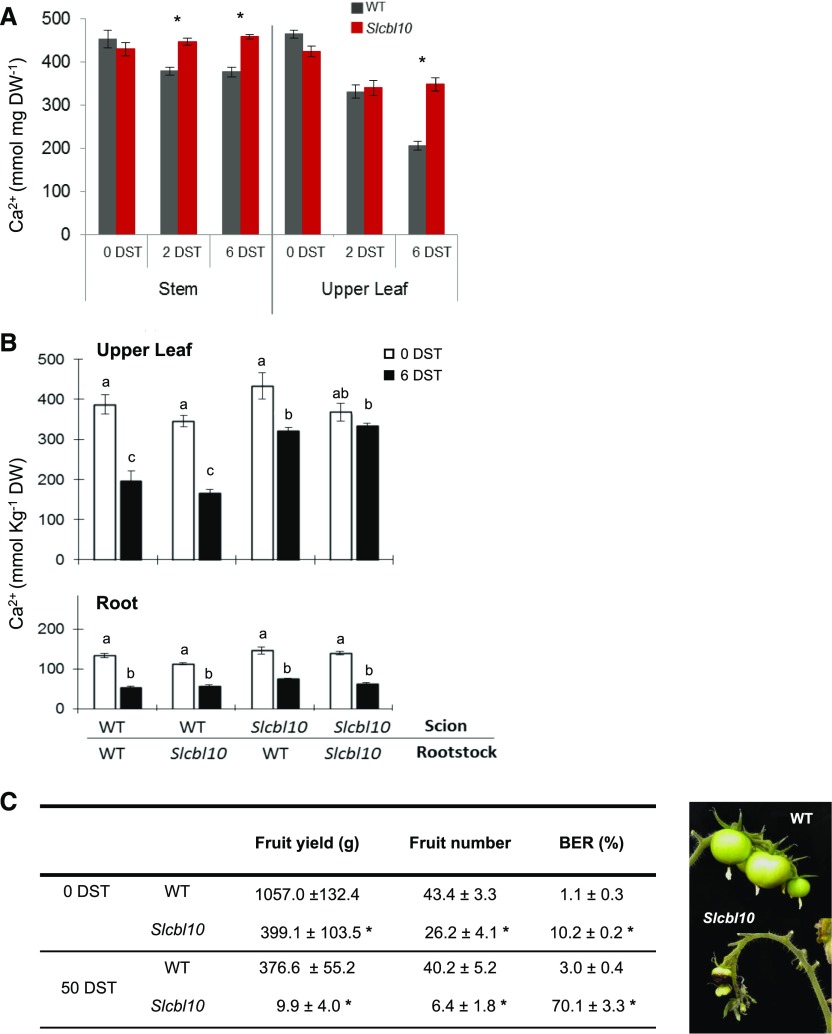
Another singular feature of Slcbl10 mutant extracted from ion analysis was an altered Ca2+ distribution pattern under salinity (Fig. 6A). Thus, while the hydroponic salt treatment (HSTa assay) caused a significant reduction of Ca2+ content in both stems and upper adult leaves of wild-type plants after 6 DST, no significant changes of Ca2+ content were observed in Slcbl10 mutant plants. Consequently, Ca2+ content was 80% and 60% higher in stem and upper adult leaves, respectively, in Slcbl10 than in wild-type plants.
Figure 6.
SlCBL10 disruption promoted the retention of Ca2+ in leaf and stem under salinity conditions. A, Wild-type and Slcbl10 mutant plants grown in a hydroponic system under salt conditions (HSTa assay). Ca2+ content was analyzed in stem (first internode) and in the first developed leaf prior to salt treatment (0 DST) and after 2 and 6 DST. B, Grafted plants between wild-type and Slcbl10 mutant were subjected to hydroponic salt treatment (HSTa assay conditions). Ca2+ content was analyzed in the first developed leaf and in root at 0 DST and 6 DST. C, Fruit yield, fruit number, and BER incidence in wild-type and mutant plants at 0 DST and after 50 DST. Values are the mean ± se of three biological replicates of five plants each. Asterisks indicate significant differences between wild type and Slcbl10 mutant (Student’s t test, P < 0.05). Different lowercase letters indicate significant differences in each tissue (root or leaf) determined by ANOVA (P < 0.05).
The higher Ca2+ levels detected in upper adult leaves and stem of Slcbl10 mutant plants could be due to Ca2+ retention or to an increased Ca2+ transport from roots to shoots during salt treatment. To check these two possibilities, reciprocal grafting experiments were performed between wild-type and Slcbl10 plants, and Ca2+ contents were measured in grafted plants grown under hydroponic salt treatment (HSTa assay conditions; Fig. 6B). The level of Ca2+ was significantly higher in leaves of grafted plants using Slcbl10 as scion independently of the rootstock background, while a similar decrease of Ca2+ content was found in leaves when wild-type scion was grafted on either wild-type or mutant rootstock. In addition, no significant differences of Ca2+ content were found in roots of the different combinations. Therefore, it is possible to conclude that the higher Ca2+ content found in Slcbl10 mutant organs was not due to a higher Ca2+ transport from root to shoot but, most likely, to retention in leaves of Ca2+ stores, which did not diminish under salinity.
Under calcium-deficit conditions, such as those promoted by salinity (White and Broadley, 2003), plants need to mobilize their calcium reservoirs to ensure fruit development and yield. Therefore, possible effects on fruit yield promoted by Ca2+ reservoirs improperly retained in leaves and stems of Slcbl110 plants were analyzed in a long-term salt treatment assay in a greenhouse (GST assay conditions; see “Materials and Methods”). Fruit yield and blossom end rot incidence, the latter being a well- known symptom of Ca2+ deficiency disorders in tomato fruit (Tonetto de Freitas et al., 2014; Zhai et al., 2015), were determined in adult plants after 50 DST (Fig. 6C). A considerable decrease of fruit production and higher blossom end rot incidence (7-fold) was detected in Slcbl10 plants with respect to wild type. In addition, other phenotypic alterations that coincided with those promoted by Ca2+ deficiency were observed in long-term salt-treated Slcbl10 and RNAi SlCBL10 plants, among others, reduced growth of the apical meristem, small leaves with evident chlorosis symptoms at their leaf edges, and thickened petioles and stems (Supplemental Figs. S4 and S5). Taken together, these results indicated that under salinity conditions, Ca2+ reservoirs are less available in Slcbl10 plants than in wild type, thus limiting their proper development and productivity.
To corroborate that the low Ca2+ availability is responsible for the growth restriction of Slcbl10 mutant in salt stress conditions, an in vitro assay was carried out using 1 mm suboptimal Ca2+ concentration. Under these conditions, the Slcbl10 mutant showed higher sensitivity to a Ca2+ deficiency condition (Supplemental Fig. S6, A and B). The first Ca2+ deficiency symptom observed was a collapse of the subapical shoot region resulting in a constriction necrosis below the shoot tip, followed by the apical meristem senescence. Light microscopic analysis also revealed significant alterations affecting SAM morphology and ground cells of Slcbl10 mutant plants (Supplemental Fig. S6, C and D). In addition, swollen cells were observed in the submeristematic region immediately below the collapsed zone leading to thickened stems (Supplemental Fig. S6E) whose appearance strongly resembled that observed in Slcbl10 young plants grown under salt conditions (Fig. 2). These results strongly support that tomato plants lacking SlCBL10 are not able to balance their development under Ca2+ deficient conditions.
Expression Pattern of Key Genes Involved in Vacuolar Fluxes of Ca2+
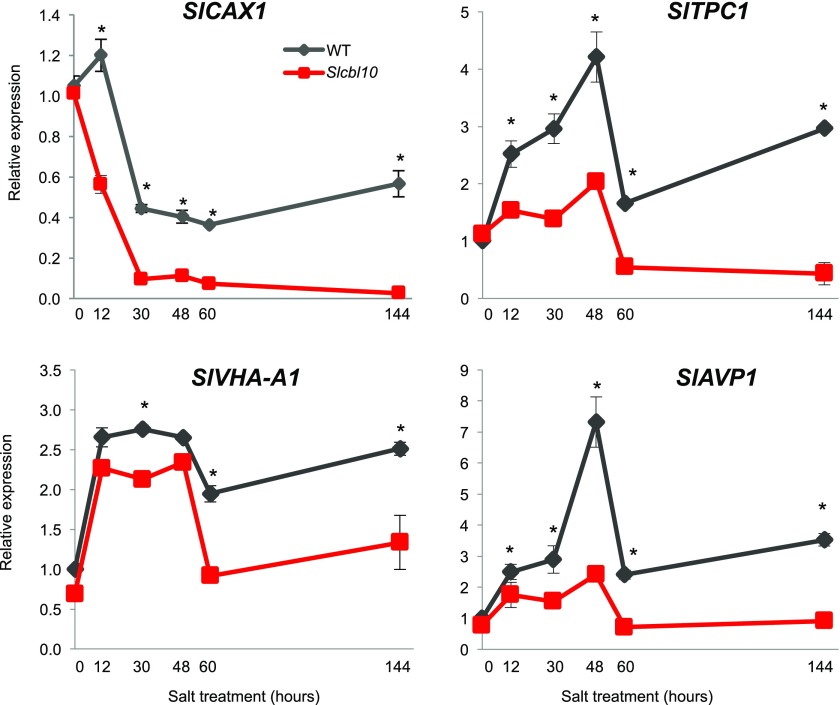
To ascertain how SlCBL10 might be involved in the mobilization of Ca2+ stores, and taking into account that the vacuole is by far the largest intracellular Ca2+ store in mature cells (Peiter, 2011), the expression profile of key genes involved in Ca2+ fluxes were analyzed. Concretely, transcript accumulation of CAX1, AVP1, and VHA-A1 genes, all required for Ca2+ compartmentalization into the vacuole (Pittman, et al., 2009), and TPC1 involved in Ca2+ release from vacuole (Hedrich and Marten, 2011), were measured in upper adult leaves of Slcbl10 mutant and wild-type plants grown under HSTa assay conditions. Given the essential role of Ca2+ as a second messenger of signaling stress, gene expression analyses were performed at early and later steps of salt treatment (Fig. 7). In wild-type plants, salinity induced a down-regulation of SlCAX1, while SlTPC1, the main responsible channel for the release of Ca2+ from vacuole, was up-regulated. The lowest levels of SlCAX1 transcripts were detected after 48 h of salt treatment, which remained low until the end of treatment. SlTPC1 registered two maximum levels of expression, one soon after 48 h and the other later, at the end of salt treatment. In Slcbl10 mutant plants, expression of SlTPC1 was not induced by salinity, but it was down-regulated (65% reduction) after 48 h of salt exposure. Moreover, SlCAX1 expression was more pronounced and earlier repressed in Slcbl10 mutant than in wild-type plants. Indeed, after 60 h of salt exposure, the level of SlCAX1 transcripts was almost null in the mutant (98% of inhibition). The expression pattern of these genes in wild type indicated that salinity induced an efflux of Ca2+ from vacuole, while in mutants this efflux may be impaired by the strong down-regulation of TPC1.
Figure 7.
SlCBL10 disruption alters the influx and efflux of Ca2+ in vacuole. Wild-type and Slcbl10 mutant plants grown in a hydroponic system under salt conditions (HSTa assay). Relative gene expression of SlCAX1, SlTPC1, SlAVP1, and SlVHA-A1 was recorded in upper adult leaf of wild type and Slcbl10 mutant during 6 d of salt stress. Values are the mean ± se of two independent assays, each with three biological replicates. Asterisks indicate significant differences (Student’s t test, P < 0.05).
Regarding the vacuolar proton-pumps coded by SlAVP1 and SlVHA-A1 genes, salinity induced a similar profile expression to that described for SlTPC1 in wild-type plants, as a peak of induction was detected at 48 h of salt treatment (Fig. 7). However, the level of transcripts of both SlAVP1 and SlVHA-A1 genes were significantly lower in Slcbl10 mutant plants than in wild type. It is interesting to highlight that the expression patterns of SlTPC1 and the two vacuolar proton pumps, SlAVP1 and SlVHA-A1, followed a very similar temporal pattern expression, suggesting that they may cooperate to regulate proper Ca2+ flux in the vacuole under salt stress in tomato.
SlCBL10 Gene Is Needed to Maintain a Proper Na+/Ca2+ Ratio in Flowers and Apex under Salinity
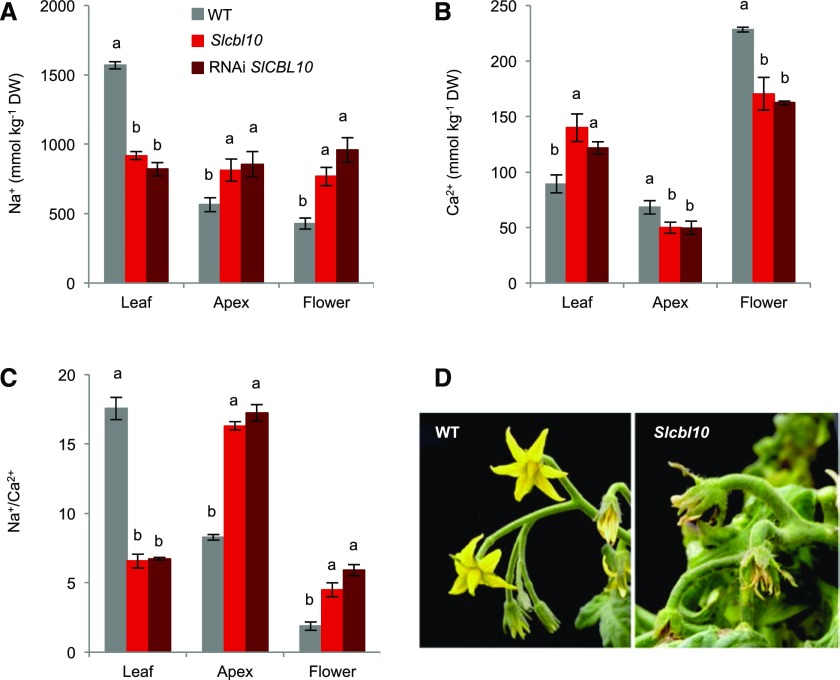
It is known that under salinity conditions, the maintenance of a proper Na+/Ca2+ low ratio in growing tissues, such as shoot apex and flowers, is essential to maintain plant growth (Manaa et al., 2013). Thus, to ascertain when the lack of SlCBL10 gene prevents the maintenance of a proper Na+/Ca2+ ratio that ensures the growth in tissues such as apex and flowers under salinity, Na+/Ca2+ ratio was assessed in wild-type, Slcbl10 mutant and RNAi SlCBL10 plants. After 6 DST, Na+ and Ca2+ contents were analyzed in upper adult leaf, apex, and flowers (first flower truss immediately below first fully developed leaf). In RNAi SlCBL10 and Slcbl10 mutant plants, a higher Ca2+ content but a lower Na+ content was registered in upper adult leaves compared to wild type (Fig. 8, A and B). Contrarily, in apex and flowers, Na+ levels increased and Ca2+ levels decreased both in Slcbl10 mutant and in RNAi plants (Fig. 8, A and B), causing the Na+/Ca2+ ratios in these tissues to be improperly high (Fig. 8C), which might be the ultimate responsible factor for the observed alterations in mutants cultivated under salinity.
Figure 8.
SlCBL10 gene is involved in maintaining a suitably low Na+/Ca2+ ratio in tomato apex and flower under salinity conditions. Wild-type, Slcbl10 mutant, and RNAi SlCBL10 plants grown in a hydroponic system and salt-treated at the fifth fully developed leaf stage (HSTa assay). Na+ (A) and Ca2+ contents (B) were analyzed in upper adult leaf, apex and flower after 6 DST, and then the Na+/Ca2+ ratio was calculated (C). Values are the mean ± se of two independent assays, each with three biological replicates. Different lowercase letters represent significant differences (P > 0.05) calculated by ANOVA. D, Representative images of wild-type and Slcbl10 mutant flowers after 10 DST.
DISCUSSION
This study reports the identification and functional characterization of the tomato Slcbl10 knock-out mutant identified from a screening of a T-DNA tomato mutant population (Pineda et al., 2012; Pérez-Martín et al., 2017). Through an anchor-PCR approach, SlCBl10 was identified as the tagged gene responsible for the salt hypersensitive mutant phenotype. Molecular complementation experiments proved that SlCBL10 is orthologous to the Arabidopsis CBL10 gene and encodes a calcium sensor CALCINEURIN B-LIKE PROTEIN 10. Although SlCBL10 was previously involved in a signaling pathway mediating tomato plant immunity (de la Torre et al., 2013), its role in regulating abiotic stress has not been studied so far in this model species.
SlCBL10 Protects Growing Tissues from Salt Stress by Na+ Retention in Adult Tissues
Results reported here indicate that the salt sensitivity phenotype of Slcbl10 mutant plants (i.e. growth inhibition, hypocotyl thickening, and apical necrosis) was similar both under in vitro and in vivo stress conditions and that such phenotype was also corroborated in RNAi SlCBL10 lines. In all cases, the lack of the SlCBL10 finally drove plants to die from apical collapse (Fig. 2). In addition, expression analysis showed that SlCBL10 was up-regulated in wild-type plants cultivated under salinity conditions, the highest expression level being detected in upper adult leaves close to the shoot apex. Together, these results indicate that transcriptional activity of SlCBL10 plays a key role in the adaptive response of tomato plants to salt stress by protecting shoot apical meristem and growing tissues from physiological damages caused by salinity.
Previous studies have shown that CBL10 is also involved in salt response in Arabidopsis (Kim et al., 2007; Quan et al., 2007) and Populus (Tang et al., 2014). Indeed, Arabidopsis cbl10 mutant plants accumulated lower Na+ and a higher K+ content as a consequence of salt treatment, being the first salt-hypersensitive mutant with a lower Na+/K+ ratio reported so far (Kim et al., 2007). Here it has been shown that tomato Slcbl10 mutant also accumulates lower Na+ and higher K+ than wild-type plants; in addition, functional complementation of the Arabidopsis cbl10 mutant by SlCBL10 leads to the recovery of Na+ levels. Under these premises, it is necessary to explain how plants accumulating less Na+, as is the case for Slcbl10 and cbl10 mutants, showed a salt-hypersensitive phenotype when the opposite would be expected. A precise dissection of Na+ relative content in young versus adult leaves as well as in leaves versus stems in tomato allowed for the elucidation of the role played by SlCBL10 in Na+ homeostasis. It is known that the mechanism of salinity tolerance in tomato involves a preferential Na+ accumulation in adult leaves and stems, which prevents Na+ from reaching the shoot apex (Cuartero et al., 2010). Our results proved that this physiological mechanism was totally altered in Slcbl10 mutant, which was not able to retain Na+ in adult tissues. This alteration leads to a 7-fold increase in Na+ content that reaches the shoot apex, which results in an inadequate high Na+/K+ ratio in growing tissues. Changes in Na+/K+ ratios are consistent with the physiological damages detected in the apical part of mutant plants and strongly support a functional role of the SlCBL10 gene in protecting shoot apex and developing tissues from salt stress conditions.
SlCBL10 Function Is Required for Na+ Compartmentalization into Vacuole
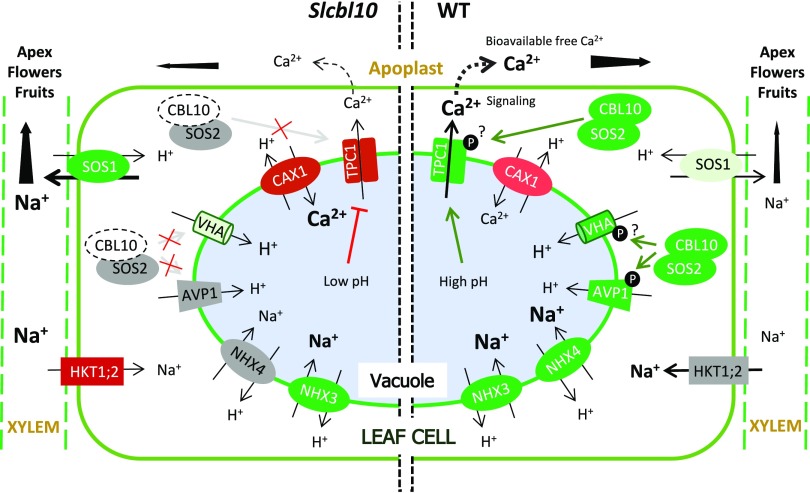
In the cytoplasm, inappropriate Na+ levels cause important metabolic alterations as this ion inhibits enzyme activity (Maathuis, 2009); for that reason, cytoplasmic Na+ content must be kept at low level by exporting Na+ into the vacuole (Albaladejo et al., 2017). Results from expression analysis revealed that the decreased capacity of Slcbl10 plants to retain Na+ in adult leaf was associated with a significant lower salt-induced expression of genes involved in Na+ compartmentalization into vacuole, such as LeNHX4, SlAVP1, SlVHA-A1, and SlSOS2. Moreover, in the leaves of Slcbl10 plants we observed both a reduced expression of SlHKT1;2, the main responsible for uploading Na+ from the xylem into the cells (Asins et al., 2013; 2015), as well as an increase of SlSOS1 gene expression, responsible for Na+ extrusion from leaf cells to xylem (Olías et al., 2009). Hence, expression analysis supports the hypothesis that Na+ compartmentalization into vacuoles as well as Na+ upload from xylem into cells were severely inhibited in Slcbl10 mutant plants, while Na+ extrusion from leaf cells to xylem was favored (Fig. 9). Such physiological changes allow the toxic ion to reach the apex and flowers promoting their collapse and the subsequent death of the plants. Therefore, results indicate that SlCBL10 is needed for regulating Na+ homeostasis through the activity of genes involved in the compartmentalization of Na+ into vacuole.
Figure 9.
Hypothetical model of the genetic and physiological mechanism proposed to explain the functional role of SlCBL10 gene in regulating Na+ and Ca2+ homeostasis under salt stress conditions. On the right side, it is indicated the activity of Na+ antiporters (SOS1, NHK, and KKT), Ca2+ antiporter (CAX), the vacuolar pumps (AVP1 and V-ATPase), and the cation vacuolar channel TPC1, all of them involved in maintaining Na+/Ca+2+ balance through the vacuolar transport. On the left side, the mechanism would be impaired due to the lack of SlCBL10 (see “Discussion” section for details). Red and green colors mean down-regulated and up-regulated genes under salt stress, respectively, while gray color represents absence of gene expression changes in salinity conditions. Different lightness of red and green colors indicates different levels of gene expression in Slcbl10 mutant (left) respect to wild-type plants (right) grown in salt conditions, where darker hues represent higher induction (green) or inhibition (red). The names of the proteins correspond to those of Arabidopsis thaliana, although results provided in this work indicate that, in general terms, this mechanism could be conserved in tomato.
In Arabidopsis, the CBL10-SOS2 complex has been proposed as a positive regulator of a still unknown vacuolar protein triggering Na+ compartmentalization into vacuoles (Kim et al., 2007; Waadt et al., 2008). Additionally, it has been determined that SOS2 regulates AtNHX1 antiporter activity (Qiu et al., 2004) and directly activates the vacuolar H+ pump V-ATPase (Batelli et al., 2007). However, a previous study in Populus suggested that CBL10 is not directly related to the function of NHX proteins (Tang et al., 2014), which points at the vacuolar H+-pumps as the potential candidates for protein targets of CBL10 (Fig. 9). In tomato, the lack of SlCBL10 function was associated with a repression of SlSOS2, and the constitutive expression of SlCBL10 rescued the phenotype of the Arabidopsis cbl10 mutant, indicating that the molecular mechanism underlying Na+ compartmentalization into vacuole mediated by SlCBL10 may be shared between Arabidopsis and tomato.
SlCBL10 Promotes Ca2+ Mobilization and Availability under Salt Stress Conditions
Ca2+ deficit usually occurs in plants growing in salinized soils, since elevated Na+ concentrations hinder Ca2+ uptake by roots (Zhai et al., 2015), which usually induces a reduction of Ca2+ content in upper leaves and stem. Decreased Ca2+ levels were detected in wild-type but not in Slcbl10 mutant plants when they grew under salinity conditions (Fig. 6). Using reciprocal grafting between wild type and the Slcbl10 mutant, it was also proven that the higher Ca2+ levels detected in Slcbl10 mutant leaves resulted from Ca2+ retention in these tissues rather than from a higher Ca2+ transport from the root to the shoot during salt treatment. Likewise, given that Ca2+ stores in apical leaves and stem could be exchanged and mobilized to other tissues (apical meristem, flower, and fruit) according to the physiological Ca2+ needs of plants (White and Broadley, 2003; Dayod et al., 2010), the retention of Ca2+ in Slcbl10 upper leaves could cause an inefficient supply of Ca2+ to other demanding tissues, such as flowers and shoot apex, thus contributing to their collapse under salinity. Several specific abnormalities have been reported due to Ca2+ deficit, mainly reduced growth of apical meristems, chlorotic leaves, tissue softening, and high BER incidence in fruits (Robertson, 2013; Uozumi et al., 2012; Tonetto de Freitas et al., 2014). Therefore, abnormal Ca2+ retention detected in upper adult leaves may also be responsible for the almost null production of fruits and high BER incidence detected in Slcbl10 plants characterized under salinity conditions.
The fact that the Slcbl10 mutant was not able to balance its development in a Ca2+ deficient medium (1 mm Ca2+) also indicates that SlCBL10 is required for an appropriate Ca2+ partitioning in tomato plants. Results showed that under suboptimal Ca2+ concentration, SlCBL10 promotes the adequate expansion and division of pith cells below the shoot apical meristem. Indeed, the truncation of SlCBL10 resulted in the collapse of subapical cells, and finally caused the death of the shoot apical meristem; said symptoms have been previously described as resulting from a suboptimal concentration of calcium reaching growing tissues (Busse et al., 2008). Therefore, under salt stress conditions, SlCBL10 gene function makes it possible that Ca2+ reservoirs can be mobilized, a feature that is essential to regulate plant growth and survival of developing tissues.
SlCBL10 and SlTPC1 Cooperate in the Proper Ca2+ Release
The vacuole is by far the largest intracellular Ca2+ store in mature cells (Peiter, 2011) and constitutes the main Ca2+ reservoirs from which Ca2+ is exchanged and mobilized according to the physiological Ca2+ needs of plant (White and Broadley, 2003; Dayod et al., 2010). The expression profile of key vacuolar genes involved in Ca2+ homeostasis suggest that the mechanism required to originate Ca2+ fluxes in vacuole was severely affected when Slcbl10 plants grew in salt stress conditions (Fig. 9). Thus, salinity did not induce the expression of SlAVP1 and SlVHA-A1 genes, which implies that the proton gradient, which is necessary to energize the Ca2+ transport toward the vacuole through CAX1 antiporters (Manohar et al., 2011), was impaired by the lack of SlCBL10 gene function. Also, the stronger inhibition of SlCAX1 observed in Slcbl10 mutant plants could contribute to altering the proton gradient. Indeed, an indirect feedback mechanism has been proposed between CAX transporters (CAX1, CAX2, CAX3) and H+-pump V-ATPase in Arabidopsis, which would generate H+ flux across the tonoplast (Cheng et al., 2003). Since TPC1 has been reported as the main factor responsible for promoting Ca2+ efflux from vacuole to cytoplasm in leaf cells (Furuichi et al., 2001), the lower H+ pump into the vacuole of Slcbl10 plants might result in an inefficient opening of the SlTPC1 channel since a low luminal pH is required to regulate the aperture of this channel (Kintzer and Stroud, 2016). Moreover, the fact that the expression of SlTPC1 was not induced by salinity in Slcbl10 mutant, as occurred in wild-type plants, together with a probable loss of efficiency for opening of SlTPC1 channel suggest that in mutant plants, the Ca2+ release from vacuole induced by salinity is disturbed, which in turn could cause a defective calcium mediated salt-stress signaling and therefore salt sensitivity (Choi et al., 2014). Indeed, it has been proven that TPC1 is involved in the generation of Ca2+ cytoplasmic concentration elevation waves directed to stress signaling purposes (Evans et al., 2016). Based on this study’s results, it seems possible to suggest that SlCBl10 plays a role in Ca2+-mediated stress signaling through direct or indirect TPC1 channel regulation (Fig. 9). Direct regulation of TPC1 by CBL10 would imply that TPC1 is a target for the CBL10-CIPK24 complex to be phosphorylated and activated. In support of that hypothesis, it has been proposed that the TPC1 channel is regulated by phosphorylation (Kintzer and Stroud, 2016). Indirect regulation of TPC1 by CBL10 could be mediated by the acidification of vacuole through the regulation of vacuolar H+-pumps, as a low pH is required to open the channel (Kintzer and Stroud, 2016). In such a way, SlAVP1 or SlV-ATPase would be the potential target for SlCBL10, which would agree with the mechanism discussed for Na+ compartmentalization (Fig. 9).
The role of SlCBL10 in Ca2+ releases from vacuole could also contribute to adaptation mechanism to salinity, allowing mobilization of Ca2+ vacuolar stores in leaf cells toward fast-growing tissues to compensate for the lower Ca2+ uptake by root under salinity. The hypothesis of a double function of SlCBL10, although associated with the same mechanism (regulation of Ca2+ fluxes in vacuole), is supported by the profile expression registered in wild-type tomato leaves during salt treatment, in which two induction phases of expression were detected. The first induction took place during short time periods (after 24 h of salt treatment), which could be involved in the signaling of the salt stress process. Later, a second increase of expression could be attributed to an increasing Ca2+ demand by fast-growing tissues as a consequence of Ca2+ deficiency caused by salinity (Fig. 9). The same expression profile was recorded by SlTPC1, SlAVP1, and SlVHA-A1, which reinforces the idea of a functional relationship between these genes.
CONCLUSIONS
This study has proven that the SlCBL10 gene function is required to maintain a proper Na+/Ca2+ ratio in growing tissues allowing plant growth under salt stress conditions. Although the functional role of CBL10 in controlling Na+ homeostasis has been previously demonstrated in Arabidopsis (Kim et al., 2007), the regulation of Ca2+ homeostasis by CBL10 has not been proposed until now. Nevertheless, Monihan et al., (2016) determined that CBL10 is critical for reproductive development under salt stress conditions and detected a higher Na+ and lower Ca2+ accumulation in Arabidopsis flowers. Such results together with the functional complementation of the Arabidopsis cbl10 mutant phenotype by SlCBL10 strongly support that SlCBL10 is a true ortholog of the Arabidopsis CBL10 and its function is conserved. Thus, it is proposed that the mechanism by which SlCBL10 participates in salt tolerance mechanism is directly related to the regulation of Na+ and Ca2+ fluxes in the vacuole of leaf cells, through the activation of a tonoplast target, being the cation channel SlTPC1 and the two vacuolar H+-pumps, SlAVP1 and SlV-ATPase, the potential targets of SlCBL10 (Fig. 9). Accordingly, under salinity conditions, CBL10 confers to adult leaves the capacity to retain Na+ avoiding toxic ion accumulation in young developing tissues as well as facilitates activation of Ca2+ release from vacuoles in leaves counterbalancing Ca2+ deficiency caused by salt stress.
MATERIALS AND METHODS
Screening and Identification of pms916 (Slcbl10) Tomato Mutant
The tomato (Solanum lycopersicum) cv Moneymaker was used to generate a collection of T-DNA mutants by means of the enhancer trap vector pD991 (Atarés et al., 2011; Pineda et al., 2012; Campos et al., 2016; Pérez-Martín et al., 2017). The in vitro screening of 1,200 T2 families of tomato T-DNA lines (10–12 plants per family) grown in basal culture medium (SCM) supplemented with 100 mm NaCl led to the detection of a mutant initially named pms916 because of its hypersensitive phenotype to salt stress. To estimate the number of inserts bearing a functional NPTII marker gene, a segregation analysis of T2 progeny in kanamycin-containing medium consisting of Murashige and Skoog (MS) salts (Murashige and Skoog, 1962), Suc (10 g L−1), and kanamycin 100 (mg L−1) was carried out. The identification of the insert responsible for the mutation and the cosegregation analysis between the insert and the mutant phenotype were performed by segregation analysis with T2 and T3 progenies in both kanamycin-containing medium and SCM. To corroborate the in vitro salt sensitivity phenotype of the pms916 mutant, two new experiments were conducted under in vivo conditions. In both experiments, pregerminated seeds of the T2 segregating progeny were sown into pots containing coconut fiber and grown under controlled climatic conditions: 26 ± 3°C day/18 ± 1°C night and extra lighting provided by wide-spectrum tubes (450 μmol s−1 m−2; Gro-lux) to expose plants to 16-h day length. To assess salt hypersensitivity, T2 plants were irrigated with half-strength Hoagland solution (Hoagland and Arnon, 1950). Salt treatment (100 mm NaCl) was initiated when the plants had developed two true leaves.
The number of T-DNA copies was determined by Southern-blot hybridization experiments. Genomic DNA was isolated from young leaves as described by Dellaporta et al. (1983). Ten µg of genomic DNA was digested with EcoRI and HindIII endonucleases, electrophoresed in 0.8% agarose gel, and blotted onto Hybond N+ membranes (GE Healthcare) as described by Ausubel et al. (1993). Hybridization was performed with a chimeric probe, fusing the complete coding sequence of the NEOMYCIN PHOSPHOTRANSFERASE II (NPTII) gene to 811 pb of coding sequence from the endogenous tomato FALSIFLORA (FA) gene, which was employed as hybridization positive control (Yuste-Lisbona et al., 2016).
Cloning of T-DNA Flanking Sequences and PCR Genotyping
The sequences flanking T-DNA were isolated by anchor-PCR according to the procedure previously established by Pérez-Martín et al. (2017). The sequences of primers used are listed in Supplemental Table S2. The cloned sequences were compared with SGN Database (http://solgenomics.net/) to assign the T-DNA insertion site on tomato genome.
Cosegregation analysis of the T-DNA insertion site with the mutant phenotype in the T2 progeny was checked by PCR using (1) the specific genomic forward (G-F) and reverse (G-R) primers to amplify the wild-type allele (without T-DNA insertion) and (2) one specific genomic primer (G-F) and the specific T-DNA border primer (T2-R) to amplify the mutant allele (carrying the T-DNA insertion). Primers were designed based on sequence information available from SGN Database (http://solgenomics.net/). The sequences of genotyping primers used are listed in Supplemental Table S2. Amplification of the genotyping primers was performed in a 30-μL volume using 25 ng of total DNA, 50 ng of each primer, 0.25 mm dNTPs, 2.5 mm MgCl2, and 1 U of REDTaq DNA polymerase (SIGMA-Aldrich) in 1x Taq buffer. DNA was amplified under the following thermal cycling conditions: 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, and a final extension of 5 min at 72°C. PCR products were analyzed in 1% agarose gels in SB buffer (10 mm sodium boric acid) and visualized with ethidium bromide.
Generation of Transgenic Tomato Lines
To generate SlCBL10 silencing lines, a RNAi approach was followed. With this aim, a 123-bp fragment of the SlCBL10 cDNA was amplified using the SlCBL10-RNAiF and SlCBL10-RNAiR primers (Supplemental Table S2), and the PCR product was cloned in sense and antisense orientation separated by intronic sequences into the pKANNIBAL vector (Wesley et al., 2001) to generate a pKANNIBAL-SlCBL10 plasmid. The resulting plasmid was digested with NotI, and the entire construct was cloned into the binary vector pART27 (Gleave, 1992). In all cases, the binary plasmids generated were electroporated into Agrobacterium tumefaciens LBA 4404 strain for further use in genetic transformation experiments. Agrobacterium-mediated transformation was performed following the protocol described by Gisbert et al. (2000). Fourteen independent diploid transgenic lines silencing SlCBL10 were generated in tomato. The SlCBL10 expression level was measured by RT-qPCR as described below. Regenerant plants (control plants) were also obtained under the same conditions except for the use of the RNAi gene construct.
Arabidopsis Transformation and Complementation Test
The Arabidopsis cbl10 mutant line (SALK_056042) was kindly donated by Professor Karen Schumaker (University of AZ). To generate transgenic lines overexpressing SlCBL10, a 774-bp fragment was cloned spanning the entire ORF of SlCBL10 (Solyc08g065330) in the vector pK7WG2D.1. The resulting construct was electroporated into A. tumefaciens GV3101 and transformed into cbl10 mutant by floral dip method (Clough and Bent, 1998).
Seeds were surface-sterilized in 100% hypochlorite sodium for 5 min, washed five times in sterilized water, and sown in petri dishes containing 0.5x MS medium supplemented with 1% Suc and 0.9% agar. Seeds were stratified for 2 d at 4°C before growth at 22°C under long-day photoperiod (16 h light/8 h dark). Five-day-old seedlings were transferred to MS medium supplemented with 100 mm NaCl. Na+ contents were determined by atomic emission spectrophotometry.
Treatment Assays
Tomato wild type (cv Moneymaker), T3 homozygous and azygous plants for the Slcbl10 mutation, RNAi SlCBL10 and regenerant plants were used for the phenotypic and physiological characterization of the Slcbl10 mutant and the functional analysis of the SlCBL10 gene. Seeds were surface-sterilized briefly with 20% (v/v) commercial bleach for 15 min and then washed with sterilized water four times and suspended in sterile water at 4°C for 72 h. Germination was performed in darkness, in a 8:3 (v/v) mixture of peat:perlite, at 28°C temperature and 90% relative humidity. Seedlings were then maintained in a controlled-environment chamber (8-h-day/16-h-night cycle at 345 µmol m−2 s−1 light, 23–25°C, 50%–60% relative humidity) until they reached the desired developmental stage for each experimental assay. During this period, plants were irrigated daily with half-strength Hoagland solution (Hoagland and Arnon, 1950).
HST and Grafting Experiments
Short-term HST assays were performed in a controlled-environment chamber (conditions above described). Tomato plants were grown hydroponically in an aerated half-strength Hoagland solution. Two types of salt treatments were performed depending on the development stage of plants: (1) young plants at cotyledon stage were treated at 50 mm NaCl for 24 h and then at 100 mm for 10 additional days (HSTy assay conditions), and (2) adult plants at the fifth fully developed leaf stage were treated at 100 mm NaCl for 24 h and then at 200 mm for 5 additional days (HSTa assay conditions). In HSTa assays, shoot and root fresh weights were taken prior to salt treatment and after 2 and 6 DST. Ions content was analyzed in root, stem (taken at the first to second leaf insertion), upper adult leaf (first fully developed leaf), and shoot apex (leaf primordial and apical meristem) at 0 DST and after 6 DST. Expression induction by salinity of interesting genes was determined in shoot apex, young leaf (not fully developed leaf), upper adult leaf, stem, and root of plants after 2 DST. Additionally, gene expressions were also determined in upper adult leaf after 12, 24, 30, 60, and 144 h of salt stress. All samples were previously frozen in LN2 and kept at −80°C until further gene expression analysis. For each genotype, three biological replicates constituted by five plants each were analyzed. Two independent assays were carried out in the same experimental hydroponic conditions described above.
An additional HSTa assay was performed using reciprocal grafting between wild-type and Slcbl10 mutant plants as well as autografting with wild-type and Slcbl10 mutant plants in which Ca2+ content was analyzed in root and upper adult leaf of grafted plants at 0 DST and after 6 DST.
Greenhouse salt treatment assay (GST)
A long-term salt experiment was conducted in a greenhouse of southeastern Spain with adult plants. At the seventh to eighth fully developed leaf stage, wild-type and Slcbl10 plants were transferred from the controlled culture chamber to a polyethylene greenhouse and grown on cocoa peat, using a drip irrigation system, as previously described (García-Abellan et al., 2014). The fertirrigation solution was prepared in 2,000-L tanks with local irrigation water (electrical conductivity = 0.9 dS m−1). Before salt treatment, plants were grown under those conditions for 21 additional days until 10 leaves were fully developed. Then, 15 plants per genotype were salt-treated (100 mm NaCl), keeping 15 additional plants per genotype growing in the absence of salt. Salt treatment was performed by adding 100 mm NaCl to the tanks under 30/15°C day/night temperatures, 40% relative humidity, and 500 µmol m−2 s−1 of natural light irradiance. After 50 d, fruits of each plant were counted (no. of fruits) and weighed (reproductive biomass) and the BER incidence calculated (percentage of fruit with BER symptoms).
Calcium Deficiency in Vitro Assay
Pregerminated wild-type and Slcb110 mutant seeds were grown on MS medium supplemented with a suboptimal calcium concentration of 1 mm using calcium chloride as a source. Culture media contained 3 g L−1 Suc, 0.1 g L−1 myo-inositol, and 0.9% agar. The pH was adjusted to 5.6 ± 0.02. Media was autoclaved at 121°C for 20 min before use. Plants were cultured in 20 x 150 mm glass capped tubes containing 10 mL of media. Culture tubes were placed under 8-h-day/16-h-night cycle light at 76 µmol m−2 s−1 photosynthetic photon flux density from cool-white fluorescent lamps measured at the top of the culture tubes. The temperature was maintained at 23 ± 2°C. Fifty plants per genotype were examined after 20 and 35 d of in vitro culture. After 20 d of culture in a suboptimal calcium concentration medium, five shoot tips were taken from each genotype and fixed with 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 m sodium phosphate buffer (pH 7.2) for 3 h at 4°C. Afterward, samples were washed thrice with phosphate buffer and then incubated with 1% osmium tetroxide in the same buffer for 2 h. Subsequently, three washes with phosphate buffer were performed. Fixed tissues were dehydrated in a graded series of ethanol (35, 50, 70, 96, and 100%), and then infiltrated with a propylene oxide and JB4 resin mixture. After that, they were immersed in JB4 resin overnight at 4°C and finally transferred to flat embedding molds filled with JB4 resin that polymerized at 68°C for 24 h. Polymerized blocks were sectioned (0.5–0.7 mm thick) with a Leica EM UC6 ultramicrotome (Leica Mikrosysteme). The sections were stained for 5 min at 60°C in 1% (w/v) toluidine blue and rinsed with de-ionized water. Finally, stained sections were observed under light microscopy and digital images were obtained.
Ion Content Analysis
Concentration of Na+, K+, and Ca2+ was measured in plant material dried for 48 h at 80°C, milled to powder, and digested in a concentrated HNO3:HClO4 (2:1 v/v) solution. Na+, K+, and Ca2+ were analyzed by inductively coupled plasma spectrometry (Ionomic Service of CEBAS-CSIC).
Gene Expression Analysis
Different vegetable tissues previously frozen in LN2 and stored at −80°C were analyzed by RT-qPCR. Total RNA was isolated using a RNeasy kit (Qiagen), contaminating DNA was removed with RNAse-free DNase (DNA-free kit, Ambion), and RNA quality was assessed by electrophoresis on a denaturing agarose gel. Total RNA was quantified in a GeneQuant II spectrophotometer (Pharmacia Biotech) and 5 μg was used for cDNA synthesis with the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). RT-qPCR was performed using 1 μL of undiluted cDNA mixed with iQSyBr Green Supermix (BioRad), and 0.45 μm of forward and reverse primers using assay conditions as previously described (Asins et al., 2013). All reactions were performed in triplicate. The presence of a single band on an agarose gel electrophoresis and of a single peak in the Tm curve confirmed the specificity of RT-qPCR amplification. Relative expression data were calculated as described by Asins et al. (2013) using the tomato elongation factor 1α (LeEF1α, acc. AB061263) as housekeeping gene. The expression level was calculated from 2–ΔΔCt (Livak and Schmittgen, 2001) using the expression level of each gene from nontreated untransformed tissue as the calibrator sample. Data were statistically analyzed using the SPSS 13.0 software package. All data are given as mean ± se of three biological replicates of five plants each. Significant differences among means were analyzed by Student’s t and ANOVA tests (P < 0.05).
Specifically, it was evaluated the expression level of SlCBL10 (Solyc08g065330), SlSOS1 (Solyc01g005020), SlSOS2 (Solyc12g009570), SlHKT1;1 (Solyc07g014690), SlHKT1;2 (Solyc07g014680), LeNHX3 (Solyc01g067710), LeNHX4 (Solyc01g098190), SlAVP1 (Solyc06g068240), SlVHA-A1 (Solyc12g055800), and SlTPC1 (Solyc07g053970) genes. Regarding tomato CAX homologs, two genes homologous to Arabidopsis CAX1 and CAX3, that is Solyc09g005260 (83.9% and 79.1% similarity to CAX3 and CAX1, respectively) and Solyc06g006110 (79.7% and 77.1% similarity to CAX3 and CAX1, respectively) were identified. Of the two CAX genes analyzed, only Solyc06g006110 was responsive to salinity, and therefore it was named as SlCAX1 and used for further analysis. Sequences of evaluated genes are available in SGN Database (ITAG 2.5; http://solgenomics.net/). All primers used for RT-qPCR are listed in Supplemental Table S2.
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Table S1. Genetic analysis of the T2 progeny of pms916 mutant.
Supplemental Table S2. Primers used for standard and RT-qPCR analyses.
Supplemental Figure S1. Phenotype of T3 azygous (wild-type phenotype, left) and homozygous (Slcbl10 mutant phenotype, right) plants for the Slcbl10 mutation grown in absence of salt stress.
Supplemental Figure S2. The lack of SlCBL10 alters K+ content in tomato plant under salt stress.
Supplemental Figure S3. Na+ extrusion is increased in Slcbl10 under salinity.
Supplemental Figure S4. Phenotype of Slcbl10 tomato mutant plants salt-treated for a long time in a greenhouse.
Supplemental Figure S5. Phenotype of RNAi SlCBL10 plants salt-treated for a long time in a greenhouse.
Supplemental Figure S6. Effects of calcium deficiency on shoot apex development.
Footnotes
This study was supported by grants from the Plant KBBE Program (EUI2009-04074), the Spanish Ministerio de Economia y Competitividad (AGL2012-40150, AGL2015-64991-C3-1-R/2-R/3-R, and BIO2016-79187-R), as well as the French National Research Agency ENDOREPIGEN project. A.O.-A. was supported by a PhD fellowship from the Ministerio de Economia y Competitividad (BIO2009-11484).
References
- Albaladejo I, Meco V, Felix Plasencia F, Flores FB, Bolarin MC, Egea I (2017) Unravelling the strategies used by the wild tomato species Solanum pennellii to confront salt stress: from leaf anatomical adaptations to molecular responses. Environ Exp Bot 135: 1–12 [Google Scholar]
- Almeida P, de Boer GJ, de Boer AH (2014) Differences in shoot Na+ accumulation between two tomato species are due to differences in ion affinity of HKT1;2. J Plant Physiol 171: 438–447 [DOI] [PubMed] [Google Scholar]
- Asins MJ, Raga V, Roca D, Belver A, Carbonell EA (2015) Genetic dissection of tomato rootstock effects on scion traits under moderate salinity. Theor Appl Genet 128: 667–679 [DOI] [PubMed] [Google Scholar]
- Asins MJ, Villalta I, Aly MM, Olías R, Álvarez DE Morales P, Huertas R, Li J, Jaime-Pérez N, Haro R, Raga V, et al. (2013) Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+ /K+ homeostasis. Plant Cell Environ 36: 1171–1191 [DOI] [PubMed] [Google Scholar]
- Atarés A, Moyano E, Morales B, Schleicher P, García-Abellán JO, Antón T, García-Sogo B, Pérez-Martín F, Lozano R, Flores FB, Moreno V, et al. (2011) An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild-related species Solanum pennellii. Plant Cell Rep 30: 1865–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1993) Current Protocols in Molecular Biology, Vol 2. Wiley Interscience, New York, pp 2.9.1–2.9.15 [Google Scholar]
- Bageshwar UK, Taneja-Bageshwar S, Moharram HM, Binzel ML (2005) Two isoforms of the A subunit of the vacuolar H(+)-ATPase in Lycopersicon esculentum: highly similar proteins but divergent patterns of tissue localization. Planta 220: 632–643 [DOI] [PubMed] [Google Scholar]
- Batelli G, Verslues PE, Agius F, Qiu Q, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu JK (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol 27: 7781–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Bonales-Alatorre E, Pottosin I, Shabala L, Chen ZH, Zeng F, Jacobsen SE, Shabala S (2013) Differential activity of plasma and vacuolar membrane transporters contributes to genotypic differences in salinity tolerance in a halophyte species, Chenopodium quinoa. Int J Mol Sci 14: 9267–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse SB, Ozgen S, Palta JP (2008) Influence of root zone calcium on subapical necrosis in potato shoot cultures: localization of injury at the tissue and cellular levels. J Am Soc Hortic Sci 133: 653–662 [Google Scholar]
- Campos JF, Cara B, Pérez-Martín F, Pineda B, Egea I, Flores FB, Fernández-García N, Capel J, Moreno V, Angosto T, et al. (2016) The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol J 14: 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD (2003) The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111: 6497–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng NH, Stancombe MA, Hirschi KD, Webb AAR, et al. (2011) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero J, Bolarin MC, Moreno V, Pineda B (2010) Molecular tools for enhancing salinity tolerance in plants. In Jain SM, Brar DS, eds, Molecular Techniques in Crop Improvement. Springer, New York, pp 373–405 [Google Scholar]
- Dayod M, Tyerman SD, Leigh RA, Gilliham M (2010) Calcium storage in plants and the implications for calcium biofortification. Protoplasma 247: 215–231 [DOI] [PubMed] [Google Scholar]
- de la Torre F, Gutiérrez-Beltrán E, Pareja-Jaime Y, Chakravarthy S, Martin GB, del Pozo O (2013) The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25: 2748–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 1: 19–21 [Google Scholar]
- Evans MJ, Choi WG, Gilroy S, Morris RJ (2016) A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol 171: 1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Galal HK, Bromham L (2010) Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol 37: 604–612 [Google Scholar]
- Furuichi T, Cunningham KW, Muto S (2001) A putative two pore channel AtTPC1 mediates Ca(2+) flux in Arabidopsis leaf cells. Plant Cell Physiol 42: 900–905 [DOI] [PubMed] [Google Scholar]
- Gálvez FJ, Baghour M, Hao G, Cagnac O, Rodríguez-Rosales MP, Venema K (2012) Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiol Biochem 51: 109–115 [DOI] [PubMed] [Google Scholar]
- García-Abellan JO, Egea I, Pineda B, Sanchez-Bel P, Belver A, García-Sogo B, Flores FB, Atarés A, Moreno V, Bolarin MC (2014) Heterologous expression of the yeast HAL5 gene in tomato enhances salt tolerance by reducing shoot Na+ accumulation in the long term. Physiol Plant 152: 700–713 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581: 2204–2214 [DOI] [PubMed] [Google Scholar]
- Gisbert C, Rus AM, Bolarín MC, López-Coronado JM, Arrillaga I, Montesinos C, Caro M, Serrano R, Moreno V (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123: 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM. (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92: 19–31 [Google Scholar]
- Hedrich R, Marten I (2011) TPC1-SV channels gain shape. Mol Plant 4: 428–441 [DOI] [PubMed] [Google Scholar]
- Hirschi KD. (1999) Expression of Arabidopsis CAX1 in tobacco: altered calcium homeostasis and increased stress sensitivity. Plant Cell 11: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water–culture method for growing plants without soil. In Agricultural Experiment Station Circular 347, Berkeley, CA, The College of Agriculture University of California, pp 1–32 [Google Scholar]
- Huertas R, Rubio L, Cagnac O, García-Sánchez MJ, Alché JdeD, Venema K, Fernández JA, Rodríguez-Rosales MP (2013) The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant Cell Environ 36: 2135–2149 [DOI] [PubMed] [Google Scholar]
- Kang HK, Nam KH (2016) Reverse function of ROS-induced CBL10 during salt and drought stress responses. Plant Sci 243: 49–55 [DOI] [PubMed] [Google Scholar]
- Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schültke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kim KN. (2013) Stress responses mediated by the CBL calcium sensors in plants. Plant Biotechnol Rep 7: 1–8 [Google Scholar]
- Kintzer AF, Stroud RM (2016) Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature 531: 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y (2009) Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. (2014) Sodium in plants: perception, signalling, and regulation of sodium fluxes. J Exp Bot 65: 849–858 [DOI] [PubMed] [Google Scholar]
- Maeshima M. (2000) Vacuolar H(+)-pyrophosphatase. Biochim Biophys Acta 1465: 37–51 [DOI] [PubMed] [Google Scholar]
- Manaa A, Faurobert M, Valot B, Bouchet JP, Grasselly D, Causse M, Ahmed HB (2013) Effect of salinity and calcium on tomato fruit proteome. OMICS 17: 338–352 [DOI] [PubMed] [Google Scholar]
- Manohar M, Shigaki T, Hirschi KD (2011) Plant cation/H+ exchangers (CAXs): biological functions and genetic manipulations. Plant Biol (Stuttg) 13: 561–569 [DOI] [PubMed] [Google Scholar]
- Monihan SM, Magness CA, Yadegari R, Smith SE, Schumaker KS (2016) Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiol 171: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Olías R, Eljakaoui Z, Li J, De Morales PA, Marín-Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32: 904–916 [DOI] [PubMed] [Google Scholar]
- Park S, Cheng NH, Pittman JK, Yoo KS, Park J, Smith RH, Hirschi KD (2005) Increased calcium levels and prolonged shelf life in tomatoes expressing Arabidopsis H+/Ca2+ transporters. Plant Physiol 139: 1194–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E. (2011) The plant vacuole: emitter and receiver of calcium signals. Cell Calcium 50: 120–128 [DOI] [PubMed] [Google Scholar]
- Pérez-Martín F, Yuste-Lisbona FJ, Pineda B, Angarita-Díaz MP, García-Sogo B, Antón T, Sánchez S, Giménez E, Atarés A, Fernández-Lozano A, et al. (2017) A collection of enhancer trap insertional mutants for functional genomics in tomato. Plant Biotechnol J 15: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda B, García–Abellán JO, Antón T, Pérez F, Moyano E, García–Sogo B, Campos JF, Angosto T, et al. (2012) Genomic approaches for salt and drought stress tolerance in tomato. In Tuteja N, Gill SS, Tiburcio AF, Tuteja R, eds, Improving Crop Resistance to Abiotic Stress. Wiley-VCH Verlag and Co. KGaA, Weinheim, pp 1085–1110 [Google Scholar]
- Pittman JK, Edmond C, Sunderland PA, Bray CM (2009) A cation-regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J Biol Chem 284: 525–533 [DOI] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, et al. (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11: 372–374 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279: 207–215 [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. (2013) Modulating Plant Calcium for Better Nutrition and Stress Tolerance. ISRN Botany Article ID 952043.
- Shabala S. (2013) Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot 112: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Shabala L, Van Volkenburgh E, Newman I (2005) Effect of divalent cations on ion fluxes and leaf photochemistry in salinized barley leaves. J Exp Bot 56: 1369–1378 [DOI] [PubMed] [Google Scholar]
- Tang RJ, Yang Y, Yang L, Liu H, Wang CT, Yu MM, Gao XS, Zhang HX (2014) Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ 37: 573–588 [DOI] [PubMed] [Google Scholar]
- Tonetto de Freitas S, McElrone AJ, Shackel KA, Mitcham EJ (2014) Calcium partitioning and allocation and blossom-end rot development in tomato plants in response to whole-plant and fruit-specific abscisic acid treatments. J Exp Bot 65: 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi A, Ikeda H, Hiraga M, Kanno H, Nanzyo M, Nishiyama M, Kanayama Y (2012) Tolerance to salt stress and blossom-end rot in an introgression line, IL8-3, of tomato. Sci Hortic (Amsterdam) 138: 1–6 [Google Scholar]
- Villalta I, Reina-Sánchez A, Bolarín MC, Cuartero J, Belver A, Venema K, Carbonell EA, Asins MJ (2008) Genetic analysis of Na(+) and K (+) concentrations in leaf and stem as physiological components of salt tolerance in tomato. Theor Appl Genet 116: 869–880 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste-Lisbona FJ, Quinet M, Fernández-Lozano A, Pineda B, Moreno V, Angosto T, Lozano R (2016) Characterization of vegetative inflorescence (mc-vin) mutant provides new insight into the role of MACROCALYX in regulating inflorescence development of tomato. Sci Rep 6: 18796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Yang Q, Hou M (2015) The effects of saline water drip irrigation on tomato yield, quality, and blossom–end rot incidence: a 3a case study in the south of China. PLoS One 10: e0142204. [DOI] [PMC free article] [PubMed] [Google Scholar]