The majority of plant species preferentially express NLRs in root tissues, but the Brassicaceae family displays consistent shoot-skewed NLR expression across different phylogenetic NLR clades.
Abstract
Nucleotide-binding site leucine-rich repeat resistance genes (NLRs) allow plants to detect microbial effectors. We hypothesized that NLR expression patterns could reflect organ-specific differences in effector challenge and tested this by carrying out a meta-analysis of expression data for 1,235 NLRs from nine plant species. We found stable NLR root/shoot expression ratios within species, suggesting organ-specific hardwiring of NLR expression patterns in anticipation of distinct challenges. Most monocot and dicot plant species preferentially expressed NLRs in roots. In contrast, Brassicaceae species, including oilseed rape (Brassica napus) and the model plant Arabidopsis (Arabidopsis thaliana), were unique in showing NLR expression skewed toward the shoot across multiple phylogenetically distinct groups of NLRs. The Brassicaceae are also outliers in the sense that they have lost the common symbiosis signaling pathway, which enables intracellular infection by root symbionts. While it is unclear if these two events are related, the NLR expression shift identified here suggests that the Brassicaceae may have evolved unique pattern-recognition receptors and antimicrobial root metabolites to substitute for NLR protection. Such innovations in root protection could potentially be exploited in crop rotation schemes or for enhancing root defense systems of non-Brassicaceae crops.
The sessile nature of vascular plants has spurred development of mechanisms for coping with biotic and abiotic stresses and for optimizing uptake of inorganic compounds under low nutrient availability. In response to these challenges, plant roots and shoots have evolved specialized functions above and below ground, where they have also adapted to interact with the distinct microbial communities of the phyllo- or rhizosphere. These diverse plant-microbe interactions range from symbiosis over parasitism to pathogenic infection (Bulgarelli et al., 2013; Fatima and Senthil-Kumar, 2015; Vandenkoornhuyse et al., 2015).
Reflecting the different characteristics of plant roots and shoots, distinct host-microbe combinations have been used to unravel the molecular components required for transspecies interaction and communication. In plant shoots, the focus has almost exclusively been on pathogenic interactions, where work in the model plant Arabidopsis (Arabidopsis thaliana) from the Brassicaceae family has provided great insight into plant immunity (Jones and Dangl, 2006; Nishimura and Dangl, 2010). Passive defenses, such as the waxy cuticle on epidermal cells, cell walls, and preformed antimicrobial chemicals form the first barriers for microbes and are often sufficient for deterring would-be pathogens (Thordal-Christensen, 2003). Microbes that successfully evade these obstacles encounter a large repertoire of resistance (R) proteins in the form of transmembrane receptor-like proteins and receptor-like kinases on the surface of plant cells, which recognize conserved microbe-associated molecular patterns. Upon activation, these pattern-recognition receptors (PRRs) trigger complex intracellular signaling cascades, such as phytohormone perturbations, accumulation of ions, MAPK activation, and production of reactive oxygen species, ultimately leading to transcriptional and translational changes that promote the production of defense compounds (Pel and Pieterse, 2012; Muthamilarasan and Prasad, 2013).
To escape this microbe-associated molecular pattern-triggered immunity, microbes have evolved effectors that are injected into the plant cell cytoplasm using specialized secretion systems that penetrate the plant cell membrane. Upon translocation, these effectors target components of the defense machinery, suppressing immune signaling and gene expression through degradation, allosteric or covalent modification of host molecules, thus adapting the local environment to be more suitable for microbial growth and improving the chances of successful tissue colonization (Jones and Dangl, 2006; Xin and He, 2013; Le Fevre et al., 2015). In response, plant cells employ a family of intracellular R proteins that recognize effectors either by direct interaction, or indirectly through detection of modifications made to host proteins (Khan et al., 2016). Effector-triggered immunity activation by an intracellular R protein leads to a stronger immune response than that of molecular pattern-triggered immunity and is often associated with localized cell death to limit the spread of biotrophic pathogens (Jones and Dangl, 2006; Hofius et al., 2007).
The majority of intracellular R proteins share a similar structure with an amino-terminal signaling domain followed by a highly conserved nucleotide binding domain (NBD) and a carboxy-terminal leucine-rich repeat (LRR) domain of variable length (van der Biezen and Jones, 1998; Takken and Goverse, 2012). This class of R proteins is referred to as nucleotide-binding site leucine-rich repeat (NLR) proteins. The NBD is shared by Apaf1, plant R proteins and CED4 (NB-ARC) and is highly conserved among all NLR proteins. It acts as a molecular switch, and cycles between active ATP-bound and inactive ADP-bound states depending on the activity of the LRR domain. The LRR domain is believed to be directly involved in protein-protein interactions with microbial effectors or host proteins and to function by auto-suppressing the NBD of the NLR (Jones and Jones, 1997; Takken et al., 2006; Marquenet and Richet, 2007; Lukasik and Takken, 2009; Takken and Goverse, 2012). The amino-terminal signaling domain is generally divided into two separate classes based on homology to either the signaling domain of Toll/IL-1 Receptors (TIR) or the presence of a coiled-coil (CC) domain. These two distinct signaling components share common downstream signaling pathways; however, both classes have also been observed to activate separate downstream components (Aarts et al., 1998; Falk et al., 1999; Meyers et al., 1999; Pan et al., 2000; Takken et al., 2006; Hofius et al., 2009). While both CC and TIR type NLRs (CNLs and TNLs, respectively) are widely distributed in dicots, canonical TNLs appear to be absent in monocots (Meyers et al., 1999; Pan et al., 2000; Meyers et al., 2002; Tarr and Alexander, 2009). In addition, variations of the signaling domain-NBD-LRR (NLR) structure can be found in most plant species, with NBD-containing proteins lacking either the amino-terminal signaling domain or the carboxy-terminal LRR domain, or having juxtaposed noncanonical domains, extending their flexibility as signaling components or effector decoys for host proteins (Bonardi et al., 2012; Kroj et al., 2016).
While many NLRs play important roles in Arabidopsis shoot immunity, little is known about how Arabidopsis roots mount immune response against microbes, or what role NLRs play. However, the PRR FLAGELLIN-SENSITIVE2 is fully functional in roots and activates similar downstream MAP-kinase cascades in both root and shoot (Millet et al., 2010). There are reported differences between roots and shoots for the phytohormone salicylic acid, which is considered a requirement for basal defense in leaves against biotrophic pathogens, but does not appear to be as important in root immune responses (Jones and Dangl, 2006; Millet et al., 2010).
Unlike the work on Arabidopsis pathogen responses, studies of root-microbe interactions have focused on endosymbiosis with nitrogen fixing rhizobia and mycorrhizal fungi. Up to 90% of all terrestrial plants are believed to associate with arbuscular mycorrhizal (am) fungi to enhance their acquisition of phosphorus and other nutrients. Plant associations with nitrogen-fixing bacteria contained within root nodules is restricted to around 10 families, including the agriculturally important Fabaceae (legume) family (Doyle, 1998; Gualtieri and Bisseling, 2000; Parniske, 2008). Arabidopsis belongs to the Brassicaceae family, which is one of the few plant families that has lost the capacity for root endosymbiosis with mycorrhizal fungi that is ancestral to the Angiospermae (flowering plants) (Gualtieri and Bisseling, 2000; Smith and Read, 2010; Delaux et al., 2014). Two model plants from the legume family, Lotus (Lotus japonicus) and Medicago (Medicago truncatula), have been extensively used for unraveling the genetic pathways required for root nodulation through their symbiotic association with nitrogen fixing rhizobia (Barker et al., 1990; Handberg and Stougaard, 1992). This work has led to the discovery of nodulation factors, key signal molecules secreted by rhizobia, and several host receptors that perceive and transduce the signal through regulatory components to modulate downstream transcriptional regulation and coordinate nodule organogenesis and infection (Long, 1989; Schauser et al., 1999; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Lévy et al., 2004; Kaló et al., 2005; Smit et al., 2005; Tirichine et al., 2006; Kouchi et al., 2010; Madsen et al., 2010). Similar to nodulation factors produced by rhizobia, AM fungi secrete Myc factors to activate symbiotic signaling in the host. Despite their distinct phenotypic characteristics, AM and nodulation pathways share a core of signaling components known as the common symbiosis signaling pathway, likely owing to their common evolutionary origin (Oldroyd and Downie, 2006; Parniske, 2008; Banba et al., 2008; Singh and Parniske, 2012; Guillotin et al., 2016).
Despite the history of focusing on pathogenic plant-microbe interactions in plant shoots and on symbiotic interactions in roots, both organs are prone to pathogen infection and would presumably be protected by NLR proteins present in cells subject to effector challenge. Currently, little is known about the expression characteristics of NLRs and, unless they are ubiquitously expressed across all plant organs, NLR gene expression patterns could provide indications about differences in pathogen effector pressures between plant tissues and across plant species. Here we present a meta-analysis of NLR gene expression data, including both monocot and dicot plant species. Our analysis revealed stable root to shoot NLR gene expression ratios within species, with all monocot and legume plant species examined predominantly expressing NLRs in roots. In contrast, members of the Brassicaceae family displayed an aberrant shoot-skewed NLR expression, which suggested an unusual mode of plant-microbe interaction for this plant family.
RESULTS
NLR Gene Expression Varies between Tissues in a Species-Specific Manner
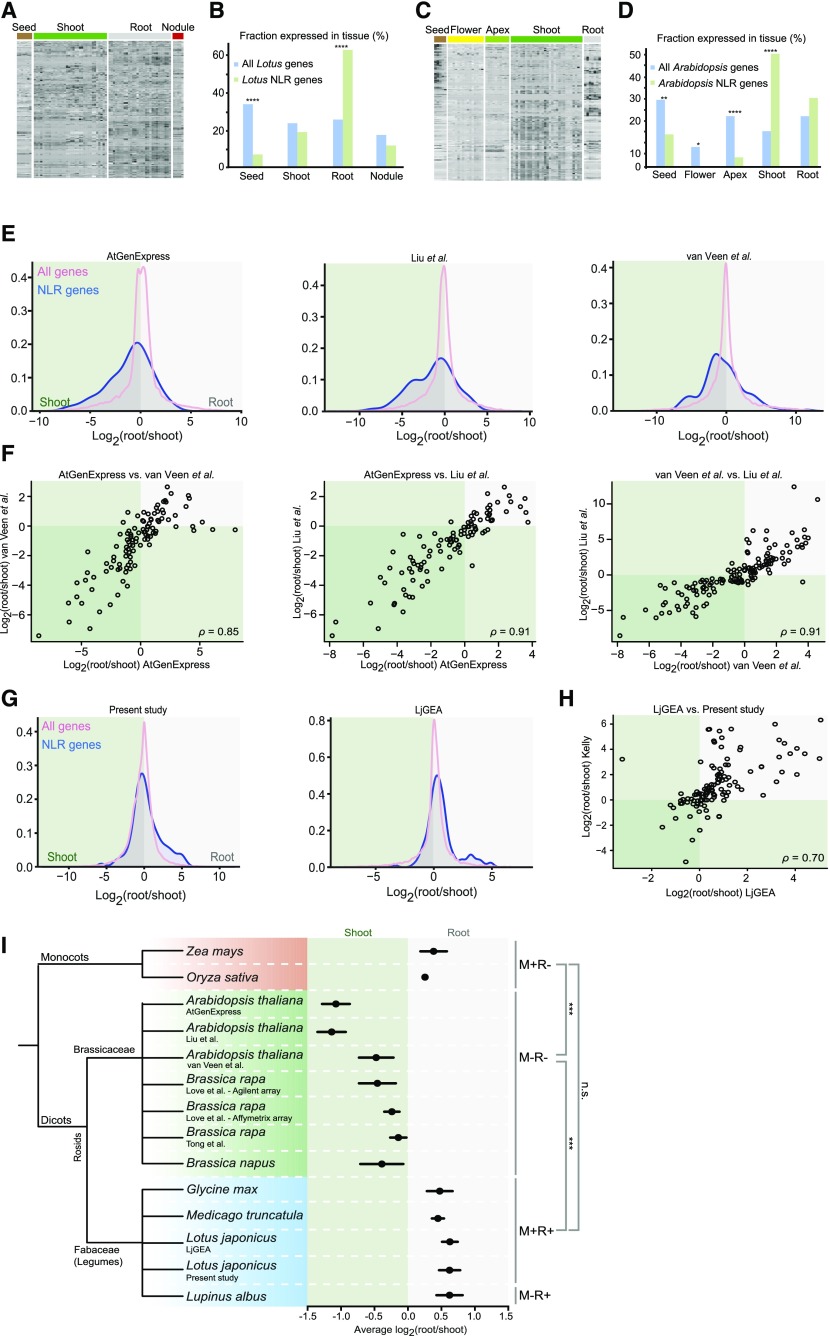
Individual plant organs have evolved to function in specific environments, where they interact with distinct microbiota (Vandenkoornhuyse et al., 2015). In addition, plant species, including Lotus, which partake in symbiosis with rhizobia and mycorrhizal fungi, harbor microbiota that diverge greatly from those of nonsymbiotic species like Arabidopsis (Zgadzaj et al., 2016). To investigate if NLR expression patterns reflected these cross-tissue and -species differences, we analyzed data from Lotus and Arabidopsis, which differ in symbiotic status and have microarray expression atlas data available for multiple tissues (Schmid et al., 2005; Høgslund et al., 2009; Verdier et al., 2013; Supplemental Table S1–2; Supplemental File S1). We identified all putative NLRs in Lotus and Arabidopsis based on the presence of an NBD and at least one other NLR-associated domain and then classified genes by enriched expression in reproductive, shoot, root, or root nodule tissues. NLR expression in Lotus shoot and nodule tissues did not show significant differences compared to overall gene expression, but reproductive tissues showed strong depletion of NLR expression and Lotus roots displayed a significant enrichment of NLR expression (Figure 1, A and B). For Arabidopsis, reproductive tissues also showed a significant depletion of expressed NLR genes, but Arabidopsis roots did not show enriched NLR gene expression. Instead, Arabidopsis shoots displayed a significant enrichment of NLR gene expression (Figure 1, C and D).
Figure 1.
NLR gene expression patterns across species. A, Expression patterns of 198 putative Lotus NLR genes. B, Enrichment of Lotus NLR genes by tissue type. The fraction of all genes and NLR genes with enriched expression in the given tissue are shown. C, Expression patterns of 160 putative Arabidopsis NLR genes. D, Enrichment of Arabidopsis NLR genes by tissue type. The fraction of all genes and NLR genes with enriched expression in the given tissue are shown. P values indicate the probability that the fraction of NLR genes showing enriched expression in a specific tissue is identical to that of all genes. E, Density plots displaying the distribution of the logarithm of the root/shoot expression ratios of all genes and NLR genes for each Arabidopsis expression data set indicated. See main text for sources. F, Root/shoot expression correlations for Arabidopsis NLR genes from the three data sets shown in Figure 1E. Each circle represents one NLR gene for which expression data are available in both of the datasets compared. G, Density plots displaying the distribution of the logarithm of the root/shoot expression ratios of all genes and NLR genes for each Lotus expression data set indicated. H, Root/shoot expression correlations for Lotus NLR genes between two datasets. Each circle represents one NLR gene for which expression data are available in both datasets. I, Phylogenetic tree of species for which both shoot and root expression data are available, along with their average NLR gene root/shoot expression values (black dots). Error bars indicate SEM. The symbiotic status of each species is indicated on the right. M, mycorrhiza; R, rhizobia; +, engages in endosymbiosis; -, does not engage in endosymbiosis. Significance levels of comparisons between species groups are indicated on the far right. ***P ≤ 0.001. n.s., not significant. Generalized linear model ANOVA was used for calculation of P values. See Supplemental Table S6 for P values for interspecies differences.
To investigate if the contrasting root/shoot NLR gene expression ratios were general for the two species, we studied additional data sets. For Arabidopsis, we examined two recent RNA-seq experiments, including both root and shoot samples in the same experimental series (van Veen et al., 2016; Liu et al., 2016). Since no equivalent data sets were available for Lotus, we carried out an RNA-seq experiment including mock and rhizobium inoculated root and shoot samples. Transcript quantification by RNA-seq is based on digital read counts, which provide a large dynamic range. The methodology can, however, cause issues with normalization between samples because of compositional bias effects, where a pronounced increase in the expression of a few genes reduces the number of reads allocated to other transcripts in the same library (Li, Witten, et al., 2012). Comparisons between root and shoot samples, which have different expression profiles, could be subject to such biases. We therefore evaluated four different normalization methods – total library read count, Trimmed Means of M-values (TMM), PoissonSeq (PSS), and voom (Robinson et al., 2010; Li, Witten et al., 2012; Ritchie et al., 2015) – for their efficacy in controlling for compositional bias effects on root/shoot gene expression ratios. To compare the methods, we simulated compositional biases in the RNA-seq data by randomly elevating the expression of 100 genes 10,000-fold, followed by scaling back expression of the remaining genes to retain the same overall library size. Normalization of the simulated data using voom or total read counts produced erratic root/shoot expression ratios, which depended on the direction of the introduced bias. In contrast, TMM and PSS normalization both performed well, with PSS-based root/shoot ratios slightly more robust to the introduced biases (Supplemental Table S3). We therefore used PSS normalized data for downstream analysis.
Both Arabidopsis RNA-seq data sets showed a clear shoot skew for NLRs relative to the average expression ratio for all genes (Fig. 1E; Supplemental Table S4), and the NLR expression ratios were strongly correlated across array and RNA-seq experiments (Fig. 1F). For Lotus, the RNA-seq data were consistent with the array data in showing a pronounced root skewed NLR expression (Fig. 1, G and H; Supplemental Table S4).
Bacterial inoculation is another factor that could potentially influence NLR root/shoot expression ratios. We compared Lotus inoculated and uninoculated samples, but found no significant differences in the root/shoot NLR expression ratios for neither the array nor the RNA-seq experiment (Supplemental Fig. S1). NLR root/shoot expression ratios thus showed clear differences between Lotus and Arabidopsis, and these differences were consistent across independent experiments carried out using either array or RNA-seq methodology for transcript quantification, indicating that regulation of NLR gene expression varied between organs in a species-specific manner.
The Brassicaceae Family Shows Aberrant Shoot-Skewed NLR Gene Expression
To determine which of these contrasting patterns of NLR gene expression was predominant among flowering plants, we analyzed additional species for which root and shoot tissues had been subjected to global expression profiling in the same experiment. These included three legume species (Medicago, Glycine max, Lupinus albus), two Brassicaceae family members (Brassica rapa ssp. Pekinensis and Brassica napus), and two monocots (Zea mays and Oryza sativa; Figure 1I; Supplemental Figs. S2 and S3). We calculated root/shoot expression ratios for all genes, including only samples where root and shoot tissues had been analyzed in the same experimental series (Supplemental Tables 1 and 2). We identified a total of 2,167 NLR genes across the selected species, and expression data were available for 1,235 of these (Supplemental Table S5).
Like Lotus, the three other dicot legumes and the two monocots displayed NLR gene expression skewed toward the root when compared to the overall gene expression pattern (Fig. 1I; Supplemental Figs. S2 and S3; Supplemental Table S4). In comparison, the three Brassicaceae species stood out by displaying shoot-skewed NLR gene expression (Fig. 1I; Supplemental Figs. S2 and S3; Supplemental Table S4). Comparisons within the legume, Brassicaceae, or monocot groups showed very few statistically significant differences. However, when we compared between species groups, many comparisons showed significant differences, with the overall differences between Brassicaceae versus both legumes and monocots highly significant (P < 1e-04; Fig. 1I; Supplemental Table S6). Among the flowering plants investigated, shoot-skewed expression of NLR genes was a feature exclusive to the dicot Brassicaceae family, while the remaining monocots and dicot species all displayed root-skewed expression.
The Brassicaceae Expression Shift Is Seen across Multiple NLR Clades
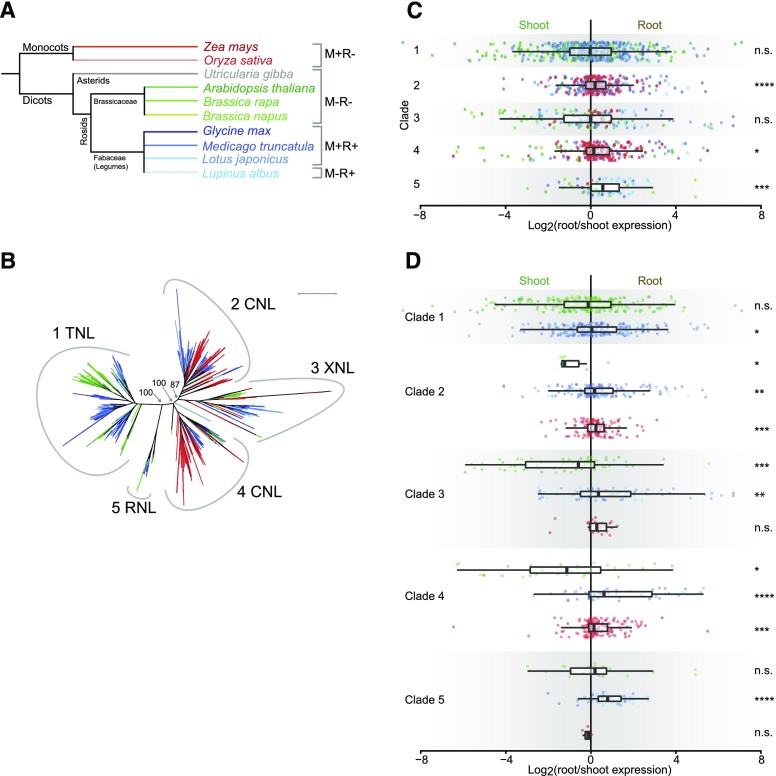
We speculated that the Brassicaceae expression shift could have been caused by the loss of a specialized set of phylogenetically related NLRs evolved specifically to guard root specific components. To test this hypothesis, we categorized all identified NLRs by aligning their NBDs and constructing a phylogenetic tree based on 2,033 sequences (Figs. 2, A and B). In addition to the previously mentioned species, we included the carnivorous and submerged aquatic bladderwort Utricularia gibba from the Asterids clade, which lacks a true root (Ibarra-Laclette et al., 2013). The phylogenetic analysis allowed us to identify five well-supported major NLR clades (Fig. 2B; Supplemental File S2; Supplemental Table S7). To investigate if the Brassicaceae expression shift was correlated with a specific NLR subtype, we also classified all NLRs according to domain composition, initially identifying TIR and CC domains, and compared these results to our phylogenetic analysis (Supplemental Fig. S4; Supplemental Table S7). We found that several of the sequences in our initially identified set of NLRs from all eight species contained a specific subtype of the CC domain, the CCR domain, which shows high sequence similarity to the amino terminal domain of the non-NBD-LRR R gene RPW8 (RESISTANCE TO POWDERY MILDEW 8; Xiao et al., 1997; Xiao et al., 2001; Collier and Moffett, 2009; Collier et al., 2011; Bonardi et al., 2011; Shao et al., 2016). We therefore included the CCR domain proteins as a separate group in the analysis (Xiao et al., 2001; Meyers et al., 2003; Shao et al., 2016). Hereafter, we refer to NLRs containing TIR, CC, and CCR domains as TNLs, CNLs, and RNLs, respectively. NLRs containing neither of the three described domains are referred to as XNLs. We performed a stringent search against the NCBI conserved domain database CDD (Marchler-Bauer et al., 2011) for identification of canonical CC and TIR domains. We did not classify as many NLRs into TNL, and especially CNL groups, as in previous studies (Supplemental Table S7; Monosi et al., 2004; Ameline-Torregrosa et al., 2008; Kim et al., 2012; Shao et al., 2014; Yu et al., 2014; Song et al., 2015; Sarris et al., 2016; Shao et al., 2016). We chose this conservative approach due to relatively poorly defined sequence composition and structure of especially the CC domains.
Figure 2.
NLR expression patterns by phylogenic clade. Colors indicate the plant species from which the NLR originates with reference to Figure 2A. A, Species-level phylogenetic tree. The symbiotic status of each species is indicated on the right. M, mycorrhiza; R, rhizobia; +, engages in endosymbiosis; -, does not engage in endosymbiosis. B, Phylogenetic tree based on the NBD protein sequence of identified NLR genes in the species indicated in Figure 2A. Numbers at branches indicate bootstrap values for the branching of the five major clades. Peripheral numbers indicate clade designation, and NLR designation indicate enrichment of the corresponding NLR type in the given clade. Scale bar indicates 1.0 average amino acid substitutions per site. See Supplemental File S2 for full bootstrap analysis of the tree. See Supplemental Table S7 for NLR distribution at the clade and species level. C, Per clade log2 root/shoot expression ratios of the NLR genes shown in B for which expression data are available. Each colored dot represents one NLR gene. Box plot bars show median with boxes indicating 25th and 75th percentiles and whiskers indicating 1.5 times the interquartile range. D, Same as C but with expression data separated into groups depending on the species evolutionary descent colored according to Figure 2A. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. n.s., not significant. Student’s t test was used for calculation of P values. See Supplemental Table S9 for P values for interclade and interspecies differences.
Clade 1 was highly enriched in TNLs (708/806), CNLs dominated clade 2 (307/510) and clade 4 (326/385), clade 5 was enriched for RNLs (62/87), and clade 3 contained mainly XNLs (211/245; Fig. 2B; Supplemental Table S7). The clear correlation between domain structure and the NBD-based phylogeny indicated that the NBD sequences contained sufficient information for inferring the evolutionary history of the plant NLR family, as previously suggested (Pan et al., 2000).
In accordance with previous studies, we did not observe any sequences from monocots in the TNL-enriched clade 1, but among all sequences analyzed we recovered 7 monocot NLRs that had an identifiable TIR-like domain, which has previously been observed to be juxtaposed irregularly compared to the normal TIR domain (Meyers et al., 2002; Nandety et al., 2013). We did not identify any monocot RNLs using our stringent filtering criteria, although at least one RNL has previously been found in both rice and maize (Shao et al., 2016). Nor did we recover any TIR or TIR-related domain containing NLR sequences from U. gibba, despite it being a dicot (Pan et al., 2000; Fluhr 2001; Tarr and Alexander, 2009; Ibarra-Laclette et al., 2013). In fact, U. gibba sequences were only found in clades 2 and 4 (Figure 2B; Supplemental Table S7).
Examining the expression characteristics of NLRs grouped by their protein domain composition, we did not observe expression bias toward either organ for TNLs and XNLs (Supplemental Fig. S4A; Supplemental Table S8). In contrast, CNLs and RNLs displayed highly significant root skews (Supplemental Fig. S4A; Supplemental Table S8). Legume and monocot TNLs and CNLs showed significant root skews, but Brassicaceae CNLs and XNLs showed significant shoot skews (Supplemental Fig. S4B; Supplemental Table S8). Reflecting the contrasting Brassicaceae and legume expression skews, XNLs, TNLs, and CNLs all showed significantly different means in Brassicaceae versus legume comparisons (Supplemental Fig. S4B; Supplemental Table S8).
We then studied NLR root/shoot ratios for the five NLR clades defined on the basis of the sequence similarities of their NBDs. Across data from all species, we observed highly significant root skews for the CNL-enriched clade 2 and for the RNL-enriched clade 5 (Fig. 2C; Supplemental Table S9). When examining the Brassicaceae, legume and monocot species groups separately, we found significant shoot skews for Brassicaceae clades 2, 3, and 4, and significant root skews for monocot clades 2 and 4 and for all legume NLR clades. The mean Brassicaceae root/shoot expression ratios deviated significantly from those of legumes for clades 1 to 4, and from monocots for clades 3 and 4 (Fig. 2D; Supplemental Table S9). In contrast, we did not find significant deviations between Brassicaceae and legumes for the RNL-enriched clade 5, where both species groups showed root-skewed expression. Since we observed significant Brassicaceae deviations for multiple NLR clades, a monophyletic group of NLRs was not responsible for the Brassicaceae expression shift. However, there were differences between the NLR clades in the severity of the shift, with the smallest effect seen for the TNL-enriched clade 1.
NLR Clade 2 Depletion Is Characteristic of the Brassicaceae Family
Comparing the species tree (Fig. 2A) to the NBD-based NLR tree (Fig. 2B), we noted that clade 1 and 5 in the NBD tree contained mainly dicot members, whereas clades 3 and 4 comprised monocot and dicot members from all species, in line with the species tree. In contrast, clade 2 from the NBD-tree was depleted in dicot Brassicaceae members, while both legume and monocot members were well represented, suggesting a possible family-specific depletion of a major NLR clade in the Brassicaceae (Fig. 2B; Supplemental Table S7).
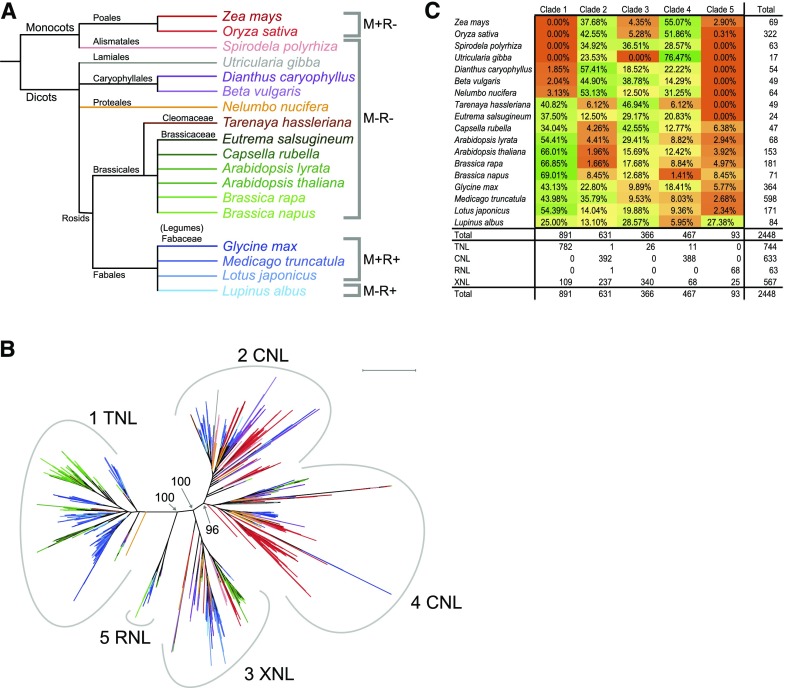
To investigate if NLR clade 2 depletion was Brassicaceae specific, we examined NLR protein sequences from 8 additional plant species, including Brassicaceae members and species from other families without a functional common symbiosis signaling pathway (Fig. 3, A and B; Supplemental Table S10; Supplemental File S3). We found that 120 of the 415 new NLR sequences were present in clade 2, indicating that clade 2 was not generally depleted in species without a common symbiosis signaling pathway (Fig. 3C; Supplemental Table S10). After including three additional Brassicaceae species, we still observed a pronounced family-specific Brassicaceae depletion in clade 2, as we only found 20 of 544 Brassicaceae NLRs belonging to this family (Supplemental Table S10), suggesting that NLR clade 2 depletion is characteristic of the Brassicaceae family.
Figure 3.
NBD phylogeny including additional nonmycorrhizal species. A, Species-level phylogenetic tree. The symbiotic status of each species is indicated on the right. M, mycorrhiza; R, rhizobia; +, engages in endosymbiosis; -, does not engage in endosymbiosis. B, Phylogenetic tree based on the NBD protein sequence of identified NLR genes in the species indicated in Figure 3A. Numbers at branches indicate bootstrap values for the branching of the five major clades. Peripheral numbers indicate clade designation. Scale bar indicates 1.0 average amino acid substitutions per site. Colors indicate the plant species from which the NBD originates with reference to Figure 3A. See Supplemental File S3 for full bootstrap analysis of the tree. See Supplemental Table S10 for NLR distribution at the clade and species level. C, Table showing the percentage of NLRs in each species, with respect to each clade shown in Figure 3B.
DISCUSSION
Cytoplasmic NLRs make up the last line of defense against potentially pathogenic microbes that have evaded physical barriers and membrane-localized PRRs to successfully deliver effectors into plant cells. The stable root/shoot NLR expression ratios observed here are consistent with a defense system in which NLR expression patterns are hardwired in anticipation of organ-specific effector challenges (Bhardwaj et al., 2011; Wang et al., 2011). Indeed, we also found that rhizobium inoculation of the nodulating legume Lotus did not alter the overall pattern of NLR expression, further underlining the stability of NLR root/shoot expression ratios within species. It was striking that we found an overall root-skew in NLR expression in the majority of plant species. This suggests that roots generally experience a higher level of effector pressure than shoots, despite that NLR function has mainly been characterized in the context of shoot-pathogen interactions (Erb et al., 2009; Nishimura and Dangl, 2010). It might not be surprising given the complexity of soil microbial communities, but our data does underline the need for establishing new root pathosystems and for understanding the role of NLRs in root-microbe interactions.
It is conceivable that a specialized set of phylogenetically related NLRs could have evolved specifically to guard the root defense machinery or other root specific pathways, and that the Brassicaceae NLR expression shift might be caused by the loss of such a group of NLRs. Here, we tested this hypothesis by grouping NLRs according to the sequence homology of their NBD domains, identifying five major clades. While the CNL-enriched NLR clade 2 was strongly depleted in the Brassicaceae, it was well-represented in all other plant species examined. In addition, we observed a Brassicaceae shoot skew for all NLR clades, with the smallest shift observed for TNLs, which are absent in the monocots rice (Oryza sativa) and maize (Zea mays) and therefore cannot be generally required for protecting root components. The shift in Brassicaceae NLR expression could thus not be attributed to the loss of a single NLR clade, and our data did not support the existence of a specific group of phylogenetically distinct NLRs guarding the root.
Brassicaceae and monocot species have previously been identified as outliers with respect to NLR regulation in the context of small RNAs. Whereas large numbers of NLR transcripts are targeted by small RNAs in legumes and other dicots (Zhai et al., 2011; Zhang et al., 2016), Brassicaceae and monocot species appear only to regulate a handful of NLRs by small RNA targeting (Zhang et al., 2016). Small RNA NLR regulation is often initiated by 22 nucleotide miRNA targeting followed by NLR transcript degradation, which leads to the production of secondary phased and transacting small interfering RNAs (Arikit et al., 2014; Axtell, 2013; Fei et al., 2015; Zhai et al., 2011). In most species, the majority of small RNA-regulated NLRs are part of large gene families targeted by the miR482/2118 superfamily of microRNAs (Li, Pignatta et al., 2012; Shivaprasad et al., 2012; Zhai et al., 2011; Zhang et al., 2016). It has been suggested that the limited miRNA targeting of NLRs in Brassicaceae and monocots could be due to absence of the large NLR gene families targeted by miR482 (Zhang et al., 2016). Although the Brassicaceae and monocots show opposite trends with respect to root/shoot NLR expression ratios, it is possible that small RNA regulation of NLR transcript levels could play a role in establishing organ-specific NLR expression patterns in legumes and other species with large numbers of NLR targeted by small RNAs.
In the Brassicaceae, the general expression shift toward the shoot across four major NLR clades suggests a reduced anticipation of effector challenge to root cells relative to shoot cells. We envisage two scenarios, which are not mutually exclusive, that could account for the shift. First, our data are consistent with a model where NLRs were randomly recruited from an expanding NLR complement, regardless of phylogenetic origin, for guarding root specific components. When the guarded pathways became defunct in the Brassicaceae family, it gradually lost the associated root-expressed NLRs across the different NLR clades, leading to the overall shoot skew in NLR expression. Second, rather than passively reducing the effector challenge level to roots by loss of a signaling pathway prone to microbial effector targeting, the Brassicaceae could have minimized their requirement for root NLR protection by developing family-specific active measures that efficiently deter putative soil pathogens before they have a chance to deploy their effectors. One possibility is that the Brassicaceae maintain high levels of antimicrobial glucosinolates in the root apoplasm, and there are indications that roots have higher constitutive glucosinolate levels than shoots (Van Dam et al., 2009). Another is that the Brassicaceae have evolved a unique set of highly efficient pattern recognition receptors that quickly eliminate putative root pathogens. For instance, the Ef-Tu and lipopolysaccharide PRRs are thought to be Brassicaceae-specific (Kunze et al., 2004; Ranf et al., 2015).
The Brassicaceae family made up a very conspicuous group of outliers that displayed shoot- rather than root-skewed NLR expression. The Brassicaceae are also outliers in the sense that they have lost the common symbiosis signaling pathway, which remains functional in 80% to 90% of land plants (Parniske, 2008; Delaux et al., 2014). It is tempting speculate if the two events are related, since the loss of the common symbiosis signaling pathway would both have removed a potentially heavily NLR-guarded pathway and eliminated constraints impeding development of more effective general root defense systems. In any case, the NLR expression shift described here suggests that the Brassicaceae may have evolved rich repertoires of unique PRRs and antimicrobial root metabolites to substitute for NLR protection. Such fundamental differences in root protection strategies could potentially be exploited in crop rotation schemes or for enhancing root defense systems of non-Brassicaceae crops.
MATERIALS AND METHODS
Identification of Putative NLR Genes
To allow identification of putative NLR genes, protein sequences were downloaded as indicated (Supplemental Table S1). Annotation versions were chosen for compatibility with the available microarray or RNA-seq data to allow subsequent expression analysis. This is why the latest versions were not used in all cases. NLR genes were then identified in a three-step procedure. First, candidate genes were selected using HMMER 3.1b1 (Eddy 2011) based on the NB-ARC PFAM protein domain PF00931. Second, the candidate list was filtered by performing a search for conserved protein domains using CDD (Marchler-Bauer et al., 2011), requiring that the selected putative NLR proteins contain, in addition to the NB-ARC domain, either LRR, TIR, PLN00113, PLN03194, or PLN03210 domains. Third, all NLR protein sequences were manually curated to identify and remove false positives. The total number of identified NLR proteins in each of the 18 species is shown in Supplemental Table S5, with sequences available in Supplemental File S4.
Lotus RNA-seq
Lotus (Lotus japonicus) ecotype Gifu (Handberg and Stougaard, 1992) seeds were surface sterilized, germinated, and grown in conditions as described previously (Kawaharada et al., 2015). Three biological replicates per sample were analyzed with each consisting of 10 seedlings grown on 1/4 B&D plates for 10 d before inoculation of the roots with 750 μL of an M. loti R7A suspension (OD600 = 0.02) or water. Three days post inoculation, roots and shoots were separated and total RNA was isolated using a NucleoSpin RNA Plant kit (Macherey-Nagel) according to the manufacturer’s instructions. RNA quality was assessed with an Agilent 2100 Bioanalyser, and samples were sent to GATC Biotech (http://gatc-biotech.com/) for library preparation and sequencing. Sequencing data have been deposited at the NCBI Short Read Archive with BioProject ID PRJNA384655 and are available for analysis on Lotus Base (Mun et al., 2016). Read counts were generated by mapping the reads to the L. japonicus v. 3.0 genome using CLC Genomics Workbench 9.5.3.
Analysis of NLR Gene Expression Data
For tissue-specific gene expression enrichment analysis (Fig. 1, A–D), we classified genes as being enriched in a specific tissue group if the average expression level in that group was higher than the average of all other tissue groups and at least two times higher than that of at least one other tissue group.
To evaluate root/shoot expression ratios, available expression data were downloaded as indicated in Supplemental Table S2. Sample IDs along with normalized expression values are available in Supplemental File S1. For Lotus and Brassica rapa, probes were reassigned to the updated annotation using BLAST to match probe and cDNA sequences (e-value cutoff 0.001), assigning only the best matching probe to a gene. For Lotus, Medicago, and soybean (Glycine max), samples representing identical or closely related plant accessions were used in the analysis. For rice (Oryza sativa) and maize (Zea mays), data from a number of different accessions were used, but only data where both root and shoot samples had been assayed within the same experiment were used to ensure the comparability of samples from the two tissues. For oilseed rape (Brassica napus), the analysis was based on raw RNA-seq reads. RNA-seq data files were downloaded from the NCBI short read archive (https://www.ncbi.nlm.nih.gov/sra), and reads from each library were assembled using Trinity (–full_cleanup) (Haas et al., 2013) followed by clustering using cd-hit-est v.4.6.6 (-M 16000 -T 8) (Fu et al., 2012). Next, the longest open reading frames were identified for each transcript, and the corresponding protein sequences were used for identification of NLRs as described. Reads were mapped back to the gene set output from cd-hit-est using STAR (–runMode genomeGenerate–genomeChrBinNbits 14) parameters for index generation and standard options for mapping (Dobin et al., 2013). Finally, reads mapping to multiple locations were filtered out followed by summarizing read counts per gene for each sample. Microarray data were analyzed directly, maintaining the original intersample normalization. For RNA-seq data, we simulated compositional bias by randomly selecting 100 genes and increasing their expression level 10,000-fold in either root or shoot samples followed by downscaling expression levels of all genes to match the original library size. RNA-seq data normalization was carried out using TMM implemented in edgeR (Robinson et al., 2010), PoissonSeq (Li, Witten, et al., 2012), voom from the limma R package (Ritchie et al., 2015), and normalization factors calculated based on total library read count.
For all species, expression data from the genes with the 15% lowest expression levels were filtered out, and the log2 NLR root/shoot expression ratios were normalized by subtracting the mean value for all genes. Expression ratios were plotted using ggplot2 in R version 3.1.2. Where multiple data sets were available for each species, RNA-seq data were used in the analysis of root/shoot expression ratios to include as many genes as possible. For Arabidopsis (Arabidopsis thaliana), average expression ratios based on the two available RNA-seq data sets were used.
The significance of differences in mean expression ratios between all genes and NLR genes were evaluated using Student’s t test (t.test) and the Kolmogorov-Smirnov test (ks.test) as implemented in R (Supplemental Table S4). Density plots were generated using the geom_density function of ggplot2 in R with standard options, and experimental cumulative distribution plots were generated using the R ecdf function with standard options. The significance levels of interspecies differences in root/shoot expression ratios (Supplemental Table S6) and differences in the average root/shoot expression by NLR gene clade or domain based on the phylogenetic tree shown in Figure 2B (Supplemental Tables S8 and S9) were evaluated by fitting a generalized linear model, for example, model <− glm(log(root/shoot) ∼ species), and subsequently using the glht function from the multcomp R package to calculate P values, for example, glht(model, linfct = mcp(Species = ”Tukey”)).
Construction of NLR Protein Phylogeny
Sequences of the NB-ARC domains of identified R genes were extracted based on domains identified by the CCD search using a python script, and aligned using Clustal Omega v1.2.3 (Sievers et al., 2011). Sequences were then filtered for low coverage positions (50% cutoff) and sequences lacking >50% of the aligned NB-ARC domain were removed. Phylogenetic trees were constructed in IQ-Tree v.1.5.2 and evaluated using the ultrafast bootstrap approximation approach (UFBoot; Minh et al., 2013; Nguyen et al., 2015). The resulting tree was colored by species using colorTree v1.1 and visualized using Dendroscope v3.5.7 (Chen and Lercher, 2009; Huson and Scornavacca, 2012). See Supplemental Files S2 and S3 for NB-ARC domain alignments of the trees described in Figures 2B and 3B, respectively, along with bootstrap analysis. See Supplemental File S4 for sequences for all NLRs used to construct the phylogenetic trees, and Supplemental File S5 for a general overview of all NLRs used in the study. All python scripts and commands for the above analyses are available in Supplemental File S6.
Accession Numbers
Sequence data from this article can be found in the NCBI Sequence Read Archive under accession number PRJNA384655.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of rhizobium inoculation on root/shoot expression ratios.
Supplemental Figure S2. Interspecies variation in root/shoot expression ratios: density plots.
Supplemental Figure S3. Interspecies variation in root/shoot expression ratios: cumulative distribution function plots.
Supplemental Figure S4. Log root/shoot expression ratios by NLR domain type.
Supplemental File S1. Normalized expression data.
Supplemental File S2. NB-ARC alignment of sequences used to construct the phylogenetic tree in Figure 2B, along with bootstrap analysis of the resulting phylogenetic tree.
Supplemental File S3. NB-ARC alignment of sequences used to construct the phylogenetic tree in Supplemental Figure S3B, along with bootstrap analysis of the resulting phylogenetic tree.
Supplemental File S4. Full-length sequences for all NLRs identified and used in this study.
Supplemental File S5. Full list of NLR genes identified, with domains, designations, and normalized log2 root/shoot expression ratios.
Supplemental File S6. Python scripts and commands used for extraction, alignment, bootstrapping, and creation of phylogenetic trees of NBD sequences.
Supplemental Table S1. Sources of the protein sequences used in the NLR analysis.
Supplemental Table S2. Expression data sources used in the NLR analysis.
Supplemental Table S3. Effects of RNA-seq data normalization methods.
Supplemental Table S4. Mean log (root/shoot) gene expression ratios for NLR and all genes.
Supplemental Table S5. Summary of NLR genes and expression data availability.
Supplemental Table S6. Cross-species comparison of normalized log2 root/shoot NLR expression ratios supporting Figure 1I.
Supplemental Table S7. Clade distribution of NLR genes for the phylogenetic tree used in Figure 2B.
Supplemental Table S8. Cross-species domain comparison of normalized log2 root/shoot NLR gene expression ratios for Supplemental Figure S4.
Supplemental Table S9. Cross-species clade comparison of normalized log2 root/shoot NLR gene expression ratios for Figure 2, C and D.
Supplemental Table S10. Clade distribution of NLR genes for the phylogenetic tree shown in Figure 3B.
Acknowledgments
The authors acknowledge all research groups contributing expression data used in our meta-analysis.
Footnotes
This work was supported by the Danish National Research Foundation grant no. DNRF79.
Articles can be viewed without a subscription.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels JM, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A 95: 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameline-Torregrosa C, Wang B-B, O'Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146: 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdés-López O, Prince S, Musket TA, Nguyen HT, et al. (2014) An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Banba M, Gutjahr C, Miyao A, Hirochika H, Paszkowski U, Kouchi H, Imaizumi-Anraku H (2008) Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol 49: 1659–1671 [DOI] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P (1990) Medicago Truncatula, a model plant for studying the molecular genetics of the rhizobium-legume symbiosis. Plant Mol Biol Rep 8: 40–49. [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC (2011) Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6: e26968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Op Immunol 24: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A 108: 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838 [DOI] [PubMed] [Google Scholar]
- Chen WH, Lercher MJ (2009) ColorTree: a batch customization tool for phylogenic trees. BMC Res Notes 2: 155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel L-P, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM (2014) Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet 10: e1004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. (1998) Phylogenetic perspectives on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci [Google Scholar]
- Eddy SR. (2011) Accelerated profile HMM searches. PLOS Comput Biol 7: e1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Lenk C, Degenhardt J, Turlings TC (2009) The underestimated role of roots in defense against leaf attackers. Trends Plant Sci 14: 653–659 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci U S A 96: 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima U, Senthil-Kumar M (2015) Plant and pathogen nutrient acquisition strategies. Front Plant Sci 6: 750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Li P, Teng C, Meyers BC (2015) Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J 83: 451–465 [DOI] [PubMed] [Google Scholar]
- Fluhr R. (2001) Sentinels of disease. Plant resistance genes. Plant Physiol 127: 1367–1374 [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualtieri G, Bisseling T (2000) The evolution of nodulation. Plant Mol Biol 42: 181–194 [PubMed] [Google Scholar]
- Guillotin B, Couzigou J-M, Combier J-P (2016) NIN is involved in the regulation of arbuscular mycorrhizal symbiosis. Front Plant Sci 7: 1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus Japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496. [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hofius D, Tsitsigiannis DI, Jones JD, Mundy J (2007) Inducible cell death in plant immunity. Semin Cancer Biol 17: 166–187 [DOI] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS One 4: e6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Lyons E, Hernández-Guzmán G, Pérez-Torres CA, Carretero-Paulet L, Chang T-H, Lan T, Welch AJ, Juárez MJ, Simpson J, et al. (2013) Architecture and evolution of a minute plant genome. Nature 498: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24: 89–167 [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D (2016) Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr Opin Microbiol 29: 49–55 [DOI] [PubMed] [Google Scholar]
- Kim J, Lim CJ, Lee B-W, Choi J-P, Oh S-K, Ahmad R, Kwon S-Y, Ahn J, Hur C-G (2012) A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant kingdom. Mol Cells 33: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel J-B (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol 210: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre R, Evangelisti E, Rey T, Schornack S (2015) Modulation of host cell biology by plant pathogenic microbes. Annu Rev Cell Dev Biol 31: 201–229 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet E-P, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Witten DM, Johnstone IM, Tibshirani R (2012) Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 13: 523–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Yang S-Y, Hong Y-T, Huang S-M, Wang F-N, Chiang S-F, Tsai S-Y, Lu W-C, Chiou T-J (2016) Identification of plant vacuolar transporters mediating phosphate storage. Nat Commun 7: 11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR. (1989) Rhizobium-legume nodulation: life together in the underground. Cell 56: 203–214 [DOI] [PubMed] [Google Scholar]
- Lukasik E, Takken FL (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquenet E, Richet E (2007) How integration of positive and negative regulatory signals by a STAND signaling protein depends on ATP hydrolysis. Mol Cell 28: 187–199 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30: 1188–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109: 1434–1447 [DOI] [PubMed] [Google Scholar]
- Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU (2016) Lotus Base: an integrated information portal for the model legume Lotus japonicus. Sci Rep 6: 39447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamilarasan M, Prasad M (2013) Plant innate immunity: an updated insight into defense mechanism. J Biosci 38: 433–449 [DOI] [PubMed] [Google Scholar]
- Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL (2010) Arabidopsis and the plant immune system. Plant J 61: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Op Plant Biol 9: 351–357. [DOI] [PubMed] [Google Scholar]
- Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50: 203–213 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pel MJC, Pieterse CMJ (2012) Microbial recognition and evasion of host immunity. J Exp Bot 64: 1237–1248. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, Sánchez-Carballo PM, Zähringer U, Hückelhoven R, Lee J, et al. (2015) A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 16: 426–433 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shao Z-Q, Zhang Y-M, Hang Y-Y, Xue J-Y, Zhou G-C, Wu P, Wu X-Y, Wu X-Z, Wang Q, Wang B et al. (2014) Long-Term Evolution of Nucleotide-Binding Site-Leucine-Rich Repeat Genes: Understanding Gained from and beyond the Legume Family. Plant Physiol 166: 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z-Q, Xue J-Y, Wu P, Zhang Y-M, Wu Y, Hang Y-Y, Wang B, Chen J-Q (2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BACM, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (2010) Mycorrhizal Symbiosis. Academic Press. [Google Scholar]
- Song H, Wang PF, Li TT, Xia H, Zhao SZ, Hou L, Zhao CZ (2015) Genome-wide identification and evolutionary analysis of nucleotide-binding site-encoding resistance genes in Lotus japonicus (Fabaceae). Genet Mol Res 14: 16024–16040 [DOI] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Takken FLW, Goverse A (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr Op Plant Biol 15: 375–384. doi:. [DOI] [PubMed] [Google Scholar]
- Tarr DE, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2: 197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H. (2003) Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Van Dam NM, Tytgat TOG, Kirkegaard JA (2009) Root and shoot glucosinolates: a comparison of their diversity, function and interactions in natural and managed ecosystems. Phytochem Rev [Google Scholar]
- van der Biezen EA, Jones JD (1998) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van Veen H, Vashisht D, Akman M, Girke T, Mustroph A, Reinen E, Hartman S, Kooiker M, van Tienderen P, Schranz ME, et al. (2016) Transcriptomes of eight Arabidopsis thaliana accessions reveal core conserved, genotype- and organ-specific responses to flooding stress. Plant Physiol 172: 668–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206: 1196–1206 [DOI] [PubMed] [Google Scholar]
- Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, Tang Y, Murray JD, Udvardi MK (2013) Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J 74: 351–362 [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee DU, Fu X-D, Dong X (2011) Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Findlay K, Oliver RP, Turner JG (1997) Characterization of three loci controlling resistance of. Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J 12: 110–768 [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Xin X-F, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Zhang F, Tong C, Huang J, Cheng X, Dong C, Zhou Y, Qin R, Hua W et al. (2014) Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana BMC genomics 15: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgadzaj R, Garrido-Oter R, Jensen DB, Koprivova A, Schulze-Lefert P, Radutoiu S (2016) Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA 113: E7996–E8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong D-H, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xia R, Kuang H, Meyers BC (2016) The diversification of plant NBS-LRR defense genes directs the evolution of MicroRNAs that target them. Mol Biol Evol 33: 2692–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]