Despite previous arguments on chloroplast lipid droplets, all lipid droplets are present in the cytosolic compartment and not in the chloroplast in Chlamydomonas reinhardtii.
Abstract
The precise localization of the lipid droplets and the metabolic pathways associated with oil production are crucial to the engineering of microalgae for biofuel production. Several studies have reported detecting lipid droplets within the chloroplast of the microalga Chlamydomonas reinhardtii, which accumulates considerable amounts of triacylglycerol and starch within the cell under nitrogen deprivation or high-light stress conditions. Starch undoubtedly accumulates within the chloroplast, but there have been debates on the localization of the lipid droplets, which are cytosolic organelles in other organisms. Although it is impossible to prove an absence, we tried to repeat experiments that previously indicated the presence of lipid droplets in chloroplasts. Here, we present microscopic results showing no evidence for the presence of lipid droplets within the chloroplast stroma, even though some lipid droplets existed in close association with the chloroplast or were largely engulfed by the chloroplasts. These lipid droplets are cytosolic structures, distinct from the plastoglobules present in the chloroplast stroma. These results not only contrast with the old ideas but also point out that what were previously thought to be chloroplast lipid droplets are likely to be embedded within chloroplast invaginations in association with the outer envelope of the chloroplast without intervention of the endoplasmic reticulum. These findings point to the intriguing possibility of a tight metabolic flow from the chloroplast to the lipid droplet through a close association rather than direct contact of both organelles.
“Il est impossible de prouver la négative,” answered Louis Pasteur (1922) to the partisans of spontaneous generation. This famous phrase was meant to express the theoretical impossibility of denying what never exists. The same argument seems to be valid for the current debate on whether lipid droplets occur within the chloroplast in the microalga Chlamydomonas reinhardtii (Chlamydomonas), a model alga that accumulates a considerable amount of triacylglycerol (TAG) and starch within the cell under nitrogen-deprived or high-light stress conditions (Siaut et al., 2011). Starch undoubtedly accumulates within the chloroplast, but there have been arguments on the localization of the lipid droplets, which are normally cytosolic organelles in plants and other organisms. Three studies on the presence of lipid droplets in the chloroplast have reported two lines of evidence: one based on starchless mutants under nitrogen-deficient conditions (Fan et al., 2011; Goodson et al., 2011) and the other based on wild-type cells under high-light stress (Goold et al., 2016). Localization of the lipid droplets is crucial to engineering microalgae for biofuel production, which relies on the biochemical and cytological understanding of TAG synthesis and accumulation (Merchant et al., 2012; Johnson and Alric, 2013; Li-Beisson et al., 2015). Thus, we sought to resolve the controversy by carefully repeating the previous electron microscopy experiments and using confocal 3D image reconstruction.
The lipid droplet is a globular organelle enclosing TAG as a major constituent and is covered by a one-half-unit membrane consisting of phospholipids and specific proteins. A homologous organelle is found in plants and algae as well as in nonphotosynthetic eukaryotes. In plants, the lipid droplet is known to emerge from the endoplasmic reticulum (ER) by budding. Many protein components of the lipid droplet membrane are known in plants and include oleosin, caleosin, and sterol dehydrogenase (Chapman et al., 2012), which are believed to stabilize the lipid droplet structure. In Chlamydomonas, different kinds of proteins are present in the limiting membrane of the lipid droplet (Moellering and Benning, 2010; Nguyen et al., 2011; Tsai et al., 2015). Lipid droplets can be small (less than 200 nm in diameter) or large (up to 1 µm in diameter). A number of microalgal biofuel production studies have focused on increasing oil production. TAG, a major component of the lipid droplet, is synthesized in the ER and on the surface of the lipid droplet, with diacylglycerol and acyl-CoA or phosphatidylcholine as major precursors (Bates, 2016; Zienkiewicz et al., 2016). The pathway of TAG synthesis in plants and microalgae is supposed to be similar to that in animal cells and yeast, with some additional pathways (Merchant et al., 2012; Johnson and Alric, 2013; Li-Beisson et al., 2015; Bates, 2016; Zienkiewicz et al., 2016).
The chloroplast is the major site of fatty acid synthesis in plants and algae. Some of the fatty acids are transported to the cytosolic compartment and used for the synthesis of phospholipids in the ER, whereas the remaining fatty acids are used to synthesize chloroplast lipids, such as galactolipids (Li-Beisson et al., 2015). Under nitrogen-deprived conditions, fatty acids are mobilized from galactolipids to synthesize TAG (Li et al., 2012; Zienkiewicz et al., 2016). This raises the naïve suggestion that TAG also could be synthesized and accumulated within the chloroplast (Fan et al., 2011; Goodson et al., 2011). An argument for this comes from the presence of TAG in the plastoglobule, another type of lipid-containing structure that is ubiquitously present in the chloroplast (Bréhélin et al., 2007; Lohscheider and Río Bártulos, 2016). Cyanobacteria, which are considered descendants of an ancestral endosymbiont that engendered chloroplasts, also contain a lipidic structure apparently similar to plastoglobules (van de Meene et al., 2006). The plastoglobule is a small globular particle (100–200 nm in diameter) present in the vicinity of the photosynthetic membranes. It contains various isoprenoids, such as plastoquinones and tocopherols, but also TAG and membrane lipids (Tevini and Steinmüller, 1985). The enzyme farnesyl ester synthetase (FES1/2) has the ability to synthesize TAG in chloroplasts (Lippold et al., 2012).
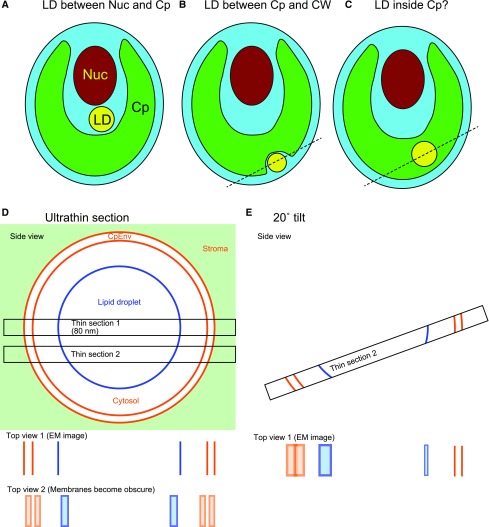
The localization of lipid droplets has been analyzed in detail by freeze-fracture electron microscopy in Chlamydomonas (Goodson et al., 2011). That study affirmed that lipid droplets are present (1) in the cytosol between the nucleus and the chloroplast, (2) in the cytosol outside the chloroplast, or (3) within the chloroplast stroma, shown as localization types A to C (Fig. 1, A–C). The first two locations are commonly found in oil-accumulating Chlamydomonas, and this is consistent with the presence of lipid droplets within the cytosol in animal cells and yeast. The third location is reportedly found only in starchless mutants under acetate-boost conditions. This type of lipid droplet is called cpst-LB (for chloroplast lipid body). Another study that also reported lipid droplets within the chloroplast examined ultrathin sections by transmission electron microscopy (Fan et al., 2011). A more recent study argues that lipid droplets accumulate in the chloroplast under saturating light stress (Goold et al., 2016).
Figure 1.
Schematic model of the relationship between the lipid droplet and the chloroplast. A to C, Schematic views of hypothetical different localizations of lipid droplets (LD). Cp, Chloroplast; CW, cell wall; Nuc, nucleus. D and E, Schematic explanation of different views of the chloroplast envelope membranes (CpEnv) in an ultrathin section. Detection of the chloroplast envelope membranes is crucial in identifying the localization of a lipid droplet. We consider the case in which a lipid droplet is present within a cytosolic compartment enclosed within the chloroplast, as shown by the broken lines in B and C. In this case, the lipid droplet must be surrounded by the chloroplast envelope membranes. If a section is cut at the center of the lipid droplet that appears to be located within the chloroplast, we can recognize that the lipid droplet is surrounded by the chloroplast envelope membranes and, therefore, is located within the cytosolic compartment (section 1). But if a section (section 2) is cut out of the center, then the chloroplast envelope membranes and the lipid droplet half-membrane become obscure, because the thickness of the section (usually 70–100 nm) is much larger than the thickness of the membranes (7–10 nm for envelope membranes and 3–4 nm for the lipid droplet half-membrane). If the section is appropriately tilted (E), then we can identify the membranes partly. EM, Electron micrograph.
The presence of lipid droplets in the chloroplast is currently being discussed by the lipid research community. There are several reasons to cast doubt about type C localization. First, all known enzymes (except FES1/2) involved in TAG synthesis are targeted to the ER. Second, there is no solid evidence that cyanobacteria have the ability to accumulate TAG as lipid droplets, as is seen in eukaryotes. In addition, the lipid droplet and plastoglobule are different entities that must be distinguished.
This study was undertaken in an effort to find evidence for type C localization. It was important for us to repeat the previous experiments that showed type C localization with great care. If we obtained a positive result for the presence of lipid droplets within the chloroplast, this could be useful in algal engineering (Misra et al., 2012; Gargouri et al., 2015; Bhowmick et al., 2016). We acknowledge that even if we could not confirm type C localization, such a localization could have been present in other experiments under different conditions, at least theoretically. Nevertheless, if we were unable to reproduce past observations under a variety of conditions, it would call into question past findings of type C localization. Here, we present various relevant observations that lead us to be quite doubtful about type C localization; however, we did notice an intriguing possibility of direct metabolic flow between the chloroplast and lipid droplets.
RESULTS
High-Light Intensity-Induced Accumulation of Lipid Droplets in Wild-Type Cells
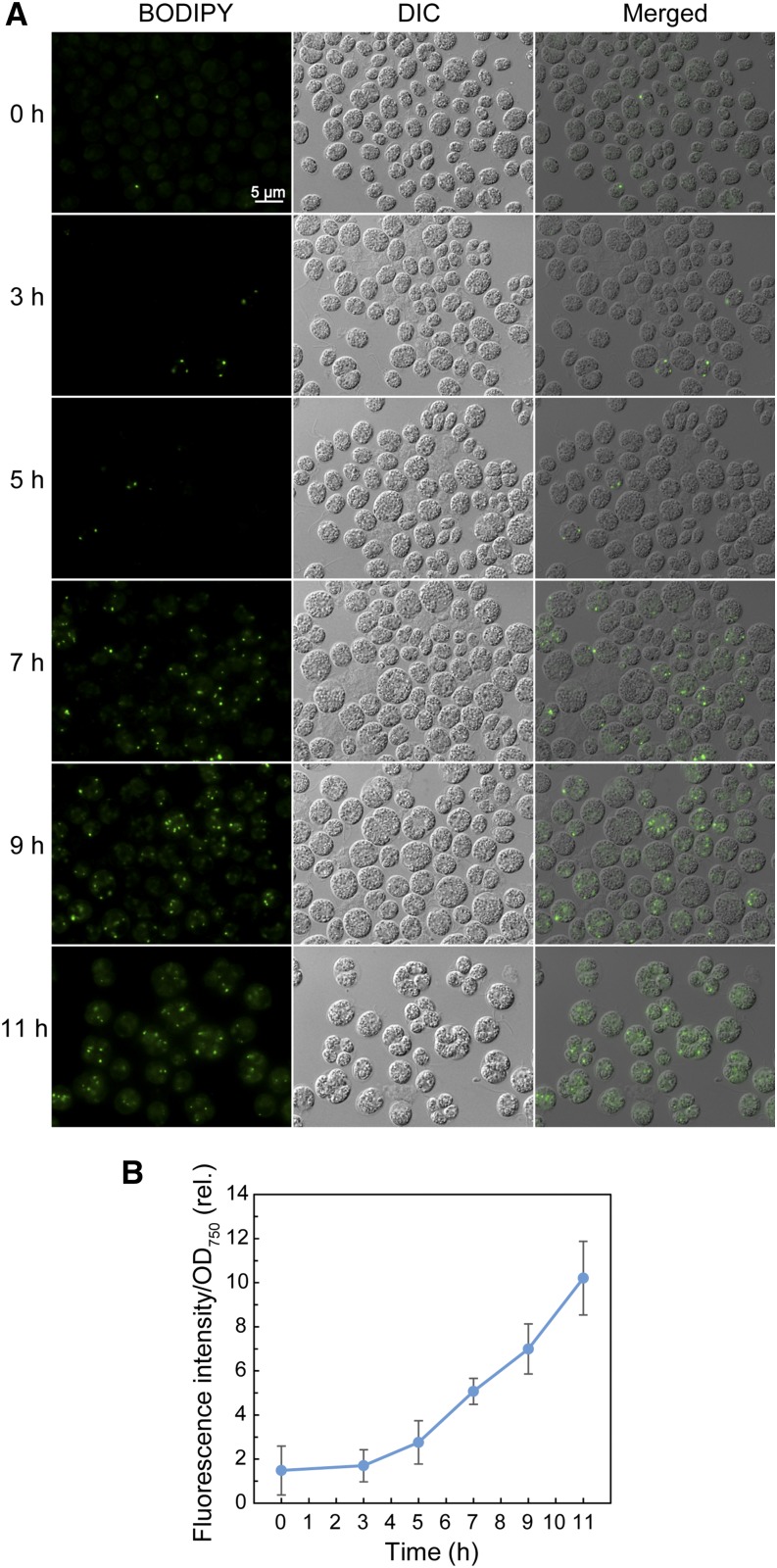
To confirm that the wild-type cells accumulate neutral lipids in response to high-light intensity, as reported in strain CC-124 (Goold et al., 2016), we used strain CC-1010 (Sakurai et al., 2014) and examined lipid droplets stained with BODIPY after a shift from low light (40 μmol m–2 s–1) to high light (200 μmol m–2 s–1; Fig. 2A). Lipid droplet accumulation was estimated by quantifying Nile Red fluorescence (Fig. 2B). The results showed that the fluorescence signals of the lipid droplets increased linearly with time under the high-light condition. As far as the microscopy images indicated, the lipid droplets detected corresponded to the plastidal lipid droplets reported by Goold et al. (2016), with respect to their size and distribution within the cell. Quantification of TAG by chemical analysis (Table I) gave consistent results with those of fluorometry, which revealed that CC-1010 cells also accumulated TAG upon transfer to the high-light condition and that the continuous culture system used by Goold et al. (2016) was not obligatory for this phenomenon. The TAG contained only a low level of 16:4 and 18:3(9,12,15) fatty acids, suggesting that the contribution of chloroplast membrane lipids to the TAG pool was not important under this condition, in contrast to the nitrogen-deficient conditions reported previously (Li et al., 2012; Sakurai et al., 2014).
Figure 2.
Neutral lipid accumulation in Chlamydomonas CC1010 cells subjected to high-light irradiation. A, Observation of BODIPY-stained cells. The cells grown under low light were shifted to high light and grown for 11 h. DIC, Differential interference contrast microscope image; Merged, merged image of BODIPY and differential interference contrast microscopy images. B, Measurement of the fluorescent signal of Nile Red. Cells grown as in A were stained with Nile Red, and the fluorescent signal was measured by a plate reader.
Table I. Fatty acid composition and accumulation of TAG in Chlamydomonas CC1010 cells subjected to high-light irradiation for 7 h (n = 3).
The values show the composition of fatty acids in percentage. The level of TAG is shown at the bottom. It was noted that the fatty acids specific to chloroplast lipids, namely 16:4 and 18:3(9,12,15), did not appreciably accumulate in TAG. As reported by Sakurai et al. (2014), 18:4 was supposed to be a mixture of 18:4(5,9,12,15) and 18:4(6,9,12,15), but, as reported by Sato et al. (2016), 18:4(6,9,12,15) was not detected in Chlamydomonas CC-1010.
| Fatty Acid | Total Lipids |
TAG |
||
|---|---|---|---|---|
| 0 h | 7 h | 0 h | 7 h | |
| 14:0 | 0.4 ± 0.1 | 0.4 ± 0.0 | 4.4 ± 0.7 | 2.2 ± 0.8 |
| 14:1 | 0.3 ± 0.1 | 0.2 ± 0.0 | 1.4 ± 0.3 | 0.9 ± 0.7 |
| 16:0 | 24.2 ± 1.8 | 24.7 ± 0.3 | 35.4 ± 2.5 | 30.6 ± 3.2 |
| 16:1(7) | 2.0 ± 0.4 | 2.5 ± 0.6 | 1.7 ± 0.1 | 2.0 ± 0.5 |
| 16:2(7,10) | 4.8 ± 1.0 | 3.3 ± 0.7 | 1.1 ± 0.3 | 2.1 ± 0.8 |
| 16:3(4,7,10) | 0.9 ± 0.2 | 0.9 ± 0.0 | 0.3 ± 0.3 | 0.4 ± 0.1 |
| 16:3(7,10,13) | 7.8 ± 0.3 | 7.7 ± 0.8 | 6.7 ± 0.8 | 6.9 ± 1.5 |
| 16:4(4,7,10,13) | 10.1 ± 2.3 | 10.0 ± 1.3 | 2.3 ± 0.2 | 3.3 ± 0.1 |
| 17:0 | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.4 ± 0.4 | 0.4 ± 0.2 |
| 18:1(9) | 4.1 ± 0.2 | 5.1 ± 1.2 | 12.6 ± 1.9 | 16.1 ± 1.7 |
| 18:1(11) | 2.9 ± 0.7 | 3.0 ± 0.3 | 4.2 ± 1.0 | 3.9 ± 0.4 |
| 18:2(9,12) | 16.1 ± 2.8 | 14.7 ± 2.3 | 13.1 ± 1.3 | 14.5 ± 1.8 |
| 18:3(5,9,12) | 8.7 ± 0.5 | 9.0 ± 0.5 | 8.8 ± 1.1 | 7.3 ± 0.5 |
| 18:3(9,12,15) | 15.9 ± 1.9 | 16.9 ± 1.1 | 6.1 ± 0.6 | 8.0 ± 0.3 |
| 18:4(5,9,12,15) | 1.7 ± 0.2 | 1.6 ± 0.2 | 1.5 ± 0.4 | 1.7 ± 0.2 |
| TAG (% of total lipids) | – | – | 3.5 ± 0.2 | 7.8 ± 1.3 |
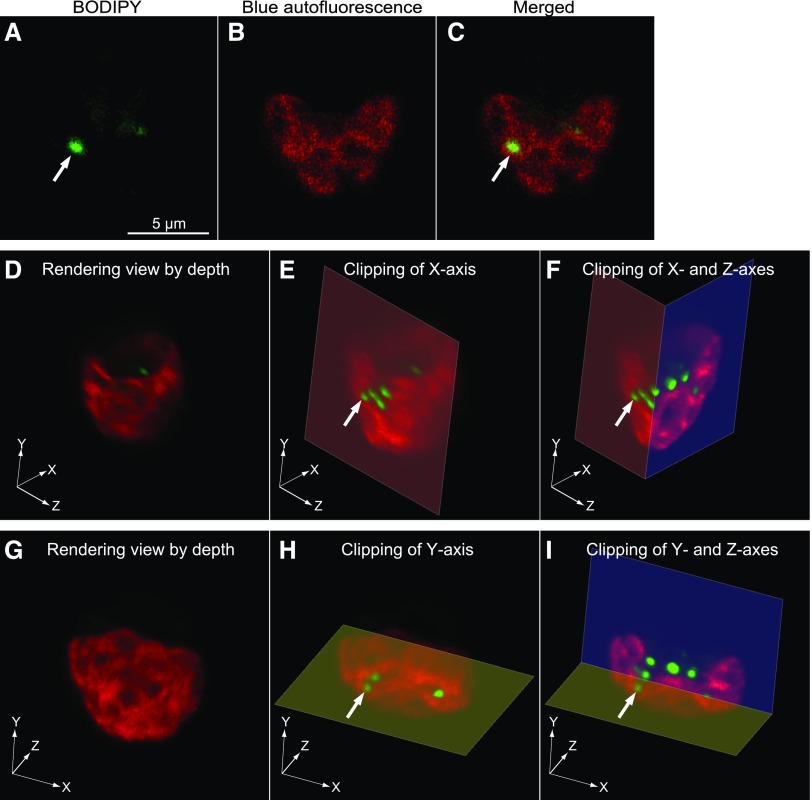
Subcellular localization of lipid droplets in cells 7 to 11 h after transfer to high light was examined by Z-stacking confocal fluorescence microscopy after staining with BODIPY or LipidTOX. The BODIPY and LipidTOX fluorescence signals coincided with each other (Supplemental Fig. S1). The great majority of lipid droplets appeared in the cytosolic compartment in the two-dimensional images. In rare cases, the lipid droplets appeared entirely embedded in the chloroplast (Fig. 3, A–C). Note that the chloroplast images were obtained from blue autofluorescence (shown in red pseudocolor) and not from chlorophyll fluorescence. This was useful for avoiding the contamination of lipid droplet fluorescence in the chloroplast image (for details, see “Materials and Methods”). The lipid droplets found in the cytosol and those apparent within the chloroplast were similar in size and appearance. We obtained Z-stacking images containing such lipid droplets at intervals of 0.2 to 0.4 μm and carefully examined the structures of the chloroplast and lipid droplets in the 3D image with clipping of the x, y, or z axis. We show three cases as examples in which the lipid droplets seemed to be present in the chloroplast. In case 1 (Fig. 3) and case 2 (Supplemental Fig. S2), the lipid droplets indicated by arrows/arrowheads were present within an invagination of the chloroplast (Fig. 3, D–I; Supplemental Fig. S2, D–F) but appeared entirely embedded within the chloroplast depending on the angle (Fig. 3; Supplemental Fig. S2, A–C). Case 3 (Supplemental Fig. S3) shows that the lipid droplet indicated by the arrowhead penetrated the chloroplast, whereas another lipid droplet (arrow) was located within an invagination of the chloroplast. Sequential TIF files for rendering the view and selected movie files of the stereo images with rolling are provided as Supplemental Data S1 to S5.
Figure 3.
3D reconstruction by confocal fluorescence microscopy of a wild-type Chlamydomonas CC-1010 cell after the transfer from low light to high light. Cells grown under high light for 7 h were stained with BODIPY and observed by confocal microscopy with Z-stacking. The fluorescence of BODIPY and blue autofluorescence are pseudocolored. A to C, 2D images of a slice in which a lipid droplet (white arrow) seemed to be entirely enclosed by the chloroplast. Images A and B show the fluorescence of BODIPY and blue autofluorescence, respectively. Merged, Merged image of BODIPY and autofluorescence images. D to I, Rendering images of the same cell as the one shown in A to C. D to F show the images viewed from an identical angle. G to I show the images viewed from a different angle. E and F are clipped images of D at the x axis (E) or the x and z axes (F). H and I are clipped images of G at the y axis (H) or the y and z axes (I). The white arrows indicate an identical lipid droplet in A, C, E, F, H, and I.
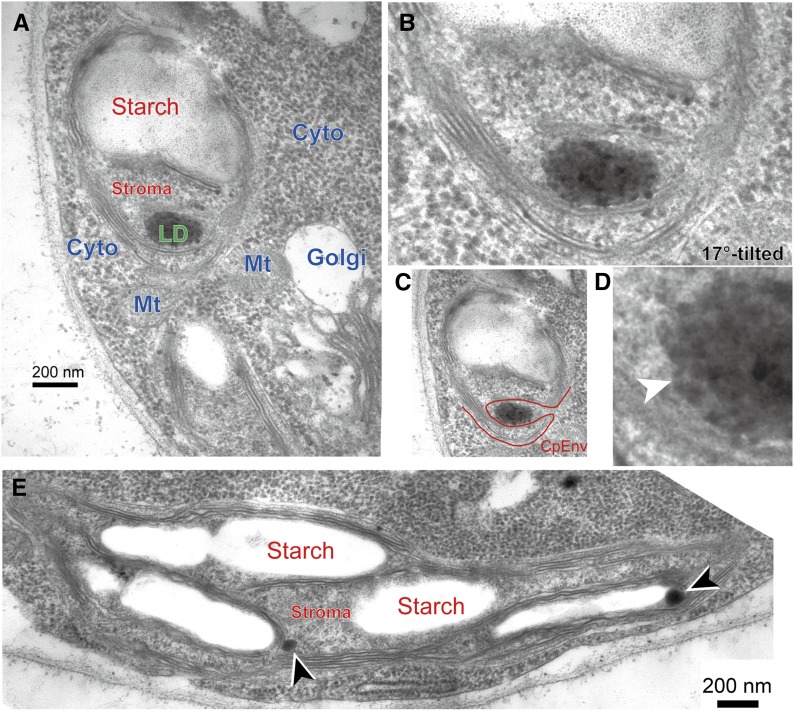
Examination by transmission electron microscopy also revealed the infrequent occurrence of lipid droplets that appeared to be within the chloroplast. The presence of chloroplast envelope membranes, which separated the inside and outside of the chloroplast, is a good criterion to judge if a lipid droplet is located within or outside a chloroplast. Because the thickness of the thin sections was about 10 times larger than the thickness of the chloroplast envelope membranes, the membranes were not always seen clearly (Fig. 1D). Proper tilting of the section in the microscope made it possible to clearly trace the envelope membranes (Fig. 1E). Figure 4A shows an example of a small lipid droplet apparently located within a chloroplast; however, an invagination was visualized by tilting (Fig. 4, B and C). This lipid droplet seemed to have a thin limiting membrane (Fig. 4D). In addition, the lipid droplet was clearly different from the plastoglobules, which were about 100-nm particles present in the stroma in close association with starch granules and thylakoid membranes (Fig. 4E). Although we examined many samples by confocal fluorescence and electron microscopy, we were unable to find any lipid droplet entirely enclosed within a chloroplast. In other words, all lipid droplets were present within the cytosolic compartment under high-light conditions.
Figure 4.
Electron micrographs of a lipid droplet entrapped by a chloroplast in wild-type Chlamydomonas CC-1010 cells 7 h after the transfer from low light to high light. A, Image of a chloroplast seemingly including a lipid droplet in its stroma. B, Image of the same chloroplast with 17° tilting. C, Same as B with tracing of the chloroplast envelope. D, Enlarged lipid droplet showing a putative limiting membrane (white arrowhead). E, Plastoglobules within the chloroplast (arrowheads). Cyto, Cytosol; Golgi, Golgi apparatus; LD, lipid droplet; Mt, mitochondrion; Starch, starch granule; Stroma, chloroplast stroma.
Starchless Mutants under Nitrogen Deprivation
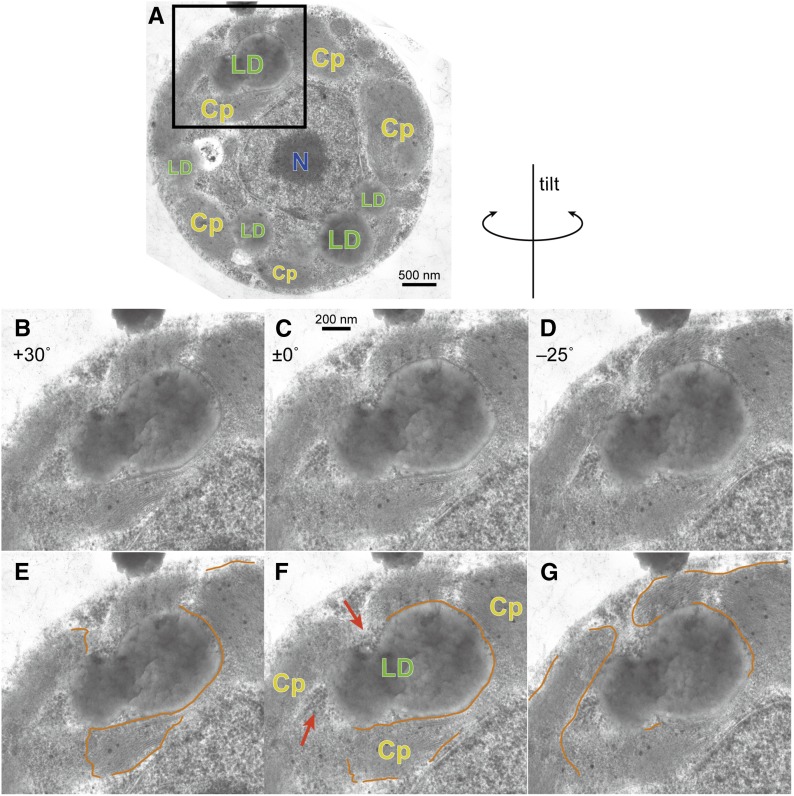
We then examined the subcellular localization of the lipid droplets in the starchless mutants cw15sta6 and cw15sta7 (Work et al., 2010; Fan et al., 2011; Goodson et al., 2011; Siaut et al., 2011), which accumulate TAG after an acetate boost under nitrogen deprivation. Figure 5A shows an electron microscopy image of a representative cw15sta6 cell containing lipid droplets, some of which were apparently present in the chloroplast (two lipid droplets were fused in this case). Tilted images of the lipid droplets (Fig. 5, B–D) allowed us to trace the envelope membranes enclosing the chloroplasts (Fig. 5, E–G, orange lines). A careful examination by connecting all traces revealed that the lipid droplets contacted with the cytosol at the sites indicated by the arrows. Supplemental Figure S4 shows the lipid droplets in cw15sta7 cells. It was clear that the lipid droplets were present in close association with the chloroplast, but they existed outside the chloroplast and were never entirely embedded in the chloroplast stroma.
Figure 5.
Electron micrographs of lipid droplets in the starchless mutant (cw15sta6) cells of Chlamydomonas, deprived of nitrogen for 24 h with supplementation of acetate. A, Whole cell. B to D, Tilted images as indicated along the directions shown above. E to G, Traces of identifiable parts of chloroplast envelope membranes (single lines for a set of inner and outer membranes). The visibility of the membranes depended on the tilt angle. Cp, Chloroplast; LD, lipid droplet; N, nucleus. Red arrows indicate the cytoplasmic ribosomes.
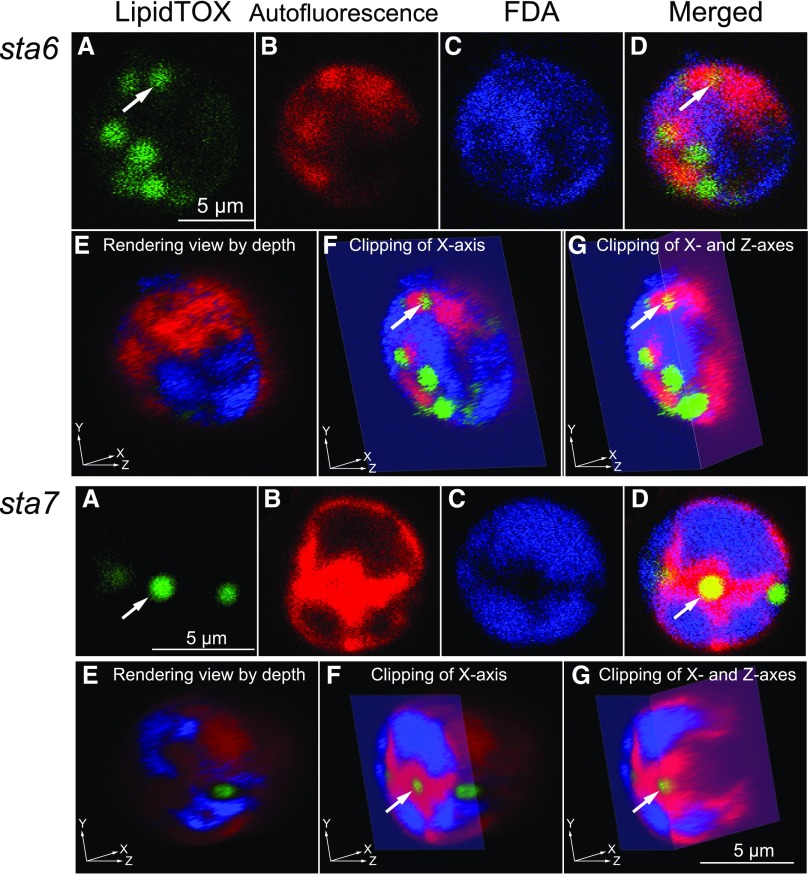
We also examined starchless mutants by confocal microscopy and 3D image reconstruction (Fig. 6). In this case, the cells were stained with visualizing reagents for lipid and cytosol without fixing the cells. Cytosolic staining with fluorescein diacetate (FDA) was possible only in living cells, because the staining depends on the cytosolic esterase that hydrolyzes FDA. Some lipid droplets indicated by the arrows were apparently located within the chloroplast, but clipping at appropriate slices showed that they were present within the cytosolic compartment, although they were deeply embedded within the invaginations. As in the case of lipid droplets that accumulate under high light, we never found any lipid droplets present in the chloroplast in the starchless mutants.
Figure 6.
3D reconstruction by confocal fluorescence microscopy of the starchless mutant cells of Chlamydomonas, deprived of nitrogen for 24 h with acetate supplementation. The top set of images shows a representative cw15sta6 cell; the bottom set of images shows a representative cw15sta7 cell. All images are pseudocolored. A, Localization of lipid droplets detected by LipidTOX fluorescence (green). B, Localization of chloroplast detected by blue autofluorescence (but shown in red). C, Localization of cytosol detected by FDA fluorescence (blue). D, Merged images of A to C. E to G, Views of 3D reconstructed images by different clipping. The staining with FDA in living cells will give uniform fluorescence after a long time due to hydrolysis of FDA by esterases leaked from broken cells. We had to work rapidly with the lowest concentration of FDA. That is why the intensity of fluorescence was weak in this observation.
DISCUSSION
Resolution of Type C Localization
We were finally led to the idea that it is impossible to prove what never exists, as Pasteur thought about 150 years ago with respect to spontaneous generation (Pasteur, 1922; the original version was published in the 1860s). He recognized the impossibility of disproving spontaneous generation but also understood the importance of checking against all possible pitfalls behind the arguments in its favor. We took the same strategy in our study on the presumptive lipid droplets in the chloroplast. We found various conditions in which lipid droplets were apparently present in the chloroplast (type C localization). We then carefully examined these lipid droplets by focusing on the presence of the chloroplast envelope membranes and the cytosol. All of our observations led us to conclude that the lipid droplets were not present within the chloroplast stroma, either under the high-light condition or under nitrogen deprivation.
We also reexamined the results of previously published studies on the type C localization of lipid droplets. Unfortunately, all previously presented data do not support type C localization. The lipid droplets in the fracture face reported by Goodson et al. (2011; Figure 11A) were clearly enclosed by double membranes, which were unambiguously identified as envelope membranes; in other words, they were located outside the chloroplast. The lipid droplets reported by Fan et al. (2011) were present in a chloroplast invagination but not entirely enclosed by a chloroplast (Supplemental Fig. S5A). In the fluorescence micrographs of a more recent report (Goold et al., 2016), fluorescence of the lipid droplets (stained with Nile Red) also appeared in the chlorophyll fluorescence channel (Supplemental Fig. S5B; in fact, each channel was a monochromatic image in the confocal microscope). As a consequence, the site where lipid droplets reside was shown as a red fluorescent area, and the lipid droplet (found in the green channel) was erroneously located within the chloroplast (but the red fluorescence in this area must be fluorescence due to the lipid droplet itself contaminating the red channel). Nile Red has a broad absorption peak with a very low absorbance, above 600 nm, and this resulted in an observable fluorescence excited at 635 nm (Supplemental Fig. S6A), which is normally used as an excitation beam for imaging chlorophylls. The same was essentially true for LipidTOX (Supplemental Figs. S6C and S7, D–F), whereas the problem was significantly alleviated with BODIPY (Supplemental Figs. S6B and S7, G–I). All of these problems were avoided by using blue autofluorescence for imaging chloroplasts or thylakoid membranes (Supplemental Fig. S7A and all other fluorescence figures in this study), although the exact origin of this fluorescence is unknown. All of these measurements suggested that the lipid droplets seemingly located within the chloroplast, as reported by Goold et al. (2016), were not really located within the chloroplast. Therefore, we conclude that there is currently no solid evidence for the presence of lipid droplets within the chloroplast in Chlamydomonas.
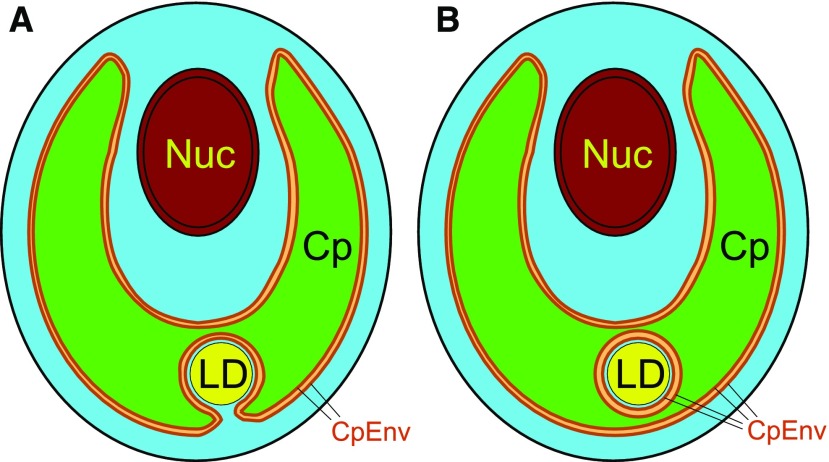
We finally propose a realistic model for the chloroplast lipid droplets in Chlamydomonas (Fig. 7). The lipid droplets seemingly present within the chloroplast stroma are actually present in the invaginations of the chloroplast (Fig. 7A). It is theoretically possible that they could be perfectly isolated from the cytosol (Fig. 7B), but we never found such lipid droplets with confidence. If the lipid droplets were actually isolated from the bulk cytosol, biochemical activities, such as protein synthesis and the supply of important metabolites such as ATP, would be quite limited.
Figure 7.
Models of the localization of lipid droplets in Chlamydomonas. A, Plausible model of a lipid droplet seemingly located within a chloroplast but localized within the cytosolic compartment, which is present in a chloroplast invagination. The close association of the chloroplast envelope and the lipid droplet half-membrane supports the transfer of fatty acids from the chloroplast to the lipid droplet. B, Hypothetical model of a lipid droplet seemingly embedded within a chloroplast. The cytosol is shown by cyan. Note that the lipid droplet is surrounded by cytosol. Cp, Chloroplast; CpEnv, chloroplast envelope; LD, lipid droplet; Nuc, nucleus. This figure shows only a lipid droplet seemingly present within the chloroplast (type C localization). Besides this type of lipid droplet, there are a number of lipid droplets located in the cytosol between the nucleus and the chloroplast (type A) or outside the chloroplast (type B).
Biochemical Data Arguments for the Localization of Lipid Droplets
In addition to electron microscopy images, Fan et al. (2011) presented biochemical data that they tried to use as evidence for the chloroplast origin of lipid droplets. They analyzed the fatty acid composition of the TAG that accumulated in starchless mutants under nitrogen deprivation and showed that C16 fatty acids were abundant. There is a prevailing belief that the C18/C16 lipid molecular species originate from the lipid biosynthetic pathway in the chloroplast and that the C18/C18 molecular species are synthesized in the ER. These two pathways are often called the prokaryotic and eukaryotic pathways, respectively. In Chlamydomonas, this tradition began with a study by Eichenberger’s group in the 1980s (Giroud et al., 1988), which was motivated by the popular paradigm of the plant lipid field of the time (Roughan and Slack, 1982). More recently, Warakanont et al. (2015) showed that the chain length is not a key to classify the origin of lipids in the context of the lipid traffic in Chlamydomonas. C16 fatty acids can be provided from the ER for the synthesis of chloroplast lipids. TAG contains both C16 and C18 fatty acids, which are provided from either ER lipids or chloroplast lipids. The 16:0 fatty acids in TAG are likely to be provided by diacylglyceryl-N,N,N-trimethylhomoserine, whereas the 16:4 fatty acid is probably provided by monogalactosyl diacylglycerol (MGDG), because MGDG is the sole lipid class that contains 16:4 as an abundant fatty acid (Sakurai et al., 2014, and many other studies). This does not mean that TAG containing 16:4 fatty acid is present in the chloroplast, as stated by Fan et al. (2011). We imagine that the 16:4 fatty acid is hydrolyzed from MGDG (Li et al., 2012), transported to the cytosolic compartment, and incorporated into TAG by acyltransfer reactions.
Another biochemical argument for localization of the lipid droplets is related to the possible use of a protein in the lipid droplet membrane as a marker for the cytosolic lipid droplet. Various proteins in the lipid droplet membrane have been characterized in plants and are used as markers for the lipid droplet membrane. In Chlamydomonas, major lipid droplet protein (MLDP) has been characterized as the major constituent of the lipid droplet membrane formed under nitrogen deprivation (Moellering and Benning, 2010; Tsai et al., 2015). We analyzed the level of the MLDP transcript in conditions known to favor the accumulation of lipid droplets (Supplemental Fig. S8). Under nitrogen deprivation, expression of the MLDP gene increased about 15-fold, which was consistent with the results of Moellering and Benning (2010). However, expression of the MLDP gene under the high-light condition did not increase above the control level. The data presented by Goold et al. (2016) were ambiguous: the MLDP signal was scarcely found on the silver-stained gel, whereas MLDP was detected as the 14th most abundant lipid droplet protein by a proteome analysis. These results suggest that it is difficult to use MLDP as a general marker of lipid droplet membranes. In addition, all of the lipid droplet analytical data were obtained for lipid droplets isolated from whole cells, including both cytosolic lipid droplets and the putative chloroplast lipid droplets (if such entities exist). It is theoretically impossible to distinguish the cytosolic lipid droplet and the hypothetical chloroplast lipid droplet by using a marker developed with the lipid droplet preparation isolated in the published way. That is why we preferred microscopy methods to biochemical methods to identify the cytosolic nature of the lipid droplets, which seemed to be chloroplast lipid droplets.
Putative Interorganellar Metabolic Flow
A remaining problem is why some lipid droplets are present in invaginations of the chloroplast. Similar invaginations are not found in the chloroplasts of land plants. This phenomenon might be specific to Chlamydomonas or related algae having a single large chloroplast. Invaginations or holes might be naturally present in such large chloroplasts, and the holes could facilitate the transport of cytosolic materials to the outer cytosolic compartment between the chloroplast and the plasma membrane. Holes and invaginations might be hardly observable without careful examination by fluorescence microscopy. A single chloroplast often appeared as several segments in the electron micrographs of wild-type cells under normal conditions without lipid droplet accumulation (Supplemental Fig. S9, arrows), and these represented holes or the reticulated or branched architecture of the chloroplast. We identified several holes within the chloroplast in the confocal images (Supplemental Fig. S7C, arrowheads). Under the conditions in which TAG accumulates, these invaginations and holes are filled with growing lipid droplets. We still do not know if this filling of the invaginations by lipid droplets occurs passively as a result of mechanical packing within the cell. Alternatively, the tight association between the lipid droplet and the chloroplast at the site of an invagination or hole might have intriguing importance in the metabolic flow of fatty acids from the chloroplast to the lipid droplet. We will discuss this possibility further.
We were impressed by the close positioning of the lipid droplets with the outer envelope membrane, which formed the inner surface of the chloroplast invagination (Fig. 5; Supplemental Figs. S4 and S10, B and C). This close relationship between the lipid droplet and the chloroplast envelope membrane could facilitate the efficient transport of fatty acids synthesized within the chloroplast to the growing lipid droplet. In general, lipid droplets emerge from the ER membrane by budding. The precursors of TAG, such as diacylglycerol, phosphatidylcholine, and acyl-CoA, are provided by enzymes localized in the ER. However, no ER membranes were detected between the lipid droplet and the chloroplast envelope at the site of their close association. The space between the chloroplast envelope membrane and the surface of the lipid droplet was very narrow. It was narrower than the thickness of a typical ER cisterna. All precursors for synthesizing TAG must be supplied directly from the chloroplast to the lipid droplet in the lipid droplets located within the invagination of the chloroplast. This was an intriguing new aspect of lipid droplet formation. The problem of the chloroplast lipid droplet will have to be studied under a different perspective, namely, lipid traffic (as in Block and Jouhet, 2015, for a somewhat different type of traffic) from the chloroplast to the lipid droplet. We still do not know how the initial lipid droplet is formed, which finally localizes to the chloroplast invagination.
Tsai et al. (2015) noted the presence of plastid proteins and the plastid lipid digalactosyl diacylglycerol in the lipid droplets of Chlamydomonas and discussed a putative origin of lipid droplets from the chloroplast envelope membranes. Many outer envelope proteins, such as TOC75, TOC34, TGD2, and MGD1, coimmunoprecipitated with MLDP. This is explicitly depicted in Figure 2 of Liu and Benning (2013), who suggested a possible flow of materials from the chloroplast envelope to the lipid droplet. An integration of the lipid droplet within the chloroplast envelope, rather than contact of the membrane and the lipid droplet, was presented in Figure 3 of Li-Beisson et al. (2015). Although Liu and Benning (2013) were very cautious about the probable nonspecific contamination, the presence of light-harvesting chlorophyll proteins and ATPase subunits, which often are detected in the proteomics of subcellular fractions, suggested that we need different types of evidence, such as fluorescence energy transfer results, to conclude the presence of envelope membrane proteins in the lipid droplet. Despite the figure in Liu and Benning (2013) that showed direct contact or complete fusion of the lipid droplet membrane and the outer envelope, no such image has ever been either presented in the literature or detected by our observations. Most electron micrographs presented in the literature were taken at low magnifications to show the entire structure of the cell (about ×10,000), but the relationship between the lipid droplet and the envelope must be observed at a higher magnification, such as ×50,000 or higher. As shown schematically in Figure 1 for the two envelope membranes, two parallel membranes can be seen as fused in the examination of thin sections. The same is true for the lipid droplet membrane and the envelope membranes. We occasionally observed a lipid droplet apparently fused with the envelope membranes in thin sections, but the two objects were separated if we observed the sections by tilting them in the manner shown in Figure 5. In this sense, no image has ever been presented showing direct contact or fusion of the lipid droplet membrane and the chloroplast envelope.
The proteomics or lipid analysis data of the isolated lipid droplet could be interpreted to show that, during the fractionation, the lipid droplet present within the chloroplast invagination was released as a complex structure consisting of the lipid droplet wrapped by the chloroplast envelope membranes, with the inner envelope outside and the outer envelope inside. This is a more realistic model of isolated lipid droplets bearing digalactosyl diacylglycerol and outer envelope proteins, which are present in a minor proportion (the majority of the lipid droplets are certainly those that originate from the cytosolic compartment). Therefore, a realistic image of the chloroplast lipid droplet is a lipid droplet, which is present within the chloroplast invagination, in close association with the chloroplast envelope membrane by keeping a very narrow layer of cytosol. We cannot exclude the possibility that some structures bridge the lipid droplet and the envelope. In this regard, it is interesting that we also found close associations between the Golgi apparatus, the lipid droplets, and the chloroplast (Fig. 4A; Supplemental Fig. S10). It appears that the Golgi apparatus provides enzymes for TAG synthesis while the chloroplast provides fatty acids.
Distinguishing the Plastoglobule from the Lipid Droplet
The plastoglobule is known to contain TAG, which is supposed to be synthesized by FES1/2 (Lippold et al., 2012), also called DGAT3/4. As clearly stated by Schimper (1885), the plastoglobule and lipid droplet must be clearly distinguished. He showed that the former is soluble in ethanol, whereas the latter is not. The plastoglobule is different from a lipid droplet in that it contains various compounds other than TAG, such as plastoquinones and tocopherols, among others. The plastoglobule contains various types of specific proteins known as fibrillins (FBNs; Lundquist et al., 2012; Lohscheider and Río Bártulos, 2016). In the list of proteins detected by the proteomic analysis of plastoglobules in Arabidopsis (Arabidopsis thaliana), we found no protein that was known to localize to the cytosolic lipid droplets. In the discussions provided by both Fan et al. (2011) and Goold et al. (2016), the ability of the chloroplast to synthesize TAG by FES1/2 was taken as evidence for the presence of a lipid droplet within the chloroplast. If a significant number of lipid droplets are present within the chloroplast, and if TAG is provided by the plastoglobules as suggested, then FBNs might be detected in the proteome of lipid droplets. However, there is no protein in plastoglobules, such as FBN, in the list of proteins detected by the proteomic analysis of Chlamydomonas lipid droplets (Moellering and Benning, 2010; Nguyen et al., 2011; Tsai et al., 2015). The only putative FBN-related protein is g7551 (530 amino acids), described by Tsai et al. (2015). It has partial homology to plant FBN, but this entry was changed to a short (130 amino acids) protein (A8JH10) and is not included in the list of FBNs of Lohscheider and Río Bártulos (2016). Therefore, no FBNs are detected in isolated lipid droplets, and it is unlikely that even a small population of lipid droplets is localized in the chloroplast and supplied with TAG synthesized by plastoglobules.
During our electron microscopy sample preparation, the cells were dehydrated with ethanol; thus, TAG remained as a liquid material mixed with the resin within the section for observation. Some of the electron micrographs showed wavy-appearing lipid droplets due to inefficient cutting of the TAG/resin mixture. The plastoglobules were seen as small particles (Fig. 4E). Goold et al. (2016) presented plastidal lipid droplets, but according to our examination, these are lipid droplets, not plastoglobules: the size of the droplets was smaller than the typical large lipid droplets accumulated in nitrogen-starved cells, but they were fairly larger (more than 200 nm in diameter) than typical plastoglobules, which were about 100 nm in diameter (Fig. 4E). The lipid droplets presumptively located within the chloroplast were similar in size to those undoubtedly present in the cytosol. There is no reason to assume that the plastidal lipid droplets were something other than typical lipid droplets. All lipid droplets, whether chloroplast lipid droplets or not, must be present within the cytosolic compartment.
We noted that lipid droplets that appear to localize within the chloroplast retain a thin limiting membrane. This is very difficult to see, but a thin membrane was visible in a limited region depending on the tilt angles (Fig. 4D; Supplemental Fig. S10B). No limiting membrane is known in the plastoglobule. This observation also is evidence that the lipid droplets are considered to be cytosolic and not located in the chloroplast.
CONCLUSION
Based on the electron microscopy examination and the 3D reconstruction by confocal fluorescence microscopy, we conclude that there is no evidence for the presence of lipid droplets in the chloroplast of Chlamydomonas. We also provide various biochemical arguments using published data on the lipid and protein compositions of the lipid droplets and plastoglobules, which are taken as convincing evidence that the lipid droplets are essentially different from plastoglobules. In addition, we found a new aspect of research on the lipid droplets, namely, the traffic from the chloroplast to the lipid droplet without an intervening ER membrane.
MATERIALS AND METHODS
Algal Strains and Growth Conditions
The green alga Chlamydomonas reinhardtii strains CC-1010, CC-4348 (cw15sta6-1; BAFJ5), and CC-4334 (cw15sta7-1) were obtained from the Chlamydomonas Resource Center. CC-1010 cells were grown in modified Bristol’s medium (Watanabe, 1960) with aeration by 2% (v/v) CO2 in air at 25°C. For lipid accumulation under high-light irradiation, the culture was first illuminated at a photon flux of 40 µmol m–2 s–1 until OD750 became 0.2 to 0.4 and then was illuminated at 200 µmol m–2 s–1 according to Goold et al. (2016). The starchless mutants, sta6 and sta7 cells, were grown in Tris-acetate phosphate medium (Gorman and Levine, 1965) at 25°C under light (50 µmol m–2 s–1) with shaking. The cells were transferred to nitrogen-depleted Tris-acetate phosphate medium containing 20 mm potassium acetate for 24 h to accumulate lipids.
Microscopic Examination
CC-1010 cells stained with BODIPY were observed with a fluorescence microscope as described previously (Sakurai et al., 2014). For confocal microscopy, CC-1010 cells were fixed in 0.25% (w/v) glutaraldehyde and stained with 10 μg mL–1 BODIPY or 0.5% (v/v) LipidTOX Red solution (Invitrogen). Unfixed sta6 and sta7 cells were stained with 0.5% (v/v) LipidTOX Red solution and 1 μm FDA cytosolic staining. The samples were placed on a glass-based dish (coverglass thickness, 0.08–0.12 mm) and were imaged with a Nikon C2+ confocal imaging system (Nikon Instech) mounted on a Nikon Eclipse Ti-E inverted microscope. Fluorescence signals of chloroplasts and LipidTOX were acquired sequentially with 408-nm excitation and 417- to 477-nm emission (see below) and with 561-nm excitation and 575- to 615-nm emission, respectively. BODIPY and FDA fluorescence signals were acquired by 488-nm excitation and 500- to 550-nm emission. Images were obtained as Z-stacks and processed with Fiji software. Reconstructed 3D images were visualized by FluoRender software (http://www.sci.utah.edu/software/fluorender.html).
Chloroplasts are usually imaged with chlorophyll fluorescence, typically using excitation at 640 nm and emission at 660 to 1,000 nm (Supplemental Fig. S7B). As pointed out in Supplemental Figure S5, fluorescence of Nile Red or LipidTOX can enter the chlorophyll channel (Supplemental Figs. S6 and S7). This is inevitable if chlorophyll fluorescence is used for imaging chloroplasts (or thylakoid membranes). We discovered that the blue region can be used to obtain images of chloroplasts (excitation at 408 nm and emission at 417–477 nm). The image obtained with blue fluorescence was identical to the image obtained with conventional red fluorescence (Supplemental Fig. S7, A–C). We still do not know the exact origin of this blue fluorescence of chloroplasts, but this technique is useful for avoiding the fluorescence of lipid stains (Nile Red, BODIPY, or LipidTOX) and misleading overlays on the chloroplast image.
Fluorometric Quantification of Neutral Lipids
The cells were fixed with 0.25% glutaraldehyde and stored at 4°C until measurement. A 2× Nile Red solution (1 μg mL–1 Nile Red and 50% (v/v) dimethyl sulfoxide) was added to each sample, and the mixture was incubated for 5 min at room temperature. Fluorescence emission at 604 nm of Nile Red bound to neutral lipid was measured under excitation at 530 nm in an EnSpire plate reader (PerkinElmer). Background fluorescence also was measured and subtracted.
Fluorescence Spectral Analysis
The fluorescence of Nile Red, LipidTOX, and BODIPY dissolved in TAG was measured using a Shimadzu RF-5300PC fluorescence spectrophotometer.
Lipid Analysis
Extraction of lipids from cells, thin-layer chromatographic separation of lipid classes, and gas chromatographic determination of fatty acid methyl esters were performed essentially as described previously (Sakurai et al., 2014).
Transmission Electron Microscopy
Cells were fixed in 0.125% (w/v) glutaraldehyde for 10 min and then with 1% (w/v) glutaraldehyde for 30 min. They were centrifuged at 400g for 10 min, and the precipitate was embedded in 1% (w/v) low-melting agarose. Dehydration through an ethanol series, embedding in Epon resin, sectioning, and staining were performed as described previously (Sato et al., 2014; Toyoshima and Sato, 2015).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of LipidTOX and BODIPY staining.
Supplemental Figure S2. 3D reconstruction by confocal fluorescence microscopy of another wild-type cell of Chlamydomonas CC-1010.
Supplemental Figure S3. 3D reconstruction by confocal fluorescence microscopy of a third wild-type cell of Chlamydomonas CC-1010.
Supplemental Figure S4. Electron micrographs of lipid droplets in the starchless mutant (cw15sta7) cells of Chlamydomonas deprived of nitrogen for 24 h with acetate supplementation.
Supplemental Figure S5. Reexamination of the published figures of chloroplast localization of lipid droplets in Chlamydomonas.
Supplemental Figure S6. Fluorescence spectra of representative pigments for lipid droplet staining in pure TAG.
Supplemental Figure S7. Confocal imaging of chloroplasts and lipid droplets.
Supplemental Figure S8. Levels of MLDP gene transcripts under various growth conditions in Chlamydomonas CC-1010.
Supplemental Figure S9. Putative holes in the chloroplast in the strain CC-1010.
Supplemental Figure S10. Close association of the Golgi apparatus, lipid droplet, and chloroplast.
Supplemental Data S1. Sequential TIF files of the 3D reconstruction of LD-accumulating CC-1010 cells for Figure 3.
Supplemental Data S2. Sequential TIF files of the 3D reconstruction of LD-accumulating CC-1010 cells for Supplemental Figure S2.
Supplemental Data S3. Sequential TIF files of the 3D reconstruction of LD-accumulating CC-1010 cells for Supplemental Figure S3.
Supplemental Data S4. Sequential TIF files of the 3D reconstruction of LD-accumulating sta6 cells for the top part of Figure 6
Supplemental Data S5. Sequential TIF files of the 3D reconstruction of LD-accumulating sta7 cells for the bottom part of Figure 6.
Acknowledgments
We are grateful to Takashi Hirashima in our laboratory for discussion on plastoglobules.
Footnotes
This work was supported in part by a KAKENHI (15K12433 and 17H03715 to N.S.) from the Japan Society for the Promotion of Science (JSPS) and a grant, Core Research for Evolutional Science and Technology, from the Japan Science and Technology Agency to N.S and H.W.
Articles can be viewed without a subscription.
References
- Bates PD. (2016) Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta 1861: 1214–1225 [DOI] [PubMed] [Google Scholar]
- Bhowmick GD, Koduru L, Sen R (2016) Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application: a review. Renew Sustain Energy Rev 50: 1239–1253 [Google Scholar]
- Block MA, Jouhet J (2015) Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr Opin Cell Biol 35: 21–29 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F, van Wijk KJ (2007) Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci 12: 260–266 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants: thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Gargouri M, Park JJ, Holguin FO, Kim MJ, Wang H, Deshpande RR, Shachar-Hill Y, Hicks LM, Gang DR (2015) Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J Exp Bot 66: 4551–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud C, Gerber A, Eichenberger W (1988) Lipids of Chlamydomonas reinhardtii: analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol 29: 587–595 [Google Scholar]
- Goodson C, Roth R, Wang ZT, Goodenough U (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell 10: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold HD, Cuiné S, Légeret B, Liang Y, Brugière S, Auroy P, Javot H, Tardif M, Jones B, Beisson F, et al. (2016) Saturating light induces sustained accumulation of oil in plastidal lipid droplets in Chlamydomonas reinhardtii. Plant Physiol 171: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12: 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C (2012) A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82: 504–522 [DOI] [PubMed] [Google Scholar]
- Lippold F, vom Dorp K, Abraham M, Hölzl G, Wewer V, Yilmaz JL, Lager I, Montandon C, Besagni C, Kessler F, et al. (2012) Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24: 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Benning C (2013) Lipid metabolism in microalgae distinguishes itself. Curr Opin Biotechnol 24: 300–309 [DOI] [PubMed] [Google Scholar]
- Lohscheider JN, Río Bártulos C (2016) Plastoglobules in algae: a comprehensive comparative study of the presence of major structural and functional components in complex plastids. Mar Genomics 28: 127–136 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23: 352–363 [DOI] [PubMed] [Google Scholar]
- Misra N, Panda PK, Parida BK, Mishra BK (2012) Phylogenomic study of lipid genes involved in microalgal biofuel production: candidate gene mining and metabolic pathway analyses. Evol Bioinform Online 8: 545–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- Pasteur L. (1922) Œuvres de Pasteur, Vol II Masson, Paris, pp 295, 459 [Google Scholar]
- Roughan PG, Slack CR (1982) Cellular organization of glycerolipid metabolism. Annu Rev Plant Physiol 33: 97–132 [Google Scholar]
- Sakurai K, Moriyama T, Sato N (2014) Detailed identification of fatty acid isomers sheds light on the probable precursors of triacylglycerol accumulation in photoautotrophically grown Chlamydomonas reinhardtii. Eukaryot Cell 13: 256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Katsumata Y, Sato K, Tajima N (2014) Cellular dynamics drives the emergence of supracellular structure in the cyanobacterium, Phormidium sp. KS. Life (Basel) 4: 819–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Mori N, Hirashima T, Moriyama T (2016) Diverse pathways of phosphatidylcholine biosynthesis in algae as estimated by labeling studies and genomic sequence analysis. Plant J 87: 281–292 [DOI] [PubMed] [Google Scholar]
- Schimper AFW. (1885) Untersuchungen über die Chlorophyllkörper und die ihnen homologen Gebilde. Jarb F Wiss Botanik 16: 1–247 [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, et al. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevini M, Steinmüller D (1985) Composition and function of plastoglobuli. II. Lipid composition of leaves and plastoglobuli during beech leaf senescence. Planta 163: 91–96 [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Sato N (2015) High-level accumulation of triacylglycerol and starch in photoautotrophically grown Chlamydomonas debaryana NIES-2212. Plant Cell Physiol 56: 2447–2456 [DOI] [PubMed] [Google Scholar]
- Tsai CH, Zienkiewicz K, Amstutz CL, Brink BG, Warakanont J, Roston R, Benning C (2015) Dynamics of protein and polar lipid recruitment during lipid droplet assembly in Chlamydomonas reinhardtii. Plant J 83: 650–660 [DOI] [PubMed] [Google Scholar]
- van de Meene AML, Hohmann-Marriott MF, Vermaas WFJ, Roberson RW (2006) The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol 184: 259–270 [DOI] [PubMed] [Google Scholar]
- Warakanont J, Tsai CH, Michel EJS, Murphy GR III, Hsueh PY, Roston RL, Sears BB, Benning C (2015) Chloroplast lipid transfer processes in Chlamydomonas reinhardtii involving a TRIGALACTOSYLDIACYLGLYCEROL 2 (TGD2) orthologue. Plant J 84: 1005–1020 [DOI] [PubMed] [Google Scholar]
- Watanabe A. (1960) List of algal strains in collection at the Institute of Applied Microbiology, University of Tokyo. J Gen Appl Microbiol 6: 283–292 [Google Scholar]
- Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LMI, Dismukes GC, Posewitz MC (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell 9: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz K, Du ZY, Ma W, Vollheyde K, Benning C (2016) Stress-induced neutral lipid biosynthesis in microalgae: molecular, cellular and physiological insights. Biochim Biophys Acta 1861: 1269–1281 [DOI] [PubMed] [Google Scholar]