The establishment of photosynthesis is a two-phase process with a clear checkpoint associated with the second regulatory phase allowing coordination of the activities of the nuclear and plastid genomes.
Abstract
Chloroplasts develop from undifferentiated proplastids present in meristematic tissue. Thus, chloroplast biogenesis is closely connected to leaf development, which restricts our ability to study the process of chloroplast biogenesis per se. As a consequence, we know relatively little about the regulatory mechanisms behind the establishment of the photosynthetic reactions and how the activities of the two genomes involved are coordinated during chloroplast development. We developed a single cell-based experimental system from Arabidopsis (Arabidopsis thaliana) with high temporal resolution allowing for investigations of the transition from proplastids to functional chloroplasts. Using this unique cell line, we could show that the establishment of photosynthesis is dependent on a regulatory mechanism involving two distinct phases. The first phase is triggered by rapid light-induced changes in gene expression and the metabolome. The second phase is dependent on the activation of the chloroplast and generates massive changes in the nuclear gene expression required for the transition to photosynthetically functional chloroplasts. The second phase also is associated with a spatial transition of the chloroplasts from clusters around the nucleus to the final position at the cell cortex. Thus, the establishment of photosynthesis is a two-phase process with a clear checkpoint associated with the second regulatory phase allowing coordination of the activities of the nuclear and plastid genomes.
In eukaryotes, photosynthesis occurs in the chloroplasts. Plastid differentiation into photosynthetically active chloroplasts follows a clear developmental program where functional chloroplasts are derived from nonphotosynthetic progenitors, either directly from proplastids present in meristematic cells or via the dark-grown intermediate form known as etioplasts (Pogson and Albrecht, 2011). Most studies on the early light response and chloroplast development have been performed on cotyledons of dark-grown seedlings shifted to light. This is not a true reflection of what occurs in the developing leaves of the plant, and genetic studies have shown that chloroplast development proceeds differently in cotyledons and true leaves (Pogson and Albrecht, 2011). In addition, chloroplast biogenesis has to be coordinated with leaf growth to enable a switch from heterotrophic to photoautotrophic metabolism. The leaf becomes photosynthetically active at the same time as it shifts from primary to secondary morphogenesis, and chloroplast differentiation is an important regulator of the simultaneous onset of cell expansion and photosynthesis (Andriankaja et al., 2012).
Chloroplasts, like mitochondria, evolved from free-living prokaryotic organisms that entered the eukaryotic cell through endosymbiosis. The division of genetic information between different compartments presents a complex problem to the eukaryotic cell, requiring the coordination of the activities of the different genomes during the process of chloroplast development. Several of the components required for the transcription and translation of the plastid-encoded photosynthesis genes are nuclearly encoded, such as the sigma factors (SIG) of the plastid-encoded RNA polymerase (PEP; Woodson et al., 2013; Chi et al., 2015). In addition, the photosynthetic apparatus is composed of proteins encoded by genes from both the nucleus and the chloroplast. In the photosynthetic electron transport complexes of the thylakoid membrane, for example, the core subunits are encoded by the plastid genome and the peripheral subunits are encoded by the nuclear genome. Thus, it is clear that the initiation of chloroplast development in the light is dependent on the expression of nucleus-encoded components but also that a tight stoichiometric assembly of nucleus-encoded and plastid-encoded proteins is essential for the establishment of photosynthesis (Nelson and Yocum, 2006). To achieve this, there must be communication between the chloroplast and the nucleus through so-called anterograde (nucleus-to-plastid) and retrograde (plastid-to-nucleus) mechanisms (Pogson et al., 2008; Barajas-López et al., 2013a).
The establishment of functional chloroplasts is a complex process involving several cellular compartments. First, chloroplast development is dependent on light, and the initial light signal triggers the activation of the phytochrome and cryptochrome photoreceptors (Waters and Langdale, 2009). These photoreceptors perceive light signals and initiate intracellular signaling pathways involving the proteolytic degradation of signaling components such as phytochrome interaction factors and a large reorganization of the transcriptional program to modulate plant growth and development (Chen et al., 2004). Second, expression of the plastid-encoded photosynthesis genes needs to be initiated, and this induction depends upon the expression and assembly of the nucleus-encoded components, such as the SIGs and polymerase-associated proteins (PAPs), required for the activity of the PEP (Kindgren and Strand, 2015). Third, there are indications from the literature that a hypothetical retrograde signaling mechanism is required for full activation of the nucleus-encoded photosynthesis genes (Woodson et al., 2013). However, it is unclear if this regulatory mechanism is separate from the initial light signal and whether this retrograde signal is positive or negative and, thus, if healthy chloroplasts stimulate or if impaired chloroplasts block nuclear gene expression.
The complexity of leaf development imposes experimental limitations on our ability to study chloroplast biogenesis, restricting the utility of traditional genetic approaches. In planta, chloroplast biogenesis is strongly connected to the development of new tissues and the inhibition of cell elongation; thus, only a few cells at any one moment are in the same state of chloroplast development. In grasses, a spatial gradient of chloroplast development can be observed in a single leaf (Li et al., 2010; Pick et al., 2011), but such developmental gradients in leaves of dicot plants are much harder to isolate. In a single cell-based system, it is possible to observe the synchronous development of many cells in parallel, facilitating the deciphering of the regulatory mechanisms at the level of the individual cell controlling the transition from a proplastid to a functional chloroplast. Thus, to address the limitations of traditional in planta dicot systems, and to develop a well-controlled experimental setup where the individual regulatory checkpoints of chloroplast development could be defined, a pluripotent inducible cell line was generated from Arabidopsis (Arabidopsis thaliana). Following light exposure, this cell line was shown to differentiate into photosynthetically active cells with functional chloroplasts, providing an experimental system with a temporal gradient of chloroplast development. Using this unique cell line from the dicot Arabidopsis in combination with the monocot maize (Zea mays), we could demonstrate that the development from a proplastid to a functional chloroplast, and thereby the establishment of photosynthesis, is dependent on a regulatory mechanism involving two distinct phases. First, light exposure triggers an initial change in gene expression, metabolite profile, chlorophyll accumulation, and plastid structure. Second, a second signal, most likely triggered by activation of the chloroplast, is required for the full transition to a functional chloroplast. A clear checkpoint is associated with the second regulatory phase, as it is dependent on a plastid signal enabling the plant to synchronize the expression of photosynthetic genes from the nuclear and chloroplast genomes during seedling establishment.
RESULTS
An Inducible Cell Line from Arabidopsis Demonstrated the Differentiation of Proplastids to Chloroplasts
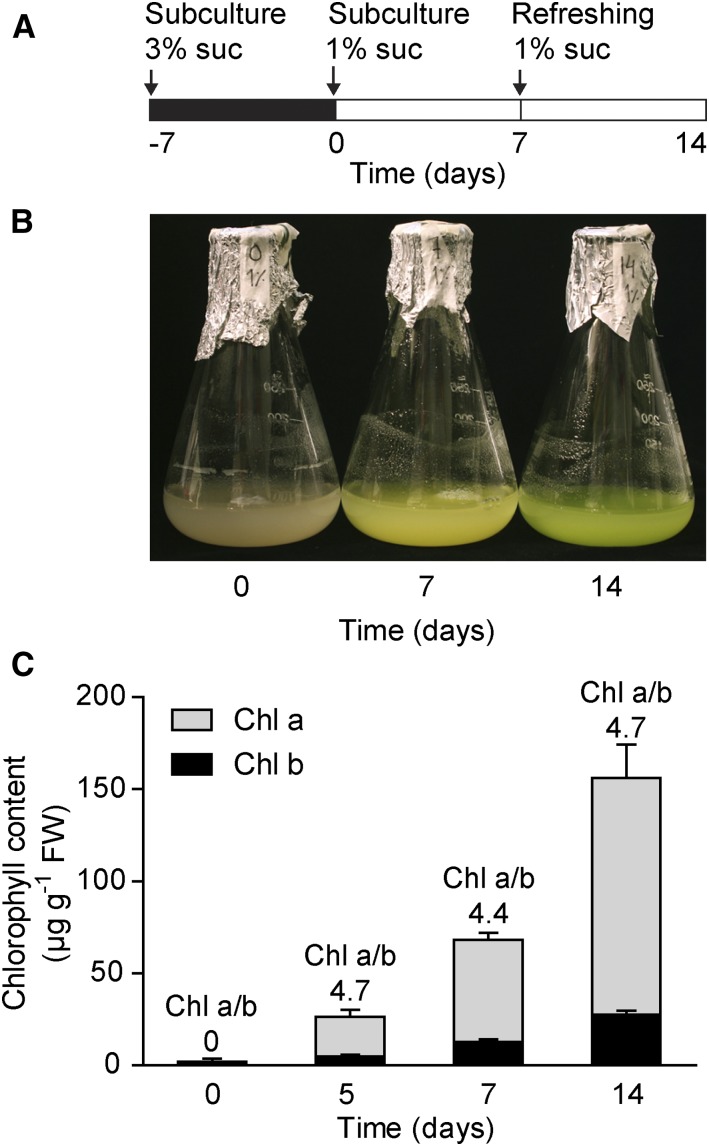
A pluripotent inducible cell line was generated from Arabidopsis (Pesquet et al., 2010). This habituated cell line can be propagated in the dark without the addition of hormones or growth factors (Pesquet et al., 2010). We here show that chloroplast development can be triggered on demand in these cells by exposing the culture to light. Green cell culture-based systems that display a photosynthetic capacity similar to the one in planta have been described previously (González-Pérez et al., 2011; Zhang et al., 2011; Hampp et al., 2012). However, in contrast to our cell line, those cell cultures are constitutively green and cannot be grown in the dark, making such systems unusable to investigate the chloroplast developmental process. The cells were grown on Murashige and Skoog (MS) medium supplied with 3% Suc in the dark, and optimal conditions for greening of the cell culture were selected following a screening procedure (methods for subculture, Suc concentrations, light conditions, etc.). A carbon source is required during chloroplast development, and during the deetiolation of dark-grown seedlings the carbon used for the establishment of photosynthesis is taken from the storage in the seed. In a single-cell system, carbon must be provided in the medium. However, to promote source activity (e.g. photosynthesis) rather than sink activity (e.g. growth and storage; Rolland et al., 2002), the amount of sugar in the medium was decreased to 1% prior to exposure to light and the medium was refreshed after 7 d in the light (Fig. 1A). The equilibrated cells at 1% Suc (T0) were exposed to constant light (Fig. 1A). This condition was maintained to allow for sampling at any time point during chloroplast biogenesis and exclude circadian or diurnal fluctuations. The green color of the cell cultures following light exposure also was confirmed by chlorophyll quantification (Fig. 1, B and C). Thus, the chronological progression of chloroplast differentiation in the light was reflected by the increase in chlorophyll content (Fig. 1C).
Figure 1.
Greening process induced by light in Arabidopsis cell culture. A, Experimental conditions used to investigate chloroplast biogenesis in Arabidopsis cell culture. Cells were subcultured in MS medium with 1% (w/v) Suc, equilibrated (T0), and then placed under continuous light. At 7 d, the MS medium was replaced with the same volume of fresh MS medium. B, Seven- and 14-d cells compared with the control (T0). C, Chlorophyll a and b contents following light exposure. Values are means ± sd of three biological replicates. Chl a/b, Chlorophyll a/b ratio; FW, fresh weight of the cell culture.
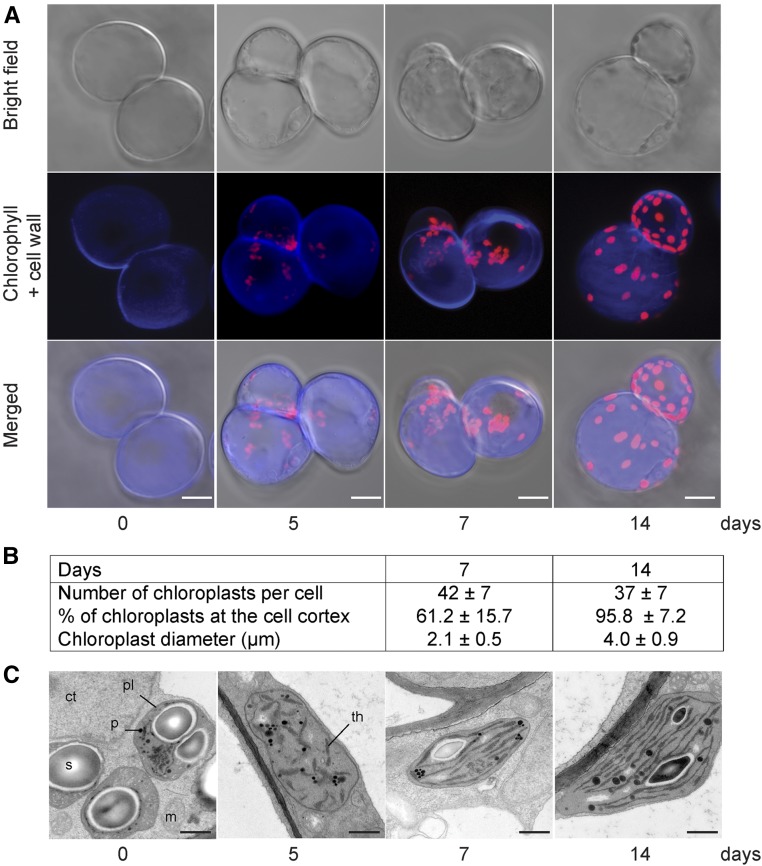
Analysis using confocal microscopy of the cells at different stages during the light exposure was conducted to investigate the changes in plastid morphology and intracellular position during chloroplast development (Fig. 2A). Combining the xyz-stack images from the confocal microscopy showed that the shape of the plastids in the 5-d cells was poorly defined and the plastids were grouped together (Fig. 2A). Thus, the structure of the 5-d plastids made it difficult to get accurate counts for plastid numbers and positions, but at 7 d, the mean number of chloroplasts per cell reached 42 ± 7. About 61% of the chloroplasts were positioned at the cell cortex, whereas the rest remained clustered around the nucleus (Fig. 2B). In addition, the morphology of the plastids changed into a more defined globular shape of 2.1 ± 0.5 µm in diameter after 7 d in the light (Fig. 2B). After 14 d in the light, the number of chloroplasts per cell remained unchanged (37 ± 7 chloroplasts per cell), but the size of the chloroplasts increased to 4 ± 0.9 µm in diameter compared with the 7-d sample, and nearly 96% of the chloroplasts were now positioned at the cell cortex, as seen in a leaf mesophyll cell (Fig. 2B).
Figure 2.
Visualization of proplastids developing into functional chloroplasts. A, Confocal laser scanning microscopy of the Arabidopsis cell line during light exposure. Confocal optical sections were combined for a three-dimensional reconstruction showing the bright field, Calcofluor White-stained cell wall, and chlorophyll autofluorescence. Bars = 20 μm. B, Plastid size, numbers, and cellular positions at 7 and 14 d. C, Electron microscopy images of proplastids developing into chloroplasts following light exposure. Representative images from at least two independent experiments for each developmental stage are shown. ct, Cytoplasm; m, mitochondrion; p, plastoglobule; pl, plastid; s, starch; th, thylakoids. Bars = 0.5 μm.
The detailed ultrastructure of the plastids during the greening process was investigated using transmission electron microscopy (Fig. 2C). No prolamellar bodies could be detected in the plastids of the dark-grown samples. Prolamellar bodies are characteristics of etioplasts; thus, the plastids of the dark-grown cell culture were similar to proplastids found in meristematic cells. In the dark, the proplastids displayed an electron-dense aggregate zone that contained large starch deposits and plastoglobuli. Following light exposure for 5 d, an internal structure with small grana of interconnected thylakoids with short intergranal thylakoids appeared in the starch-free plastids (Fig. 2C). After 7 d in the light, the chloroplasts contained a well-developed structure with numerous grana of four to five thylakoids and intergranal thylakoids (Fig. 2C). At 14 d, the thylakoid structure was complete and the size and the morphology of the cell culture chloroplasts were similar to those observed in Arabidopsis and pea (Pisum sativum) plants, where the chloroplasts typically are 5 to 10 µm long and have five to seven thylakoids per grana with intergranal thylakoids (Lopez-Juez and Pyke, 2005; Barajas-López et al., 2013b; Myouga et al., 2013).
Photosynthesis Was Established in the Differentiated Chloroplasts
So far, we have shown that chlorophyll accumulates following light exposure and that the structure and the organization of the chloroplasts in the green cell culture were similar to what was seen in a leaf mesophyll cell. The critical question is whether cultured Arabidopsis cells are capable of performing photosynthesis. First, we looked at the assembly of the photosynthetic protein complexes by analyzing the thylakoid membrane proteins by blue-native PAGE (Fig. 3A; Järvi et al., 2011). Seven major complexes were defined according to Arabidopsis plant thylakoid membrane preparations (Fig. 3A, right lane). We were able to distinguish the known protein complexes following light exposure of the cell culture (Fig. 3A). Most of the photosynthetic protein complexes were already present in the 5-d cells, although at low concentrations, and by 7 d, chlorophyll accumulated in the LHCII trimer (Fig. 3A). Photosystem assembly was completed in 14-d cells, where the protein bands corresponding to the PSII-LHCII supercomplex, PSI-PSII dimer, ATP synthase, PSII monomer, and cytochrome b6f were clearly visible (Fig. 3A). Thus, the protein complex profile of 14-d cells was similar to the profile from leaves of 3-week-old Arabidopsis plants (Fig. 3A). Western-blot analysis confirmed the accumulation of the photosynthetic proteins during the chloroplast differentiation process (Fig. 3B).
Figure 3.
Photosynthetic activity established in the Arabidopsis cell culture. A, Blue-native PAGE analysis of thylakoid protein complexes from 5-, 7-, and 14-d cells and from 3-week-old Arabidopsis Columbia-0 (Col-0) plants. The gel shown is representative of two independent experiments. The adjusted loading was according to protein content. B, Western blot against photosynthetic protein in samples from 0-, 1-, 4-, 5-, 7-, and 14-d cells. C, Chlorophyll fluorescence variation at 7 and 14 d. The graph is representative of three independent experiments. AL, Actinic light; ML, measuring light. D, The chlorophyll fluorescence parameter Fv/Fm measured at 7 and 14 d. E, Oxygen evolution rate measured in 5-, 7-, and 14-d cells. Values represent means ± sd of three independent experiments with three biological replicates per experiment. FW, Fresh weight of the cell culture.
To evaluate the activity of photosynthetic electron transport, we measured chlorophyll fluorescence parameters (Fig. 3, C and D). Electron transport was already efficient in 7-d cells, and it increased further in 14-d cells (Fig. 3C). The ratio of variable fluorescence to maximum fluorescence (Fv/Fm), representing the maximum photochemical efficiency of PSII, is 0.73 in the 14-d cells (Fig. 3D). This Fv/Fm was similar to what was frequently reported for 3- to 4-week-old Arabidopsis plants (Hou et al., 2015; Vercruyssen et al., 2015). To confirm photosynthetic activities, oxygen evolution also was determined in 5-, 7-, and 14-d cells (Fig. 3E). By 5 d in the light, the oxygen produced by photosynthesis was already higher than the amounts consumed by respiration. Oxygen evolution increased considerably in the following days. Oxygen evolution in the 7- and 14-d cells was found to be very stable, as the cells could be submitted to many (more than eight) 2-min light/dark cycles without losing oxygen-evolving activity, suggesting that the chloroplasts of the cell culture were very robust. Taken together, these results demonstrate that the cells in this inducible cell culture system behave in many ways like leaf mesophyll cells and can develop functional photosynthetic chloroplasts on demand.
The Establishment of Photosynthesis Was Associated with a Metabolic Change
We investigated the changes in the metabolome during the transition from proplastids to a functional chloroplast and, consequently, the shift from heterotrophic to photoautotrophic metabolism. Relative changes of known metabolites were determined from six biological replicates at each time point. Using this method, 47 metabolites were identified (Supplemental Table S1), and the samples were separated by a principal component analysis (PCA; Fig. 4A). The first two principal components accounted for 63.5% of the total variance of the metabolite data, and the groups differentiated mainly according to the developmental stage (principal component 1). However, two phases during chloroplast development were clearly identified in the metabolome. First, in the response to light, the day-1 to day-4 cells cluster together (constituting phase 1). The metabolome of the 5-d cells was clearly discriminated from the cluster of the day-1 to day-4 cells (principal component 2), suggesting that a critical developmental switch takes place between day 4 and day 5 (initiation of phase 2). Consecutively, 7-, 10-, and 14-d cells were clustered tightly together, suggesting that the cells had switched to photosynthetic metabolism before 7 d. We clustered the metabolite levels (Supplemental Fig. S1) and found that organic acids became less abundant during the transition from proplastid to a functional chloroplast (Fig. 4B). This group was composed mainly of tricarboxylic acid cycle intermediates such as citric acid, succinic acid, fumaric acid, and malic acid (Supplemental Fig. S1). Amino acids cluster in two separate groups, one group that gradually increased and a second that varied in amounts during the developmental process (Supplemental Fig. S1). In the same group as the increasing amino acids, we found other nitrogen-rich metabolites such as allantoin and urea (Fig. 4C; Supplemental Fig. S1). Chloroplast development in this system correlates with large modifications of metabolism, with a depletion of carbon-rich metabolites and an accumulation of nitrogen-rich metabolites. However, at this point, how these metabolite concentrations relate to fluxes through carbon and nitrogen metabolism is difficult to say.
Figure 4.
Evolution of the metabolome during chloroplast development. A, PCA plot for the metabolite profile at different time points of chloroplast development. The two first components (PC1 and PC2) are plotted proportionally. The different colors represent days in light (0–14 d), with six independent biological replicates per day; the percentages shown on the axes indicate the proportion of variance for each principal component. B and C, Box-plot diagrams showing the accumulation of metabolites down- and up-regulated during the chloroplast development process, respectively. Colors are not connected to T0 and T14 in A.
Changes in the Global Expression Profiles of the Nuclear and Plastid Genes Required for the Establishment of Photosynthesis
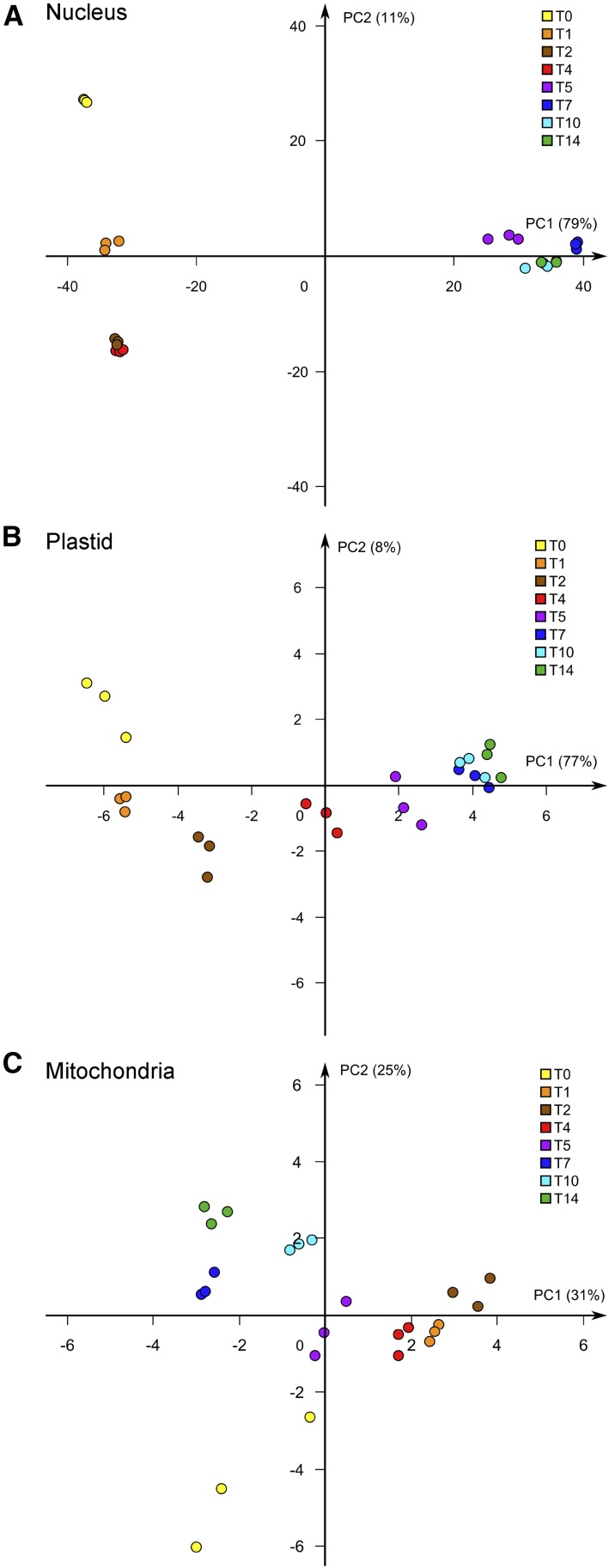
Changes in nuclear, plastid, and mitochondrial gene expression during the chloroplast development process were determined by RNA sequencing analysis. PCA analysis (Fig. 5) showed that there is a large distance between the clusters of the nuclear genes for day 0 and day 1 (Fig. 5A), indicating that a large shift in gene expression occurred during the first day of light exposure. This agrees with the report that when dark-grown seedlings were exposed to light, as much as one-third of the nuclear genes showed transcript changes (Chen et al., 2010). Another large distance between the clusters was observed between days 4 and 5 (Fig. 5A). After day 5, there was a very small separation of the consecutive clusters for days 7 to 14. This expression pattern of the nuclear genes indicates, similar to what was observed for the metabolome (Fig. 4A), two distinct phases of the regulation of gene expression, with differential expression accentuated between days 0 and 1 and days 4 and 5. Testing for differential expression of the nuclear genes revealed that 3,084 genes were differentially expressed between days 0 and 1 (P < 0.01), whereas 12,411 genes were differentially expressed when day 4 was compared with day 5 (P < 0.01).
Figure 5.
Global analysis of gene expression in response to light. PCA plots are shown for nuclear gene expression (A), plastid gene expression (B), and mitochondrial gene expression (C) in response to light. The different colors represent days in light (0–14 d), with three independent biological replicates per day; the percentages shown on the axes indicate the proportion of variance for each principal component (PC1 and PC2).
The plastid genes also showed a strong change in expression following exposure to light, and the distance between the clusters for day 0 and day 1 was similar to that observed for the nucleus-encoded genes (Fig. 5B). However, the two-phase pattern of gene expression seen for the nuclear genes was not as apparent for the plastid genes, where the separation between the clusters for day 4 and day 5 was less pronounced. The difference in gene expression was most pronounced during the first 5 d and less so from day 5 onward. Expression of the mitochondrially encoded genes changed less in response to light compared with the nucleus- and plastid-encoded genes (Fig. 5C).
Gene Ontology (GO) analyses were performed on the genes that showed a change in expression during the two highly differentiated time points, day 0 versus day 1 (D 0-1) and day 4 versus day 5 (D 4-5). Tables I and II show the top 20 terms within the two ontologies cellular components and biological processes at D 0-1 and D 4-5, respectively. The expression data confirm that chloroplast biogenesis is initiated during the first day of light exposure (D 0-1) and established during the second phase of regulation (D 4-5). Genes categorized under the terms chloroplast (GO:0009507), chloroplast thylakoid membrane (GO:0009535), chloroplast envelope (GO:0009941), and chloroplast stroma (GO:0009570) are found in large numbers among the top-ranked genes for both time points. As many as ∼2,400 differentially expressed genes are tagged with the term chloroplast (GO:0009507) at D 4-5 (Table II).
Table I. Top 20 GO terms for biological processes and cellular components ontologies for the genes differentially expressed between day 0 and day 1.
| Biological Process |
Cellular Component |
||||
|---|---|---|---|---|---|
| GO Identifier | Term | No. of Genes | GO Identifier | Term | No. of Genes |
| GO:0006412 | Translation | 291 | GO:0022625 | Cytosolic large ribosomal subunit | 100 |
| GO:0001510 | RNA methylation | 143 | GO:0022627 | Cytosolic small ribosomal subunit | 83 |
| GO:0006364 | rRNA processing | 151 | GO:0009535 | Chloroplast thylakoid membrane | 154 |
| GO:0009220 | Pyrimidine ribonucleotide biosynthesis | 95 | GO:0005730 | Nucleolus | 122 |
| GO:0042254 | Ribosome biogenesis | 229 | GO:0009941 | Chloroplast envelope | 178 |
| GO:0010207 | PSII assembly | 82 | GO:0009570 | Chloroplast stroma | 203 |
| GO:0006098 | Pentose-phosphate shunt | 92 | GO:0048046 | Apoplast | 107 |
| GO:0016036 | Cellular response to phosphate starvation | 69 | GO:0009506 | Plasmodesma | 199 |
| GO:0010200 | Response to chitin | 137 | GO:0005618 | Cell wall | 137 |
| GO:0009657 | Plastid organization | 158 | GO:0022626 | Cytosolic ribosome | 219 |
| GO:0006612 | Protein targeting to membrane | 127 | GO:0009522 | PSI | 14 |
| GO:0010363 | Regulation of plant-type hypersensitive response | 124 | GO:0009579 | Thylakoid | 219 |
| GO:0019288 | Isopentenyl diphosphate biosynthetic | 92 | GO:0010287 | Plastoglobule | 28 |
| GO:0019375 | Galactolipid biosynthetic process | 52 | GO:0005840 | Ribosome | 267 |
| GO:0009773 | Photosynthetic electron transport | 33 | GO:0009543 | Chloroplast thylakoid lumen | 31 |
| GO:0010218 | Response to far-red light | 50 | GO:0030076 | Light-harvesting complex | 16 |
| GO:0019344 | Cys biosynthetic process | 80 | GO:0009507 | Chloroplast | 770 |
| GO:0009744 | Response to Suc | 79 | GO:0010319 | Stromule | 18 |
| GO:0009862 | Systemic acquired resistance | 85 | GO:0030095 | Chloroplast PSII | 12 |
| GO:0009637 | Response to blue light | 56 | GO:0005773 | Vacuole | 195 |
Table II. Top 20 GO terms for biological processes and cellular components ontologies for the genes differentially expressed between day 4 and day 5.
| Biological Process |
Cellular Component |
||||
|---|---|---|---|---|---|
| GO Identifier | Term | No. of Genes | GO Identifier | Term | No. of Genes |
| GO:0006412 | Translation | 477 | GO:0009506 | Plasmodesma | 621 |
| GO:0001510 | RNA methylation | 169 | GO:0009570 | Chloroplast stroma | 503 |
| GO:0046686 | Response to cadmium | 374 | GO:0009535 | Chloroplast thylakoid membrane | 246 |
| GO:0051567 | Histone H3-K9 methylation | 168 | GO:0005829 | Cytosol | 1,213 |
| GO:0006364 | rRNA processing | 222 | GO:0005886 | Plasma membrane | 1,929 |
| GO:0006364 | Response to salt stress | 555 | GO:0022625 | Cytosolic large ribosomal subunit | 110 |
| GO:0006098 | Pentose-phosphate shunt | 174 | GO:0022627 | Cytosolic small ribosomal subunit | 97 |
| GO:0006275 | Regulation of DNA replication | 125 | GO:0009941 | Chloroplast envelope | 422 |
| GO:0042254 | Ribosome biogenesis | 327 | GO:0005774 | Vacuolar membrane | 357 |
| GO:0009220 | Pyrimidine ribonucleotide biosynthesis | 128 | GO:0005730 | Nucleolus | 234 |
| GO:0009853 | Photorespiration | 142 | GO:0005773 | Vacuole | 623 |
| GO:0019288 | Isopentenyl diphosphate biosynthesis | 191 | GO:0048046 | Apoplast | 234 |
| GO:0006094 | Gluconeogenesis | 147 | GO:0009507 | Chloroplast | 2,385 |
| GO:0006096 | Glycolytic process | 182 | GO:0009543 | Chloroplast thylakoid lumen | 64 |
| GO:0008283 | Cell proliferation | 190 | GO:0010287 | Plastoglobule | 54 |
| GO:0006270 | DNA replication initiation | 65 | GO:0005618 | Cell wall | 381 |
| GO:0010207 | PSII assembly | 134 | GO:0005747 | Mitochondrial respiratory chain complex | 50 |
| GO:0009902 | Chloroplast relocation | 94 | GO:0005840 | Ribosome | 375 |
| GO:0010389 | Regulation of G2/M transition | 66 | GO:0005794 | Golgi apparatus | 599 |
| GO:0010027 | Thylakoid membrane organization | 160 | GO:0022626 | Cytosolic ribosome | 268 |
Regarding the ontology biological processes, the categories response to blue light (GO:0009367) and response to far-red light (GO:0010218) involved in photomorphogenesis were found among the genes changing in expression during the first day of light exposure (D 0-1). The categories photosynthetic electron transport (GO:0009773) and plastid organization (GO:0009657) also were found in the day-0 versus day-1 comparison (Table I). In the day-4 versus day-5 comparison, the terms PSII assembly (GO:0006098), thylakoid membrane organization (GO:0010027), chloroplast relocation (GO:0009902), and photorespiration (GO:0009853) appear in the top 20 GO terms (Table II). Interestingly, the group histone H3-K9 methylation (GO:0051567) including 168 genes appeared in the comparison between the 4- and 5-d samples, suggesting that chromatin modification is involved in the second phase of regulation of the nuclear genes (Table II; Fig. 5A).
A Two-Phase Expression Pattern of Nucleus-Encoded Photosynthesis Components
The expression of genes encoding the components building the photosynthetic machinery was investigated in detail. Heat maps were generated to visualize the expression levels over the time period of chloroplast development and the establishment of photosynthesis (Fig. 6). The expression profiles of the nuclear PSII- and PSI-, cytochrome b6f-, and ATPase-associated genes displayed a very strong induction of expression in response to light when the levels at time 0 were compared with those of the 1-d cells (Fig. 6, A and B). Following the strong light induction, the expression levels were stationary until day 5, when a very strong induction of the expression levels was observed again. This two-phase expression profile also was clear from the PCA plots that showed differential gene expression between days 0 and 1 and days 4 and 5, and this profile appears universal for all the nuclear PSII- and PSI-associated genes. The plastid-encoded photosynthesis-associated genes showed a more gradual increase of expression levels in response to light (Figs. 5B and 6, A and B).
Figure 6.
Expression of the components required for the photosynthetic light reaction. Gene expression heat maps are shown for PSII components (A) and PSI components (B) of nucleus-encoded genes (top) and plastid-encoded genes (bottom), photosystem assembly factors (C), and chlorophyll biosynthesis enzymes (D). Each column represents gene expression at a time point compared with the previous one [log2(T/T-1)]. Red color indicates genes that were up-regulated, and yellow color indicates genes that were down-regulated, compared with the previous time point.
In contrast to the nuclear genes encoding structural components of the photosystems, the nuclear genes encoding photosystem assembly factors (Plöchinger et al., 2016) did not uniformly display the two-phase expression profile. Notably, the expression of ALB3, LPA1, and PSBN was not increased strongly until day 5 (Fig. 6C). The expression profile of the assembly factors correlates with the results from blue-native PAGE (Fig. 3A), where the photosynthetic protein complexes were fully in place first after 7 d in the light and the assembly was completed in 14-d cells. The majority of the nuclear genes encoding the enzymes required for chlorophyll biosynthesis showed the strongest induction in expression levels when day 1 was compared with the control T0 (Fig. 6D), which also coincides with the rapid induction of chlorophyll biosynthesis in response to light (Fig. 1). Interestingly, CAO and HEMA showed a delayed induction of expression compared with the other genes encoding enzymes required for chlorophyll biosynthesis (Fig. 6D).
We also analyzed the expression of nuclear transcription factors known to be involved in chloroplast biogenesis, LONG HYPOCOTYL5 (HY5) and GOLDEN2-LIKE PROTEIN2 (GLK2; Waters et al., 2009; Kobayashi et al., 2012). HY5 activity is correlated directly with the stage of photomorphogenesis (Osterlund et al., 2000). HY5 expression level displayed a strong induction following light exposure in the 1-d sample, and then the expression levels declined (Supplemental Fig. S2). In contrast, GLK2 expression showed an induction first after 4 d in the light (Supplemental Fig. S2). Thus, HY5 and GLK2 showed contrasting expression patterns during the chloroplast developmental process, suggesting different roles. This was also supported by the analysis of hy5 and glk1glk2 mutants (Supplemental Fig. S2). The induction of LHCB expression in response to light was severely impaired in the hy5 mutant but not significantly altered in the glk1glk2 mutant compared with the wild type. In contrast, the more gradual induction of CAO was impaired in glk1glk2 but not in the hy5 mutant (Supplemental Fig. S2).
A Two-Phase Expression Profile for LHCB Was Observed Also in the Spatial Developmental Gradient of a Maize Leaf
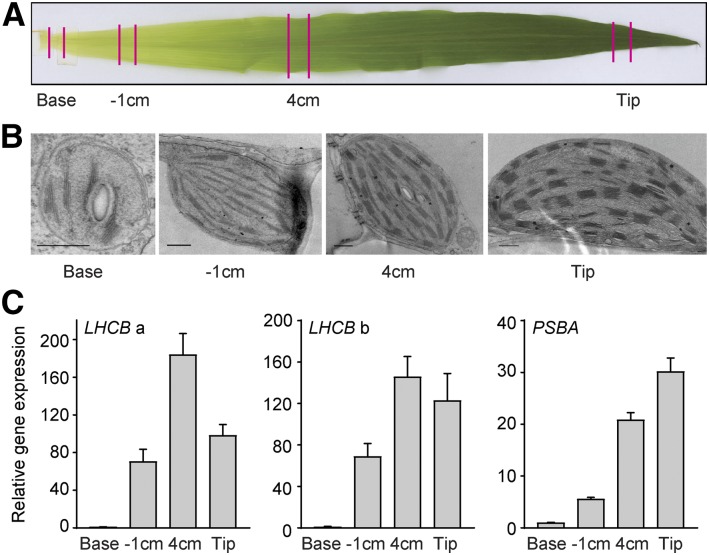
In our cell culture system, it is clear that the expression of the key components required for the establishment of photosynthesis occurs in two distinct phases. To test if this regulation also is the case in monocot plants, we used maize leaves, where a gradient of chloroplast development can be observed in a single leaf. Although it is important to emphasize that, in maize, the photosynthetic processes are partitioned between the mesophyll and the bundle sheath chloroplasts, the chloroplast developmental process has been well documented in maize (Li et al., 2010; Pick et al., 2011). The developmental stage of the chloroplasts in the maize mesophyll cells was investigated by transmission electron microscopy at different positions of the leaf (Fig. 7). At the base of the leaf, where the cells are exposed to only limited if any light, the plastids showed a structure indicative of a proplastid, like that observed for the dark-grown Arabidopsis cells (Figs. 2C and 7B). Following the leaf developmental gradient, an internal structure with small grana of interconnected thylakoids was observed 1 cm from the base. At the next sample point, the chloroplasts had a well-developed structure with numerous grana and intergranal thylakoids, and at the tip leaf, the thylakoid structure was complete (Fig. 7B). The expression levels of two LHCB genes were investigated in the maize samples (Supplemental Fig. S3). In the 1-cm sample, a strong induction of LHCB expression compared with the base sample was observed (Fig. 7C). This was similar to what was observed in the 1-d sample from our Arabidopsis cell culture. At the next sample point, 4 cm, a further induction of LHCB expression was observed (Fig. 7C). The expression levels were now stationary, and no further induction was observed at the tip of the leaf (Fig. 7C). Thus, the spatial developmental gradient from the maize leaf showed the same two-phase expression profile as was observed from the temporal developmental gradient in our Arabidopsis cell culture (Fig. 7). In contrast, a gradual increase in plastid PSBA expression along the maize leaf gradient was observed, similar to what was observed in the Arabidopsis cells (Figs. 6A and 7C).
Figure 7.
Expression of photosynthetic components along the chloroplast developmental gradient of a maize leaf. A, Illustration of the samples collected from the leaf gradient. B, Electron microscopy images of proplastids developing into chloroplasts following the leaf gradient. Images were chosen from at least two independent experiments for each developmental stage. Bars = 0.5 μm. C, LHCB and PSBA gene expression. LHCBa and LHCBb represent GRMZM2G351977 and GRMZM2G120619, respectively. Gene expression was normalized to UBIQUITIN-CONJUGATING ENZYME E2 (ZmUBI; qGRMZM2G102421) and related to the amount present in the base sample. Each data point represents the mean ± se of at least three independent replicates.
Phase 2 of LHCB Expression Can Be Blocked by the Inhibition of Chloroplast Development
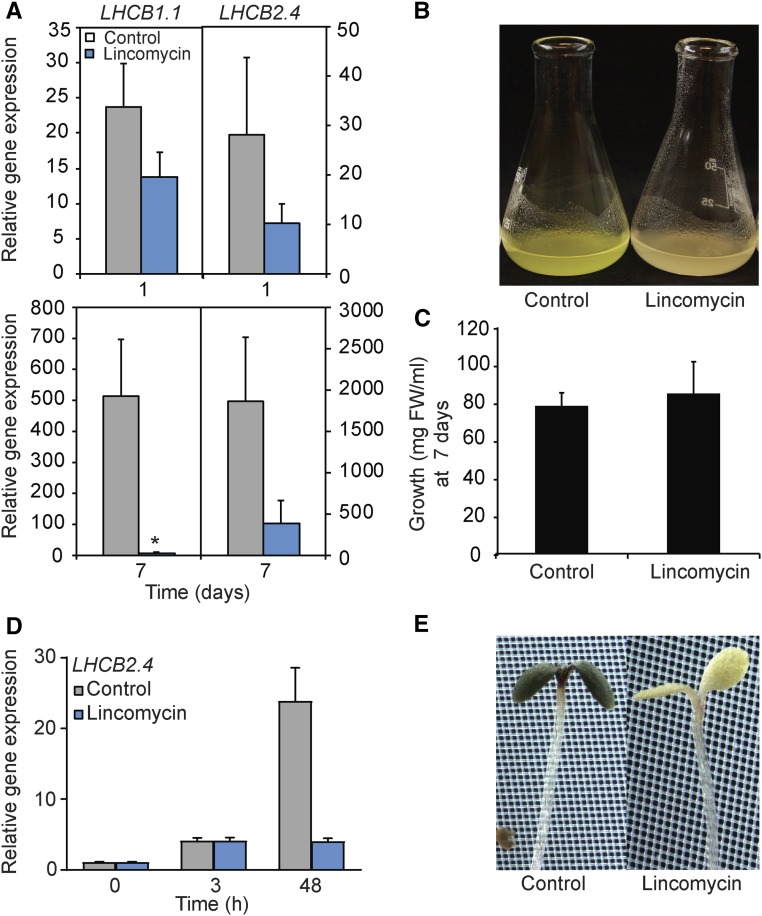
The two-phase expression profile of LHCB genes was observed in two independent greening systems: Arabidopsis cell cultures and maize leaves. In addition, following 3 h of light exposure of etiolated seedlings, a strong induction of LHCB expression was observed. The expression levels were then stationary for the first 12 h of light exposure, but between 12 and 24 h, a second significant induction in gene expression was observed, and between 24 and 48 h of light exposure, no further induction was observed (Supplemental Fig. S4). The collected data suggest that the second phase of induction of the nucleus-encoded photosynthesis genes is linked to a certain developmental stage and/or activation of the chloroplasts. To test if the induction of gene expression is linked to the status of the chloroplast, we used inhibitory agents to block proper chloroplast development. We exposed the cells to high Suc concentrations (Supplemental Fig. S5). Chloroplast development was blocked by high Suc concentrations, and the cells were then unable to perform photosynthesis (Supplemental Fig. S5). As an alternative condition, we used lincomycin, an inhibitor of plastid protein translation (Fig. 8). The initial induction of the LHCB expression triggered by light exposure was not affected following the lincomycin or high Suc concentrations (Fig. 8; Supplemental Fig. S5). However, the second phase of induction was absent following the Suc and lincomycin treatments when chloroplast development was blocked. Thus, phase 2 of the regulatory mechanism controlling LHCB expression was shown to be distinct from the initial light response and clearly linked to proper chloroplast development, as was shown both in cell cultures and Arabidopsis seedlings (Fig. 8; Supplemental Fig. S5).
Figure 8.
Lincomycin treatment blocks phase 2 in the regulation of LHCB expression in cell culture and Arabidopsis seedlings. A, Gene expression of nucleus-encoded LHCB1.1 and LHCB2.4 in the cell culture 1 and 7 d following the addition of 500 μm lincomycin. Gene expression was normalized to ubiquitin-like protein (At4g36800) and related to the amount present in the dark. Each data point represents the mean ± sd of at least three independent replicates. The asterisk indicates a significant difference between the control and lincomycin conditions (Student’s t test: *, P < 0.05). B, Representative photograph of 7-d-old control and lincomycin-treated cells. C, Growth parameters of 7-d-old control and lincomycin-treated cells. FW, Fresh weight of the cell culture. D, LHCB2.4 expression in Arabidopsis seedlings 3 and 48 h following a shift to 1 mm lincomycin. Gene expression was normalized to ubiquitin-like protein (At4g36800) and related to the amount present in the dark. Each data point represents the mean ± se of at least three independent replicates. E, Representative photographs of control and lincomycin-treated Arabidopsis seedlings following a 48-h light exposure.
Mathematical Calculations Suggest a Feedback-Controlled Coordination of Nuclear and Plastid Gene Expression
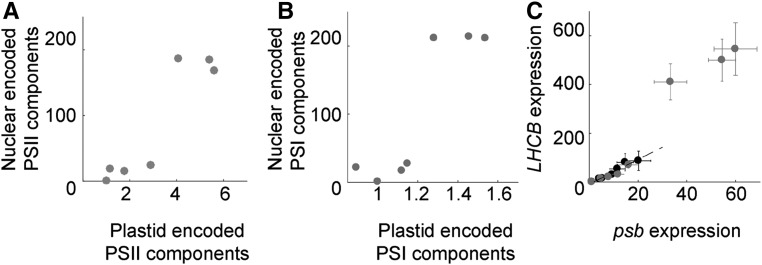
The initial light response is mediated via nuclear components, but our results (Figs. 6–8) strongly suggest that a retrograde signaling mechanism is required for the full expression of photosynthesis-associated nuclear genes (PhANGs). To test this conclusion, we mathematically assessed if the nuclear and plastid genomes are connected via positive feedback (Mitrophanov and Groisman, 2008). Positive feedback has been demonstrated to contribute to the efficiency of transcriptional regulatory systems (Mitrophanov and Groisman, 2008). To convert our experimental data to mathematical parameters, the expression of PSII and PSI components was recalculated to a geometric mean (Fig. 9). Nuclear gene expression (y axis) displayed a large dynamic range, with two distinct phases, where low and high expression clustered together. The calculations displayed a linear coordination of expression from the nucleus and the plastids for low expression levels (Fig. 9, A and B). This linear coordination could be explained by the fact that the nucleus-encoded SIGs and PAPs are required for PEP activity and the expression of PSII and PSI components in the plastid. Following this initial linear phase, a dramatic increase in expression levels occurred and the nuclear expression levels were elevated to a second phase. However, when chloroplast development was inhibited by high Suc or lincomycin treatment, nuclear gene expression followed the initial linear response of the two genomes (Fig. 9C, black circles) but did not enter the second high-expression phase. This suggests that the second phase is due to a positive retrograde feedback, linked to plastid activities, regulating nuclear gene expression. Mathematically, when two components display an initial linear response for small values with an increasing nonlinear response and a distinct jump to a second phase, it is demonstrated that the two components are mutually connected through a positive feedback mechanism (Supplemental Data S1). Thus, the calculations support our conclusions that the two components, nucleus and chloroplast, are coordinated by positive feedback during chloroplast development and that the activation of chloroplast transcription is the trigger for the feedback signal required for the full expression of PhANGs.
Figure 9.
Coordination of plastid and nuclear expression. Expression is shown for PSII and PSI components encoded in the nucleus (y axis) plotted versus components encoded in the plastids (x axis). A, Geometric mean expression of nucleus- versus plastid-encoded PSII components displayed in Figure 6A. B, Geometric mean expression of PSI components displayed in Figure 6B. C, Geometric mean of LHCB1.1 and LHCB2.4 (y axis) and psbA and psbD (x axis). Black and gray circles are expression levels measured for 3% and 1% Suc, respectively.
DISCUSSION
Using a single cell culture from Arabidopsis, we could show that the establishment of photosynthesis through the development from proplastids to functional chloroplasts occurs in two distinct phases (Fig. 10). During the initial phase, light exposure triggers a significant change in gene expression (Fig. 5) and the metabolite profile changes in response to light (Fig. 4). As many as 3,084 genes were differentially expressed between day 0 and day 1, and genes within the categories response to blue light (GO:0009367) and response to far-red light (GO:0010218) were highly represented among those genes (Table I), confirming the important role of the photoreceptors during the early light response (Strasser et al., 2010). Following the initial response to light exposure, the expression levels remained rather stationary until day 5, when the second phase of the developmental process was triggered. During phase 2, the final transition to photosynthetically functional chloroplasts occurs, and this transition requires a major reorganization of cellular metabolic activity. Such a complete reorganization of the metabolism will naturally require the expression of novel components, and as many as 12,411 genes were differentially expressed when day 4 was compared with day 5 (P < 0.01; Fig. 5). The GO enrichment also showed that a more diverse set of genes was triggered in the second expression phase, although a high percentage of the genes would fall within the category chloroplast (GO:0009507; Table II). Interestingly, the group histone H3-K9 methylation including 168 genes suggested that chromatin modification could be involved in the second phase of regulation of the nuclear genes (Table II; Fig. 5A). In mammalian cells, it was shown that changes to intracellular metabolism alter the expression of specific histone methyltransferases and acetyltransferases, conferring widespread variations in epigenetic patterns (Keating and El-Osta, 2015). Thus, the large number of genes changing in expression between day 4 and day 5 also would argue for a regulation of gene expression at a higher level of organization. Associated with the massive change in gene expression during phase 2 was a change in the metabolome, supporting a critical metabolic shift between day 4 and day 5 (Fig. 4).
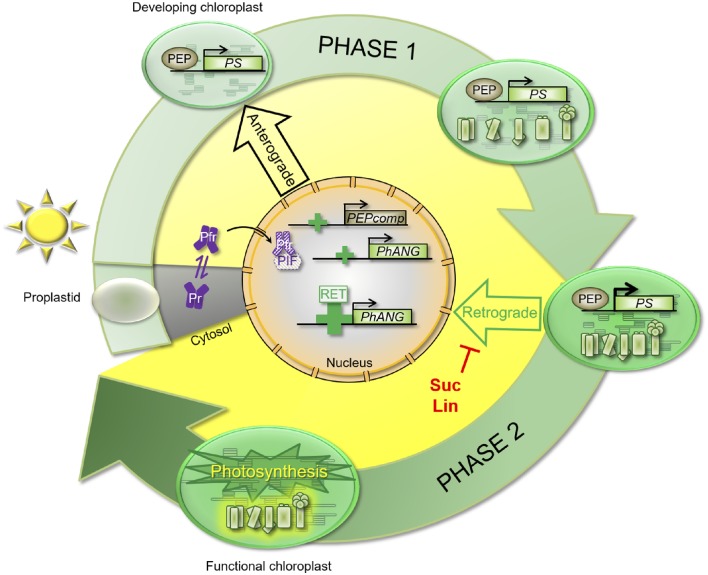
Figure 10.
Model for the two distinct regulatory phases required for the full expression of nucleus-encoded photosynthesis genes. In response to light, a rapid induction of nucleus-encoded photosynthesis-associated genes is observed (PEPcomp and PhANG). This induction was assigned previously the response of the photoreceptors (Pfr and Pr). Once a certain developmental stage has been reached, the first thylakoid membranes have been formed, some photosynthetic activity detected, and expression of the plastid-encoded photosynthesis genes (PS) activated, a second strong induction of gene expression was observed. This second induction was inhibited by Suc or lincomycin (Lin) treatments, whereas the inhibitors did not affect the first light-triggered induction of gene expression. This suggests that the two regulatory phases are controlled by different mechanisms and that the second phase is dependent on a positive retrograde signal (Retrograde). PIF, Phytochrome-interacting factor.
In contrast to the nucleus-encoded genes, the plastid and mitochondrial genes did not show the same two-phase expression profile, and the increase in photosynthetic gene expression was more gradual in response to light. PEP represents the major transcription machinery in mature chloroplasts, and over 80% of all primary plastid transcripts are transcribed by PEP (Zhelyazkova et al., 2012). The initiation of chloroplast development in the light and the activation of the photosynthetic reactions are believed to be accompanied by a repression of NEP activity and an increase of PEP-mediated plastid transcription (Liere et al., 2011; Börner et al., 2015). However, the mechanisms underlying this change in major RNA polymerase activity and the division of labor between NEP and PEP in the chloroplast are unknown (Zhelyazkova et al., 2012). The transcript levels for the plastid-encoded photosynthesis components were relatively high compared with the nucleus-encoded components at T0, suggesting that the transcription of plastid-encoded photosynthesis genes occurs to some degree already in the proplastids in the dark. Possibly, this indicates the NEP-driven expression of those components already in the dark.
The second phase of nuclear gene expression coincided with the appearance of the first chloroplast-like structures. At day 5, an internal structure with interconnected thylakoids appeared in the plastids (Fig. 2), and some photosynthetic activity could be detected (Fig. 3). During phase 2, the photosynthetic complexes are assembled and finalized. More than 40 proteins are associated with PSII, either stably or transiently (Järvi et al., 2011). Due to such complexity, the photosystem apparatus must be assembled in an orderly manner, ensuring that the components are inserted in the correct sequence. It appears unlikely that this is controlled by the order in which different components are expressed, as the data showed that they are all expressed simultaneously following illumination. The photosystem assembly factors, such as SRP54, ALB3, LPA1, and LPA2, play a key role in the assembly of a functional PSII and PSI, and full expression of these factors was not observed until day 5 (Fig. 6C). The expression profile of the assembly factors correlates with the results from blue-native PAGE (Fig. 3A), where the photosynthetic protein complexes only appeared after 7 d in the light. The assembly of PSII and PSI was completed in 14-d cells, when all the photosynthetic protein complexes, such as the PSII-LHCII supercomplex, PSI-PSII dimer, ATP synthase, PSII monomer, and cytochrome b6f, were clearly visible by green-native PAGE (Fig. 3A).
During chloroplast formation, distinct morphological changes to the plastids were observed, including a gradual change in chloroplast shape from elongated tubes to globular organelles (Fig. 2). The morphological changes were accompanied by a clear positional shift from the nuclear vicinity to the cellular cortex (Fig. 2). Thus, we have demonstrated that the shape and the cellular position of the plastids are highly dynamic during the transition from proplastid to functional chloroplast. A critical developmental switch occurred between day 4 and day 5, as indicated by the massive change in gene expression and the change in the metabolome. At this time point, the plastids appeared clustered around the nucleus (Fig. 2A), possibly to facilitate the information flow from the nucleus to the chloroplasts. As the development into a functional chloroplast proceeded, the plastids moved toward the plasma membrane. In the 7-d cells, some plastids appeared near the cell cortex, whereas following 14 d in the light, the chloroplasts were clearly separated from each other and positioned at the cell cortex, as seen in leaf mesophyll cells (Fig. 2A). The final position of the chloroplasts at the cell cortex also was associated with fully functional chloroplasts and the establishment of true photosynthetic activity. Thus, the development of functional chloroplast also is associated with major cytological changes, where the cellular position of the plastids is very dynamic. Chloroplasts were shown to move from the cell cortex to the nucleus upon illumination as a mechanism of photoavoidance in the centric diatom Pleurosira leavis (Furukawa et al., 1998), but the movement of plastid during chloroplast development, to our knowledge, has not been demonstrated previously.
The second phase of expression of the nucleus-encoded photosynthesis genes was clearly linked to the activity of the chloroplast. Correlated with the large change in nuclear gene expression between day 4 and day 5 was the establishment of a significant expression of the plastid-encoded photosynthesis genes (Figs. 6 and 9). When the expression profile of the temporal chloroplast differentiation gradient in our Arabidopsis cell line was compared with the gradient described in a single leaf of maize, the spatial developmental gradient from the maize leaf showed the same two-phase expression profile (Fig. 7). The maize leaves contain the two cell types, mesophyll and bundle sheath cells, with different roles in the photosynthetic process; thus, the samples for the expression data come from a mix of the two cell types. However, the second induction of LHCB expression was correlated with the formation of plastid structures in the mesophyll cells and the expression of the plastid-encoded photosynthesis genes (Fig. 7). In addition, published expression profiles from maize showed that samples from the midpoint of the leaves showed a strong induction of photosynthesis genes whereas the genes encoding the enzymes in tetrapyrrole biosynthesis were expressed closer to the base of the leaf, similar to the profile detected in the Arabidopsis cell line (Fig. 6; Li et al., 2010; Pick et al., 2011). A good correlation between expression levels and protein levels of photosynthetic components has also been demonstrated (Majeran et al., 2010). Thus, gene expression profiles of those different categories of photosynthesis-related genes were linked to the same developmental stages of the plastids in the two different systems representing dicots and monocots, respectively (Figs. 6 and 7).
The initial induction of LHCB expression triggered by light exposure was not affected by the lincomycin treatment or the high Suc concentrations (Figs. 8 and 9; Supplemental Fig. S5). In contrast, the second phase of induction was absent when chloroplast development was blocked. Thus, the mechanism behind phase 2 of PhANG expression is separate from the initial light signal (Fig. 8). The second phase is most likely induced by a retrograde signal dependent on normal chloroplast development (Fig. 9). Our mathematical calculations suggest that the nucleus and the plastids are mutually connected by a positive feedback mechanism (Fig. 9). Once PEP is activated and the expression of the plastid-encoded photosynthesis genes is induced, we postulate that a positive plastid signal is generated through a retrograde mechanism. The calculations and the expression profiles show that this retrograde signal is most likely linked to the transcription of plastid-encoded photosynthesis genes. The status of the PEP complex links the functional state of the chloroplast to the nucleus, enabling the plant to synchronize the expression of photosynthetic genes from the nuclear and chloroplast genomes during seedling establishment. The plastid signals operating during chloroplast biogenesis have not yet been discovered, but the lack of PhANG expression when chloroplast development is blocked by mutations or chemical treatments has usually been interpreted as being due to a repressive signal emanating from damaged or abnormal plastids. However, the possibility was recently presented in which lincomycin would either (1) disrupt a positive plastid-emitted signal, which acts in a GENOME UNCOUPLED1 (GUN1)-regulated manner, or (2) induce a negative plastid-emitted signal, which acts to repress nuclear transcription in a GUN1-mediated manner (Martín et al., 2016). Our results conclude that PhANG expression is dependent on a positive signal from healthy developing plastids. In this new understanding of the coupling of nuclear and plastid gene expression, the gun mutations could be interpreted as allowing a positive signal to proceed that would normally have been shut down. In this case, the mutated components would be expected to be part of a gating machinery that controls whether the signal is produced. This new interpretation may help us reinterpret puzzling old data or generate new hypotheses to test.
MATERIALS AND METHODS
Cell Culture, Growth Conditions, and Cell Sampling
Arabidopsis (Arabidopsis thaliana) Col-0 cell lines were grown in MS medium supplied with 3% (w/v) Suc, pH 5.7, in the dark at 25°C and shaken at 140 rpm (Pesquet et al., 2010). Cells were subcultured weekly by a 1:10 dilution. For all experiments, 7-d-old cells from dark conditions were subcultured in a 1:10 ratio in MS medium with 1% (w/v) Suc, equilibrated, and placed in a growth cabinet under continuous light (150 µmol photons m−2 s−1) and constant rotary agitation. After 7 d in the light, cells were pelleted without centrifugation and the medium was replaced. Cells were placed back in the light until 14 d for the final sample. Cells were collected from the culture by filtration, immediately frozen in liquid nitrogen, and ground in a mortar. For metabolomics analysis, a washing step with ice-cold distilled sterile water of the filtered cells was added before freezing the samples. The oxygen evolution rate and chlorophyll fluorescence parameters were determined with 1- and 2-mL aliquots of cell culture, respectively.
Plant Growth Conditions and Tissue Collection
All experiments were performed with Arabidopsis ecotype Col-0. The glk1glk2 double mutant and hy5-1 mutant lines were described elsewhere (Fitter et al., 2002; Maxwell et al., 2003; Kleine et al., 2007). For the etiolated seedlings, seeds were surface sterilized and cultured on phytagel-solidified (2.7 g L−1; Sigma-Aldrich) MS medium (2.2 g L−1; Duchefa) including Suc (10 g L−1; VWR) and MES buffer (0.5 g L−1; Sigma-Aldrich), pH 5.7. The MS plates were covered with aluminum foil and kept for 3 d at 4°C in darkness and then moved into continuous white light (150 µmol m−2 s−1) for 3 h at 22°C to potentiate germination. MS plates were kept in total darkness at 22°C for 5 d. After 5 d, the etiolated seedlings were exposed in continuous white light (550 µmol m−2 s−1) and 22°C. For lincomycin treatment, the etiolated seedlings were transferred onto MS plates containing 1 mm lincomycin (Sigma-Aldrich) or distilled, deionized water as a mock treatment and then exposed to continuous white light (150 µmol m−2 s−1) and 22°C.
For maize (Zea mays) samples, tissue was collected from the third leaf of 9-d-old maize B73 seedlings according to Li et al. (2010). Briefly, seeds were sown in the soil directly and grown under light of 150 µmol m−2 s−1, 12/12-h light/dark, 31°C light/22°C dark, and 50% relative humidity. Four different segments from each leaf were collected: basal (1 cm above the third leaf ligule), transitional (1 cm below the second leaf ligule), maturing (4 cm above the second leaf ligule), and mature (1 cm below the third leaf tip).
Chlorophyll Analysis
Chlorophyll was extracted by adding 1 mL of buffered acetone (80% acetone and 0.2 m Tris-HCl, pH 7) to 80 mg fresh weight of cells. Samples were incubated overnight at 4°C and centrifuged for 10 min at 15,000g. Chlorophyll content was measured and expressed according to Porra et al. (1989).
Confocal and Transmission Electron Microscopy
Analysis of chlorophyll autofluorescence was performed by confocal laser scanning microscopy (Zeiss LSM 780). The cell wall was stained by incubating samples with Calcofluor White (0.002% final concentration) for 5 min. Cells were scanned sequentially to prevent any cross talk between fluorescence channels. Confocal 0.37-µm-thick optical sections of bright-field, Calcofluor White staining, and chlorophyll autofluorescence signals were combined for a three-dimensional reconstruction of cells using ImageJ. Chloroplast density, diameter, and cortical positioning were determined using ImageJ. As the chlorophyll intensity is low in 5-d cell samples compared with 7- and 14-d cell samples, the level of chlorophyll intensity was adjusted to correctly visualize the plastids in the 5-d cell sample. For transmission electron microscopy, the samples were fixed using 2.5% (v/v) glutaraldehyde in 0.1 m cacodylate buffer overnight at 4°C. After washing three times in buffer, the specimens were postfixed with 1% (v/v) osmium tetroxide in the medium buffer for 1 h and washed twice in distilled water. Samples were dehydrated with 50%, 70%, 95%, and 100% ethanol and infiltrated and embedded in Spurr’s resin. Using a Diatome diamond knife on a Leica EM UC7 device, thin sections (60–90 nm) were collected onto copper grids, treated with 5% uranyl acetate in water for 20 min, followed by Sato’s lead staining for 5 min. Sections were examined in a JEOL 1230 transmission electron microscope, and digital images were captured using a Gatan MSC 600CW camera.
Isolation of Thylakoid Protein Complexes and Blue-Native PAGE
Thylakoid membrane purification was done according to Hall et al. (2011) with modifications. Briefly, cells were pelleted by centrifugation (200g, 2 min), incubated with cellulase (1%, w/v) and macerozyme (0.2%, w/v), and shaken in the dark for 4 h to digest the cell wall. After homogenization, successive centrifugation, and washing steps, thylakoid membranes were resuspended in a thylakoid wash buffer (Hall et al., 2011). Protein quantification was done with the Pierce BCA Protein Assay Kit (Thermo Scientific) after precipitation of the proteins in chlorophyll extraction buffer and resuspension of the pellet in 160 mm Tris-HCl and 2% SDS. A total of 50 µg of protein complexes from isolated thylakoids was diluted in 25 mm Bis-Tris-HCl, pH 7, and 2% glycerol and solubilized with 3% β-dodecylmaltoside (Sigma-Aldrich) for 10 min at 4°C in the dark. Unsolubilized material was removed by centrifugation at 18,000g for 20 min. A total of 35 μg of protein was loaded with BN loading buffer on a 4-12% Bis-Tris gel (NuPAGE Novex 1.0 mm; Invitrogen).
Protein Gel Electrophoresis and Immunoblotting
Total proteins were extracted from 100 mg fresh weight of cells with 200 µL of buffer (65 mm Tris-HCl, pH 6.8, 10% glycerol, 3% SDS, 0.005% Bromophenol Blue, and 5% β-mercaptoethanol), heated at 95°C for 5 min, and spun down for 10 min at 14,000 rpm. A total of 15 µL of protein extract was loaded, and proteins were separated on a 10% acrylamide gel for SDS-PAGE analysis and then transferred on a nitrocellulose membrane for western-blot analysis. Proteins LHCB1, LHCB2, psaC, psbD, and RBCL were detected with primary rabbit antibodies at dilutions of 1:5,000 (LHCB1 and LHCB2), 1:100 (psaC), 1:2,000 (psbD), and 1:2,500 (RBCL; Agrisera) and a secondary donkey anti-rabbit IgG antibody conjugated to horseradish peroxidase (1:50,000). For α-tubulin detection, a primary mouse antibody (1:1,000) and a secondary goat anti-mouse IgG antibody conjugated to horseradish peroxidase (1:10,000) were used. Luminescence was detected using the ECL Prime Western Blotting Detection Reagent (Amersham).
Oxygen Evolution Analysis and Chlorophyll Fluorescence
Oxygen evolution was measured using a 1-mL cell suspension and an oxygen electrode (Hansatech) at 20°C (no external electron acceptors were added). Samples were dark incubated on the electrode with slow steering for 2 min and then illuminated with saturating white light (2,000 µmol photons m−2 s−1) for 2 min. Oxygen evolution was recorded and evaluated by Oxygraph version 1.15 (Hansatech), and the rates were converted into nanomoles of oxygen produced per minute and standardized to total fresh weight of cells. Chlorophyll fluorescence was measured using a Dual-PAM-100 (Walz) on a 2-mL aliquot of cell culture in a glass cuvette under agitation. Before each measurement, samples were dark adapted for 20 min. The F0 (minimum fluorescence yield) was measured under weak modulated measuring light (9 µmol photons m−2 s−1) and the Fm (maximum fluorescence yield) was measured by applying a saturating pulse of white light (3,000 µmol photons m−2 s−1 for 0.6 s). The actinic light was 125 µmol photons m−2 s−1. Fv/Fm was calculated from the ratio (Fm – F0)/Fm.
Metabolite Analysis
A total of 30 mg of frozen cell powder from six independent cell cultures (biological replicates) was submitted to metabolite analysis according to Kusano et al. (2011), except for the statistical analysis. Metabolites present in the UPSC library were automatically investigated in the chromatograms, and corresponding peaks were integrated and quantified. Obtained peaks were subsequently confirmed by manual curation against the UPSC library and the Golm library (Schauer et al., 2005). A matrix was then generated containing the peak areas (normalized using the internal standards and the fresh weight) and the samples and then was used for statistical analysis performed with SIMCA13.0.3 software.
RNA Isolation and RNA Sequencing
Total RNA was isolated using the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. RNA was quantified with a Nanodrop ND-100 spectrophotometer, and RNA quality was checked by electrophoresis on a 1% agarose gel. RNA samples were ethanol precipitated and diluted to 500 ng μL−1. RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies), and the RNA was quantified with a Qubit 2.0 fluorometer (Thermo Fisher Scientific), both according to the manufacturer’s instructions.
RNA sequencing libraries were prepared using the TruSeq Stranded Total RNA with Ribo-Zero Plant Kit (Illumina), according to the manufacturer’s instructions, using half-reactions and 200 ng of RNA. Successful rRNA depletion was verified by electrophoresis using the High Sensitivity RNA ScreenTape System on the Agilent 2200 TapeStation (Agilent Technologies), according to the manufacturer’s instructions. Enrichment of cDNA fragments was performed using 11 cycles generating cDNA fragments of ∼270 bp. Prior to sequencing, single-molecule DNA templates were bridge amplified on the cBot (Illumina) to form clonal clusters inside the flow cell using the TruSeq SR Cluster Kit v3-cBot-HS (Illumina), according to the manufacturer’s instructions. Single-read sequencing was carried out for 51 cycles on the HiSeq 1500 (Illumina) using the TruSeq SBS Kit v3-HS (Illumina), following the manufacturer’s instructions. An average cluster density of ∼850 K mm−2 was recorded.
Computational Analysis
Raw read data were converted to fastq files using the Casava program and assessed for quality using the program FastQC version 0.11.3 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then filtered using the program Trimmomatic version 0.33 (Bolger et al., 2014), removing the Illumina adapters and reads with quality below 20. Raw read count numbers per gene were obtained via pseudoalignment using the Kallisto program (Bray et al., 2015). TAIR10 genome and accompanying gene annotation file were used as references (www.arabidopsis.org). A correction was first made to the YCF3 gene annotation (ATCG00360) in accordance with de Longevialle et al. (2008). Raw counts produced by Kallisto were processed using a custom Java script that took the maximum isoform count for each gene and rounded it to an integer value for compatibility with downstream analysis tools. The resulting single matrix of raw count data was split into three separate count matrices based on the encoding compartment (nucleus, plastid, and mitochondrion). Each of these data sets was normalized separately using the DESeq2 package (Love et al., 2014). This was performed using the DESeq likelihood ratio test. PCA was carried out within DESeq2 using a variance-stabilizing transformation on count data. The reads from RNA sequencing have been uploaded to the European Nucleotide Archive database (accession no. E-MTAB-5777).
GO Term Enrichment Analysis
Differential gene expression results of nuclear genes for D 0-1 and D 4-5 were obtained from normalized count data in DESeq2 using a Wald test with a log-fold change threshold of 1 and an adjusted P value cutoff of less than 0.01. Annotation of differentially expressed genes was provided by the R package GO.db (bioconductor.org/packages/GO.db). Background genes were determined using the Manhattan method of genefinder in the R package Genefilter (bioconductor.org/packages/genefilter). Top GO terms were compiled with the R package topGO (Alexa and Rahnenfuhrer, 2010) using the Fisher test statistic.
Gene Expression Analyses
Total RNA was extracted from Arabidopsis seedlings or segments of maize leaf using the E.Z.N.A.Plant RNA Kit (VWR). The extracted total RNA was quantified with an ND-1000 spectrophotometer (NanoDrop Technologies). One microgram of total RNA was used as a template for reverse transcription with the iScript cDNA Synthesis Kit (Bio-Rad) before the possible genomic DNA was eliminated with DNase I (Thermo Fisher Scientific). Equal amounts of first-strand cDNAs were used as templates for real-time PCR amplification using the following primer combinations (Supplemental Table S2): qLHCB1.1_F/qLHCB1.1_R, qLHCB2.4_F/qLHCB2.4_R, qCAO_F/qCAO_R, qZm2G351977_F/qZm2G351977_R, qZm2G120619_F/qZM2G120619_R, and qZMPSBA_F/qZmPSBA_R. For Arabidopsis, PROTEIN PHOSPHATASE2A SUBUNIT A3 (PP2A; AT1G13320) was amplified using the primer combination qPP2A_F/qPP2A_R. For maize, ZmUBI (qGRMZM2G102421) was amplified using the primer combination qZm2G102421_F/qZm2G102421_R. Quantitative real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) with a CFX96 Real-Time System (Bio-Rad) quantitative PCR machine. LHCB1.1, LHCB2.4, and CAO transcript levels were quantified in relation to Arabidopsis PP2A levels, and GRMZM2G351977 and GRMZM2G120619 transcript levels were quantified in relation to ZmUBI levels.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Heat map and cluster representation of the metabolite profiles in the different samples during chloroplast development.
Supplemental Figure S2. Gene expression of HY5, GLK2, LHCB1.1, and CAO in Arabidopsis cell culture and seedlings.
Supplemental Figure S3. Phylogenetic analysis of the maize family genes, showing the phylogenetic relationships among LHCBs and PSBAs in maize and Arabidopsis.
Supplemental Figure S4. Deetiolation of Arabidopsis seedlings.
Supplemental Figure S5. High Suc concentration inhibits chloroplast development.
Supplemental Table S1. List of identified metabolites with their normalized peak areas in every sample.
Supplemental Table S2. List of primers used for the RT-PCR analysis.
Supplemental Data S1. Theory section with details of the mathematical calculations.
Acknowledgments
We thank Dr. Jane Langdale for the glk1glk2 seeds.
Glossary
- MS
Murashige and Skoog
- PCA
principal component analysis
- GO
Gene Ontology
- Col-0
Columbia-0
Footnotes
This work was supported by grants from the Swedish Research Council (to Å.S. and E.P.) and Solar Fuels/Artificial Leaf grants (to Å.S. and W.P.S.), the Kempe Foundation (to J.H. and T.D.), the Carl Tryggers Foundation (to W.P.S.), and the Australian Research Council (grant no. CE140100008 to I.S.).
References
- Alexa A, Rahnenfuhrer J (2010) topGO: Enrichment Analysis for Gene Ontology. R package version 2.22.0
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Barajas-López JdeD, Blanco NE, Strand Å (2013a) Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833: 425–437 [DOI] [PubMed] [Google Scholar]
- Barajas-López JdeD, Kremnev D, Shaikhali J, Piñas-Fernández A, Strand A (2013b) PAPP5 is involved in the tetrapyrrole mediated plastid signalling during chloroplast development. PLoS ONE 8: e60305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, Pachter L (2015) Near-optimal RNA-Seq quantification. Cornell University Library [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Jiang J, Zhang L (2015) Plastid sigma factors: their individual functions and regulation in transcription. Biochim Biophys Acta 1847: 770–778 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Watanabe M, Shihira-Ishikawa I (1998) Green- and blue-light mediate chloroplast migration in the centric diatom Pleurosira laevis. Protoplasma 203: 214–220 [Google Scholar]
- González-Pérez S, Gutiérrez J, García-García F, Osuna D, Dopazo J, Lorenzo Ó, Revuelta JL, Arellano JB (2011) Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol 156: 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Mishra Y, Schröder WP (2011) Preparation of stroma, thylakoid membrane, and lumen fractions from Arabidopsis thaliana chloroplasts for proteomic analysis. Methods Mol Biol 775: 207–222 [DOI] [PubMed] [Google Scholar]
- Hampp C, Richter A, Osorio S, Zellnig G, Sinha AK, Jammer A, Fernie AR, Grimm B, Roitsch T (2012) Establishment of a photoautotrophic cell suspension culture of Arabidopsis thaliana for photosynthetic, metabolic, and signaling studies. Mol Plant 5: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Fu A, Garcia VJ, Buchanan BB, Luan S (2015) PSB27: a thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc Natl Acad Sci USA 112: 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Keating ST, El-Osta A (2015) Epigenetics and metabolism. Circ Res 116: 715–736 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Strand Å (2015) Chloroplast transcription, untangling the Gordian knot. New Phytol 206: 889–891 [DOI] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keränen M, Aro EM, Fukaki H, Ohta H, Sugimoto K, et al. (2012) Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Jonsson P, Fukushima A, Gullberg J, Sjöström M, Trygg J, Moritz T (2011) Metabolite signature during short-day induced growth cessation in Populus. Front Plant Sci 2: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, et al (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell BB, Andersson CR, Poole DS, Kay SA, Chory J (2003) HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiol 133: 1565–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA (2008) Positive feedback in cellular control systems. BioEssays 30: 542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Akiyama K, Tomonaga Y, Kato A, Sato Y, Kobayashi M, Nagata N, Sakurai T, Shinozaki K (2013) The Chloroplast Function Database II: a comprehensive collection of homozygous mutants and their phenotypic/genotypic traits for nuclear-encoded chloroplast proteins. Plant Cell Physiol 54: e2. [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW (2010) The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr Biol 20: 744–749 [DOI] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, et al. (2011) Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23: 4208–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöchinger M, Schwenkert S, von Sydow L, Meuer J, Schröder W (2016) Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, Terc and ALB3 in maintenance and assembly of PSII. Front Plant Sci 7: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Tognetti VB, Gonzalez N, Van Dingenen J, De Milde L, Bielach A, De Rycke R, Van Breusegem F, Inzé D (2015) GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol 167: 817–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T (2012) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]