Recent discoveries focus on the central phytochrome signaling mechanisms that have profound impact on plant growth and development in response to light.
Abstract
The basic helix-loop-helix domain-containing transcription factors that interact physically with the red and far-red light photoreceptors, phytochromes, are called PHYTOCHROME INTERACTING FACTORS (PIFs). In the last two decades, the phytochrome-PIF signaling module has been shown to be conserved from Physcomitrella patens to higher plants. Exciting recent studies highlight the discovery of at least four distinct kinases (PPKs, CK2, BIN2, and phytochrome itself) and four families of ubiquitin ligases (SCFEBF1/2, CUL3LRB, CUL3BOP, and CUL4COP1-SPA) that regulate PIF abundance both in dark and light conditions. This review discusses these recent discoveries with a focus on the central phytochrome signaling mechanisms that have a profound impact on plant growth and development in response to light.
Plants undergo two contrasting developmental programs: skotomorphogenesis in the absence of light and photomorphogenesis in the presence of light. Dark-grown plants display etiolated phenotypes such as longer hypocotyls, apical hooks, and closed and yellowish cotyledons. In contrast, light-grown plants display photomorphogenic phenotypes characterized by short hypocotyls and open and expanded green cotyledons. Light also modulates other growth and developmental programs by regulating directional growth, shade avoidance, and photoperiodic flowering.
One of the most important photoreceptors that perceives and responds to the red and far-red light spectrum is the phytochrome (phy) family (Bae and Choi, 2008). Phytochromes are chromoproteins where the apoprotein is attached to a billin chromophore, forming a holoprotein. Upon red light exposure, phytochromes allosterically change the conformation from the inactive Pr to the active Pfr form. The inactive Pr form resides in the cytosol, while the active Pfr form of all phytochromes translocates into the nucleus. (Van Buskirk et al., 2012; Klose et al., 2015). In the cytosol, the Pfr forms of phytochromes regulate the translation of mRNA (Paik et al., 2012). However, in the nucleus, phytochromes interact with multiple partners to modulate the transcription of downstream target genes to mediate light responses (Huq and Quail, 2005). One of the pivotal interacting partners is PHYTOCHROME INTERACTING FACTORs (PIFs), which act as a key regulator of the transition from skotomorphogenesis to photomorphogenesis (Castillon et al., 2007; Leivar and Quail, 2011). PIFs are encoded by a subset of the basic helix-loop-helix (bHLH) transcription factor superfamily. PIFs function as negative regulators of light responses by repressing photomorphogenesis and maintaining the skotomorphogenic state of the etiolated seedlings in darkness (Leivar and Quail, 2011; Leivar and Monte, 2014). Upon exposure to light, phytochromes promote the turnover of PIFs through rapid phosphorylation, ubiquitination, and proteasome-mediated degradation, which inhibits PIF function to induce transcriptional reprogramming, resulting in photomorphogenic development (Xu et al., 2015). Rapid progress has been made in recent years in defining the phytochrome-PIF signaling module that underlies the biochemical mechanism of phytochrome function. In this Update, we review the diverse regulatory mechanisms between phytochromes and PIFs from nonvascular to vascular plants, the diversity of PIF functions in plant growth and development, as well as the updated mechanistic details of the posttranslational regulation of PIF abundance by phytochromes, various kinases, and E3 ligase complexes. We also highlight the fact that PIF degradation occurs not only under light but also in darkness. Finally, we discuss the dynamic regulation of PIF DNA-binding and transcriptional activities through PIF-interacting partners. For details on the roles of phytochromes and PIFs in other pathways, readers are directed to recently published review articles (Legris et al., 2017; Paik et al., 2017).
THE PHYTOCHROME-PIF SIGNALING MODULE IS EVOLUTIONARILY CONSERVED FROM BRYOPHYTES TO ANGIOSPERMS
Phytochromes are present in all land plants and most green algal lineages, except in the chlorophytes (Li et al., 2015). The function of phytochrome as red/far-red light sensors has been investigated extensively in terms of photochemistry, protein structures, and transduction of light signal in seed plants (Rockwell et al., 2006; Bae and Choi, 2008). In angiosperms, the phytochrome family consists of two types, a light-labile type I and a light-stable type II, based on physiological and spectroscopic analyses (Li et al., 2015). The divergence of two types of phytochromes occurred immediately after the branching of ferns and seed plants. In nonseed plants, although phytochromes are well documented, the molecular mechanism of phytochrome signaling is beginning to be understood.
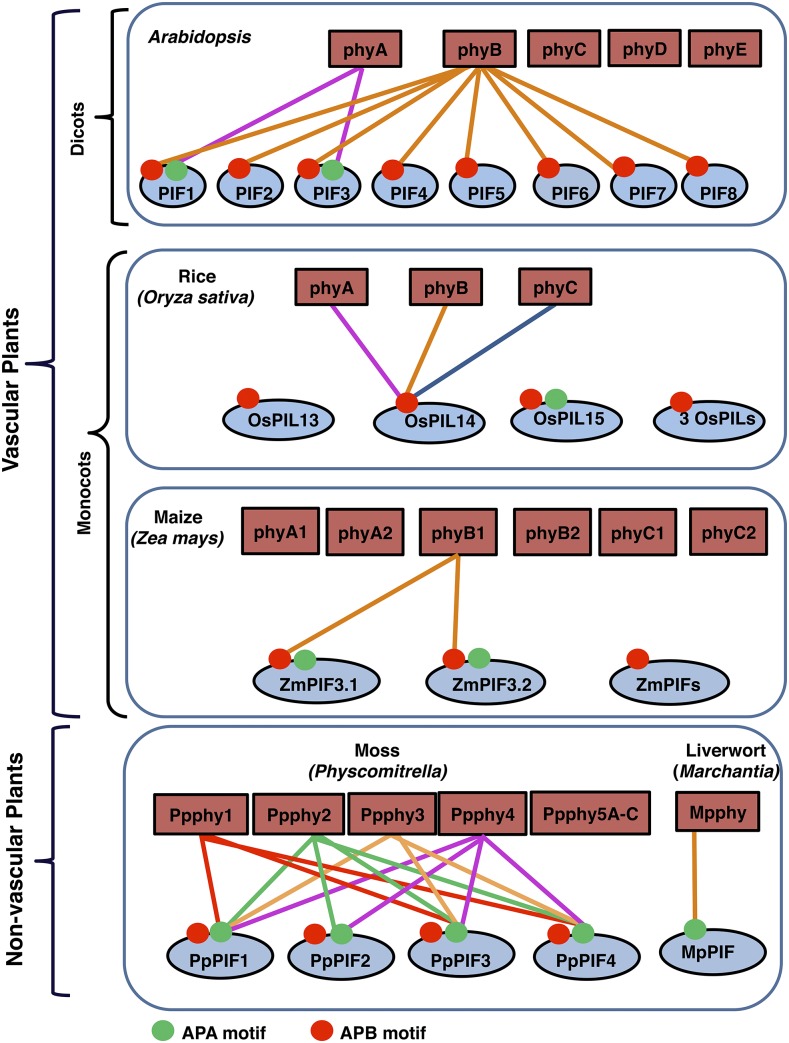
In Arabidopsis (Arabidopsis thaliana), the phytochrome family consists of five members designated as phyA to phyE (Fig. 1). Unlike the dicots, the phytochrome family in monocots contains three different phytochromes designated as phyA, phyB, and phyC (Mathews and Sharrock, 1997). Phytochromes in rice (Oryza sativa) are present as single-copy genes (Li et al., 2015). However, in maize (Zea mays), due to the gene duplication from the ancient tetraploidization in the maize ancestral lineage, phytochrome genes are present as homolog pairs for each of the three genes, known as phyA1 and phyA2, phyB1 and phyB2, and phyC1 and phyC2 (Sheehan et al., 2004). Sharing three classes of phytochromes (phyA/B/C) and missing two counterparts (phyD/E) in monocots suggest unique roles of phyD/E in dicots and also highlight the fundamental differences between the monocot and dicot phytochrome signaling pathways.
Figure 1.
The phytochrome-PIF signaling module in land plants. Schematic illustration shows the interactions between various phytochromes and PIFs from nonvascular plants to vascular plants. The schematic is based on available data on the diversification of phytochromes and PIFs in land plants. Connecting lines with the same color illustrate the interaction of each phytochrome with different PIFs. The red and green circles on different PIFs indicate the active phyB-binding (APB) and active phyA-binding (APA) motifs, respectively.
Interestingly, in Marchantia polymorpha, a liverwort, the genome contains only one phytochrome (Mpphy) gene (Inoue et al., 2016). Mpphy is unique: it is very similar to the light-stable type II phytochromes in angiosperms in terms of light stability in vivo; however, the light-dependent nuclear localization is similar to that of type I phytochrome in Arabidopsis (Inoue et al., 2016). Furthermore, similar to Arabidopsis, Mpphy regulates LHCB (LIGHT HARVESTING COMPLEX PHOTOSYSTEM II SUBUNITS) and POR (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE) gene expression reversibly under red and far-red light. In contrast to M. polymorpha, Physcomitrella patens exhibits one of the most diverse phytochrome families, with seven different phytochromes (PpPHY1, PpPHY2, PpPHY3, PpPHY4, and PpPHY5A–PpPHY5C), essentially due to three gene duplication events. These phytochromes can be divided into four distinct clades (Jaedicke et al., 2012; Possart and Hiltbrunner, 2013; Li et al., 2015). Thus, the phytochrome gene family diverged early in land plant evolution.
Similar to phytochromes, the primary signaling partners for phytochromes, PIFs, have been discovered in a variety of plant lineages from bryophytes to angiosperms (Lee and Choi, 2017). Arabidopsis has eight PIFs (PIF1, PIF3–PIF8, and PHYTOCHROME INTERACTING FACTOR3-LIKE1 [PIL1], renamed as PIF2; Fig. 1; Lee and Choi, 2017). All Arabidopsis PIFs have an APB motif; only PIF1 and PIF3 have both an APB and an APA motif. The light-dependent interactions between phyB and all of the PIFs, and between phyA and PIF1/PIF3 are well documented (Fig. 1; Huq et al., 2004; Lee and Choi, 2017). The importance of the APA and APB sequence motifs in providing the Pfr specificity of phy-PIF physical interactions has been shown by mutational analyses (Leivar and Quail, 2011).
In contrast to Arabidopsis, the functional significance of the phytochrome-PIF relationship is not fully understood in rice and maize. In rice, there are six putative PHYTOCHROME-INTERACTING FACTOR-LIKE proteins (OsPIL11–OsPIL16) with the conserved APB motif; however, only OsPIL15 contains an APA motif. Among rice PILs, only OsPIL14 has been shown to interact with OsphyA, OsphyB, and OsphyC, but it interacts preferentially with OsphyB in vitro (Fig. 1; Cordeiro et al., 2016). Similar to Arabidopsis PIFs, the overexpression of OsPILs also promotes hypocotyl and internode elongation in Arabidopsis and rice, respectively (Nakamura et al., 2007; Todaka et al., 2012). Interestingly, the overexpression of OsPIL15 in rice inhibits seedling growth in the dark, while both red and far-red light suppress its activity (Zhou et al., 2014). In tobacco (Nicotiana tabacum), overexpression of OsPIL11 results in the inhibition of hypocotyl elongation under red light (Li et al., 2012b). Thus, OsPIL11 functions similarly to Arabidopsis PIF6 in tobacco.
In maize, the analysis of bHLH sequences illustrates the presence of at least seven putative PHYTOCHROME-INTERACTING FACTORs (ZmPIFs) with a conserved APB motif, of which two (ZmPIF3.1 and ZmPIF3.2) also contain the APA motif. Recently, it was shown that ZmPIF3.1 and ZmPIF3.2 interact with the Pfr form of ZmphyB1, but not ZmphyB2. None of the other known ZmPIFs interact with either of ZmphyB1 or ZmphyB2 (Kumar et al., 2016). Furthermore, the function of ZmPIFs in light signaling pathways has not been demonstrated yet.
While the molecular mechanism of the phytochrome-PIF signaling module is well studied in higher plants, the mechanistic details of phytochrome-PIF in basal land plants is beginning to be unraveled. Recently, a single PIF gene was discovered in M. polymorpha (Inoue et al., 2016). Sequence analysis of MpPIF demonstrates the presence of a highly conserved C-terminal domain and a putative APA motif at its N terminus. Knocking out MpPIF by homologous recombination shows that MpPIF is necessary for light-mediated responses, such as the inhibition of gemma germination and the repression of LHCB and POR mRNA expression in the dark. In addition, Mpphy induces a rapid degradation of MpPIF in response to red light. These data clearly demonstrate the conservation of the phytochrome-PIF signaling module in M. polymorpha and Arabidopsis (Inoue et al., 2016).
Sequence analysis of the P. patens genome identifies four putative PIF orthologs (PpPIF1–PpPIF4; Possart et al., 2017). PpPIFs contain an APA motif and a potential APB-like motif similar to those found in Arabidopsis and other angiosperm PIFs. However, only APA, but not the APB-like sequence motif, is necessary for the light-dependent interactions between PpPIFs and Ppphys. Site-directed mutagenesis of critical amino acid residues in the APA motif significantly reduces or eliminates the interaction between PpPIFs and all Ppphys, suggesting that the APA motif is critical for the interaction. Overexpression of PpPIF1/PpPIF2 in wild-type Arabidopsis induces a hyposensitive phenotype in response to red light, while overexpression of PpPIFs in the pif quadruple mutant (pif1 pif3 pif4 pif5, termed pifQ) background complements, and largely rescues, the constitutively photomorphogenic (cop-like) morphological phenotypes in the dark. Furthermore, overexpression of PpPIFs also rescues the PIF-dependent gene expression in the pifQ background. Taken together, these data illustrate that PpPIFs from P. patens are functional in Arabidopsis. However, in contrast to Arabidopsis and M. polymorpha PIFs, PpPIFs are not degraded in response to red light exposure when expressed in Arabidopsis. These data suggest that either PpPIFs are not degraded like Arabidopsis PIF7 or PpPIF degradation might be eluded in the heterologous system tested. Thus, the light regulation of PpPIFs awaits further studies in P. patens. Overall, the identification and characterization of phytochromes and PIFs from lower to higher plants as discussed above highlight a few salient features of the phytochrome-PIF signaling module, as outlined in Box 1.
DISTINCT AND SHARED BIOLOGICAL FUNCTIONS OF PIFS IN ARABIDOPSIS
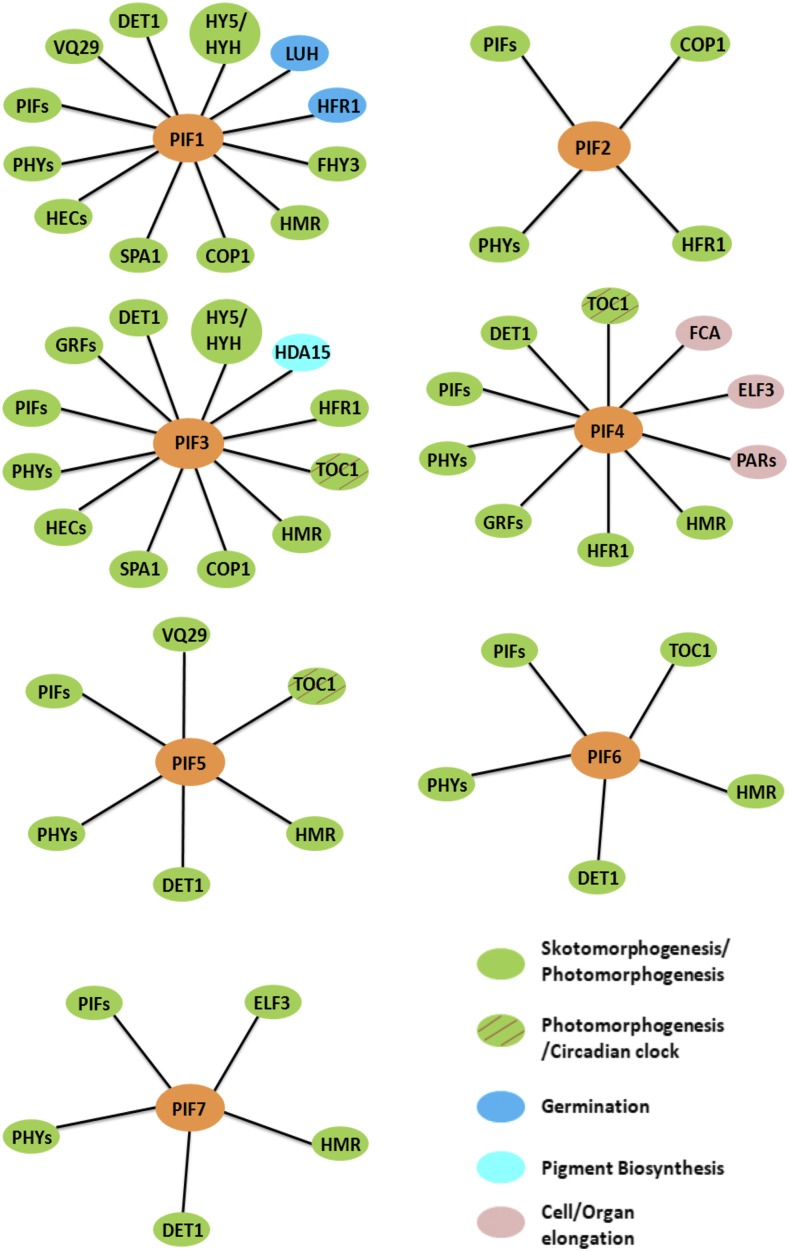
The functional importance of PIFs has been investigated extensively through genetic, biochemical, and physiological studies. PIFs function primarily as negative regulators of photomorphogenesis. Thus, pifQ exhibits constitutive photomorphogenic phenotypes (Leivar et al., 2008b; Shin et al., 2009). Conversely, overexpression of PIFs results in hyposensitive phenotypes in response to light (Huq and Quail, 2002; Kim et al., 2003; Khanna et al., 2004; Leivar et al., 2008a). In addition to their prominent role in regulating photomorphogenesis, PIFs have been shown to regulate many other pathways, including mediating metabolic signals to the circadian clock (Shor et al., 2017), thermomorphogenesis (Quint et al., 2016), hormone signaling, and biotic and abiotic responses (Paik et al., 2017). They do so by interacting with a host of other cellular signaling molecules (Fig. 2).
Figure 2.
Distinct and shared biological functions of PIFs in Arabidopsis. PIFs function as negative regulators of light signaling as well as important modulators of plant growth and development. Different PIFs interact with a number of common but also distinct interacting partners to regulate seed germination, cell and organ elongation, photosynthesis, pigment biosynthesis, and also the integration of light and circadian clock signaling. Contrary to the generally accepted functions of PIFs as negative regulators of photomorphogenesis, PIF6 functions as a positive regulator of photomorphogenesis. The distinct and shared biological functions of PIF-interacting proteins are shown in solid color ovals and/or hatched ovals.
Whereas the regulation of photomorphogenesis reflects the shared or overlapping function of PIFs, some PIFs also function distinctly to modulate certain physiological responses (i.e. some responses are controlled by a single PIF, while others by two or more PIFs; Jeong and Choi, 2013). For example, PIF1 plays a major role in inhibiting light-dependent seed germination (Oh et al., 2004). Two important regulatory proteins, LEUNIG_HOMOLOG and HFR1 (LONG HYPOCOTYL IN FAR-RED1), are known to interact with PIF1 and to coordinate seed germination by regulating the expression of abscisic acid- and GA-related genes (Fig. 2; Shi et al., 2013; Lee et al., 2015). PIF1 also regulates chlorophyll biosynthesis and plastid development by modulating the expression of a number of genes (Huq et al., 2004; Moon et al., 2008; Kim et al., 2016c). PIF2 (more commonly known as PIL1) also interacts with COP1 and phyB in vivo (Fig. 2; Luo et al., 2014). COP1 promotes the degradation of PIF2 in the dark, while phyB stabilizes PIF2 in response to light. Unlike other PIFs, PIF2 positively regulates seedling deetiolation in response to blue, red, and far-red light. PIF2 also interacts with PIF1, PIF3, PIF4, and PIF5 and prevents PIF target gene expression. In this regard, PIF2 functions similar to HFR1. In fact, PIF2 also heterodimerizes with HFR1 (Fig. 2) and promotes photomorphogenesis in an additive manner with HFR1. Further studies are necessary to determine how PIF2 and HFR1 function together to inhibit the DNA-binding activity of PIFs. PIF2 has all the necessary amino acids in the basic domain and is predicted to be a DNA-binding bHLH protein. Thus, PIF2 might recruit other PIFs to different DNA-binding sites by heterodimerization.
As a founding member of the PIFs, PIF3 functions predominantly as a negative regulator of seedling deetiolation along with other PIFs, primarily by regulating the abundance of phyB levels (Kim et al., 2003; Monte et al., 2004; Leivar et al., 2008a). PIF3 also interacts with TOC1, and this interaction optimizes the temporal regulation of diurnal growth of hypocotyl elongation (Soy et al., 2016). Similar to PIF1, PIF3 represses chlorophyll biosynthesis and photosynthesis in etiolated seedlings (Stephenson et al., 2009). PIF3 interacts with HISTONE DEACETYLASE15 (Fig. 2), which together repress the expression of target genes by concomitant decreases in histone acetylation and RNA polymerase II-associated transcription (Liu et al., 2013). Furthermore, PIF3 regulates the ethylene-induced hypocotyl elongation in light, and very recently, it was shown that PIF3 modulates the freezing tolerance by negatively regulating the expression of CBF (C-REPEAT BINDING FACTOR) genes (Zhong et al., 2012; Jiang et al., 2017).
Among all known PIFs, PIF4 uniquely regulates hypocotyl elongation in response to light, shade, temperature, and diurnal conditions (Huq and Quail, 2002; Lorrain et al., 2008; Franklin et al., 2011; Kumar et al., 2012). It does so by binding to the promoter sequences and activating the expression of target genes, including regulatory genes involved in auxin biosynthesis. Whereas the elongation of hypocotyl by PIF4 under different conditions provides competitive advantage toward overall fitness and survival, the persistent activation of PIF4 might cause a physiological imbalance due to the possible hyperelongation of hypocotyls. To prevent such events, plants employ multiple regulatory mechanisms, including inactivation of PIF4 transcriptional activity. For example, EARLY FLOWERING3, a component of an evening complex, interacts with PIF4 in an evening complex-independent manner and reduces PIF4 activity (Nieto et al., 2015). Similarly, at higher temperature, FLOWERING TIME CONTROL PROTEIN, an RNA-binding protein, interacts with PIF4 and induces its dissociation from the YUCCA8 promoter to balance hypocotyl growth (Lee et al., 2014). While in shade, HFR1, PAR1 (PHYTOCHROME RAPIDLY REGULATED1), and PAR2 interact with PIF4 and subsequently prevent PIF4 from activating its target genes (Fig. 2; Hornitschek et al., 2009; Hao et al., 2012; Lee et al., 2014; Nieto et al., 2015). Besides hypocotyl elongation, PIF4 also controls several physiological and development aspects, such as stomatal development in response to light quality, circadian gating, chlorophyll degradation and leaf senescence in darkness, freezing tolerance, and anthocyanin biosynthesis under red-light conditions (Lorrain et al., 2008; Casson et al., 2009; Lee and Thomashow, 2012; Sakuraba et al., 2014; Song et al., 2014; Liu et al., 2015; Zhang et al., 2015; Zhu et al., 2016a).
PIF5 regulates many of the same pathways regulated by PIF4, albeit in an overlapping manner. PIF5 (and PIF4) functions as a positive regulator of leaf senescence through chlorophyll degradation in darkness (Sakuraba et al., 2014; Song et al., 2014; Zhang et al., 2015) and as a negative regulator of red light-induced anthocyanin biosynthesis (Liu et al., 2015). PIF5 in collaboration with PIF4 and PIF7 functions as an important regulator of shade avoidance in Arabidopsis (Lorrain et al., 2008; Hornitschek et al., 2009; Li et al., 2012a). PIF5 also functions at the interface between red light signaling and the circadian clock through its direct interaction with TOC1 (TIMING OF CAB EXPRESSION1; Fig. 2; Fujimori et al., 2004).
PIF6 produces two splice variants, α and β, of which the β-form regulates seed dormancy (Penfield et al., 2010). Contrary to the general role of PIFs as negative regulators of photomorphogenesis, PIF6 functions as a positive regulator, as ectopically overexpressed PIF6 inhibits hypocotyl elongation under continuous red light. Interestingly, PIF6 also interacts with TOC1 (Fig. 2); however, the functional importance of this interaction has yet to be determined (Fujimori et al., 2004). Interestingly, the APB domain of PIF6 and PIF3 along with the N-terminal domain of phyB have been used to develop a light-switchable gene regulatory module for studying cellular signaling pathways in metazoans (Shimizu-Sato et al., 2002; Levskaya et al., 2009; Toettcher et al., 2011a, 2011b, 2013).
PIF7 acts as a negative regulator of seedling deetiolation under red light (Leivar et al., 2008a). PIF7, along with PIF3 and PIF4, functions additively to promote hypocotyl elongation under continuous red light by suppressing phyB levels. PIF7 also regulates shade avoidance responses by directly controlling auxin biosynthetic genes under shade conditions (Li et al., 2012a). In addition, PIF7 and PIF4 reduce freezing tolerance responses by directly down-regulating CBF gene expression in a photoperiod-dependent manner (Lee and Thomashow, 2012). Finally, the functional importance of PIF8 is largely unknown; therefore, further efforts are necessary to examine the potential role of PIF8 in light signaling pathways.
POSTTRANSLATIONAL REGULATION OF PIF ABUNDANCE
PIFs are known to be stable in the dark to promote skotomorphogenesis, and the light-induced degradation of PIFs promotes photomorphogenesis. However, in recent years, PIFs have been shown to be degraded under both dark and light conditions. An optimum level of PIFs is necessary for the proper transition from skotomorphogenesis to photomorphogenic development.
FACTORS CONTROLLING PIF ABUNDANCE UNDER DARKNESS
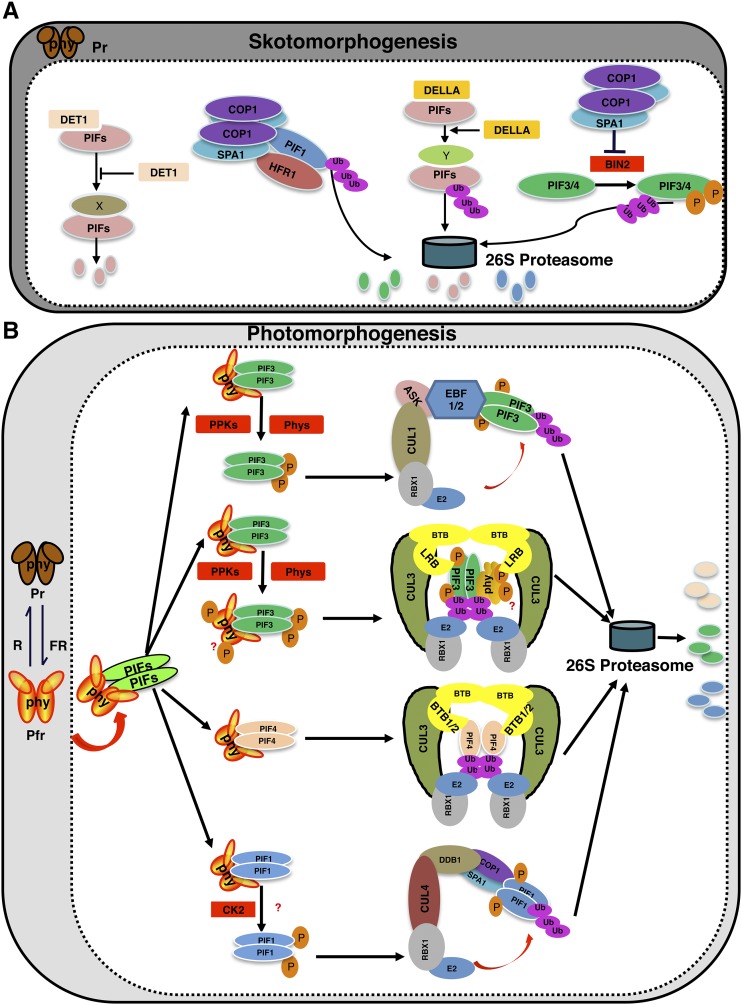
Several factors, including helix-loop-helix (HLH) proteins, kinases, and E3 ubiquitin ligases, have been shown to regulate PIF abundance in the dark. Many of these factors (e.g. DET1, HECATE2 [HEC2], and the COP1/SPA complex) stabilize PIFs in the dark to promote skotomorphogenesis, while others (e.g. BIN2, DELLA proteins, HFR1, and COP1) promote the degradation of PIFs in darkness (Fig. 3A).
Figure 3.
Dynamic regulation of PIF levels in dark and light. Model shows the mechanisms of light-dependent phosphorylation and degradation of PIFs by various kinases and E3 ubiquitin ligases. A, In the dark, DET1 interacts with PIFs and stabilizes them by an unknown mechanism. PIF1 also forms a heterodimer with HFR1, thereby triggering the codegradation of both HFR1 and PIF1. COP1 function is necessary for this codegradation of PIF1 and HFR1. Moreover, DELLAs negatively regulate PIF abundance by promoting PIF degradation through the 26S proteasome-dependent pathway both in dark and light conditions. In addition, the COP1-SPA complex promotes the stability of PIF3 by inhibiting the BIN2-mediated phosphorylation and degradation of PIF3 and PIF4. X and Y indicate unknown factors necessary for DET1- and DELLA-induced degradation of PIFs in the dark, respectively. B, Upon light exposure, different PIFs are phosphorylated by different protein kinases, including PPKs, phytochromes, and possibly other kinases, which triggers ubiquitination by different E3 ubiquitin ligase complexes followed by degradation through the 26S proteasome pathway. The ubiquitination and degradation of PIF1, PIF3, and PIF4 involve CUL4, CUL1, and CUL3-based E3 ubiquitin ligase complexes, respectively.
DET1 functions as a repressor of photomorphogenesis in part by positively regulating PIF abundance in the dark. DET1 interacts with PIFs both in vitro and in vivo (Fig. 3) and stabilizes PIFs. Strikingly, the degradation of PIFs in the det1 background is not mediated by the 26S proteasome pathway, as the inhibitors of the 26S proteasome failed to block PIF degradation in the det1 background (Dong et al., 2014). Further studies are necessary to understand how PIFs are degraded in the det1 background through the proteasome-independent pathway.
The GSK3-like kinase BRASSINOSTEROID-INSENSITIVE (BIN2) has been shown to phosphorylate both PIF3 and PIF4 in vitro (Bernardo-García et al., 2014; Ling et al., 2017). BIN2-mediated phosphorylation promotes the degradation of PIF3 and PIF4 in the dark. Mutation in the BIN2 phosphorylation sites reduces the degradation of both PIF3 and PIF4 in vivo. Strikingly, the COP1/SPA1 complex prevents BIN2-mediated phosphorylation and proteasomal degradation of PIF3 in the dark. COP1 interacts with and sequesters BIN2 from PIF3, while SPA1 interacts with PIF3 in the same binding region of PIF3 with BIN2, thus preventing BIN2 binding (Ling et al., 2017). Therefore, BIN2 promotes the degradation of PIFs, while COP1 and SPA1 stabilize them in the dark.
DELLA proteins have been shown to interact directly with PIFs and inhibit their activity to regulate hypocotyl elongation in response to light and GA (de Lucas et al., 2008; Feng et al., 2008). In a recent study, DELLA proteins also were shown to promote the degradation of all four PIFs through the 26S proteasome pathway under both dark and light conditions (Li et al., 2016). Thus, DELLAs inhibit PIF activity not only by sequestration but also by PIF degradation to regulate hypocotyl elongation. DELLA proteins are degraded in response to GA through the SLEEPY (SLY) F-box protein (McGinnis et al., 2003). It will be interesting to examine if DELLAs and PIFs are codegraded through SLY in the dark.
Similar to DELLA, HFR1 has been shown to sequester PIFs by heterodimerization to promote seed germination and seedling deetiolation (Hornitschek et al., 2009; Shi et al., 2013). Strikingly, HFR1 also promotes the degradation of PIF1 and PIF5 in the dark in a heterodimerization-dependent manner (Xu et al., 2017). Conversely, PIF1 promotes the degradation of HFR1 in darkness by enhancing the ubiquitination of HFR1 by COP1. Therefore, HFR1 not only sequesters PIFs but also negatively regulates their abundance in the dark. This regulation is important for the rapid transition from skotomorphogenesis to photomorphogenesis.
FACTORS CONTROLLING PIF ABUNDANCE UNDER LIGHT
In general, phytochromes induce the rapid degradation of PIFs in response to light signals via direct physical interactions with PIFs (Al-Sady et al., 2008; Shen et al., 2008), although phyB has been shown to induce PIF1 degradation non cell autonomously (Kim et al., 2016b). DNA binding is not necessary for the light-induced degradation of PIFs (Al-Sady et al., 2008; Shen et al., 2008). However, HEMERA has been shown to couple PIF degradation with PIF transactivation of their target genes (Qiu et al., 2015). HEMERA is necessary for PIF degradation only under light-grown seedlings (Chen et al., 2010). Two distinct biochemical events precede the light-induced degradation of PIFs. First, PIFs are phosphorylated rapidly in response to light. Second, PIFs are ubiquitinated followed by rapid degradation through the 26S proteasome pathway (Leivar and Quail, 2011). Kinetic and mutational analyses have shown that the phosphorylation precedes the ubiquitination and degradation (Al-Sady et al., 2006; Shen et al., 2008). Thus, a major focus in the field is to identify the kinases and E3 ubiquitin ligases for PIFs. Tremendous progress has been made in this area in recent years, as discussed below.
Kinases That Phosphorylate PIFs
Several kinases have been shown to phosphorylate PIFs in recent years (Fig. 3B). These include Casein Kinase II (CK2), BIN2, phytochrome itself, and four members of a recently discovered Ser/Thr kinase family called Photoregulatory Protein Kinases (PPK1–PPK4). Among these, phytochrome is perhaps the best candidate for several reasons. First, PIFs interact physically with phytochromes in response to light (Ni et al., 1999; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004). Second, direct physical interaction with phytochrome is necessary for the light-induced phosphorylation and degradation of PIFs (Al-Sady et al., 2008; Shen et al., 2008). Third, phytochromes have been shown to function as a Ser/Thr kinase in vitro (Yeh and Lagarias, 1998). However, direct in vivo evidence showing phytochrome function as a kinase was lacking until recently. Interestingly, a recent study showed that both oat (Avena sativa) and Arabidopsis phytochromes function as Ser/Thr kinases for PIFs in vitro (Shin et al., 2016). These authors have mapped the kinase domain within the PAS-GAF-PHY domains located at the N terminus of phytochromes and identified critical residues that are necessary for ATP binding. Transgenic plants expressing the mutant oat phyA impaired in the kinase activity show a hyposensitive phenotype compared with the wild type under far-red light. The light-induced phosphorylation and degradation of PIF3 is reduced significantly in the kinase mutants compared with control plants in vivo, which is consistent with the defects in ATP-binding activity and mutant phenotypes. These data strongly support that plant phytochrome functions as a kinase for PIF3 and possibly other PIFs. However, several questions still remain unanswered. First, PIF3 phosphorylation is light inducible in vivo. However, the in vitro data presented in this study did not show any light-induced phosphorylation of PIF3 by phytochromes, suggesting that other factors might be necessary in vitro to fully capture the light-regulated phosphorylation events. Second, PIF3 is phosphorylated at multiple sites in response to light and displayed a characteristic band shift in vivo (Ni et al., 2013). However, the in vitro data presented in this study did not show any band shift, suggesting that phytochrome alone is not sufficient to mediate multiple phosphorylations of PIF3. Third, an independent study from another laboratory failed to reproduce the Arabidopsis phyB kinase activity toward PIF3 (Ni et al., 2017), arguing that Arabidopsis phytochromes may not function as protein kinases. However, these differences might be due to the use of different buffers and/or preparation of the phytochrome molecule used for the kinase assays. Thus, additional experiments are necessary to establish whether Arabidopsis phytochromes indeed function as bona fide protein kinases.
Recently, four members of a protein kinase family called PPK (PPK1–PPK4) were shown to phosphorylate PIF3 in vitro (Ni et al., 2017). The PPKs display sequence similarity to CK1 and have been described previously as MUT9-Like Ser/Thr Kinases (MLKs; Wang et al., 2015; Huang et al., 2016). The addition of phyB enhances the phosphorylation of PIF3 by PPK in vitro; however, this enhancement is light independent, suggesting that both the Pr and Pfr forms of phyB enhance the phosphorylation of PIF3 by PPK. The light-induced phosphorylation and degradation of PIF3 is defective in the ppk mutants compared with the wild type, suggesting that PPKs function as a protein kinase for PIF3 both in vitro and in vivo. Although phyB is not phosphorylated directly by PPK, PPKs also induce the codegradation of PIF3 and phyB in vivo. Hence, both PIF3 and phyB are more abundant in the ppk mutant under continuous red light compared with the wild type. Consistently, ppk mutants display a hypersensitive phenotype in response to red light. This phenotype is not consistent with PIF3 degradation; rather, it is consistent with the higher level of phyB due to the reduced phyB degradation in these mutants. Although this study provides strong evidence that PPKs function as protein kinases for PIF3, PPKs do not appear to function as the primary light-regulated kinase that phosphorylates PIFs in response to light for several reasons. The biologically active Pfr form of phyB does not enhance the kinase activity of PPKs toward PIF3 (Ni et al., 2017). PPKs appear to function as a general kinase for many pathways. A companion study shows that PPKs also phosphorylate CRY2 in response to blue light (Liu et al., 2017). In addition, PPKs/MLKs function as a kinase for histone and are involved in the epigenetic regulation of gene expression (Wang et al., 2015; Su et al., 2017). Phosphorylation of Ser-95 of H2A is necessary for the regulation of FT expression and flowering time under long days. Thus, PPKs might be a general kinase functioning in many pathways. This notion is further supported by the lethality of the ppk1234 mutant, as these authors failed to isolate the quadruple mutant (Liu et al., 2017; Ni et al., 2017).
Two other kinases, CK2 and BIN2, have been shown to phosphorylate PIFs. CK2 phosphorylates PIF1 (Bu et al., 2011b), while BIN2 phosphorylates PIF3 and PIF4 (Ling et al., 2017; Bernardo-García et al., 2014). However, these kinases are not necessary for the light-induced phosphorylation of PIFs in vivo. Thus, the identification and characterization of the light-regulated protein kinase that phosphorylates PIFs await further study.
E3 Ligases for PIFs
As discussed above, PIFs are degraded through the 26S proteasome pathway in response to light. In Arabidopsis, there are three main classes of CULLIN (CUL) RING UBIQUITIN LIGASEs (CRLs) that are involved in the ubiquitination of substrates (Vierstra, 2009): CUL1, CUL3, and CUL4. Although the substrate specificity components for each of these CRLs are different, all three CRLs have been shown to be involved in the light-induced degradation of PIFs (Fig. 3B)
Genetic analysis shows that cul1-6 is hyposensitive to red light (Moon et al., 2007). However, the substrate for CUL1-based E3 ligase for regulating photomorphogenesis is not known. A recent study shows that PIF3 is ubiquitinated and degraded by the SCFEBF1/EBF2 ubiquitin ligase (Dong et al., 2017). In this case, two previously described F-box proteins called EBF1 and EBF2 act as substrate recognition components (Guo and Ecker, 2003; Potuschak et al., 2003). Consistently, the ebf1ebf2 double mutant displayed long hypocotyls under red light, similar to those of cul1-6. This study also provides evidence of a novel mechanism of SCF formation. In general, a substrate is phosphorylated by a stimulus, and the phosphorylated substrate is then recognized by the E3 ubiquitin ligase for the ubiquitination followed by degradation (Pickart, 2001). However, these authors show that PIF3 is constitutively bound to EBF1 and EBF2 in the dark, but PIF3 is not ubiquitinated by EBF1 and EBF2 in darkness. Light-induced phosphorylation of PIF3 triggers the formation of the SCFEBF1/EBF2 complex, resulting in the ubiquitination and subsequent degradation of PIF3. This study also shows that EBF1 and EBF2 induce the degradation of PIF3 under a broad range of red light conditions compared with the previously described CUL3LRB E3 ubiquitin ligase complex, which degrades PIF3 under strong red light conditions (Ni et al., 2014). However, PIF3 is still degraded in the ein3eil1ebf1ebf2 mutant background compared with the ein3eil1 control, suggesting that additional E3 ubiquitin ligases are involved in the light-induced degradation of PIF3.
As discussed above, CUL3-based E3 ubiquitin ligase plays an important role in PIF3 degradation in response to light (Ni et al., 2014). In this case, LIGHT-RESPONSE Bric-a-Brack/Tramtrack/Broad (LRB1, LRB2, and LRB3) act as substrate recognition components in the CUL3LRB complex. However, LRBs not only recruit PIF3 but also phyB in response to light signal and promote the ubiquitination and subsequent codegradation of both substrates in vivo by the 26S proteasome pathway. As a primary signaling partner acting downstream of phyB, a defect in PIF3 degradation is expected to result in a hyposensitive phenotype under red light. However, the lrb123 triple mutant displays a hypersensitive phenotype under continuous red light, consistent with the higher abundance of phyB in the triple mutant compared with the wild type. In addition, both SCFEBF1/2 and CUL3LRB display a modest effect on PIF3 degradation, suggesting that additional E3 ubiquitin ligases still might be degrading PIF3 in response to light.
Very recently, a CUL3-based E3 ubiquitin ligase was shown to promote the ubiquitination and degradation of PIF4 (Zhang et al., 2017). In this case, two BLADE ON PETIOLE (BOP1/2) proteins among six members of the BTB-ankyrin protein family act as a substrate recognition component for PIF4. The ubiquitination and degradation of PIF4 are reduced in the bop1bop2 mutant background compared with the wild type not only under red light but also under elevated temperature, suggesting that BOP proteins control PIF4 protein abundance under red light as well as under elevated temperature-mediated growth responses. Consistent with an elevated level of PIF4, the bop mutant displays a long hypocotyl under red light as well as at high ambient temperature. Interestingly, the phosphorylation of PIF4 is not required for the binding between PIF4 and BOP. This is different from PIF3, where phosphorylation is absolutely necessary for the PIF3-LRB interaction or SCFEBF1/2-PIF3 complex formation (Ni et al., 2014; Dong et al., 2017). Furthermore, a weak level of PIF4 ubiquitination is observed in the dark, which is strongly enhanced under light conditions in vivo (Zhang et al., 2017). The ubiquitination level of PIF4 is much reduced in the bop1/2 mutant both in the dark and light conditions, suggesting that BOP proteins are necessary for PIF4 ubiquitination. Although the authors concluded that the light-induced phosphorylation of PIF4 might not be necessary for the degradation of PIF4 via BOP1/2, the role of light in mediating the interaction between BOP1/2 and PIF4 was not examined extensively in this study. The authors used a transient protoplast system and a bimolecular fluorescence complementation assay as in vivo interaction assays without any dark and light controls, which may not recapture the true dark/light conditions for seedlings. Further experiments are necessary to examine if the light-induced phosphorylation of PIF4 is necessary for enhanced binding between PIF4 and BOP1/2.
CUL4-based E3 ubiquitin ligase has been shown to promote the light-induced ubiquitination and degradation of PIF1 (Zhu et al., 2015). In this case, COP1 and SPA proteins act as substrate adaptor components in recruiting the phosphorylated form of PIF1 for the light-induced ubiquitination and subsequent degradation. Both cop1-4 and spaQ display strong hyposensitive phenotypes in seed germination in response to red and far-red light, consistent with the PIF1 protein abundance at the seed and seedling stages. This study highlights a novel function of the COP1/SPA complex from a well-established negative regulator to a crucial positive regulator complex during the early transition from dark to light. It will be interesting to examine whether other PIFs also are regulated by the CUL4-based E3 ligase complex. In addition, phytochromes inhibit COP1-SPA function to promote photomorphogenesis (Lu et al., 2015; Sheerin et al., 2015; Xu et al., 2015). However, in this case, the COP1-SPA complex functions in cooperation with phytochromes to promote the degradation of PIFs. Further studies are necessary to understand the precise timing when phytochromes and the COP1-SPA complex act together and when phytochromes inhibit COP1-SPA function to promote photomorphogenesis in response to light.
From the discussion above, one wonders why there are so many kinases and E3 ubiquitin ligases regulating PIF abundance. Several speculations might help answer this question. First, the kinetics and the effects of different light quality and quantity on PIF degradation are different for different PIFs. Thus, having multiple kinases and E3 ubiquitin ligases might allow fine-tuning the rate of degradation and the sensitivity to different fluences. Second, PIFs are not involved only in light signaling pathways, but recent studies highlight the importance of PIFs in multiple aspects of plant growth and development and response pathways (Leivar and Monte, 2014; Paik et al., 2017). Thus, PIFs act as a central hub in many cellular signaling pathways (e.g. GA, brassinosteroid [BR], auxin, ethylene, JA, circadian, and many others; Leivar and Monte, 2014; Paik et al., 2017). Therefore, signaling factors from these pathways (e.g. DELLA, BIN2, EBF1/2, and possibly others) intersect with photomorphogenesis by regulating PIF levels.
POSTTRANSLATIONAL REGULATION OF PIF ACTIVITY IN THE DARK AND LIGHT
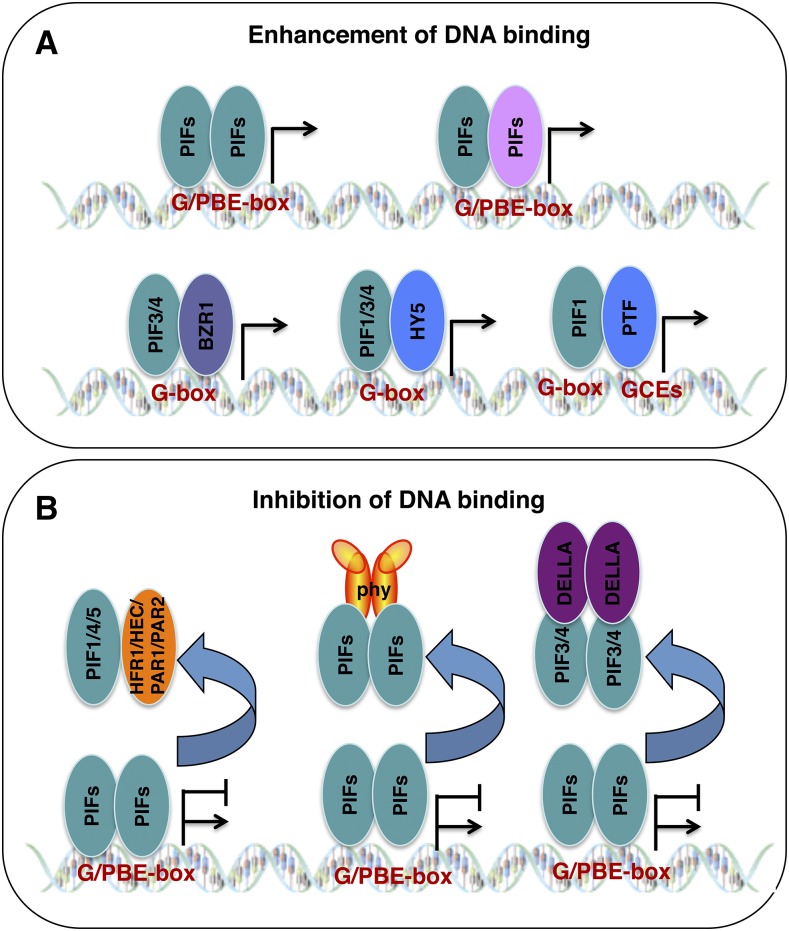
Being members of the bHLH superfamily, PIFs are predicted to bind to DNA. In vitro gel-shift, random DNA-binding selection, and in vivo chromatin immunoprecipitation (ChIP) assays show that four major PIFs (PIF1, PIF3, PIF4, and PIF5) bound to either the G-box (CACGTG) and/or the E box (CANNTG), which are called PIF-binding E boxes (Martínez-Garcia et al., 2000; Huq and Quail, 2002; Huq et al., 2004; Hornitschek et al., 2012). A number of proteins that interact directly with PIFs modulate the DNA binding and, consequently, the transcriptional activity of PIFs (Fig. 4; Paik et al., 2017). These are divided into two classes: factors enhancing PIF DNA-binding activity (Fig. 4A) and factors inhibiting PIF DNA-binding activity (Fig. 4B). This topic has been reviewed in recent years (Leivar and Monte, 2014). However, there has been new progress in this area, which will be emphasized in the discussion below.
Figure 4.
Modulation of the DNA-binding and transcriptional activities of PIFs. PIF-interacting proteins control the transcriptional activities of PIFs either by activating or inhibiting PIF’s capacity to bind directly to DNA. A, PIFs can bind to the G/PBE-box as a homodimer and/or a heterodimer with other PIFs. PIF interactions with other proteins, including BZR1 and HY5, regulate the DNA-binding and transcriptional activities of PIFs. In addition, PIF-interacting transcription factors (PTFs; e.g. group A bZIP proteins) modulate the DNA-binding and transcriptional activities of PIFs by binding to nearby G-box coupling elements (GCEs). B, Various factors inhibit the DNA-binding and subsequent transcriptional activities of PIFs either by forming heterodimers (e.g. HFR1/HEC2/PAR1/PAR2) or by direct physical interactions with PIFs followed by sequestration (e.g. phytochromes and DELLA proteins).
Factors Enhancing the DNA-Binding Ability of PIFs
One of the hallmark features of the bHLH family of proteins is that they bind to DNA as both homodimers and heterodimers. In fact, the crystal structure of MyoD shows that the basic domain binds to the DNA, while the HLH domain forms homodimers and heterodimers (Ma et al., 1994; Littlewood and Evans, 1998). PIFs also form homodimers and heterodimers, and these dimeric forms also bind to the DNA in vitro (Toledo-Ortiz et al., 2003; Bu et al., 2011a).
PIFs also interact with other transcription factors, and these interactions modulate the PIF DNA-binding activity. For example, PIF3 and PIF4 interact with BZR1 and bind to the same G-box DNA sequence element to regulate genes involved in the light and BR pathways (Oh et al., 2012; Zhang et al., 2014). PIF1 and PIF3 interact with HY5, a bZIP protein, and bind to DNA as the PIF1/PIF3-HY5 complex (Chen et al., 2013). In this case, HY5 promotes the DNA-binding ability of PIF1 and PIF3 and regulates genes involved in anthocyanin biosynthesis and reactive oxygen response pathways. Conversely, HY5 also functions antagonistically by binding to the same binding sites targeted by PIF1/3 and inhibits PIF1/3 function in regulating chlorophyll and flavonoid biosynthetic pathways (Toledo-Ortiz et al., 2014).
Interestingly, a recent study performed ChIP followed by microarray (ChIP-chip) to identify in vivo binding sites of PIF1 (Kim et al., 2016b). This study showed that PIF1 DNA-binding sites (PBSs) are enriched with not only G-boxes but also with other hexameric sequence elements named GCEs. This study showed that ∼59% of the PBSs contain at least one G-box element and 41% lack G-box elements. They proposed that PIF1 binding to PBSs might be stabilized in association with other PTFs that bind to other nearby PIF1-binding sites called GCEs. These authors identified group A bZIP transcription factors as PTFs and showed that ABI5, a member of the group A bZIP family, binds to the ACGT-containing GCE sequences. ABI5 and other group A bZIP transcription factors interact with PIF1 and, consequently, facilitate PIF1-binding activity to the DNA target gene promoters. Although these authors demonstrated the enhancement of the DNA-binding ability of PIF1 by PTFs, no biological significance of this enhanced DNA-binding activity was presented in their study. Further experiments are necessary to understand whether PTFs and PIF1 coordinately regulate any biological processes.
Factors Inhibiting the DNA-Binding Ability of PIFs
Three groups of factors inhibit the DNA-binding ability of PIFs: the well-known HLH classes of transcription factors, phyB, and DELLA proteins.
The first class encodes HLH proteins that include HFR1, PAR1, PAR2, and HEC proteins. As HLH proteins, they do not bind to DNA directly by themselves; however, they sequester PIFs by heterodimerizing with them (Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013; Zhu et al., 2016b). HFR1 and HEC1/HEC2 interact with PIF1 and inhibit seed germination in response to red and far-red light (Shi et al., 2013; Zhu et al., 2016b). HFR1, PAR1, and PAR2 interact with PIF4/PIF5 and inhibit shade avoidance responses (Lorrain et al., 2008; Hao et al., 2012).
Second, the light-activated phytochromes can interact physically with PIFs and not only induce their degradation but also sequester them from binding to DNA. It has been shown that phyB binds to PIF1/PIF3 and inhibits PIF1/PIF3-binding activity to their target promoters using in vivo ChIP assays (Park et al., 2012). Thus, phytochromes inhibit PIF function using two distinct mechanisms (i.e. light-induced degradation and light-induced sequestration).
The third class of PIF inhibitor is DELLA proteins, which interact with PIF3 and PIF4 and block the DNA-binding ability of these PIFs (de Lucas et al., 2008; Feng et al., 2008). Strikingly, a recent study showed that DELLA proteins not only sequester PIFs but also induce their degradation to regulate target gene expression (Li et al., 2016). Thus, DELLA proteins and phytochromes regulate PIFs in a similar fashion. However, the distinction is that phytochromes inhibit PIF activities in response to light, while DELLA proteins inhibit PIF activities irrespective of any light signal. The DELLA-mediated degradation and sequestration of PIFs results in optimum regulation of hypocotyl elongation in response to both light and GA signals.
CONCLUSIONS AND FUTURE PERSPECTIVES
Although PIFs are known to be degraded only under light, recent results show that PIFs also are actively degraded in darkness. Thus, a homeostasis of PIF level is necessary for the proper transition from skotomorphogenesis to photomorphogenesis. This is also important for the coordination with other factors that interact with PIFs to regulate similar processes (e.g. cell elongation). Collectively, all four major PIFs function to regulate the transition from skotomorphogenesis to photomorphogenesis. However, genetic analyses of the factors degrading PIFs also shed light on the distinct signaling roles of individual PIFs. For example, PPKs and CUL3LRBs induce the codegradation of PIF3 and phyB in response to light (Ni et al., 2014, 2017). PIF3 is more abundant in the ppk and lrb mutants compared with the wild type under red light. However, the phenotypes of the ppk and lrb mutants are not consistent with the PIF3 abundance; rather, they correlate with the phyB levels in these mutant backgrounds. Thus, it appears that the major function of PIF3 is to attenuate phyB signaling by inducing phyB degradation under light. Although light-regulated gene expression is defective in the pif3 mutant at an early time, pif3 displays a phyB overexpression phenotype under continuous red light (Kim et al., 2003; Monte et al., 2004). This conclusion also is supported by the fact that the PIF3 gene encoding a mutant PIF3 protein defective in DNA binding still rescues the pif3 mutant phenotype under continuous red light (Al-Sady et al., 2008). In contrast, the light-induced degradation of PIF1 is defective in the cop1 and spaQ mutants (Zhu et al., 2015). Both mutants display reduced seed germination in response to light, consistent with the higher abundance of PIF1 in these mutants compared with the wild type. Despite similar biochemical properties of PIF1 and PIF3, PIF3 appears to be more dedicated to signal attenuation, while PIF1 is dedicated to signal transduction. This distinction is not clear for the other PIFs based on the available data. In summary, PIFs took center stage in the last two decades, since their initial discovery in 1998 (Ni et al., 1998); much remains to be learned (see Outstanding Questions box). Further studies are necessary to answer these and many other questions.
Acknowledgments
We thank Dr. Inyup Paik for critical reading of this article. Due to space constraints, we apologize that many recent articles from other colleagues could not be discussed.
Footnotes
We acknowledge support by grants from the National Institutes of Health (NIH) (1R01 GM-114297), National Science Foundation (MCB-1543813), and U.S.-Israel Binational Science Foundation (BSF#2015316) to E.H.
Articles can be viewed without a subscription.
References
- Al-Sady B, Kikis EA, Monte E, Quail PH (2008) Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S (2014) BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev 28: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Castillon A, Chen F, Zhu L, Huq E (2011a) Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol Biol 77: 501–511 [DOI] [PubMed] [Google Scholar]
- Bu Q, Zhu L, Yu L, Dennis M, Lu X, Person M, Tobin E, Browning K, Huq E (2011b) Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biol Chem 286: 12066–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM (2009) Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol 19: 229–234 [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro AM, Figueiredo DD, Tepperman J, Borba AR, Lourenço T, Abreu IA, Ouwerkerk PB, Quail PH, Margarida Oliveira M, Saibo NJ (2016) Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim Biophys Acta 1859: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dong J, Ni W, Yu R, Deng XW, Chen H, Wei N (2017) Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr Biol 27: 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H (2014) Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T (2004) Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hao Y, Oh E, Choi G, Liang Z, Wang ZY (2012) Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant 5: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA (2016) Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2005) Phytochrome signaling. In Briggs WR, Spudich JL, eds, Handbook of Photosensory Receptors. Wiley-VCH, Weinheim, Germany, pp 151–170 [Google Scholar]
- Inoue K, Nishihama R, Kataoka H, Hosaka M, Manabe R, Nomoto M, Tada Y, Ishizaki K, Kohchi T (2016) Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28: 1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaedicke K, Lichtenthäler AL, Meyberg R, Zeidler M, Hughes J (2012) A phytochrome-phototropin light signaling complex at the plasma membrane. Proc Natl Acad Sci USA 109: 12231–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Choi G (2013) Phytochrome-interacting factors have both shared and distinct biological roles. Mol Cells 35: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci USA 114: 6695–6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang H, Park J, Kim W, Yoo J, Lee N, Kim J, Yoon TY, Choi G (2016a) PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28: 1388–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Song K, Park E, Kim K, Bae G, Choi G (2016b) Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell 28: 2770–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jeong J, Kim J, Lee N, Kim ME, Lee S, Chang Kim S, Choi G (2016c) PIF1 regulates plastid development by repressing photosynthetic genes in the endodermis. Mol Plant 9: 1415–1427 [DOI] [PubMed] [Google Scholar]
- Klose C, Viczián A, Kircher S, Schäfer E, Nagy F (2015) Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol 206: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar I, Swaminathan K, Hudson K, Hudson ME (2016) Evolutionary divergence of phytochrome protein function in Zea mays PIF3 signaling. J Exp Bot 67: 4231–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jung JH, Cortés Llorca L, Kim SG, Lee S, Baldwin IT, Park CM (2014) FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat Commun 5: 5473. [DOI] [PubMed] [Google Scholar]
- Lee N, Choi G (2017) Phytochrome-interacting factor from Arabidopsis to liverwort. Curr Opin Plant Biol 35: 54–60 [DOI] [PubMed] [Google Scholar]
- Lee N, Park J, Kim K, Choi G (2015) The transcriptional coregulator LEUNIG_HOMOLOG inhibits light-dependent seed germination in Arabidopsis. Plant Cell 27: 2301–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Nieto C, Sellaro R, Prat S, Casal JJ (2017) Perception and signalling of light and temperature cues in plants. Plant J 90: 683–697 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461: 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S (2015) Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat Commun 6: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW (2016) DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012a) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peng WF, Liu QQ, Zhou JJ, Liang WH, Xie XZ (2012b) Expression patterns of OsPIL11, a phytochrome-interacting factor in rice, and preliminary analysis of its roles in light signal transduction. Rice Sci 19: 263–268 [Google Scholar]
- Ling JJ, Li J, Zhu D, Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci USA 114: 3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T, Evans GI (1998) Helix-Loop-Helix Transcription Factors, Ed 3 Oxford University Press, New York [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. (2017) Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat Commun 8: 15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen CY, Wang KC, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, et al. (2013) PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang Y, Wang J, Li P, Zhao C, Chen Y, Bi Y (2015) Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci 238: 64–72 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ (2014) COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PCM, Rould MA, Weintraub H, Pabo CO (1994) Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20: 666–671 [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhao Y, Dai X, Zhang W, Gray WM, Huq E, Estelle M (2007) A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol 143: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T (2007) Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci Biotechnol Biochem 71: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH (2013) Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, Quail PH (2017) PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat Commun 8: 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, Huq E (2017) Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G (2012) Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci USA 109: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Halliday KJ (2010) A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol 73: 89–95 [DOI] [PubMed] [Google Scholar]
- Pickart CM. (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Possart A, Hiltbrunner A (2013) An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell 25: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A, Xu T, Paik I, Hanke S, Keim S, Hermann HM, Wolf L, Hiß M, Becker C, Huq E, et al. (2017) Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 29: 310–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Li M, Pasoreck EK, Long L, Shi Y, Galvão RM, Chou CL, Wang H, Sun AY, Zhang YC, et al. (2015) HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27: 1409–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH (2002) A light-switchable gene promoter system. Nat Biotechnol 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Sheehan MJ, Farmer PR, Brutnell TP (2004) Structure and expression of maize phytochrome family homeologs. Genetics 167: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW (2013) HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin AY, Han YJ, Baek A, Ahn T, Kim SY, Nguyen TS, Son M, Lee KW, Shen Y, Song PS, et al. (2016) Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun 7: 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, Huq E (2017) PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol 215: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E (2016) Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci USA 113: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Wang S, Zhang F, Zheng H, Liu Y, Huang T, Ding Y (2017) Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell 29: 2197–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, et al. (2012) Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci USA 109: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD (2011a) Light control of plasma membrane recruitment using the Phy-PIF system. Methods Enzymol 497: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD (2011b) Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 8: 837–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155: 1422–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Decker PV, Chen M (2012) Photobodies in light signaling. Plant Physiol 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang Z, Casas-Mollano JA, Xu J, Riethoven JJM, Zhang C, Cerutti H (2015) Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kathare PK, Pham VN, Bu Q, Nguyen A, Huq E (2017) Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in Arabidopsis. Development 144: 1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E (2015) Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci 20: 641–650 [DOI] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Holmlund M, Lorrain S, Norberg M, Bakó L, Fankhauser C, Nilsson O (2017) BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 6: e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R (2014) The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Chen Y, He JX, Bi Y (2015) PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci 237: 57–68 [DOI] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu Q, Zhang F, Wang Y, Zhang S, Cheng H, Yan L, Li L, Chen F, Xie X (2014) Overexpression of OsPIL15, a phytochrome-interacting factor-like protein gene, represses etiolated seedling growth in rice. J Integr Plant Biol 56: 373–387 [DOI] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY (2016a) TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Bu Q, Xu X, Paik I, Huang X, Hoecker U, Deng XW, Huq E (2015) CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat Commun 6: 7245. [DOI] [PubMed] [Google Scholar]
- Zhu L, Xin R, Bu Q, Shen H, Dang J, Huq E (2016b) A negative feedback loop between PHYTOCHROME INTERACTING FACTORs and HECATE proteins fine tunes photomorphogenesis in Arabidopsis. Plant Cell 28: 855–874 [DOI] [PMC free article] [PubMed] [Google Scholar]