DNA methylation differences between isogenic parental lines can directly or indirectly trigger heterosis in Arabidopsis hybrids.
Abstract
Despite the importance and wide exploitation of heterosis in commercial crop breeding, the molecular mechanisms behind this phenomenon are not completely understood. Recent studies have implicated changes in DNA methylation and small RNAs in hybrid performance; however, it remains unclear whether epigenetic changes are a cause or a consequence of heterosis. Here, we analyze a large panel of over 500 Arabidopsis (Arabidopsis thaliana) epigenetic hybrid plants (epiHybrids), which we derived from near-isogenic but epigenetically divergent parents. This proof-of-principle experimental system allowed us to quantify the contribution of parental methylation differences to heterosis. We measured traits such as leaf area, growth rate, flowering time, main stem branching, rosette branching, and final plant height and observed several strong positive and negative heterotic phenotypes among the epiHybrids. Using an epigenetic quantitative trait locus mapping approach, we were able to identify specific differentially methylated regions in the parental genomes that are associated with hybrid performance. Sequencing of methylomes, transcriptomes, and genomes of selected parent-epiHybrid combinations further showed that these parental differentially methylated regions most likely mediate the remodeling of methylation and transcriptional states at specific loci in the hybrids. Taken together, our data suggest that locus-specific epigenetic divergence between the parental lines can directly or indirectly trigger heterosis in Arabidopsis hybrids independent of genetic changes. These results add to a growing body of evidence that points to epigenetic factors as one of the key determinants of hybrid performance.
Heterosis describes an F1 hybrid phenotype that is superior compared with the phenotype of its parents. The phenomenon has been exploited extensively in agricultural breeding for decades and has improved crop performance tremendously (Chen, 2010; Schnable and Springer, 2013). Despite its commercial impact, knowledge of the molecular basis underlying heterosis remains incomplete. Most studies have focused on finding genetic explanations, resulting in the classical dominance (Jones, 1917; Crow, 1998; Schnable and Springer, 2013) and overdominance (Crow, 1948, 1998) models of heterosis. In line with genetic explanations, interspecies hybrids have been observed frequently to show a higher degree of heterosis than intraspecies hybrids, indicating that genetic distance correlates with the extent of heterosis (East, 1936; Chen, 2010).
Genetic explanations, however, do not sufficiently explain or predict heterosis. There is growing evidence that epigenetic factors also play a role in heterosis (Groszmann et al., 2013; Springer, 2013; Dapp et al., 2015; Zhu et al., 2017). For example, it has been shown that altered epigenetic profiles at genes regulating circadian rhythm play an important role in heterotic Arabidopsis (Arabidopsis thaliana) hybrids (Ni et al., 2009). Moreover, heterotic hybrids of Arabidopsis, maize (Zea mays), and tomato (Solanum lycopersicum) are shown to differ in levels of small regulatory RNAs and/or DNA methylation (5mC) relative to their parental lines (Groszmann et al., 2011; Barber et al., 2012; Shen et al., 2012; Shivaprasad et al., 2012; Zhang et al., 2016b).
Remodeling of the methylome during hybridization has been proposed to be involved in heterosis in genetic hybrids and was implicated in the formation of novel epialleles in a met1-derived epigenetic hybrid plant (epiHybrid; Groszmann et al., 2011; Greaves et al., 2012; Shen et al., 2012; Rigal et al., 2016). Processes such as the transfer of 5mC between alleles (trans-chromosomal methylation [TCM]) or the loss of 5mC at one of the alleles (trans-chromosomal demethylation [TCdM]) have been indicated to contribute to the observed remodeling of the epigenome (Greaves et al., 2012; Shivaprasad et al., 2012; Groszmann et al., 2013). These trans-chromosomal (de)methylation [TC(d)M] events occur between homologous sequences and have been shown to require the RNA-directed DNA methylation (RdDM) pathway, which involves small regulatory RNAs (Zhang et al., 2016b). Strikingly, some of these changes in 5mC levels have been shown to be stable over multiple generations (Greaves et al., 2012, 2014). However, parental lines used for these studies differed in both their genetic and epigenetic profiles, making it challenging to disentangle genetic from epigenetic effects.
It has been shown in particular isogenic epigenetic recombinant inbred lines (epiRILs) that heritable morphological variation in Arabidopsis plants can be caused exclusively by epigenetic factors (Johannes et al., 2009; Roux et al., 2011; Cortijo et al., 2014; Kooke and Keurentjes, 2015). To specifically address the contribution of parental epigenetic variation to F1 heterosis, we made use of the same epiRILs (Johannes et al., 2009; Reinders et al., 2009) to generate F1 epigenetic hybrids, hereafter called epiHybrids (Dapp et al., 2015). EpiRILs are near isogenic but display mosaic patterns in terms of their epigenomes. The two Arabidopsis epiRIL populations reported have been created by crossing the wild-type Columbia accession (Col-wt) with Col-wt lines carrying a mutation in either METHYLTRANSFERASE1 (MET1-3) or DECREASE IN DNA METHYLATION1 (DDM1-2; Johannes et al., 2009; Reinders et al., 2009). MET1 maintains DNA methylation at cytosines in a CG sequence context (mCG) but also affects non-CG methylation (Finnegan et al., 1996; Mathieu et al., 2007; Stroud et al., 2013). Loss of MET1 causes an almost complete elimination of mCG genome wide, which is associated with misregulated gene expression and transcriptional activation of transposable elements (TEs; Zhang et al., 2006; Cokus et al., 2008; Lister et al., 2008). DDM1 is a nucleosome remodeler, and the ddm1-2 mutation leads to an ∼70% reduction in DNA methylation (Kakutani et al., 1995), predominantly affecting mCG and mCHG (where H represents A, C, or T) and, to a lesser extent, mCHH (Zemach et al., 2013), in primarily long transposable elements (Zemach et al., 2013). Loss of DDM1 also affects genic loci where it reduces mCG and causes CHG hypermethylation in gene bodies (Zemach et al., 2013).
Recently, epiHybrids have been generated by crossing a met1-derived epiRIL with Col-wt, and heterosis for biomass was reported for one of the epiHybrids (Dapp et al., 2015). The effect was observed only with the epiRIL as maternal parent, suggesting a strong maternal effect (Dapp et al., 2015). In this study, we created epiHybrids by crossing Col-wt as a maternal parent to 19 different near-isogenic but epigenetically divergent ddm1-derived epiRILs, allowing the assessment of epigenetic variation in identical maternal backgrounds. Using high-throughput phenotyping, we observed various positive and negative heterotic effects in one or more of the six traits monitored among the 19 epiHybrids, indicating that epigenetic divergence among parents has a direct or indirect role in triggering heterosis. Furthermore, in contrast to previous studies, our experimental design allowed us to employ an epigenetic quantitative trait locus (QTL) mapping approach that, in combination with methylome and transcriptome profiling, allowed quantifying and characterizing the contribution of parental methylation differences to heterosis. Using this approach, we were able to identify specific differentially methylated regions (DMRs) in the parental genomes that are associated with heterotic phenotypes in the epiHybrids. We provide evidence that these parental DMRs mediate local methylome and transcriptome remodeling at putatively causative genes, thus providing molecular mechanisms underlying the observed heterotic phenotypes in the epiHybrids.
RESULTS
Construction of epiHybrids
Hybrids are usually generated from parental lines that vary at both the genomic and epigenomic levels, and disentangling those two sources of variation is challenging. To overcome this limitation, we generated epigenetic Arabidopsis F1 hybrids (epiHybrids) from near-isogenic but epigenetically divergent parental lines by crossing Col-wt as the maternal parent to near-isogenic ddm1-2-derived epiRILs (Johannes et al., 2009) as the paternal parent (Fig. 1A). EpiRILs carry chromosomes that are a mosaic of Col-wt and hypomethylated ddm1-2-derived genomic regions (Johannes et al., 2009; Colomé-Tatché et al., 2012; Cortijo et al., 2014; Fig. 1A). Nineteen epiRIL parental lines were selected that sample a broad range of 5mC divergence from the Col-wt reference methylome (Fig. 1B; Supplemental Table S1). Lines were chosen that have a wild-type methylation profile at FLOWERING LOCUS WAGENINGEN (FWA; Supplemental Fig. S1; Supplemental Table S1), as loss of DNA methylation at the FWA locus is known to affect flowering time (FT) and, as such, many related developmental traits (Soppe et al., 2000). Furthermore, we selected epiRILs covering a wide range of phenotypic variation in FT and root length, two traits that have been monitored previously (Supplemental Table S1; Johannes et al., 2009).
Figure 1.
Heterosis occurs in epiHybrids. A, Experimental setup. Lines are depicted schematically as one chromosome with the numbers indicating the epiRIL identifier (e.g. 371 and 492) and the respective epiHybrid (e.g. 371H and 492H). B, Genome-wide 5mC levels (y axis) of the Col-wt line in green and the epiRIL parental lines in salmon. Numbers indicate the epiRIL identifiers. The 5mC levels were calculated as the proportion of methylated DNA immunoprecipitation probes with respect to the total amount of probes. C, Col-wt, epiHybrid 232H, and epiRIL 232 at 13 DAS as an example for high-parent heterosis. D to F, Three classes of phenotypic effects monitored in the epiHybrids. The black dashed lines indicate the MPV. The green and salmon peaks indicate the mean performance of the parental lines. The white dashed lines indicate the mean performance of the epiHybrids. G, Phenotypic effects in six traits monitored across the 19 epiHybrids. The columns at the right summarize positive and negative heterotic effects per trait. H to J, Examples of epiHybrids exhibiting high-parent heterosis in LA and HT (H and I) and low-parent heterosis in FT (J) Error bars represent ±1 se. Deviation from high parent or low parent is shown in percentage.
Heterotic Phenotypes Occur in the epiHybrids
The phenotypic performance of the 19 epiHybrid lines and their 20 parental lines was assessed. In total, we monitored about 1,090 plants (∼28 replicate plants per epiHybrid or parental line) for a range of quantitative traits: leaf area (LA), growth rate (GR), FT, main stem branching (MSB), rosette branching (RB), plant height (HT), and seed yield (SY; Supplemental Tables S2–S7). The hybrids and parental lines were grown in parallel in a climate-controlled chamber with automated watering. The plants were randomized throughout the chamber to level out phenotypic effects caused by plant position. Leaf area was measured up to 14 d after sowing (DAS) using an automated camera system (Fig. 1C), and GR was determined based on these data (Supplemental Methods S1). Flowering time was scored manually as the day of opening of the first flower. After all plants started flowering, they were transferred to the greenhouse and grown to maturity. MSB, RB, and HT were scored manually after harvesting of the plants. The phenotypic observations for SY were inconsistent in a replication experiment; therefore, those data sets were excluded from further analysis.
The extent of heterosis was evaluated by comparing the phenotypic performance of the hybrids with that of their parental lines. We distinguished five effects: additivity, positive midparent heterosis, negative midparent heterosis, high-parent heterosis, and low-parent heterosis (Fig. 1, D–F). Briefly, an additive effect is defined by a hybrid mean phenotype that is equal or close to the average phenotype of the two parents (the midparent value [MPV]). Midparent heterosis, by contrast, refers to positive or negative deviations of the hybrid mean phenotype from the MPV. High-parent heterosis and low-parent heterosis are important special cases of midparent heterosis in which the hybrid mean phenotype either exceeds the mean phenotype of the high parent or falls below that of the lowest performing parent. In crop breeding, the focus is usually on obtaining high-parent heterosis and low-parent heterosis, as these present novel phenotypes that are outside the parental range. Depending on the trait and commercial application, either high-parent heterosis or low-parent heterosis can be considered superior. For instance, early flowering may be preferable over late flowering; in such cases, maximizing low-parent heterosis may be desirable. For other traits, such as yield or biomass, it is more important to maximize high-parent heterosis. However, in order to obtain a comprehensive view of hybrid performance, it is informative to also monitor midparent heterosis, as many traits of mature plants are affected by other traits that may not display fully penetrant heterotic effects.
We observed a remarkably wide range of heterotic phenotypes among the epiHybrids (Fig. 1G; Supplemental Tables S2–S7). The magnitude of these phenotypic effects was substantial (Fig. 1, H–J; Supplemental Fig. S2; Supplemental Tables S8–S19) and similar to that typically seen in hybrids of Arabidopsis natural accessions (Groszmann et al., 2014; Wang et al., 2015). Many epiHybrids (16 of 19) exhibited significant midparent heterosis in at least one of the six monitored traits (false discovery rate = 0.05; Fig. 1G). Across all epiHybrids and traits, we identified 30 cases of positive midparent heterosis and negative midparent heterosis. Among those, four cases show low-parent heterosis and nine cases show high-parent heterosis (Fig. 1G). Interestingly, in 11 out of the 17 remaining cases of midparent heterosis, the phenotypic means of the epiHybrids were in the direction of the phenotypic means of the epiRIL parent rather than in the direction of the Col-wt parent (Fig. 1G; Supplemental Tables S2–S7, F1 trend). Also, all four low-parent heterosis and two of the high-parent heterosis cases were in the direction of the epiRIL parent (Fig. 1, G, I, and J; Supplemental Fig. S2). This observation illustrates that ddm1-2-derived hypomethylated epialleles are often (partially) dominant over wild-type epialleles.
We observed cases of high-parent heterosis for LA, HT, and MSB and cases of low-parent heterosis for FT and MSB. High-parent heterosis for LA occurred in three out of 19 epiHybrids, namely 232H, 195H, and 193H (the number in the identifier refers to the epiRIL parent, and the H stands for F1 epiHybrid line). The epiHybrids 232H, 195H, and 193H significantly exceeded their best parent (Col-wt) by 17%, 18%, and 15%, respectively (Fig. 1H; Supplemental Table S19). Interestingly, although GR is developmentally related to LA, hybrid effects in GR were only moderately, albeit positively, correlated with LA (ρ = 0.57, P = 0.02), which implies that LA heterosis is determined by other traits besides GR.
For HT, we detected five cases of significant high-parent heterosis with up to 6% increases in HT (Fig. 1I; Supplemental Table S14). One may expect LA high-parent heterosis to strongly correlate with HT high-parent heterosis, as the rosette provides nutrients for the developing shoot (Bennett et al., 2012). However, high-parent heterosis for both LA and HT occurred only in one epiHybrid (193H; Fig. 1, G–I). For MSB, we detected one case of high-parent heterosis (64H; Fig. 1G; Supplemental Fig. S2).
Besides positive heterosis, our phenotypic screen revealed strong negative heterotic effects for FT (earlier flowering) and MSB (less MSB). Significant low-parent heterosis occurred in epiHybrids 232H, 208H, 344H (FT), and 438H (MSB; Fig. 1J; Supplemental Fig. S2; Supplemental Tables S15 and S17). In the most prominent case for FT, the epiHybrid (232H) flowered about 10% earlier than the earliest flowering parent. The epiHybrids 208H and 344H flowered 3% and 4% earlier than their earliest parent (epiRIL 208 and epiRIL 344), respectively. The epiHybrid 438H showed 14% less MSB than its least branched parent (Supplemental Fig. S2).
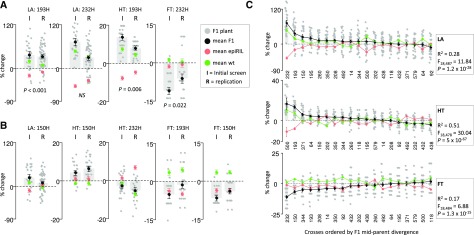
The reproducibility of our findings was tested by performing replicate experiments under the same growth conditions as before, using seeds from newly performed crosses. We focused on epiHybrids that exhibited relatively strong positive or negative heterotic phenotypes in the initial screen (193H, 150H, and 232H; Fig. 1G) and measured LA, FT, and HT in the epiHybrids and their parents. We monitored about 540 plants for LA (∼60 replicates per line) and 270 plants for FT and HT (∼30 replicates per line). The direction of the heterotic effects in LA, FT, and HT was reproducible in all tested cases (Fig. 2, A and B). Importantly, the LA and HT high-parent heterosis observed for 193H, and the strong FT low-parent heterosis for 232H, were perfectly reproducible, while LA high-parent heterosis observed for 232H was reduced to midparent heterosis (Fig. 2A). Taken together, these results show that the heterotic effects observed in the epiHybrids are relatively stable for LA, HT, and FT even across fresh seed batches, which is not always the case for Arabidopsis phenotypes (Massonnet et al., 2010).
Figure 2.
Confirmation of midparent (MP) divergence in the initial screen and replicate experiment for epiHybrids 150H, 193H, and 232H. A, Results for cases of high-parent heterosis and low-parent heterosis for LA, HT, and FT in the initial and replicate experiments. B, Results for cases showing less prominent phenotypic effects for LA, HT, and FT. The MPV is shown as dashed horizontal lines, and the MP divergence is shown as change from MPV in percentage. To illustrate the F1 epiHybrid distribution for each trait, the individual replicate plants are depicted as dots. C, F1 MP divergence for LA, HT, and FT for all epiHybrids. The MPV is shown as horizontal dashed lines, and MP divergence is shown as change from MPV in percentage. The epiHybrids are ordered from highest (left) to lowest (right) F1 MP divergence. To illustrate the F1 epiHybrid distribution for each trait, the individual replicate plants are depicted as dots. Variance component analysis was used to estimate how much of the total variation in MP divergence can be explained by between-cross variation. The F-statistic from this analysis is shown in the boxes.
Parental DMRs Are Associated with epiHybrid Performance
In order to quantify the proportion of variation in LA, HT, and FT heterosis that can be attributed to differences between the epiRIL parents, we calculated the phenotypic divergence of each of the 530 epiHybrid plants (∼28 plants from each of 19 epiHybrid lines) from their respective MPVs (Fig. 2C). Treating the degree of divergence as a quantitative trait, our goal was to estimate the within- and between-cross variance components (Supplemental Methods S1). For LA, HT, and FT, we found that 28% (F18,487 = 11.84, P = 1.2 × 10−28), 51% (F18,478 = 30.04, P = 5 × 10−67), and 17% (F18,484 = 6.88, P = 1.3 × 10−15) of the total variation in midparent divergence could be attributed to the between-cross variance component, respectively, indicating that (epi)genomic differences between the 19 paternal epiRILs are important determinants of hybrid performance (Fig. 2C; Supplemental Table S20; Supplemental Methods S1).
In an effort to identify specific methylome features that could account for these differences, we made use of previously published Col-0 and epiRIL methylated DNA immunoprecipitation tiling array data (Colomé-Tatché et al., 2012). We previously showed that the epiRILs differ in total 5mC content as a result of the stochastic fixation of ddm1-derived hypomethylated epihaplotypes during inbreeding (Colomé-Tatché et al., 2012; Fig. 3A). Therefore, we asked whether genome-wide 5mC levels is predictive of hybrid performance. To do this, we used tiling-resolution methylation calls and calculated the methylated proportion of each epiRIL genome (Supplemental Methods S1). Our analysis revealed no significant correlation between paternal 5mC content and average hybrid performance in LA, HT, and FT among the 19 crosses (Supplemental Fig. S3).
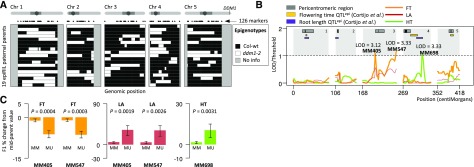
Figure 3.
Parental epigenotypes are associated with heterotic phenotypes in the F1 epiHybrids. A, Genome-wide patterns of Col-wt- and ddm1-2-inherited epihaplotypes in the (epi)genomes of the parental epiRILs used in this study. The epiRILs depicted are 14, 64, 92, 118, 150, 193, 195, 202, 208, 232, 260, 344, 350, 371, 432, 438, 492, 500, and 579 (from top to bottom). B, The parental epigenotypes were associated with F1 phenotypes in a genome-wide QTL scan. QTL peaks indicate that specific differentially methylated regions in the parental methylomes contribute to heterosis in the F1 epiHybrids. Shown are the QTL profiles for FT, HT, and LA. Published QTLsepi for root length and FT are shown as well. C, Effect direction of the peak QTL markers (MM405, MM547, and MM698). The y axis shows the mean phenotypic divergence of epiHybrids from their MPVs. MM refers to epiHybrids whose parents were both wild-type methylated at the peak QTL markers (MM epihomozygous in Col-wt female and MM epihomozygous in epiRIL male); MU refers to epiHybrids whose parents were differentially methylated (MM epihomozygous in Col-wt female and UU epihomozygous in epiRIL male). Error bars represent ±1 se of the estimate.
We reasoned that variation in hybrid phenotypes could be associated with more localized methylation differences between the parents rather than the result of global differences in 5mC content. To test this hypothesis, we made use of 126 epiRIL DMRs that were shown previously to mark ddm1-derived hypomethylated epihaplotypes (Colomé-Tatché et al., 2012; Cortijo et al., 2014). At each of these DMRs, a given epiRIL parent is either epihomozygous for the wild-type-like methylated state or epihomozygous for the ddm1-like hypomethylated state. In Col-0, the maternal parent for the epiHybrids, these loci are homozygously methylated. Therefore, the parents of a given cross can either be differentially methylated at these loci (i.e. the paternal epiRIL locus is homozygously hypomethylated and the maternal Col-0 locus is homozygously methylated) or, alternatively, the two parents have the same methylation state (i.e. the locus is homozygously methylated in both).
We used this information in a QTL scan and tested whether parental DMRs were associated with the (average) phenotypic performance of the different epiHybrid lines (Fig. 3A; Supplemental Fig. S4; Supplemental Methods S1). Despite the relatively small sample size (n = 19), our scan revealed two genome-wide significant QTLs on chromosome 3 that contributed to the between-cross variation in midparent heterosis in FT (QTL 1: logarithm of the odds [LOD] = 3.12, 37.62 centiMorgan [cM]; QTL 2: LOD = 3.33, 101.44 cM; Fig. 3B; Supplemental Table S21). The epiHybrids whose parents were differentially methylated at these loci (homozygously methylated versus hypomethylated) showed significant negative midparent heterosis compared with the epiHybrids whose parents both had the wild-type state (homozygously methylated in both parents; Fig. 3C). While not significant at the genome-wide scale (Fig. 3B), the same two QTLs had suggestive effects on LA heterosis in the opposite direction than FT (Fig. 3, B and C), indicating that both QTLs act pleiotropically. We also detected a single significant QTL on chromosome 4 (LOD = 3.33, 56 cM) that contributed to the between-cross variation in midparent heterosis for HT (Fig. 3B; Supplemental Table S21). In this case, epiHybrids whose parents were differentially methylated showed positive midparent heterosis compared with epiHybrids whose parents were not (Fig. 3C). Interestingly, the HT QTL overlaps with a previously identified QTLepi for root length in a panel of 123 epiRILs (Cortijo et al., 2014). The same study identified QTLsepi associated with variation in FT (Cortijo et al., 2014) that we did not detect here (Fig. 3B), implying that other regions may play a role in FT trait variation than in FT heterosis. Taken together, our QTL mapping results show that local methylation differences between the parents have a direct or indirect impact on hybrid performance in our experimental system.
Methylome and Transcriptome Remodeling in epiHybrids
A number of plant studies have shown that, when homologous methylated and hypomethylated regions come together during hybridization, the methylation state of one region can be acquired by its homologous (allelic) counterpart (Greaves et al., 2012; Rigal et al., 2016; Zhang et al., 2016b). This can involve the acquisition of both lower as well as higher DNA methylation levels by TC(d)M events, resulting in methylation levels that diverge from the MPV. Nonadditive methylation levels can drive nonadditive expression changes at proximal genes (Greaves et al., 2012; Rigal et al., 2016) and could provide a molecular basis for the heterotic effects observed in this study.
We assessed the extent of methylome and transcriptome remodeling in four of the epiHybrids (92H, 150H, 193H, and 232H) and their parental lines by performing whole-genome bisulfite sequencing (BS-seq) and RNA sequencing (RNA-seq) on 21-DAS rosette leaf tissue (see “Materials and Methods”). Biological replicates of the BS-seq data sets (methylation frequency) and those of the gene expression data sets (reads per kilobase of transcript per million mapped reads [RPKM]) were compared and revealed high correlations among the replicates (BS-seq rs > 0.85 [Supplemental Fig. S5A] and RNA-seq rs > 0.9 [Supplemental Fig. S5B; Supplemental Tables S22 and S23]). For further analysis, BS-seq reads from biological replicates were pooled, and RNA-seq replicates were used to estimate the variance among the replicates to calculate the final expression levels in each line.
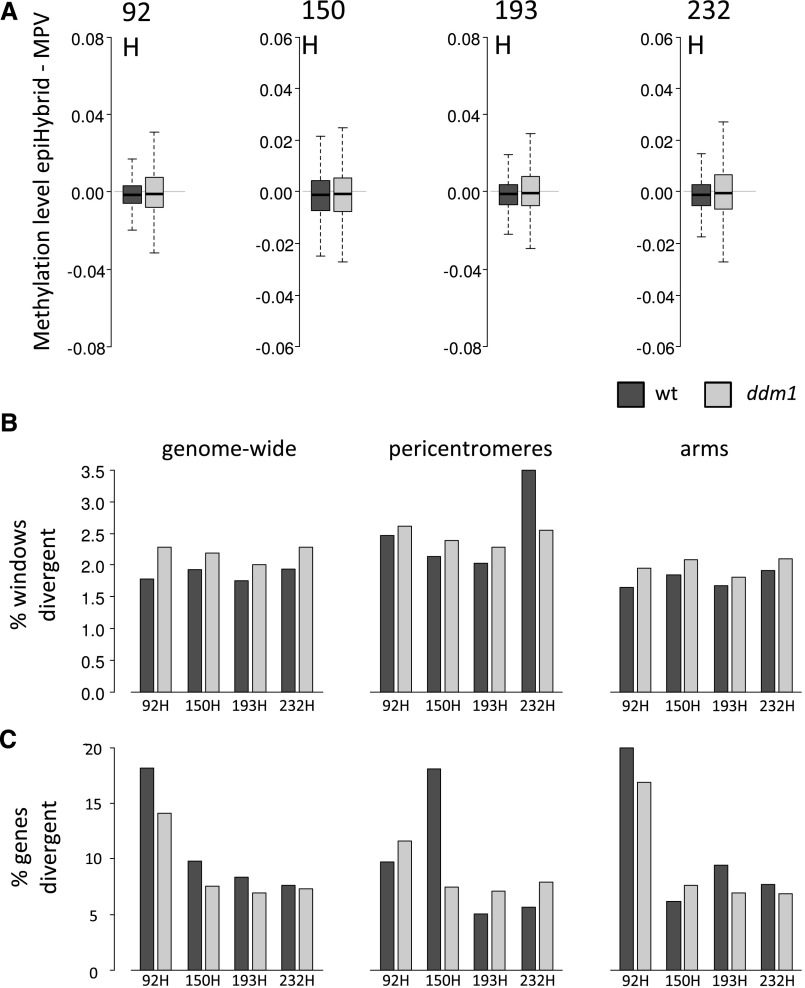
Overall, DNA methylation levels were higher in Col-wt than in any epiRIL parent. Globally, DNA methylation levels of epiHybrids were between those of their parental lines (Supplemental Fig. S6). In order to identify specific nonadditively methylated regions in the epiHybrids, a genome-wide sliding window approach was used (window size of 100 bp, step size of 50 bp). We found that nonadditively methylated regions occurred throughout the genome in all four epiHybrids but were more prevalent in regions where a given epiRIL parental line carried a hypomethylated ddm1-derived epihaplotype, compared with windows where both parents were Col-wt (Fig. 4, A and B). The nonadditively methylated regions deviated in the positive and negative directions from MPV for both Col-wt/Col-wt and ddm1/Col-wt regions (Supplemental Fig. S7).
Figure 4.
Methylome and transcriptome remodeling at wild-type (wt)- and ddm1-derived regions in the epiHybrids. A, Divergence in DNA methylation from MPV of all wild-type- (black) and ddm1-derived windows (gray) for the four epiHybrids indicated. DNA methylation level is determined as methylated reads per cytosine/total reads per cytosine, averaged over entire windows. Windows refer to 100-bp sliding windows, with a step size of 50 bp. Results are shown for methylation levels based on all cytosines regardless of sequence context. B, Percentage of wild-type- and ddm1-derived windows with methylation divergence from MPV across the four epiHybrids. The first graph displays genome-wide results, the second graph displays pericentromeric regions only, and the third graph displays euchromatic chromosome arms only. Windows refer to 100-bp sliding windows, with a step size of 50 bp (see “Materials and Methods”). C, Percentage of genes within wild-type- and ddm1-derived regions with expression divergence from MPV across the four epiHybrids. The first graph displays genome-wide results, the second graph displays pericentromeric regions only, and the third graph displays euchromatic chromosome arms only.
Trends became more apparent when filtering for clear cases of nonadditively methylated regions (Table I), which we defined as those windows that showed a fold change from MPV of at least 1.5 and an absolute difference in methylation level of at least 0.05. Genome wide, about 2.1% of the 100-bp windows across the four epiHybrids showed negative deviation from MPV (methylation loss), while about 2.4% showed positive deviation from MPV (methylation gain; Table I). Interestingly, a substantial portion of the nonadditively methylated regions (48%–71%) showing negative deviation from MPV occurred in regions where the two parental lines differed in their methylation levels, suggesting that these nonadditively methylated regions are possibly the result of TCdM events. Also nonadditively methylated regions with positive midparent deviation were detected in the regions that were differentially methylated between the parents (25%–32%), suggesting that these are the result of TCM events. Most of these latter events, however, occurred in regions that were not differentially methylated (Table I), suggesting de novo DNA methylation by small RNAs derived from nonallelic homologous sequences. These results are consistent with a recent report on a genetic Arabidopsis hybrid that found most TCdM events at loci that are differentially methylated in the parents and TCM events at similarly methylated loci (Zhang et al., 2016b).
Table I. Genome-wide nonadditively methylated regions in the epiHybrids.
Reported are the number of windows (sliding window size of 100 bp, step size of 50 bp) that displayed divergence from MPV with a fold change of at least 1.5 and an absolute difference in methylation level of at least 0.05. Divergence in the positive direction indicates a methylation gain (Signif. higher), and divergence in the negative direction indicates a loss (Signif. lower). The last three columns report the number of nonadditively methylated regions at regions that were differentially methylated in the parents. Also reported are the number of windows for which none of the cytosines contained sufficient coverage (Insuff. coverage) as well as the number of windows that did not show a significant difference from the MPV (Not signif.). Win. #, Number of windows; Win. %, percentage of windows with respect to all windows that are also differentially methylated between the parents; Div. %, percentage of windows that are differentially methylated between the parents with respect to all divergent windows of that category.
| Hybrid | Description | Coveragea |
Divergent Windowsb |
Differentially Methylated between Parentsc |
||||
|---|---|---|---|---|---|---|---|---|
| Win. # | Win. % | Win. # | Win. % | Win. # | Win. % | Div. % | ||
| 92H | Insuff. coverage | 186,792 | 7.84 | |||||
| Suff. coverage | 2,196,138 | 92.16 | ||||||
| Not signif. | 2,096,616 | 95.47 | 135,247 | 74.43 | 6.45 | |||
| Signif. lower | 48,270 | 2.20 | 31,993 | 17.61 | 66.28 | |||
| Signif. higher | 51,252 | 2.33 | 14,476 | 7.97 | 28.24 | |||
| 150H | Insuff. coverage | 190,152 | 7.98 | |||||
| Suff. coverage | 2,192,778 | 92.02 | ||||||
| Not signif. | 2,095,298 | 95.55 | 80,006 | 68.80 | 3.82 | |||
| Signif. lower | 52,269 | 2.38 | 25,041 | 21.53 | 47.91 | |||
| Signif. higher | 45,211 | 2.06 | 11,240 | 9.67 | 24.86 | |||
| 193H | Insuff. coverage | 192,708 | 8.09 | |||||
| Suff. coverage | 2,190,222 | 91.91 | ||||||
| Not signif. | 2,099,032 | 95.84 | 103,926 | 75.04 | 4.95 | |||
| Signif. lower | 39,065 | 1.78 | 20,051 | 14.48 | 51.33 | |||
| Signif. higher | 52,125 | 2.38 | 14,512 | 10.48 | 27.84 | |||
| 232H | Insuff. coverage | 177,413 | 7.45 | |||||
| Suff. coverage | 2,205,517 | 92.55 | ||||||
| Not signif. | 2,098,056 | 95.13 | 164,309 | 75.62 | 7.83 | |||
| Signif. lower | 48,647 | 2.21 | 34,396 | 15.83 | 70.71 | |||
| Signif. higher | 58,814 | 2.67 | 18,579 | 8.55 | 31.59 | |||
Sufficient coverage: a minimum read coverage of three. bNonadditively methylated region criteria: fold change of at least 1.5 from MPV and an absolute difference in methylation level of at least 0.05. cDifferential methylation between parents: difference in methylation level should be equal to or lower than −0.1 (see “Materials and Methods”).
Genome-wide analysis performed on the transcriptome data revealed that both ddm1 and Col-wt-derived regions were also divergent from the MPV at the gene expression level, but here, the Col-wt regions had a (slightly) higher percentage of genes displaying expression divergence (Fig. 4C). For three of the epiHybrids this was true for the euchromatic chromosome arms, but not for the pericentromeric regions, where ddm1-derived regions had a higher percentage of divergently expressed genes than wild-type regions. The only exception to this trend was epiHybrid 150H, which showed slightly more divergently expressed genes in pericentromeric wild-type regions. Similar to identifying nonadditively methylated regions with clear divergence from the MPV, using the four parent-epiHybrid combinations, the number of genome-wide nonadditively expressed genes was identified with a minimum RPKM of 2 and a fold change of at least 1.4. Across the four epiHybrids, we found 738 to 2,535 nonadditively expressed genes (2.4%–8.3% of all genes) that showed a negative deviation from MPV (expression loss), while 1,245 to 2,623 nonadditively expressed genes (4.1%–8.6% of all genes) showed a positive expression deviation (expression gain) from the MPV (Supplemental Table S24).
Taken together, these results show that methylomes are partially remodeled in the epiHybrids, not only in regions that are differentially methylated in the parents but also in nondifferentially methylated regions. Remodeling in differentially methylated regions is probably mediated by allelic TC(d)M events. Remodeling in regions where both parents were similarly methylated suggests nonallelic TC(d)M events. Transcriptional remodeling also occurs both in ddm1- and Col-wt-derived regions, and we hypothesize that these are driven by allelic and nonallelic mechanisms as well.
Identification of Putative Loci Mediating Hybrid Performance
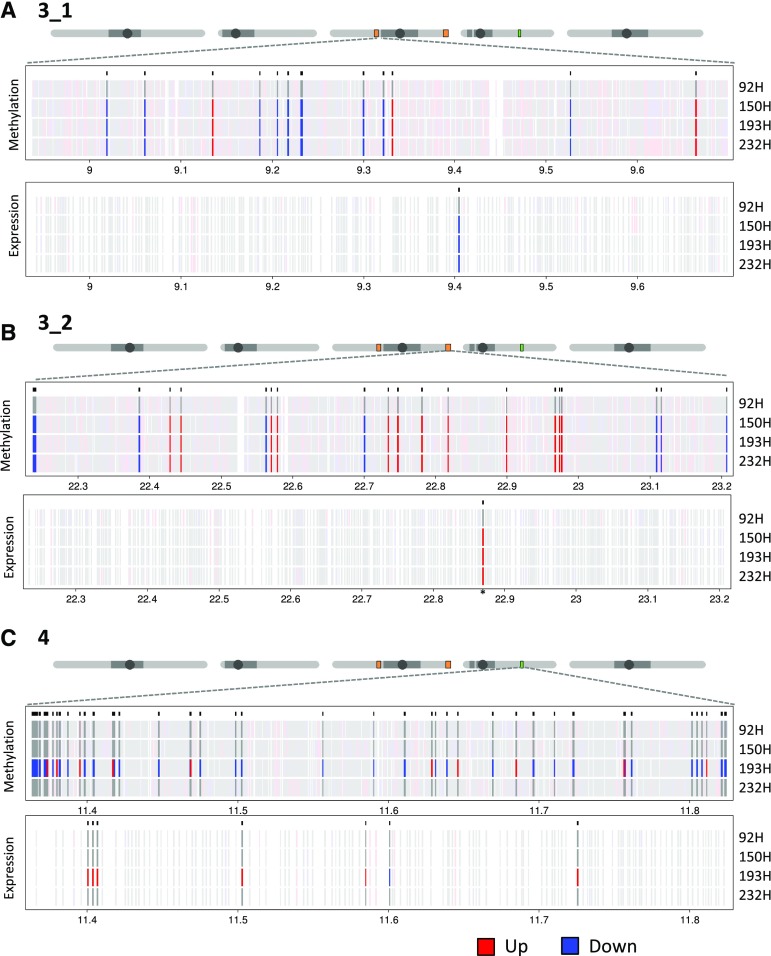
Our QTL analysis indicated that parental methylation differences that are in linkage with specific parental DMRs are associated with hybrid performance for FT, LA, and HT. In line with recent molecular epigenetic studies in other hybrid systems (Rigal et al., 2016; Zhang et al., 2016b), we argued that the heterotic effects may be due to allelic (Greaves et al., 2012; Rigal et al., 2016; Zhang et al., 2016b) and nonallelic transfer of 5mC, specifically at phenotypically relevant loci within the QTL intervals. To assess this possibility, all nonadditively methylated regions and nonadditively expressed genes within the QTL intervals in the four parent-epiHybrid combinations were identified. Candidate nonadditively methylated regions and nonadditively expressed genes were required to be consistent in direction (i.e. negative or positive divergence from MPV) as well as consistent with the parental methylation state at the peak QTL markers. Only 155 of the nonadditively methylated regions and nine of the nonadditively expressed genes met these criteria in the four parent-epiHybrid combinations, and these represent a conservative set of candidates that might explain the QTL effects (Fig. 5; Supplemental Table S25). Of these 155, QTL intervals 3_1, 3_2, and 4 contained 14, 30, and 111 nonadditively methylated regions and one, one, and seven nonadditively expressed genes, respectively.
Figure 5.
Nonadditively methylated regions (naMRs) and nonadditively expressed genes (naEGs) within QTL intervals 3_1 (A), 3_2 (B), and 4 (C) in the four epiHybrids. On top of each section, a schematic depiction of the five Arabidopsis chromosomes with the approximate position of the QTL interval is shown. Criteria for naMR selection were a fold change of at least 1.5 from MPV, an absolute divergence in methylation level of 0.05, and consistency with the parental methylation state at the peak QTL marker (Supplemental Table S25). Criteria for naEG selection were an RPKM of 2, a fold change of at least 1.4 from MPV, consistency with the haplotype at the peak marker (Supplemental Table S25), and consistency in direction. Significant increases (blue) and decreases (red) in DNA methylation and expression in the right direction are indicated. Gray indicates the naMRs and naEGs that are not significantly different from MPV. Red and blue in the background indicate naMRs and naEGs that are significantly different from MPV but not consistent with the haplotype and/or not in the right direction.
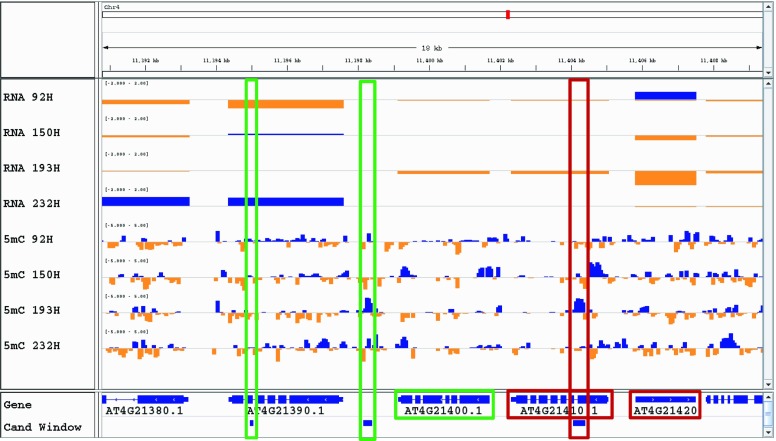
In FT QTL interval 3_1, we found one gene (AT3G25760) whose expression was up-regulated in the three epiHybrids whose parents were differentially methylated at the peak QTL marker (150H, 193H, and 232H; Supplemental Tables S25 and S26). The gene encodes an allene oxide cyclase, which is involved in jasmonic acid biosynthesis, and was observed to be down-regulated in C24 × Landsberg erecta genetic hybrids displaying heterotic growth of the rosette (Groszmann et al., 2015). The direction of the change in expression is opposite. We hypothesize that, depending on the parent combinations, the level of a particular gene product associated with heterosis may be different (Zhang et al., 2016a). In FT QTL interval 3_2, we identified a down-regulated candidate gene, encoding EMBRYO DEFECTIVE1703, in the same three epiHybrids.
In the HT QTL interval on chromosome 4, we identified seven nonadditively expressed genes, including a TE (AT4G21420). Five of the six genes (AT4G21400, AT4G21410, AT4G21650, AT4G21830, and AT4G22130), and the TE, were down-regulated in epiHybrid 193H, whose parents were differentially methylated at the peak QTL marker. In addition, another candidate gene (AT4G21860) was up-regulated in the same epiHybrid. Five of the six genes (AT4G21400, AT4G21410, AT4G21650, AT4G21830, and AT4G21860) were interesting in terms of their functionality, as they were shown to be involved in stress or defense responses (Rizhsky et al., 2004; Laugier et al., 2010; Li et al., 2012; Yadeta et al., 2017). The down-regulation of two (AT4G21650 and AT4G21830) of these genes was observed previously in heterotic hybrids between different Arabidopsis accessions (Groszmann et al., 2015). It has been proposed that a suppressed defense response could be relevant for mediating heterosis (Groszmann et al., 2015), in line with the known tradeoff between plant defense and growth or yield (Denancé et al., 2013; Huot et al., 2014). A change in expression of the same genes in both genetic and epigenetic heterotic hybrids could point toward such genes being, at least in part, controlled by epigenetic variation.
The precise mechanisms through which epigenetic variation affects nonadditive gene expression states in the epiHybrids remain unclear. Interestingly, of the seven nonadditively expressed genes in QTL interval 4, six (AT4G21400, AT4G21410, AT4G21420, AT4G21650, AT4G21830, and AT4G22130) were colocating with 21 nonadditively methylated regions within a distance of 5 kb, indicating a cis-regulatory effect for these candidates (Fig. 5; Supplemental Table S27). All six nonadditively expressed genes were down-regulated in 193H, while all associated nonadditively methylated regions showed an increase in DNA methylation level. A subset of nonadditively methylated regions was located within nonadditively expressed genes, while others were flanking them (Fig. 6; Supplemental Fig. S8). We hypothesize that the remaining putatively causal nonadditively methylated regions identified in this study may affect target genes within or outside the QTL intervals from distances larger than 5 kb, for example by regulating the expression of enhancer elements (Shlyueva et al., 2014; Weber et al., 2016). In addition, the relatively stringent filtering for candidate nonadditively methylated regions and nonadditively expressed genes may have missed potentially colocating pairs. Many more epiHybrid lines would be needed to explore this possibility. Nonetheless, the candidate genes identified here provide excellent targets for follow-up studies.
Figure 6.
Example of fold changes from MPV in gene expression and DNA methylation at candidate windows in epiHybrids where nonadditively expressed genes and nonadditively methylated regions are within 5 kb of each other. RNA tracks show gene expression-level fold changes, 5mC tracks show DNA methylation-level fold changes, the Gene track displays the gene annotation from TAIR10, and Cand Window presents the identified candidate windows. Blue and orange bars indicate positive and negative deviation from MPVs. Green and red boxes highlight the candidate windows and their associated genes.
Whole-Genome Sequencing Rules Out Genetic Variants as a Cause for Hybrid Performance
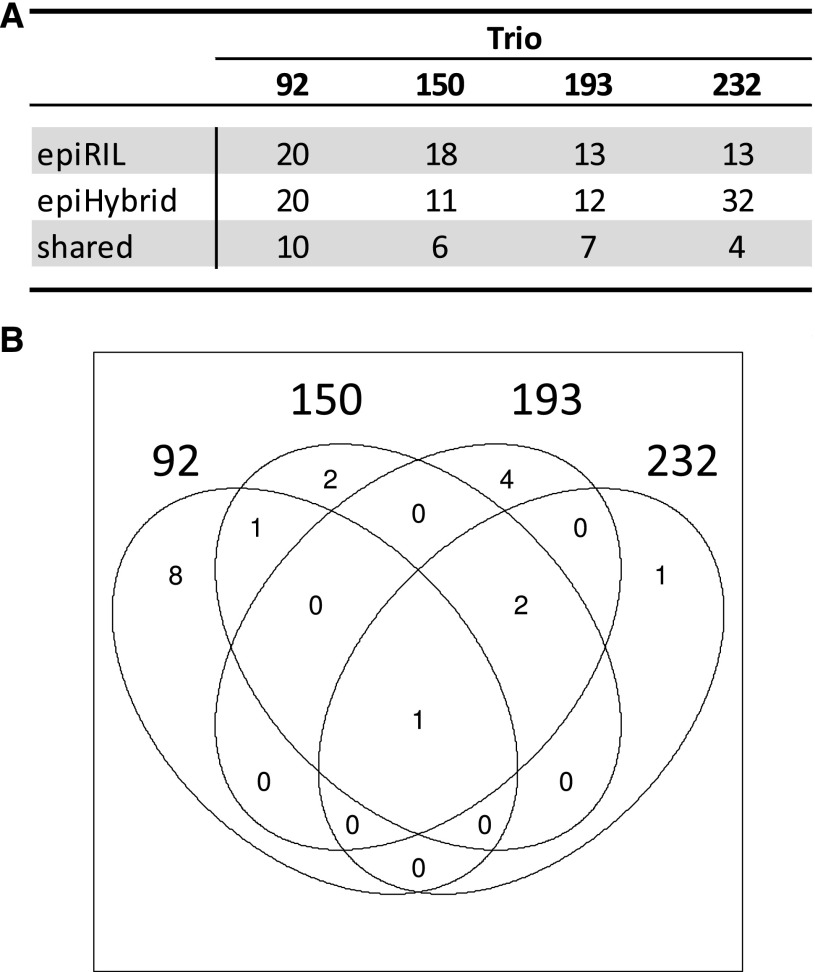
Previous mate-pair resequencing of over 100 epiRILs identified a number of TE mobilization events in this experimental system (Cortijo et al., 2014). While the majority of these events occurred in a line-specific manner during inbreeding, a subset of the insertions was shared among the epiRILs and appeared to have been derived from the genome of the original ddm1 founder line. It is plausible that our detected heterosis QTLs are the outcome of classical genetic (over)dominance effects resulting from ddm1-inherited TEs that are in linkage disequilibrium with the peak QTL markers. To assess this possibility, we resequenced the genomes of the four parent-epiHybrid combinations (in replicates; each replicate consists of a pool of five plants) that were used for methylome and transcriptome analysis by Illumina paired-end sequencing (see “Materials and Methods”). The replicate design enabled us to identify high-confidence TE insertions, which we defined as those insertions that were detectable both in the epiHybrids as well as in their epiRIL parental lines. Genome wide, we detected ten, six, seven, and four high-confidence shared TE insertion events that were detected in the epiHybrids and their respective epiRIL parents (Fig. 7). Of the detected TEs, only four were shared between at least two epiRILs, indicating that most TEs are unlikely to be derived from the ddm1 founder line (Fig. 7). None of the QTL intervals harbored any of the above-mentioned high-confidence TE insertions (Supplemental Table S28). We identified one TE insertion in QTL interval 3_1 for FT and LA in two independent F1 replicates of epiHybrid 150H (Supplemental Table S28). However, the epiRIL did not carry this insertion, indicating that this TE is either a false positive/negative or that the TE genotypes of the epiHybrid parents used for crossing differed from those used for sequencing, possibly because of incomplete homozygosity. We also detected two TE insertions in QTL interval 3_2 in only one of two replicates of epiHybrid 232H (Supplemental Table S28). Again, the epiRIL parent did not carry the insertion, thus rendering its origin ambiguous. To support our conclusion that TEs inherited from the ddm1 founder line are not the cause for the detected heterosis QTL, we also reanalyzed the mate-pair sequencing data of the 19 epiRILs (Cortijo et al., 2014) used as paternal parents for the initial phenotypic screen, but again we were unable to identify shared events in the QTL intervals (Supplemental Methods S1).
Figure 7.
Genome-wide shared high-confidence TE insertions. A, Table depicting the number of unique, high-confidence TEs observed in the four epiHybrids and their respective epiRIL parental lines. Trio, epiHybrid-parent combination. The TE insertions that were shared between the epiHybrid and respective epiRIL parental lines are indicated as well. B, Venn diagram depicting the TE insertions shared between the four epiRIL-epiHybrid combinations.
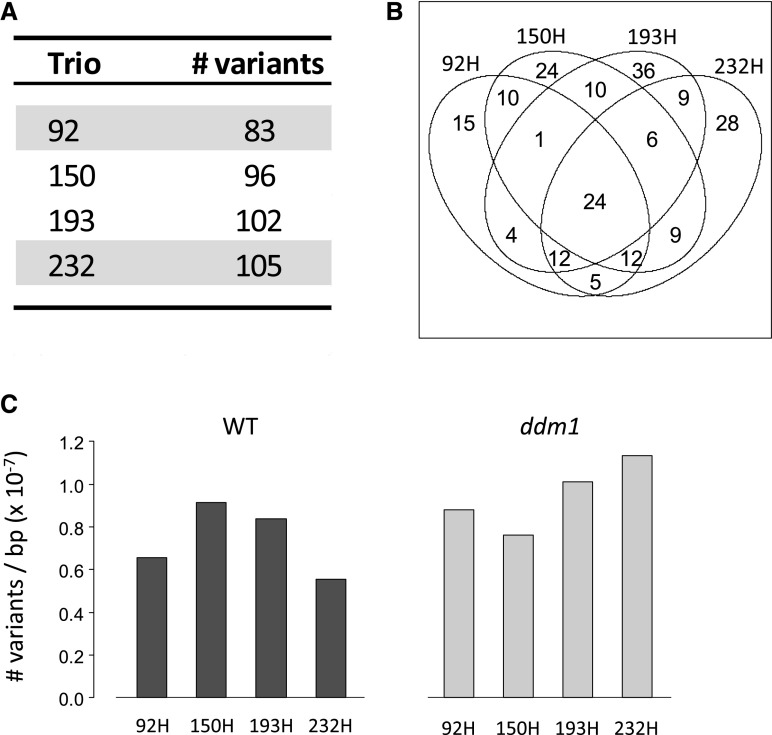
Apart from facilitating TE mobilization, hypomethylated sequences in the epiRIL genomes may promote higher mutation rates in general, perhaps as a result of a more accessible chromatin structure. Therefore, we used the replicate genome-wide sequencing data of our four epiHybrids and their parental lines and searched for single-nucleotide polymorphisms (SNPs) and small insertions and deletions (INDELs; see “Materials and Methods”). Genome wide, we identified 83, 96, 102, and 105 homozygote SNPs/INDELs in epiRILs 92, 150, 193, and 232, respectively, that were polymorphic with respect to the sequenced Col-wt parental line and heterozygote in the F1 offspring (i.e. in epiHybrids 92H, 150H, 193H, and 232H; Fig. 8A). Interestingly, a large percentage of these detected variants (73%; 283 out of 386) were shared between at least two epiRILs (Fig. 8B). One interpretation of this observation is that the epiRIL ddm1 founder line had a relatively high mutation load, which is supported by the fact that the ddm1-derived regions in the epiRILs on average contain more SNP/INDEL polymorphisms per base pair than the wild-type-inherited genomic regions (Fig. 8C). However, the Col-0 sequence of the epiRIL ddm1 founder line may have differed from that of the epiRIL wild-type founder line, and each may have differed to a varying extent from that of the sequence of the Col-wt used in our study. While the origin of shared variants in the epiRILs is difficult to establish, the remaining 27% (103 out of 386) of high-confidence variants were nonshared and most likely originated during the six generations of inbreeding of the epiRILs. We detected 15, 24, 36, and 28 nonshared variants in epiRILs 92, 150, 193, and 232, respectively (Fig. 8B). These numbers indicate that between 2.5 and six variants arose per generation, corresponding to a lower-bound estimate of the mutation rate of about 9.9 × 10−9 to 2.4 × 10−8 per generation per haplotype genome. This rate is close to the 7.1 × 10−9 ± 0.7 × 10−9 estimate in Arabidopsis mutation accumulation lines (Ossowski et al., 2010), arguing against extensive hypermutability in the epiRILs due to segregating ddm1-derived hypomethylated sequences. Focusing on the QTL intervals specifically, we only found three SNPs/INDELs that were homozygous in the epiRIL and heterozygous in the F1. Two of these variants were identified in epiHybrid 150H and one in 193H, neither of which was shared (Supplemental Fig. S9).
Figure 8.
Analysis of genome-wide SNPs and INDELs in four epiHybrids and parental lines. A, Number of high-confidence variants detected in each trio (epiHybrid-parent combination). High-confidence variants were defined as those for which one parental line was homozygous (AA), the other parental line was homozygous (BB), and the epiHybrid was heterozygous (AB). B, Number of high-confidence variants that are shared or not shared across the four trios. C, Total number of variants detected in the epiHybrids for wild-type (WT)- and ddm1-derived regions separately.
Taken together, our TE and SNP/INDEL results argue against a genetic basis underlying the detected QTL.
DISCUSSION
Heterotic hybrids have been shown to display widespread changes in DNA methylation and small RNAs relative to their parental lines (Groszmann et al., 2011; Barber et al., 2012; Shen et al., 2012; Shivaprasad et al., 2012). It remains unclear whether these changes are a cause or a consequence of heterosis and the extent to which they are determined by the genomes and/or epigenomes of the parents. Here, we presented the construction and analysis of a large panel of Arabidopsis epiHybrids that we derived from near-isogenic but epigenetically divergent parents. This proof-of-principle experimental system allowed us to test whether epigenetic divergences between the parental lines are associated with heterosis in F1 hybrids independently of genetic differences. Phenotypic analysis uncovered a wide range of heterotic effects in most of the epiHybrid lines. For GR, RB, LA, and FT, heterotic effects were observed in only one direction, while for HT and MSB, depending on the epiHybrid, positive and negative heterosis was detected. This suggests that the covariation in the different traits measured in our study is epigenotype dependent and, thus, hybrid specific. Much larger panels of epiHybrids would be needed to assess this hypothesis systematically.
Unlike previous epigenetic studies of hybrids, our experimental design allowed us to employ an epigenetic QTL mapping approach to associate the heterotic phenotypes observed in the epiHybrids with specific DMRs in the parental genomes. Using this approach, we identified several heterosis QTLs, while genomic resequencing did not detect any shared TE insertions, SNPs, or INDELs in the QTL confidence intervals, indicating that these QTLs have an epigenetic rather than a genetic basis. Interestingly, methylome and transcriptome sequencing of selected epiHybrids uncovered a number of regions and genes that showed nonadditive methylation and expression changes within the QTL intervals. Several of these methylation and transcript-level changes co-occurred within 5 kb from each other, suggesting a possible cis-regulatory link.
Prior studies on Arabidopsis heterotic hybrids identified candidate genes underlying heterotic phenotypes in pathways of the circadian clock, flavonoid biosynthesis, auxin transport, or salicylic acid metabolism and response (Ni et al., 2009; Shen et al., 2012; Groszmann et al., 2015; Zhang et al., 2016a). A Gene Ontology analysis on differentially expressed genes in a met1-derived epiHybrid did not show an overrepresentation of particular gene categories; however, the study proposed a gene (RECOGNITION OF PERONOSPORA PARASITICA5) within an epigenetically regulated resistance gene cluster to be a possible candidate (Dapp et al., 2015). Most studies focused on a biomass-related trait in the vegetative growth phase (i.e. plant fresh/dry weight, leaf width, LA; Ni et al., 2009; Shen et al., 2012; Groszmann et al., 2015; Zhang et al., 2016a), which can be compared with the LA trait measured here. Nonetheless, we did not identify an epigenetic QTL for LA, apart from the suggestive pleiotropic effect of the FT heterosis QTLs on LA. Even though none of the previously suggested genes involved in pathways leading to heterosis resided in our QTL intervals, we do not rule out a possible role of these genes in mediating heterotic phenotypes in our study, particularly because our QTLs cannot explain all the between-line phenotypic variation (Fig. 2C; Supplemental Table S20; Supplemental Methods S1). One reason for not detecting these genes in our QTL approach, besides the relatively small population size and inherent low mapping power, may be that DNA methylation is not a major factor for the regulation of these genes. That said, candidate genes explaining heterosis should also be viewed with respect to the specific phenotypes that are monitored; genes involved in biomass heterosis may not necessarily be relevant for heterosis in FT or final HT. Moreover, the extent of heterosis may vary depending on the direction of the cross (Ni et al., 2009). In our approach, parent-of-origin effects between the F1 lines were deliberately excluded by performing all crosses in the same direction using Col-wt as the maternal line. An example of a parent-of-origin effect is illustrated with the CCA1 gene, where, in F1 hybrids, increased CHH methylation in the promoter region is associated with lower CCA1 expression and an increased biomass heterosis (Ng et al., 2014). To identify such cases, reciprocal crosses should be included in future study designs.
In addition to the nonadditively methylated regions and nonadditively expressed genes within the QTL support intervals, we also observed substantial methylome and transcriptome remodeling elsewhere in the epiHybrid genomes. Genome wide, nonadditively methylated regions occurred in regions of the epiHybrid genomes that were differentially methylated between the parents but also in regions that were similarly methylated. The nonadditively methylated regions in the epiHybrids displayed primarily significantly lower methylation levels when they were differentially methylated in the parents, while these regions showed mainly increased methylation levels when they were similarly methylated in the parents. Although the origin of these nonadditive changes remains unclear, our data are consistent with the notion that nonadditive methylation changes in hybrids are the outcome of allelic and nonallelic TCM or TCdM events (Greaves et al., 2012; Shivaprasad et al., 2012; Groszmann et al., 2013; Zhang et al., 2016b). As observed for hybrids between genetically divergent parents (Zhang et al., 2016b), TCM occurred primarily at similarly methylated regions, suggesting de novo DNA methylation mediated by small RNAs from nonallelic homologous regions. In line with this, in genetic hybrids, the majority of TCM events in similarly methylated regions are located in repetitive heterochromatic pericentromeric sequences (Zhang et al., 2016b).
A study of genetic hybrids also reported that TCdM preferentially occurs in small interfering RNA-producing regions showing high levels of sequence polymorphisms between the parents (Zhang et al., 2016b). Indeed, small RNAs need to have sufficient homology with the target sequence for RdDM to occur. Hence, sequence polymorphisms at small RNA target sites can hamper de novo DNA methylation between allelic sequences in the hybrid (Zhang et al., 2016b). In heterozygotes, the effective small interfering RNA concentration in the nucleus is decreased, resulting in a loss of de novo DNA methylation at RdDM target loci (Zhang et al., 2016b). In our study, genetic variation between the parental genomes is virtually absent; therefore, the high incidence rate of TCdM indicates that this can occur independently of DNA sequence polymorphisms at small RNA target sites.
There is ample evidence that changes in DNA methylation can alter transcriptional states (Law and Jacobsen, 2010), and we hypothesize that nonadditive methylation levels in the QTL intervals establish nonadditive expression states of phenotypically relevant genes. However, we cannot discriminate cause from consequences in this study and do not exclude the possibility that changes in DNA methylation are triggered by changes in gene transcription. In support of the latter, a recent study provides compelling evidence that increased gene transcription, induced by phosphate starvation, led to increased DNA methylation of TEs flanking those genes. The increase in DNA methylation was postulated to play a role in maintaining the repression of TEs. However, contrary to this observation, the six candidate nonadditively expressed genes that colocated with nonadditively methylated regions in our study displayed a negative association (i.e. a decrease in expression was associated with an increase in DNA methylation). Moreover, five of the six colocating nonadditively methylated regions did not include TE sequences. It remains possible, however, that the nonadditive methylation changes are (partly) a consequence of transcription changes in our setup. Parental expression and DNA methylation profiles may have the potential to predict those regions in hybrids in either case.
In our study, heterotic effects were observed in epiHybrids produced by crossing Col-wt and ddm1-derived epiRILs (Johannes et al., 2009). In a recent study, heterosis for rosette area was reported in an epiHybrid generated by crossing a met1-derived epiRIL with Col-wt (Dapp et al., 2015). MET1 is involved in the maintenance of DNA methylation at cytosines in a CG sequence context, and a mutation in this gene causes severe and abundant losses of CG and CHH methylation, respectively (Stroud et al., 2014). Heterosis was observed in a parent-of-origin manner; the reciprocal cross did not result in heterosis (Dapp et al., 2015). This suggests a maternal effect (Park et al., 2016). In our design, we used Col-wt as the female parent and various ddm1-derived epiRILs as the male parent. While there could have been maternal contributions to heterotic phenotypes observed in our study, these effects cannot account for the variation in heterotic phenotypes seen between the 19 different epiHybrid lines, as the maternal parental line was invariant. It is plausible, nonetheless, that paternal effects could have contributed to between-cross variation in hybrid performance, for example via the cytoplasmic transmission of differential small RNAs in the pollen (Martínez et al., 2016). However, these paternally inherited effects, if they occurred, cannot explain the epigenetic QTL associations detected in our study. In fact, the QTL effects in our study provide the most compelling evidence to suggest that methylome divergence between the parental lines can trigger heterosis in F1 hybrids, independent of genetic differences and any other types of parental effects.
A more detailed insight into the molecular mechanisms underlying the QTL effects would require scaling up our study design to hundreds of epiHybrid families. This would provide a way to systematically study how parental methylation differences determine hybrid epigenomes in both cis as well as trans.
CONCLUSION
Here, we reported on the observation of extensive heterosis for several traits in a collection of Arabidopsis epiHybrids. These results could be confirmed in two independent experiments. We could exclude maternal parent-of-origin effects and genetic causes as the source of this variation. Three epigenetic QTLs were detected that explained variation in hybrid performance by differences in the methylation state of genomic loci, thus in part ruling out paternal effects. Substantial epigenetic and transcriptional remodeling in the epiHybrids was observed at genomic loci that were differentially methylated in the parental lines. Most striking was a strong enrichment for trans-chromosomal demethylation events. Finally, this study identified a number of candidate genes coupling the decrease in transcript levels with increased methylation levels, strongly suggesting a cis-regulatory role in controlling heterosis in epiHybrids. Taken together, our epigenetic QTL mapping results indicate that local methylation differences between parents can have a direct or indirect impact on hybrid performance, independent of genetic differences and other types of parental effects. Future research needs to address the causal relationship between the observed changes in methylome and transcript levels, in cis as well as in trans, and hybrid performance. This can be achieved by applying integrative QTL mapping approaches to phenome, methylome, and transcriptome data collected from much larger and more diverse panels of epiHybrid families. Our proof-of-principle study provides the needed rationale to initiate such efforts in crop species.
MATERIALS AND METHODS
Plant Material
The epiRILs in our study were generated by Johannes et al. (2009). The epiRILs were constructed as follows. An Arabidopsis (Arabidopsis thaliana) Col-0 line deficient for ddm1-2 was crossed to an isogenic Col-wt line, and the resulting F1 was backcrossed as the female parent to Col-wt. Subsequently, about 500 progeny plants with a wild-type DDM1 allele were selected and propagated through six more rounds of selfing, generating a population of 500 different epiRILs. We selected 19 different epiRILs as paternal plants for generating epiHybrids (line identifiers 14, 232, 92, 208, 438, 195, 350, 500, 150, 118, 432, 202, 344, 64, 492, 193, 260, 579, and 371). Our selection criteria were as follows: (1) a wide range of DNA methylation divergence from Col-wt and among the selected lines; (2) a wild-type DNA methylation state at the FWA locus in order to avoid differences in DNA methylation at this locus giving rise to differences in FT (Soppe et al., 2000) in the hybrids; and (3) a wide range of phenotypic variation in FT and root length among the selected lines. The epiRILs were purchased from the Arabidopsis stock center of the Institut National de la Recherche Agronomique in Versailles (http://publiclines.versailles.inra.fr/).
Crosses
To generate F1 hybrids from the selected epiRILs and Col-wt, all parental plants were grown in parallel in soil (Jongkind 7 from Jongkind) in pots (Danish size 40 cell; Desch Plantpak). The plants were grown at 20°C, 60% humidity, in long-day conditions (16 h of light, 8 h of dark), and were watered three times per week. All crosses were performed in parallel in a time frame of 2 weeks to avoid phenotypic effects in the F1 progeny due to differences in growing conditions. To exclude differences in maternal cytoplasm affecting the phenotypes of the F1 plants, Col-wt plants were used as the maternal parent and the epiRILs as paternal parents. In parallel, all parental lines, Col-wt and epiRILs, were propagated by manual selfing. This was done to (1) ensure that parental and F1 hybrid seeds were generated under the same growing conditions and (2) exclude potential phenotypic effects derived from hand pollination (Meyer et al., 2004).
Phenotypic Screen
The seeds were stratified at 4°C for 3 d on petri dishes containing filter paper and water before transferring them onto Rockwool/Grodan blocks (soaked in Hyponex NPK: 6.5–6.19 medium) in a climate-controlled chamber (20°C, 70% humidity, and long-day conditions [as above]). The transfer of the seeds onto the Rockwool blocks is defined as time point 0 DAS. Seeds from each parental and hybrid line were sown in 28 replicates, and their positions were randomized throughout the growth chamber to level out phenotypic effects caused by plant position. The plants were watered two or three times per week depending on their size. After the plants started flowering, they were transferred to the greenhouse (20°C, 60% humidity, and long-day conditions [as above]). In the greenhouse, the plants were watered three times per week and stabilized by binding them to wooden sticks at later developmental stages. The plants were harvested once the siliques of the main inflorescence and its side branches were ripe.
Rosette LA
LA was monitored by an automated camera system (Kooke et al., 2016) from 4 DAS. The system consists of 14 fixed cameras that can take photographs of up to 2,145 plants daily, every 2 h. We monitored LA until 14 DAS, since at later time points leaves start overlapping, hampering the correct detection of LA. Leaf area in mm2 was calculated by an ImageJ-based measurement setup (http://edepot.wur.nl/169770).
FT
FT was defined as the DAS at which the first flower opened. FT was scored manually each day before 12 am.
HT
HT was scored manually in cm on dried plants. The measurement was taken at the main inflorescence, from the rosette to the highest flower head.
Branching
Branching was scored on the dried plants by counting the branches emerging from the rosette (RB) and from the main stem (MSB).
Total SY
Seeds were harvested from the dried plants, cleaned by filtering, and SY was determined subsequently by weighing (resulting in mg of seeds per plant).
Variance Component Analysis and QTL Mapping Approach
For the QTL mapping approach and variance component analysis, see Supplemental Methods S1.
Replication Experiment with Selected Hybrids
Freshly ordered seeds of epiRILs (line identifiers 92, 150, 193, and 232) from the Arabidopsis stock center at Versailles were used for the replication experiment with the hybrids selected. The crosses with the epiRILs and the phenotypic screen were performed as described above, with the exception that more replicates were monitored for each parental and hybrid line: 60 replicates for LA and 30 replicates for the traits FT and HT. Furthermore, branching was not examined in the replication experiment.
DNA Sample Preparation for Whole-Genome-seq and Whole-Genome BS-seq
Plant material for sequencing was grown in parallel with the second phenotypic screen, under the exact same controlled environmental conditions as described above. Aerial rosette tissue at 21/22 DAS was harvested before noon on two consecutive days and snap frozen immediately in liquid nitrogen. Material was stored at –80°C until processing. Genomic DNA from two biological replicates (2 × 6 rosettes) was extracted using a standard cetyl-trimethyl-ammonium bromide-based extraction protocol followed by an RNase digest. For both whole-genome-seq and whole-genome BS-seq, at least 5 μg of DNA per sample were submitted to the Beijing Genome Institute. For whole-genome BS-seq, DNA was treated with bisulfite. Whole-genome-seq and whole-genome BS-seq libraries were made with an insert size of 500 and 200 bp, followed by sequencing, generating 90-bp and 150-bp paired-end reads, respectively. Sequencing was performed on an Illumina HiSeq4000 instrument.
Alignment of Bisulfite-Treated Reads and Construction of DNA Methylomes
Read sequences were quality trimmed, and adapter sequences were removed with the use of Cutadapt (version 1.9; Python version 2.7.9; Martin, 2011). Trimming was performed using the paired-end mode, and the quality threshold was set to a phred score of 20 (q = 20). We applied the default error rate of 10% in the adapter sequence for their removal. Afterward, we discarded reads that were shorter than 40 bp.
Reads were subsequently mapped to the TAIR10 Arabidopsis reference genome with the use of BS-Seeker2 (version 2.0.10; Guo et al., 2013). The maximum allowed proportion of mismatches was set to 0.05 (m = 0.05), and the maximum insert size was set to 1,000 bp (X = 1,000). Bowtie2 (version 2.2.2; Langmead and Salzberg, 2012) was used for the alignment of the reads. Mapping was done in two steps. Read pairs were first aligned in paired-end mode. Reads that could not be aligned concordantly were aligned subsequently in single-end mode as described on the Web site of BS-seeker2 (http://pellegrini.mcdb.ucla.edu/BS_Seeker2/). Mate 2 reads were first reverse complemented before alignment.
Duplicate read pairs were removed in paired mode using in-house scripts by comparing start positions (5′ ends) of the mates and the orientation (mapped to forward or reverse strand). Samtools (version 1.2; using htslib 1.2.1; Li et al., 2009) was used for sorting the mapped read pairs in this procedure. When only one mate of the pair was aligned, duplicate reads were removed by comparing only with read pairs for which only one mate was aligned (same strategy; comparing start of the alignment and orientation). In both cases (paired and single mode), we kept one read pair (randomly chosen). Also, when both mates of one pair were overlapping with each other, we corrected for the overlap by trimming the reads until the middle of the overlapping part. When one mate contained a mismatch at a cytosine position and the other mate contained a methylation call (methylated or unmethylated), we kept the call even if it came from the mate for which this overlapping part was discarded (or trimmed).
After the removal of duplicate read pairs, the sam files were sorted using Samtools (Li et al., 2009), and methylomes were subsequently reconstructed. With the use of the information in the sam files that indicates which reference cytosines are unmethylated (a T mapped to a C; bisulfite conversion) or methylated (a C mapped to a C; no conversion), the number of reads indicating methylation and the total number of reads were determined for each reference cytosine. After the determination of these frequencies, the methylation level of each cytosine was determined. This methylation level is defined as the proportion of reads that indicate that the cytosine is methylated (proportion of nonconverted cytosines).
Identification of Nonadditively Methylated Regions
After the construction of the methylomes, average methylation levels were calculated for genomic windows with a size of 100 bp and a step size 50 bp. For this analysis, only cytosines with a coverage of three or more reads were selected (sufficient read coverage). The methylation level of each cytosine was calculated as the number of reads that indicated that the cytosine was methylated (a C in the read sequence; nonconverted cytosine) divided by the total number of reads covering the cytosine. The midparent methylation level was calculated as the average of both parents (average methylation level of the wild type and epiRIL). Midparent divergence was calculated by subtracting the midparent methylation level from the methylation level of the F1 hybrid. Windows were classified as nonadditively methylated regions when they met the following criteria: (1) the fold change from MPV should be higher than or equal to 1.5 and (2) the absolute difference in methylation level should be equal to or higher than 0.05.
RNA Sample Preparation for RNA-seq
Plant material for sequencing was grown in parallel with the second phenotypic screen under the exact same controlled environmental conditions as described above. Aerial rosette tissue at 21/22 DAS was harvested before noon on two consecutive days and snap frozen immediately in liquid nitrogen. Material was stored at –80°C until processing. RNA was prepared for two or three biological replicates (five rosettes per replicate). Two biological replicates were prepared for lines 92, 92H, and 232. Three biological replicates were prepared for lines 150, 150H, 193, 193H, 232H, and Col-wt.
RNA was extracted using a protocol combining Trizol-based (TRI Reagent; Sigma; T9424) extraction with the RNeasy Plant Mini Kit (Qiagen; catalog no. 74903). Frozen plant material was ground in liquid nitrogen, and TRI Reagent was added to the frozen powder (∼1 mL per 100 mg of tissue). After an incubation of 5 to 30 min at room temperature, 0.2 mL of chloroform:isoamyl alcohol (ratio of 24:1)/mL TRI Reagant was added, mixed, and incubated for another 3 min at room temperature. Next, the samples were centrifuged for 15 min at 12,000g at 4°C, and the upper phase was transferred into a new tube. One volume of isopropanol was added, and samples were incubated at room temperature for 10 min before transferring them to an RNeasy MINI spin column (supplied in the RNeasy Plant Mini Kit). A centrifugation step for 15 s at 9,000g was performed, flow through was discarded, and 700 µL of buffer RW1 (supplied in the RNeasy Plant Mini Kit) was added to the column, followed by another centrifugation step for 15 s at 9,000g. A total of 500 µL of buffer RPE (supplied in the RNeasy Plant Mini Kit) was added to the column, and the sample was centrifuged again for 15 s at 9,000g. Again, 500 µL of buffer RPE was added to the column, and the sample was centrifuged for 2 min at 9,000g. Next, the column was transferred to a new 2-mL collection tube followed by a centrifugation step for 1 min at high speed to dry the membrane. Then, the column was transferred to a new 1.5-mL collection tube, and 50 μL of RNase-free water was added directly on the column membrane. The RNA was eluted by centrifugation for 1 min at 9,000g. To remove traces of DNA, a DNase I digest was performed (DNA-free DNA removal kit; Ambion; AM1906) according to the manufacturer’s instructions.
One to 2 μg of RNA per sample was submitted for library preparation and sequencing on an Illumina HiSeq2500 instrument to Wageningen University and Research. Libraries were prepared using the Illumina TruSeq RNA stranded library preparation kit and poly(A) selection. Either 50-bp single-read or 125-bp paired-end-read sequencing was performed.
RNA-seq
The 125-bp paired-end reads were trimmed to 50 bp using the Fastx toolkit (Gordon, 2016), and only one end was used for read mapping to make the analysis comparable to the other libraries with 50-bp single-end reads. The sequenced reads were trimmed at both ends based on sequencing quality (Q20), and the remaining Illumina adaptor sequences were removed using Trimmomatic (Bolger et al., 2014). When the remaining read length was less than 35 bp, the read was removed from the analysis. The reads were aligned, allowing one mismatch, to the reference genome, Arabidopsis TAIR10 assembly obtained from Ensembl Plants (Kersey et al., 2016), using TopHat2 (Kim et al., 2013) and Bowtie (Langmead et al., 2009). The transcript assembly and gene expression level calculation for each replicate were performed with a guided reference (TAIR10; Kersey et al., 2016) using the Cufflinks pipeline (Cufflink, Cuffquant, and Cuffnorm; Trapnell et al., 2010). Only uniquely mapping reads were retained for further analysis. To determine the reproducibility, the RPKM values for each gene in every pair of replicates were plotted against each other, and Spearman’s rank correlations were calculated in R (R Development Core Team, 2008). The RPKM values for each sample were calculated with multiread correction and with the variance information among the two or three replicates using Cuffdiff (Trapnell et al., 2013). The cutoffs for significantly nonadditively expressed genes were an RPKM of 2 and a fold change of 1.4 from MPV.
Whole-Genome Sequencing Analysis to Detect SNPs, Small INDELs, and TEs
Read sequences were quality trimmed, and adapter sequences were removed with the use of Cutadapt (version 1.9; Python version 2.7.9; Martin, 2011). Trimming was performed using the paired-end mode, and the quality threshold was set to a phred score of 20 (q = 20). We applied the default error rate of 10% in the adapter sequence for their removal. Afterward, we discarded reads that were shorter than 40 bp. Reads were subsequently mapped to the TAIR10 Arabidopsis reference genome with the use of BWA (BWA-MEM algorithm [default settings]; Li and Durbin, 2009). Read pairs with a mapping quality lower than 10 and (remaining) duplicate read pairs were removed in paired mode using in-house scripts by comparing start positions (5′ ends) of the mates and the orientation (mapped to forward or reverse strand). Samtools (version 1.2; using htslib 1.2.1; Li et al., 2009) was used for sorting the mapped read pairs in this procedure. When only one mate of the pair was aligned, duplicate reads were removed by comparing only with read pairs for which only one mate was aligned (same strategy; comparing the start of the alignment and orientation). In both cases (paired and single mode), we kept one read pair (randomly chosen).
SNPs and INDELs were called for each trio (Col-wt, epiRIL, and hybrid) using the GATK Haplotypecaller program (settings: –genotyping_mode DISCOVERY -stand_emit_conf 10 -stand_call_conf 30 -mmq 20; version 3.4-46-gbc02625; McKenna et al., 2010). Samtools (version 1.2; using htslib 1.2.1; Li et al., 2009) was used for merging the bam files of the parents and hybrid. After SNP and INDEL calling, the raw variants were filtered using the values of six annotations, FS, MQ, MQRankSum, QD, ReadPosRankSum, and SOR, that were most informative according to GATK (https://software.broadinstitute.org/gatk/documentation/article.php?id=6925). We considered the values of the variants with genotype combination AA (Col-wt)|BB (epiRIL)|AB (hybrid) as a true positive set and used this distribution as a reference for filtering the remaining variants. It is unlikely that this combination of genotypes occurred by chance or due to technical reasons. The cutoffs that were determined for each annotation correspond to a false negative rate of 5% of the AA|BB|AB variants. SNPs and INDELs were filtered separately.
For the detection of new TE insertion events, discordant read pairs were used. Read pairs for which the mates were mapped at least 10 kb apart from each other were extracted from the alignment files (Supplemental Fig. S10A) to reduce possible noise. A smaller distance also could be due to rare long fragments in the sequencing library. Mates of discordant read pairs were merged subsequently when the mates of both pairs were overlapping each other and were mapped to the same strand. In the next step, we searched for possible read-pair groups that could explain a TE insertion event. At least two read pairs were required to form a group as evidence for a TE insertion event. One of the mates of a read pair was mapped to one end of the TE, and the other mate was mapped to the genomic region that is flanking the insertion point (Supplemental Fig. S10B). In order to find such combinations of read pairs, we required a maximum distance of 1 kb between the mates outside of the TE and a maximum distance of 30 kb between the other mates that map inside the TE (position of the annotated TE). The 1-kb limit outside of TEs was selected due to the fragment lengths being approximately 500 bp. The mate that is flanking the insertion point cannot be farther away from the insertion point than 500 bp; otherwise, the other mate of the sequenced fragment would also flank the insertion point and not map to the TE. Therefore, the maximum distance between the mates that flank the insertion point was set to 1 kb (2 × 500 bp). The maximum distance of 30 kb between mates inside the TE corresponds to the maximum length of annotated TEs. In the next step, read pairs for which at least one of the mates was mapped to an annotated TE were kept. For this purpose, TE annotations from TAIR as well as from Quesneville were used. In the final step, the read-pair groups were filtered using several criteria: (1) one of the mates of every read pair in a given group must overlap the same TE; (2) for the same reason as explained above, the distances between all the combinations of the mates outside of the TE must be less than 1 kb (Supplemental Fig. S10B); (3) the mates in the TE must be within 500 bp from either end of the TE (if this distance were larger, than both mates of a pair would be mapped to the TE); and (4) all the mates must be mapped to chromosome sequences and not to the chloroplast genome. To accommodate alignment errors, a maximum of 10-bp protrusion of the TE mates from the TE was allowed. When one or more read pairs in a group did not fulfill one of the criteria above, the group was removed.
Accession Number
The accession number is GSE99482.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Methylation profile at the FWA locus.
Supplemental Figure S2. Detected cases of high-parent heterosis and low-parent heterosis for MSB.
Supplemental Figure S3. Relationship between genome-wide methylation level of paternal epiRILs (x axis) and level of midparent heterosis in F1 epiHybrids derived from these epiRILs (y axis).
Supplemental Figure S4. Frequency histograms of the percentage change from MPV for the 19 epiHybrid crosses.
Supplemental Figure S5. Spearman correlation analysis between BS-seq replicates and between RNA-seq replicates.
Supplemental Figure S6. Global DNA methylation levels in the epiHybrids and their parental lines.
Supplemental Figure S7. Methylome remodeling in epiHybrids.
Supplemental Figure S8. Example of fold changes from MPV in gene expression and DNA methylation at candidate windows in epiHybrids where nonadditively expressed genes and nonadditively methylated regions are within 5 kb of each other.
Supplemental Figure S9. Analysis of SNPs and INDELs (variants) in four epiHybrids, genome wide and within the QTL intervals.
Supplemental Figure S10. TE insertion analysis.
Supplemental Table S1. Selection of epiRILs.
Supplemental Table S2. Phenotype summary for HT.
Supplemental Table S3. Phenotype summary for MSB.
Supplemental Table S4. Phenotype summary for RB.
Supplemental Table S5. Phenotype summary for FT.
Supplemental Table S6. Phenotype summary for GR.
Supplemental Table S7. Phenotype summary for LA.
Supplemental Table S8. Test for midparent heterosis in HT.
Supplemental Table S9. Test for midparent heterosis in MSB.
Supplemental Table S10. Test for midparent heterosis in RB.
Supplemental Table S11. Test for midparent heterosis in FT.
Supplemental Table S12. Test for midparent heterosis in GR.
Supplemental Table S13. Test for midparent heterosis in LA.
Supplemental Table S14. Test for high(low)-parent heterosis in HT.
Supplemental Table S15. Test for high(low)-parent heterosis in MSB.
Supplemental Table S16. Test for high(low)-parent heterosis in RB.
Supplemental Table S17. Test for high(low)-parent heterosis in FT.
Supplemental Table S18. Test for high(low)-parent heterosis in GR.
Supplemental Table S19. Test for high(low)-parent heterosis in LA.
Supplemental Table S20. Variance component analysis for midparent heterosis.
Supplemental Table S21. Summary of interval mapping results.
Supplemental Table S22. Coverage for BS-seq.
Supplemental Table S23. Number of reads that were sequenced in our RNA-seq experiments.
Supplemental Table S24. List of nonadditively expressed genes and their expression levels in the studied lines.
Supplemental Table S25. List of differentially expressed genes in our QTL regions.
Supplemental Table S26. Epihaplotypes at the markers within the QTL intervals in the respective epiRILs.
Supplemental Table S27. Distances between candidate windows and genes.
Supplemental Table S28. Transposon insertions in the epiHybrids and their respective epiRIL parents compared with Col-wt.
Supplemental Methods S1. Variance component analysis and QTL mapping approach.
Acknowledgments
We thank F. Becker, I. Hövel, D. Angorro, R. Kooke, J.A. Bac-Molenaar, M. Tark-Dame, P. Sanderson, M. Koini, T. Bey, B. Weber, L. Tikovsky, and Unifarm Wageningen for technical support during sowing or phenotyping. We thank Elio Schijlen and Bas te Lintel Hekkert for generating RNA-seq libraries and transcriptome sequencing. We thank H. Westerhoff for discussion and critically reading the article and D. Vreugdenhil for support with the plant phenotypic screen.
Footnotes
We acknowledge the support of the Centre for Improving Plant Yield (CIPY; part of the Netherlands Genomics Initiative and the Netherlands Organization for Scientific Research) for K.L. The first phenotypic screen was supported by the Enabling Technologies Hotels (ETH) Programme from CIPY. We gratefully acknowledge the financial support of the European Commission 7th Framework-People-2012-ITN Project EpiTRAITS (Epigenetic Regulation of Economically Important Plant Traits; 316965) to R.O. and M.S. F.J. acknowledges support from the Technical University of Munich-Institute for Advanced Study funded by the German Excellence Initiative and the European Union Seventh Framework Programme under grant agreement #291763 and the SFB924/Sonderforschungsbereich924 of the Deutsche Forschungsgemeinschaft (DFG).
These authors contributed equally to the article.
Articles can be viewed without a subscription.
References
- Barber WT, Zhang W, Win H, Varala KK, Dorweiler JE, Hudson ME, Moose SP (2012) Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA 109: 10444–10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E, Roberts JA, Wagstaff C (2012) Manipulating resource allocation in plants. J Exp Bot 63: 3391–3400 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomé-Tatché M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, Etcheverry M, Martin A, Feng S, Duvernois-Berthet E, et al. (2012) Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci USA 109: 16240–16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo S, Wardenaar R, Colomé-Tatché M, Gilly A, Etcheverry M, Labadie K, Caillieux E, Hospital F, Aury JM, Wincker P, et al. (2014) Mapping the epigenetic basis of complex traits. Science 343: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Crow JF. (1948) Alternative hypotheses of hybrid vigor. Genetics 33: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. (1998) 90 years ago: the beginning of hybrid maize. Genetics 148: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapp M, Reinders J, Bédiée A, Balsera C, Bucher E, Theiler G, Granier C, Paszkowski J (2015) Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants 1: 15092. [DOI] [PubMed] [Google Scholar]
- Denancé N, Sánchez-Vallet A, Goffner D, Molina A (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM. (1936) Heterosis. Genetics 21: 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. (2016) FASTX-Toolkit. http://hannonlab.cshl.edu/fastx_toolkit/index.html (January 19, 2016)
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES (2014) Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 111: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA 109: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Gonzalez-Bayon R, Greaves IK, Wang L, Huen AK, Peacock WJ, Dennis ES (2014) Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol 166: 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Gonzalez-Bayon R, Lyons RL, Greaves IK, Kazan K, Peacock WJ, Dennis ES (2015) Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 112: E6397–E6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M (2013) BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7: 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, et al. (2009) Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DF. (1917) Dominance of linked factors as a means of accounting for heterosis. Genetics 2: 466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Richards EJ (1995) Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res 23: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, et al. (2016) Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 44: D574–D580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Keurentjes JJB (2015) Epigenetic variation contributes to environmental adaptation of Arabidopsis thaliana. Plant Signal Behav 10: e1057368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Kruijer W, Bours R, Becker F, Kuhn A, van de Geest H, Buntjer J, Doeswijk T, Guerra J, Bouwmeester H, et al. (2016) Genome-wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis. Plant Physiol 170: 2187–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugier E, Tarrago L, Vieira Dos Santos C, Eymery F, Havaux M, Rey P (2010) Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. Plant J 61: 271–282 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Lee SH, Chieh PS, Lin CS, Wang YC, Chan MT (2012) Arabidopsis root-abundant cytosolic methionine sulfoxide reductase B genes MsrB7 and MsrB8 are involved in tolerance to oxidative stress. Plant Cell Physiol 53: 1707–1719 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK (2016) Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Čaikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Törjék O, Becher M, Altmann T (2004) Heterosis of biomass production in Arabidopsis: establishment during early development. Plant Physiol 134: 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DWK, Miller M, Yu HH, Huang TY, Kim ED, Lu J, Xie Q, McClung CR, Chen ZJ (2014) A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell 26: 2430–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim MY, Vickers M, Park JS, Hyun Y, Okamoto T, Zilberman D, Fischer RL, Feng X, Choi Y, et al. (2016) DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA 113: 15138–15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Reinders J, Wulff BBH, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M, Becker C, Pélissier T, Pogorelcnik R, Devos J, Ikeda Y, Weigel D, Mathieu O (2016) Epigenome confrontation triggers immediate reprogramming of DNA methylation and transposon silencing in Arabidopsis thaliana F1 epihybrids. Proc Natl Acad Sci USA 113: E2083–E2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Colomé-Tatché M, Edelist C, Wardenaar R, Guerche P, Hospital F, Colot V, Jansen RC, Johannes F (2011) Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 188: 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Springer NM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64: 71–88 [DOI] [PubMed] [Google Scholar]
- Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, et al. (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15: 272–286 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Springer NM. (2013) Epigenetics and crop improvement. Trends Genet 29: 241–247 [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Greaves IK, Groszmann M, Wu LM, Dennis ES, Peacock WJ (2015) Hybrid mimics and hybrid vigor in Arabidopsis. Proc Natl Acad Sci USA 112: E4959–E4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Zicola J, Oka R, Stam M (2016) Plant enhancers: a call for discovery. Trends Plant Sci 21: 974–987 [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Elmore JM, Creer AY, Feng B, Franco JY, Rufian JS, He P, Phinney B, Coaker G (2017) A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol 173: 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Xu T, Srivastava AK, Wang D, Zeng L, Yang L, He L, Zhang H, Zheng Z, et al. (2016a) The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov 2: 16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C (2016b) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 113: E4248–E4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Chen L, Zhang C, Hao P, Jing X, Li X (2017) Global transcriptome analysis reveals extensive gene remodeling, alternative splicing and differential transcription profiles in non-seed vascular plant Selaginella moellendorffii. BMC Genomics (Suppl 1) 18: 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]