Abstract
The de novo generation of hematopoietic stem and progenitor cells (HSPC) occurs solely during embryogenesis from a population of epithelial cells called hemogenic endothelium (HE). During midgestation HE cells in multiple intra- and extraembryonic vascular beds leave the vessel wall as they transition into HSPCs in a process termed the endothelial to hematopoietic transition (EHT). Runx1 expression in HE cells orchestrates the transcriptional switch necessary for the transdifferentiation of endothelial cells to functional HSPCs. Runx1 is widely considered the master regulator of developmental hematopoiesis because it plays an essential function during specification of the hematopoietic lineage during embryogenesis. Here we review the role of Runx1 in embryonic HSPC formation, with a particular focus on its role in hemogenic endothelium.
Keywords: Runx1, hemogenic endothelium, hematopoiesis, aorta-gonad-mesonephros region, hematopoietic stem cell
Almost all blood cells in the adult mammal differentiate from hematopoietic stem cells (HSCs) in the bone marrow. However HSCs do not originate in the bone marrow, and instead differentiate in the embryo before bone or bone marrow forms (Müller et al., 1994). The majority of HSCs, defined as cells that can engraft adult transplant recipients, differentiate from immature HSC precursors called pre-HSCs (Rybtsov et al., 2011, Taoudi et al., 2008). Pre-HSCs, in turn, differentiate from a small population of hemogenic endothelial cells (Zovein et al., 2008, Chen et al., 2009). The maturation of pre-HSCs into HSCs predominantly takes place in the fetal liver, which is colonized by pre-HSCs via the circulation (Rybtsov et al., 2016, Kieusseian et al., 2012). Following birth, HSCs leave the fetal liver and settle in the bone marrow, where they remain for the rest of adult life.
HSCs develop at midgestation in the mouse embryo, and at 1 month of gestation in the human embryo (Ivanovs et al., 2011, Müller et al., 1994). However, before HSCs are present, several other primitive types of blood cells emerge that are essential for embryonic viability. Hematopoietic progenitors (cells that can produce differentiated blood cells, but do not possess long-term multilineage reconstitution potential) and HSCs form in three waves, as described below. Runx1 is important for the differentiation of all embryonic blood cell lineages, and is particularly essential for the differentiation of blood cells in the second two waves from hemogenic endothelium.
Primitive hematopoiesis-the first wave
Primitive hematopoietic cells are one of the earliest functional cell populations to appear during embryogenesis. They emerge in the extraembryonic yolk sac shortly after gastrulation, and prior to the onset of circulation, a functional vascular system, or the development of HSCs (Palis et al., 1999, Ferkowicz and Yoder, 2005). Primitive hematopoietic cells in this first wave include unipotent primitive erythrocyte progenitors, bi-potent erythrocyte/megakaryocyte progenitors, and primitive macrophages (Xu M et al., 2001, Tober et al., 2007, Palis et al., 1999, Moore and Metcalf, 1970, Haar and Ackerman, 1971, Tracey et al., 1998). These primitive blood cells have distinct morphological and functional features compared to their “definitive” counterparts that form during the second and third waves of hematopoiesis. For example, primitive erythrocytes are larger than definitive erythrocytes, they express embryonic and adult globins, and they retain their nucleus when entering the circulation (Palis et al., 1999, Palis, 2014, Kingsley et al., 2004). Primitive megakaryocytes have a lower ploidy class than definitive megakaryocytes, and more rapidly produce platelets that prevent hemorrhaging in the primitive vascular plexus as it develops into a functional cardiovascular system (Xu M et al., 2001, Potts et al., 2014).
The mesodermal cells that give rise to primitive hematopoietic cells originate from a population of proximal epiblasts that migrate through the primitive streak and into the extraembryonic yolk sac early during gastrulation (Lawson et al., 1991). The mesoderm accumulates to form thickened regions called mesodermal masses that then differentiate into blood islands consisting of primitive erythroblasts, and into angioblasts that will form the vascular plexus of the yolk sac (Ferkowicz and Yoder, 2005, Haar and Ackerman, 1971). Due to their parallel development and close physical association, it was initially hypothesized that blood and endothelial cells in the yolk sac shared a common progenitor called the hemangioblast (Murray, 1932, Sabin, 1920). This idea was supported by the demonstration that hemangioblast-like progenitors that gave rise to both blood and endothelial cells could be isolated from embryonic stem (ES) cell cultures (Choi et al., 1998, Zambidis et al., 2005). It later became apparent that the putative bi-potent hemangioblast was actually a tri-lineage progenitor that could also give rise to smooth muscle cells (Ema et al., 2003). However, in vivo clonal analyses provided evidence against the existence of a bi-potential hemangioblast in the yolk sac, and instead suggested that yolk sac endothelium and hematopoietic cells are derived from adjacent but independent regions of the epiblast, and are thus specified prior to entering the primitive streak (Padron-Barthe et al., 2014, Ueno and Weissman, 2006).
Runx1 is expressed in the mesodermal mass in the yolk sac, and in the progenitors of primitive hematopoietic cells in the mouse embryo with the exception of primitive erythrocytes that initially express Runx1 but rapidly downregulate its expression shortly after emergence (North et al., 1999, Zeigler et al., 2006, Lacaud et al., 2002). Two of the three primitive hematopoietic lineages, primitive erythrocytes and megakaryocytes can form in the absence of Runx1, however their normal development is affected by Runx1 loss. Runx1-deficient embryos produce numbers of primitive erythroid colonies comparable to littermate controls and do not appear anemic (Yokomizo et al., 2008, Lacaud et al., 2002). However, more detailed analysis revealed reduced expression of cell surface Ter119 and the hematopoietic transcription factors KLF1 and GATA1, and defective maturation of Runx1-deficient erythrocytes (Yokomizo et al., 2008, Castilla et al., 1996). Furthermore, about 30% of primitive erythrocytes derived from Runx1−/− embryos displayed a deformed shape characterized by a rough punctate surface (Yokomizo et al., 2008). Despite these abnormalities Runx1−/− primitive erythrocytes are functional, as indicated by normal levels of benzidine staining (hemoglobinization) and the fact that Runx1−/− embryos survive until E12.5, which is longer than GATA1-deficient embryos, which die by E10.5 with severe anemia due to the lack of functional primitive erythrocytes (Yokomizo et al., 2008, Fujiwara et al., 1996, Okuda et al., 1996, Wang et al., 1996a). Runx1 is not required for the formation of primitive diploid megakaryocytes, although their numbers were lower in Runx1 deficient yolk sacs (Potts et al., 2014). Primitive macrophages, on the other hand, absolutely require Runx1, as they are lacking in Runx1−/− embryonic stem cell differentiation cultures (Lacaud et al., 2002) and embryos (Li et al., 2006). In summary, in the absence of Runx1, primitive macrophages are absent, diploid megakaryocytes are reduced in number, and primitive erythropoiesis is abnormal. Although it is often stated that Runx1 is required for definitive, but not primitive hematopoiesis, this is inaccurate as Runx1 is strictly required for the development of one primitive blood cell lineage, and important for the normal development of two others.
Runx1 has also been shown to play a role during primitive hematopoiesis in zebrafish and Xenopus embryos. In Xenopus embryos, Runx1 is expressed in the ventral blood island (VBI), which is analogous to mouse yolk sac blood islands (Tracey et al., 1998). Inhibiting Runx1 function via the injection of a dominant negative form of Runx1 mRNA prior to the VBI stage drastically reduced the number of Benzidine+ primitive erythrocytes (Tracey et al., 1998). Similarly, in zebrafish embryos, morpholino knockdown of Runx1 expression at the one to eight cell stage resulted in fewer primitive erythrocytes (Kalev-Zylinska et al., 2002). The primitive macrophage and megakaryocyte populations were not examined in either species. The decrease in primitive erythrocytes in both zebrafish and Xenopus embryos is contrary to what is observed in the mouse and suggests that Runx1 plays a more essential role in primitive erythropoiesis during zebrafish and Xenopus development.
Definitive hematopoiesis-the second and third waves
The term “definitive” in the context of developmental hematopoiesis has several meanings, but was originally used to describe adult erythrocytes, which unlike primitive erythrocytes are small and concave, lose their nuclei before entering the circulation, and do not express embryonic globin (Palis et al., 1999, Palis, 2014, Kingsley et al., 2004). Defined this way, definitive hematopoiesis encompasses two overlapping waves of blood development. Wave 2 is characterized by the generation of erythro-myeloid progenitors (EMPs) and lymphoid progenitors in the yolk sac and embryo proper (Yoder, 2014). EMPs can be found as early as E8.25 in the murine yolk sac (Palis et al., 1999, McGrath et al., 2015) and heart (Nakano et al., 2013). The next wave 2 progenitor to appear are lymphoid progenitors, which are found at E9.5 in the yolk sac and the paired dorsal aorta, and by E10.5 in the umbilical artery (UA) and vitelline artery (VA) (Yoshimoto et al., 2011, Yoshimoto et al., 2012). Adult repopulating HSCs (wave 3) do not appear until E10.5; they are generated initially in the dorsal aorta (DA), UA, and VA, and can subsequently be found in the yolk sac, head and placental vasculature (Li et al., 2012, Li et al., 2016, Rhodes et al., 2008, Gordon-Keylock et al., 2013, de Bruijn et al., 2000, Gekas et al., 2005). They are thought to arrive via circulation in these latter sites, instead of being generated in situ (Dieterlen-Lievre, 1975, Cumano et al., 2001, Medvinsky and Dzierzak, 1996).
Definitive hematopoietic cells are derived from a population of epithelial cells called hemogenic endothelium (HE) that are part of the interior lining of specific blood vessels in the embryo (Swiers et al., 2013b). HE is a transient population that gives rise to hematopoietic progenitors and stem cells in a process termed the endothelial to hematopoietic transition (EHT) (Kissa and Herbomel, 2010). Live-imaging studies of HE cells in vitro and in vivo have captured this dynamic process (Kissa and Herbomel, 2010, Boisset et al., 2010, Bertrand et al., 2010, Lancrin et al., 2009). Initially, HE cells appear flat in images generated by confocal microscopy, and integrated in the endothelial monolayer. Scanning electron microscopy of mouse HE cells revealed them to be more oblong, with rounded cell bodies and filopodia-like protrusions of the membrane as compared to non-hemogenic endothelial cells (Bos et al., 2015). As the EHT progresses, the HE cell bends away from the vessel wall until it rounds up and detaches from the endothelial layer becoming a mobile hematopoietic cell (Kissa and Herbomel, 2010, Boisset et al., 2010, Bertrand et al., 2010, Lancrin et al., 2009, Eilken et al., 2009).
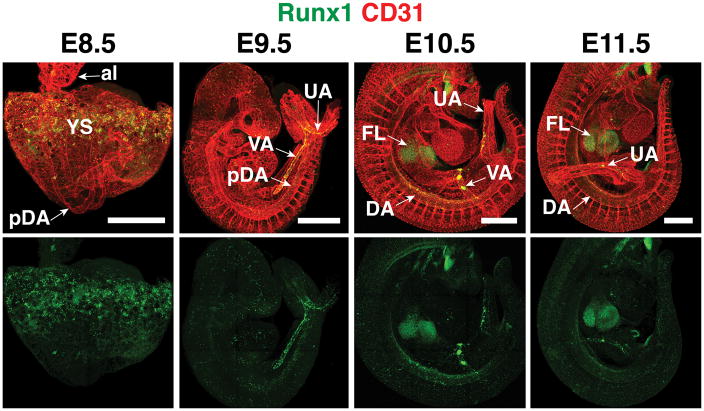
In mouse embryos, HE is localized in the yolk sac, the large arteries of the embryo proper, the heart, and the chorionic plexus (Rhodes et al., 2008, Li et al., 2012, Nakano et al., 2013, Yzaguirre and Speck, 2016). HE cells are identified based on Runx1 expression (North et al., 1999) (Figure 1). Runx1 is a critical regulator of the EHT and as such, suppresses an endothelial transcriptional program and initiates a hematopoietic program in HE allowing the EHT to occur (Lancrin et al., 2012, Chen et al., 2009, North et al., 1999, Yokomizo et al., 2001). Transcriptional and functional analyses demonstrated that HE cells derived from E8.5 mouse embryos preferentially form endothelial tubules in culture conditions that support both endothelial and hematopoietic cells (Swiers et al., 2013a). In contrast, E10.5 HE preferentially forms hematopoietic cells in vitro. The functional change that occurs between E8.5 and E10.5 was accompanied by a transcriptional shift characterized by the upregulation of hematopoietic factors such as Runx1, Meis1, Gata2, Gata3 and Myb suggesting that initially HE cells are functional endothelial cells, but as the hematopoietic program ramps up during midgestation HE loses endothelial function and gains hematopoietic potential (Swiers et al., 2013a).
Figure 1. Location of Runx1 expression and hemogenic endothelium in the mouse embryo.
Confocal Z-projections of mouse embryos between embryonic day (E) 8.5 and E11.5 immunostained for the endothelial and hematopoietic marker CD31 (red) and Runx1 (green). Runx1 is expressed in hemogenic endothelium in the yolk sac (YS) at E8.5. At E9.5, Runx1 expression is prominent in the vitelline artery (VA) and umbilical artery (UA). An E10.5 embryo (head removed) shows Runx1 protein in the vitelline artery, umbilical artery, dorsal aorta (DA), and the site of colonization, the fetal liver (FL). al, allantois ; pDA, paired dorsal aorta. Scale bar = 500μm.
In mammalian embryos, after the EHT occurs, newborn hematopoietic cells adhere to the vessel wall within the lumen forming clusters of hematopoietic cells. The peak of EHT in the mouse embryo (E10.5) is marked by the formation of hundreds of Kit+ hematopoietic clusters within the lumens of the DA, VA and UA and dozens residing within the vascular plexus of the yolk sac (Yokomizo and Dzierzak, 2010, Frame et al., 2015, Yzaguirre and Speck, 2016). Analysis of the Kit+ hematopoietic clusters within the embryo proper between E10.5 and E11.5 has revealed that they consist of lymphoid progenitors, a small number of myeloid progenitors, and pre-HSCs that can mature into HSCs capable of long-term multilineage reconstitution (Boisset et al., 2015, Li et al., 2014, Taoudi et al., 2008, Rybtsov et al., 2011). By E12.5 most hematopoietic cluster cells have entered the circulation and made their way to the fetal liver where they undergo maturation and proliferation, expanding the pool of HSCs and hematopoietic progenitors (Ema and Nakauchi, 2000, Kieusseian et al., 2012, Rybtsov et al., 2016). Beginning at E17.5 HSCs migrate to the bone marrow where they will reside throughout the lifetime of the animal (Christensen et al., 2004).
In zebrafish embryos the EHT occurs away from the lumen of the dorsal aorta, and the newly formed hematopoietic cells must traverse through the subaortic space and enter circulation via the axial vein (Kissa et al., 2008). Once in circulation hematopoietic cells migrate to the caudal hematopoietic tissue that is akin to mammalian fetal liver where they differentiate and expand before traveling to definitive hematopoietic organs (Murayama et al., 2006).
Runx1 is required during definitive hematopoiesis
Runx1 is expressed in all sites of blood formation. During gastrulation Runx1 is expressed in the extraembryonic mesoderm that gives rise to primitive hematopoietic cells (Swiers et al., 2013a, Lacaud et al., 2002, Zeigler et al., 2006). During definitive hematopoiesis Runx1 is the most reliable marker of hemogenic endothelium and is expressed by all hematopoietic cells with the exception of erythrocytes (North et al., 2004, North et al., 1999, North et al., 2002, Lorsbach et al., 2004). In addition to hematopoietic tissues, Runx1 is expressed in the olfactory epithelium, spinal ganglia, maxillary processes and the mesenchyme that flanks the ventral length of the dorsal aorta (North et al., 1999, Levanon et al., 2001a). Germline deletion of Runx1 results in the elimination of all definitive hematopoietic cells and embryonic lethality by E12.5 (Wang et al., 1996a, Okuda et al., 1996). Embryonic lethality of Runx1 deficient embryos is due to hemorrhaging within the ventricle of the central nervous system, the pericardial space, and the peritoneal cavity (Okuda et al., 1996, Wang et al., 1996a). The hemorrhaging is likely secondary to the lack of definitive hematopoietic cells because hematopoietic cells are involved in vascular remodeling during embryogenesis. For example, hematopoietic cells express angiopoietin-1 (Ang-1), a chemoattractant that promotes blood vessel sprouting (Witzenbichler et al., 1998). Analysis of the vasculature of Runx1 deficient embryos revealed decreased branching in the head, pericardium and vitelline artery in the yolk sac (Takakura et al., 2000). When Runx1 deficient explants were supplemented with hematopoietic cells or Ang-1 the vascular defects were rescued, suggesting that the vascular defects that cause hemorrhaging in Runx1 deficient embryos are due to the loss of Ang-1 expressing definitive hematopoietic cells (Takakura et al., 2000).
Runx1 can bind DNA as a monomer in vitro, but when Runx1 heterodimerizes with its non-DNA binding subunit CBFβ, flexible DNA-recognition loops in Runx1 are stabilized and its binding affinity for DNA increases (see Bushweller and Tahirov, this volume). Embryos deficient for CBFβ died by E12.5 with hemorrhaging akin to Runx1 deficient embryos and had significantly fewer definitive hematopoietic progenitors in their fetal livers when compared to littermate controls (Sasaki et al., 1996, Bresciani et al., 2014, Wang et al., 1996b, Niki et al., 1997). Similar results were obtained in CBFβ deficient zebrafish, confirming that CBFβ is required for Runx1 to function during definitive hematopoiesis (Sasaki et al., 1996, Bresciani et al., 2014). Interestingly, definitive hematopoiesis is not completely blocked in CBFβ deficient embryos to the same extent as in Runx1 deficient embryos. For example, the hematopoietic-specific transcription factor c-Myb is not expressed at sites of definitive hematopoiesis in Runx1 deficient zebrafish embryos, but it is expressed in the dorsal aorta of CBFβ deficient zebrafish embryos (Bresciani et al., 2014). Furthermore, definitive erythroid and myeloid progenitors are never found within the fetal livers of Runx1 deficient embryos but small numbers (approximately 40-fold less than wildtype controls) are present in CBFβ deficient fetal livers (Wang et al., 1996b). These studies suggest that the low-affinity binding of Runx1 to DNA in the absence of CBFβ is enough to initiate definitive hematopoiesis but is not sufficient to supply enough definitive hematopoietic cells to prevent embryonic lethality.
Unlike Runx1 and CBFβ deficient embryos, Runx1 heterozygous embryos survive well into adulthood and have relatively minor defects in hematopoietic development. There are fewer erythroid/myeloid progenitors in the yolk sacs, fetal livers and aorta/gonad/mesonephros regions of Runx1+/− embryos compared to wild type littermate controls (Wang et al., 1996b, Wang et al., 1996a, Mukouyama et al., 2000). Unexpectedly, the development of HSCs in Runx1+/− embryos is accelerated and spatially shifted (Cai et al., 2000). Specifically, HSCs were readily detected in the E10.5 AGM, and could also be detected in the yolk sacs of Runx1+/− embryos at E10.5 (Cai et al., 2000). This is in contrast to wild type embryos in which very few HSCs are present in the E10.5 dorsal aorta and are found in the yolk sac approximately 24 hours later (Müller et al., 1994). Therefore, reduced Runx1 dosage suppresses definitive hematopoiesis (wave 2) and changes the spatial and temporal development of HSCs (wave 3). The mechanism behind the temporal and spatial shift in HSC development associated with Runx1 heterozygosity is not known, but a subsequent study on the differentiation of Runx1 heterozygous embryonic stem (ES) cells provided a clue. The commitment of Runx1+/− ES cells to hemangioblasts, and subsequently to hematopoietic lineages was found to be accelerated by approximately 12 hours compared to that of wildtype ES cells (Lacaud et al., 2004). Therefore the acceleration in HSC formation may originate at a very early stage in hematopoietic development, in the formation of the tri-lineage hemangioblast, in which Runx1 is expressed (Lacaud et al., 2002).
Runx1 is required in hemogenic endothelium for the development of definitive hematopoietic cells
The studies of Runx1 and CBFβ knockout mice demonstrated that Runx1 and CBFβ are essential for definitive hematopoiesis but did not pinpoint when and in which cell population Runx1 is necessary. The observation that Runx1 is expressed in endothelial cells at all sites of hematopoietic cluster formation led to the hypothesis that Runx1 is required for the transition from endothelial to definitive hematopoietic cells. To test this hypothesis Runx1 was ablated in endothelial cells via endothelial specific Cre-recombinase mediated excision, which led to the complete abrogation of definitive hematopoiesis and embryonic lethality by E13.5 (Li et al., 2006, Chen et al., 2009). Also, endothelium sorted from the yolk sac and embryo proper of E10.5 Runx1−/− mice could not generate hematopoietic cells when plated on an OP9 stromal cell layer in conditions that support EHT (Yokomizo et al., 2001). These findings suggest that Runx1 expression is required in endothelial cells for the de novo generation of definitive hematopoietic cells. This point was further supported by a study that took the reverse approach by restoring endogenous Runx1 expression in Tek-expressing endothelial cells in Runx1 reversible knockout mouse embryos (Liakhovitskaia et al., 2009). Restoration of Runx1 expression in endothelial cells was sufficient to rescue lymphoid lineages, myeloid lineages and HSCs, and prolonged the life of the embryos up until birth (Liakhovitskaia et al., 2009). Postnatal lethality of these mice likely resulted from the loss of Runx1 expression in non-hematopoietic tissues. In fact, Runx1 null mice in which Runx1 expression is restored in only endothelial/hematopoietic cells have defects in neuronal differentiation and mineralization of the skull and sternum, demonstrating additional roles of Runx1 during development (Kobayashi et al., 2012, Liakhovitskaia et al., 2010).
After HE cells transition into hematopoietic cells they continue to express Runx1, which led to the hypothesis that Runx1 remains essential even after the EHT. However, conditional deletion of Runx1 in hematopoietic cells via Vav1-Cre, did not result in the ablation of EMPs or HSCs, nor did it affect embryonic or adult viability, indicating that Runx1 is not required in Vav1 expressing hematopoietic cells (Chen et al., 2009). However, Runx1 deletion in hematopoietic cells does cause defects that include thrombocytopenia and defective lymphopoiesis in adult mice (Chen et al., 2009, Ichikawa et al., 2004, Growney et al., 2005, Putz et al., 2006). Therefore, although Runx1 expression in hematopoietic cells is not essential for the generation and survival of definitive hematopoietic cells, it is required for lineage-specific differentiation and homeostasis. One caveat of this study is that Vav1-Cre is active in circulating and fetal liver hematopoietic cells but not in hematopoietic cluster cells within the dorsal aorta, leaving open the possibility that Runx1 is required for a short period after the EHT and before fetal liver colonization.
To more precisely determine the temporal requirement of Runx1 expression in hemogenic endothelium, Tober et al. conditionally deleted Runx1 during 24-hour intervals between E7.5 and E11.5 using a tamoxifen-inducible endothelial-specific Cre driven from vascular endothelial cadherin (Cdh5) regulatory sequences (Cdh5-CreERT) then assessed hematopoiesis (Tober et al., 2013). They found that when Runx1 was deleted between E8.25 –E9.25 that EMP numbers were dramatically reduced, indicating that Runx1 is critical in that time frame for the formation of EMPs from hemogenic endothelium. On the other hand deletion between E9.0 – E10.0 had no effect on EMP numbers, indicating that by E10.0 the requirement for Runx1 in hemogenic endothelium for the majority of EMP formation has ended. In contrast, the de novo development of HSCs was dependent on Runx1 expression in the endothelium up until E11.5. Thus, the requirement for Runx1 expression in HE for the development of EMPs and HSCs is temporally uncoupled, which is consistent with the sequential development of EMPs and HSCs during embryogenesis. This study however, did not determine if Runx1 was required in hematopoietic cluster cells because although vascular endothelial cadherin protein (CD144) is expressed on the surface of hematopoietic cluster cells, Cdh5 mRNA is downregulated 6-fold in hematopoietic cluster cells (Tober et al., 2013, North et al., 2002, Jaffredo et al., 2005, Fraser et al., 2003). It was unclear whether CreERT protein levels correlated with Cdh5 mRNA or vascular endothelial cadherin protein levels, and was present and active in hematopoietic cluster cells.
The molecular basis for the transient requirement for Runx1 was explored in a mouse embryonic stem (ES) cell model (Hoogenkamp et al., 2009, Lichtinger et al., 2012). Using a Runx1−/− mouse ES cell line expressing inducible Runx1, Hoogenkamp et al. demonstrated that Runx1 bound to an upstream regulatory element (URE) of the Spi1, encoding the hematopoietic transcription factor Pu.1. Spi1 is a downstream Runx1 target that is required for myelopoiesis. Runx1 initiated chromatin unfolding in the Spi1 URE at the onset of hematopoietic development (Hoogenkamp et al., 2009). Furthermore, using ChIP and in vivo footprinting they found that weak and transient binding of Runx1 to the URE was sufficient to establish stable transcription factor complexes at cis-regulatory elements that could sustain Spi1 expression even after removal of Runx1 (Hoogenkamp et al., 2009).
Genome-wide analysis by the same group using the same ES cell differentiation model compared ES-derived HE cells before and after the induction of Runx1 (Lichtinger et al., 2012). They found that after Runx1 induction in HE, Runx1 bound to sites that contained little or no H3K9Ac and subsequently strongly increased H3K9Ac levels, illustrating that Runx1 does not require high levels of active chromatin marks to bind to its target sites, but once bound can induce chromatin activation. Furthermore, Runx1 was shown to recruit hematopoietic regulators, SCL/TAL1 and FLI1 to target sites in HE cells to activate a hematopoietic transcriptional program (Lichtinger et al., 2012). This study illustrates Runx1’s ability to orchestrate a hematopoietic-specific program in HE by changing the binding profiles of hematopoietic regulators and insuring proper progression through the EHT.
As hemogenic endothelial cells begin to transition into hematopoietic cells, one of the earliest hematopoietic markers to be expressed is the αIIb integrin subunit CD41 (Mikkola et al., 2003). A subset of endothelial cells in the dorsal aorta of Runx1 deficient embryos express CD41, suggesting that in the absence of Runx1, hemogenic endothelium is at least partially specified and can switch on hematopoietic gene expression (Liakhovitskaia et al., 2014). To determine if Runx1 expression close to the onset of EHT is sufficient for generating definitive hematopoietic cells, Liakhovitskaia et al., restored Runx1 expression in CD41+ cells in Runx1 deficient embryos via CD41 (Itga2b)-Cre (Liakhovitskaia et al., 2014). Restoring Runx1 expression in CD41+ cells rescued the generation of HSCs, and the embryos survived until birth, suggesting that Runx1 is required and sufficient for the progression of CD41+ cells into HSCs (Liakhovitskaia et al., 2014). CD41+ cells isolated from wild type mouse embryos or embryonic stem cell cultures can give rise to hematopoietic cells but cannot generate endothelial progenitors, indicating that CD41+ cells are committed to the hematopoietic lineage (Hashimoto et al., 2007, Li et al., 2005). The finding that restoring Runx1 expression in CD41+ (Itga2b-Cre expressing) cells can rescue HSCs suggests that Runx1 is not required until the endothelial to hematopoietic transition is initiated and hematopoietic fate has been cemented. However, transcriptional analysis of hemogenic and non-hemogenic endothelial cells isolated from E8.5 embryos revealed that while CD41 protein at the surface of either cell population is low to non-existent at E8.5, both hemogenic and non-hemogenic endothelial cells express Itga2b mRNA (Swiers et al., 2013a). Therefore the Itga2b-Cre used by Liakhovitskaia et al. (Liakhovitskaia et al., 2014) may have restored Runx1 expression in all endothelium at E8.5 rather than specifically in HE cells initiating the EHT. Thus, it is formally possible that Runx1 expression in Itga2b-expressing endothelial cells earlier in development, prior to the onset of EHT, is necessary for the de novo generation of definitive hematopoietic cells.
Regulation of Runx1 expression during the specification of hemogenic endothelium
Although Runx1 is required for the successful transition of HE cells into hematopoietic cells it is not required for the specification of hemogenic endothelium. This was perhaps best illustrated in live-imaging studies of EHT in Runx1 deficient zebrafish embryos. In Runx1 morphant zebrafish embryos, HE cells bend away from the endothelial monolayer, initiating the EHT, but fragment before forming a hematopoietic cell (Kissa and Herbomel, 2010, Zhen et al., 2013), a phenomenon that was also observed in Runx1−/− mouse ES-derived HE cells (Lancrin et al., 2009, Eilken et al., 2009). Furthermore, as mentioned above CD41 is expressed by HE cells in the DA of E10.5 Runx1−/− mouse embryos, indicating that the hematopoietic program is at least partially initiated in the absence of Runx1 expression (Liakhovitskaia et al., 2014).
Although Runx1 is not required for the specification of HE it was proposed to play a role in determining cell fate in mesoderm-derived progenitors. Etv2+ Flk1+ mesodermal cells give rise to both endothelial cells and blood (Kataoka et al., 2011, Wareing et al., 2012). Whether the Etv2+ Flk1+ mesodermal progenitor gives rise to a non-hemogenic endothelial cell or a HE cell was recently reported to be controlled, at least in part, by Runx1 (Eliades et al., 2016). At E7.5, Runx1+ Etv2+ Flk1+ cells reside within the extraembryonic yolk sac and co-express mesodermal and endothelial specific markers. At E8.5, a subset of Etv2+ cells migrate from the area at the boundary of the yolk sac and embryo proper into the embryo proper and downregulate mesoderm-specific genes (Eliades et al., 2016). A similar observation was made by Tanaka et al., who reported that between E7.5 and E8.5 Runx1+ Gata1− cells located at the boundary between the extraembryonic yolk sac and the embryo proper, migrate to the embryo proper where they contribute to the intraembryonic vasculature and blood (Tanaka et al., 2014). Interestingly, the Etv2+ population at E7.5 expresses Runx1 and has hemogenic potential, likely representing at least in part the yolk sac blood island cells. At E8.5, in contrast most Etv2+ cells do not express Runx1, and lack hematopoietic potential, from which it was suggested that Runx1 is silenced in the majority of Etv2+ cells between E7.5 and E8.5 (Eliades et al., 2016). The mechanism of silencing involves Bmi1, a member of the Polycomb Repressive Complex 1 (PRC1) (Eliades et al., 2016), which physically interacts with Runx1 (Yu et al., 2012). Ectopic expression of Runx1, or inhibition of PRC1 conferred hemogenic potential to the E8.5 Etv2+ population, suggesting that the hemogenic potential of the E8.5 Etv2+ population is restricted through Runx1 silencing (Eliades et al., 2016). These results demonstrate that the default program in Etv2+ Flk1+ progenitors may be the hematopoietic program, initiated by Runx1. Bmi1 then represses Runx1 expression at E8.5 to promote a vascular fate.
Silencing of Runx1 expression in endothelium is also mediated through the homeobox protein, HoxA3. During hematopoietic development the expression of Runx1 and HoxA3 in the endothelium is mutually exclusive, in part because HoxA3 directly interacts with and represses Runx1 expression (Iacovino et al., 2011). Ectopic expression of HoxA3 during ES cell differentiation and in cells derived from E10.5 mouse embryos resulted in the downregulation of hematopoietic markers and inhibited hematopoietic specification, and increased the expression of endothelial-specific genes, suggesting that HoxA3 reinforces an endothelial fate while suppressing the hematopoietic potential of endothelial progenitors (Iacovino et al., 2011). Interestingly, when Runx1 is ectopically expressed in HoxA3-induced ES-derived endothelial progenitor cells the expression of hematopoietic genes is rescued, indicating that high levels of Runx1 can override HoxA3 activity (Iacovino et al., 2011).
Does Runx1 function as a master regulator of hematopoiesis?
The term “master regulator” is often used to describe a gene that sits at the very top of a regulatory hierarchy. However a stringent test of a master regulator is whether it can reprogram one cell type into another (Chan and Kyba, 2013). Logically, the most likely cells that would respond to direct reprogramming by Runx1 are endothelial cells. However direct reprogramming studies have shown that Runx1 alone is not sufficient to reprogram either human umbilical vein endothelial cells (HUVECs) or human adult dermal endothelial cells (hDMECs) into hematopoietic progenitor cells (Sandler et al., 2014). Only when Runx1 was combined with Spi1, Fosb and Gfi1 could relatively efficient reprogramming of endothelial cells be achieved (Sandler et al., 2014). Interestingly, both Spi1 (Pu.1) and Gfi1 are direct downstream targets of Runx1 (Lancrin et al., 2012, Huang et al., 2008, Hoogenkamp et al., 2009), but when they were individually removed from a transduction cocktail containing all four transcription factors the efficiency of reprogramming significantly decreased, suggesting that ectopic Runx1 alone was unable to efficiently drive their expression (Sandler et al., 2014). Therefore, by this strict definition Runx1 is not a master regulator, as it is not by itself sufficient to reprogram HUVECs or hDMECs into blood cells. The reason for this is unclear, but may be because Runx1 cannot access various downstream targets in specific endothelial subtypes. Endothelial cells of different tissues and developmental stages are diverse in function, phenotype, transcription and chromatin state (Nolan et al., 2013, Aird, 2012, Chi et al., 2003, Casanello et al., 2014), therefore it would be interesting to determine if other endothelial subtypes are more permissive to respecification by Runx1. Runx1 can induce a hematopoietic program in E8.5 Etv2+ endothelial cells, therefore the ability of endothelial cells to respond to Runx1 activity may be lost as development proceeds (Eliades et al., 2016).
Downstream targets of Runx1 that regulate the EHT
In order to transition morphologically and functionally into hematopoietic cells, HE cells must extinguish their endothelial-specific transcriptional program and upregulate a hematopoietic program; a transcriptional switch that is largely orchestrated by Runx1. Two direct targets of Runx1, Gfi1 and Gfi1b, encode nuclear zinc finger transcriptional repressors that inhibit expression of endothelial genes in HE during the EHT (Lancrin et al., 2012). In Gfi1/Gfi1b deficient mouse embryos, HE cells in the yolk sac fail to transition morphologically into hematopoietic cells and remain locked in the endothelial layer (Lancrin et al., 2012). However, dissociation of Gfi1/Gfi1b deficient yolk sac frees the hematopoietic cells, which can then form hematopoietic colonies in clonogenic assays, suggesting that Gfi1/Gfi1b deficient HE cells can form functional hematopoietic progenitors but are unable to physically transition into a morphological hematopoietic cell (Lancrin et al., 2012). Conversely, when Gfi1 and Gfi1b expression was induced in Runx1−/− embryonic stem cell derived-HE, the HE cells could undergo the morphological transition into rounded cells but the round cells could not form colonies in hematopoietic clonogenic assays, thus illustrating that during the EHT Gfi1 and Gfi1b repress an endothelial fate allowing for the morphological transition of flat HE cells into rounded hematopoietic cells (Lancrin et al., 2012). Interestingly, a subsequent study found that hematopoietic clusters did not form in the arteries of Gfi1/Gfi1b deficient embryos, and Gfi1 expressing cells remained within the endothelial layer. However, unlike in the yolk sac, dissociated cells from the arteries could not differentiate into hematopoietic colonies, indicating that Gfi1 and Gfi1b have functions in blood cell formation in the major arteries in addition to their requirement for the EHT (Thambyrajah et al., 2016).
Identifying the transcriptional program regulated by Runx1 in hemogenic endothelium is challenging because HE is a rare population that exists only transiently during midgestation. To overcome these challenges Lie-A-ling et al. (Lie-A-Ling et al., 2014) used an alternative technique to chromatin immunoprecipitation called DNA adenine methyltransferase identification (DamID). DamID relies on the fusion of a transcription factor (such as Runx1) to the Escherichia coli DNA adenine methyltransferase (Dam). When the transcription factor binds DNA the fused Dam protein adds stable methylation tags to adenines within nearby GATC sequences allowing for identification of transcription factor binding sites without the need for antibodies (Lie-A-Ling et al., 2014). To identify Runx1 targets in HE, Lie-A-Ling et al. established Runx1−/− ES cell lines containing doxycycline inducible Runx1-Dam, and then differentiated the ES cells into HE. Fortuitously, the inducible system was leaky, allowing for low levels of Runx1 expression in the absence of doxycycline that were not sufficient for EHT, but were sufficient for the detection of Runx1 occupancy by DamID (Lie-A-Ling et al., 2014). Comparison of the Runx1-DamID methylation and RNA-Seq datasets led to the identification of 235 genes that were both bound by Runx1 and differential expressed in HE cells generated from wild type and Runx1−/− ES cells (Lie-A-Ling et al., 2014). The expression of 80 of the genes was negatively correlated with Runx1 occupancy and 155 genes were positively correlated (Lie-A-Ling et al., 2014), consistent with Runx1’s ability to function as a transcriptional repressor or activator in the same cell type (Canon and Banerjee, 2003). The target genes that were positively correlated with Runx1 expression were associated with cell adhesion, integrin signaling, cellular movement and interaction with the extracellular matrix (Lie-A-Ling et al., 2014). Interestingly, very few hematopoietic genes were identified as Runx1 targets, suggesting that the HE was in an early stage of differentiation and had not yet initiated a hematopoietic-specific program. Thus an early function of Runx1 in HE is to regulate the expression of genes involved in the activation of migration and adhesion of HE cells prior to the EHT.
Transcriptional and translation regulation of Runx1 expression during embryonic hematopoiesis
The spatio-temporal specific expression pattern of Runx1 during embryonic hematopoiesis is controlled, in part, through transcriptional regulation. In vertebrates, Runx1 is transcribed from two alternative promoters, the distal (P1) promoter and the proximal (P2) promoter (Ghozi et al., 1996, Rennert et al., 2003, Levanon et al., 2001b, Bee et al., 2009b, Telfer and Rothenberg, 2001). The P2 promoter differs from the P1 promoter in that it is associated with a large CpG island that may influence differential regulation of P1 versus P2 transcription (Levanon et al., 2001b, Bee et al., 2009b). Furthermore, the conserved binding sites associated with each promoter are different; P1 contains a cAMP-responsive element, a CCAAT box, GATA, SMAD and RUNX motifs whereas P2 contains CCATT boxes, initiator sequences, a GC-box, OCT and ETS motifs (Ghozi et al., 1996, Bee et al., 2009a, Bee et al., 2010, Martinez et al., 2016). Differential promoter usage in addition to RNA splicing leads to a vast array of Runx1 isoforms. The full-length isoforms generated from the P1 and P2 promoters are referred to as Runx1c and Runx1b, respectively. Runx1c (465aa) is larger than Runx1b (451aa) due to a difference of 19aa at their N-termini, but there are no data that suggest these differences confer distinct properties to the Runx1c and Runx1b proteins (Fujita et al., 2001, Challen and Goodell, 2010).
Differential promoter usage during hematopoiesis does, however, control the timing and level of Runx1 expression. Analysis of Runx1 promoter activity in mouse embryos and ES cell differentiation models revealed that the P2 promoter is dominant early during primitive hematopoiesis and at the onset of definitive hematopoiesis, whereas P1 activity ramps up later in development during fetal liver and bone marrow hematopoiesis (Bee et al., 2009b, Bee et al., 2010, Fujita et al., 2001, Pozner et al., 2007, Sroczynska et al., 2009). In mice, abrogation of the P2 promoter via insertion of a neomycin resistance gene resulted in fewer hematopoietic clusters in the large arteries of the embryo proper, significantly fewer hematopoietic progenitors in the fetal liver and yolk sac, reduced thymopoiesis, and perinatal lethality (Pozner et al., 2007, Bee et al., 2010). The prolonged survival of P2-attenuated mice compared to Runx1 null mice (E12.5 lethality), is likely due to the overlap of P1 and P2 activity in hemogenic endothelium, and therefore P1 promoter activity alone promotes the de novo generation of sufficient numbers of definitive hematopoietic cells to prevent embryonic lethality (Sroczynska et al., 2009, Bee et al., 2009b). Loss of P1 promoter activity, on the other hand, is less detrimental than P2 loss. P1-null mouse embryos have fewer hematopoietic clusters and produce fewer hematopoietic progenitors in the yolk sac and large arteries of the embryo proper compared to littermate controls, but the decrease in hematopoietic cells is not as severe as that caused by P2 attenuation, and loss of P1 is not lethal (Bee et al., 2010). However, the bone marrow and peripheral blood of P1-null adult mice does exhibit a significant decrease in white blood cells and platelets and an increase in the percentage of bone marrow HSCs and hematopoietic progenitors (Bee et al., 2010). Interestingly, one functional P2-deleted Runx1 allele in the absence of P1-activity was sufficient to rescue embryonic lethality, but one functional P1 allele in the absence of P2 was not, suggesting that the dosage and timing of Runx1 expression is critical for the generation of definitive hematopoietic cells (Bee et al., 2010, Pozner et al., 2007).
The P1 and P2 promoters regulate the timing and dosage of Runx1 during hematopoiesis but they do not confer tissue specificity in mammalian embryos (Ghozi et al., 1996, Bee et al., 2009a). Hematopoietic specific expression is mediated by enhancers located within and upstream of the Runx1 gene locus (Schutte et al., 2016). The best known of these is a 531 bp enhancer located between P1 and P2, 23.4kb downstream of the ATG in exon 1 of Runx1 (Nottingham et al., 2007, Ng et al., 2010). The +23 enhancer drives reporter expression at all sites of hematopoiesis in mouse embryos (Nottingham et al., 2007, Ng et al., 2010). Specifically, the +23 enhancer is active in hemogenic endothelium, hematopoietic clusters and fetal liver hematopoietic cells. It is not, however, active in non-hematopoietic tissues that express Runx1, such as the mesenchyme beneath the dorsal aorta (Nottingham et al., 2007, Ng et al., 2010).. ChIP analysis of the +23 enhancer demonstrated association with Gata2, Runx1, Ets transcription factors, and the SCL/Lmo2/Lbd-1 complex in a myeloid progenitor cell line (Nottingham et al., 2007). To determine if transcription factor binding was necessary for activity of the +23 enhancer in mouse embryos, Nottingham et al. assessed whether or not activity of the enhancer was disrupted after mutating the RUNX, ETS or GATA motifs. They found that the RUNX motif was not required for +23 enhancer activity but the ETS and GATA motifs were required, therefore the +23 enhancer confers hematopoietic specific expression of Runx1 and is regulated, in part, through interaction with Gata and Ets transcription factors (Nottingham et al., 2007).
Post-transcriptional control of Runx1 occurs through variations in translational efficiency and transcript attenuation via miRNAs. The translational efficiencies of P1 and P2-derived transcripts differ due to distinct 5′ untranslated regions (UTR). P1-derived transcripts have a relatively short 5′ UTR (452 bp) that directs efficient cap-dependent translation (Pozner et al., 2000). In contrast, P2-derived transcripts have a long 5′ UTR (1,631bp) containing an internal ribosomal entry site (IRES), which mediates cap-independent translation (Pozner et al., 2000). It has been proposed that P2-derived transcripts are poorly transcribed due to the length of the UTR and cis-acting elements within it, including the IRES as well as multiple upstream AUG codons and GC-rich islands (Pozner et al., 2000, Levanon et al., 1996). A possible explanation for the presence of both IRES and cap-dependent translation of Runx1 mRNA is that IRES-containing transcripts are translated during mitosis and under stress conditions when cap-dependent translation is impaired (Levanon and Groner, 2004). Further post-transcriptional regulation of Runx1 occurs through miRNA transcript attenuation. In addition to distinct 5′UTRs, Runx1 mRNA isoforms have different 3′ UTRs that range in size from 150 to 4,000 bp (Levanon et al., 2001b). Several putative miRNA binding sites were identified in the 3′ UTR of Runx1, and the length of the 3′UTR was shown to change the susceptibility to miRNA targeting and attenuation (Ben-Ami et al., 2009). Although the role that translational regulation of Runx1 plays during embryonic hematopoiesis has not been elucidated, it is plausible that it influences isoform, dose, timing and cell specific expression of Runx1 during development.
The past 10 years have seen the shaping of the roadmap of HSPC development from hemogenic endothelium: discrete cellular intermediates of the HSC lineage have been identified, along with the identification of distinct populations of HE giving rise to HPCs and HSCs. In addition, our understanding of the critical role Runx1 plays in this process has deepened with the identification of new target genes. The rapid developments in imaging and expression profiling technologies now enable taking the study of de novo HSPC generation to the single cell level, the level at which cell fate decisions are made. This will no doubt lead to more exciting insights into the role of the master regulator Runx1 in blood stem and progenitor cell generation.
References
- AIRD WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. doi: 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEE T, ASHLEY EL, BICKLEY SR, JARRATT A, LI PS, SLOANE-STANLEY J, GOTTGENS B, DE BRUIJN MF. The mouse Runx1 +23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood. 2009a;113:5121–4. doi: 10.1182/blood-2008-12-193003. [DOI] [PubMed] [Google Scholar]
- BEE T, LIDDIARD K, SWIERS G, BICKLEY SR, VINK CS, JARRATT A, HUGHES JR, MEDVINSKY A, DE BRUIJN MF. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol Dis. 2009b;43:35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- BEE T, SWIERS G, MUROI S, POZNER A, NOTTINGHAM W, SANTOS AC, LI PS, TANIUCHI I, DE BRUIJN MF. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–50. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- BEN-AMI O, PENCOVICH N, LOTEM J, LEVANON D, GRONER Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–43. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTRAND JY, CHI NC, SANTOSO B, TENG S, STAINIER DY, TRAVER D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–11. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOISSET JC, CLAPES T, KLAUS A, PAPAZIAN N, ONDERWATER J, MOMMAAS-KIENHUIS M, CUPEDO T, ROBIN C. Progressive maturation toward hematopoietic stem cells in the mouse embryo aorta. Blood. 2015;125:465–9. doi: 10.1182/blood-2014-07-588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOISSET JC, VAN CAPPELLEN W, ANDRIEU-SOLER C, GALJART N, DZIERZAK E, ROBIN C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–20. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- BOS FL, HAWKINS JS, ZOVEIN AC. Single-cell resolution of morphological changes in hemogenic endothelium. Development. 2015;142:2719–24. doi: 10.1242/dev.121350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRESCIANI E, CARRINGTON B, WINCOVITCH S, JONES M, GORE AV, WEINSTEIN BM, SOOD R, LIU PP. CBFβ and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood. 2014;124:70–8. doi: 10.1182/blood-2013-10-531988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAI Z, DE BRUIJN M, MA X, DORTLAND B, LUTEIJN T, DOWNING RJ, DZIERZAK E. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000;13:423–31. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- CANON J, BANERJEE U. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev. 2003;17:838–43. doi: 10.1101/gad.1064803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASANELLO P, SCHNEIDER D, HERRERA EA, UAUY R, KRAUSE BJ. Endothelial heterogeneity in the umbilico-placental unit: DNA methylation as an innuendo of epigenetic diversity. Front Pharmacol. 2014;5:49. doi: 10.3389/fphar.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASTILLA LH, WIJMENGA C, WANG Q, STACY T, SPECK NA, ECKHAUS M, MARÍN-PADILLA M, COLLINS FS, WYNSHAW-BORIS A, LIU PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–96. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- CHALLEN GA, GOODELL MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol. 2010;38:403–16. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAN SS, KYBA M. What is a Master Regulator? J Stem Cell Res Ther. 2013:3. doi: 10.4172/2157-7633.1000e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN MJ, YOKOMIZO T, ZEIGLER BM, DZIERZAK E, SPECK NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–91. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHI JT, CHANG HY, HARALDSEN G, JAHNSEN FL, TROYANSKAYA OG, CHANG DS, WANG Z, ROCKSON SG, VAN DE RIJN M, BOTSTEIN D, BROWN PO. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003;100:10623–8. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI K, KENNEDY M, KAZAROV A, PAPADIMITRIOU JC, KELLER G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–32. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- CHRISTENSEN JL, WRIGHT DE, WAGERS AJ, WEISSMAN IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2:E75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUMANO A, FERRAZ JC, KLAINE M, DI SANTO JP, GODIN I. Intraembryonic, but not yolk sac hematopoietic precursors, isolated before circulation, provide long-term multilineage reconstitution. Immunity. 2001;15:477–85. doi: 10.1016/s1074-7613(01)00190-x. [DOI] [PubMed] [Google Scholar]
- DE BRUIJN MF, SPECK NA, PEETERS MC, DZIERZAK E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–74. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIETERLEN-LIEVRE F. On the origin of haemopoietic stem cells in the avian embryo: an experimental approach. J Embryol Exp Morphol. 1975;33:607–19. [PubMed] [Google Scholar]
- EILKEN HM, NISHIKAWA S, SCHROEDER T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- ELIADES A, WAREING S, MARINOPOULOU E, FADLULLAH MZ, PATEL R, GRABAREK JB, PLUSA B, LACAUD G, KOUSKOFF V. The Hemogenic Competence of Endothelial Progenitors Is Restricted by Runx1 Silencing during Embryonic Development. Cell Rep. 2016;15:2185–99. doi: 10.1016/j.celrep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA H, NAKAUCHI H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–8. [PubMed] [Google Scholar]
- EMA M, FALOON P, ZHANG WJ, HIRASHIMA M, REID T, STANFORD WL, ORKIN S, CHOI K, ROSSANT J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–93. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERKOWICZ MJ, YODER MC. Blood island formation: longstanding observations and modern interpretations. Exp Hematol. 2005;33:1041–7. doi: 10.1016/j.exphem.2005.06.006. [DOI] [PubMed] [Google Scholar]
- FRAME JM, FEGAN KH, CONWAY SJ, MCGRATH KE, PALIS J. Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells. 2015 doi: 10.1002/stem.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRASER ST, OGAWA M, YOKOMIZO T, ITO Y, NISHIKAWA S. Putative intermediate precursor between hematogenic endothelial cells and blood cells in the developing embryo. Dev Growth Differ. 2003;45:63–75. doi: 10.1046/j.1440-169x.2003.00675.x. [DOI] [PubMed] [Google Scholar]
- FUJITA Y, NISHIMURA M, TANIWAKI M, ABE T, OKUDA T. Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem Biophys Res Commun. 2001;281:1248–55. doi: 10.1006/bbrc.2001.4513. [DOI] [PubMed] [Google Scholar]
- FUJIWARA Y, BROWNE CP, CUNNIFF K, GOFF SC, ORKIN SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–8. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEKAS C, DIETERLEN-LIEVRE F, ORKIN SH, MIKKOLA HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- GHOZI MC, BERNSTEIN Y, NEGREANU V, LEVANON D, GRONER Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci U S A. 1996;93:1935–40. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORDON-KEYLOCK S, SOBIESIAK M, RYBTSOV S, MOORE K, MEDVINSKY A. Mouse extraembryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood. 2013;122:2338–45. doi: 10.1182/blood-2012-12-470971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROWNEY JD, SHIGEMATSU H, LI Z, LEE BH, ADELSPERGER J, ROWAN R, CURLEY DP, KUTOK JL, AKASHI K, WILLIAMS IR, SPECK NA, GILLILAND DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAAR JL, ACKERMAN GA. A phase and electron microscopic study of vasculogenesis and erythropoiesis in the yolk sac of the mouse. Anat Rec. 1971;170:199–223. doi: 10.1002/ar.1091700206. [DOI] [PubMed] [Google Scholar]
- HASHIMOTO K, FUJIMOTO T, SHIMODA Y, HUANG X, SAKAMOTO H, OGAWA M. Distinct hemogenic potential of endothelial cells and CD41+ cells in mouse embryos. Dev Growth Differ. 2007;49:287–300. doi: 10.1111/j.1440-169X.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- HOOGENKAMP M, LICHTINGER M, KRYSINSKA H, LANCRIN C, CLARKE D, WILLIAMSON A, MAZZARELLA L, INGRAM R, JORGENSEN H, FISHER A, TENEN DG, KOUSKOFF V, LACAUD G, BONIFER C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG G, ZHANG P, HIRAI H, ELF S, YAN X, CHEN Z, KOSCHMIEDER S, OKUNO Y, DAYARAM T, GROWNEY JD, SHIVDASANI RA, GILLILAND DG, SPECK NA, NIMER SD, TENEN DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- IACOVINO M, CHONG D, SZATMARI I, HARTWECK L, RUX D, CAPRIOLI A, CLEAVER O, KYBA M. HoxA3 is an apical regulator of haemogenic endothelium. Nat Cell Biol. 2011;13:72–8. doi: 10.1038/ncb2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICHIKAWA M, ASAI T, SAITO T, SEO S, YAMAZAKI I, YAMAGATA T, MITANI K, CHIBA S, OGAWA S, KUROKAWA M, HIRAI H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- IVANOVS A, RYBTSOV S, WELCH L, ANDERSON RA, TURNER ML, MEDVINSKY A. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J Exp Med. 2011;208:2417–27. doi: 10.1084/jem.20111688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAFFREDO T, BOLLEROT K, SUGIYAMA D, GAUTIER R, DREVON C. Tracing the hemangioblast during embryogenesis: developmental relationships between endothelial and hematopoietic cells. Int J Dev Biol. 2005;49:269–77. doi: 10.1387/ijdb.041948tj. [DOI] [PubMed] [Google Scholar]
- KALEV-ZYLINSKA ML, HORSFIELD JA, FLORES MV, POSTLETHWAIT JH, VITAS MR, BAAS AM, CROSIER PS, CROSIER KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–30. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- KATAOKA H, HAYASHI M, NAKAGAWA R, TANAKA Y, IZUMI N, NISHIKAWA S, JAKT ML, TARUI H. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood. 2011;118:6975–86. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- KIEUSSEIAN A, BRUNET DE LA GRANGE P, BURLEN-DEFRANOUX O, GODIN I, CUMANO A. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 2012;139:3521–30. doi: 10.1242/dev.079210. [DOI] [PubMed] [Google Scholar]
- KINGSLEY PD, MALIK J, FANTAUZZO KA, PALIS J. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood. 2004;104:19–25. doi: 10.1182/blood-2003-12-4162. [DOI] [PubMed] [Google Scholar]
- KISSA K, HERBOMEL P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–5. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- KISSA K, MURAYAMA E, ZAPATA A, CORTES A, PERRET E, MACHU C, HERBOMEL P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–56. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- KOBAYASHI A, SENZAKI K, OZAKI S, YOSHIKAWA M, SHIGA T. Runx1 promotes neuronal differentiation in dorsal root ganglion. Mol Cell Neurosci. 2012;49:23–31. doi: 10.1016/j.mcn.2011.08.009. [DOI] [PubMed] [Google Scholar]
- LACAUD G, GORE L, KENNEDY M, KOUSKOFF V, KINGSLEY P, HOGAN C, CARLSSON L, SPECK N, PALIS J, KELLER G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100:458–66. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- LACAUD G, KOUSKOFF V, TRUMBLE A, SCHWANTZ S, KELLER G. Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood. 2004;103:886–9. doi: 10.1182/blood-2003-06-2149. [DOI] [PubMed] [Google Scholar]
- LANCRIN C, MAZAN M, STEFANSKA M, PATEL R, LICHTINGER M, COSTA G, VARGEL O, WILSON NK, MOROY T, BONIFER C, GOTTGENS B, KOUSKOFF V, LACAUD G. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–22. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- LANCRIN C, SROCZYNSKA P, STEPHENSON C, ALLEN T, KOUSKOFF V, LACAUD G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–5. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWSON KA, MENESES JJ, PEDERSEN RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- LEVANON D, BERNSTEIN Y, NEGREANU V, GHOZI MC, BAR-AM I, ALOYA R, GOLDENBERG D, LOTEM J, GRONER Y. A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol. 1996;15:175–85. doi: 10.1089/dna.1996.15.175. [DOI] [PubMed] [Google Scholar]
- LEVANON D, BRENNER O, NEGREANU V, BETTOUN D, WOOLF E, EILAM R, LOTEM J, GAT U, OTTO F, SPECK N, GRONER Y. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech Dev. 2001a;109:413–7. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- LEVANON D, GLUSMAN G, BANGSOW T, BEN-ASHER E, MALE DA, AVIDAN N, BANGSOW C, HATTORI M, TAYLOR TD, TAUDIEN S, BLECHSCHMIDT K, SHIMIZU N, ROSENTHAL A, SAKAKI Y, LANCET D, GRONER Y. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001b;262:23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- LEVANON D, GRONER Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–9. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- LI W, FERKOWICZ MJ, JOHNSON SA, SHELLEY WC, YODER MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- LI Y, ESAIN V, TENG L, XU J, KWAN W, FROST IM, YZAGUIRRE AD, CAI X, CORTES M, MAIJENBURG MW, TOBER J, DZIERZAK E, ORKIN SH, TAN K, NORTH TE, SPECK NA. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 2014;28:2597–612. doi: 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z, CHEN MJ, STACY T, SPECK NA. Runx1 function in hematopoiesis is required in cells that express Tek. Blood. 2006;107:106–10. doi: 10.1182/blood-2005-05-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z, LAN Y, HE W, CHEN D, WANG J, ZHOU F, WANG Y, SUN H, CHEN X, XU C, LI S, PANG Y, ZHANG G, YANG L, ZHU L, FAN M, SHANG A, JU Z, LUO L, DING Y, GUO W, YUAN W, YANG X, LIU B. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell. 2012;11:663–75. doi: 10.1016/j.stem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- LI Z, VINK CS, MARIANI SA, DZIERZAK E. Subregional localization and characterization of Ly6aGFP-expressing hematopoietic cells in the mouse embryonic head. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAKHOVITSKAIA A, GRIBI R, STAMATERIS E, VILLAIN G, JAFFREDO T, WILKIE R, GILCHRIST D, YANG J, URE J, MEDVINSKY A. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009;27:1616–24. doi: 10.1002/stem.71. [DOI] [PubMed] [Google Scholar]
- LIAKHOVITSKAIA A, LANA-ELOLA E, STAMATERIS E, RICE DP, VAN’T HOF RJ, MEDVINSKY A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010;340:539–46. doi: 10.1016/j.ydbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- LIAKHOVITSKAIA A, RYBTSOV S, SMITH T, BATSIVARI A, RYBTSOVA N, RODE C, DE BRUIJN M, BUCHHOLZ F, GORDON-KEYLOCK S, ZHAO S, MEDVINSKY A. Runx1 is required for progression of CD41+ embryonic precursors into HSCs but not prior to this. Development. 2014;141:3319–23. doi: 10.1242/dev.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LICHTINGER M, INGRAM R, HANNAH R, MÜLLER D, CLARKE D, ASSI SA, LIE-A-LING M, NOAILLES L, VIJAYABASKAR MS, WU M, TENEN DG, WESTHEAD DR, KOUSKOFF V, LACAUD G, GÖTTGENS B, BONIFER C. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–33. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIE-A-LING M, MARINOPOULOU E, LI Y, PATEL R, STEFANSKA M, BONIFER C, MILLER C, KOUSKOFF V, LACAUD G. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood. 2014;124:e11–20. doi: 10.1182/blood-2014-04-572958. [DOI] [PubMed] [Google Scholar]
- LORSBACH RB, MOORE J, ANG SO, SUN W, LENNY N, DOWNING JR. Role of Runx1 in adult hematopoiesis: analysis of Runx1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103:2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- MARTINEZ M, HINOJOSA M, TROMBLY D, MORIN V, STEIN J, STEIN G, JAVED A, GUTIERREZ SE. Transcriptional Auto-Regulation of RUNX1 P1 Promoter. PLoS One. 2016;11:e0149119. doi: 10.1371/journal.pone.0149119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGRATH KE, FRAME JM, FEGAN KH, BOWEN JR, CONWAY SJ, CATHERMAN SC, KINGSLEY PD, KONISKI AD, PALIS J. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 2015;11:1892–904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEDVINSKY A, DZIERZAK E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- MIKKOLA HK, FUJIWARA Y, SCHLAEGER TM, TRAVER D, ORKIN SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–16. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- MOORE MA, METCALF D. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br J Haematol. 1970;18:279–96. doi: 10.1111/j.1365-2141.1970.tb01443.x. [DOI] [PubMed] [Google Scholar]
- MUKOUYAMA Y, CHIBA N, HARA T, OKADA H, ITO Y, KANAMARU R, MIYAJIMA A, SATAKE M, WATANABE T. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonad-mesonephros region. Dev Biol. 2000;220:27–36. doi: 10.1006/dbio.2000.9617. [DOI] [PubMed] [Google Scholar]
- MÜLLER AM, MEDVINSKY A, STROUBOULIS J, GROSVELD F, DZIERZAK E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- MURAYAMA E, KISSA K, ZAPATA A, MORDELET E, BRIOLAT V, LIN HF, HANDIN RI, HERBOMEL P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–75. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- MURRAY P. The Development in vitro of the Blood of the Early Chick Embryo. London. Proceedings of the Royal Society.1932. [Google Scholar]
- NAKANO H, LIU X, ARSHI A, NAKASHIMA Y, VAN HANDEL B, SASIDHARAN R, HARMON AW, SHIN JH, SCHWARTZ RJ, CONWAY SJ, HARVEY RP, PASHMFOROUSH M, MIKKOLA HK, NAKANO A. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun. 2013;4:1564. doi: 10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NG CE, YOKOMIZO T, YAMASHITA N, CIROVIC B, JIN H, WEN Z, ITO Y, OSATO M. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells. 2010;28:1869–81. doi: 10.1002/stem.507. [DOI] [PubMed] [Google Scholar]
- NIKI M, OKADA H, TAKANO H, KUNO J, TANI K, HIBINO H, ASANO S, ITO Y, SATAKE M, NODA T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci U S A. 1997;94:5697–702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOLAN DJ, GINSBERG M, ISRAELY E, PALIKUQI B, POULOS MG, JAMES D, DING BS, SCHACHTERLE W, LIU Y, ROSENWAKS Z, BUTLER JM, XIANG J, RAFII A, SHIDO K, RABBANY SY, ELEMENTO O, RAFII S. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–19. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORTH T, GU TL, STACY T, WANG Q, HOWARD L, BINDER M, MARÍN-PADILLA M, SPECK NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–75. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- NORTH TE, DE BRUIJN MF, STACY T, TALEBIAN L, LIND E, ROBIN C, BINDER M, DZIERZAK E, SPECK NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–72. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- NORTH TE, STACY T, MATHENY CJ, SPECK NA, DE BRUIJN MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22:158–68. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- NOTTINGHAM WT, JARRATT A, BURGESS M, SPECK CL, CHENG JF, PRABHAKAR S, RUBIN EM, LI PS, SLOANE-STANLEY J, KONG-A-SAN J, DE BRUIJN MF. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–97. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKUDA T, VAN DEURSEN J, HIEBERT SW, GROSVELD G, DOWNING JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–30. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- PADRON-BARTHE L, TEMINO S, VILLA DEL CAMPO C, CARRAMOLINO L, ISERN J, TORRES M. Clonal analysis identifies hemogenic endothelium as the source of the blood-endothelial common lineage in the mouse embryo. Blood. 2014;124:2523–32. doi: 10.1182/blood-2013-12-545939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALIS J. Primitive and definitive erythropoiesis in mammals. Front Physiol. 2014;5:3. doi: 10.3389/fphys.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALIS J, ROBERTSON S, KENNEDY M, WALL C, KELLER G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–84. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- POTTS KS, SARGEANT TJ, MARKHAM JF, SHI W, BIBEN C, JOSEFSSON EC, WHITEHEAD LW, ROGERS KL, LIAKHOVITSKAIA A, SMYTH GK, KILE BT, MEDVINSKY A, ALEXANDER WS, HILTON DJ, TAOUDI S. A lineage of diploid platelet-forming cells precedes polyploid megakaryocyte formation in the mouse embryo. Blood. 2014;124:2725–9. doi: 10.1182/blood-2014-02-559468. [DOI] [PubMed] [Google Scholar]
- POZNER A, GOLDENBERG D, NEGREANU V, LE SY, ELROY-STEIN O, LEVANON D, GRONER Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol Cell Biol. 2000;20:2297–307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POZNER A, LOTEM J, XIAO C, GOLDENBERG D, BRENNER O, NEGREANU V, LEVANON D, GRONER Y. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev Biol. 2007;7:84. doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUTZ G, ROSNER A, NUESSLEIN I, SCHMITZ N, BUCHHOLZ F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–39. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- RENNERT J, COFFMAN JA, MUSHEGIAN AR, ROBERTSON AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHODES KE, GEKAS C, WANG Y, LUX CT, FRANCIS CS, CHAN DN, CONWAY S, ORKIN SH, YODER MC, MIKKOLA HK. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–63. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYBTSOV S, IV, ANOVS A, ZHAO S, MEDVINSKY A. Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development. 2016;143:1284–9. doi: 10.1242/dev.131193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYBTSOV S, SOBIESIAK M, TAOUDI S, SOUILHOL C, SENSERRICH J, LIAKHOVITSKAIA A, IV, ANOVS A, FRAMPTON J, ZHAO S, MEDVINSKY A. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med. 2011;208:1305–15. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SABIN FR. Studies on the origin of blood-vessels and of red blood-corpuscles as seen in the living blastoderm of chicks during the second day of incubation. Contributions to Embryology 1920 [Google Scholar]
- SANDLER VM, LIS R, LIU Y, KEDEM A, JAMES D, ELEMENTO O, BUTLER JM, SCANDURA JM, RAFII S. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511:312–8. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASAKI K, YAGI H, BRONSON RT, TOMINAGA K, MATSUNASHI T, DEGUCHI K, TANI Y, KISHIMOTO T, KOMORI T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93:12359–63. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SROCZYNSKA P, LANCRIN C, KOUSKOFF V, LACAUD G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009;114:5279–89. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- SWIERS G, BAUMANN C, O’ROURKE J, GIANNOULATOU E, TAYLOR S, JOSHI A, MOIGNARD V, PINA C, BEE T, KOKKALIARIS KD, YOSHIMOTO M, YODER MC, FRAMPTON J, SCHROEDER T, ENVER T, GOTTGENS B, DE BRUIJN MF. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat Commun. 2013a;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWIERS G, RODE C, AZZONI E, DE BRUIJN MF. A short history of hemogenic endothelium. Blood Cells Mol Dis. 2013b;51:206–12. doi: 10.1016/j.bcmd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAKURA N, WATANABE T, SUENOBU S, YAMADA Y, NODA T, ITO Y, SATAKE M, SUDA T. A role for hematopoietic stem cells in promoting angiogenesis. Cell. 2000;102:199–209. doi: 10.1016/s0092-8674(00)00025-8. [DOI] [PubMed] [Google Scholar]
- TANAKA Y, SANCHEZ V, TAKATA N, YOKOMIZO T, YAMANAKA Y, KATAOKA H, HOPPE PS, SCHROEDER T, NISHIKAWA S. Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep. 2014;8:31–9. doi: 10.1016/j.celrep.2014.05.055. [DOI] [PubMed] [Google Scholar]
- TAOUDI S, GONNEAU C, MOORE K, SHERIDAN JM, BLACKBURN CC, TAYLOR E, MEDVINSKY A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3:99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- TELFER JC, ROTHENBERG EV. Expression and function of a stem cell promoter for the murine CBFalpha2 gene: distinct roles and regulation in natural killer and T cell development. Dev Biol. 2001;229:363–82. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- THAMBYRAJAH R, MAZAN M, PATEL R, MOIGNARD V, STEFANSKA M, MARINOPOULOU E, LI Y, LANCRIN C, CLAPES T, MÖRÖY T, ROBIN C, MILLER C, COWLEY S, GÖTTGENS B, KOUSKOFF V, LACAUD G. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. 2016;18:21–32. doi: 10.1038/ncb3276. [DOI] [PubMed] [Google Scholar]
- TOBER J, KONISKI A, MCGRATH KE, VEMISHETTI R, EMERSON R, DE MESY-BENTLEY KK, WAUGH R, PALIS J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109:1433–41. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOBER J, YZAGUIRRE AD, PIWARZYK E, SPECK NA. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development. 2013;140:3765–76. doi: 10.1242/dev.094961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRACEY WD, PEPLING ME, HORB ME, THOMSEN GH, GERGEN JP. A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development. 1998;125:1371–80. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- UENO H, WEISSMAN IL. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev Cell. 2006;11:519–33. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- WANG Q, STACY T, BINDER M, MARIN-PADILLA M, SHARPE AH, SPECK NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996a;93:3444–9. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Q, STACY T, MILLER JD, LEWIS AF, GU TL, HUANG X, BUSHWELLER JH, BORIES JC, ALT FW, RYAN G, LIU PP, WYNSHAW-BORIS A, BINDER M, MARÍN-PADILLA M, SHARPE AH, SPECK NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- WAREING S, ELIADES A, LACAUD G, KOUSKOFF V. ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev Dyn. 2012;241:1454–64. doi: 10.1002/dvdy.23825. [DOI] [PubMed] [Google Scholar]
- WITZENBICHLER B, MAISONPIERRE PC, JONES P, YANCOPOULOS GD, ISNER JM. Chemotactic properties of angiopoietin-1 and -2, ligands for the endothelial-specific receptor tyrosine kinase Tie2. J Biol Chem. 1998;273:18514–21. doi: 10.1074/jbc.273.29.18514. [DOI] [PubMed] [Google Scholar]
- XU MJ, MATSUOKA S, YANG FC, EBIHARA Y, MANABE A, TANAKA R, EGUCHI M, ASANO S, NAKAHATA T, TSUJI K. Evidence for the presence of murine primitive megakaryocytopoiesis in the early yolk sac. Blood. 2001;97:2016–22. doi: 10.1182/blood.v97.7.2016. [DOI] [PubMed] [Google Scholar]
- YODER MC. Inducing definitive hematopoiesis in a dish. Nat Biotechnol. 2014;32:539–41. doi: 10.1038/nbt.2929. [DOI] [PubMed] [Google Scholar]
- YOKOMIZO T, DZIERZAK E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–61. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOMIZO T, HASEGAWA K, ISHITOBI H, OSATO M, EMA M, ITO Y, YAMAMOTO M, TAKAHASHI S. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111:4075–80. doi: 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- YOKOMIZO T, OGAWA M, OSATO M, KANNO T, YOSHIDA H, FUJIMOTO T, FRASER S, NISHIKAWA S, OKADA H, SATAKE M, NODA T, ITO Y. Requirement of Runx1/AML1/PEBP2alphaB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- YOSHIMOTO M, MONTECINO-RODRIGUEZ E, FERKOWICZ MJ, PORAYETTE P, SHELLEY WC, CONWAY SJ, DORSHKIND K, YODER MC. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A. 2011;108:1468–73. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIMOTO M, PORAYETTE P, GLOSSON NL, CONWAY SJ, CARLESSO N, CARDOSO AA, KAPLAN MH, YODER MC. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–14. doi: 10.1182/blood-2011-12-397489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU M, MAZOR T, HUANG H, HUANG HT, KATHREIN KL, WOO AJ, CHOUINARD CR, LABADORF A, AKIE TE, MORAN TB, XIE H, ZACHAREK S, TANIUCHI I, ROEDER RG, KIM CF, ZON LI, FRAENKEL E, CANTOR AB. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–43. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YZAGUIRRE AD, SPECK NA. Insights into blood cell formation from hemogenic endothelium in lesser-known anatomic sites. Dev Dyn. 2016 doi: 10.1002/dvdy.24430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAMBIDIS ET, PEAULT B, PARK TS, BUNZ F, CIVIN CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–70. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEIGLER BM, SUGIYAMA D, CHEN M, GUO Y, DOWNS KM, SPECK NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–92. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- ZHEN F, LAN Y, YAN B, ZHANG W, WEN Z. Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development. 2013;140:3977–85. doi: 10.1242/dev.097071. [DOI] [PubMed] [Google Scholar]
- ZOVEIN AC, HOFMANN JJ, LYNCH M, FRENCH WJ, TURLO KA, YANG Y, BECKER MS, ZANETTA L, DEJANA E, GASSON JC, TALLQUIST MD, IRUELA-ARISPE ML. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–36. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]