SYNOPSIS
Prenatal whole exome sequencing (WES) has the potential to increase the ability to provide more diagnostic capabilities in fetuses with sonographic abnormalities which will then improve the ability to counsel families. It is also often the first step in improving the path towards informed diagnosis and treatment, which is especially important in the era of advancing in utero fetal therapy. In this review, we will discuss the current literature regarding prenatal WES, clinical indications for WES, challenges with interpretation/counseling (variants of unknown significance), research priorities, ethical issues, and potential future advances.
Keywords: Fetal, exome, sequencing, prenatal ultrasound, abnormalities
Background
Ultrasound detected fetal sonographic abnormalities are identified in 2–3% of pregnancies.1 Genetic diagnosis with amniocentesis or chorionic villus sampling with chromosomal microarray (CMA) and karyotype are routinely offered in these cases. Of cases that undergo diagnostic testing, a karyotype abnormality is found in 8–10% of cases, whereas a micro-deletion/duplication is identified in another 6% leaving the majority of families without a specific genetic diagnosis.2 These families must therefore be counseled based on ultrasound findings alone. Management decisions thus need to be made on the basis of limited information and counseling is challenging because of the broad differential diagnosis and large range of prognoses and expectations. Whole exome sequencing (WES), rather than targeted disease specific gene panels, is now being studied and used to improve prenatal diagnosis in cases where structural abnormalities are identified sonographically. Initial studies show that prenatal WES can elucidate the responsible pathogenic variants in an additional 203 –804% of cases when standard genetic testing (karyotype and CMA) is normal. The diagnostic yield of prenatal WES is known to be highly dependent on the indication for WES.
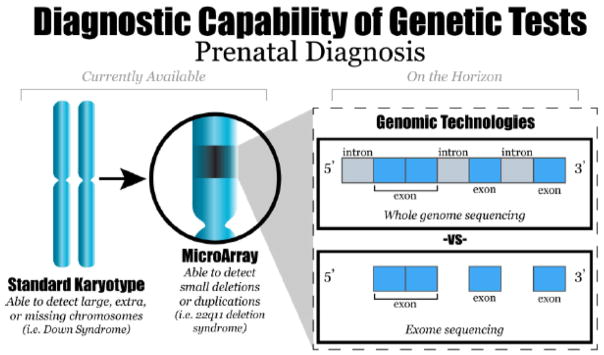
WES, unlike whole genome sequencing (WGS), is currently clinically available and focuses on the exons or protein coding regions of the genome only (Figure 1). Exons account for 1.5%5 of the DNA in the genome, comprising approximately 22,000 genes. Most identified genes implicated in Mendelian disease involve the exons.6 Thus, WES is more cost-effective than WGS. In addition, WES is preferred because the ability to interpret intronic regions of the genome is currently extremely limited. Prenatal WES has the ability to increase diagnostic rates in cases where fetal anomalies are present and enhance our understanding regarding pathogenic variants that are developmentally lethal.7 WES also has the potential to expand known disease phenotypes to the prenatal period. Multiple challenges of prenatal WES include the following: 1) interpreting the vast amount of data in a timely manner; 2) identifying pathogenic variants in diseases with reduced penetrance and variable expressivity; and 3) providing adequate pre- and post- test counseling particularly in regards to the stress/uncertainly associated with discovering variants of unknown significance (VUS).8
Figure 1.
New genomic technologies such as whole genome and whole exome sequencing have the ability to interrogate the fetal genome more comprehensively than currently available tests.
From Hardisty EE, Vora NL. Advances in genetic prenatal diagnosis and screening. Curr Opin Pediatr 2014;26(6):634–8; with permission.
Prenatal WES has the potential to increase the ability to provide a more precise diagnosis which will then improve our ability to counsel families. It is also often the first step in improving the path towards informed diagnosis and treatment, which is especially important in the era of advancing in utero fetal therapy. In this review, we will discuss the current literature regarding prenatal WES, clinical indications for WES, challenges with interpretation/counseling (VUS), research priorities, ethical issues, and potential future advances.
Prenatal
As of July 2017, the prenatal data for exome sequencing currently includes 16 case series with 5 or more fetuses (7 articles and 9 conference abstracts) and several additional case reports.9 The overall diagnostic rate ranges from 6.210 to 57.1% (Table 1)11 in published articles and up to 80% when abstracts were included.4,9 The diagnostic rate is dependent on the indication for WES and varies based on multiple factors including the following: single vs multiple organ system affected, specific organ system affected, and proband vs trio sequencing. The lowest reported rate of 6.2% was obtained by performing WES on all fetuses with any anomaly visualized on ultrasound, while the highest rates were in small studies with carefully chosen cases.11,12,13 WES was successful in determining the diagnosis in 3.6–6.2% of isolated anomalies and 14.3–16% of fetuses with ultrasound findings affecting multiple organ systems.10 Certain organ systems appear to have a higher yield of reported pathogenic variants. For example, in a cohort of 84 deceased fetuses, a pathogenic variant was identified by WES more frequently in fetuses with a isolated neurologic abnormalities compared to those with isolated cardiovascular findings (37 vs 31%).14 The yield is also increased when WES is performed on a trio (maternal, paternal, proband).15 The use of trios allows for prioritization of review of variants that have an increased likelihood of being pathogenic using a standard analytic framework that takes into account potential de novo variants (present in fetus but absent from the parents) or recessively inherited variants (homozygous or compound heterozygous in the fetus and heterozygous in parents).
Table 1.
Fetal exome sequencing published case series
| First Author | Number of Cases | Cohort description | Proband vs trio | Pathogenic variant | Likely pathogenic variant |
|---|---|---|---|---|---|
| Yang12 | 11 | Terminated anomalous fetuses | Trio | 6/11 (54%) | -- |
| Carss15 | 30 | Prenatal sonographic anomalies | Trio | 3/30 (10%) | 5/30 (16.7%) |
| Drury3 | 24 | Prenatal sonographic anomalies including NT ≥3.5 | 14 Proband 10 trio |
5/24 (20.8%) | 1/24 (4.2%) |
| Alamillo11 | 7 | Multiple sonographic anomalies termination or demise | Trio | 3/7 (42.9%) | 1/7 (14.3%) |
| Pangalos13 | 14 | Prenatal sonographic anomalies | Proband only | 6/14 (42.9%) | -- |
| Yates14 | 84 | Demise or termination | 33 Proband/duo 51 Trio/quad |
17/84 (20%) | 38/84 (45%) |
| Vora26 | 15 | Multiple sonographic anomalies | Trio | 7/15 (46.7%) | 1/15 (6.7%) |
The outcomes of a prenatal diagnosis can be extreme, and require significant counseling with regard to reproductive decision-making16. When the pregnancy is continued the diagnosis can assist with palliative care decisions including comfort care or a diagnosis can inform the pediatricians about management or possible treatments that may be offered postnatally. A diagnosis with WES may also reduce hospital costs by decreasing the number of tests performed postnatally, thereby avoiding the ‘diagnostic odyssey’ and potentially decreasing the length of the hospital stay.17,18 When performed clinically in the prenatal or neonatal period, a positive result can have implications for other family members and/or for future pregnancies. In a subsequent pregnancy, preimplantation genetic diagnosis (PGD) could be performed or the patient could have diagnostic testing by chorionic villus sampling (CVS) or amniocentesis. Understanding the genetic etiology can also provide closure for the family and help with the grieving process in the case of fetal or neonatal loss.19
Many Mendelian disorders may not have a known prenatal phenotype and the molecular diagnosis may not be suspected because the phenotype may be atypical from what is described postnatally. Use of prenatal WES has the ability to expand phenotypes to the prenatal period and increase our understanding of genes that may be critical to human development.
Patient preferences and understanding of prenatal genetic diagnosis are extremely variable and likely affect the psychosocial impact of the relayed results. Preference to undergo testing and desire to make decisions based on the results vary by ethnicity,20,21,22 socioeconomic status, cultural and religious beliefs, acceptability of termination of pregnancy, and experiences with disability.23, 24,25 Vora et al., found in a pilot study on prenatal WES that women with a lower family income scored significantly lower on the genetics literacy assessment compared with women with a higher family income. They also found that women with a lower family income (<$50,000 vs >89,999) had increased expectations that WES would provide a reason for the fetal abnormalities, (Likert scale: 5.2 out of 10) despite appropriate pretest counseling by a genetic counselor about an approximate 30% diagnostic yield. Further research on this critical topic of patient expectations and understanding is needed to ensure that patients’ needs are being met as new technologies inevitably become implemented into clinical practice.26
Therapy
The use of fetal intervention to treat genetic disease has been in use for over 50 decades. One of the earliest uses of in utero therapy is the use of intrauterine transfusions for fetal Rh isoimmunization27. In the era of advanced fetal therapy, including: in utero myelomenigocele repair, fetal cardiac intervention28, fetal endotracheal occlusion for congenital diaphragmatic hernia, and potentially in utero amnioinfusion for bilateral renal agenesis29; additional knowledge of the underlying genetic etiology of the defect is invaluable.30 For example, an in utero CDH repair may be less effective in fetuses with syndromic CDH than isolated CDH. It has been proposed31 that in utero fetal intracerebral shunts could effectively treat severe fetal ventriculomegaly.32,33 In males, isolated severe fetal ventriculomegaly is secondary to an L1CAM mutation in 2–15% of cases.34 These infants are known to have more severe neurodevelopmental outcomes than other cases of apparently isolated severe ventriculomegaly.35 It may therefore be reasonable to exclude fetuses with L1CAM mutations or perform subgroup analyses on these infants when trials evaluating the utility of fetal intervention for this condition are performed. This example can be extrapolated to other inherited etiologies of fetal anomalies. It has yet to be determined whether or not certain underlying genetic conditions may benefit more or less from fetal intervention. Prenatal WES should be studied in cases where fetal intervention is possible or planned so that further information about best surgical candidates can be obtained.
Until recently, there were few recognizable fetal phenotypes for which evaluation of single gene mutations were routinely performed. Prenatal molecular diagnosis of specific skeletal dysplasias can often be made with targeted panel testing.36 However, due to the broad phenotypic spectrum of skeletal and other conditions and the limited information obtained with prenatal ultrasound, WES may be more efficient and cost effective. New fetal therapies are currently emerging, and demonstrating improved outcomes in specific skeletal dysplasias.37 Particularly promising is teriparatide for treatment of Osteogenesis Imperfecta (OI).38 The only current in utero therapy for OI is in utero mesenchymal stem cell transplantation via the fetal umbilical vein.39 A transient decrease in fractures has been demonstrated in case series.40 The BOOSTB4 study is a feasibility study that is underway to study the use of stem cell transplantation in 15 cases of OI with in utero diagnosis.41 As additional postnatal therapy for genetic syndromes evolves, their application to fetal life will need to be explored. Thus, application of prenatal WES to such cases may help identify which pregnancies would benefit most from in utero therapies.
Clinical Indications
WES is recommended in pediatric and adult medicine for clinical indications such as multiple birth defects or neurodevelopmental delay when other tests have been uninformative. Postnatal data shows an overall diagnostic yield of 25–30%, depending on the affected organ system.12,42 The American College of Medical Genetics and Genomics (ACMG) recommends WES when a genetic disorder is suggested by phenotype and family history and either targeted genetic testing is not available or available testing performed is not diagnostic.43 It is both current practice, and cost effective to perform a microarray prior to whole exome sequencing. Experts also suggest performing targeted testing prior to WES when a specific syndrome is suspected. If the presenting disorder is highly genetically heterogenous, WES is potentially more cost effective than sequencing individual genes.44
ACMG and the Society of Maternal Fetal Medicine (SMFM) recommend that all patients considering WES receive counseling from a provider with genetics expertise. However, given that WES is in its infancy and studies are ongoing to determine its clinical utility, ACMG and SMFM do not recommend WES for routine use for prenatal diagnosis.45
In select cases where other approaches to diagnosis have been uninformative, it may be appropriate to offer WES. Examples of such cases include recurrent or multiple congenital anomalies, heterotaxy, and undiagnosed skeletal dysplasias. Prenatal WES also has a role in cases where a fetus has structural abnormalities with reported consanguinity or homozygosity indicative of relatedness on microarray.
ACMG reportable variants
The current ACMG reporting recommendations include not only reporting the clinically relevant findings that could be contributing to the primary phenotype for which the testing was requested, but also offering reports of “secondary findings” for medical conditions that are potentially medically actionable. Secondary findings include pathogenic variants in genes responsible for conditions that are unrelated to the indication for which testing was initially performed. The initial ACMG recommendation included mandatory reporting of specified variants in 56 genes.46 ACMG recently modified the recommendations to optional reporting, removing 4 genes and adding an additional gene, resulting in reporting recommendations for 59 genes.47 ACMG recommends reporting only known pathogenic or disease causing variants within the genes and not benign variants. Literature suggests WES will identify a secondary finding in 1% of cases.48 The majority of reportable incidental variants occur in genes responsible for a predisposition to cancer or a cardiac event. The clinical treatment modalities for these variants have variable efficacy and are currently far from curative.
Prior to these ACMGs guidelines, societies did not recommend testing children for adult onset diseases until they reached sufficient maturity to provide informed consent.49 The incidental variant guidelines created a paradigm shift, recommending parents opt out of receiving information about secondary findings identified on WES of their child or fetus.
Interpretation/Counseling
Pre- and post-test counseling is recommended for all prenatal genetic screening and diagnostic testing. Ideally, both the mother and father are present for the counseling session. A trained professional with genetics expertise should obtain complete clinical information along with a three-generation pedigree. Specifically when taking a pedigree in a prenatal genetics clinic, the fetus and any siblings, miscarriages or fetal losses should be included in one of these three generations. Testing should be ordered only by specialists who are comfortable with the interpretation and explanation of results. Information should include options for reproductive decision making, pregnancy and perinatal management. 50
There are risks, benefits and limitations to every form of prenatal screening or diagnosis. There are 5 specific items that should be mentioned during pre-test counseling for prenatal WES. The results of the testing could provide the following information:
A primary finding could be identified or suggested to explain the fetal phenotype. This includes variants in genes that could explain the medical conditions for which the testing is being performed.
A negative result for which a diagnosis is not available for the phenotype in question.
A variant of unknown significance (VOUS), which may or may not be related to the condition for which the testing is obtained.
A secondary or incidental finding could be identified. This refers to a pathogenic variant in a gene that causes a genetic condition, unrelated to the indication for testing.
Genetic relationships could be identified including the identification of false paternity or consanguinity.
The patient undergoing prenatal WES needs appropriate informed consent to make decisions about opting in or out of receiving information on VOUS and secondary/incidental findings. In addition to the 5 statements above, patients should also be informed that the knowledge of variants is continuously evolving so that an unreported variant or a variant that is coded as unknown in significance at the time WES is initially performed, may be found to be pathogenic in the future. Primary or secondary gene mutations identified in the fetus could also have impacts on the patient’s health, other family members’ health, and future pregnancies.
Variants of unknown significance (VOUS) are particularly problematic in the prenatal setting.51 Providers who offer and counsel about prenatal microarray have experience with counseling patients regarding uncertain results. Laboratories performing WES often provide limited reports to decrease the number of VOUS reported to the clinician. Herein lies the ethical balance of reporting and counseling. Current databases such as ClinVar52 and Human Genetics Mutation Database53, and Exome Aggregation Consortium54 include limited fetal phenotypic data, making fetal phenotypes especially challenging to interpret.
The interpretation of WES is simple when a previously reported pathogenic mutation with a well-documented phenotype is identified. The most critical data to arrive at the correct diagnosis involves phenotypic data from ultrasound, fetal MRI (if performed), and fetal autopsy. However, detailed phenotypic information is not often available prenatally. Thus, a definitive diagnosis cannot be made in many cases. Instead multiple variants are identified and must be filtered using pipelines specific to trios along with whatever phenotypic data is available to determine possible pathogenicity. Laboratories and clinics should have clear guidelines and policies related to reporting of findings and all reported variants should be confirmed with Sanger sequencing as false positive results can occur with WES given that sequencing of many fragments occurs simultaneously. Databases with phenotypic and molecular data (ClinGen/ClinVar) currently do not have a prenatal component. However, given that WES is starting to be used prenatally in select cases, it is critical that a shared prenatal database become available to further understand genotype/phenotype correlations prenatally and to enable clinicians and researchers to determine if the variants found in their patients were seen in other patients with the same phenotype.
Limitations
While WES is performed to evaluate single nucleotide variants in expressed areas of the genome, it has many limitations. Some of the limitations can be overcome by increasing the depth of sequencing, whereas others are inherent defects of the current technology. WES does not detect differences in copy number variation, for which microarray technology is required. It is not designed to detect aneuploidy (i.e. trisomy 21), polyploidy (i.e. triploidy, tetraploidy), nor does it detect translocations, trinucleotide repeats (repeat expansions, tandem repeat size), or low level mosaicism. There are several regions in the genome with poor depth/coverage, particularly GC rich areas,55 that are not adequately sequenced with WES. Sanger sequencing is recommended to verify all reported results.
In addition to the overall challenges of WES, the long turnaround time is especially problematic in the prenatal setting. Limited anatomy can be visualized on sonogram prior to the anatomical survey performed at 18–22 weeks gestation. In addition, primary testing with microarray or panel testing is generally performed prior to sending WES. When performed rapidly, results can be obtained within 2–3 weeks. It is anticipated that the turnaround will become faster and more cost-effective over time. 56
Research implications
Several NIH funded centers are currently performing whole exome sequencing for the purpose of identification of novel genes. Initiation of WES was quickly followed by a rise in the discovery of the known function of genes.57 The majority of novel genes are identified postnatally, however genes responsible for lethal fetal conditions or recurrent miscarriages rely on prenatal WES and WGS to provide insight into the function of genes that may be critical to human development
It is critical that researchers performing prenatal WES be committed to sharing data. A common database for prenatal phenotypes with molecular findings by WES is essential to optimizing clinical care. Data sharing is recommended for clinical laboratories as well as researchers. The NIH requires grant submitters to supply information on their current plans, including patient consent, to participate in genome wide data sharing. Centers are often willing to perform WES on patients with specific fetal defects, for example the DHREAMS study58 based out of Columbia will perform WES on CDH patients seen at other centers.
Ethics
Multiple ethical issues associated with prenatal WES have been brought to attention.59 Of primary concern to the patient is adequate interpretation and disclosure of appropriate variants. This is particularly important in the prenatal period where the decision to continue a pregnancy could be determined by the reported variant. In addition, patient counseling requires appropriate consent and disclosure of secondary and incidental variants, re-contact for reclassification of variants, discussion of informing affected family members, loss of privacy associated with data-sharing, as well as potential discrimination if a genetic diagnosis is made.
Genetic professionals have an ethical obligation to follow appropriate protocols for patient consent60 and interpretation of variants. It is particularly important to discuss the options of opting in or out of receiving information on secondary/incidental findings and VOUS.61 In the case of trio analysis, it is essential to consent the mother separately from the father to discuss that non-paternity may be disclosed from prenatal WES. She would then have an option to opt out of testing. The patient should consider all testing options available including panel testing when appropriate. In the future, and potentially even now, WES could be more cost effective than panel testing, however WES can result in a significant amount of unintended information.
An ethical dilemma inherent in genetic testing is ensuring that genetic information remains confidential.62 This is confounded by the importance of data sharing in the understanding and classification of variants. Efforts have been made to limit potential discrimination secondary to results. The Genetic Information Non-discrimination Act (GINA) of 2008 prevents utilization of genetic information for health insurance. It does not, however, apply to life, disability, long-term-care, or certain federal employee health care programs.
Challenges of interpretation and reporting remain barriers to widespread use of prenatal WES. Molecular geneticists and molecular variant analysts are responsible for maximizing reporting of positives while minimizing the reporting of potentially benign variants. Regardless of this balance, until comprehensive WES data is deposited in publicly available databases, false negatives and false positives will be reported.
Costs
The majority of the cost of performing WES is due to costs of variant analysis. A well-trained molecular variant analyst or molecular geneticist is essential to providing accurate results. Most clinical and research laboratories have committees made of multidisciplinary teams to review the findings from prenatal WES. These may include molecular geneticists, cytogeneticists, clinical geneticists, genetic counselors, bioethicists, and bioinformaticians. In addition to accurately interpreting and reporting primary findings which explain the fetal phenotype, it is essential to interpret and report secondary findings presuming consent was obtained from the patients to receive such findings. A system for re-analysis and re-evaluation of variants over time is also necessary because genes whose functions were previously unknown are being discovered and categorized at a rapid rate. Sanger sequencing is also recommended for confirmation of variants for the patient and for the family members who receive results about medically actionable findings that were unrelated to the fetal phenotype such as any of the ACMG specified variants in 59 genes.45 Costs will presumably decrease as technology advances and interpretation algorithms improve.
Although a diagnosis is thought to be cost effective, it often does incur medical costs including additional recommended medical screening, evaluation, and or therapy related to the diagnosis for either the patient and/or family members. There is also an emotional cost associated with having a test performed whether it is resulted as positive, negative, or VOUS.63
Future advances
WES and WGS are rapidly evolving with advances in research and technology. The ability to perform WGS has quickly followed WES and is currently being studied on a research basis. The limitations of interpretation of WES seem minor compared to interpretation challenges of WGS data.64 There will likely be a pivotal point in the future where cost and knowledge will make WGS more cost-effective and widely performed.
In addition, advances in prenatal screening, including cell free DNA (cfDNA), have reduced the utilization of chorionic villus sampling and amniocentesis. CfDNA technology allows for detection of cell free placental DNA in maternal blood and is a clinically recommended screening method for women who are at high risk for having a pregnancy affected with a common aneuploidy.65 Capabilities of performing WES on cell free DNA currently exist.66,67 As technology improves and cost decreases it is likely that WES using cell free DNA will be clinically available. Some of the limitations of this technology will persist, including the fact that the cells in the maternal system are derived from trophoblasts and not a pure representation of fetal DNA. When screening tests using cell free DNA show an increased risk for a specific diagnosis, a diagnostic test will continue to be necessary to rule out placental mosaicism or other etiologies of a false positive screen.
Single cell technology has been proposed as a more accurate technology than the current cell free platforms68,69 Rather than relying on maternal/fetal ratios, single cell technology allows for a single fetal cell to be specifically evaluated. If utilized, several cell wills need to be evaluated to evaluate for sequencing errors or mosaicism. This technology potentially has more promise for cell free whole exome sequencing than the current non-invasive cell free technologies secondary to the lack of maternal contamination.
In summary, further research is needed to determine best practices for clinical implementation of WES specifically related to interpretation, turn-around time, and optimal ways to counsel patients given that it is only a matter of time before WES becomes available using noninvasive technologies.
KEY POINTS.
Prenatal whole exome sequencing is emerging as a valuable tool for fetal diagnosis in the setting of sonographic abnormalities.
Diagnostic rates are variable across studies with improved rates when trio (proband, mother father) whole exome sequencing is performed.
Prenatal genetic counseling is crucial for appropriate parental consent for whole exome sequencing.
There are many ethical considerations including risks of discrimination that must be considered when whole exome sequencing is performed.
Footnotes
DISCLOSURE STATEMENT
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Update on overall prevalence of major birth defects--Atlanta, Georgia, 1978–2005. MMWR Morb Mortal Wkly Rep. 2008;57:1–5. [PubMed] [Google Scholar]
- 2.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367:2175–84. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drury S, Williams H, Trump N, Boustred C, Lench N, Scott RH, et al. Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn. 2015;35:1010–7. doi: 10.1002/pd.4675. [DOI] [PubMed] [Google Scholar]
- 4.Yadava SM, Ashkinadze E. 125: Whole exome sequencing (WES) in prenatal diagnosis for carefully selected cases. Am J Obstet Gynecol. 2017;216:S87–S88. [Google Scholar]
- 5.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? J Med Genet. 2011;48:580–9. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- 7.Medeira A, Norman A, Haslam J, Clayton-Smith J, Donnai D. Examination of fetuses after induced abortion for fetal abnormality—a follow-up study. Prenat Diagn. 1994;14:381–5. doi: 10.1002/pd.1970140507. [DOI] [PubMed] [Google Scholar]
- 8.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004 Aug 6;305(5685):869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 9.Best S, Wou K, Vora N, Van den Veyver IB, Wapner R, Chitty LS. Promises, Pitfalls and Practicalities of Prenatal Whole ExomeSequencing. Prenat Diagn. 2017 Jun 27; doi: 10.1002/pd.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wapner RPs, Brennan K, Bier L, Wou K, Goldstein D. Whole exome sequencing in the evaluation of fetal structural anomalies: A prospective study of sequential patients. Am J Obstet Gynecol. 2017;216:S5–6. [Google Scholar]
- 11.Alamillo CL, Powis Z, Farwell K, Shahmirzadi L, Weltmer EC, Turocy J, et al. Exome sequencing positively identified relevant alterations in more than half of cases with an indication of prenatal ultrasound anomalies. Prenat Diagn. 2015;35:1073–8. doi: 10.1002/pd.4648. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1807–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pangalos C, Hagnefelt B, Lilakos K, Konialis C. First applications of a targeted exome sequencing approach in fetuses with ultrasound abnormalities reveals an important fraction of cases with associated gene defects. PeerJ. 2016;34:e1955. doi: 10.7717/peerj.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yates CL, Monaghan KG, Copenheaver D, Retterer K, Scuffins J, Kucera CR, Friedman B, Richard G, Juusola J. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med. 2017 Apr 20; doi: 10.1038/gim.2017.31. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Carss KJ, Hillman SC, Parthiban V, McMullan DJ, Maher ER, Kilby MD, Hurles ME. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet. 2014 Jun 15;23(12):3269–77. doi: 10.1093/hmg/ddu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhardt BA, Soucier D, Hanson K, Savage MS, Jackson L, Wapner RJ. Women’s experiences receiving abnormal prenatal chromosomal microarray testing results. Genet Med. 2013 Feb;15(2):139–45. doi: 10.1038/gim.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia CA, Husami A, Holle J, Johnson JA, Qian Y, Mathur A, Wei C, Indugula SR, Zou F, Meng H, Wang L, Li X, Fisher R, Tan T, Hogart Begtrup A, Collins K, Wusik KA, Neilson D, Burrow T, Schorry E, Hopkin R, Keddache M, Harley JB, Kaufman KM, Zhang K. Clinical Impact and Cost-Effectiveness of Whole Exome Sequencing as a Diagnostic Tool: A Pediatric Center’s Experience. Front Pediatr. 2015 Aug 3;3:67. doi: 10.3389/fped.2015.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willing LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–87. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguire M, Light A, Kuppermann M, Dalton VK, Steinauer JE, Kerns JL. Grief after second-trimester termination for fetal anomaly: a qualitative study. Contraception. 2015 Mar;91(3):234–9. doi: 10.1016/j.contraception.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tschudin S, Huang D, Mor-Gültekin H, Alder J, Bitzer J, Tercanli S. Prenatal counseling--implications of the cultural background of pregnant women on information processing, emotional response and acceptance. Ultraschall Med. 2011 Dec;32(Suppl 2):E100–7. doi: 10.1055/s-0031-1281665. Epub 2011 Dec 20. [DOI] [PubMed] [Google Scholar]
- 21.Muhsen K, Na’amnah W, Lesser Y, Volovik I, Cohen D, Shohat T. Determinates of underutilization of amniocentesis among Israeli Arab women. Prenat Diagn. 2010 Feb;30(2):138–43. doi: 10.1002/pd.2428. [DOI] [PubMed] [Google Scholar]
- 22.Kuppermann M, Learman LA, Gates E, et al. Beyond race or ethnicity and socioeconomic status: predictors of prenatal testing for Down syndrome. Obstet Gynecol. 2006;107:1087–1097. doi: 10.1097/01.AOG.0000214953.90248.db. [DOI] [PubMed] [Google Scholar]
- 23.Case AP, Ramadhani TA, Canfield MA, Wicklund CA. Awareness and attitudes regarding prenatal testing among Texas women of childbearing age. Journal of genetic counseling. 2007 Oct;16(5):655–661. doi: 10.1007/s10897-007-9103-6. [DOI] [PubMed] [Google Scholar]
- 24.Kuppermann M, Gates E, Washington AE. Racial-ethnic differences in prenatal diagnostic test use and outcomes: preferences, socioeconomics, or patient knowledge? Obstetrics and gynecology. 1996 May;87(5 Pt 1):675–682. doi: 10.1016/0029-7844(96)00017-8. [DOI] [PubMed] [Google Scholar]
- 25.Kuppermann M, Nakagawa S, Cohen SR, Dominguez-Pareto I, Shaffer BL, Holloway SD. Attitudes toward prenatal testing and pregnancy termination among a diverse population of parents of children with intellectual disabilities. Prenatal diagnosis. 2011 Dec;31(13):1251–1258. doi: 10.1002/pd.2880. [DOI] [PubMed] [Google Scholar]
- 26.Vora NL, Powell B, Brandt A, Strande N, Hardisty E, Gilmore K, Foreman AKM, Wilhelmsen K, Bizon C, Reilly J, Owen P, Powell CM, Skinner D, Rini C, Lyerly AD, Boggess KA, Weck K, Berg JS, Evans JP. Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet Med. 2017 May 18; doi: 10.1038/gim.2017.33. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattison NS, Roberts AB, Mantell N. Intrauterine fetal transfusion, 1963–90. Ultrasound Obstet Gynecol. 1992 Sep 1;2(5):329–32. doi: 10.1046/j.1469-0705.1992.02050329.x. [DOI] [PubMed] [Google Scholar]
- 28.Moon-Grady AJ, Morris SA, Belfort M, et al. International Fetal Cardiac Intervention Registry: A Worldwide Collaborative Description and Preliminary Outcomes. J Am Coll Cardiol. 2015 Jul 28;66(4):388–99. doi: 10.1016/j.jacc.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 29.Goebel J. New Nephrological Frontiers: Opportunities and Challenges Created by Fetal Care Centers. Adv Pediatr. 2017 Aug;64(1):73–86. doi: 10.1016/j.yapd.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 30.McGivern MR, Best KE, Rankin J, Wellesley D, Greenlees R, Addor MC, et al. Epidemiology of congenital diaphragmatic hernia in Europe: a register-based study. Arch Dis Child Fetal Neonatal Ed. 2015;100:F137–44. doi: 10.1136/archdischild-2014-306174. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Emery SP, Maxey AP, Gu X, Wagner WR, Chun Y. A novel low-profile ventriculoamniotic shunt for foetal aqueductal stenosis. J Med Eng Technol. 2016;40(4):186–98. doi: 10.3109/03091902.2016.1154617. [DOI] [PubMed] [Google Scholar]
- 32.Cavalheiro S, da Costa MDS, Mendonça JN, Dastoli PA, Suriano IC, Barbosa MM, Moron AF. Antenatal management of fetal neurosurgical diseases. Child’s Nervous System. 33(7):1125–1141. doi: 10.1007/s00381-017-3442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavalheiro S, Moron AF, Zymberg ST, Dastoli P. Fetal hydrocephalus—prenatal treatment. Childs Nerv Syst. 2003;19:561–573. doi: 10.1007/s00381-003-0772-7. [DOI] [PubMed] [Google Scholar]
- 34.Kenwrick S, Jouet M, Donnai D. X linked hydrocephalus and MASA syndrome. J Med Genet. 1996 Jan;33(1):59–65. doi: 10.1136/jmg.33.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stumpel C, Vos YJ. L1 Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993–2017. [PubMed] [Google Scholar]
- 36.Chitty LS, Mason S, Barrett AN, McKay F, Lench N, Daley R, et al. Non-invasive prenatal diagnosis of achondroplasia and thanatophoric dysplasia: Next generation sequencing allows for a safer, more accurate, and comprehensive approach. Prenat Diagn. 2015;35:656–62. doi: 10.1002/pd.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jelin AC, O’Hare E, Blakemore K, Jelin EB, Valle D, Hoover-Fong J. Skeletal Dysplasias: Growing Therapy for Growing Bones. Front Pharmacol. 2017 Mar 6;8:79. doi: 10.3389/fphar.2017.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orwoll ES, Shapiro J, Veith S, Wang Y, Lapidus J, Vanek C, et al. Evaluation of teriparatide treatment in adults with osteogenesis imperfecta. J Clin Invest. 2014;124:491–498. doi: 10.1172/JCI71101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Blanc K, Götherström C, Ringdén O, Hassan M, McMahon R, Horwitz E, Anneren G, Axelsson O, Nunn J, Ewald U, Nordén-Lindeberg S, Jansson M, Dalton A, Aström E, Westgren M. Fetal mesenchymal stem-cell engraftment in bone after in utero transplantation in a patient with severe osteogenesis imperfecta. Transplantation. 2005 Jun 15;79(11):1607–14. doi: 10.1097/01.tp.0000159029.48678.93. [DOI] [PubMed] [Google Scholar]
- 40.Chan JK, Götherström C. Prenatal transplantation of mesenchymal stem cells to treat osteogenesis imperfecta. Front Pharmacol. 2014;5:223. doi: 10.3389/fphar.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chitty LS, David AL, Gottschalk I, Oepkes D, Westgren M, Götherström C, et al. EP21.04: BOOSTB4: a clinical study to determine safety and efficacy of pre- and/or postnatal stem cell transplantation for treatment of osteogenesis imperfecta. Ultrasound Obstet Gynecol. 2016;48(Suppl 1):356. [Google Scholar]
- 42.Deciphering Developmental Disorders Study. Large-Scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ACMG Board of Directors. Points to consider in the clinical application of genomic sequencing. Genet Med. 2012 Aug;14(8):759–61. doi: 10.1038/gim.2012.74. [DOI] [PubMed] [Google Scholar]
- 44.Stark Z, Schofield D, Alam K, Wilson W, Mupfeki N, Macciocca I, Shrestha R, White SM, Gaff C. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017 Jan 26; doi: 10.1038/gim.2016.221. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 45.Committee Opinion No682: Microarrays. Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics Gynecology Committee on Genetics and the Society for Maternal-Fetal Medicine. Obstet Gynecol. 2016 Dec;128(6):e262–e268. doi: 10.1097/AOG.0000000000001817. [DOI] [PubMed] [Google Scholar]
- 46.Green RC1, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013 Jul;15(7):565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 48.Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: Screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. American Journal of Human Genetics. 2012 Jul 13;91(1):97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borry P, Evers-Kiebooms G, Cornel MC, Clarke A, Dierickx K. Genetic testing in asymptomatic minors. Background considerations towards ESHG Recommendations. Eur J Hum Genet. 2009;17:711–9. doi: 10.1038/ejhg.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skirton H, Goldsmith L, Jackson L, Lewis C, Chitty L. Offering prenatal diagnostic tests: European guidelines for clinical practice. Eur J Hum Genet. 2014;22:580–6. doi: 10.1038/ejhg.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westerfield L, Darilek S, van den Veyver IB. Counseling challenges with Variants of Uncertain Significance and Incidental Findings in Prenatal Genetic Screening and Diagnosis. J Clin Med. 2014;3:1018–32. doi: 10.3390/jcm3031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Gennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40(10):e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012 Oct 3;4(154):154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson D, et al. Exome Sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2911;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 58.Longoni M, High FA, Qi H, Joy MP, Hila R, Coletti CM, Wynn J, Loscertales M, Shan L, Bult CJ, Wilson JM, Shen Y, Chung WK, Donahoe PK. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet. 2017 Jun;136(6):679–691. doi: 10.1007/s00439-017-1774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horn R, Parker M. Opening Pandora’s box?: ethical issues in prenatal whole genome and exome sequencing. Prenat Diagn. 2017 Jul 10; doi: 10.1002/pd.5114. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bunnik EM1, de Jong A, Nijsingh N, de Wert GM. The new genetics and informed consent: differentiating choice to preserve autonomy. Bioethics. 2013 Jul;27(6):348–55. doi: 10.1111/bioe.12030. [DOI] [PubMed] [Google Scholar]
- 61.Pinxten W, Howard HC. Ethical issues raised by whole genome sequencing. Best Pract Res Clin Gastroenterol. 2014;28(2):269–79. doi: 10.1016/j.bpg.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Gymrek Melissa, McGuire Amy L, Golan David, Halperin Eran, Erlich Yaniv. Identifying Personal Genomes by Surname Inference. Science. 2013;339:321–4. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 63.Botkin JR, Belmont JW, Berg JS, Berkman BE, Bombard Y, Holm IA, et al. Points to Consider: Ethical, Legal and Psychosocial Implications of Genetic Testing in Children and Adolescents. Am J Hum Genet. 2015;97:6–21. doi: 10.1016/j.ajhg.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lacey S, Chung JY, Lin H. A compariso of whole genome sequencing with exome sequencing for family-based association studies. BMC Proc. 2014 Jun 17;8(Suppl 1 Genetic Analysis Workshop 18Vanessa Olmo):S3. doi: 10.1186/1753-6561-8-S1-S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Committee Opinion No 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstet Gynecol. 2015 Sep;126(3):e31–7. doi: 10.1097/AOG.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 66.Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Wuake SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487(7407):320–4. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4(137):137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bi W, Breman A, Shaw CA, Stankiewicz P, Gambin T, Lu X, Cheung SW, Jackson LG, Lupski JR, Van den Veyver IB, Beaudet AL. Detection of ≥1Mb microdeletions and microduplications in a single cell using custom oligonucleotide arrays. Prenat Diagn. 2012 Jan;32(1):10–20. doi: 10.1002/pd.2855. [DOI] [PubMed] [Google Scholar]
- 69.Breman AM, Chow JC, U’Ren L, Normand EA, Qdaisat S, Zhao L, Henke DM, Chen R, Shaw CA, Jackson L, Yang Y, Vossaert L, Needham RH, Chang EJ, Campton D, Werbin JL, Seubert RC, Van den Veyver IB, Stilwell JL, Kaldjian EP, Beaudet AL. Evidence for feasibility of fetal trophoblastic cell-based noninvasive prenatal testing. Prenat Diagn. 2016 Nov;36(11):1009–1019. doi: 10.1002/pd.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]