Two carcinoma cases with ERBB2 transmembrane domain mutation, that showed clinical improvement with afatinib, are presented. The treatment strategy for the second patient was based on the experience of afatinib treatment for the first patient and was accomplished by sharing information of such rare mutations interinstitutionally. This article highlights the importance of establishing an integrated database of genomic and clinical information, including therapeutic outcomes, to implement precision oncology medicine.
Abstract
We previously reported on a family with hereditary lung cancer, in which a germline mutation in the transmembrane domain (G660D) of avian erythroblastic leukemia viral oncogene homolog 2 (erb‐b2 receptor tyrosine kinase 2) (ERBB2; human epidermal growth factor receptor 2 [HER2]) seemed to be responsible for the cancer predisposition. Although few data are available on treatment, anti‐ERBB2 therapeutic agents may be effective for ERBB2‐mutant cancers. The familial lung cancer patient in one of the authors’ institutes developed bone metastasis with enlarging lung tumors and was treated with the ERBB2 inhibitor afatinib. We also encountered a patient with ampullary adenocarcinoma with ERBB2 G660D and S310F comutations in another institute of the authors’, revealed by comprehensive genomic profiling. This patient was then treated with afatinib and also achieved transitory response. We also searched for ERBB2 transmembrane mutations in various types of cancers in PubMed, The Cancer Genome Atlas (TCGA), and the Memorial Sloan Kettering‐Integrated Mutation Profiling of Actionable Cancer Targets (MSK‐IMPACT) database. Besides our two cases, two patients with V659E mutations were found via PubMed. Three potential patients were found in TCGA. In addition, MSK‐IMPACT allowed identification of three additional urothelial carcinomas with G660D mutations and two lung adenocarcinomas with V659E mutations. Our experience suggests that establishing a database of integrated information regarding the clinical genome and therapeutic outcome of patients with recurrent but less common mutations is essential to implement precision oncology.
Key Points.
Rare but targetable mutations such as avian erythroblastic leukemia viral oncogene homolog 2 (erb‐b2 receptor tyrosine kinase 2) (ERBB2; human epidermal growth factor receptor 2 [HER2]) transmembrane domain (TMD) mutations can be detected by comprehensive genomic profiling.
Afatinib may be effective for patients with cancer with ERBB2 (HER2) TMD mutations.
In order to implement precision oncology, it is important to establish a database of integrated information regarding the clinical genomes and therapeutic outcomes of patients with recurrent but less common mutations.
摘要
我们既往报道过一个有遗传性肺癌的家族, 其中鸟类成红细胞白血病病毒致癌基因同源物2(erb‐b2受体酪氨酸激酶2)[ERBB2;人表皮生长因子受体2(HER2)]跨膜结构域(G660D)中的种系突变似乎是造成癌症倾向的原因。虽然与治疗有关的数据非常少, 但抗ERBB2治疗剂可能对ERBB2突变型癌症有效。其中一位作者所在研究机构的家族性肺癌患者发生了骨转移, 并伴有肺肿瘤扩大, 接受了ERBB2抑制剂阿法替尼的治疗。在作者的另一个研究机构中, 我们还发现了一例经全面的基因组分析显示具ERBB2 G660D和S310F突变的壶腹部腺癌患者。该患者随后接受了阿法替尼治疗, 并实现了短暂的反应。我们还在PubMed、肿瘤基因图谱(TCGA)以及Memorial Sloan Kettering‐可操作肿瘤靶标综合突变分析序列(MSK‐IMPACT)数据库中搜索了ERBB2跨膜突变。除我们的两个病例外, 通过PubMed还发现了两例具V659E突变的患者。在TCGA中发现了三例潜在的患者。另外, 通过 MSK‐IMPACT 发现了三例具有G660D突变的泌尿道上皮癌和两例具有V659E突变的肺腺癌。我们的经验表明, 要实现肿瘤精准治疗, 必须建立一个综合信息数据库, 其中包括具有频发但不常见突变的患者的临床基因组和治疗结果信息。
要点:
• 罕见但可靶向的突变, 如鸟类成红细胞白血病病毒致癌基因同源物2(erb‐b2受体酪氨酸激酶2)[ERBB2;人表皮生长因子受体2(HER2)]跨膜结构域(TMD)突变可通过全面的基因组分析进行检测。
• 阿法替尼对具有ERBB2(HER2)跨膜结构域突变的癌症患者可能有效。
• 为了实现肿瘤精准治疗, 必须建立一个综合信息数据库, 其中包括具有频发但不常见突变的患者的临床基因组和治疗结果信息。
Patient Stories
Patient 1
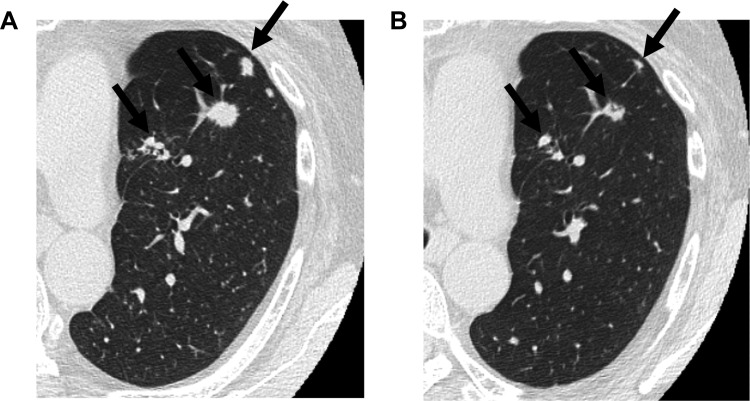
Patient 1, the familial lung cancer patient in Okayama University Hospital, was a 56‐year‐old woman with the avian erythroblastic leukemia viral oncogene homolog 2 (erb‐b2 receptor tyrosine kinase 2) (ERBB2; human epidermal growth factor receptor 2 [HER2]) G660D mutation described in our previous report [1]. She developed bone metastasis with enlarging multiple lung tumors. She was a light smoker with a 1.2‐pack‐year smoking history. She received carboplatin and pemetrexed but developed progressive disease. We examined the effect of afatinib on ERBB2 G660D‐transfected cells, leading to the inhibition of phosphorylation of ERBB2 G660D protein [2]. Based on our in vitro findings, we administered afatinib (40 mg per day) to Patient 1 using Health Insurance Claims Review and Reimbursement Service Board. Informed consent was obtained from the patient. Although a grade 3 adverse effect of an acne‐like rash appeared, the lesions in lung have shown a partial response (Fig. 1), and those in the bone have remained stable for the past 16 months with a reduced afatinib dose (20 mg per day). Clinical responses were based on Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 criteria.
Figure 1.
Effect of afatinib in Patient 1 with an ERBB2 G660D mutation. Computed tomography scan of the chest before (A) and after (B) treatment with afatinib. Shrinkage of multiple lung nodules (arrows) was observed.
Patient 2
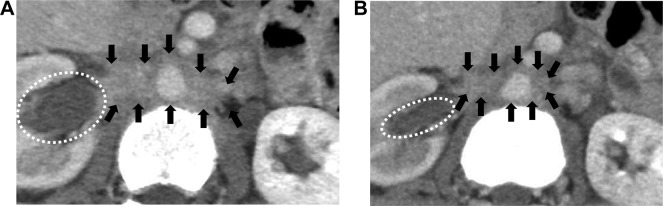
In Kyoto University Hospital, a 52‐year‐old woman, Patient 2, developed a postoperative metastatic lung nodule originating from adenocarcinoma of the ampulla of Vater. She had no smoking history. She had undergone curative resection followed by adjuvant chemotherapy. She developed recurrent disease in the lung 15 months after surgery and received gemcitabine and cisplatin because of progressive disease. Finally, she became resistant to the standard regimen for biliary tract cancer, and comprehensive genomic profiling, as described below, was performed on the resected primary tumor. Because we identified the same mutation, ERBB2 G660D, in Patient 2, as described below, we treated her with afatinib after the approval of the Ethics Committee. The treatment regimens were approved by the Institutional Review Board, and informed consent was obtained from the patient. The results of afatinib therapy are described in the Patient Update section, and the imaging findings are shown in Figure 2.
Figure 2.
Effect of afatinib in Patient 2 with ERBB2 G660D and S310F comutations. Computed tomography scan of the abdomen before (A) and after (B) treatment with afatinib. Shrinkage of the metastatic region of the para‐aorta (region circled with arrows) and improvement of hydronephrosis (region circled with dots) were observed.
Molecular Tumor Board
Comprehensive genomic profiling using the OncoPrime panel test, which sequences the entire coding region of 215 genes and examines the rearrangement of 17 frequently rearranged genes with clinical and preclinical relevance in human cancers [3], is performed for cancer patients in both Okayama University Hospital and Kyoto University Hospital, and the results are discussed at each institute. Patients are presented by the physician, and the board members, including doctors and bioinformaticians, discuss the results of the OncoPrime panel test and suggest any applicable drugs.
Genotyping Results and Interpretation of the Molecular Results
The OncoPrime [3] panel test was performed on the resected primary tumor in Patient 2. Detailed method was shown in the previous report [3]. OncoPrime identified ERBB2 G660D and S310F mutations in Patient 2. As the allele frequency result from OncoPrime for ERBB2‐S310F was 0.568, there was a possibility of germline mutation for S310F. We performed Sanger sequencing for both codons G660 and S310 using tumor and peripheral blood specimens. The ratio of the heights of mutant and wild type waves in the sequencing electropherogram for S310F in the tumor specimen was almost 1:1, whereas the variant allele was not detected in the specimen of peripheral blood for either G660 or S310, confirming that both G660D and S310F were somatic mutations, which suggested the presence of focal amplification in the locus of S310F. OncoPrime also reported a CTNNB1 T41A variant in this patient. At a joint conference of the members from both institutions, we shared the information that afatinib had been effective for Patient 1. In addition, a previous report strongly suggested that afatinib might be effective for disease with ERBB2 S310F mutations [4]. As described in the Patient Stories section, we treated Patient 2 with afatinib after the approval of the Ethics Committee.
Functional and Clinical Significance of ERBB2 Transmembrane Domain Mutations
Genomic and functional analyses suggested that the germline mutation at the transmembrane domain (TMD) of ERBB2 (G660D) in Patient 1 was responsible for familial lung cancer [1], [2]. We also found one patient with sporadic lung adenocarcinoma with an ERBB2 V659E mutation [1]. Furthermore, afatinib inhibited the phosphorylation of ERBB2 G660D or V659E protein in ERBB2 G660D‐ or V659E‐transfected cells [2]. TMD mutations V659E and G660D are located within the glycine zipper motif The652‐X3‐Ser656‐X3‐Gly660, a tandem variant of the GG4‐like motif, at the N‐terminal portion of the transmembrane domain, and it is reported that this motif is critically related to the dimerization of ERBB2 [5, 6].
We searched for ERBB2 TMD mutations in various type of cancers in PubMed and The Cancer Genome Atlas (TCGA) via cBioPortal [7]. We also performed a search for the Memorial Sloan Kettering‐Integrated Mutation Profiling of Actionable Cancer Targets (MSK‐IMPACT) clinical sequencing cohort [8]. Rare but recurrent mutations of the ERBB2 TMD occurred at residues G660 and V659, as summarized in Table 1. Besides our patient in the previous report [1], only two patients with V659E mutation were found via the PubMed database [9], [10]. Out of 147 studies and datasets, three potential patients were found in TCGA. In addition, five patients with ERBB2 TMD mutations were identified in the MSK‐IMPACT database. We have no detailed data on afatinib treatment for the ERBB2 TMD‐mutant cases shown in Table 1.
Table 1. Characteristics of patients with tumors harboring mutations in ERBB2 residues G660 and V659.
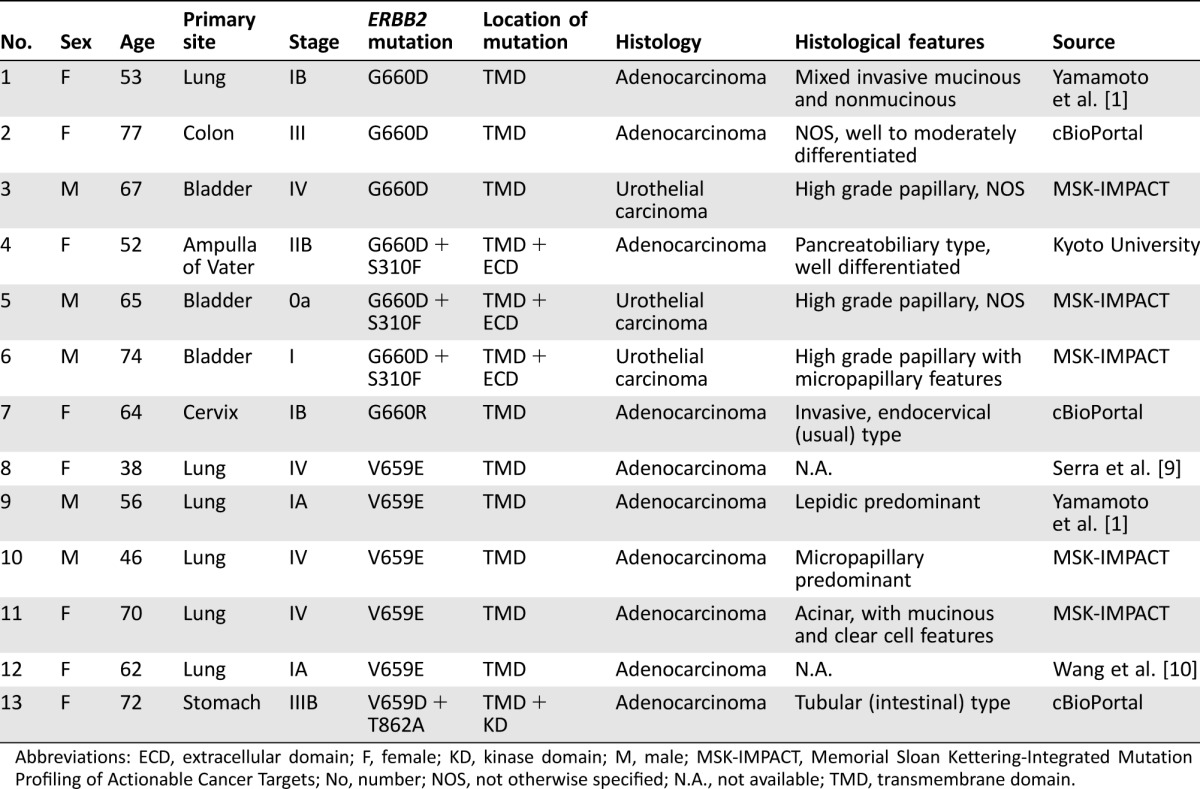
Abbreviations: ECD, extracellular domain; F, female; KD, kinase domain; M, male; MSK‐IMPACT, Memorial Sloan Kettering‐Integrated Mutation Profiling of Actionable Cancer Targets; No, number; NOS, not otherwise specified; N.A., not available; TMD, transmembrane domain.
A schematic figure of ERBB2 with the locations of other missense or in‐frame mutations in lung cancer via cBioPortal is shown in Figure 3A. ERBB2 mutations exist in the extracellular domain (ECD), TMD, and tail domain, in addition to the kinase domain (KD). Although little is known of the significance of the ERBB2 mutations in the ECD and tail domain, there is a possibility of conformational changes via these mutations like those in the KD and TMD, resulting in the increased activity of ERBB2. The MEK/ERK signaling pathway is stimulated by dimerization of ERBB2 [11]. ERBB2 amplification may also increase kinase activity. However, conformational changes do not occur via amplification itself, and increased kinase activity is dependent on the extent of the amplification. In non‐small cell lung cancer, ERBB2 mutation and amplification are mutually exclusive [12].
Figure 3.
Schema of ERBB2. (A): The G660D mutation is located in the ERBB2 transmembrane domain. Other missense or in‐frame mutations are also shown. The MEK/ERK signaling pathway is stimulated by dimerization of ERBB2. (B): Mechanisms of monoclonal antibodies, such as trastuzumab, and tyrosine kinase inhibitors (TKIs), such as afatinib, for ERBB2. Trastuzumab and other monoclonal antibodies bind to the ECD and prevent dimerization of ERBB2, whereas afatinib and other TKIs bind to the kinase domain (KD) and inhibit ERBB2. The KD is activated in ERBB2 KD‐ or TMD‐mutant tumors regardless of ECD status.
Abbreviations: ECD, extracellular domain; ERBB2, avian erythroblastic leukemia viral oncogene homolog 2 (erb‐b2 receptor tyrosine kinase 2); ERK, extracellular signal‐regulated kinases; MEK, MAPK‐ERK kinase; Mut, mutation; RAF, rapidly accelerated fibrosarcoma proto‐oncogene, serine/threonine kinase; RAS, rat sarcoma viral oncogene homolog; TMD, transmembrane domain.
Potential Strategies to Target the Pathway and Implications for Clinical Practice
Recurrent but less common mutations, such as ERBB2 TMD mutations, have potential for targeting therapy. However, it may be difficult to develop a treatment strategy, as the functional data on rare mutations are limited. In the current study, it was useful to share information interinstitutionally regarding the clinical genomes and therapeutic outcomes of patients with these rare mutations in order to perform the appropriate targeting therapy.
The development of comprehensive genomic profiling has opened the door for precision medicine. Our findings demonstrate that ERBB2 G660D is an actionable mutation for afatinib therapy. Although clinical response to lapatinib has been reported in a patient with ERBB2 V659E‐mutant lung cancer [9], our data suggest that responses to ERBB2‐targeted agents are not uniform across all patients with these ERBB2 mutations. Regarding Patient 1, the candidate alterations were restricted to 29 variants by comparing the results of whole‐exome sequencing, and the ERBB2 TMD mutation was picked up using five types of standardized deleteriousness scores (SIFT [13], PolyPhen‐2 [14], LRT [15], MutationTaster [16], and phyloP [17]). According to these scores, SRP54, MIPOL1, SLC26A1, IDUA, and SYTL5 were considered possible causative genes other than ERBB2, although the impact of these variants seems to be limited. As for Patient 2, the CTNNB1 T41A variant, which was mainly reported in soft tissue tumors, was identified in addition to ERBB2. CTNNB1 is responsible for Wnt signaling, suggesting that another pathway was also activated in this case. Identified mutations other than ERBB2 may affect the variable responsiveness to afatinib. Additionally, skin toxicity was observed in Patient 1 and not in Patient 2. As there was an association between the therapeutic effect of afatinib and skin toxicity in EGFR‐mutant lung cancer [18], we consider that there may also be a relationship between them in ERBB2 TMD‐mutant tumors.
As for ERBB2‐targeting therapy, possible options are small‐molecule inhibitors and monoclonal antibodies (Fig. 3B). There are several ERBB2‐targeting tyrosine kinase inhibitors (TKIs), such as lapatinib, neratinib, pyrotinib, and poziotinib, in addition to afatinib. Those TKIs have a potential to inhibit ERBB2 KD‐mutant tumors, as they directly bind to the KD of ERBB2. ERBB2 TMD‐mutant tumors also respond to those TKIs, as TMD mutations favor a kinase‐active conformation [5], [6], [19]. On the other hand, trastuzumab and other monoclonal antibodies target the ECD, preventing the dimerization of ERBB2. Because the KD is constitutively activated in ERBB2 KD‐mutant tumors [20] even if dimerization of ERBB2 is inhibited by trastuzumab or other monoclonal antibodies, their antiproliferative effect may be limited. In ERBB2 TMD‐mutant tumors, the effect of trastuzumab and other antibodies may also be limited, as dimerization of ERBB2 is predicted to be stable even if trastuzumab or other antibodies bind to the ECD [19]. Therefore, ERBB2‐targeting TKIs such as afatinib have a therapeutic advantage over trastuzumab and other monoclonal antibodies.
Patient Update
For Patient 2, there was a tendency of slight size reduction of the metastatic para‐aortic lymph node at 2.4 months after afatinib introduction that resulted in obvious clinical improvement of stenotic hydronephrosis (Fig. 2). Although the effect of afatinib was observed, the evaluation of the lesions was a stable disease, according to RECIST version 1.1 criteria. Patient 2 finally experienced progression of disease 4 months later. Skin rash was not observed.
Conclusion
We experienced two cases of patients with carcinoma with ERBB2 TMD mutation who showed clinical improvement with afatinib. The treatment strategy for Patient 2 was based on our experience of afatinib treatment for Patient 1, which was accomplished by sharing information of such rare mutations interinstitutionally. Our experience of sharing treatment outcomes highlights the importance of establishing an integrated database of genomic and clinical information, including therapeutic outcomes, to implement precision oncology medicine.
Acknowledgments
This study was supported by a Management Expenses Grant for National Universities in Japan, and NIH/NCI Cancer Center Support Grant P30 CA008748 (authors from Memorial Sloan Kettering Cancer Center)
Author Contributions
Conception/design: Shinichi Toyooka, Katsuyuki Kiura, Manabu Muto, Marc Ladanyi
Provision of study material or patients: Hiromasa Yamamoto, Takashi Ninomiya, Shigemi Matsumoto, Masashi Kanai
Collection and/or assembly of data: Shuta Tomida, Ken Suzawa
Data analysis and interpretation: Shuta Tomida, Ken Suzawa, Patrice Desmeules, Mark G. Kris, Bob T. Li
Manuscript writing: Hiromasa Yamamoto, Shinichi Toyooka, Marc Ladanyi
Final approval of manuscript: Shinichi Toyooka, Marc Ladanyi
Disclosures
Shinichi Toyooka: Boehringer‐Ingelheim (RF); Katsuyuki Kiura: Chugai Pharmaceutical, Pfizer, Novaltis Pharma, Taiho Pharmaceutical, Eli Lilly, Ono Pharmaceutical (RF); AstraZeneca, Boehringer‐Ingelheim, Nippon Kayaku, Daiichi Sankyo, Shionogi, Taiho Pharmaceutical, Ono Pharmaceutical (H); Marc Ladanyi: Boehringer‐Ingelheim, AstraZeneca (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Yamamoto H, Higasa K, Sakaguchi M et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst 2014;106:djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suzawa K, Toyooka S, Sakaguchi M et al. Antitumor effect of afatinib, as a human epidermal growth factor receptor 2‐targeted therapy, in lung cancers harboring HER2 oncogene alterations. Cancer Sci 2016;107:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kou T, Kanai M, Matsumoto S et al. The possibility of clinical sequencing in the management of cancer. Jpn J Clin Oncol 2016;46:399–406. [DOI] [PubMed] [Google Scholar]
- 4. Greulich H, Kaplan B, Mertins P et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci USA 2012;109:14476–14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bocharov EV, Mineev KS, Volynsky PE et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem 2008;283:6950–6956. [DOI] [PubMed] [Google Scholar]
- 6. Mineev KS, Bocharov EV, Pustovalova YE et al. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J Mol Biol 2010;400:231–243. [DOI] [PubMed] [Google Scholar]
- 7. Cerami E, Gao J, Dogrusoz U et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng DT, Mitchell TN, Zehir A et al. Memorial Sloan Kettering‐Integrated Mutation Profiling of Actionable Cancer Targets (MSK‐IMPACT): A hybridization capture‐based next‐generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serra V, Vivancos A, Puente XS et al. Clinical response to a lapatinib‐based therapy for a Li‐Fraumeni syndrome patient with a novel HER2V659E mutation. Cancer Discov 2013;3:1238–1244. [DOI] [PubMed] [Google Scholar]
- 10. Wang R, Zhang Y, Pan Y et al. Comprehensive investigation of oncogenic driver mutations in Chinese non‐small cell lung cancer patients. Oncotarget 2015;6:34300–34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohtsuka T, Sakaguchi M, Yamamoto H et al. Interaction of cytokeratin 19 head domain and HER2 in the cytoplasm leads to activation of HER2‐Erk pathway. Sci Rep 2016;6:39557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arcila ME, Chaft JE, Nafa K et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the sift algorithm. Nat Protoc 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 14. Adzhubei IA, Schmidt S, Peshkin L et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res 2009;19:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarz JM, Rodelsperger C, Schuelke M et al. Mutationtaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- 17. Siepel A, Pollard KS, Haussler D. New methods for detecting lineage-specific selection. In: Apostolico A, Guerra C, Istrail S, Pevzner PA, Waterman M, eds. Research in computational molecular biology, Berlin: Springer Berlin Heidelberg, 2006;190–205. [Google Scholar]
- 18. Kudo K, Hotta K, Bessho A et al. Development of a skin rash within the first week and the therapeutic effect in afatinib monotherapy for EGFR‐mutant non‐small cell lung cancer (NSCLC): Okayama Lung Cancer Study Group experience. Cancer Chemother Pharmacol 2016;77:1005–1009. [DOI] [PubMed] [Google Scholar]
- 19. Ou SI, Schrock AB, Bocharov EV et al. HER2 transmembrane domain (TMD) mutations (V659/G660) that stabilize homo‐ and heterodimerization are rare oncogenic drivers in lung adenocarcinoma that respond to afatinib. J Thorac Oncol 2017;12:446–457. [DOI] [PubMed] [Google Scholar]
- 20. Wang SE, Narasanna A, Perez‐Torres M et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 2006;10:25–38. [DOI] [PubMed] [Google Scholar]