Abstract
Thousands of long noncoding RNAs (lncRNAs) have been identified in eukaryotic genomes, many of which are expressed in spatially and temporally restricted patterns. Nonetheless, the roles of the majority of these transcripts are still unknown. One of the mechanisms by which lncRNAs function is through the modulation of chromatin state. To assess the functions of lncRNAs we developed RNA-DamID, a novel approach that detects lncRNA-genome interactions in a cell-type specific manner in vivo with high sensitivity and accuracy. Identifying the cell-type-specific genome occupancy of lncRNAs is key to understanding their mechanisms of action in development and disease. We used RNA-DamID to investigate targeting of the lncRNAs in the Drosophila dosage compensation complex (DCC) and show that initial targeting is cell-type-specific.
Introduction
Despite the potential importance of lncRNA-chromatin interactions in development and disease the function of the majority of lncRNAs in vivo is still unknown. One class of lncRNAs are found in the nucleus where they may function to regulate gene expression by facilitating the assembly of protein complexes at specific genomic loci 1–4. We developed RNA-DamID (RNA DNA adenine methylase identification) to map cell-type-specific lncRNA binding sites in vivo. RNA-DamID is based on Targeted DamID (TaDa) 5,6 in which DNA or chromatin binding proteins are fused to the E.coli adenine methylase Dam (DNA adenine methylase) and expressed at low levels under the spatio-temporal control of the GAL4 system 7.
We used RNA-DamID to investigate targeting of the Drosophila dosage compensation complex (DCC), which is comprised of lncRNAs roX1 and roX2 and the male-specific lethal proteins MSL1, 2 and 3, MOF and MLE. The roX RNAs are paradigms of lncRNA biology and are critical for assembly of the DCC on the male X chromosome. Here we show that the roX lncRNAs bind to DCC assembly sites in a cell-type-specific fashion. Surprisingly, we found that roX2 can also bind to a subset of target sites in females. Previously it was thought that Msl2 is not expressed in females. However, we found that roX2 binding is abolished in Msl2 mutant females, demonstrating that Msl2 is expressed in females at levels that are sufficient to localise roX2 to a subset of high-affinity chromatin entry sites (CES). roX2 is critically dependent on Msl2 for its recruitment.
Results
RNA-DamID a system for detecting cell-type specific lncRNA-chromatin interactions
We adapted TaDa to detect RNA-chromatin interactions using the bacteriophage MCP-MS2 system. First we fused the MS2 coat protein (MCP) tandem dimer to the bacterial Dam methylase (Supplementary Fig. 1a). Next we tagged the lncRNA of interest with three MS2 RNA stem loops (Supplementary Fig. 1b). MCP is able to bind to the MS2 tag with nanomolar affinity 8. If the tagged lncRNA binds, or is recruited to, genomic DNA or chromatin then the Dam-MCP fusion should recognise the tagged RNA and methylate adenine residues in the sequence GATC in the vicinity of the RNA-chromatin interaction (Fig. 1a). As a negative control we expressed three MS2 RNA loops lacking a fused lncRNA (Supplementary Fig. 1c, 2a). Spatio-temporal control of RNA-DamID expression by GAL4 and GAL80ts enables the detection of RNA-chromatin interactions in any cell type of interest. Digestion with the methylation-specific restriction enzyme, DpnI, followed by adaptor ligation, PCR and deep sequencing allows genome-wide detection of RNA occupancy. RNA-DamID works in intact tissues and does not require cell sorting, crosslinking or immunoprecipitation.
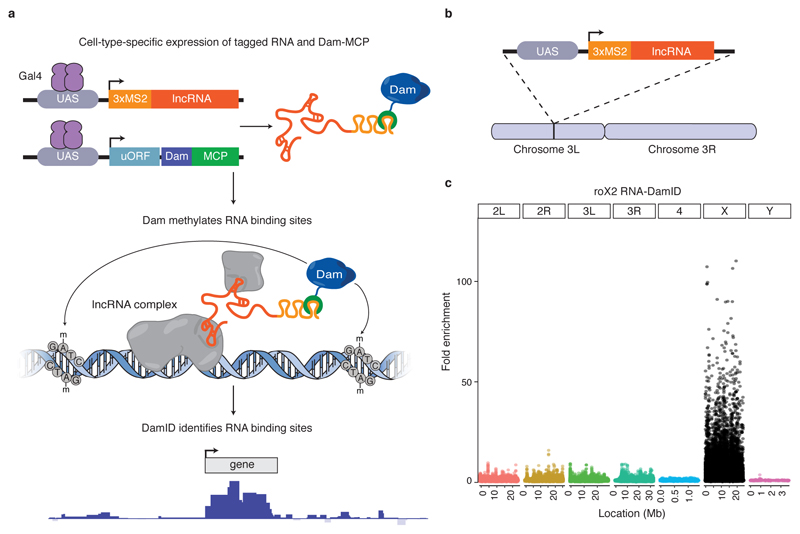
Figure 1. RNA-DamID accurately detects lncRNA-chromatin interactions in vivo.
(a) Schematic representation of RNA-DamID. A lncRNA of interest (red) tagged with 3xMS2 stem loops is co-expressed with a Dam-MCP fusion protein (blue and green, respectively) under the spatial and temporal control of GAL4 (purple). The Dam-MCP fusion is recruited to sites of lncRNA-chromatin interaction and methylates adenines within the sequence GATC. lncRNA binding sites are identified genome-wide by DamID. (b) A UAS-3xMS2-roX2 transgene was inserted on chromosome 3L. (c) Fold enrichment of roX2 RNA-DamID binding in whole male larvae, normalised to the negative control, reveals binding exclusively to the X chromosome (average of two biological replicates).
RNA-DamID detects lncRNA-chromatin interaction in vivo with high sensitivity
The lncRNAs roX1 and roX2, which regulate Drosophila dosage compensation, are among the best-understood examples of lncRNA function. The roX RNAs assemble into a complex with the male-specific lethal (MSL) proteins at specific sites on the male X chromosome 4,9. The MSL complex hyperactivates genes on the male X chromosome to equalise gene expression between males and females. We generated a UAS-3xMS2-roX2 transgene integrated on chromosome 3L (Fig. 1b). Driving expression with ubiquitously expressed GAL4 resulted in a strong enrichment of methylation on the X chromosome in male larvae (Fig. 1c). We normalised the RNA-DamID signal to the negative control, 3xMS2 stem loops co-expressed with MCP-Dam (Supplementary Fig. 2a-b). The result is similar to normalisation to Dam-alone (Supplementary Fig. 2c).
771/779 (99%) of binding peaks localise to the X chromosome while only 8 are detected on autosomes (Supplementary Data Set 1). Biological replicates show high signal and correlation on the X chromosome (Spearman’s correlation=0.805, R2=0.648), but not on the autosomes (Spearman’s correlation=0.0142, R2=0.002) (Supplementary Fig. 3). Our results for roX2 occupancy, determined by RNA-DamID from only 4 larvae, agreed closely with profiles generated previously with ChIRP from 300-1500 larvae 10. Therefore, RNA-DamID is able to profile accurately RNA-genome interactions with high sensitivity.
RNA-DamID has higher accuracy and sensitivity than ChIRP-seq
We also carried out TaDa of the MSL complex protein, Msl3, and compared our results with H4K16ac ChIP 11 (Fig. 2a-b). RNA-DamID detected 244/267 (91%) of peaks on the X identified by ChIRP in larvae 10 (Fig. 2b). However, none of the 26 autosomal peaks identified by ChIRP were detected using roX2 RNA-DamID, nor were these sites occupied by Msl3 or marked by H4K16ac, suggesting that they are likely to be false positives.
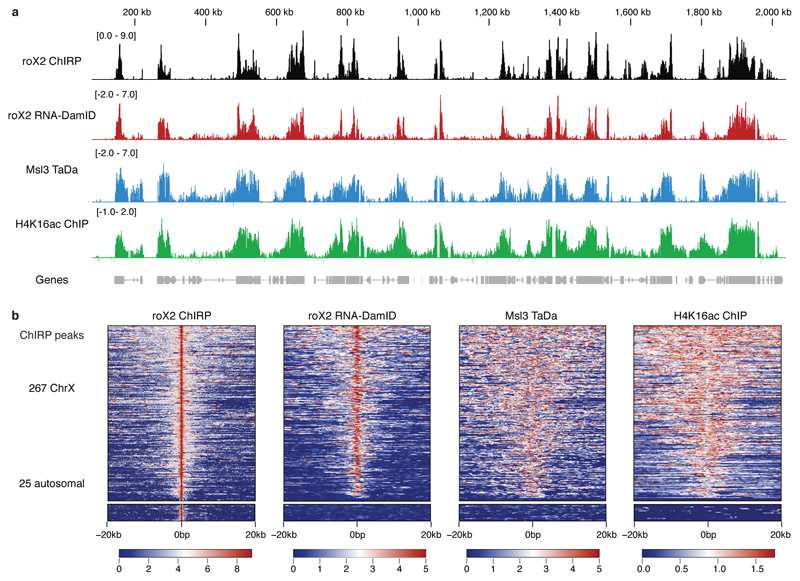
Figure 2. roX2 co-localises with the male-specific lethal complex.
(a) Peaks identified by roX2 RNA-DamID co-localise with roX2 ChIRP 10, Msl3 TaDa and H4K16ac ChIP 11. RNA-DamID scale represents log2 fold change of 3xMS2-roX2 and Dam-MCP compared to 3xMS2 and Dam-MCP (average of two biological replicates). Msl3 TaDa scale is a log2 fold change of Msl3-Dam fusion compared to Dam-alone. Chirp signal is log2 transformed. H4K16ac represents log2 fold change of H4K16ac ChIP over input. (b) Heat map of roX2 RNA-DamID, roX2 ChIRP, Msl3 and H4K16 ChIP signal plotted with over a 20kb window either side of roX2 ChIRP peaks. The majority of ChIRP X chromosome peaks, but not autosomal ChIRP peaks, show an enrichment of RNA-DamID, Msl3 TaDa and H4K16ac ChIP signal.
The results obtained with RNA-DamID of a roX2 transgene are highly similar to those obtained by ChIRP of endogenous roX2, but RNA-DamID is at least 75 fold more sensitive than ChIRP and shows greater specificity (Fig. 2b). In addition, we found 563 novel roX2 binding sites, also co-occupied by Msl3 and H4K16ac, that had not previously been detected (Supplementary Fig. 4). Interestingly, we did not observe spreading of roX2 in cis from the transgene insertion site (Supplementary Fig. 5). This supports the surprising observation that low level transcription of roX transgenes results in extensive spreading in cis, whereas robust induction of UAS-driven roX RNAs with GAL4 results in localisation exclusively on the X chromosome 12. Our results demonstrate that roX2 binding to the male X chromosome is independent of the genomic locus from which it is transcribed and that roX2 can truly act in trans.
roX RNA binding to chromatin entry sites is cell-type-specific
Dosage compensation occurs in two stages. First, the MSL/roX complex assembles in a sequence-dependent fashion at ~150 “chromatin entry sites” (CES) 13,14. Second, the processive MSL complex spreads to active genes on the X chromosome, which are recognised by Msl3 through binding to the histone mark H3K36me315.. This step is sequence-independent and depends upon gene expression. As yet it is not known whether the DCC binds initially to all CES in every cell, or whether its binding pattern is unique to each cell type
To assess whether initial targeting of the MSL complex to CES is cell-type-specific, we took advantage of an endogenously tagged allele of roX1 (6xMS2-roX1)16. Although roX1 and roX2 are significantly different in length (0.6 and 3.7 kb respectively) and exhibit little sequence homology 17, they are functionally redundant. The rox16xMS2 allele was generated in a roX2 mutant background, eliminating competition between the two RNAs for complex formation.
To assess roX1 occupancy in different tissues of the Drosophila larva, we analysed neural stem cells (NSCs), salivary glands (driving MCP-Dam with inscuteable-GAL4) and ubiquitous expression (tubulin-GAL4). We found that RNA-DamID is able to detect genome-wide binding of lncRNAs in different cell types in vivo. roX1 binding was enriched on the X chromosome in whole larvae, in NSCs and in salivary gland cells (Fig. 3a). While the average levels of roX occupancy are identical, we observed cell-type-specific binding of roX1 to CES (Fig. 3b-c): 73.3 % (110/150) of CES are bound by roX1 in all cell-types, while 20% (30/150) are cell-type-specific (Supplementary Data Set 1). In addition, 6.66% of CES (10/150) are not bound at detectable levels in any of the three tissues. The salivary gland and NSCs show the highest number of unbound CES (22 and 20 respectively), while the highest number of bound CES are found in whole larvae (139/150). We propose that the initial binding of the dosage compensation complex to CES is cell-type-specific, enabling the complex to spread efficiently to expressed genes.
Figure 3. roX1 binding to CES is cell-type specific.
(a) roX1 RNA-DamID signal from NSCs, salivary glands and whole larvae is enriched on the X chromosome (average of two biological replicates). Signal is plotted as fold enrichment over negative control. (b) Heat map of cell-type-specific binding to CES. Quantile-normalised log2 RNA-DamID is plotted over each CES (c) Example of cell-type-specific CES. log2 fold change of 3xMS2-roX2, Dam-MCP compared to 3xMS2, Dam-MCP.
Our results demonstrate that RNA-DamID was able to profile accurately, and with high sensitivity (~30,000 NSCs), the binding of a lncRNA expressed at its own endogenous levels: roX1 occupancy correlated very highly with roX2 occupancy in whole larvae (X chromosome Spearman’s correlation = 0.904, R2 = 0.817, Supplementary Fig. 6)
Targeting of the roX RNAs to CES requires Msl2
Some lncRNAs interact with chromatin through an RNA-intrinsic mechanism while proteins recruit others. It has been suggested that the roX RNAs are recruited to CES on the X chromosome by the core protein Msl2, which can bind the roX RNAs directly 18,19. Nonetheless, it is possible that the roX RNAs also have an intrinsic affinity for the X chromosome. It has been reported that the roX RNAs, and Msl2, are expressed exclusively in males 9 . To test if roX2 has an intrinsic ability to bind to chromosomal entry sites, we expressed the roX2 transgene ectopically in female larvae, where Msl2 expression is repressed by Sex lethal 20. If roX2 binding depends on Msl2, we expected to see no binding. However, if roX2 has an intrinsic ability to bind chromatin, then we should see binding.
We performed roX2 RNA-DamID and found that roX2 was specifically enriched on the X chromosome in females, suggesting that roX2 could bind chromatin independently of Msl2 (Fig. 4A). roX2 bound to a cluster of 14 CES in female larvae (Fig. 4D, Supplementary Data Set 1). Five of these sites correspond to the recently described “pioneering on X” regions, which are the first sites of MSL complex assembly19. roX2 was not enriched at most male binding sites (Fig. 4B), but 10/10 of the top female roX2 peaks were found within 1kb of a CES (Supplementary Data Set 1).
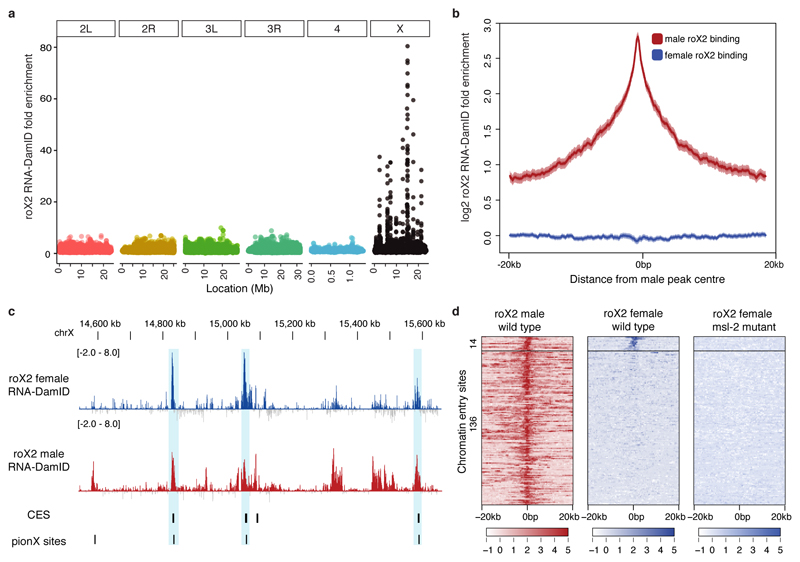
Figure 4. roX2 targets a subset of chromatin entry sites in females.
(a) roX2 is enriched on the X chromosome when expressed in female larvae. Data is plotted as fold enrichment over negative control. (b) roX2 occupancy is not enriched over most male binding sites. Data is plotted as the average log2 fold enrichment over the midpoint of each male peak. (c) Examples of female-bound chromatin entry sites. (d) Heatmap of roX2 occupancy on a subset of CES. Binding is abolished in msl-2 mutants 22. Data are represented as log2 transformed RNA-DamID signal. CES are k-means clustered according to roX2 signal in wild-type female larvae.
To confirm that roX2 binding in females was Msl2-independent, we performed RNA-DamID on female larvae homozygous null for Msl2 21. To our surprise, roX2 binding at the female-bound CES was abolished, demonstrating that binding is Msl2-dependent. We conclude that, contrary to previous reports, Msl2 is expressed in females at sufficiently high levels to localise ectopic roX2 RNA to a subset of CES. We conclude that roX RNA binding to the X chromosome is critically dependent on Msl2 protein, and that roX2 has no intrinsic affinity for chromatin.
Discussion
Here we have developed RNA-DamID, a powerful new approach for cell-type-specific analysis of the mechanism of action of lncRNAs. RNA-DamID can detect genome-wide lncRNA-chromatin interactions in a cell-type specific manner in vivo with high sensitivity and accuracy. We used RNA-DamID to assay binding of the lncRNAs in the Drosophila dosage compensation complex. Our results demonstrate that the initial targeting of the dosage compensation lncRNA, roX1, to CES on the X chromosome is cell-type-specific, differing in neural stem cells and salivary glands. We propose that cell type-specific targeting may increase the efficiency with which the DCC spreads to active genes.
Surprisingly, and contrary to previous reports, we found that females express Msl2, When we express roX2 ectopically in females, Msl2 is able to direct binding to a subset of dosage compensation complex assembly sites. We saw no binding of roX2 in the absence of Msl2, demonstrating the absolute dependence on Msl2 of roX2 recruitment.
Previous studies observed variation in CES occupancy between embryos and cultured cells, but it was unclear whether this resulted from the use of different technical approaches 13. Here we demonstrate context-dependent targeting of the roX RNAs in vivo ,suggesting that the extent of dosage compensation, and the genes escaping compensation, may vary between cell-types. Our results extend the current model of roX targeting to CES, which depends upon cell-identity, topology23 and sequence 13,14.
The interaction of other lncRNAs with chromatin may also be spatially and temporally regulated, and altered dynamically in development and disease. RNA-DamID enables the elucidation of lncRNA function in vivo and the discovery of the roles of lncRNAs, some of which that are precisely expressed throughout development. We have recently adapted TaDa for use in mammalian cells (Cheetham and Brand, unpublished) and anticipate that the application of RNA-DamID in other model systems will be straightforward and effective.
Online Methods
Cloning
To clone pUAST-LT3-Dam-NLS-MCPx2, NLS-MCPx2 was amplified and inserted into pUAST-LT3-Dam with XhoI and NotI. To clone pUAST_3xMS2, 3xMS2 was amplified from pCDNA3-3xMS2 (gift of P. Amaral) and inserted into pUAST-attb linearised with BglII and NotI by Gibson Assembly (NEB #E2611L) as per manufacturer’s instructions. To make the 5’ 3xMS2 tagged roX2, roX2 was amplified from pYP137 (gift of M. Kuroda) and inserted into pUAST_3xMS2 linearised with XhoI using Gibson Assembly. MSL3 was amplified from an embryonic cDNA library and inserted into pUAST-LT3-Dam with NotI and XbaI.
Primers
A full list of primers used in this study can be found in Supplementary Table 1.
Fly lines
Drosophila melanogaster stocks were raised at 25°C in a humidified incubator. tubulin-GAL4 24 and inscuteable-GAL4 25 were used in combination with tubulin-GAL80ts 26 for DamID experiments. The stocks UAS-Dam-NLS-MCPx2 [attp154], UAS-Dam-MSL3 [attp154], UAS-3xMS2-roX2 [attp2] were generated by coinjection of the relevant plasmid with the phiC31 integrase plasmid pBS130 into attp lines 27. roX16xMS2, roX2Δ was a kind gift of V. Meller. w;;tubulin-GAL4, tubulin-GAL80ts was kindly provided by T. Megraw. A full list of genotypes used in this study can be found in Supplementary Table 2.
Sample collection
DamID construct containing flies were crossed to a GAL4 driver with tubulin-GAL80ts. Embryos were collected on apple juice plates with yeast for 4 hrs at 25°C. Plates were then transferred to 18°C until larval hatching (~44 hrs). All larvae that hatched in a 3 hr collection window were transferred to a food plate with yeast. After 4 days at 18°C plates were shifted to 29°C and larvae were dissected after 2 days. For the NSC and salivary gland experiments 30 central nervous systems and salivary glands respectively were dissected. Males and females were separated by gonadal presence. Balancers were eliminated by fluorescence (CyO, act>GFP) or phenotype (tubby). Two biological replicates collected on separate days from separate crosses were analysed for each experiment.
DamID-seq
DamID was performed as previously described 28. Briefly, Genomic DNA was extracted using the QIAamp DNA Micro Kit (Qiagen, cat. no. 56304). The gDNA was eluted in 50 μl of H2O. 44 μl of DNA was incubated with 1μl of DpnI and 5μl of Cutsmart buffer overnight at 37° C. The DNA was purified using a Qiagen PCR purification kit and eluted in 32 μl. 15 μl of DNA was ligated to the 0.8 μl of adaptors (50 μM) with 2 μl of T4 Ligase Buffer 1.2 μl H2O and 1 μl of T4 DNA ligase. The ligation was incubated for 2 hrs at 16° and then heat inactivated at 65° for 20 minutes. 4 μl of DpnII buffer and 15 μl of H2O and 1μl of DpnII were added to the reaction. After 2 hrs incubation at 37° 16 μl of cDNA PCR buffer, 2.5 µl of DamID_PCR primer (50 µM), 3.2 µl 10 mM dNTPs, 96.3 µl of H2O and 2 µl of Advantage 2 cDNA polymerase enzyme were added and split into 4 PCR reactions. The following PCR program was run:
| Cycle number | Denaturation | Annealing | Extension |
|---|---|---|---|
| 0 | 68 °C, 10 min | ||
| 1 | 94 °C, 30 sec | 65 °C, 5 min | 68 °C, 15 min |
| 2-4 | 94 °C, 30 sec | 65 °C, 1 min | 68 °C, 10 min |
| 5-21 | 94 °C, 30 sec | 65 °C, 1 min | 68 °C, 2 min |
| 22 | 68 °C, 5 min |
Subsequently the DNA was purified using a Qiagen PCR Purification column and eluted in the 30 µl of H2O. 2 µg of DNA was added to 10 µl of Cutsmart buffer and made up to 100 µl with H2O. The solution was sonicated on high intensity for 6 cycles (30 secs on, 30 secs off) on a Diagenode Bioruptor® Plus. 1 µl of AwlI was added to cleave off the DamID adaptors and then library was prepared for sequencing using the modified TruSeq protocol elaborated upon in 28.
Data analysis and visualisation
DamID reads were aligned to Drosophila genome release dm6 and normalised as described previously 29. Briefly, reads for controls and the lncRNA-MS2, Dam-MCP libraries were binned into GATC fragments. The ratio of test over control was normalised by a kernel density estimate using readcounts from the accessible regions of the genome (assessed by the methylation pattern of the control). To reduce noise generated from ratios between regions with low signal, pseudocounts are added proportionally to the number of mapped reads. This normalisation process is described in further detail in 29. ChIRP bedgraph files were downloaded from the Gene Expression Omnibus (GSE69208) and converted to dm6 by UCSC liftover. ChIRP peaks, Chromatin Entry Sites 14 and pionX sites 19 were converted from dm3 to dm6 using UCSC Liftover. H4K16ac fastq files were downloaded from modENCODE and mapped to dm6 with Bowtie2 30. Seqplots 31 was used to generate heatmaps and density plots. Fold enrichment plotted over a 20kb window (Fig. 2b and 3d) or 1kb window (Fig. 3b) either side of the peak midpoint.
Peak calling
Peaks were called using a simple FDR peak caller 29. In brief, binding intensity thresholds are calculated from the normalised bedgraph file and compared to a randomly shuffled version of the dataset. The frequency of adjoining GATC fragments with intensity higher than the threshold is calculated. As the number of adjoining GATC fragments and the random observed frequency follows a logarithmic relationship it can be modelled by linear regression for any number of fragments. The FDR represents the ratio of observed consecutive fragments over the threshold compared to that expected over a threshold. Peaks with FDR<0.01 common to both biological replicates were considered.
Supplementary Material
Acknowledgements
We thank M. Kuroda (Harvard University), V. Meller (Wayne State University), T. Megraw (Florida State University) and P. Amaral (Gurdon Institute) for reagents; T. Leonardi for advice on data visualisation; P. Amaral, T. Southall, O. Marshall and members of the Brand Lab for advice and discussion. This work was funded by Wellcome Trust Senior Investigator Award 103792 and Wellcome Trust Programme Grant 092545 to AHB. S.W.C. was funded by a Herchel Smith Research Studentship. A.H.B acknowledges core funding to the Gurdon Institute from the Wellcome Trust (092096) and CRUK (C6946/A14492).
Footnotes
Author Contributions
S.W.C and A.H.B designed the experiments. S.W.C performed the experiments. S.W.C and A.H.B analysed the data. S.W.C and A.H.B wrote the manuscript. All authors reviewed the manuscript prior to submission.
Competing financial interests
The authors declare no competing financial interests
Data availability
DamID-Seq data is deposited in the Gene Expression Omnibus under the accession number GSE97456.
Life Sciences Reporting Summary
A Life Sciences Reporting Summary for this article is available
References
- 1.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 2.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 5.Southall TD, et al. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev Cell. 2013;26:101–12. doi: 10.1016/j.devcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–8. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 7.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 8.Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–57. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- 10.Quinn JJ, et al. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016;30:191–207. doi: 10.1101/gad.272187.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho JWK, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–52. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- 13.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The Chromosomal High-Affinity Binding Sites for the Drosophila Dosage Compensation Complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alekseyenko AA, et al. A Sequence Motif within Chromatin Entry Sites Directs MSL Establishment on the Drosophila X Chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller CI, Akhtar A. The MSL complex: Juggling RNA-protein interactions for dosage compensation and beyond. Curr Opin Genet Dev. 2015;31:1–11. doi: 10.1016/j.gde.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Apte MS, et al. Generation of a useful roX1 allele by targeted gene conversion. G3. 2014;4:155–62. doi: 10.1534/g3.113.008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 6:113–21. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 18.Ilik IA, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–73. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villa R, Schauer T, Smialowski P, Straub T, Becker PB. PionX sites mark the X chromosome for dosage compensation. Nature. 2016;537:244–248. doi: 10.1038/nature19338. [DOI] [PubMed] [Google Scholar]
- 20.Bashaw GJ, Baker BS. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995;121:3245–3258. doi: 10.1242/dev.121.10.3245. [DOI] [PubMed] [Google Scholar]
- 21.Zhou S, et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 1995;14:2884–95. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S, et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 1995;14:2884–95. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez F, et al. High-Affinity Sites Form an Interaction Network to Facilitate Spreading of the MSL Complex across the X Chromosome in Drosophila. Mol Cell. 2015;60:146–162. doi: 10.1016/j.molcel.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 25.Luo L, Joyce Liao Y, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 26.McGuire SE. Spatiotemporal Rescue of Memory Dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 27.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–83. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall OJ, Southall TD, Cheetham SW, Brand AH. Cell-type-specific profiling of protein–DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat Protoc. 2016;11:1586–1598. doi: 10.1038/nprot.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall OJ, Brand AH. Damidseq-pipeline: An automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015;31:3371–3373. doi: 10.1093/bioinformatics/btv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stempor P, Ahringer J. SeqPlots - Interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 2016;1:14. doi: 10.12688/wellcomeopenres.10004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.