Abstract
To fully achieve the goals of a genomics‐enabled learning health care system, purposeful efforts to understand and reduce health disparities and improve equity of care are essential. This paper highlights 3 major challenges facing genomics‐enabled learning health care systems, as they pertain to ancestrally diverse populations: inequality in the utility of genomic medicine; lack of access to pharmacogenomics in clinical care; and inadequate incorporation of social and environmental data into the electronic health care record. We advance a framework that cannot only be used to directly improve care for all within the learning health system but can also be used to focus on the needs to address racial and ethnic health disparities and improve health equity.
Keywords: health equity, health disparities, genomic medicine, precision medicine, learning health care system
1. INTRODUCTION
Electronic health records (EHRs) are revolutionizing the practice of medicine. Not only does the digitalization and standardization of medical records help to improve patient outcomes by facilitating integrated care within complex medical settings,1 these properties also enable the improvement of the health care system itself2, 3, 4 by simplifying the periodic assessment of system function and quality,5, 6 the so‐called learning health system model. With the adoption of EHRs also comes changes to the content of the medical record. For example, the burgeoning ubiquity of genomic information in the clinical setting, from the increased use of genetic tests as an element of clinical care to direct to consumer testing results provided by patients, has galvanized efforts to integrate these data into the medical record.7, 8 This surge has also spurred the development of means of providing of relevant clinical knowledge and patient‐specific information related to medical genomics, known as genomic clinical decision support (CDS).9, 10, 11 However, the foundations for most integration and genomic CDS efforts, and indeed for the basis for many clinical genetic tests, rests in a body of work severely limited by the diversity of its participant populations.12 In some cases, these shortcomings have been identified; yet without adequate data on diverse populations, we cannot truly know the extent of these limitations, potentially compounding already gaping disparities in health.
Genomic integration into EHRs therefore comes with both significant caveats and substantial opportunity. On the one hand, care must be taken to assess the utility of genomic testing and the meaning of genomic results in specific contexts.11, 13, 14, 15, 16 On the other, the nature of EHRs and the facility with which they can be analyzed as a dataset allows health care systems to evaluate the effects of genetically enabled care on health outcomes, which have the potential to add not only to the improvement of patient care directly, through the refinement of clinical practices, but also indirectly through enhancement of the body of knowledge upon which biomedical discovery and development is based.17 In so doing, these health care systems can shed light on gaps in our scientific knowledge and stimulate research that will help our medical advances serve all populations equitably.
In this paper, we examine the conceptual foundations of EHRs and the learning health care model. We also explore the major challenges to health equity presented by the current state of EHRs and genomic and precision medicine, focusing on issues related to ancestral diversity. Finally, we take stock of these challenges and discuss a framework to aid in addressing them through research, technology, and learning health care.
2. THE LEARNING HEALTH CARE MODEL
Many aspects of the modern practice of medicine we owe to the scientific method. Aseptic technique, vaccines, CPR: these cornerstones of clinical care arose via a process of hypothesis‐driven, empirical determination of optimal methodology.18, 19, 20 This process is routinely applied to the tools of medicine, resulting in new treatment strategies and technologies that promise the radical improvement of clinical outcomes. However, these new tools are only as useful as their successful deployment. Outcomes may differ based on distinct factors affecting certain populations or individual patients, which must be considered in the care strategy. Moreover, the blanket application of a specific treatment for a given condition may drive up costs with limited medical benefits across the board.21 Proper implementation, therefore, requires its own application of the scientific method, embodied by the continuously learning health care system (LHS), to determine the ideal mechanism and context for use.
2.1. Assumptions of a Continuous Learning Health Care System
In 2012, the National Academy of Medicine, then the Institute of Medicine, published a roadmap to guide the development of LHSs.22 The guide assumed several characteristics of LHSs that touch on capabilities science and informatics, the patient‐clinician relationship, incentives, and culture (Table 1). A tacit assumption of the framework is the commitment to quality care for an increasingly diverse patient population. The human body, while generally universal among human beings, is subject to environmental impingements, both positive and negative, that result in a constellation of characteristics affecting individual patient care. The LHS model acknowledges the many forces acting on a patient's condition and demands that the information available for clinical decision making reflect the individual patient's situation to the greatest extent possible. The derivation of this information comes from the systematic reappraisal of action‐outcome relationships in the context of multiple factors and the integration of these findings into clinical practice.
Table 1.
Characteristics of a Continuously Learning Health Care System
| Science and Informatics |
| • Real‐time access to knowledge – A learning health care system continuously and reliably captures, curates, and delivers the best available evidence to guide, support, tailor, and improve clinical decision making and care safety and quality. |
| • Digital capture of the care experience – A learning health care system captures the care experience on digital platforms for real‐time generation and application of knowledge for care improvement. |
| Patient‐Clinician Relationships |
| • Engaged, empowered patients – A learning health care system is anchored on patient needs and perspectives and promotes the inclusion of patients, families, and other caregivers as vital members of the continuously learning care team. |
| Incentives |
| • Incentives aligned for value – In a learning health care system, incentives are actively aligned to encourage continuous improvement, identify and reduce waste, and reward high‐value care. |
| • Full transparency – A learning health care system systematically monitors the safety, quality, processes, prices, costs, and outcomes of care, and makes information available for care improvement and informed choices and decision making by clinicians, patients, and their families. |
| Culture |
| • Leadership‐instilled culture of learning – A learning health care system is stewarded by leadership committed to a culture of teamwork, collaboration, and adaptability in support of continuous learning as a core aim. |
| • Supportive system competencies – In a learning health care system, complex care operations and processes are constantly refined through ongoing team training and skill building, systems analysis and information development, and creation of the feedback loops for continuous learning and system improvement. |
Adapted from IOM, 2012 22
2.2. Benefits of a Learning Health Care System
The LHS model offers myriad prospective benefits over the classic model of clinical medicine. First and foremost is the potential for improvement in patient care. In 2012 to 2013, there were an estimated 265 860 premature deaths that could have been prevented with effective and timely health care.18, 22 Of those, over 83 000 deaths could have been avoided if all states improved their care to the level of the best‐performing state. Health disparities are dramatically impacted by social determinants of health like socioeconomic status, urbanicity/rurality, sexual orientation, and gender identity.23, 24, 25, 26, 27, 28, 29 Inequality in care in the United States is also disproportionately detrimental to racial and ethnic minority groups.30 This inequality has worsened for certain key measures—eg, at risk adults without a doctor visit; adults without a usual source of care; older adults without recommended preventive care—in the recent past.18 The LHS model offers the means by which the utility of particular care strategies can be dissected according to situation to give practitioners a better understanding of what works and when. Implementation of that knowledge in the care setting allows for the improved delivery of appropriate, often life‐saving treatment that is sensitive to individual needs.
Knowing what is likely to work in a given care scenario minimizes unnecessary procedures and care that drive up cost without improving benefits by identifying the most probabilistically effective courses of treatment. According to recent figures, wasteful spending constitutes up to a third of the money spent annually in the United States on health care.31 In 2011, wasteful health spending, excluding fraud, may have amounted to nearly $1 trillion (Table 2).32 The LHS model is uniquely poised to address wasteful health spending across the board. When information is leveraged to improve care, not only do patients receive better care tailored to their individual needs, treatment costs are also more likely to represent well‐informed medical decisions, improving overall value of clinical care.
Table 2.
Estimates of Waste in U.S. Health Care Spending in 2011, by Category
| Cost to Medicare and Medicaida (In billions) | Total Cost to U.S. Health careb (In billions) | |||||
|---|---|---|---|---|---|---|
| Low | Midpoint | High | Low | Midpoint | High | |
| Failures of Care Delivery | $26 | $36 | $45 | $102 | $128 | $154 |
| Failures of Care Coordination | $21 | $30 | $39 | $25 | $35 | $45 |
| Overtreatment | $67 | $77 | $87 | $158 | $192 | $226 |
| Administrative Complexity | $16 | $36 | $56 | $107 | $248 | $389 |
| Pricing Failures | $36 | $56 | $77 | $84 | $131 | $178 |
| Subtotal (%) | $166 (6%) | $253 (9%) | $304 (11%) | $476 (18%) | $734 (27%) | $992 (37%) |
| Fraud and Abuse | $30 | $64 | $98 | $82 | $177 | $272 |
| Total (%) | $197 | $300 | $402 | $558 (21%) | $910 (34%) | $1,263 (47%) |
Adapted from Lallemand, 2012.32 Totals may not match sum of components, due to rounding. a Includes state portion of Medicaid. b Total U.S. health care spending estimated at $2.687 trillion.
3. EHR TECHNOLOGY
The explosion of techniques, tools, and treatments, while aiding in the advancement of our clinical capabilities, have also exponentially complicated medical practice. To draw meaningful links between patient conditions, patient‐provider interactions, numerous intervention strategies, and health outcomes, and to distribute that information in a way that reaches the point of care, LHSs need a means of recording the relevant information that lends itself to data capture, curation, and delivery assumed by the model.22 The EHRs standardize the collection of information about patients, their care, and their health outcomes and facilitate the serial revision of practices according to system‐wide syntheses of available information. Without this standardization, connecting outcomes to practices would require great effort, and the undertaking on a large scale would not be feasible. Today, the EHR plays an essential role within a complex and rapidly evolving health IT ecosystem, consisting of technological infrastructure and methodology to aggregate, store, distribute, analyze, and extract information from the health record. However, in leveraging health IT, information fed back into the system is only as good as the information collected at the front end. To meet the needs of diverse individuals in an increasingly complex clinical environment, that diversity and complexity must be adequately reflected in the EHR as a first step towards effectively leveraging this information throughout the health IT ecosystem.
Adequate reflection of diversity not only means that EHRs must support inclusion of relevant data but also means that efforts must be made to record as much information from each patient as possible to capture their unique condition and context. For patients who receive routine care at a limited number of sites, information acquisition is a relatively simple task. But many patients have limited access to care, primarily seek out care in settings where the focus is on urgent issue remediation, and rely on care from multiple disparate settings, not all of which may share information or even utilize EHR technology. The eMERGE research study found that general IT implementation was the primary challenge in the integration of pharmacogenomics in the CDS tools in the EHR.11 The challenges of general information technology implementation will adversely impact federally qualified health clinics, community hospitals, and rural health care systems. These patients may be far more likely to bear the brunt of health disparities, in part because the seamless development of a complete electronic medical record is much more difficult.33 For these patients, providers must be conscientious about using every patient‐provider interaction as an opportunity to enhance the comprehensiveness of the EHR.
3.1. The Genomic Medicine‐Enabled EHR
Changes to the practice of medicine are changing medical record content. With the advent of precision medicine, health care has seen a rapid increase of the use of genomic information in the clinical setting.34, 35 This information may be derived from clinical tests for disease diagnosis or risk, as well as testing for pharmacological response. However, genomic information may also enter the clinical setting via the patient directly. Direct‐to‐consumer genetic testing platforms and research use of genomic sequencing have made personal genomic information readily accessible to public citizens, and patients may share this information with their health care providers.
Genomic information of all kinds is making its way into the EHR.36 Genomic information and interpretation in the EHR must be not only clinically relevant, it must also signify the most advanced information available. The rapidly expanding body of knowledge pertaining to genetic variation and gene‐environment interactions that shape health add a dimension of complexity to this effort. It also presents a growing burden on practitioners who, with relatively little formal training in genomics,37 must stay abreast of major developments in the field and to apply relevant information at the point of encounter. As such, there is a growing need for genomic CDS integrated into the EHR that both are resilient to consistent updates and provides information with adequate ease and depth to benefit clinical care.
Although the needs relating to genomic data integration and CDS have been widely recognized,13, 36 complications have stymied most efforts to broadly address these needs. Foremost, among these complications are the absence of standardized genomic data models, exchange formats, and representations of knowledge that would facilitate integration across systems in a scalable and consistent fashion. Such efforts are further hampered by lack of platform interoperability and widespread data sharing. Thus, genomic data and CDS integration projects have largely been platform‐ or site‐specific, drawing on different methodology and genomic information and data sources, a recipe for considerable variation in the utility of the final product.
4. HEALTH EQUITY CHALLENGES OF GENOMICS‐ENABLED LEARNING HEALTH SYSTEMS
The promise and productivity generated by the completion of the human genome project in 2003 have propelled genomic and precision medicine, as well as the biomedical research agenda, into the future. A proponent of this new era of precision medicine, President Barack Obama asked:
“Instead of trying a one‐size‐fits‐all treatment, what if medical experts could tailor one specifically for everyone's body? By bringing together doctors and data like never before, precision medicine aims to deliver the right treatments in the right dosage at the right time—every time.”38
While precision medicine and its genomic foundations do harbor great potential, neither can be embraced without caveat. The latent shortcomings of these approaches were presaged in a cautionary perspective published in Nature Genetics only a year after the completion of the human genome project. The article by Tate and Goldstein―entitled “Will tomorrow's medicines work for everyone?”―grappled with the precise challenges we face today, more than a decade later:
“If genetics does eventually prove relevant to the treatment of common diseases, then to the extent that genetic advances are uneven among racial and ethnic groups, disparities may result.”39
The medical benefits of genomics are beginning to be realized. Yet researchers have already observed that a lack of inclusion of ancestrally diverse populations is undermining genomic and precision medicine.40 Thus, not only are we are faced with the challenge of incorporating genomic information into clinical care, we do so with the full knowledge that this information is woefully incomplete. The lack of publicly available data on clinically relevant variants in diverse ancestral populations adversely affects the utility of new genomic knowledge for all patients for both diagnosis and treatment. To fully realize the benefits of precision medicine, EHRs must also be able to support the integration of relevant social and environmental, as well as genomic, information, and this information must reflect the broad diversity of humanity. The LHS model is uniquely poised not only to help address these challenges to health equity but also to evaluate and implement refined clinical tools.
4.1. Challenge I: Clinical Utility of Genomic Medicine
The results of genome‐wide association studies (GWAS) that examine the association between disease incidence and genomic variation comprise a large portion of the medical genetic literature. In 2010, Need and Goldstein published an article in Trends in Genetics that first highlighted the lack of ancestral diversity in GWAS available in public databases.41 A year later, Bustamante, Burchard, and De La Vega further enunciated the numerous barriers in place for truly diverse genomics research.42 In their comment in Nature, they reported that, as of 2010, 96% of GWAS were performed in populations of exclusively European descent. “Such challenges,” they maintained, “do not justify restricting the beneficiaries of medical genomic research to a small subset of humanity. Population‐based studies must be carried out on a global scale.”
Popejoy and Fullerton's 2016 update on Bustamante et al original analysis indicated a nearly 300% increase in the proportion of GWAS participants of non‐European descent.43 While promising, that still means non‐European individuals constitute only 19% of GWAS participants overall. Almost two‐thirds of non‐European GWAS participants are Asian, primarily the result of an increase in studies conducted in Asia, leaving individuals of African, Hispanic and Latin, Pacific Islander, Arab and Middle Eastern, Native, and mixed ancestry to comprise only 5% of the overall GWAS population.
Although the inclusion of diverse ancestral populations in such genomic databases as OMIM begin to address the issue of data availability, considerably more effort is needed to address the staggering dearth of information in non‐European ancestral populations. It is estimated that 25% of GWAS findings identified in primarily European ancestral populations have significantly different effect sizes in non‐European ancestral groups.44 Moreover, GWAS homogeneity and bias towards inclusion of subjects with European ancestry may miss key associations due to low frequency of variants in these populations.45 “Irrespective of what's driving it,” Popejoy and Fullerton note, “the continued under‐representation of populations of mixed ancestry or of people whose ancestry is not European is a problem. Until researchers are able to conduct amply powered GWAS on each major ancestral population across the world, scientists will continue to miss important information about disease biology.”
The GWAS is not the only category of genetic and genomic research lacking study population diversity, and this lack of diversity could have dire consequences for minority patients. Nowhere was this more clearly illustrated than in a 2016 study by Manrai et al detailing the validity of clinical genetic tests for hypertrophic cardiomyopathy risk when applied in ancestrally diverse patient populations.46 The study found that misclassification of benign variants as pathogenic occurred in multiple patients all of whom were of African or unspecified ancestry. The authors further articulated the importance of these findings:
“Such misclassifications invalidate risk assessments undertaken in relatives, requiring a chain of amended reports and management plans. Our findings suggest that false positive reports are an important and perhaps underappreciated component of the “genotype‐positive –phenotype‐negative” subgroup of tested persons. These findings show how health disparities may arise from genomic misdiagnosis.”
The future of genomic and precision medicine is contingent on the success of genomics research in understanding genetic variation in diverse ancestral populations.47 To adequately meet their mission of service to their entire patient population, genomic‐enabled LHSs must acknowledge this problem and commit to playing an essential role in resolving it.
4.2. Challenge II: Pharmacogenomics and Health Equity
Lack of diversity in research populations affects not only diagnostic and risk assessment tools but also treatment strategies. For example, the prescription drug clopidogrel (known by the brand name Plavix) is an inhibitor of platelet aggregation; its action is contingent of the metabolism of the drug in the liver into its active form, a process that depends on the CYP2C19 enzyme.48 Mutations in the CYP2C19 gene can significantly reduce drug metabolism, and thus drug efficacy.49 Clopidogrel was originally approved by the FDA in 1997 as an alternative to aspirin for preventing acute myocardial infarction.50 The FDA issued a black box warning in 2010 stating that poor metabolizers may not receive the full benefit of the drug treatment and may remain at risk for heart attack, stroke, and cardiovascular death.51
In March 2014, David Louie, District Attorney for the State of Hawaii, filed a lawsuit against the manufacturers of clopidogrel, Bristol‐Myers Squibb Company and Sanofi‐Aventis U.S. The complaint asserted claims against the Defendants for violation of Hawaii state laws for false, deceptive, and unfair marketing of clopidogrel.52 The Complaint asserts that the drug “has diminished or no effect on approximately 30% of the patient population” of the State of Hawaii. The Attorney General, on behalf of the State, contends that the pharmaceutical companies knew and failed to disclose, prior to the FDA's 2010 warning, that the drug did not benefit everyone within the State equally, and that individuals of certain ancestral background―Pacific Islanders and East Asians, ancestral groups that make up a significant proportion of the Hawaiian population―were more likely to harbor CYP2C19 mutations that made them poor responders to clopidogrel's active ingredient.53, 54, 55 Not all physicians, however, agree that CYP2CI9 testing should be routine in Hawaii.56
We are continuously learning about interindividual differences in drug exposure and/or drug response. Some interindividual differences are associated with individual patients’ ancestral backgrounds, and the drug development pipeline is beginning to warm to that concept.57 Today, race and ethnicity are often used as medical surrogates for biological ancestry, even in prescribing recommendations.12 There is a growing number of drugs with FDA‐approved product labeling directed at specific races and ethnicities.58 While standard GWAS analytic techniques do take hidden population structure into consideration, this information may not make its way into the interpretation and implementation of these results in clinical applications. The genomics‐enabled LHS has the potential to hasten our move beyond the crude proxies of racial and ethnic categories, measures that are fluid and change over time, to looking at the genotypes that may influence drug exposure and response.12 This would require LHSs to embrace genetic and genomic information more broadly as a component of patient care, including such information as ancestral markers in the EHR as well as the routine analysis of EHR data. Nevertheless, even understanding not only the self‐identified race and ethnicity of the patient but also the ancestral background, would augment the LHS by getting us closer to the end goal of geographically derived genetic differences.
4.3. Challenge III: Social and Environmental Determinants of Interindividual Health Differences
The statement “your zip code is a better predictor of your health than your genetic code” has been used for many years to emphasize the limitation of genomics.59 It also highlights the fact that social and environmental factors play an important role in interindividual differences in health outcomes.60 For example, it is now commonly accepted that smoking cessation can drastically reduce risk of cardiovascular disease.61, 62 More recent research has linked such properties as neighborhood ethnic density,63 childhood trauma,64, 65 and poverty66, 67 to health outcomes. Thus, true implementation of precision medicine requires the consideration of these factors as part of clinical care. The National Academy of Science in 2013 convened a committee to recommend core social and behavioral domains for inclusion in all EHRs (Table 3).68 As outlined in the report:
“It [the Committee] identified a parsimonious panel of measures that is comprehensive, interoperable and efficient… While recognizing the additional time needed to collect such data and act upon it, the committee concluded that the health benefits of addressing these determinants outweigh the added burden to providers, patients, and health care systems.”
Table 3.
Candidate Set of Domains for Consideration for the Inclusion in All Electronic Health Records
| Sociodemographic Domains |
| • Sexual orientation Race/ethnicity Country of origin/U.S. versus foreign born Education Employment Financial resource strain (food and housing insecurity) |
| Psychological Domains |
| • Health literacy Stress Negative mood and affect (depression, anxiety) Psychological assets (conscientiousness, patient engagement/activation, optimism, self‐efficacy) |
| Behavioral Domains |
| • Dietary patterns Physical activity Tobacco use and exposure Alcohol use |
| Individual‐Level Social Relationships and Living Conditions Domains |
| • Social connections and social isolation Exposure to violence |
| Neighborhoods and Communities |
| • Neighborhood and community compositional characteristics (socioeconomic and racial/ethnic characteristics) |
Adapted from IOM, 2014 68
Understanding the social and behavioral environment of patients, in addition to genomic information, can greatly inform clinical decision making. Social determinants of health―such as income, financial resource strain, education, and health literacy―are especially important in providing a social context for understanding a patient's clinical phenotype. Some EHRs capture such data as geolocation and employment status, as well as alcohol and smoking history, which may be used as proxies to measure the effects of certain social determinants on health and wellness.69 Income and education are strongly correlated with health and life expectancy.70 More than the effect such factors may have on health directly, they may also impinge on patients’ access to genomic and precision medicine and to new medical technologies. Therefore, potential health benefits of genomics‐enabled LHSs require the inclusion of genomic data and social and environmental EHR data to “harness the full potential of the information to provide better patient care,” as well as the recognition that social and environmental barriers to care exist and must be overcome.36 Not the least of these barriers is inconsistent appreciation for the import of such information by health care providers. Educational initiatives may be required to enhance providers’ recognition and use of social and environmental information as elements of clinical care.
5. CONCEPTUAL FRAMEWORK
It is not enough to acknowledge the limitations of our present understanding regarding the biological underpinnings of disease in diverse ancestral and social backgrounds. Nor is it prudent to allow the biomedical research community to shoulder the full burden of generating more complete data when a well‐suited mechanism for informing the biomedical research process exists at our fingertips. Inherent in the LHS model is the capacity for routine evaluation and adaptation of care practices. As more and, a wider variety of information, including genomic and social data, are incorporated into the EHR, there arises the potential for the LHS to inform not only care practices for diverse patient populations but also the biomedical research guiding precision medicine (Figure 1). Initial efforts to provide integrated CDS may be limited in its broad utility, due to the constraints mentioned previously. Yet the full extent of these limitations will remain unrealized until tested, which is precisely what can be accomplished through evaluation of health outcomes in diverse contexts.
Figure 1.
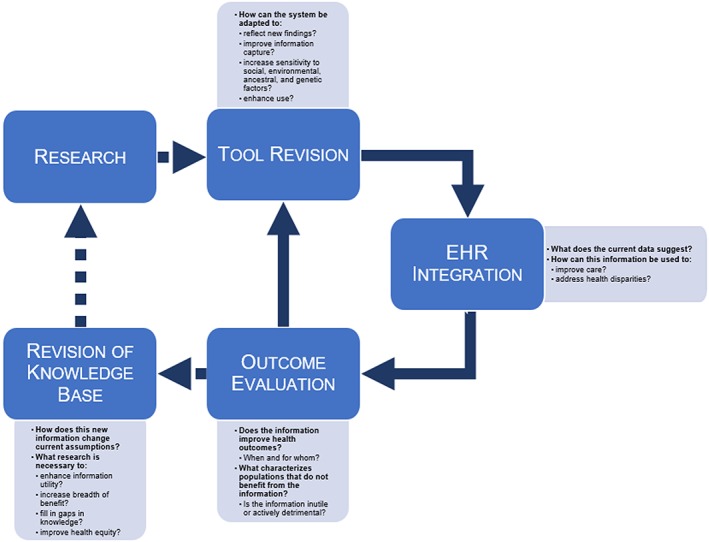
Improving Health Equity through Learning Health Care: A Framework.
The current LHS model is extremely proficient at asking the question of when information improves health outcomes; what is critical for ameliorating health disparities is the question of for whom does the information confer improvement. Attempts by LHSs to approach this question has largely been hampered by the content available in the EHR. Thus, leveraging the LHS model for health equity purposes is contingent on capturing an expanded set of data within the health record that allows for evaluation based on sociodemographic, environmental, and genomic factors. Similarly crucial for this approach is the bidirectional flow of information between the biomedical research community and the LHS. By feeding evaluations of clinical utility in the context of diverse biological, social, and environmental factors back into the research environment, we can revise the body of knowledge upon which CDS tools are based, in the interests of greater equity across the patient spectrum, and identify knowledge gaps requiring further experimental investigation.
Leaders in the field have set out principles for integrating personalized medicine into health care practice. These principles include establishing best practices for the collection and dissemination of evidence needed to demonstrate clinical utility and recognition of its value of care and to provide effective healthcare delivery infrastructure and data management systems to guide clinical decisions, so that individual patient and clinical support information is comprehensive, useful, and user friendly.71 The framework we propose can be construed as an expansion of these principles to incorporate the concept of health equity. These efforts will likely require steps be taken by the larger health care community (Table 4).
Table 4.
Steps to Improving Equity in Genomics‐Enabled Learning Health Care Systems
| Methods, Measures, and Models |
| • Develop international standards for the classification of populations to support global implementation of LHSs Develop new models to enhance the use of de‐identified clinical data from diverse populations |
| Monitoring |
| • Publicly monitor archived genotype and phenotype data available for genomic research by ancestral population |
| Patient Engagement |
| • Develop sustained and respectful relationships with ancestrally underrepresented communities to encourage participation in genomic research that may benefit clinical care Develop new methods for recruitment of ancestrally underrepresented communities for research participation within the genomics‐enabled LHS |
| EHR Infrastructure |
| • Enhance EHR information capture to improve resolution of population diversity Develop CDS that is sensitive to social, environmental, and ancestral factors, as well as genetic features Improve interoperability of EHR platforms to allow for information transfer between disparate points of care |
6. CONCLUSION
In 2015, the National Academy of Medicine held a workshop on genomics‐enabled LHSs; the workshop report included only a brief discussion of health care disparities, which referenced Charles Friedman's comments at the workshop:
“…the problems that get attention are those around which communities of interest form and generate enthusiasm for solutions. If communities of interest form around reducing disparities, learning cycles could take shape around those issues. ‘Let's look at ourselves and decide what's important.’”36
We agree that those guiding the use of the learning health care model have a responsibility to focus specifically on the question of how we can use the genomics‐enabled LHS to reduce health disparities an improve equity of care in the age of precision medicine. Moreover, in this paper, we have highlighted several major challenges facing such an enterprise: inequality in the utility of genomic medicine; lack of access to pharmacogenomics in clinical care; and inadequate incorporation of social and environmental data into the EHR. This discussion by no means encompasses the full scope of complexities facing the fields of genomic and precision medicine, which also includes such issues as inequality in genomic platforms, inconsistencies and methodological problems in older studies, often low prevalence of high‐risk pharmacogenomic variants, and a widespread need for genomics training amongst clinical providers. Nor do we attempt to put forward a solution that would comprehensively address the challenges presented in this paper. Nevertheless, we posit a framework that can not only be used to directly improve care for the full spectrum of patient diversity but can also be intentionally leveraged to address health disparities and improve health equity writ large. Adoption of this framework, in conjunction with solutions addressing the broader health IT and clinical ecosystems, as well as the research community, could usher in a new chapter in health. Precision medicine may yet revolutionize care across all patient populations, and the genomics‐enabled learning health care model is a powerful instrument we can use to ensure the revolution is equitable. By raising consciousness around the issues of health equity in the context of learning health care, and by outlining an approach to address some of the major challenges faced by precision health care, we hope to advance current efforts and stimulate dialog and positive change.
ACKNOWLEDGEMENT
This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. The opinions expressed in this article are those of the authors and do not necessarily reflect those of the National Human Genome Research Institute, the National Institutes of Health, or the Department of Health and Human Services.
Blizinsky KD, Bonham VL. Leveraging the Learning Health Care Model to Improve Equity in the Age of Genomic Medicine. Learn Health Sys. 2018;2:e10046 10.1002/lrh2.10046
REFERENCES
- 1. Shekelle PG, Morton SC, Keeler EB. Costs and benefits of health information technology. Evid Rep Technol Assess (Full Rep). Agency for Healthcare Research and Quality (US). 2006. Apr;(132):1‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowes LP, Noritz GH, Newmeyer A, et al. “Learn From Every Patient”: Implementation and early results of a learning health system. Dev Med Child Neurol. 2017;59(2):183‐191. [DOI] [PubMed] [Google Scholar]
- 3. Hessels A, Flynn L, Cimiotti JP, Bakken S, Gershon R. Impact of heath information technology on the quality of patient care. Online J Nurs Inform. NIH Public Access; 2015;19. [PMC free article] [PubMed] [Google Scholar]
- 4. Jarvis B, Johnson T, Butler P, et al. Assessing the impact of electronic health records as an enabler of hospital quality and patient satisfaction. Acad Med. 2013;88(10):1471‐1477. [DOI] [PubMed] [Google Scholar]
- 5. Etheredge LM. A rapid‐learning health system. Health Aff. 2007;(2):w107‐w118. [DOI] [PubMed] [Google Scholar]
- 6. Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2(57):57cm29 10.1126/scitranslmed.3001456 [DOI] [PubMed] [Google Scholar]
- 7. Sitapati A, Kim H, Berkovich B, et al. Integrated precision medicine: The role of electronic health records in delivering personalized treatment. Wiley Interdiscip Rev Syst Biol Med. 2017;9(3):e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warner JL, Jain SK, Levy MA. Integrating cancer genomic data into electronic health records. Genome Med. 2016;26;8(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teich JM, Osheroff JA, Pifer EA, Sittig DF, Jenders RA, Expert Review Panel CDS. Clinical decision support in electronic prescribing: Recommendations and an action plan: Report of the joint clinical decision support workgroup. J Am Med Inform Assoc. 2005;12(4):365‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Overby CL, Kohane I, Kannry JL, et al. Opportunities for genomic clinical decision support interventions. Genet Med. 2013;15(10):817‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herr TM, Bielinski SJ, Bottinger E, et al. Practical considerations in genomic decision support: The eMERGE experience. J Pathol Inform. 2015;6(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonham VL, Callier SL, Royal CD. Will precision medicine move us beyond race? N Engl J Med. 2016;374(21):2003‐2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shirts BH, Salama JS, Aronson SJ, et al. CSER and eMERGE: Current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015;22(6):1231‐1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaye JB, Schultz LE, Steiner HE, Kittles RA, Cavallari LH, Karnes JH. Warfarin pharmacogenomics in diverse populations. Pharmacotherapy. 2017;37(9):1150‐1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drozda K, Wong S, Patel SR, et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenetics and Genomics. 2015;25(2):73‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bentley AR, Callier S, Rotimi CN. Diversity and inclusion in genomic research: Why the uneven progress? J Community Genet. 1st ed. Springer: Berlin; 2017;6(Suppl 2):335–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: Past, present, and future. Genet Med. 2013;15(10):761‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCarthy D, Radley D, Hayes S. Aiming higher: Results from a scorecard on state health system performance. 2015. ed. The Commonwealth Fund; 2015.
- 19. Stern AM, Markel H. The history of vaccines and immunization: Familiar patterns, new challenges. Health Aff. 2005;24(3):611‐621. [DOI] [PubMed] [Google Scholar]
- 20. Lenzer J. Peter Joseph Safar. BMJ. 2003;327(7415):624. [Google Scholar]
- 21. Chandra A, Garthwaite C. The economics of indication‐based drug pricing. N Engl J Med. 2017;377(2):103‐106. [DOI] [PubMed] [Google Scholar]
- 22. Best Care at Lower Cost: The Path to Continuously Learning Health Care in America [Internet]. Washington, D.C: National Academies Press; 2013. Available from: https://www.nap.edu/read/13444/chapter/1. [PubMed] [Google Scholar]
- 23. Andrews AL, Kazley AS, Basco WT, Teufel RJ. Lower rates of EMR use in rural hospitals represent a previously unexplored child health disparity. Hosp Pediatr. 2014;4(4):211‐216. [DOI] [PubMed] [Google Scholar]
- 24. Butler MJ, Harootunian G, Johnson WG. Are low income patients receiving the benefits of electronic health records? A statewide survey. Health Informatics J. 2013;19(2):91‐100. [DOI] [PubMed] [Google Scholar]
- 25. 2016. National Healthcare Quality and Disparities Report [Internet] . AHRQ Pub. Rockville, MD; 2017 Jul. Report No.: AHRQ Pub. No. 17‐0001. Available from:http://www.ahrq.gov/research/findings/nhqrdr/nhqdr16/index.html
- 26. Thornton RLJ, Glover CM, Cené CW, Glik DC, Henderson JA, Williams DR. Evaluating strategies for reducing health disparities by addressing the social determinants of health. Health Aff. 2016;35(8):1416‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woolf SH. Progress in achieving health equity requires attention to root causes. Health Aff. 2017;36(6):984‐991. [DOI] [PubMed] [Google Scholar]
- 28. Braveman PA, Cubbin C, Egerter S, et al. Socioeconomic status in health research: One size does not fit all. JAMA. 2005;294(22):2879‐2888. [DOI] [PubMed] [Google Scholar]
- 29. Collins TW, Grineski SE, Morales DX. Environmental injustice and sexual minority health disparities: A national study of inequitable health risks from air pollution among same‐sex partners. Soc Sci Med. 2017;191:38‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williams DR, Purdie‐Vaughns V. Needed interventions to reduce racial/ethnic disparities in health. J Health Polit Policy Law. 2016;41(4):627‐651. 10.1215/03616878-3620857 [DOI] [PubMed] [Google Scholar]
- 31. Berwick DM, Hackbarth AD. Eliminating waste in US health care. JAMA. 2012;307(14):1513‐1516. [DOI] [PubMed] [Google Scholar]
- 32. Lallemand NC. Health Policy Brief: Reducing Waste in Health Care [Internet]. Health Affairs. 2012. Dec.; Available from: http://www.healthaffairs.org/healthpolicybriefs/brief.php?brief_id=82 [Google Scholar]
- 33. Sperber NR, Carpenter JS, Cavallari LH, et al. Challenges and strategies for implementing genomic services in diverse settings: Experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med Genomics. 2017;10(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature. 2015;526(7573):336‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abul‐Husn NS, Owusu Obeng A, Sanderson SC, Gottesman O, Scott SA. Implementation and utilization of genetic testing in personalized medicine. Pharmgenomics Pers Med. 2014;7:227‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roundtable on Translating Genomic‐Based Research for Health, Board on Health Sciences Policy, Institute of Medicine . Genomics‐Enabled Learning Health Care Systems: Gathering and Using Genomic Information to Improve Patient Care and Research: Workshop Summary. Washington (DC): National Academies Press (US); 2015 Jul 8. [PubMed]
- 37. Plunkett‐Rondeau J, Hyland K, Dasgupta S. Training future physicians in the era of genomic medicine: Trends in undergraduate medical genetics education. Genet Med. 2015;17(11):927‐934. [DOI] [PubMed] [Google Scholar]
- 38. Obama B. Medicine's next step. Boston Globe [Internet]. Boston, MA; 2016. Jul 7. Available from: https://www.bostonglobe.com/opinion/2016/07/06/medicine‐next‐step/tPdgf4XfOHvUckHpTTbuvN/story.html
- 39. Tate SK, Goldstein DB. Will tomorrow's medicines work for everyone? Nat Genet. 2004;36(11s):S34‐S42. [DOI] [PubMed] [Google Scholar]
- 40. Ramos E, Callier SL, Rotimi CN. Genetic misdiagnoses and the potential for health disparities. Per Med. 9(8):839‐847.23543886 [Google Scholar]
- 41. Need AC, Goldstein DB. Next generation disparities in human genomics: Concerns and remedies. Trends Genet. 2009;25(11):489‐494. [DOI] [PubMed] [Google Scholar]
- 42. Bustamante CD, Burchard EG, la Vega De FM. Genomics for the world. Nature. 2011;475(7355):163‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carlson CS, Matise TC, North KE, et al. Generalization and dilution of association results from European GWAS in populations of non‐European ancestry: The PAGE study. PLoS Biol. 2013;11(9):e1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fumagalli M, Moltke I, Grarup N, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349(6254):1343‐1347. [DOI] [PubMed] [Google Scholar]
- 46. Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cornel MC, Bonham VL. Genomics for all in the 21st century? J Community Genet. 2017;475:163‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenetics and Genomics. 2010;20(7):463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. De Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S‐mephenytoin metabolism in humans. J Biol Chem. 1994;269(22):15419‐15422. [PubMed] [Google Scholar]
- 50. Wu AH, White MJ, Oh S, Burchard E. The Hawaii clopidogrel lawsuit: The possible effect on clinical laboratory testing. Future Med. 2015; 12(3):179–181. https://doi.org/102217/pme154 [DOI] [PubMed] [Google Scholar]
- 51. Administration FAD . FDA Announces New Boxed Warning on Plavix: Alerts patients, health care professionals to potential for reduced effectiveness. Vol. Sandy Walsh, U.S. Food and Drug Administration. Press release. Retrieved March; 2010.
- 52. Department of the Attorney General of the State of Hawaii . Attorney General Files Suit Against Manufacturers and Distributors of the Prescription Drug Plavix [Internet]. Vol. Anne Lopez, Special Assistant to the Attorney General, Department of the Attorney General. Honolulu, HI; 2014. Available from: http://ag.hawaii.gov/wp‐content/uploads/2014/01/News‐Release‐2014‐09.pdf
- 53. Nakamura K, Goto F, Ray WA, et al. Interethnic differences in genetic polymorphism of debrisoquin and mephenytoin hydroxylation between Japanese and Caucasian populations. Clin Pharmacol Ther. 1985;38(4):402‐408. [DOI] [PubMed] [Google Scholar]
- 54. Kaneko A, Lum JK, Yaviong J, et al. High and variable frequencies of CYP2C19 mutations: Medical consequences of poor drug metabolism in vanuatu and other pacific islands. Pharmacogenetics and Genomics. 1999;9(5):581. [PubMed] [Google Scholar]
- 55. 2010 Census Interactive Population Search [Internet] . 2010 Census Interactive Population Search ‐ Hawaii. Washington, DC; [cited 2017. Mar 4]. Available from: https://www.census.gov/2010census/popmap/ipmtext.php?fl=15
- 56. Bhopalwala AM, Hong RA, Khan ZR, Valentin MR, Badawi RA. Routine screening for CYP2C19 polymorphisms for patients being treated with clopidogrel is not recommended. Hawaii J Med Public Health. 2015;74(1):16‐20. [PMC free article] [PubMed] [Google Scholar]
- 57. Administration UFAD . FDA Report: Action plan to enhance the collection and availability of demographic subgroup data. 2015.
- 58. Ramamoorthy A, Pacanowski MA, Bull J, Zhang L. Racial/ethnic differences in drug disposition and response: Review of recently approved drugs. Clin Pharmacol Ther. 2015;97(3):263‐273. [DOI] [PubMed] [Google Scholar]
- 59. Gordy C. The Root: The Shaky Future Of Health Care For Blacks [Internet]. NPR. 2011 [cited 2017. Mar 4]. Available from: http://www.npr.org/2011/04/06/135172202/the‐root‐the‐shaky‐future‐of‐health‐care‐for‐blacks
- 60. Hirsch A, Schwartz BS. The key to your health could be in your ZIP code [Internet]. The Conversation. 2015 [cited 2017. Mar 4]. Available from: https://theconversation.com/the‐key‐to‐your‐health‐could‐be‐in‐your‐zip‐code‐46304
- 61. Mensah GA, Wei GS, Sorlie PD, et al. Decline in cardiovascular mortality: Possible causes and implications. Circ Res. 2017;120(2):366‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Joseph P, Leong D, McKee M, et al. Reducing the global burden of cardiovascular disease, Part 1: The epidemiology and risk factors. Circ Res. 2017;121(6):677‐694. [DOI] [PubMed] [Google Scholar]
- 63. Mensah GA, Goff DC, Gibbons GH. Cardiovascular mortality differences‐place matters. JAMA. 2017;317(19):1955–1957. [DOI] [PubMed] [Google Scholar]
- 64. Oral R, Ramirez M, Coohey C, et al. Adverse childhood experiences and trauma informed care: The future of health care. Pediatr Res. 2016;79(1‐2):227–33. [DOI] [PubMed] [Google Scholar]
- 65. López‐Martínez AE, Serrano‐Ibáñez ER, Ruiz‐Párraga GT, Gómez‐Pérez L, Ramírez‐Maestre C, Esteve R. Physical health consequences of interpersonal trauma: A systematic review of the role of psychological variables. Trauma Violence Abuse. 10.1177/1524838016659488 [DOI] [PubMed] [Google Scholar]
- 66. COUNCIL ON COMMUNITY PEDIATRICS . Poverty and child health in the United States. Pediatrics. 2016;137(4):e20160339–9. [DOI] [PubMed] [Google Scholar]
- 67. Halfon N, Larson K, Son J, Lu M, Bethell C. Income inequality and the differential effect of adverse childhood experiences in US children. Acad Pediatr. 2017;17(7S):S70‐S78. [DOI] [PubMed] [Google Scholar]
- 68. Committee on the Recommended Social and Behavioral Domains and Measures for Electronic Health Records , Board on Population Health and Public Health Practice, Institute of Medicine Capturing Social and Behavioral Domains and Measures in Electronic Health Records: Phase 2. Washington (DC): National Academies Press (US); 2015. [PubMed] [Google Scholar]
- 69. Hollister BM, Restrepo NA, Farber‐Eger E, Crawford DC, Aldrich MC, Non A. Development and performance of text‐mining algorithms to extract socioeconomic status from de‐identified electronic health records. Pac Symp Biocomput. NIH Public Access. 2016;22:230‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Woolf SH, Braveman P. Where health disparities begin: The role of social and economic determinants‐‐and why current policies may make matters worse. Health Aff. 2011;30(10):1852‐1859. [DOI] [PubMed] [Google Scholar]
- 71. Pritchard DE, Moeckel F, Villa MS, Housman LT, Ca McCarty, Hl McLeod. Strategies for integrating personalized medicine into healthcare practice. Future Med. 2017;14(2):141–152. http://dxdoiorg/102217/pme‐2016‐0064. [DOI] [PubMed] [Google Scholar]


