Abstract
During embryonic development, cells become progressively restricted in their differentiation potential. This is thought to be regulated by dynamic changes in chromatin structure and associated modifications, which act together to stabilize distinct specialized cell lineages. Remarkably, differentiated cells can be experimentally reprogrammed to a stem cell-like state or to alternative lineages. Thus, cellular reprogramming provides a valuable platform to study the mechanisms that normally safeguard cell identity and identify factors whose manipulation facilitates cell fate transitions. Recent work has uncovered the chromatin assembly factor complex CAF-1 as a potent barrier to cellular reprogramming. In addition, CAF-1 has been implicated in the reversion of pluripotent cells to a totipotent-like state and in various lineage conversion paradigms, suggesting that modulation of CAF-1 levels may endow cells with a developmentally more plastic state. Here, we review these exciting results, discuss potential mechanisms and speculate on the possibility of exploiting chromatin assembly pathways to manipulate cell identity.
Introduction
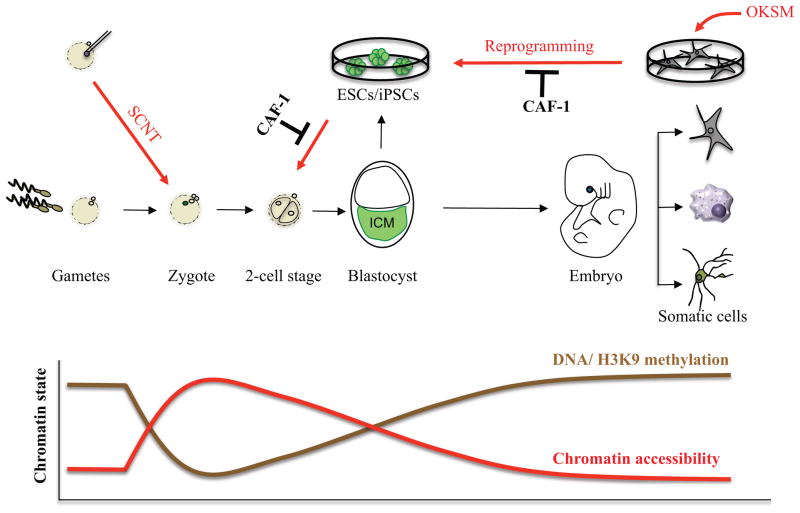
Development of multicellular organisms encompasses discrete stages of patterning and lineage specification, resulting in the production of all specialized cell types of the adult body. In mammals, fertilized zygotes and blastomeres of the cleavage stage embryo represent the developmentally most plastic state and are thus coined “totipotent,” which defines the ability of cells to produce all embryonic and extra-embryonic lineages of the developing organism [1] (Fig. 1). Fertilization represents a natural reprogramming process whereby the gametes’ chromatin undergoes dramatic chromatin reorganization in order to prepare the genome for embryonic development [2–5]. Seminal work by John Gurdon, Ian Wilmut and others demonstrated that the transfer of differentiated nuclei into enucleated oocytes using somatic cell nuclear transfer (SCNT) recapitulates this process and endows the somatic nucleus with a zygote-like state capable of supporting the development of a cloned animal [6,7].
Figure 1. Chromatin accessibility and modifications during development and nuclear reprogramming.
Development is accompanied by a gradual increase in chromatin compaction and the acquisition of repressive histone and DNA methylation patterns, which stabilize somatic cell fate and function as barriers to cellular reprogramming. Reprogramming to pluripotency and totipotency reverses these processes by chromatin decompaction (red arrows) and loss of silencing marks. The overexpression of transcription factors in somatic cells yields induced pluripotent stem cells (iPSCs) while the injection of somatic nuclei into oocytes by somatic cell nuclear transfer (SCNT) yields totipotent cells. CAF-1 suppression enhances the reprogramming of somatic cells to iPSCs and of ESCs/iPSCs to a totipotent-like state.
Following cleavage divisions and formation of the blastocyst embryo in mammals, cells residing within the inner cell mass (ICM) give rise to all embryonic lineages including the germ line. ICM cells are therefore called “pluripotent” [1]. Pluripotency can be captured in vitro by explanting blastocysts and deriving embryonic stem cells (ESCs)[8,9], which self-renew indefinitely in culture while retaining the potential to give rise to an entire animal upon reintroduction into host blastocysts [10,11]. Similar to zygotes, pluripotent cells are characterized by an open chromatin structure that reflects their potential to give rise to all embryonic cell types in vitro or in vivo. By contrast, differentiated cells exhibit a much more closed chromatin structure that correlates with their limited developmental potential and the establishment of specialized transcriptional programs [12](Fig. 1). Remarkably, Takahashi and Yamanaka demonstrated that pluripotency can be re-established in somatic cells by the ectopic expression of ESC-associated transcription factors such as Oct4, Klf4, Sox2 and c-Myc (OKSM), giving rise to induced pluripotent stem cells (iPSCs) [13]. The generation of iPSCs also reinvigorated earlier attempts of transcription factor-mediated cell fate change within somatic lineages [14,15] and led to more recent efforts to directly convert one mature cell type into another mature cell type using lineage-specific transcription factors or small molecules, a process termed direct lineage conversion or transdifferentiation [16].
The reprogramming of somatic cells to pluripotency or totipotency remains an ineffective process, suggesting that epigenetic barriers are established during development to safeguard somatic cell identity and resist cell fate change. Over the past decade, several chromatin pathways such as DNA methylation and histone H3K9 methylation, which are typically enriched within differentiated cells (Fig. 1), have been recognized as major roadblocks to nuclear reprogramming in the context of SCNT and iPSC generation [17,18]. However, the mechanisms by which chromatin modifications and overall chromatin accessibility act together to impede reprogramming and therefore protect cell identity remain incompletely understood. Here we review recent studies that have uncovered the chromatin assembly factor-1 (CAF-1) as a barrier to iPSCs generation and alternative cell fate transitions and thus implicate this essential complex in the regulation of cellular plasticity [19,20]. CAF-1 was identified almost three decades ago as a catalyzer of nucleosome assembly during DNA replication and repair [21,22]. Biochemical analyses further showed that CAF-1 is composed of three major subunits, p150, p60 and RbAp48 (also known as p48) that orchestrate complex interactions with histones and chromatin-modifying enzymes [23].
CAF-1 and cell fate control
The silencing of the somatic program occurs efficiently and rapidly in differentiated cells expressing the Yamanaka factors, OKSM [24]. By contrast, activation of the pluripotency-associated program is inefficient and slow. ChIP-seq analyses during early stages of human reprogramming revealed that transcription factors predominantly bind to distal elements that correspond to DNase I-resistant enhancers [25]. This observation is consistent with the idea of “pioneer factors”, which bind to closed chromatin and gradually recruit additional cofactors and remodeling enzymes to activate silenced target genes [26]. A subsequent study explored transcription factor occupancy during early stages of mouse reprogramming and confirmed binding of OKSM to enhancer elements, although no preferential binding to inaccessible enhancers was observed [27]. Specifically, this study found that OKSM facilitate the rapid silencing of somatic enhancers by physically associating with somatic transcription factors, leading to their redistribution to sites elsewhere engaged by OKSM. In addition, OKSM bind to pluripotency enhancers in a step-wise and collaborative manner, resulting in their gradual activation. The discrepancy between these studies with regards to the pioneering activity of OKSM could be due epigenetic differences between the somatic starting cells that were used, differences between the analyzed stages of reprogramming or species-specific differences. Despite these differences, a common conclusion is that the early engagement of transcription factors with chromatin fails to immediately activate a pluripotency-specific transcriptional program, suggesting that the pre-existing chromatin structure and associated modifications provide a profound impediment to effective transcriptional activation. In support of this notion, the manipulation of repressive and activating chromatin factors such as Setdb1, Dnmt1, PRC2 and SWI/SNF synergizes with OKSM to activate pluripotency-associated genes [28].
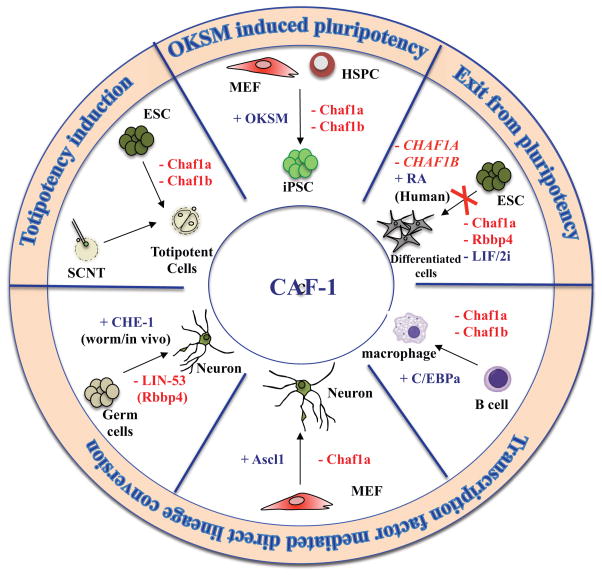
Additionally, several unbiased loss-of-function screens have been conducted during iPSCs generation to identify novel chromatin-associated factors that resist reprogramming [20,29–33]. In one such effort, our lab uncovered the two largest subunits of the chromatin assembly factor complex CAF-1, Chaf1a (p150) and Chaf1b (p60), as major roadblocks to iPSCs derivation (Fig. 2) [20]. Specifically, suppression of Chaf1a or Chaf1b enhanced the efficiency and speed of reprogramming by several orders of magnitude compared to other factors involved in heterochromatin maintenance or DNA methylation such as Dnmt1 and Setdb1. As CAF-1 is essential for cellular viability, optimal suppression was necessary to detect enhanced reprogramming and preserve a normal proliferative potential. This observation may also explain why previously identified chromatin barriers to reprogramming that are essential in somatic cells such as Mbd3 did not score prominently in our screen. Importantly, iPSCs generated with CAF-1 knockdown supported the development of high-grade germ-line chimeras, suggesting that transient CAF-1 suppression does not compromise the genomic or epigenetic integrity of cells. However, given that CAF-1 has previously been implicated in DNA damage [23], and genome stability is reportedly challenged during cellular reprogramming [34], additional experiments are warranted to rule out subtle changes to the genomic stability of iPSCs following CAF-1 depletion. Together these observations implicated CAF-1 for the first time in somatic cell fate control and raised the question of whether this complex may also be involved in other cell fate transitions.
Figure 2. Roles of CAF-1 in controlling cellular plasticity.
CAF-1 suppression facilitates cell fate change in different cellular systems with or without ectopic expression of transcription factors. Most cell fate switches were performed ex vivo using mouse cells unless noted. MEF (Mouse Embryonic Fibroblast), HSPC (Hematopoetic Stem and Progenitor Cell)
Indeed, Torres-Padilla and colleagues recently found that CAF-1 suppression endows ESCs with a more primitive developmental state that resembles totipotent 2-cell (2C) stage blastomeres of the cleavage stage embryo (Fig. 2)[19]. Interestingly, CAF-1 depleted ESCs were also more efficient at generating cloned blastocysts following SCNT, supporting the idea that hypomorphic CAF-1 expression endows ESCs with a chromatin state that is more amenable to reprogramming [19]. However, as blastocyst formation is not a very stringent readout for cloning efficiency [35], it will be informative to assess whether CAF-1 depleted nuclei also promote the postnatal survival of cloned mice. It will be equally interesting to assess the ability of CAF-1 depleted 2C-like cells to contribute to the extra-embryonic lineage upon injection into cleavage stage embryos, as has been shown for other cell populations with totipotent-like features [36,37]. Another question of interest is why only a small fraction of ESCs converted to the 2C-like state upon CAF-1 knockdown and whether this reflects different levels of CAF-1 depletion, a differential intrinsic susceptibility of some ESCs to revert or the acquisition of alternative cell states.
Interestingly, depletion of CAF-1 in ESCs not only endows them with a more primitive state but also appears to prevent their differentiation upon withdrawal from culture conditions that support self-renewal (Fig. 2). This conclusion is based on preliminary data from several recent RNAi screens aimed at identifying factors whose suppression delays or blocks ESCs differentiation [38–41]. Although no mechanism was provided on how CAF-1 loss impacts differentiation, it is tempting to speculate that CAF-1 depleted ESCs differentiate less due to the acquisition of a totipotent-like state, which may be less responsive to differentiation-inducing cues.
Notably, the loss of CAF-1 also facilitates other induced cell fate transitions that do not involve a pluripotent or totipotent state. For example, our lab recently showed that CAF-1 suppression enhances the conversion of B cells into macrophages upon overexpression of the myeloid transcription factor C/ebpa and of fibroblasts into neurons upon overexpression of the neuronal transcription factor Ascl1, representing two well-established transdifferentiation paradigms [20](Fig. 2). In addition, the smallest subunit of CAF-1, Rbbp4, scored as an epigenetic barrier in a chromatin-focused in vivo reprogramming screen in C. elegans (Fig. 2)[42]. Specifically, loss of the Rbbp4 ortholog Lin-53, together with overexpression of the neuronal-specific transcription factor CHE-1, led to the direct conversion of germ cells into neurons, indicating that Lin-53 normally safeguards germ cell fate. However, since Rbbp4 is not an exclusive component of the CAF-1 complex, it cannot be ruled out that the observed phenotypes were due to other chromatin complexes containing Rbbp4 such as the NURD and NURF nucleosome remodeling complexes and the SIN3A transcriptional repressor complex [23]. It should further be interesting to repeat this in vivo lineage conversion assay by knocking down the worm orthologs of the Chaf1a and Chaf1b subunits.
Altogether, these results are consistent with the notion that hypomorphic CAF-1 expression increases the epigenetic and developmental plasticity of cells in different cell lineages and across multiple model organisms (Fig. 2). A fundamental question emerging from these observations is how manipulation of CAF-1 facilitates cellular plasticity, which will be discussed in the following two sections.
Roles of CAF-1 in nucleosome assembly, heterochromatin maintenance and epigenetic memory
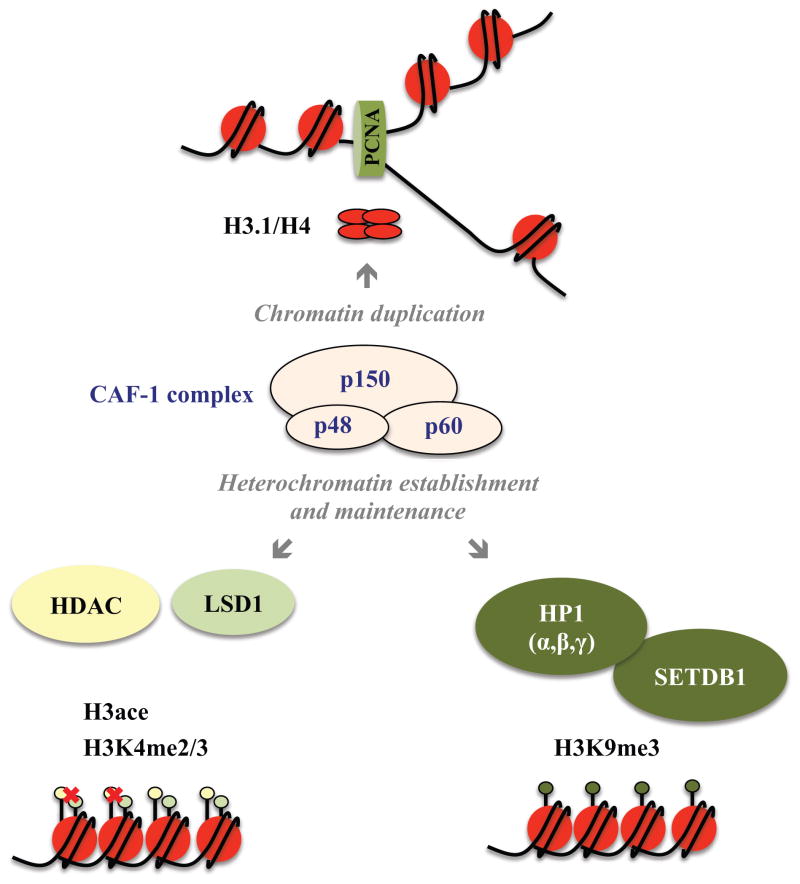
CAF-1 was originally characterized as a heterotrimeric complex that promotes the assembly of nucleosomes on replicating Simian Virus 40 (SV40) plasmid DNA using a cell-free replication system (Fig. 3) [21]. Subsequent complementation assays with human and Xenopus cell extracts demonstrated that CAF-1 is also involved in restoring nucleosome assembly after DNA repair [22]. CAF-1 directly binds to newly synthesized histones H3.1 and H4, supporting its replication-dependent function during nucleosome assembly [43,44]. In addition, CAF-1 localizes to replication foci through p150’s interaction with Proliferating Cell Nuclear Antigen (PCNA), which forms the replication clamp [45,46]. Notably, RNAi-mediated depletion of CAF-1 or overexpression of dominant negative mutants in immortalized cell lines decreases the assembly of newly replicated DNA into nucleosomes, stalling the replication process, activating DNA damage checkpoints and resulting in S phase arrest during cell division [47–49].
Figure 3. Functional diversity of the CAF-1 complex and its influence on chromatin structure and histone modifications.
Depiction of CAF-1 complex composition, highlighting its function as (1) a replication-dependent histone chaperone via its interaction with PCNA and association with H3/H4 histone tetramers, (2) heterochromatin silencing factor via recruitment of silencing complexes such as HP1/Sedtb1, which influence H3K9me3 deposition and LSD1 and HDAC, which influence erasure of H3K4 di- and tri-methylation and H3 acetylation.
In addition to its conserved function in nucleosome assembly, the CAF-1 complex also contributes to heterochromatin maintenance by forming distinct chromatin silencing complexes (Fig. 3). The p150 subunit contains domains that interact with the heterochromatin reader proteins HP1α and HP1γ (MOD1) and the H3K9 methyltransferase Setdb1 [50,51]. Mutational analyses of these HP1 interaction domains in mouse cells confirm their role in the maintenance of heterochromatin and this function also appears to be essential for cell viability [52]. Importantly, defective HP1 recruitment in these CAF-1 mutants does not seem to alter global nucleosome assembly. Thus, CAF-1 participates in at least two crucial and separable functions, nucleosome assembly and heterochromatin maintenance. To understand CAF-1’s molecular role in a given biological system, including reprogramming and transdifferentiation, it is therefore important to dissect the individual contribution of both processes. A case in point is the drosophila system where loss of CAF-1 causes defects in proliferation of mitotic and endocycling cells during larval development [53,54]. While these phenotypes were initially attributed to CAF-1’s classical chromatin assembly function, defects in heterochromatin maintenance could not be ruled out initially. Indeed, a more recent study showed that expression of a newly identified and evolutionarily conserved HP1 interacting domain within the large CAF-1 subunit rescues the embryo-lethal null phenotype, unmasking an unprecedented role of CAF-1 during oogenesis [55].
Recent evidence suggests that maintenance of gene silencing at heterochromatin loci and Polycomb targets is maintained through self-propagation of the respective histone marks or sequence-specific recruitment of silencing factors to replicated chromatin [56–59]. Considering the dual effect of CAF-1 in nucleosome assembly and heterochromatin maintenance, it is tempting to speculate that the complex contributes to this process by propagating silenced gene expression patterns through cell division and thus perpetuating an epigenetic memory. In support of this hypothesis, reduction of CAF-1 in Drosophila suppresses heterochromatin-dependent silencing and Polycomb-depedent gene repression [54]. It will certainly be interesting to test the role of CAF-1 in epigenetic memory in the context of cellular reprogramming [60].
Possible mechanisms by which CAF-1 depletion may induce cellular plasticity
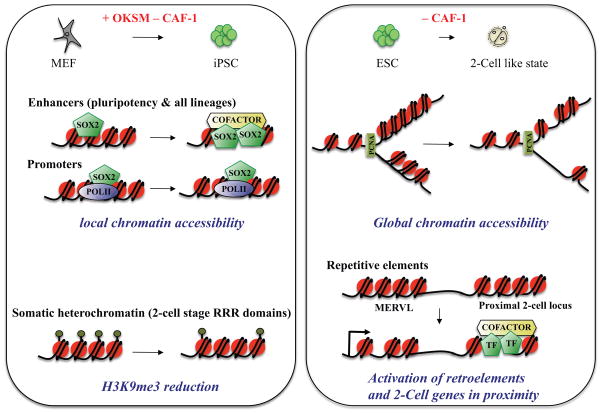
Analysis of genome-wide chromatin accessibility and Sox2 binding during iPSC reprogramming revealed that CAF-1 loss acts locally by facilitating the opening of chromatin and binding of Sox2 to enhancer elements (Fig. 4). The examination of somatic heterochromatin domains indicated that CAF-1 depletion also leads to the dilution of H3K9 trimethylation across so-called Reprogramming-Resistant Regions (RRRs). These heterochromatic RRR domains are normally active at the 2C stage in fertilization-derived embryos, yet remain silenced in SCNT embryos and thus provide an impediment to the efficient generation of cloned mice [61]. However, CAF-1 suppression did not affect H3K9me3 deposition elsewhere in the genome, nor the expression of transposable elements during the generation of iPSCs. This observation is consistent with previous CAF-1 knockdown experiments in fibroblasts [62] and argues that alternative mechanisms are in place in somatic cells to keep retro-elements silent. By contrast, acute CAF-1 suppression in undifferentiated ESCs does perturb heterochromatin organization. Specifically, repressive H3K9me3 and H4K20me3 histone marks as well as HP1α are lost from pericentric heterochromatin in CAF-1 depleted ESCs, while overall nucleosome organization remains unperturbed [62]. These results suggest that at least some of the phenotypes observed in CAF-1 depleted ESCs (e.g., differentiation block) are due to perturbed heterochromatin maintenance rather than chromatin assembly.
Figure 4. Mechanisms by which suppression of CAF-1 facilitates acquisition of a pluripotent or totipotent-like state.
Shown are models of how CAF-1 modulation may influence chromatin accessibility and histone modifications over distinct chromatin domains. During the reprogramming of somatic cells to iPSCs, suppression of CAF-1 acts locally at enhancer elements, making them more accessible to transcription factor binding. CAF-1 suppression also results in a local reduction of the H3K9me3 silencing mark at 2-cell (2C) stage-associated “reprogramming resistant regions” (RRRs), which are normally repressed in somatic cells. During the conversion of ESCs to a 2C-like state upon CAF-1 suppression, chromatin becomes more accessible globally, resulting in activation of endogenous retro-elements, such as MERVL transcripts, and neighboring genes.
Two recent studies established an intriguing link between CAF-1-dependent chromatin decondensation, repetitive element regulation and the ectopic expression of a 2C-like transcriptional program [19,63](Fig. 4). The activation of transposable elements in cleavage stage embryos has been proposed as a mechanism to rewire transcriptional networks and thus regulate stem cell identity [37,64,65]. Supporting this idea, Torres-Padilla and colleagues discovered that loss of CAF-1 in ESCs activates a subset of transposable elements that influence the expression of neighboring 2C stage-associated loci. The authors of that study further proposed that the reversion of ESCs to a 2C-like state depends on the nucleosome assembly function of CAF-1 during cell division. In further agreement with CAF-1’s role in repressing transposable elements and thus preserving the ESC state, Loh and colleagues identified CAF-1 as a major player in a genome-wide RNAi screen for regulators of retroviral silencing in ESCs [63]. CAF-1 depletion led to the de-repression of newly integrated proviruses and the reactivation of several endogenous retroviruses (ERVs) in that study. Expression analysis of CAF-1 knockdown ESCs as well as functional and biochemical characterization of CAF-1-associated complexes suggest that both CAF-1’s histone deposition function as well as its role in the recruitment of chromatin silencing complexes are involved in retro-element silencing. Strikingly, CAF-1 recruitment to different classes of ERVs seems to be regulated by co-binding to distinct cofactors such as Setdb1, Kdm1a and HDACs. However, the effect of CAF-1 on retro-element silencing in ESCs does not appear to be global as other subtypes such as intra-cisternal A particles (IAP) are not affected [19,63]. Interestingly, a recent report implicated H3.3 replication-independent histone chaperones, including a-thalassaemia/mental retardation syndrome X-linked (ATRX) and death-domain-associated protein (DAXX), in the silencing of IAP elements [66].
In summary, given the functional diversity of the CAF-1 complex and the recent characterization of additional domains within its subunits [23,55], it will be important to dissect the interdependence between nucleosome assembly and heterochromatin regulation in the context of reprogramming, transdifferentiation and pluripotency maintenance in the future. For instance, it should be informative to assess whether mutations within the newly identified HP1-interacting domain of CAF-1, discussed above, or within regions affecting binding to Kdm1a and HDACs affect the reversion of ESCs to a totipotent state, which has thus far been ascribed to CAF-1’s nucleosome assembly function [19].
Developmental roles of CAF-1
The composition and biochemical activities of the CAF-1 complex are evolutionarily conserved across human, mouse, amphibian, chicken, drosophila and yeast [23,67]. A pertinent question is therefore whether CAF-1 may function as a stabilizer of cell identity during normal development and tissue homeostasis in different multicellular organisms. However, addressing this question is challenging due to the early embryonic lethality of CAF-1 mutant animals. For example, CAF-1 knockout mice arrest between the 8- and 16-cell stage of pre-implantation development and no conditional allele has yet been reported [62]. Cytological analysis of CAF-1 mutant embryos using DAPI and HP1 staining implies a defect in constitutive heterochromatin domains, which are normally established after the second cleavage division. In support of this observation, two recent studies reported defects in heterochromatin organization and the repression of transposable elements upon CAF-1 knockdown during mouse pre-implantation development, possibly due to impaired distribution of histone H3 variants [68,69]. Interestingly these studies suggest that alternative deposition of the replication-independent histone variant H3.3 on chromatin is responsible for the perturbation of heterochromatin organization and activation of transposable elements upon CAF-1 knockdown. Strikingly, the embryonic lethality could be partially rescued by inhibition of reverse transcriptase activity, supporting the important role of CAF-1 in safeguarding the integrity of transcriptional networks during pre-implantation development by maintaining heterochromatin domains [69].
CAF-1 may also be required to maintain cell identity at later stages of development and in the adult. For example, CAF-1 is more abundantly expressed in stem cells compared to differentiated cells, suggesting that its downregulation may be important for cellular differentiation and tissue regeneration [63,70]. In support of this notion, CAF-1 scored as one of the top hits in a chromatin-focused RNAi screen for factors that prevent planarian regeneration by neoblasts, which serve as a model system for adult stem cells [70]. However, it remains to be determined whether the observed regeneration defect of CAF-1-depleted animals is due to a change of neoblast identity. Moreover, CAF-1 reportedly shuttles from the nucleus to the cytoplasm in developing mouse germ cells [71]. Since germ cells undergo major chromatin changes during gametogenesis, it is plausible that exclusion of CAF-1 from the nucleus facilitates epigenetic reprogramming through the deposition of histone variants by alternative histone chaperones. Further functional analysis of CAF-1 subunits during development using conditional and tunable genetic perturbation systems that circumvent early lethality will be critical to understand how this and other chromatin assembly pathways contribute to cellular plasticity during mammalian development.
Analysis of CAF-1 mutants in zebrafish and worms also points to a role in cellular differentiation and lineage specification [72–74]. Despite the embryonic lethality of CAF-1 mutants in both species, embryos develop to a stage that allowed probing the function of CAF-1 in early cell fate decisions. In zebrafish, CAF-1 loss leads to cell cycle arrest and differentiation defects in several organs including the retina, pectoral fins and head skeleton [72]. It is unclear at this point whether these phenotypes are due to CAF-1’s role in chromatin assembly or heterochromatin regulation. By contrast, the phenotypic similarity of mutants within histone H3 and CAF-1 in the worm points to an unprecedented role for CAF-1 histone deposition activity in generating bilateral asymmetry during embryonic development [73]. More recently, this lineage determination process has been ascribed to CAF-1’s role in suppressing Notch signaling [74]. Notably, CAF-1 may also promote Notch signaling during fly development where it reportedly collaborates with the transcriptional activator Suppressor of Hairless Su(H) [75].
Taken together, the loss-of-function phenotypes of this conserved and essential molecule across different animal models indicates that the action of CAF-1 as a transcriptional activator or repressor is highly context-dependent and points to additional nuclear functions for CAF-1 beyond histone deposition and heterochromatin maintenance.
Role of other histone chaperones in cellular plasticity and outlook
A number of additional histone chaperones have evolved to control the deposition of a wide repertoire of histone variants, raising the question of whether they might also influence reprogramming, development and cellular plasticity in mammals [76–78]. Indeed, histone chaperones are important for different developmental processes such as gastrulation, myogenesis and neurogenesis [76,79]. Surprisingly, these alternative histone chaperones did not score prominently in unbiased screens of reprogramming, although the manipulation of individual members such as HIRA reportedly influences SCNT [80,81]. It is plausible that CAF-1 scores more frequently and strongly in cell transition assays because it acts as a general chromatin factor affecting both nucleosome assembly and heterochromatin organization. Moreover, CAF-1 is proposed to affect a wide range of activating and repressive histone marks including H3K56ace, H3K9ace, H3K27ace, H3K9me3 and H3K4me2/3 [62,63,82]. It also remains poorly understood how CAF-1 interacts with alternative histone deposition pathways. For example, analysis of histone deposition in CAF-1 depleted HeLa cells indicates that alternate deposition of the histone variant H3.3 by the histone chaperone HIRA provides a gap-filling mechanism to compensate for CAF-1 loss [83]. Interestingly, previous work suggested that replacement of H3.3 and the concomitant activation of pluripotency genes during amphibian SCNT are, in part, mediated by the histone chaperone HIRA [80]. More recently, H3.3 deposition within the donor nucleus was also shown to facilitate mouse SCNT, indicating functional conservation of this mechanism across species [84]. It will be important to test whether CAF-1 depletion in somatic donor cells or in cells undergoing reprogramming into iPSCs leads to elevated H3.3 incorporation into chromatin and whether this underlies the increase in reprogramming efficiency.
In addition to CAF-1, the histone chaperone Asf1a has been implicated in cellular reprogramming and the maintenance of ESCs in human [81]. Asf1a acts upstream of CAF-1 by transferring newly synthesized and acetylated histones to CAF-1 [85]. In contrast to CAF-1, Asf1a overexpression rather than downregulation enhances human iPSCs formation. It is tempting to speculate that this phenotype is mediated by increased deposition of acetylated histones, thus inducing a more accessible chromatin state. It would further be interesting to test whether Asf1a overexpression overrides the repressive function of CAF-1 during iPSCs reprogramming.
Accumulating evidence suggests that common epigenetic mechanisms control cell fate change in the context of reprogramming, carcinogenesis and aging [28,86,87]. Given the profound effect of CAF-1 loss on the speed and efficiency of iPSCs formation, it will be interesting to test whether this complex also plays a role in cancer and aging. In support of this notion, recent studies found a connection between histone chaperone pathways and different types of cancers [77,79,88,89]. For example, the H3 chaperones ATRX and DAAX are mutated in pediatric glioblastoma, and CAF-1 levels are dysregulated in different solid tumors. As these findings are purely correlative, further studies are warranted to test whether alteration in histone chaperone pathways functionally contribute to tumor initiation, progression or maintenance. The observation that CAF-1 dosage is critical for reprogramming raises the intriguing possibility that CAF-1 may either promote or suppress tumor formation depending on cellular context. Indeed, while CAF-1 is typically overexpressed in solid tumors [90], a recent study found that its depletion endows epithelial cells with increased motility and invasive-like properties [91]. Finally, CAF-1 may play a role in ageing since both CAF-1 and histone levels decrease with cellular age. While a functional role for CAF-1 in ageing remains to be established, the recent observation that H3K9me3 levels also decrease with age suggests that CAF-1 may counter ageing by maintaining heterochromatin and thus safeguarding cell identity [92].
Altogether, these observations suggest that the effect CAF-1 perturbation on cell fate is context-dependent and influenced by several factors including the chromatin state of the target cell, environmental signals and chronological age. Although CAF-1 was discovered nearly three decades ago, its function in maintaining and safeguarding cell fate is only beginning to be recognized. Dissecting the mechanisms by which CAF-1 controls chromatin structure and function will be instrumental for a better understanding of the principles of cellular plasticity in health and disease.
Highlights.
Recent RNAi screens have identified CAF-1 as a barrier to cell fate change in various cellular and developmental systems
Effects of CAF-1 suppression are cell context-dependent, facilitating either differentiation, dedifferentiation or lineage conversion
CAF-1 suppression influences cellular plasticity by altering local or global chromatin states
Acknowledgments
We thank Raul Mostoslavsky and Michelle Carmell for useful discussions and comments. K.H. was supported by funds from the MGH, NIH (R01 HD058013-06) and the Gerald and Darlene Jordan Chair in Regenerative Medicine. S.C. was supported by funds from the Department of Defense Peer-Reviewed Cancer Research Program visionary postdoctoral fellowship (CA120212) and the Massachusetts General Hospital ECOR Tosteson Fund for Medical Discovery fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, Lensch MW, Lujan E, Pei D, Rossant J, Wernig M, Park PJ, Daley GQ. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 2.Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol. 2013;25:281–288. doi: 10.1016/j.ceb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Ishiuchi T, Torres-Padilla ME. Towards an understanding of the regulatory mechanisms of totipotency. Curr Opin Genet Dev. 2013;23:512–518. doi: 10.1016/j.gde.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, Li W, Li Y, Ma J, Peng X, Zheng H, Ming J, Zhang W, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang X, Xie W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. Using an improved version of ATAC-seq technology, the authors provide evidence for a unique chromatin state at repetitive elements and transcriptional end sites during the first three cleavage divisions. They further identify broad ATAC-seq domains over MERVL retro-elements and non-repeat early 2-cell associated genes. [DOI] [PubMed] [Google Scholar]
- 6.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 7.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 8.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 9.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 11.Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, 3rd, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 15.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 16.Hochedlinger K, Jaenisch R. Induced Pluripotency and Epigenetic Reprogramming. Cold Spring Harb Perspect Biol. 2015:7. doi: 10.1101/cshperspect.a019448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. This study reports that CAF-1 represses dinstinct transposable elements in ESCs and its loss induces a totipotent-like cell state, which requires CAF-1’s nucleosome assembly activity. [DOI] [PubMed] [Google Scholar]
- 20••.Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, Wesche DJ, Abazova N, Hogue M, Tasdemir N, Brumbaugh J, Rathert P, Jude J, Ferrari F, Blanco A, Fellner M, Wenzel D, Zinner M, Vidal SE, Bell O, Stadtfeld M, Chang HY, Almouzni G, Lowe SW, Rinn J, Wernig M, Aravin A, Shi Y, Park PJ, Penninger JM, Zuber J, Hochedlinger K. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–224. doi: 10.1038/nature15749. This paper implicates CAF-1 for the first time in transcription factor mediated reprogramming and transdifferentiation. The authors show that CAF-1 safeguards somatic cell identity during nuclear reprogramming by preserving chromatin accessibility and H3K9me3 at enhancer regions and specific heterochromatin areas, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. Oiginal paper defining CAF-1 complex composition and characterizing its function in nucleosome assembly during DNA replication. [DOI] [PubMed] [Google Scholar]
- 22.Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 23.Ridgway P, Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J Cell Sci. 2000;113(Pt 15):2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 24.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017;168:442–459. e420. doi: 10.1016/j.cell.2016.12.016. This is the first comprehensive chromatin map of early stages of mouse reprogramming, including binding patterns for 10 transcription factors, three chromatin factors and a broad range of histone marks, in addition to chromatin accessibility. The study provides valuable mechanistic insights into the silencing of somatic and the activation of pluripotency enhancer elements over time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang CS, Chang KY, Rana TM. Genome-wide functional analysis reveals factors needed at the transition steps of induced reprogramming. Cell Rep. 2014;8:327–337. doi: 10.1016/j.celrep.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, Mor N, Baran D, Weinberger L, Jaitin DA, Lara-Astiaso D, Blecher-Gonen R, Shipony Z, Mukamel Z, Hagai T, Gilad S, Amann-Zalcenstein D, Tanay A, Amit I, Novershtern N, Hanna JH. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 31.Qin H, Diaz A, Blouin L, Lebbink RJ, Patena W, Tanbun P, LeProust EM, McManus MT, Song JS, Ramalho-Santos M. Systematic identification of barriers to human iPSC generation. Cell. 2014;158:449–461. doi: 10.1016/j.cell.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dejosez M, Ura H, Brandt VL, Zwaka TP. Safeguards for cell cooperation in mouse embryogenesis shown by genome-wide cheater screen. Science. 2013;341:1511–1514. doi: 10.1126/science.1241628. [DOI] [PubMed] [Google Scholar]
- 34.von Joest M, Bua Aguin S, Li H. Genomic stability during cellular reprogramming: Mission impossible? Mutat Res. 2016;788:12–16. doi: 10.1016/j.mrfmmm.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 36•.Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, Macfarlan T, Peng W, Lloyd KC, Kim SY, Speed TP, He L. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017:355. doi: 10.1126/science.aag1927. This paper shows how suppression of single microRNA relieves the repression of a MERVL endogenous retroelement in ESC, inducing a totipotent-like state. The authors provide a mechanism whereby miR-34a represses GATA2, which contributes to MERVL silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzales KA, Liang H, Lim YS, Chan YS, Yeo JC, Tan CP, Gao B, Le B, Tan ZY, Low KY, Liou YC, Bard F, Ng HH. Deterministic Restriction on Pluripotent State Dissolution by Cell-Cycle Pathways. Cell. 2015;162:564–579. doi: 10.1016/j.cell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang SH, Kalkan T, Morrisroe C, Smith A, Sharrocks AD. A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet. 2012;8:e1003112. doi: 10.1371/journal.pgen.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3. 3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 44.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 45.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 46.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 47.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 49.Nabatiyan A, Krude T. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol Cell Biol. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 51.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 52.Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 53.Klapholz B, Dietrich BH, Schaffner C, Heredia F, Quivy JP, Almouzni G, Dostatni N. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118:235–248. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- 54.Song Y, He F, Xie G, Guo X, Xu Y, Chen Y, Liang X, Stagljar I, Egli D, Ma J, Jiao R. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev Biol. 2007;311:213–222. doi: 10.1016/j.ydbio.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 55••.Roelens B, Clemot M, Leroux-Coyau M, Klapholz B, Dostatni N. Maintenance of Heterochromatin by the Large Subunit of the CAF-1 Replication-Coupled Histone Chaperone Requires Its Interaction with HP1a Through a Conserved Motif. Genetics. 2017;205:125–137. doi: 10.1534/genetics.116.190785. This paper characterizes a novel evolutionary conserved HP1a interacting domain in the largest CAF-1 subunit in Drosophila. Deletion of this domain in the fly rescues the reported larval lethality and reveal heterochromatin associated functions of CAF-1 in position effect variegation and homologous chromosome pairing during oocyte meiosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De S, Kassis JA. Passing epigenetic silence to the next generation. Science. 2017;356:28–29. doi: 10.1126/science.aan1493. [DOI] [PubMed] [Google Scholar]
- 57.Laprell F, Finkl K, Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 58.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017:356. doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356:88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan GJ, Bai Y, Fletcher J, Wilmut I. Induced pluripotent stem cells: epigenetic memories and practical implications. Mol Hum Reprod. 2010;16:880–885. doi: 10.1093/molehr/gaq091. [DOI] [PubMed] [Google Scholar]
- 61••.Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. This paper identifies a subset of somatic heterochromatin demains that are marked by H3K9me3 and show resistance to reprogramming in SCNT embryos. Overexpression of the H3K9 demethylase Kdm4d in SCNT embryos dissolves these domains and enhances cloning efficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, Li Y, Lorincz MC, Tergaonkar V, Lim TM, Chen L, Gunaratne J, Collins JJ, Goff SP, Daley GQ, Li H, Bard FA, Loh YH. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. This paper is the first unbiased genome-wide loss-of-function screen for regulators of retrovirus silencing in ESCs. The authors identify components of the sumoylation and CAF-1 complexes as crucial mediators of retroviral silencing and provide insights into their mechanisms of action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, Duc J, Cohen MA, Wert KJ, Castanon R, Zhang Z, Huang Y, Nery JR, Drotar J, Lungjangwa T, Trono D, Ecker JR, Jaenisch R. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. This paper suggests that the expression of transposable elements is a more sensitive readout than the expression of coding genes when distinguishing human naïve and primed pluripotent stem cells. The authors further show that the transposable element signature of naïve hESCs mirrors that of late morulae/early blastocysts and implicate the KRAB-ZFP/KAP1 transcriptional regulators in their control. The authors further support the idea that active transposable elements influence proximal gene expression acting as promoters or enhancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. This paper provides a detailed map and classification of human endogenous retroviruses (HERVs) during pre-implantation development and in human embryonic stem cells using single-cell RNA-Seq analysis. This study proposes a mechanism by which HERVs regulate stage-specfic transcriptional signatures through LTR elements and preserved splice donor sites. [DOI] [PubMed] [Google Scholar]
- 66•.Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3. 3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. This paper describes how nucleosome diversity contributes to the silencing of different types of transposable elements in ESCs. The authors establish a link between histone H3.3 deposition and a silent chromatin state that is dependent on the histone chaperone complexes ATRX and DAXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Z, Liu J, Deng WM, Jiao R. Histone chaperone CAF-1: essential roles in multicellular organism development. Cell Mol Life Sci. 2015;72:327–337. doi: 10.1007/s00018-014-1748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, Ohkawa Y, Tsukada Y, Ogura A. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci U S A. 2015;112:14641–14646. doi: 10.1073/pnas.1512775112. The authors’ data suggest that CAF-1 is involved in the exchange of the histone variant H3.3 with histones H3.1/H3.2 in fertilized mouse occytes. This ensures the establishment of repressive histone marks around transposable elements to keep them silenced. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng A, Li YQ, Wang C, Han XS, Li G, Wang JY, Li DS, Qin YW, Shi Y, Brewer G, Jing Q. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J Cell Biol. 2013;201:409–425. doi: 10.1083/jcb.201207172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fischer S, Prykhozhij S, Rau MJ, Neumann CJ. Mutation of zebrafish caf-1b results in S phase arrest, defective differentiation, and p53-mediated apoptosis during organogenesis. Cell Cycle. 2007;6:2962–2969. doi: 10.4161/cc.6.23.4950. [DOI] [PubMed] [Google Scholar]
- 73.Nakano S, Stillman B, Horvitz HR. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 2011;147:1525–1536. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du Z, Santella A, He F, Shah PK, Kamikawa Y, Bao Z. The Regulatory Landscape of Lineage Differentiation in a Metazoan Embryo. Dev Cell. 2015;34:592–607. doi: 10.1016/j.devcel.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu Z, Wu H, Chen H, Wang R, Liang X, Liu J, Li C, Deng WM, Jiao R. CAF-1 promotes Notch signaling through epigenetic control of target gene expression during Drosophila development. Development. 2013;140:3635–3644. doi: 10.1242/dev.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filipescu D, Szenker E, Almouzni G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 2013;29:630–640. doi: 10.1016/j.tig.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santoro SW, Dulac C. Histone variants and cellular plasticity. Trends Genet. 2015;31:516–527. doi: 10.1016/j.tig.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 80.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3. 3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5:17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gonzalez-Munoz E, Arboleda-Estudillo Y, Otu HH, Cibelli JB. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 82.Marquardt S, Escalante-Chong R, Pho N, Wang J, Churchman LS, Springer M, Buratowski S. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, Almouzni G. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3. 3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 84.Wen D, Banaszynski LA, Rosenwaks Z, Allis CD, Rafii S. H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus. 2014;5:369–375. doi: 10.4161/nucl.36231. [DOI] [PubMed] [Google Scholar]
- 85.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soria-Valles C, Lopez-Otin C. iPSCs. On the Road to Reprogramming Aging. Trends Mol Med. 2016;22:713–724. doi: 10.1016/j.molmed.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 88.Volk A, Crispino JD. The role of the chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in homeostasis and disease. Biochim Biophys Acta. 2015;1849:979–986. doi: 10.1016/j.bbagrm.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zink LM, Hake SB. Histone variants: nuclear function and disease. Curr Opin Genet Dev. 2016;37:82–89. doi: 10.1016/j.gde.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 90.Polo SE, Theocharis SE, Grandin L, Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E, Almouzni G. Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology. 2010;57:716–724. doi: 10.1111/j.1365-2559.2010.03681.x. [DOI] [PubMed] [Google Scholar]
- 91.Endo A, Ly T, Pippa R, Bensaddek D, Nicolas A, Lamond AI. The Chromatin Assembly Factor Complex 1 (CAF1) and 5-Azacytidine (5-AzaC) Affect Cell Motility in Src-transformed Human Epithelial Cells. J Biol Chem. 2017;292:172–184. doi: 10.1074/jbc.M116.751024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JC. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]