SUMMARY
Nitrogen is one of the most important essential nutrient sources for biogenic activities. Regulation of nitrogen metabolism in microorganisms is complicated and elaborate. For this review, the yeast Saccharomyces cerevisiae was chosen to demonstrate the regulatory mechanism of nitrogen metabolism because of its relative clear genetic background. Current opinions on the regulation processes of nitrogen metabolism in S. cerevisiae, including nitrogen sensing, transport, and catabolism, are systematically reviewed. Two major upstream signaling pathways, the Ssy1-Ptr3-Ssy5 sensor system and the target of rapamycin pathway, which are responsible for sensing extracellular and intracellular nitrogen, respectively, are discussed. The ubiquitination of nitrogen transporters, which is the most general and efficient means for controlling nitrogen transport, is also summarized. The following metabolic step, nitrogen catabolism, is demonstrated at two levels: the transcriptional regulation process related to GATA transcriptional factors and the translational regulation process related to the general amino acid control pathway. The interplay between nitrogen regulation and carbon regulation is also discussed. As a model system, understanding the meticulous process by which nitrogen metabolism is regulated in S. cerevisiae not only could facilitate research on global regulation mechanisms and yeast metabolic engineering but also could provide important insights and inspiration for future studies of other common microorganisms and higher eukaryotic cells.
KEYWORDS: Agp1, GAAC pathway, Gap1, NCR, nitrogen regulation, RTG pathway, Saccharomyces cerevisiae, SPS sensor system, TOR pathway, ubiquitination
INTRODUCTION
To survive under different environmental conditions, microbial cells extensively regulate DNA duplication, chromatin remodeling, transcription, translation, and metabolism (1). Carbon metabolism and nitrogen metabolism are fundamental for producing cellular components and supplying energy metabolism. Previous work has extensively reported that the budding yeast Saccharomyces cerevisiae can preferentially use different kinds of carbon sources. Glucose is generally easier to be utilized than galactose, which is regarded as a nonfermentative carbon source, because it enters into the glycolytic pathway more easily and generates a higher carbon flux for tricarboxylic acid (TCA). The central metabolism pathway can then produce energy and metabolic intermediates for the downstream biosynthetic pathways (2, 3). This process is termed glucose repression. In the process of culture of S. cerevisiae, researchers also noticed that the yeast could preferentially utilize different nitrogen sources, such as glutamine and asparagine. During the process, these preferred sources could further repress the utilization of nonpreferred ones. This process is termed nitrogen catabolite repression (NCR) (4, 5). Accordingly, S. cerevisiae has evolved a series of mechanisms for adapting to different environments, which enables it to use the proper metabolic pathways that ensure optimal cell survival and proliferation.
Nitrogen sources are essential for life, and their metabolism is regulated precisely. In industrial biotechnology processes, nitrogen sources play an essential role in the production of various products, e.g., antibiotics (6–10), amino acids (11), and enzymes (12). In addition, some hazardous nitrogen compounds that accumulate during the production of fermented food, such as ethyl carbamate (EC) during alcoholic beverage production (13–15) and soy sauce production (16–18), are also related to the preferential utilization of nitrogen sources. Therefore, nitrogen catabolism could also seriously impact the quality of different fermented foods, such as Chinese rice wine (19) and cheese (20). Since S. cerevisiae is the one of most investigated eukaryotic model organisms, understanding the mechanisms of nitrogen catabolite repression in S. cerevisiae could also provide useful clues for the investigation of nitrogen metabolism in other similar eukaryotic microorganisms and even higher organisms (21).
In S. cerevisiae, nitrogenous materials for amino acid biosynthesis are mainly converted from glutamate and glutamine. The amino nitrogen from glutamate and the amide group from glutamine account for 85% and 15%, respectively, of the total cellular nitrogen (4, 22). Meanwhile, glutamate and glutamine work with α-ketoglutarate to connect the TCA cycle and nitrogen metabolism via NADPH-dependent glutamate dehydrogenase (GDH1) (23), glutamine synthetase (GLN1) (24, 25), NADH-dependent glutamate synthase (GLT1) (26, 27), and NAD+-linked glutamate dehydrogenase (GDH2) (26, 28). Nitrogen metabolism and its regulation have been proven to be critical in the catabolism and anabolism of proteins, amino acids, and other nitrogenous compounds; thus, they play a dominant role in general metabolism.
Nitrogen metabolism regulation involves a set of interconnected processes, such as the Ssy1-Ptr3-Ssy5 signaling sensor system (SPS sensor system) (29), the target of rapamycin (TOR) regulatory pathway (30), NCR (31), the general amino acid control (GAAC) pathway (32, 33), and other related regulatory mechanisms. All of these regulatory pathways comprise a complicated and sophisticated system for mediating nitrogen flow originating from nitrogen sensing, including nitrogen transportation, to nitrogen catabolism. Based on the sophisticated understanding of nitrogen regulation in S. cerevisiae, more precise control can be performed during industrial biotechnology processes to improve the efficiency of utilization of nitrogen sources and avoid the accumulation of hazardous nitrogenous compounds in fermented foods. Moreover, details of nitrogen regulation in S. cerevisiae could also provide important clues and inspiration for future studies of other common microorganisms and higher eukaryotic cells (21).
REGULATION OF NITROGEN SENSING
Yeast cells grow with various nitrogen sources in different growth environments. Therefore, the sensing of nitrogen sources is essential for cells to rewire cellular metabolism for optimal growth and proliferation (34). Two major regulatory pathways are responsible for the sensing of nitrogen sources in the environment: the SPS sensor system and the TOR pathway. The SPS sensor system is responsible for sensing extracellular amino acids, while the TOR pathway is mainly involved in sensing intracellular amino acids.
External Amino Acid Sensing by the SPS Sensor System
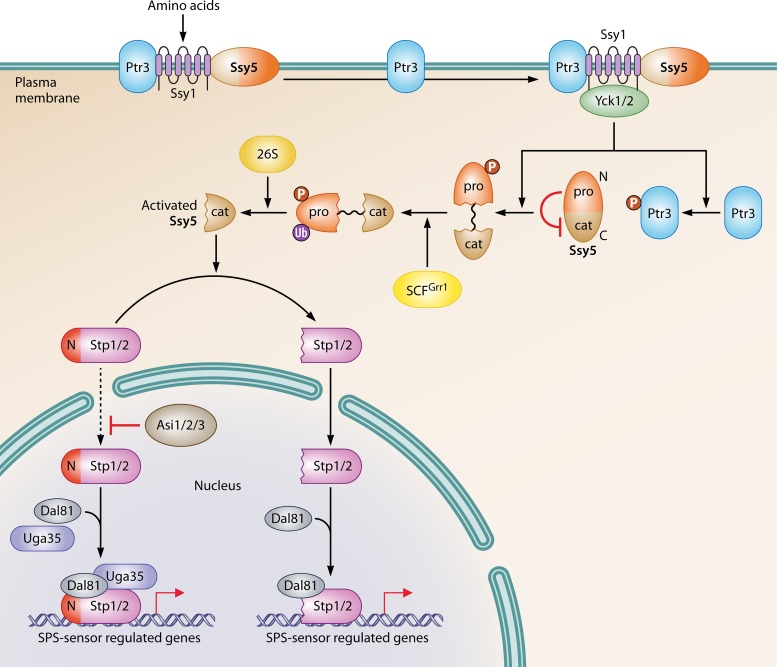
In yeast, there is a series of transporter-like membrane proteins which are responsible for sensing rather than transporting extracellular nutrients. For example, in S. cerevisiae, Ssy1 shows high sequence similarity with other amino acid permeases (AAPs), while it has no transport activity for any amino acid. Ssy1 has a long, cytoplasmically oriented amino-terminal (N-terminal) extension, which is significantly different from that of other AAPs. The sensor function of Ssy1 is dependent on its unique N-terminal region (35, 36). Ssy1 interacts physically with two other membrane proteins, Ptr3 and Ssy5, through its N-terminal domain and forms the SPS sensor system (29, 30). In yeast, the SPS sensor system is crucial for regulation of the expression of genes involved in amino acid catabolism in response to extracellular amino acid conditions (Fig. 1) (37). Mutations of Ssy1 could change its signaling conformation in the cytomembrane, which alters the binding affinity with amino acids, to form hyperresponsive or hyporesponsive alleles (38, 39). However, details of the interactions between Ssy1 and different substrates still need further studies, including sensing dynamics and protein structure analysis.
FIG 1.
Regulation of the SPS sensor system. The SPS sensor system is composed of three membrane proteins: Ssy1, Ptr3, and Ssy5. In the system, Ssy1 senses and is stimulated by extracellular amino acids. The signal transduces via Ptr3 and promotes the interaction between Ssy1 and Yck1/2. As a result, Ptr3 and the inhibitory prodomain of Ssy5 are hyperphosphorylated by Yck1/2. Next, the phosphorylated prodomain of Ssy5 is further ubiquitylated with the help of SCFGrr1, which, consequently, induces its degradation by the 26S proteasome. The resulting cat domain of Ssy5 activates Stp1 and Stp2 by digesting their negative regulatory domains at their N termini. Processed Stp1 and Stp2 translocate into the nucleus and activate their target genes by binding their promoters. Another group of transcriptional activators, Uga35/Dal81, which are also activated by the SPS sensor system, facilitate the binding of Stp1 and Stp2 to the promoters of related genes. However, the SPS sensor-dependent nuclear translocation of Stp1 and Stp2 is not strict. Low levels of preprocessed Stp1 and Stp2 can still “leak” into the nucleus (dashed line) and activate SPS-regulated genes with the help of Dal81. However, this pathway is blocked by the Asi protein.
There are two downstream effectors of SPS sensor system, Stp1 and Stp2 (Fig. 1) (40). They have redundant and overlapping abilities to activate transcription by binding to certain promoter regions of SPS sensor-regulated genes with coactivators, such as the Cyc8/Tup1 and Uga35/Dal81 complexes (41, 42). Originally, there are inhibitory domains in the N termini of both Stp1 and Stp2, which prevent them from migrating into the nucleus to function as a transcriptional activator of relevant genes. Therefore, the cleavage of N-terminal inhibitory domains is required before nuclear translocation of Stp1 and Stp2 under certain amino acid conditions (40, 43, 44). The endoproteolytic process induced by the SPS sensor system to remove negative domains of Stp1 and Stp2, which is termed receptor-activated proteolysis (RAP), is dependent on one component of the SPS sensor system, Ssy5 (45).
Ssy5 is an activating endoprotease, and it consists of a large N-terminal prodomain and a carboxyl-terminal (C-terminal) chymotrypsin-like catalytic (Cat) domain. The activation of Ssy5 is inhibited by its prodomain. The prodomain is spontaneously and incompletely cleaved from the Cat domain after Ssy5 synthesis. However, these domains remain associated with each other before the degradation of the prodomain. In response to certain amino acid conditions, the prodomain of Ssy5 is degraded via three steps. First, a conformational change of a conserved phosphodegron of the prodomain, which consists of phosphoacceptor sites and ubiquitin (Ub)-accepting lysine residues, is induced by extracellular amino acids. This conformational signal is transduced by Ptr3 to trigger the interaction of Ssy1 and the casein kinase (Yck1/2), which is constitutively activated and localized on plasma membrane. This results in the hyperphosphorylation of both Ptr3 and the prodomain of Ssy5, which is mediated by the phosphorylation of the phosphodegron. Next, the prodomain is polyubiquitylated by the Skp1/Cullin/Grr1 E3 ubiquitin ligase complex (SCFGrr1) (46). Lastly, the polyubiquitylated prodomain is degraded by the 26S proteasome (47). Under amino acid starvation conditions, Rst1, a regulatory component of protein phosphatase type 2A (PP2A) triggers the dephosphorylation of the prodomain to inhibit the activation of Ssy5 (48). PP2A is a major effector of intracellular amino acid signaling sensed by the TOR pathway. The PP2A-dependent negative regulation of Ssy5 indicates that intracellular amino acid signaling is prior to extracellular signaling to regulate the metabolism of amino acids.
The Ssy5-dependent cleavage of the inhibitory N-terminal fragments of Stp1 and Stp2 is highly dependent on the SPS sensor system, and it generates activated Stp1 and Stp2. The activated Stp1 and Stp2 consist of only the DNA-binding and transactivation domains, respectively. Both of them can be translocated into the nucleus (48). In addition to Stp1 and Stp2, another Uga35/Dal81-dependent transcriptional circuit is also activated by the SPS sensor system (42). Uga35/Dal81 facilitates the binding of the processed Stp1 and Stp2 with the promoters of SPS-regulated genes to amplify their transactivation effects. However, the translocation of Stp1 and Stp2 into the nucleus is not absolutely dependent on the SPS sensor system. In fact, a small amount of preprocessed Stp1 and Stp2 can still “leak” into the nucleus. They can also activate SPS-regulated genes with the help of Dal81, while this function is abolished by Asi proteins (Asi1, Asi2, and Asi3), the integral components of the inner nuclear membrane (49–51). Therefore, based on the identification of other components on the nuclear membrane involved in the relocation of transcription factors into the nucleus, modification of these components on the nuclear membrane could change the nuclear translocation of transcription factors to being nitrogen insensitive. Stp1 and Stp2 have different regulation and localization patterns, although they have redundant functions (46, 52). The E2 ubiquitin-conjugating enzyme Cdc34 is required for the degradation of both unprocessed and processed Stp1 but not for that of processed Stp2. In addition to the cytoplasm, the unprocessed Stp1 also localizes to the cell periphery, whereas unprocessed Stp2 does not (46).
Intracellular Nitrogen Sensing by the TOR Pathway
Tor proteins have been found in all eukaryotes, and they have conserved structures and functions (53, 54). They have been proven to be essential for cells to rewire metabolism for optimal growth in response to nutrient availability (53, 54). S. cerevisiae possesses two TOR genes, which is different from the case for many other eukaryotes, which usually have only one. The two TOR genes, TOR1 and TOR2, encode the basic members of the phosphatidylinositol protein kinase family (also called phosphatidylinositol 3′-kinase-related kinases [PIKKs]) (55). Some essential parts are conserved in all Tor proteins. The first one is the HEAT (Huntingtin elongation factor 3, regulatory subunit A of PP2A, TOR1) repeats, which contain approximately 20 HEAT motifs and function to interact with other proteins (56). The second is the FAT (FRAP, ATM, TTRAP) and FATC (FAT C-terminal) domains, which are conserved in PIKKs (57). The third is the FRB (FKBP12-rapamycin-binding) domain, the binding loci of FK506-binding protein (FKBP)-rapamycin (53). The last part is the kinase domain.
In S. cerevisiae, Tor1 and Tor2 can assemble with distinct subunits to build different regulatory protein complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2). In detail, TORC1 consists of four components. The major one could be either Tor1 or Tor2, which associates with the other three subunits, Kog1, Lst8, and Tco89. In contrast, TORC2 is made up of Tor2, Lst8, Avo1 to -3, and Bit6. The different components of TORC1 and TORC2 endow them with distinct characteristics (58–60). First, TORC1 takes part in many cell processes, including nutrient sensing, uptake, metabolism, translation initiation and ribosome biogenesis, mitochondrial function, and longevity pathways, as well as autophagy (61, 62). In contrast, TORC2 is involved mainly in regulating cell wall synthesis and the actin cytoskeleton to control the spatial bud growth of yeast (62). In addition, TORC1 is sensitive to rapamycin, while TORC2 is not. Rapamycin can hijack the cytosolic peptidyl-prolyl cis-trans-isomerase FKBP12, Fpr1 (FK506-binding protein 12), to bind the FRB domain of TORC1, resulting in the inhibition of TORC1 (63). While in TORC2, Avo1 and Avo3 can prevent rapamycin from binding the FRB domain, which accounts for its insensitivity to rapamycin (56, 58, 64).
EGOC is one of the upstream regulatory modules of the TOR pathway.
The TOR pathway was thought to detect amino acid levels inside rather than outside cells, particularly those from vacuoles and lysosomes (65–67). In yeast, in vivo localization studies suggested that TORC1 is activated and functions in the vacuolar membrane (66), while in mammalian cells, mTORC1 works in the lysosomal membrane in response to intracellular amino acid levels (68). In both yeast and mammalian cells, signals of intracellular amino acid changes stimulate the TOR pathway via an upstream regulator, the EGO complex (EGOC) (Fig. 2A).
FIG 2.
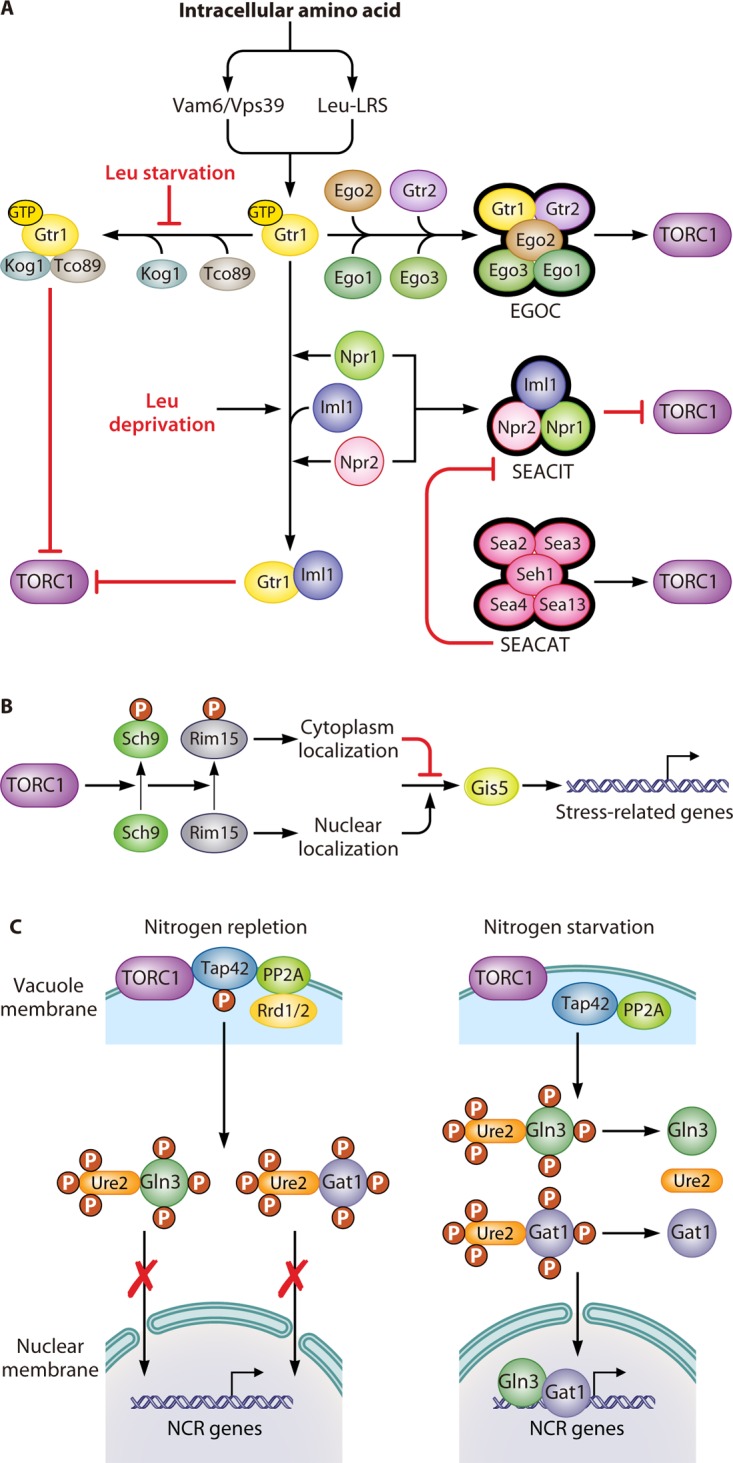
Regulation of the TOR pathway. (A) Upstream of the TOR pathway. The guanine nucleotide-binding status of Gtr1 plays an essential role in the sensing of intracellular amino acids by the TOR pathway. When sufficient levels of amino acids are present in the cytoplasm, this signal transduces through two factors, Vam6/Vps39 and LRS. They induce Gtr1 to recruit four other proteins, Gtr2, Ego1, Ego2, and Ego3, which comprise the EGOC, which activates the TORC1 complex. The GTP-bound format of Gtr1 can physically interact with Tco89 and Kog1, resulting in the inhibition of the TORC1 complex. However, this inhibition can be prevented by leucine starvation. Additionally, the SEACIT, which consists of Iml1, Npr2, and Npr3, inhibits TORC1 activity through an Npr2- and Npr3-dependent transient interaction between Iml1 and Gtr1. Leucine deprivation can promote this kind of transient interaction. Furthermore, the SEACAT activates the TORC1 complex by inhibiting the activity of the SEACIT. (B) Sch9, a downstream effector of the TOR pathway, controls the expression of stress-related genes. The activation of TORC1 promotes the phosphorylation of Sch9, which results in the subsequent phosphorylation of Rim15. Phosphorylated Rim15 is sequestrated in the cytosol, where it prevents Gis1 from activating stress-related genes. (C) The Tap42-PPase complex, another downstream effector of the TOR pathway, controls the expression of NCR-related genes. The activation of the TORC1 complex mediates the phosphorylation of Tap42 under nitrogen repletion conditions. Phosphorylated Tap42 binds with PP2A, along with either one of the regulatory proteins Rrd1 or Rrd2, and localizes on the vacuolar membrane, which facilitates their interaction with the TORC1 complex. The sequestration of the Tap42-PP2A complex on the vacuolar membrane prevents them from dephosphorylating the transcriptional activators Gln3 and Gat1, which prevents them from leaving the cytoplasm. In contrast, the inhibition of the TORC1 complex by nitrogen starvation or rapamycin treatment promotes the dephosphorylation of Tap42 and the release of Tap42-PP2A from the vacuolar membrane. Free Tap42-PP2A leads to the dephosphorylation of Gln3 and Gat1, thereby facilitating their translocation into the nucleus, where they activate NCR genes.
The regulatory subunits of the EGOC in S. cerevisiae are different from those of mammals, which are Gtr1/2 and the small GTPases RagA/B/C/D, respectively. However, they have same structural subunit Rag/Ego complex, which consists of Ego1 and Ego3, and the novel subunit Ego2 (69–71). In yeast, the EGOC is anchored to the vacuolar membrane by Ego1 (67, 72–74). Ego3 is recruited to Ego1 via binding with its extreme C terminus. This interaction is stabilized by Ego2 through its association with the α-helix of the Ego1 C terminus (75, 76). The regulatory subunit Gtr1 can form a heterodimer with Gtr2 (77). The Rag heterodimers are completely activated after GTP and GDP binding to Gtr1 and Gtr2, respectively (67, 78, 79). However, when both Gtr1 and Gtr2 are loaded with GTP, the heterodimer has only partial activity (80, 81). Both the completely and partially active forms of the Rag heterodimer are able to bind to TORC1 via Kog1 to stimulate TORC1 activity, although the completely active form does so to a greater degree (67, 79, 80).
Several kinds of regulators are involved in modulating the nucleotide-binding state of the Rag heterodimers to regulate the amino acid-dependent activity of TORC1. The first one is the guanine nucleotide exchange factor (GEF), which facilitates the conversion from GDP to GTP. In yeast, Vam6/Vps39, one of the GEFs of Gtr1, has been proposed to be sensitive to the perturbation of intracellular amino acid levels (67, 78). Furthermore, a vam6 mutant abrogates the interaction between Gtr1 and Ego1 and renders TORC1 insensitive to the increase of free intracellular amino acids through suppressing translation elongation (82). The second one is the GTPase-activating protein (GAP), which promotes the conversion from GTP to GDP (76). l-Leucyl-tRNA synthetase (LRS) is a conserved GAP in yeast as well as in mammalian cells. In yeast, when leucine is abundant, a leucine-binding LRS, Cdc60, favors the GTP-bound state of Gtr1 and activates TORC1 signaling. In contrast, after leucine is depleted, LRS secedes from Gtr1 and turns to correct mistakenly charged tRNALeu. As a result, GTP releases from Gtr1, and the activity of TORC1 is repressed (83, 84). There is another GAP that is conserved between yeast and mammalian cells, which is known as the Lst4-Lst7 complex in yeast (85) and the FNIP-FLCN complex in mammals (86, 87). In yeast, the Lst4-Lst7 complex will be recruited to the vacuolar membrane under amino acid depletion conditions. However, refeeding of amino acids will induce the transient interaction between the Lst4-Lst7 complex and Gtr2GTP and the release of the Lst4-Lst7 complex from the vacuolar membrane. As a result, GTP loaded on Gtr2 will be hydrolyzed into GDP, and the TORC1 signaling pathway will be completely activated (85). In addition, the Seh1-associated complex (SEAC) proteins Npr2 and Npr3 are another kind of mediator of TORC1 in response to changes in intracellular amino acid concentration (88). SEAC is conserved in mammals and yeast, and it consists of eight proteins that can be divided into two subcomplexes. In yeast, one is SEACIT (for SEAC inhibiting TORC1), consisting of Iml1/Sea1, Npr2, and Npr3, which negatively regulates TORC1. Leucine deprivation can promote a transient interaction between Iml1 and Gtr1 supported by Npr1 and Npr2. Consequently, Gtr1GTP could be converted to Gtr1GDP under the function of Iml1, as a GAP, resulting in the inhibition of TORC1 function (89). The other is SEACAT (for SEAC activating TORC1), consisting of Seh1, Sec13, Sea2, Sea3, and Sea4, which positively regulates TORC1 (89). This complex can inhibit SEACIT-mediated TORC1 inhibition (90, 91). Both SEACIT and SEACAT have corresponding mammalian homologs, which are termed GATOR1 and GATOR2, respectively (89).
tRNA is another upstream regulatory module of the TOR pathway.
Amino acids are fundamental nutrients. S. cerevisiae has the ability to remodel cellular metabolism in response to different kinds of amino acids. This suggests that there should be a precise sensing mechanism in cells to detect each intracellular amino acid signaling individually. However, the above-mentioned Rag factor-mediated TORC1 sensor mechanism cannot meet a such demand. Moreover, recent studies found that cellular TORC1 activity was not affected by the lack of Rag factors (92, 93). Taking the data together, there should be some other regulatory modules of the TOR pathway which sense intracellular amino acids independent of the Rag factor.
A recent study suggested that tRNAs played an important role in intracellular amino acid sensing by the TOR pathway (92). tRNAs can accurately bind with each of 20 proteogenic amino acids to synthesize aminoacyl-tRNAs (aa-tRNAs), with the help of aminoacyl-tRNA synthetases. Under conditions of nitrogen supply including amino acids, TORC1 was inhibited in aminoacyl-tRNA synthetase mutants, which suggested that amino acid sensing by TORC1 requires the participation of aminoacyl-tRNA synthetases and/or their substrate tRNAs and aa-tRNAs, but not amino acids (61). The addition of free tRNA or aa-tRNA drastically inhibited the activity of TORC1 in vitro. Moreover, mutation of RPC34, which is responsible for transcribing tRNA genes, significantly attenuated TORC1 inhibition in response to nutrient limitation conditions. These results indicated that tRNAs serve as a direct inhibitor in intracellular amino acid sensing by the TOR pathway (92). Hence, a model of a tRNA-mediated TORC1 regulatory mechanism was built. Under amino acid-rich conditions, most free tRNAs are charged with amino acids to synthesize aa-tRNAs. Most aa-tRNAs are further consumed by protein synthesis, which makes them have less chance to interact with TORC1. As a result, TORC1 is activated under such conditions. However, under amino acid-limited conditions, most tRNAs have a low probability of combining with amino acids, which results in the accumulation of free tRNAs. Thus, free tRNAs are able to interact with TORC1 to repress its activity (92).
Although these studies supported the hypothesis that tRNA is the direct regulator of TORC1, there is no direct evidence to explain how the specific tRNAs could regulate the TORC1 activity according to corresponding intracellular amino acids. Moreover, how the TOR pathway responses to each kind of amino acid remains unclear. There may be a potential TORC1-dependent pathway to regulate the consumption and/or transportation of each amino acid in response to its intracellular level.
The Sch9 and Tap42-PPase effectors are downstream of the TOR pathway.
The intracellular amino acid signaling sensed by TORC1 is mostly transduced to its two important effectors to regulate cellular activities, which are the AGC kinase Sch9 (Fig. 2B) and the Tap42-protein phosphatase (Tap42-PPase) complex (Fig. 2C) (53, 94). Sch9 is homologous to mammalian S6 kinase (S6K) and has four domains: a central kinase catalytic domain, an activation loop, a turn motif, and a C-terminal regulatory domain. The regulatory domain contains a hydrophobic motif that has six amino acid residues, which are latently phosphorylated by activated TORC1 directly (62). Activation of Sch9 is dependent on such TORC1-mediated phosphorylation, and replacing those residues with Asp/Glu releases the dependence of Sch9 activity on TORC1. Activated Sch9 regulates many genes participating in mitochondrial function (95), sphingolipid homeostasis and signaling (96), autophagy and longevity (97), and entry into the G0 phase of the cell cycle (98). For instance, activated Sch9 phosphorylates Ser1061 of Rim15 to block its nuclear translocation. The cytosolic sequestration of Rim15 prevents Gis1 from activating the expression of stress response genes (98). Of note, Sch9-dependent TORC1 activity does not regulate the expression of Gln3-dependent genes (65). When yeast cells are subjected to rapamycin treatment or nutrient starvation, Sch9 is rapidly dephosphorylated to repress TORC1 (65, 67).
Tap42 is another essential downstream regulatory protein of the TOR pathway. TORC1 activation leads to the phosphorylation of Tap42, which promotes its interaction with PP2A or PP2A-like complexes, as well as either one of the regulators Rrd1 or Rrd2, to inhibit PP2A (99–102). The PP2A phosphatase is a heterotrimer that consists of three subunits: the scaffolding subunit Tpd3, the catalytic subunit (either Pph21, Pph22, or Pph3), and the regulatory subunit (either Cdc55 or Rts1) (103, 104). The PP2A-like phosphatase is made up of the catalytic subunit, Sit4 or Ppg1, and the regulatory subunit, either Sap4, Sap155, Sap185, or Sap190 (105). The Tap42-PP2A and Tap42-PP2A-like phosphatase complexes are located mainly on vacuolar membranes, which facilitates their interaction with TORC1 (106, 107). Under rapamycin treatment or nitrogen starvation conditions, the Tap42-PP2A (and Tap42-PP2A-like) complexes are released from the vacuolar membrane to the cytosol, and Tap42 is dephosphorylated (108). The cytosolic Tap42-PP2A (and Tap42-PP2A-like) phosphatase complexes activate the expression of genes related to nitrogen catabolite repression and stress response by dephosphorylating special regulators, such as Gln3 (109, 110). Tip4 appears to be part of a feedback loop in the TOR pathway because its binding with Tap42 inhibits Tap42 to activate Sit4, which in turn dephosphorylates Tip4. Furthermore, the dephosphorylation of Tip4 by Sit4 directly or indirectly enhances its association with Tap42 (111).
In addition to downregulating Sch9 and upregulating the Tap42-PP2A phosphatase complex, inhibition of the TOR pathway also activates the cell wall integrity (CWI) pathway. When cells grow under stress conditions, Rho1, the major sensor element of the CWI pathway, will bind to Kog1 to repress TORC1 activity (112). Moreover, the interaction between Rho1 and Kog1 will release TORC1 from the vacuolar membrane and activate the Tap42-PP2A phosphatase (113). Remarkably, in addition to nitrogen repletion, the increase of intracellular S-adenosylmethionine levels could also activate TORC1 (114). S-Adenosylmethionine is the major methyl donor and comes mainly from methionine. The catalytic subunit of the PP2A complex will be methylated by the methyltransferase, Ppm1, in response to intracellular S-adenosylmethionine abundance. After that, Npr2 will be dephosphorylated by the methylated PP2A complex, leading to inhibition of SEACIT and activation of TORC1 (114).
REGULATION OF NITROGEN TRANSPORTATION
The yeast S. cerevisiae contains 24 amino acid transporters according to the current available literature (Table 1). Each of them presents in a similar conformation, which consists of 12 transmembrane domains and the cytoplasmically oriented N and C termini. They work mainly to transport amino acids as well as other amines through the plasma membrane (115–117). However, some of them, such as the general amino acid permease (Gap1) (118), also work as a sensor to detect the abundance of substrates to regulate their activity. Essentially, S. cerevisiae controls transporters at three levels: gene transcription, intracellular membrane trafficking of the synthesized proteins, and modulation of their intrinsic activity (119). In the last 10 years, a series of studies have focused on the regulation of yeast transporters at the level of membrane trafficking, in which ubiquitin is well established as a central player (Fig. 3) (120, 121). Since the regulation of these transporters exists at different levels, the regulation of amino acid transporters by ubiquitination is discussed in the following sections, while the transcriptional regulation process is covered in later sections.
TABLE 1.
Permeases of amino acids and ammonium in S. cerevisiae
| Permease | Substrate(s) | Affinity | Reference(s) |
|---|---|---|---|
| Agp1 | Asparagine, glutamine, other amino acids | Low | 224 |
| Agp3 | Glutamine | High | 225 |
| Alp1 | Arginine | —a | 157 |
| Bap2 | Leucine | High | 226 |
| Bap3 | Cysteine, leucine, isoleucine, valine | High | 227 |
| Can1 | Arginine | High | 228, 229 |
| Dip5 | Dicarboxylic amino acids | High | 230 |
| Gap1 | l-Amino acids | High | 146 |
| Gnp1 | Glutamine | High | 231 |
| Hip1 | Histidine | High | 232 |
| Lyp1 | Lysine | High | 157, 233 |
| Mep1 | Ammonium | High | 234 |
| Mep2 | Ammonium | High | 234 |
| Mep3 | Ammonium | Low | 234 |
| Mmp1 | S-Methylmethionine | High | 235 |
| Mup1 | Methionine | High | 236, 237 |
| Mup3 | Methionine | Low | 238 |
| Ort1 | Ornithine | — | 239 |
| Put4 | Proline | High | 238 |
| Sam3 | S-Adenosylmethionine | High | 235 |
| Tat1 | Valine, leucine, isoleucine, tyrosine, tryptophan, histidine | Low | 240 |
| Tat2 | Tryptophan, tyrosine | High | 240 |
| Uga4 | γ-Aminobutyrate | — | 241 |
| Yct1 | Cysteine | High | 242 |
—, not reported.
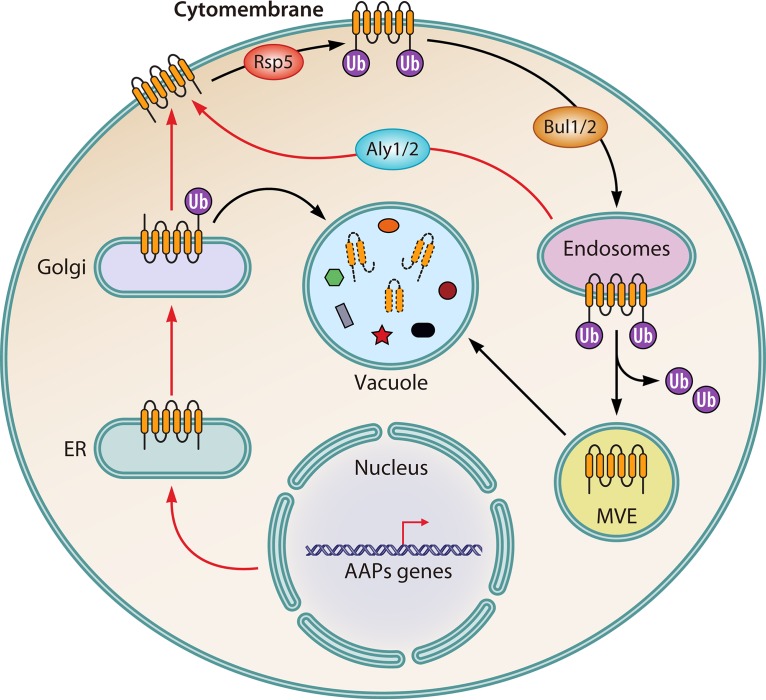
FIG 3.
Ubiquitylation regulation of AAPs. Under nitrogen starvation conditions (red arrows), genes encoding amino acid permeases (AAPs) are activated. The AAPs are first translocated into the endoplasmic reticulum for further procession and modification. They then are transported to Golgi apparatus, followed by direction to the plasma membrane, where they fulfill their functions. Under nitrogen repletion conditions (blue arrows), the Golgi apparatus-located AAPs are ubiquitylated by Rap5, which results in their vacuolar translocation and degradation. The plasma membrane-located AAPs are also ubiquitylated by Rap5, which promotes the Bul1/2-mediated endocytosis of AAPs, followed by their transportation to endosomes. When present on multivesicular endosomes, AAPs are finally targeted to the vacuole and degraded. However, there is a recycling pathway when nitrogen is depleted or cells are transferred from nitrogen repletion to starvation conditions. Through the recycling pathway, AAPs that are located on endosomes can be retargeted to the plasma membrane, which is dependent on Aly1/2.
Ubiquitination-Mediated Degradation of Nitrogen Transporters
Ubiquitin (Ub) consists of 76 amino acid residues that can link to target proteins via binding the glycine carboxyl group in its termini to the ε-amino group of a lysine residue of the target protein (122, 123). Three enzymatic reactions that are responsible for Ub conjugation, or ubiquitination, are sequentially catalyzed by the Ub-activating enzyme E1, the Ub-conjugating enzyme E2, and the Ub ligase E3 (124–126). Because there are seven lysine residues on Ub, ubiquitination could recur to target proteins via the binding of Ub to either its own lysine residues or the amino group in its N terminus. This recursive ubiquitination can generate polyubiquitin chains, which can assemble in different ways and result in distinguishing structures and properties (122, 127). The conjugated Ub can be cleaved from substrates by deubiquitinating enzymes (128).
In S. cerevisiae, nitrogen permeases need to be trafficked to different organelles for modification, degradation, or activation (Fig. 3) (21). Generally, mRNAs of genes encoding nitrogen permeases are translated in the endoplasmic reticulum. The translational products are then translocated to the Golgi apparatus (129). There are two possible fates of those permeases in the Golgi apparatus: to be trafficked to the plasma membrane to activate their transportation function (130) or to be trafficked to the vacuole for degradation, either through endosomes (131) or directly (132). Furthermore, those permeases that locate in the plasma membrane can be internalized via endocytosis (133). The internalized unit will be either redirected into the plasma membrane through the Golgi apparatus or finally degraded in the vacuole (134). Generally, endocytosis is triggered by ubiquitination modification of the target protein. In S. cerevisiae, Rsp5, one type of Ub ligase, is responsible for such ubiquitination modification (135, 136). Rsp5 consists of several functional domains, which include one C2 domain anchored to the membrane, three WW domains responsible for recognition and binding with PY motifs (XPXY) of the substrate, and one HECT domain responsible for ligating Ub to the target protein (137).
Gap1 was chosen here as a model to elucidate the Ub-mediated trafficking trajectory of nitrogen transporters in response to nitrogen sources (Fig. 3) (120, 138). Under nitrogen starvation conditions, Gap1 is recycled from the endosome with the help of two arrestin-like proteins, Aly1 and Aly2. The recycled Gap1 will be relocated to the plasma membrane and activated (139). In contrast, after being transferred to nitrogen repletion conditions, Gap1 is rapidly ubiquitinated by Rsp5. Ubiquitinated Gap1 will be internalized from the plasma membrane, then sorted into multivesicular endosomes (MVEs) and vacuoles, and finally degraded (140, 141). Remarkably, because there are no PY motifs in Gap1, the interaction between Gap1 and Rsp5 is indirect and is dependent on special PY motifs containing adaptors (142, 143). During the nitrogen repletion-induced ubiquitination of Gap1, Rsp5 is recruited to Gap1 with the help of two arrestin-like proteins, Bul1 and Bul2 (Bul1/2), which contain PY motifs (142). Furthermore, ubiquitination of Gap1 is influenced by the phosphorylation status of Bul1/2. Bul1/2 can be phosphorylated in the Npr1-dependent way, leading to its interaction with 14-3-3 protein (144). In addition, the Thr357 residue of Rsp5 would be phosphorylated, which prevents Gap1 from Rsp5-mediated ubiquitination under nitrogen starvation conditions (145).
Substrate-Regulated Ubiquitination and Trafficking of Gap1
Gap1 is well known as an amino acid transporter with broad substrate range as well as high substrate affinity (146). It also plays role in transduction of nitrogen signals to the protein kinase A pathway (147). It is regulated in response to intracellular amino acid abundance through oligo- and polyubiquitination-guided endocytosis (148). As discussed above, the intracellular substrate-induced endocytosis of Gap1 is dependent on the TORC1-mediated dephosphorylation of Bul1/2. However, the ubiquitination and endocytosis of Gap1 are also regulated by the abundance of extracellular amino acids independent of the phosphorylation of Bul1/2 (148). This kind of endocytosis of Gap1 is based on its conformation change induced by binding of extracellular substrates. Such an extracellular substrate-induced conformation change has also been found in the endocytosis of other yeast amino acid permeases, e.g., Can1 (149), Lyp1 (143), Dip5 (150), and Tat2 (151), as well as the uracil permease Fur4. Previous studies revealed that a conformational change of Gap1 is essential to its endocytosis in response to extracellular substrate availability before releasing conjoint substrate into the cytosol (148). In addition, such a conformational state should be stable enough to promote its interaction with the arrestin-like adaptors (148). Moreover, the binding of substrate to Gap1 may contribute to disrupt the interaction between its N-terminal tail with internal loops, which has been proven in other conserved permeases, such as Fur4 (151) and AdiC (152). The conformational change induced by substrate binding would facilitate the interaction between the candidate ubiquitination sites on its N-terminal tail with ubiquitination factors. Furthermore, Gap1 mutants that have totally lost their transportation capability are not endocytosed in such a substrate-induced way, whether they are able to bind with these substrates or not (116, 148). This indicated that the binding of substrates to Gap1 is an essential but not sufficient condition for its substrate-induced degradation. In other words, only when the orientation of substrate-binding Gap1 changes to the cytoplasmic state from the exoplasmic state will its N terminus be released freely and ubiquitination happen.
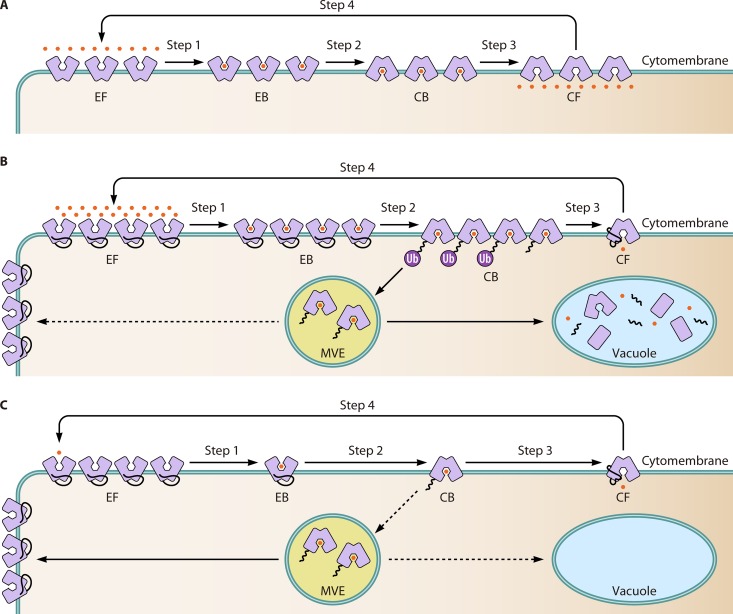
The mechanism of extracellular substrate-induced endocytosis of Gap1 could be explained based on the previously proposed transport model (153) (Fig. 4A). This transport model suggested that there are four conformational states of permeases on the plasma membrane during the transport cycle, including exoplasmically oriented free (EF), exoplasmically oriented bound (EB), cytoplasmically oriented bound (CB), and cytoplasmically oriented free (CF) (153). This model has been almost proven by the crystallizing the proteins of the 5+5 superfamily in different conformations, including the EF, EB, and CB conformations (152). Based on this transport model, a sufficient amount of amino acids will increase the amount of substrate-binding Gap1, including that in the changed conformation. The accumulation of conformation-changed Gap1 with free N termini facilitates the recognition of lysine residues by ubiquitination effectors and promotes the ubiquitination and degradation of Gap1 (Fig. 4B). However, the exhaustion of extracellular amino acids decreases the efficiency of interaction between Gap1 and amino acids as well as the abundance of conformation-changed Gap1, which hinders the access of ubiquitination effectors to the N-terminal lysine residues before the dissociation of amino acids from Gap1. As a result, Gap1 escapes from ubiquitination degradation and changes to its original state at the beginning of the transport cycle to active the transportation of amino acids (Fig. 4C). Furthermore, those Gap1 molecules that have been ubiquitinated and sorted into MVEs will be recycled and redirected to the cytomembrane to activate the uptake of amino acids under conditions of amino acid scarcity (154). In contrast, such recycling of Gap1 from MVEs could be blocked by inducing the overproduction of intercellular amino acids. The phenomenon could be observed in mutants such as mks1 and lst8 mutants (155, 156). It should be noted is that there are no more details about the regulatory mechanism of recycling Gap1 from MVEs to the cytomembrane.
FIG 4.
Substrate-regulated ubiquitination and trafficking of Gap1. (A) The general transport cycle of the permease-mediated transportation process. Generally, there are four face states of the permease during the process of its transportation of substrate from an extracellular to an intracellular location, which are exoplasmically oriented free (EF), exoplasmically oriented bound (EB), cytoplasmically oriented bound (CB), and cytoplasmically oriented free (CF). Originally, the permeases is in the EF state. After substrate binding, it changes to the EB state (step 1). It then transforms to the CB state (step 2) and subsequently to the CF state (step 3) after releasing its substrate into the cytoplasm. Finally, it reverts to the EF state (step 4) to begin another cycle. (B) In an amino acid-replete environment, many Gap1 molecules are loaded with amino acid substrates. As a result, the abundance of conformation-changed Gap1 (in the CB state) is increased. The accumulation of conformation-changed Gap1 facilities the ubiquitination adaptor to recognize the lysine residues on free N termini of Gap1. The ubiquitination signal triggers the endocytosis of cytomembrane Gap1. The endocytosis leads to direct Gap1 to the vacuole through multivesicular endosomes. Under this condition, the Gap1 in multivesicular endosomes will not be recycled back to the cytomembrane. (C) When cells are amino acid exhausted or cultured under amino acid-scarce conditions, a low substrate concentration reduces the abundance of cytoplasmically oriented Gap1. As a result, the substrate is released immediately from Gap1, which prevents the ubiquitination and endocytosis of Gap1. In addition, the preexisting ubiquitinated Gap1 can be recycled and trafficked to the cytomembrane, regulated by an unknown mechanism.
External Amino Acids Induce Transcriptional Activation of Agp1
AGP1 encodes an amino acid permease with a broad substrate range but low substrate affinity. There are several 5′-GATA-3′ motifs in its upstream region, which are common binding sites of the GATA family transcription factors. The binding of GATA factor Gln3 to these motifs could upregulate the transcription of AGP1 approximately 10-fold under nitrogen starvation conditions (157). However, the transcriptional activation of AGP1 strictly depends on external amino acids (such as phenylalanine and citrulline) (158). In addition to Gln3, one effector of the SPS sensor system, Stp1, also significantly activates transcription of AGP1 under amino acid depletion conditions, while the other effector, Stp2, could not. This differs from the effects of Stp1 and Stp2 on the transcriptional activation of the high-affinity leucine permease-encoding genes BAP2 and BAP3, where Stp1 and Stp2 perform redundant activation roles (159, 160).
Stp1 attempts to bind with a cis upstream activating sequence of AGP1, called UASAA, with help of another regulator, Uga35/Dal80. There are two inversely repetitive 5′-CGGC-3′ motifs which are separated by six nucleotides (5′-CGGCN6GCCG–3′) in the UASAA region (44). Such an arrangement of these units has been proven to be optimal for the amino acid-induced activation of AGP1 (44). The amino acid-induced activation of AGP1 could be weakened to some extent, though not completely, by mutating its UASAA region, such as by rearranging the orientation of the repetitive 5′-CGGC-3′ motifs from inverse to direct, mutating the nucleotides of the central six nucleotides, or changing the length of the central nucleotides (44). Interestingly, the expression level of a lacZ reporter gene constructed by inserting a UASAA element of GAP1 into its upstream region is insensitive to nitrogen sources, despite the presence of the GATA factor Gln3, Ure2, or Gzf3 (44), which suggests that the UASAA element is necessary but not sufficient for amino acid-induced activation of GAP1.
In addition to the Gap1 and Agp1 permeases, which have a broad substrate range, there are many other specific permeases in S. cerevisiae (Table 1). When cells grow with preferred nitrogen sources, these specific permeases, such as the glutamine transporter Gnp1, transport the preferred nitrogen sources into the cell, whereas other permeases that are responsible for transporting nonpreferred nitrogen sources are repressed. Upon the depletion of preferred nitrogen sources, nonpreferred nitrogen permeases are induced to ensure the intracellular transport of the corresponding nitrogen source (160). Importantly, most of these specific permeases are NCR sensitive.
TRANSCRIPTIONAL REGULATION OF NITROGEN CATABOLISM
Transcriptional Regulation via NCR
Genes which regulated by NCR are called NCR genes. There are four transcription factors involved in regulating the expression of NCR genes, including two activators, Gln3 and Gat1, and two repressors, Gzf3 and Dal80. The transcriptional regulation of NCR genes by these transcription factors is achieved via their interactions with GATA sequences in the promoters of NCR genes. More details about the process are summarized in several earlier reviews (4, 31, 161). In contrast to Gln3, which is constitutively expressed, the transcription of the other three regulators is controlled by NCR. The activation of NCR genes, induced by Gat1, requires the Gln3-mediated activation of Gat1. In contrast, Gln3 can activate the transcription of NCR genes independently (162). Gln3 and Gat1 will be translocated into the nucleus under nitrogen starvation conditions, while under nitrogen repletion conditions, they are sequestered in the cytoplasm by TOR-mediated phosphorylation (14, 163). In addition, the transcriptional regulator Ure2 works as another anchor to prevent the translocation of Gln3 into the nucleus. Dephosphorylation of Gln3 and Gat1, which is induced by phosphatase Sit4 or Pph3, promotes their translocation into the nucleus to activate NCR genes under nitrogen limitation or rapamycin treatment conditions (164).
In addition to the four global transcription factors, other transcription regulators are needed in regulation of some genes in a specific metabolism pathway for a particular nitrogen source. For example, Aro80 and Dal81 are required to activate the transcription of genes responsible for metabolism of γ-aminobutyric acid, urea, arginine, and allantoin (165). In many cases, the global regulators Gln3 and Gat1 interact with these regulators to regulate the transcription of NCR genes (166, 167).
Regulation of Gln3 and Gat1
Gln3 and Gat1 play important roles in transcriptional activation of NCR genes. The expression of GLN3 is simply constitutive, and its transcription is not affected by NCR (162), whereas the expression of GAT1 is much more complicated. The alternative transcriptional initiation of GAT1, which could begin from either methionine 40, 95, or 102, is insensitive to nitrogen sources, and it results in a low level of transcription (168, 169). In contrast, the premature transcriptional termination of GAT1, which is terminated at the Ser-233 site, is highly controlled by the nitrogen source, and the transcriptional level is higher in proline-containing than in glutamine-containing medium. Nevertheless, both expression patterns depend on both Gln3 and UASGATA elements in the promoter region, which are generally essential for the transcriptional activation of NCR genes (168, 169). In the NCR-sensitive pattern, the expression of GAT1 is repressed by two negative NCR regulators, Dal80 and Gzf3, which competitively bind to its promoter and repress its transcription (162).
In addition to transcriptional regulation, posttranscriptional modification of Gln3 and Gat1 is also essential for repressing Gln3- and Gat1-mediated activation of NCR genes (170). The intracellular localization of Gln3 and Gat1 is similar in response to preferred and nonpreferred nitrogen sources during steady-state growth (171). Gln3 and Gat1 are bound by Ure2 and incarcerated in the cytosol with optimal nitrogen sources, while they are translocated into the nucleus under nitrogen-poor conditions (172). However, their intracellular localization is quite different during nutritional transitions. Gat1 responds to such transitions more rapidly than Gln3 (171). Unlike the sequestration of Gln3 in the cytoplasm, which absolutely requires Ure2 under nitrogen repletion conditions, intracellular Gat1 localization is largely independent of Ure2 (170). In addition, the translocation into and out of the nucleus, as well as the dephosphorylation of Gln3 and Gat1, is regulated separately by rapamycin- and nitrogen limitation-induced inhibition of TORC1, and they have different requirements for glutamine tRNACUG. For instance, Gat1 is translocated into the nucleus after rapamycin treatment, with only a limited requirement for Sit4 and no requirement for glutamine tRNACUG, whereas the effect of rapamycin on the localization of Gln3 absolutely requires both Sit4 and glutamine tRNACUG (170, 173, 174). Furthermore, when cells are grown in proline-containing medium, the nuclear localization of Gln3 does not depends on PP2A, whereas that of Gat1 is absolutely dependent on PP2A (175). In addition, Gln3, but not Gat1, is translocated from the cytosol to the nucleus in a Sit4-independent way under treatment with methionine sulfoximine (Msx), which serves as an inhibitor of glutamine synthase, and such translocation of Gln3 depends partially on glutamine tRNACUG (173, 174). Previous studies suggested that the association of Gln3 and light membrane could facilitate TORC1-dependent phosphorylation/dephosphorylation on Gln3, which finally controls the localization of Gln3 (176). After Gln3 is translocated into the nucleus, two phenomena regulate its export. When the glutamine level is high, Gln3 exits the nucleus even when its DNA-binding residues 64 to 73 are altered, while when the glutamine level is lowered, the export of Gln3 from the nucleus is dependent on its DNA-binding residues (177). Although residues 332 to 345 of Gln3 have been proven to be essential for its export from the nucleus (177), the clear function of glutamine and its DNA-binding residues in nuclear Gln3 exportation is still unknown.
Although the TOR pathway has a strong influence on the phosphorylation of Gln3, the phosphorylation status of Gln3 is not only controlled by TORC1. For instance, rapamycin treatment, which represses TORC1, can reduce the phosphorylation of Gln3 by activating the dephosphorylation function of the Tap42-PP2A and Tap42-Sit4 complexes (178), while the reduction of Gln3 phosphorylation is not detected under nitrogen limitation conditions. Moreover, many studies suggested that nitrogen starvation serves as an independent inducer, while not a part of TORC1, of derepression of the transcription of NCR genes. Under Tap42 inactivation conditions, rapamycin cannot activate NCR, while the activation of nitrogen limitation still works (110). The localization of a modified Gln3, Gln3656–666, which lacks residues 656 to 666 responsible for TORC1-interacting and cannot associate with TORC1, is not affected by rapamycin, but it responds to nitrogen limitation (179). Above all, there must be an unknown regulatory pathway involved in the Gln3-dependent activation of NCR genes in addition to the TOR pathway.
In addition to the phosphorylation status of Gln3 and Gat1, phosphorylation modification of Ure2 also influenced their translocation from the cytosol into the nucleus (180). Ure2 is active as a homodimer, and each monomer consists of two functional regions. One is the N terminus (Ure21–93), which has rich glutamine-asparagine adjacent residues and endows Ure2 with a pre-prion-like character. The other is the C terminus (Ure294–354) and is required for the cytoplasmic retention of Gln3 and Gat1 in response to nitrogen repletion (181, 182). In the C-terminal domain, there is a flexible protruding α-cap (Ure2267–298), which is important for the distinguishing performance of Gln3 and Gat1 in response to rapamycin treatment as well as nitrogen starvation. Mutating serine residue 283 or the serine/threonine 286 to 292 repeats in this α-cap leads to repression of the rapamycin-mediated nuclear translocation of Gat1, and to a lesser extent of Gln3, but nitrogen limitation-dependent nuclear translocation remains unaffected (180). Furthermore, rapamycin treatment has a strong effect on inducing dephosphorylation of Ure2, while nitrogen starvation has just a slight or even no effect (180). Furthermore, rapamycin-induced Ure2 dephosphorylation, which can be prevented by mutating the α-cap, is independent of Sit4 and PP2A, which is in contrast to the dephosphorylation of Gln3.
Interestingly, carbon starvation is also involved in regulating the translocation of Gln3 and Gat1 in to the nucleus via Snf1-mediated hyperphosphorylation of Gln3 and Gat1 (171). Furthermore, the carbon starvation-induced translocation of Gln3 into the nucleus is in opposition to TORC1 activity (183). This suggests that Gln3 is the convergence point of the TOR-nitrogen and Snf1-glucose signaling pathways.
TRANSLATIONAL REGULATION OF NITROGEN CATABOLISM
S. cerevisiae regulates the translation of genes related to nitrogen metabolism to adapt to various nitrogen sources. The regulation of translation occurs at different steps, such as translational initiation and posttranslational modification. Posttranslational modifications involve mainly phosphorylation and ubiquitination, as has been discussed in Regulation of Nitrogen Sensing and in Regulation of Nitrogen Transportation above. Thus, the following paragraphs focus on regulation of the translational initiation of nitrogen metabolism-related genes.
The GAAC Pathway Regulates Translational Processes
In S. cerevisiae, the GAAC pathway is activated under amino acid starvation conditions. The activation of the GAAC pathway globally inhibits the translational initiation of many genes, including the Gcn4 gene (Fig. 5A). Gcn4 activates 57 genes, which participate in amino acid biosynthesis, nitrogen utilization, and signaling through interacting with their promoter regions (184).
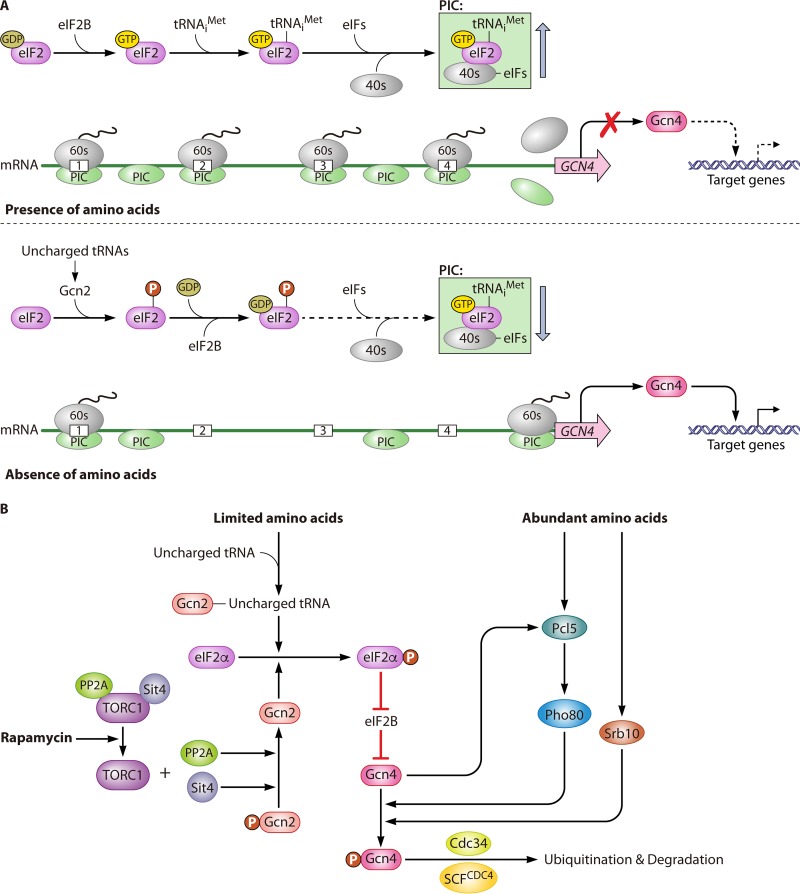
FIG 5.
Nitrogen regulation of the GAAC pathway. (A) Translational regulation of nitrogen catabolite genes by the GAAC pathway with different nitrogen sources. When amino acids are present, the α subunit of the eukaryotic translation initiation factor eIF2 binds with GDP, which then transforms into a GTP-bound state with the help of the GEF eIF2B. The GTP-bound form of eIF2 recruits a charged methionyl initiator tRNA to form the TC. The subsequent recruitment of the 40S small ribosomal subunit and other eIFs by the TC generates the 43S PIC. The PIC scans the mRNA to find an AUG start codon. Once the start codon is found, eIF2-GTP is hydrolyzed to eIF2-GDP and released from the TC. The modified PIC then recruits the 60S ribosomal unit to form the 80S initiation complex. Because there are four μORFs in the 5′-UTR of GCN4 mRNA, the 80S initiation complex has difficulty reaching the true start codon of the mRNA. Thus, under this condition, the translation of GCN4 mRNA is very limited. In the absence of amino acids, the levels of uncharged tRNAs increases, leading to the activation of the Gcn2 protein kinase. The activation of Gcn2 mediates the phosphorylation of eIF2, which results in it tightly binding with GDP and eIF2B to prevent the exchange of GDP for GTP on eIF2. The GDP-bound state of eIF2 decreases the TC level, resulting in a significant reduction in general protein synthesis. However, the activation of the translation of GCN4 under this condition benefits from the presence of four μORFs in its 5′-UTR. The lack of translational initiation from the TC leads the only existing TC to read through the μORFs until it reaches the true AUG site of the GCN4 mRNA and initiates its translation, thereby generating Gcn4, which finally activates its target genes. (B) Regulation of the intracellular abundance of Gcn4. In addition to nitrogen sources, Gcn4 is also regulated by the TOR pathway. Rapamycin treatment inhibits the TORC1 complex, which releases PP2A and Sit4 from the complex. Free PP2A and Sit4 dephosphorylate Gcn2, which results in the enhanced phosphorylation of eIF2α and finally represses the translation of GCN4 mRNA. Alternatively, abundant amino acid conditions lead to a rapid burst of Pcl5 production, which activates Pho85 to phosphorylate Gcn4. Meanwhile, another regulator, Srb10, is also responsible for the phosphorylation of Gcn4. Phosphorylated Gcn4 is then ubiquitylated by Cdc34 and SCFCDC4, which finally leads to the degradation of Gcn4.
Eukaryotic translation initiation factor 2 (eIF2) is a heterotrimer made up of three subunits, i.e., the α subunit (eIF2α), β subunit (eIF2β), and γ subunit (eIF2γ). eIF2 is activated after GTP binding to eIF2α to initiate the formation of the ternary complex (TC). The TC is made up of three elements, which are eIF2, GTP, and the charged methionyl initiator tRNA. The association of the TC along with the 40S small ribosomal subunit, as well as other eIFs, generates the 43S preinitiation complex (PIC). The PIC can bind to mRNA and scan the mRNA to find an AUG start codon. After start codon recognition, GTP is hydrolyzed to GDP, leading to the release of the eIF2-GDP heterodimer from the PIC. Following this, the 60S ribosomal subunit is recruited by the modified PIC to generate the 80S initiation complex, and then translation starts (185). The free eIF2-GDP must be converted into eIF2-GTP to reform the TC, which depends on the GEF eIF2B. Normally, amino acid starvation can increase uncharged tRNAs levels to allosterically activate the phosphorylase Gcn2. Gcn2 can bind to uncharged tRNAs via its histidyl-tRNA synthetase-like domain. Aminoacyl-tRNA synthetases catalyze the reaction of amino acids with cognate uncharged tRNAs. Their proofreading function is essential for minimizing mistranslation through hydrolysis of misactivated aminoacyl adenylates (pretransfer editing) and hydrolysis of misaminoacylated aa-tRNA (posttransfer editing) (186). The absence of editing causes accumulation of misaminoacylated tRNA, but not deacylated tRNA, under amino acid starvation conditions, which represses the transcription of Gcn2 (186). The activation of Gcn2 induces the phosphorylation of Ser51 of eIF2α (187–191) to enhance its affinity for the GEF eIF2B, which inhibits the exchange rate from the GDP- to GTP-bound status of eIF2 and finally reduces the TC formation rate (33, 192).
The decreasing level of the TC reduces the formation of the PIC, which lowers the efficiency of ribosome scanning that reinitiates the translation of most mRNAs, except for GCN4, which encodes the general control nonderepressible 4 protein (Gcn4). It is a basic leucine zipper transcription factor, which could bind to the UASGCRE (GA[C/G]TCA) motifs of GAAC-responsive genes, leading to their transcriptional activation (33). In the 5′ untranslated region (5′-UTR) of GCN4 mRNA, there are four short open reading frames (μORFs), which typically function as translational barriers. In the translational process of GCN4 mRNA, each of the four upstream AUGs of these μORFs will be mistakenly recognized as an initiation site instead of the real start codon of the Gcn4 ORF. However, the reinitiation efficiency of such a mistaken translation process is generally low, but it is under the control of nitrogen sources. With optimal nitrogen sources, high levels of the TC facilitate the reinitiation of translation after the first μORF, which dramatically decreases the probability of proper translation of GCN4 mRNA. However, under amino acid starvation conditions, low levels of the TC strongly reduce the reinitiation efficiency after the translation of the first μORF, which enables the 40S ribosomal subunit to keep scanning along the mRNA and finally move to the actual distant start codon of the Gcn4 ORF prior to its release from the mRNA (Fig. 5A).
Regulation of Gcn4
GCN4 mRNA abundance is modulated according to amino acid availability. Amino acid starvation rapidly (within 20 min) induces the translational activation of GCN4, even though a change in the GCN4 mRNA abundance is not detectable within this time frame. After 3 to 4 h under starvation conditions, an approximately 2-fold increase of GCN4 mRNA can be detected, which seems to be another (slower) way of accumulating Gcn4 (193). Moreover, mutating GCN2, GCN3, or GCN4 further promotes the accumulation of steady-state GCN4 mRNA in comparison to wild-type cells under infertile conditions (194, 195). This indicates that Gcn2, Gcn3, and Gcn4 could be involved in regulation of stability or synthesis of GCN4 mRNA. However, there is no currently available report to demonstrate the details of the mechanism.
In addition to translational regulation of GCN4, its cellular concentration is also regulated and is controlled by the level of protein degradation (196). The Gcn4 degradation rate under nutrient repletion conditions is high, with a short half-life of approximately 2 to 3 min. However, the Gcn4 degradation rate is four to five times lower when an auxotroph grows under severe starvation conditions with no supplement of an essential amino acid. Translation repression caused by cycloheximide treatment helps to stabilize Gcn4 (197, 198), which indicates that the rapid degradation of Gcn4 in a nitrogen-replete is intended to prevent Gcn4-mediated overtranslation to ensure cost-optimal cell survival. Under nonstarvation conditions, the rapid degradation of Gcn4 is induced by phosphorylation in its transcriptional activation domain. This phosphorylation is catalyzed by the cyclin-dependent protein kinases Pho85 and Srb10 (199, 200) and promotes the ubiquitination of Gcn4 mediated by Cdc34 and SCFCDC4 (Fig. 5B) (198, 200). In contrast, under amino acid starvation conditions, the disappearance of Pho85 cyclin Pcl5 protects Gcn4 from being phosphorylated by Pho85 and enhances its stability (197, 199).
Nitrogen starvation conditions activate the transcription of PLC5 through Gcn4, whereas the protein concentration of Plc5 does not increase for two reasons. The first is because starvation conditions lead to a general reduction in protein synthesis. The second is because Pcl5 is inherently unstable under starvation or nonstarvation conditions. Under nutrition repletion conditions that result in higher translation ratios, the transcription of PCL5 is activated, which results in a rapid burst of Pcl5 production. As a result, Pho85 is activated, which then accelerates the rate of degradation of preexisting Gcn4 (197). Above all, once the amino acid level is replenished, Gcn4 stimulates the rapid clearance of itself by activating and repressing the translation of PCL5 and GCN4, respectively.
In addition to the aforementioned Pcl5-dependent manner, Gcn4 could also be degraded in an Srb10-dependent manner (Fig. 5B). The kinase Srb10, termed the Srb mediator, is one of the transcriptional coactivators, and it is essential for Gcn4-mediated transcriptional activation (201, 202). It is hypothesized that Srb10 is first recruited by Gcn4 and then induces the phosphorylation of Gcn4, which triggers the subsequent ubiquitination and final degradation of Gcn4 (200). Mutating the nuclear localization sequence of Gcn4 sequesters Gcn4 in the cytosol and protects it from degradation, which suggests that Gcn4 is degraded mainly in the nucleus. Considering the cellular localization of Pho85, which is primarily in the nucleus, the degradation of Gcn4 in both Srb10- and Ph85-mediated manners seems to be restricted in the nucleus. Moreover, it has been reported that the translocation of Gcn4 in the nucleus is not influenced by amino acid availability (203). In conclusion, the expression of Gcn4 is regulated on two levels, i.e., the translational regulation of GCN4 mRNA in the cytoplasm and the posttranslational modification and degradation of Gcn4 in the nucleus. This raises an interesting question of why the degradation of Gcn4 is restricted in the nucleus, where it exerts its activation function.
Connection between the GAAC and TOR Pathways
The GAAC pathway has a strong connection with the TORC1 pathway (Fig. 5B). Rapamycin treatment abrogates the TORC1-dependent, Tap42-mediated inhibition of PP2A and Sit4, leading to the dephosphorylation of Gcn2, which results in enhanced eIF2α phosphorylation. In addition to the dephosphorylation induced by TORC1 inhibition, Gcn2 is simultaneously activated by the accumulation of uncharged tRNA, while the activation signaling from TORC1 inhibition occurs faster than that from uncharged tRNA accumulation (184). However, nitrogen (such as histidine) starvation-induced activation of TORC1 results only in phosphorylation of eIF2α and does not reduce the phosphorylation of the Gcn2 Ser577 residue (204), which suggests that sufficiently high levels of uncharged tRNAs can overcome the negative effect of Ser577 phosphorylation on Gcn2 (205).
In addition to functioning as a target of the TOR pathway, a recent study found that Gcn2 is also essential for TORC1 signaling. During amino acid starvation, Gcn2 downregulated TORC1 activity through directly phosphorylating the N-terminal region of Kog1 (206) in a Gcn1- and Gcn20-dependent manner. Structural analysis revealed that the N-terminal region of Kog1 is responsible for lining up with the kinase domain of TORC1, which indicates that Kog1 is responsible for targeting the catalytic domain of TORC1 to its substrates (207). In the absence of Gcn2, TORC1 would stay activated even under amino acid starvation conditions (206). The Gcn2-directed phosphorylation site of Kog1 has not been identified. This raises a possibility that there is an unknown effector that could mediate the Gcn2-dependent phosphorylation of Kog1. Interestingly, while Gcn2 is essential for the inhibition of TORC1 under amino acid starvation condition, it is not involved in the downregulation of TORC1 resulting from rapamycin or nitrogen deprivation.
REGULATION OF NITROGEN METABOLISM IN RESPONSE TO CARBON SOURCES IN S. CEREVISIAE
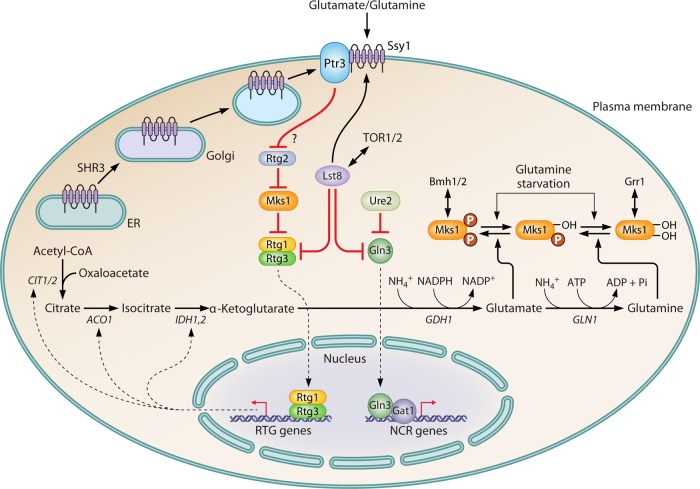
Nitrogen regulatory pathways control nitrogen metabolism by sensing the internal concentrations of ammonia, glutamate, and glutamine, which are the major precursors of amino acid biosynthesis (208). Glutamate is synthesized mainly by a reaction between α-ketoglutarate and ammonia, which is catalyzed by Gdh1, and glutamate could further be used to synthesize glutamine with ammonia, which is catalyzed by Gln1. Meanwhile, α-ketoglutarate also serves as an important intermediate in the TCA cycle of carbon metabolism. Thus, α-ketoglutarate is considered a bridge between nitrogen and carbon metabolism (Fig. 6). The expression of GDH1 and GLN1 is under the control of NCR regulatory pathways, while the expression of genes involved in converting oxaloacetate to α-ketoglutarate in the TCA cycle is instead controlled by the retrograde (RTG) pathway (209). Four activators, i.e., Rtg1 to -3 and Grr1, along with four repressors, i.e., Lst8, Mks1, Bmh1, and Bmh2, work together to regulate the expression of RTG target genes. Similar to the case for the NCR activators Gln3 and Gat1, translocation of the RTG activators Rtg1 and Rgt3 from the cytosol into the nucleus is regulated by their phosphorylation status as well as a repressor, Mks1, which induces the sequestration of Rtg1 and Rtg3 in cytosol, similar to the effect of Ure2 on Gln3 and Gat1. A complex that consists of hyperphosphorylated Mks1, 14-3-3 proteins, Bmh1, and Bmh2 is required for Mks1-induced repression of Rtg1 and Rtg3 to translocate into the nucleus. Mks1 binds and interacts with Rtg2 in such an inhibition process. In addition, Grr1 mediates the ubiquitination and degradation of Mks1 after it is released from Bmh1 and Bmh2 (1).
FIG 6.
Influence of the RTG pathway on nitrogen metabolism. When preferred nitrogen sources, such as glutamate or glutamine, are present in the environment, the inhibitory signal of the RTG pathway is generated from the interaction between glutamine or glutamate and Ssy1. The inhibitory signal is transduced to Rtg2 via Ptr3 in an unknown manner, thereby preventing Rtg2 from binding to Mks1, which allows Mks1 to interact with Rtg1/3 and retain them in the cytoplasm. The cytosolic sequestration of Rtg1/3 represses the transcription of RTG genes, including genes such as CIT1/2, ACO1, and IDH1,2, which are involved in converting oxaloacetate to α-ketoglutarate in the TCA cycle. This repression decreases the concentration of intracellular α-ketoglutarate, the precursor of glutamate and glutamine, and it finally decreases the levels of intracellular amino acids. The presence of glutamate or glutamine activates the TOR pathway, which interplays with Lst8 to regulate both the RTG and NCR pathways. Lst8 represses the RTG pathway at two levels. The first is by influencing the targeting or assembly of the SPS sensor system or its signal transduction function. The second is by inhibiting Rtg1/3 from translocating to the nucleus to activate the transcription of RTG genes. For the NCR pathway, Lst8 prevents Gln3 from translocating into the nucleus to activate the transcription of NCR genes. Mks1 is a multiply phosphorylated protein. Glutamate promotes the hyperphosphorylation of Mks1, which correlates with strong repression of the RTG pathway. When ammonia or glutamine is provided, Mks1 shifts to an intermediate state of phosphorylation. In the presence of the former, rapamycin treatment, which causes only partial dephosphorylation of Mks1, cannot dephosphorylate Mks1 to the point that the dephosphorylation level crosses the threshold required for the derepression of the RTG pathway. Additionally, Mks1 still binds with Bmh1/2 to repress Rtg1/3. In the presence of glutamine, rapamycin treatment dephosphorylates Mks1, which is sufficient to release it from the Mks1-Bmh1/2 complex to derepress the RTG pathway and to interact with Grr1, which enables it to enter the degradation pathway.
The RTG pathway is generally activated by rapamycin inhibition of TORC1, while its performance differs under different nutrient conditions. For example, when glutamate serves as the sole nitrogen source, RTG target genes are strongly repressed after rapamycin treatment. In contrast, after feeding ammonia, which promotes synthesis of glutamine via reaction with preexisting glutamate, rapamycin treatment changes to derepress the RTG pathway (210, 211). The depression of the RTG pathway is also seen after feeding glutamine or using glutamine as the sole nitrogen source. These differences may perhaps be caused by different degrees of phosphorylation of Msk1 with different nitrogen sources (211). In addition to the linkage with the TOR pathway, the RTG pathway is also connected with the SPS sensor system. When using glutamate or glutamine as the sole nitrogen source, the inhibitory signal to the RTG pathway is generated from a combination of Ssy1 and glutamate or glutamine and is transduced to Rtg2 through Ptr3. The inhibitory effect on the RTG pathway is mediated by one of the four repressors, Lst8 (53). Mutant lst8 alleles activate the RTG pathway, which increases intracellular amino acid levels via promoting the synthesis of α-ketoglutarate. As a result, Gap1 is delivered into the vacuole regardless of the extracellular nitrogen source in an lst8 mutant (156, 212). Lst8 achieves its inhibitory effect on the RTG pathway at different levels. The first occurs upstream of Rtg2, which suggests that Lst8 might function during the targeting, assembly, or signal transduction process of the SPS sensor system (213). The other occurs downstream of Rtg2, which suggests that Lst8 negatively regulates the Rtg1 and Rtg3 activators in the RTG pathway (156).
CONCLUSIONS AND PERSPECTIVES
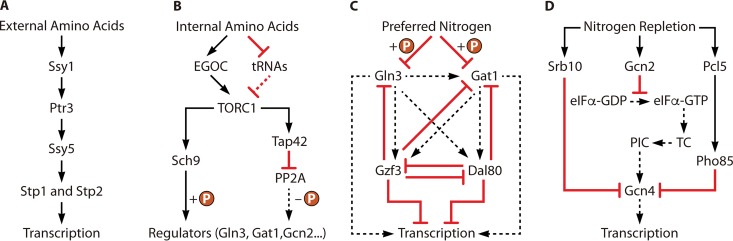
To adapt its growth to various nitrogen sources in the environment, S. cerevisiae has evolved many regulatory processes that are related to nitrogen metabolism. This review summarizes these regulatory processes on different levels, including the sensing, transportation, and catabolism of nitrogen sources, as well as their interaction with carbon metabolism. Brief outlines including the input and output of each regulatory process are shown in Fig. 7. First, nitrogen is sensed before it is metabolized. In this step, the TOR pathway and SPS sensor system play essential roles in detecting intracellular and extracellular nitrogen, respectively. Simultaneously, these two pathways are also regulated by different nitrogen signals. After receiving nitrogen signals, the TOR pathway and SPS sensor system regulate the subsequent transportation step by controlling the expression of genes that encode nitrogen transporters, as well as the degradation of nitrogen transporters via the autophagy pathway. Furthermore, cells orchestrate their nitrogen metabolism response to nitrogen signals by regulating the expression of NCR genes, which is achieved via NCR transcriptional factor-mediated transcription regulation and GAAC pathway-mediated translation regulation. In addition to nitrogen sources, carbon sources also affect nitrogen metabolism in S. cerevisiae, which occurs via the RTG pathway. Knowledge of these regulatory pathways could guide the modification of strains that are used in industrial fermentation. For example, increasing the nuclear localization of Gln3 by mutating potential phosphorylation sites on its nuclear localization signal, truncating its nuclear localization regulation signal, and disrupting URE2 significantly activated the expression of genes in the urea metabolism pathway (DUR1,2 and DUR3), which ultimately reduced the levels of urea and EC by 63% and 72%, respectively, during rice wine fermentation (14).
FIG 7.
Outline of pathways involved in nitrogen regulation in S. cerevisiae. (A) Pathway of the SPS sensor system mediated by sensing of external amino acids. Ssy1 is activated by external amino acids. The signal is transduced by Ptr3 and activates Ssy5. The activated Ssy5 is responsible for activating two transcriptional factors, Stp1 and Stp2. Finally, Stp1 and Stp2 activate the transcription of relevant genes. (B) Pathway of TOR pathway mediated by sensing of internal amino acids. The accumulation of internal amino acids promotes the establishment of the EGOC and decreases the internal abundance of tRNAs. As a result, TORC1 is activated by the EGOC and derepressed by the reduction of internal tRNAs. Next, the downstream effectors Sch9 and Tap42 are activation by activated TORC1. Finally, the phosphorylation of regulators, such as Gln3, Gat1, and Gcn2, is induced by Sch9. In addition, activated Tap42 promotes its interaction with PP2A, which represses the PP2A-mediated dephosphorylation of regulators. (C) Transcriptional regulatory pathway mediated by NCR. The major activators of NCR, Gln3 and Gat1, are phosphorylated in the presence of preferred nitrogen sources. As a result, the transcriptional activation mediated by these two activators is repressed. (D) Regulation of the GAAC pathway responds to nitrogen sources. Gcn2 is activated under nitrogen repletion conditions, which then represses the transformation of eIF2α from a GDP- to a GTP-bound status. The reduction of eIF2α-GTP decreases the TC, which then inhibits the formation of the PIC. As a result, the translation of GCN4 mRNA is repressed, leading to a decreased intracellular concentration of Gcn4. On the other hand, nitrogen repletion conditions also induce the degradation of the Gcn4 through activation of Srb10. In addition, nitrogen repletion also induces the activation of Pcl5, which then activates Pho85 and finally induces the degradation of Gcn4. The decrease of Gcn4 finally represses the transcriptional activation of relevant genes dependent on Gcn4.
Although remarkable progress has been made in the last few decades on investigating the regulatory mechanism of nitrogen metabolism, many details remain unclear. In the TOR pathway, the mechanism of alteration of TORC1 activity in response to environmental cues is yet to be revealed. Recently, studies demonstrated that Gtr-Ego-mediated activation of TORC1 is not required to prevent Gln3 translocating into the nucleus under nitrogen-rich conditions. This indicates that there is an unidentified TORC1-independent approach for sequestering Gln3 in the cytosol (214). Furthermore, the events that occur downstream of the TOR pathway are also not completely understood. Recent phospho-proteomics studies suggest that there are novel growth-related effectors that act downstream of the TOR pathway, but the identification of these effectors still needs further study. In the SPS sensor system, the molecular mechanism by which this sensor system senses extracellular amino acids, such as interactions between Ssy1 and some amino acids, remains unclear. Furthermore, as discussed above, the reason for the different degrees of accumulation of GCN4 mRNA in response to short- and long-term amino acid starvation remains unknown. Additional details, such as how NCR transcriptional factors interact with NCR genes as well as with each other, are far from completely understood. In addition, some recent studies suggested that regulation of mRNA stability via posttranscriptional modification, may have a crucial effect on changing the gene expression profile according to environmental nitrogen perturbation, which also has been seen in adaptation of cells to alteration of environmental carbon (215, 216), while the mechanism is still unclear. In addition to these details of the specific nitrogen regulatory pathway, many more questions need to be evaluated with respect to a global view of cellular growth and proliferation processes. Although links between the regulation of nitrogen and carbon catabolism have been revealed as in the tip of an iceberg, there is still a long way to go to completely interpret the complicated molecular mechanism between these two major essential nutrients. Furthermore, there is the question of influences from other macro- or micronutrients, such as phosphorus, sulfur, and metal ions, as well as vitamins, on cellular responses to nitrogen. In addition, there are different responses to short- and long-term nitrogen stimulation. There are still many challenges in understanding more details about how cells could alter their life activities in response to nitrogen stimulation to adapt these diverse environments.
As new omics technologies, such as chromatin immunoprecipitation sequencing (ChIP-seq) (217), chromatin precipitation-exo (218), nucleosome-positioning sequencing (219, 220), and phospho- and glycol-proteomics (221, 222), are developed, the regulatory mechanisms involved in nitrogen catabolism will be interpreted in greater detail. In the foreseeable future, the panorama of how a global transcriptional factor, such as Gln3, interacts with genes in the whole genome, as well as with other regulators, will be revealed with the help of ChIP-seq or ChIP-exo assays. ChIP-seq and nucleosome-positioning sequencing have been used together in a study to investigate how nucleosome positioning regulates the expression of 200 Gcn4-controlled genes. The results revealed that nucleosome eviction is crucial for the robust transcription of highly expressed genes. In contrast, the recruitment of Pol II has been shown to be the rate-limiting step during expression of weakly expressed genes (223). In addition, how the regulation of nucleosome positions responds to different nitrogen sources has been revealed primarily with the help of nucleosome-positioning sequencing (220). Using phospho- and glycol-proteomics methods, the complicated posttranslational modification regulation of nitrogen catabolism will be uncovered in detail. With the help of systematic biology, global interactions between different nitrogen regulatory pathways as well as between regulatory pathways for nitrogen and other nutrients will be explained further. The discovery of more details of the mechanism of nitrogen metabolism regulation in S. cerevisiae will greatly improve the production efficiency in the yeast fermentation industry. Furthermore, because of the similarity between yeast and humans in terms of nutrient metabolism, such discoveries may help to establish better approaches to cure human diseases, such as cancer and obesity.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2017YFC1600403), the National Natural Science Foundation of China (21390204), the Key Research and Development Program of Jiangsu Province (BE2016689), the Fundamental Research Funds for the Central Universities (JUSRP51701A), the Six Talent Peaks Project in Jiangsu Province (2015-JY-005), the Distinguished Professor Project of Jiangsu Province, and the 111 Project (111-2-06).
Biographies

Weiping Zhang is studying for his Ph.D. in fermentation engineering in the School of Biotechnology, Jiangnan University. His Ph.D. topic is the global study and regulation of nitrogen metabolism in S. cerevisiae. He began to work on this topic in 2011. His advisors, Jian Chen and Jingwen Zhou, encouraged him to write this review article to have deep knowledge of the complicated regulatory mechanism of nitrogen metabolism in S. cerevisiae. He is working on how to elucidate the nitrogen regulation mechanism through comparative genomics and transcriptional analysis of different industrial strains and epigenomic analysis of global transcriptional factors involved in nitrogen metabolism regulation, based on the understanding of the complicated regulation systems involved in nitrogen metabolism in S. cerevisiae.

Guocheng Du obtained his Ph.D. from Jiangnan University (formerly Wuxi University of Light Industry) in 1997. Since then, he became Assistant Professor, Associate Professor, and Full Professor in the School of Biotechnology, Jiangnan University. He was the Dean of the School of Biotechnology, Jiangnan University, from 2008 to 2017. He is the Distinguished Professor of Changjiang Scholars, Ministry of Education, China. His current main research focus is on bioprocess engineering, metabolic engineering, and food fermentation. He began to work on this topic in 2008 and wishes to decrease the accumulation of harmful nitrogenous compounds during Chinese traditional food fermentation processes through regulating the nitrogen metabolism pathways of fermentation strains.

Jingwen Zhou obtained his Ph.D. degree in fermentation engineering at Jiangnan University in 2009. After that, he became Assistant Professor, Associate Professor, and Full Professor in the School of Biotechnology, Jiangnan University. He finished his postdoctoral training in the Department of Chemistry and Chemical Biology at Harvard University from 2012 to 2013. His current research focuses mainly on the metabolic engineering of microorganisms to produce organic acids and plant natural products, especially α-ketoglutarate, l-ascorbic acid, and flavonoids. He began to work on this topic in 2011 and aims to decrease or eliminate the accumulation of ethyl carbamate during the fermentation of Chinese rice wine by metabolic engineering and high-throughput strategies.

Jian Chen obtained his Ph.D. degree in fermentation engineering from Jiangnan University (formerly Wuxi University of Light Industry) in 1990. Since then, he became Assistant Professor, Associate Professor, and Full Professor in the School of Biotechnology, Jiangnan University. He is the president of Jiangnan University. He was voted as Academician of the Chinese Academy of Engineering in 2017. His research interests include (i) stress tolerance and response of food microorganisms, (ii) production of food additives by use of biotechnology, and (iii) food safety issues in fermented foods. Since 2011, he has focused on studying the mechanisms of the accumulation of harmful nitrogen-containing small-molecule components during the production of fermented foods, such as ethyl carbamate and bioamines.
REFERENCES
- 1.Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu Rev Genet 42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 2.Turcotte B, Liang XB, Robert F, Soontorngun N. 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 10:2–13. doi: 10.1111/j.1567-1364.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripodi F, Nicastro R, Reghellin V, Coccetti P. 2015. Post-translational modifications on yeast carbon metabolism: regulatory mechanisms beyond transcriptional control. Biochim Biophys Acta 1850:620–627. doi: 10.1016/j.bbagen.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Magasanik B, Kaiser CA. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18. doi: 10.1016/S0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- 5.Tate JJ, Buford D, Rai R, Cooper TG. 2017. General amino acid control and 14-3-3 proteins Bmh1/2 are required for nitrogen catabolite repression-sensitive regulation of Gln3 and Gat1 localization. Genetics 205:633–655. doi: 10.1534/genetics.116.195800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haas H, Marzluf GA. 1995. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr Genet 28:177–183. doi: 10.1007/BF00315785. [DOI] [PubMed] [Google Scholar]
- 7.Wiemann P, Willmann A, Straeten M, Kleigrewe K, Beyer M, Humpf HU, Tudzynski B. 2009. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol Microbiol 72:931–946. doi: 10.1111/j.1365-2958.2009.06695.x. [DOI] [PubMed] [Google Scholar]
- 8.Studt L, Wiemann P, Kleigrewe K, Humpf HU, Tudzynski B. 2012. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl Environ Microbiol 78:4468–4480. doi: 10.1128/AEM.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Pan Y, Liu G. 2013. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet Biol 61:69–79. doi: 10.1016/j.fgb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Niehaus EM, Kleigrewe K, Wiemann P, Studt L, Sieber CMK, Connolly LR, Freitag M, Guldener U, Tudzynski B, Humpf HU. 2013. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem Biol 20:1055–1066. doi: 10.1016/j.chembiol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Brosnan JT, Brosnan ME. 2013. Glutamate: a truly functional amino acid. Amino Acids 45:413–418. doi: 10.1007/s00726-012-1280-4. [DOI] [PubMed] [Google Scholar]
- 12.Yasumura A, Abe S, Tanaka T. 2008. Involvement of nitrogen regulation in Bacillus subtilis degU expression. J Bacteriol 190:5162–5171. doi: 10.1128/JB.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Zou H, Fu J, Chen J, Zhou J, Du G. 2013. Nitrogen regulation involved in the accumulation of urea in Saccharomyces cerevisiae. Yeast 30:437–447. doi: 10.1002/yea.2980. [DOI] [PubMed] [Google Scholar]
- 14.Zhao XR, Zou HJ, Fu JW, Zhou JW, Du GC, Chen J. 2014. Metabolic engineering of the regulators in nitrogen catabolite repression to reduce the production of ethyl carbamate in a model rice wine system. Appl Environ Microbiol 80:392–398. doi: 10.1128/AEM.03055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Du G, Zou H, Fu J, Zhou J, Chen J. 2013. Progress in preventing the accumulation of ethyl carbamate in alcoholic beverages. Trends Food Sci Technol 32:97–107. doi: 10.1016/j.tifs.2013.05.009. [DOI] [Google Scholar]
- 16.Park SK, Kim CT, Lee JW, Jhee OH, Om AS, Kang JS, Moon TW. 2007. Analysis of ethyl carbamate in Korean soy sauce using high-performance liquid chromatography with fluorescence detection or tandem mass spectrometry and gas chromatography with mass spectrometry. Food Control 18:975–982. doi: 10.1016/j.foodcont.2006.05.013. [DOI] [Google Scholar]
- 17.Weber JV, Sharypov VI. 2008. Ethyl carbamate in foods and beverages: a review. Environ Chem Lett 7:233–247. doi: 10.1007/s10311-008-0168-8. [DOI] [Google Scholar]
- 18.Huang Z, Pan XD, Wu PG, Chen Q, Han JL, Shen XH. 2013. Validation (in-house and collaboratory) of the quantification method for ethyl carbamate in alcoholic beverages and soy sauce by GC-MS. Food Chem 141:4161–4165. doi: 10.1016/j.foodchem.2013.06.128. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Zou H, Du G, Chen J, Zhou J. 2015. Effects of nitrogen catabolite repression-related amino acids on the flavour of rice wine. J Inst Brew 121:581–588. doi: 10.1002/jib.269. [DOI] [Google Scholar]
- 20.Landaud S, Helinck S, Bonnarme P. 2008. Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl Microbiol Biotechnol 77:1191–1205. doi: 10.1007/s00253-007-1288-y. [DOI] [PubMed] [Google Scholar]
- 21.Feyder S, De Craene J-O, Baer S, Bertazzi DL, Friant S. 2015. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci 16:1509–1525. doi: 10.3390/ijms16011509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chisholm G, Cooper TG. 1982. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol 2:1088–1095. doi: 10.1128/MCB.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenson M, Dubois E, Piotrowska M, Drillien R, Aigle M. 1974. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol Gen Genet 128:73–85. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell AP, Magasanik B. 1983. Purification and properties of glutamine synthetase from Saccharomyces cerevisiae. J Biol Chem 258:119–124. [PubMed] [Google Scholar]
- 25.Mitchell AP. 1985. The GLN1 locus of Saccharomyces cerevisiae encodes glutamine synthetase. Genetics 111:243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller SM, Magasanik B. 1990. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol 172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cogoni C, Valenzuela L, Gonzalez-Halphen D, Olivera H, Macino G, Ballario P, Gonzalez A. 1995. Saccharomyces cerevisiae has a single glutamate synthase gene coding for a plant-like high-molecular-weight polypeptide. J Bacteriol 177:792–798. doi: 10.1128/jb.177.3.792-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ljungdahl PO, Daignan-Fornier B. 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsberg H, Ljungdahl PO. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol Cell Biol 21:814–826. doi: 10.1128/MCB.21.3.814-826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofman-Bang J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol 12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 32.Hinnebusch AG, Natarajan K. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell 1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 34.Rodkaer SV, Faergeman NJ. 2014. Glucose- and nitrogen sensing and regulatory mechanisms in Saccharomyces cerevisiae. FEMS Yeast Res 14:683–696. doi: 10.1111/1567-1364.12157. [DOI] [PubMed] [Google Scholar]
- 35.Klasson H, Fink GR, Ljungdahl PO. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol 19:5405–5416. doi: 10.1128/MCB.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souciet JL, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, Durrens P, Gaillardin C, Lepingle A, Llorente B, Malpertuy A, Neuveglise C, Ozier-Kalogeropoulos O, Potier S, Saurin W, Tekaia F, Toffano-Nioche C, Wesolowski-Louvel M, Wincker P, Weissenbach J. 2000. Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett 487:3–12. [DOI] [PubMed] [Google Scholar]
- 37.Ljungdahl PO. 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem Soc T 37:242–247. doi: 10.1042/BST0370242. [DOI] [PubMed] [Google Scholar]
- 38.Gaber RF, Ottow K, Andersen HA, Kielland-Brandt MC. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot Cell 2:922–929. doi: 10.1128/EC.2.5.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulsen P, Gaber RF, Kielland-Brandt MC. 2008. Hyper- and hyporesponsive mutant forms of the Saccharomyces cerevisiae Ssy1 amino acid sensor. Mol Membr Biol 25:164–176. doi: 10.1080/09687680701771917. [DOI] [PubMed] [Google Scholar]
- 40.Andreasson C, Ljungdahl PO. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Gene Dev 16:3158–3172. doi: 10.1101/gad.239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka N, Mukai Y. 2015. Yeast Cyc8p and Tup1p proteins function as coactivators for transcription of Stp1/2p-dependent amino acid transporter genes. Biochem Biophys Res Commun 468:32–38. doi: 10.1016/j.bbrc.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Abdel-Sater F, Bakkoury ME, Urrestarazu A, Vissers S, Andre B. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol Cell Biol 24:9771–9785. doi: 10.1128/MCB.24.22.9771-9785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreasson C, Heessen S, Ljungdahl PO. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Gene Dev 20:1563–1568. doi: 10.1101/gad.374206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdel-Sater F, Iraqui I, Urrestarazu A, Andre B. 2004. The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166:1727–1739. doi: 10.1534/genetics.166.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfirrmann T, Heessen S, Omnus DJ, Andreasson C, Ljungdahl PO. 2010. The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol Cell Biol 30:3299–3309. doi: 10.1128/MCB.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumusiime S, Zhang C, Overstreet MS, Liu Z. 2011. Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J Biol Chem 286:4620–4631. doi: 10.1074/jbc.M110.195313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omnus DJ, Pfirrmann T, Andreasson C, Ljungdahl PO. 2011. A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol Biol Cell 22:2754–2765. doi: 10.1091/mbc.E11-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omnus DJ, Ljungdahl PO. 2013. Rts1-protein phosphatase 2A antagonizes Ptr3-mediated activation of the signaling protease Ssy5 by casein kinase I. Mol Biol Cell 24:1480–1492. doi: 10.1091/mbc.E13-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boban M, Ljungdahl PO. 2007. Dal81 enhances Stp1- and Stp2-dependent transcription necessitating negative modulation by inner nuclear membrane protein asil in Saccharomyces cerevisiae. Genetics 176:2087–2097. doi: 10.1534/genetics.107.075077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boban M, Zargari A, Andreasson C, Heessen S, Thyberg J, Ljungdahl PO. 2006. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J Cell Biol 173:695–707. doi: 10.1083/jcb.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zargari A, Boban M, Heessen S, Andreasson C, Thyberg J, Ljungdahl PO. 2007. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J Biol Chem 282:594–605. doi: 10.1074/jbc.M609201200. [DOI] [PubMed] [Google Scholar]
- 52.Wielemans K, Jean C, Vissers S, Andre B. 2010. Amino acid signaling in yeast: post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J Biol Chem 285:855–865. doi: 10.1074/jbc.M109.015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Guan KL. 2011. Amino acid signaling in TOR activation. Annu Rev Biochem 80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 55.Keith CT, Schreiber SL. 1995. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 56.Wullschleger S, Loewith R, Oppliger W, Hall MN. 2005. Molecular organization of target of rapamycin complex 2. J Biol Chem 280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 57.Dames SA, Mulet JM, Rathgeb-Szabo K, Hall MN, Grzesiek S. 2005. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J Biol Chem 280:20558–20564. doi: 10.1074/jbc.M501116200. [DOI] [PubMed] [Google Scholar]
- 58.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10:457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 59.Reinke A, Anderson S, McCaffery JM, Yates J, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 60.Wedaman KP, Reinke A, Anderson S, Yates J, McCaffery JM, Powers T. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell 14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yerlikaya S, Meusburger M, Kumari R, Huber A, Anrather D, Costanzo M, Boone C, Ammerer G, Baranov PV, Loewith R. 2016. TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol Biol Cell 27:397–409. doi: 10.1091/mbc.E15-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teixeira V, Medeiros TC, Vilaca R, Ferreira J, Moradas-Ferreira P, Costa V. 2016. Ceramide signaling targets the PP2A-like protein phosphatase Sit4p to impair vacuolar function, vesicular trafficking and autophagy in Isc1p deficient cells. Biochim Biophys Acta 1861:21–33. doi: 10.1016/j.bbalip.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Schreiber SL. 1991. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 64.Gaubitz C, Oliveira TM, Prouteau M, Leitner A, Karuppasamy M, Konstantinidou G, Rispal D, Eltschinger S, Robinson GC, Thore S, Aebersold R, Schaffitzel C, Loewith R. 2015. Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol Cell 58:977–988. doi: 10.1016/j.molcel.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 65.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 66.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. 2008. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 7:1819–1830. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 68.Zoncu R, Bar-Peled L, Efeyan A, Wang SY, Sancak Y, Sabatini DM. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubouloz F, Deloche O, Wanke V, Cameron E, De Virgillo C. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 70.Stauffer B, Powers T. 2017. Target of rapamycin signaling mediates vacuolar fragmentation. Curr Genet 63:35–42. doi: 10.1007/s00294-016-0616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kira S, Kumano Y, Ukai H, Takeda E, Matsuura A, Noda T. 2016. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol Biol Cell 27:382–396. doi: 10.1091/mbc.E15-07-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashrafi K, Farazi TA, Gordon JI. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem 273:25864–25874. doi: 10.1074/jbc.273.40.25864. [DOI] [PubMed] [Google Scholar]
- 73.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR III, Davis NG. 2006. Global analysis of protein palmitoylation in yeast. Cell 125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nadolski MJ, Linder ME. 2009. Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3. J Biol Chem 284:17720–17730. doi: 10.1074/jbc.M109.005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powis K, Zhang T, Panchaud N, Wang R, De Virgilio C, Ding J. 2015. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res 25:1043–1059. doi: 10.1038/cr.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Powis K, De Virgilio C. 2016. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov 2:15049–15065. doi: 10.1038/celldisc.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakashima N, Noguchi E, Nishimoto T. 1999. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152:853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y. 2011. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev 25:1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeong JH, Lee KH, Kim YM, Kim DH, Oh BH, Kim YG. 2012. Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem 287:29648–29653. doi: 10.1074/jbc.C112.384420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valbuena N, Guan KL, Moreno S. 2012. The Vam6 and Gtr1-Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J Cell Sci 125:1920–1928. doi: 10.1242/jcs.094219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. 2012. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell 46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 84.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. 2012. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 85.Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. 2015. Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Rep 13:1–7. doi: 10.1016/j.celrep.2015.08.059. [DOI] [PubMed] [Google Scholar]
- 86.Petit CS, Roczniak-Ferguson A, Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neklesa TK, Davis RW. 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet 5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Panchaud N, Peli-Gulli M-P, De Virgilio C. 2013. SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 12:2948–2952. doi: 10.4161/cc.26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Panchaud N, Peli-Gulli M-P, De Virgilio C. 2013. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- 92.Kamada Y. 2017. Novel tRNA function in amino acid sensing of yeast Tor complex 1. Genes Cells 22:135–147. doi: 10.1111/gtc.12462. [DOI] [PubMed] [Google Scholar]
- 93.Kira S, Tabata K, Shirahama-Noda K, Nozoe A, Yoshimori T, Noda T. 2014. Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy 10:1565–1578. doi: 10.4161/auto.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broach JR. 2012. Nutritional control of growth and development in yeast. Genetics 192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet 56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 96.Swinnen E, Wilms T, Idkowiak-Baldys J, Smets B, De Snijder P, Accardo S, Ghillebert R, Thevissen K, Cammue B, De Vos D, Bielawski J, Hannun YA, Winderickx J. 2014. The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol Biol Cell 25:196–211. doi: 10.1091/mbc.E13-06-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues F, Ludovico P. 2011. Yeast chronological lifespan and proteotoxic stress: is autophagy good or bad? Biochem Soc T 39:1466–1470. doi: 10.1042/BST0391466. [DOI] [PubMed] [Google Scholar]
- 98.Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol 69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- 99.DiComo CJ, Arndt KT. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Gene Dev 10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 100.Jiang Y, Broach JR. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J 18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell 3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao Y, Boguslawski G, Zitomer RS, DePaoliRoach AA. 1997. Saccharomyces cerevisiae homologs of mammalian B and B' subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J Biol Chem 272:8256–8262. doi: 10.1074/jbc.272.13.8256. [DOI] [PubMed] [Google Scholar]
- 103.van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR. 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol 12:4946–4959. doi: 10.1128/MCB.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Healy AM, Zolnierowicz S, Stapleton AE, Goebl M, Depaoliroach AA, Pringle JR. 1991. Cdc55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: Identification, characterization, and homology to the b-subunit of mammalian type-2A protein phosphatase. Mol Cell Biol 11:5767–5780. doi: 10.1128/MCB.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luke MM, DellaSeta F, DiComo CJ, Sugimoto H, Kobayashi R, Arndt KT. 1996. The SAPs, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol 16:2744–2755. doi: 10.1128/MCB.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. 2000. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem 275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 107.Aronova S, Wedaman K, Anderson S, Yates J III, Powers T. 2007. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell 18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan G, Shen X, Jiang Y. 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J 25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shamji AF, Kuruvilla FG, Schreiber SL. 2000. Partitioning the transcriptional program induced by rapamycin among the effecters of the Tor proteins. Curr Biol 10:1574–1581. doi: 10.1016/S0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 110.Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell 11:1467–1478. doi: 10.1016/S1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 111.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell 8:1017–1026. doi: 10.1016/S1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 112.Philip B, Levin DE. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol Cell Biol 21:271–280. doi: 10.1128/MCB.21.1.271-280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan G, Lai Y, Jiang Y. 2012. The TOR complex 1 is a direct target of Rho1 GTPase. Mol Cell 45:743–753. doi: 10.1016/j.molcel.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sutter BM, Wu X, Laxman S, Tu BP. 2013. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jack DL, Paulsen IT, Saier MH. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 116.Cain NE, Kaiser CA. 2011. Transport activity-dependent intracellular sorting of the yeast general amino acid permease. Mol Biol Cell 22:1919–1929. doi: 10.1091/mbc.E10-10-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wong FH, Chen JS, Reddy V, Day JL, Shlykov MA, Wakabayashi ST, Saier MH Jr. 2012. The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol 22:105–113. doi: 10.1159/000338542. [DOI] [PubMed] [Google Scholar]
- 118.Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci 29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 119.Crapeau M, Merhi A, Andre B. 2014. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J Biol Chem 289:22103–22116. doi: 10.1074/jbc.M114.582320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. 2010. The ubiquitin code of yeast permease trafficking. Trends Cell Biol 20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 121.Zhang P, Du G, Zou H, Xie G, Chen J, Shi Z, Zhou J. 2017. Mutant potential ubiquitination sites in Dur3p enhance the urea and ethyl carbamate reduction in a model rice wine system. J Agric Food Chem 65:1641–1648. doi: 10.1021/acs.jafc.6b05348. [DOI] [PubMed] [Google Scholar]
- 122.Komander D. 2009. The emerging complexity of protein ubiquitination. Biochem Soc Trans 37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 123.Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wenzel DM, Stoll KE, Klevit RE. 2011. E2s: structurally economical and functionally replete. Biochem J 433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nagy V, Dikic I. 2010. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem 391:163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 126.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 127.Behrends C, Harper JW. 2011. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol 18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 128.Komander D, Clague MJ, Urbe S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 129.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. 2004. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 130.Harsay E, Bretscher A. 1995. Parallel secretory pathways to the cell surface in yeast. J Cell Biol 131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bowers K, Stevens TH. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 132.Cowles CR, Snyder WB, Burd CG, Emr SD. 1997. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J 16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Conibear E. 2010. Converging views of endocytosis in yeast and mammals. Curr Opin Cell Biol 22:513–518. doi: 10.1016/j.ceb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 134.Kondo Y, Hanai A, Nakai W, Katoh Y, Nakayama K, Shin HW. 2012. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct Funct 37:141–154. doi: 10.1247/csf.12015. [DOI] [PubMed] [Google Scholar]
- 135.Dupre S, Urban-Grimal D, Haguenauer-Tsapis R. 2004. Ubiquitin and endocytic intemalization in yeast and animal cells. Biochim Biophys Acta 1695:89–111. doi: 10.1016/j.bbamcr.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 136.Wang Y, Dohlman HG. 2006. Regulation of G protein and mitogen-activated protein kinase signaling by ubiquitination: insights from model organisms. Circ Res 99:1305–1314. doi: 10.1161/01.RES.0000251641.57410.81. [DOI] [PubMed] [Google Scholar]
- 137.Rotin D, Kumar S. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 138.Lv Y, Zhao X, Liu L, Du G, Zhou J, Chen J. 2013. A simple procedure for protein ubiquitination detection in Saccharomyces cerevisiae: Gap1p as an example. J Microbiol Methods 94:25–29. doi: 10.1016/j.mimet.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 139.O'Donnell AF, Apffel A, Gardner RG, Cyert MS. 2010. α-Arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell 21:3552–3566. doi: 10.1091/mbc.E10-07-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Springael JY, Andre B. 1998. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell 9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lauwers E, Jacob C, Andre B. 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soetens O, De Craene JO, Andre B. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J Biol Chem 276:43949–43957. doi: 10.1074/jbc.M102945200. [DOI] [PubMed] [Google Scholar]
- 143.Lin CH, MacGum JA, Chu T, Stefan CJ, Emr SD. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 144.Merhi A, Andre B. 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul Arrestin-like adaptors. Mol Cell Biol 32:4510–4522. doi: 10.1128/MCB.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sasaki T, Takagi H. 2013. Phosphorylation of a conserved Thr357 in yeast Nedd4-like ubiquitin ligase Rsp5 is involved in down-regulation of the general amino acid permease Gap1. Genes Cells 18:459–475. doi: 10.1111/gtc.12049. [DOI] [PubMed] [Google Scholar]
- 146.Jauniaux JC, Grenson M. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae: nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem 190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- 147.Peter GJ, During L, Ahmed A. 2006. Carbon catabolite repression regulates amino acid permeases in Saccharomyces cerevisiae via the TOR signaling pathway. J Biol Chem 281:5546–5552. doi: 10.1074/jbc.M513842200. [DOI] [PubMed] [Google Scholar]
- 148.Ghaddar K, Merhi A, Saliba E, Krammer EM, Prevost M, Andre B. 2014. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol Cell Biol 34:4447–4463. doi: 10.1128/MCB.00699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Opekarova M, Caspari T, Pinson B, Brethes D, Tanner W. 1998. Post-translational fate of CAN1 permease of Saccharomyces cerevisiae. Yeast 14:215–224. doi:. [DOI] [PubMed] [Google Scholar]
- 150.Hatakeyama R, Kamiya M, Takahara T, Maeda T. 2010. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol 30:5598–5607. doi: 10.1128/MCB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nikko E, Pelham HRB. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jeschke G. 2013. A comparative study of structures and structural transitions of secondary transporters with the LeuT fold. Eur Biophys J 42:181–197. doi: 10.1007/s00249-012-0802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jardetzky O. 1966. Simple allosteric model for membrane pumps. Nature 211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 154.Rubio-Texeira M, Kaiser CA. 2006. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol Biol Cell 17:3031–3050. doi: 10.1091/mbc.E05-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen EJ, Kaiser CA. 2002. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99:14837–14842. doi: 10.1073/pnas.232591899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen EJ, Kaiser CA. 2003. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol 161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr Genet 36:317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- 158.Iraqui I, Vissers S, Bernard F, De Craene JO, Boles E, Urrestarazu A, Andre B. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol 19:989–1001. doi: 10.1128/MCB.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Usami Y, Uemura S, Mochizuki T, Morita A, Shishido F, Inokuchi J, Abe F. 2014. Functional mapping and implications of substrate specificity of the yeast high-affinity leucine permease Bap2. Biochim Biophys Acta 1838:1719–1729. doi: 10.1016/j.bbamem.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 160.Nielsen PS, van den Hazel B, Didion T, de Boer M, Jorgensen M, Planta RJ, Kielland-Brandt MC, Andersen HA. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol Gen Genet 264:613–622. doi: 10.1007/s004380000347. [DOI] [PubMed] [Google Scholar]
- 161.Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Georis I, Feller A, Vierendeels F, Dubois E. 2009. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol Cell Biol 29:3803–3815. doi: 10.1128/MCB.00399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. 2000. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J Biol Chem 275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem 275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 165.Coornaert D, Vissers S, Andre B. 1991. The pleiotropic Uga35(Durl) regulatory gene of Saccharomyces cerevisiae cloning, sequence and identity with the Dal81 gene. Gene 97:163–171. doi: 10.1016/0378-1119(91)90048-G. [DOI] [PubMed] [Google Scholar]
- 166.Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cardillo SB, Levi CE, Bermudez Moretti M, Correa Garcia S. 2012. Interplay between the transcription factors acting on the GATA- and GABA-responsive elements of Saccharomyces cerevisiae UGA promoters. Microbiology 158:925–935. doi: 10.1099/mic.0.051235-0. [DOI] [PubMed] [Google Scholar]
- 168.Rai R, Tate JJ, Georis I, Dubois E, Cooper TG. 2014. Constitutive and nitrogen catabolite repression-sensitive production of Gat1 isoforms. J Biol Chem 289:2918–2933. doi: 10.1074/jbc.M113.516740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Georis I, Tate JJ, Vierendeels F, Cooper TG, Dubois E. 2015. Premature termination of GAT1 transcription explains paradoxical negative correlation between nitrogen-responsive mRNA, but constitutive low-level protein production. RNA Biol 12:824–837. doi: 10.1080/15476286.2015.1058476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Georis I, Tate JJ, Cooper TG, Dubois E. 2008. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J Biol Chem 283:8919–8929. doi: 10.1074/jbc.M708811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kulkarni A, Buford TD, Rai R, Cooper TG. 2006. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res 6:218–229. doi: 10.1111/j.1567-1364.2006.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Blinder D, Coschigano PW, Magasanik B. 1996. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J Bacteriol 178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tate JJ, Rai R, Cooper TG. 2015. Nitrogen starvation and TorC1 inhibition differentially affect nuclear localization of the Gln3 and Gat1 transcription factors through the rare glutamine tRNACUG in Saccharomyces cerevisiae. Genetics 199:455–474. doi: 10.1534/genetics.114.173831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Georis I, Tate JJ, Cooper TG, Dubois E. 2011. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem 286:44897–44912. doi: 10.1074/jbc.M111.290577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tate JJ, Georis I, Dubois E, Cooper TG. 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J Biol Chem 285:17880–17895. doi: 10.1074/jbc.M109.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Puria R, Zurita-Martinez SA, Cardenas ME. 2008. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 105:7194–7199. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rai R, Tate JJ, Shanmuganatham K, Howe MM, Nelson D, Cooper TG. 2015. Nuclear Gln3 import is regulated by nitrogen catabolite repression whereas export is specifically regulated by glutamine. Genetics 201:989–1016. doi: 10.1534/genetics.115.177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 179.Rai R, Tate JJ, Nelson DR, Cooper TG. 2013. gln3 mutations dissociate responses to nitrogen limitation (nitrogen catabolite repression) and rapamycin inhibition of TorC1. J Biol Chem 288:2789–2804. doi: 10.1074/jbc.M112.421826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Feller A, Georis I, Tate JJ, Cooper TG, Dubois E. 2013. Alterations in the Ure2 αcap domain elicit different GATA factor responses to rapamycin treatment and nitrogen limitation. J Biol Chem 288:1841–1855. doi: 10.1074/jbc.M112.385054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Masison DC, Wickner RB. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p prion-containing cells. Science 270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 182.Masison DC, Maddelein ML, Wickner RB. 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci U S A 94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol 22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. 2010. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem 285:16893–16911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jackson RJ, Hellen CUT, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mohler K, Mann R, Bullwinkle TJ, Hopkins K, Hwang L, Reynolds NM, Gassaway B, Aerni HR, Rinehart J, Polymenis M, Faull K, Ibba M. 2017. Editing of misaminoacylated tRNA controls the sensitivity of amino acid stress responses in Saccharomyces cerevisiae. Nucleic Acids Res 45:3985–3996. doi: 10.1093/nar/gkx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. 1992. Phosphorylation of initiation factor 2α by protein-kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585–596. doi: 10.1016/0092-8674(92)90193-G. [DOI] [PubMed] [Google Scholar]
- 188.Gomez E, Mohammad SS, Pavitt GD. 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J 21:5292–5301. doi: 10.1093/emboj/cdf515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dey M, Velyvis A, Li JJ, Chiu E, Chiovitti D, Kay LE, Sicheri F, Dever TE. 2011. Requirement for kinase-induced conformational change in eukaryotic initiation factor 2α (eIF2α) restricts phosphorylation of Ser51. Proc Natl Acad Sci U S A 108:4316–4321. doi: 10.1073/pnas.1014872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dong JS, Qiu HF, Garcia-Barrio M, Anderson J, Hinnebusch AG. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6:269–279. doi: 10.1016/S1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 191.Qiu H, Dong J, Hu C, Francklyn CS, Hinnebusch AG. 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J 20:1425–1438. doi: 10.1093/emboj/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dever TE, Chen JJ, Barber GN, Cigan AM, Feng L, Donahue TF, London IM, Katze MG, Hinnebusch AG. 1993. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 protein-kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci U S A 90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Albrecht G, Mosch HU, Hoffmann B, Reusser U, Braus GH. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J Biol Chem 273:12696–12702. doi: 10.1074/jbc.273.21.12696. [DOI] [PubMed] [Google Scholar]
- 194.Hinnebusch AG. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5:2349–2360. doi: 10.1128/MCB.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hinnebusch AG. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A 81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Irniger S, Braus GH. 2003. Controlling transcription by destruction: the regulation of yeast Gcn4p stability. Curr Genet 44:8–18. doi: 10.1007/s00294-003-0422-3. [DOI] [PubMed] [Google Scholar]
- 197.Shemer R, Meimoun A, Holtzman T, Kornitzer D. 2002. Regulation of the transcription factor Gcn4 by Pho85 cyclin Pcl5. Mol Cell Biol 22:5395–5404. doi: 10.1128/MCB.22.15.5395-5404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kornitzer D, Raboy B, Kulka RG, Fink GR. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J 13:6021–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol Biol Cell 11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Gene Dev 15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rawal Y, Qiu H, Hinnebusch AG. 2014. Accumulation of a threonine biosynthetic intermediate attenuates general amino acid control by accelerating degradation of Gcn4 via Pho85 and Cdk8. PLoS Genet 10:1–20. doi: 10.1371/journal.pgen.1004534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pries R, Bomeke K, Irniger S, Grundmann O, Braus GH. 2002. Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot Cell 1:663–672. doi: 10.1128/EC.1.5.663-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cherkasova VA, Hinnebusch AG. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Gene Dev 17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Garcia-Barrio M, Dong JS, Cherkasova VA, Zhang XL, Zhang F, Ufano S, Lai R, Qin J, Hinnebusch AG. 2002. Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2α kinase activities of GCN2. J Biol Chem 277:30675–30683. doi: 10.1074/jbc.M203187200. [DOI] [PubMed] [Google Scholar]
- 206.Yuan W, Guo S, Gao J, Zhong M, Yan G, Wu W, Chao Y, Jiang Y. 2017. General control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J Biol Chem 292:2660–2669. doi: 10.1074/jbc.M116.772194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O. 2007. Structure of TOR and its complex with KOG1. Mol Cell 27:509–516. doi: 10.1016/j.molcel.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 208.Liu Z, Butow RA. 2006. Mitochondrial retrograde signaling. Annu Rev Genet 40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 209.Giannattasio S, Liu Z, Thornton J, Butow RA. 2005. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem 280:42528–42535. doi: 10.1074/jbc.M509187200. [DOI] [PubMed] [Google Scholar]
- 210.Dilova I, Chen CY, Powers T. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol 12:389–395. doi: 10.1016/S0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- 211.Dilova I, Aronova S, Chen JCY, Powers T. 2004. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1p.Rtg3p-dependent target genes. J Biol Chem 279:46527–46535. doi: 10.1074/jbc.M409012200. [DOI] [PubMed] [Google Scholar]
- 212.Roberg KJ, Bickel S, Rowley N, Kaiser CA. 1997. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics 147:1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu ZC, Sekito T, Epstein CB, Butow RA. 2001. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J 20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tate JJ, Georis I, Rai R, Vierendeels F, Dubois E, Cooper TG. 2015. GATA factor regulation in excess nitrogen occurs independently of Gtr-Ego complex-dependent TorC1 activation. G3 5:1625–1638. doi: 10.1534/g3.115.019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Airoldi EM, Miller D, Athanasiadou R, Brandt N, Abdul-Rahman F, Neymotin B, Hashimoto T, Bahmani T, Gresham D. 2016. Steady-state and dynamic gene expression programs in Saccharomyces cerevisiae in response to variation in environmental nitrogen. Mol Biol Cell 27:1383–1396. doi: 10.1091/mbc.E14-05-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kresnowati MT, van Winden WA, Almering MJ, ten Pierick A, Ras C, Knijnenburg TA, Daran-Lapujade P, Pronk JT, Heijnen JJ, Daran JM. 2006. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol Syst Biol 2:49. doi: 10.1038/msb4100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, Mackiewicz M, Mendenhall EM, Myers RM. 2015. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 25:1581–1589. doi: 10.1101/gr.193540.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung HR, Vingron M, Thomas-Chollier M, Meijsing SH. 2015. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res 25:825–835. doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Parmar JJ, Das D, Padinhateeri R. 2016. Theoretical estimates of exposure timescales of protein binding sites on DNA regulated by nucleosome kinetics. Nucleic Acids Res 44:1630–1641. doi: 10.1093/nar/gkv1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang P, Du G, Zou H, Xie G, Chen J, Shi Z, Zhou J. 2016. Genome-wide mapping of nucleosome positions in Saccharomyces cerevisiae in response to different nitrogen conditions. Sci Rep 6:2016. doi: 10.1038/srep33970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Seeree P, Pearngam P, Kumkate S, Janvilisri T. 2015. An omics perspective on molecular biomarkers for diagnosis, prognosis, and therapeutics of cholangiocarcinoma. Int J Genomics 2015:179528. doi: 10.1155/2015/179528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhao S, Zhao X, Zou H, Fu J, Du G, Zhou J, Chen J. 2014. Comparative proteomic analysis of Saccharomyces cerevisiae under different nitrogen sources. J Proteomics 101:102–112. doi: 10.1016/j.jprot.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 223.Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. 2016. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 26:211–225. doi: 10.1101/gr.196337.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schreve J, Sin J, Garrett J. 1998. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J Bacteriol 180:2556–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Schreve JL, Garrett JM. 2004. Yeast Agp2p and Agp3p function as amino acid permeases in poor nutrient conditions. Biochem Biophys Res Commun 313:745–751. doi: 10.1016/j.bbrc.2003.11.172. [DOI] [PubMed] [Google Scholar]
- 226.Schreve J, Garrett JM. 1997. The branched-chain amino acid permease gene of Saccharomyces cerevisiae, BAP2, encodes the high-affinity leucine permease (S1). Yeast 13:435–439. doi:. [DOI] [PubMed] [Google Scholar]
- 227.De Boer M, Bebelman JP, Gonçalves PM, Maat J, Van Heerikhuizen H, Planta RJ. 1998. Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae. Mol Microbiol 30:603–613. doi: 10.1046/j.1365-2958.1998.01094.x. [DOI] [PubMed] [Google Scholar]
- 228.Ahmad M, Bussey H. 1986. Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr Genet 10:587–592. doi: 10.1007/BF00418125. [DOI] [PubMed] [Google Scholar]
- 229.Opekarova M, Kubin J. 1997. On the unidirectionality of arginine uptake in the yeast Saccharomyces cerevisiae. FEMS Microbiol Lett 152:261–267. doi: 10.1111/j.1574-6968.1997.tb10437.x. [DOI] [PubMed] [Google Scholar]
- 230.Regenberg B, Holmberg S, Olsen LD, Kielland-Brandt MC. 1998. Dip5p mediates high-affinity and high-capacity transport of l-glutamate and l-aspartate in Saccharomyces cerevisiae. Curr Genet 33:171–177. doi: 10.1007/s002940050324. [DOI] [PubMed] [Google Scholar]
- 231.Zhu X, Garrett J, Schreve J, Michaeli T. 1996. GNP1, the high-affinity glutamine permease of S cerevisiae. Curr Genet 30:107–114. doi: 10.1007/s002940050108. [DOI] [PubMed] [Google Scholar]
- 232.Tanaka J, Fink GR. 1985. The histidine permease gene (HIP1) of Saccharomyces cerevisiae. Gene 38:205–214. doi: 10.1016/0378-1119(85)90219-7. [DOI] [PubMed] [Google Scholar]
- 233.Sychrova H, Chevallier MR. 1993. Cloning and sequencing of the Saccharomyces cerevisiae gene LYP1 coding for a lysine-specific permease. Yeast 9:771–782. doi: 10.1002/yea.320090711. [DOI] [PubMed] [Google Scholar]
- 234.Marini AM, Soussi-Boudekou S, Vissers S, Andre B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol 17:4282–4293. doi: 10.1128/MCB.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rouillon A, Surdin-Kerjan Y, Thomas D. 1999. Transport of sulfonium compounds. Characterization of the S-adenosylmethionine and S-methylmethionine permeases from the yeast Saccharomyces cerevisiae. J Biol Chem 274:28096–28105. [DOI] [PubMed] [Google Scholar]
- 236.Kosugi A, Koizumi Y, Yanagida F, Udaka S. 2001. MUP1, high affinity methionine permease, is involved in cysteine uptake by Saccharomyces cerevisiae. Biosci Biotech Biochem 65:728–731. doi: 10.1271/bbb.65.728. [DOI] [PubMed] [Google Scholar]
- 237.Isnard A-D, Thomas D, Surdin-Kerjan Y. 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J Mol Biol 262:473–484. doi: 10.1006/jmbi.1996.0529. [DOI] [PubMed] [Google Scholar]
- 238.Vandenbol M, Jauniaux J-C, Grenson M. 1989. Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: similarities between CAN1, HIP1 and PUT4 permeases. Gene 83:153–159. doi: 10.1016/0378-1119(89)90413-7. [DOI] [PubMed] [Google Scholar]
- 239.Palmieri L, Lasorsa FM, Vozza A, Agrimi G, Fiermonte G, Runswick MJ, Walker JE, Palmieri F. 2000. Identification and functions of new transporters in yeast mitochondria. Biochim Biophys Acta 1459:363–369. doi: 10.1016/S0005-2728(00)00173-0. [DOI] [PubMed] [Google Scholar]
- 240.Schmidt A, Hall MN, Koller A. 1994. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol 14:6597–6606. doi: 10.1128/MCB.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.André B, Hein C, Grenson M, Jauniaux J-C. 1993. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol Gen Genet 237:17–25. [DOI] [PubMed] [Google Scholar]
- 242.Kaur J, Bachhawat AK. 2007. Yct1p, a novel, high-affinity, cysteine-specific transporter from the yeast Saccharomyces cerevisiae. Genetics 176:877–890. doi: 10.1534/genetics.107.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]