SUMMARY
Cellular aggregation is an essential step in the formation of biofilms, which promote fungal survival and persistence in hosts. In many of the known yeast cell adhesion proteins, there are amino acid sequences predicted to form amyloid-like β-aggregates. These sequences mediate amyloid formation in vitro. In vivo, these sequences mediate a phase transition from a disordered state to a partially ordered state to create patches of adhesins on the cell surface. These β-aggregated protein patches are called adhesin nanodomains, and their presence greatly increases and strengthens cell-cell interactions in fungal cell aggregation. Nanodomain formation is slow (with molecular response in minutes and the consequences being evident for hours), and strong interactions lead to enhanced biofilm formation. Unique among functional amyloids, fungal adhesin β-aggregation can be triggered by the application of physical shear force, leading to cellular responses to flow-induced stress and the formation of robust biofilms that persist under flow. Bioinformatics analysis suggests that this phenomenon may be widespread. Analysis of fungal abscesses shows the presence of surface amyloids in situ, a finding which supports the idea that phase changes to an amyloid-like state occur in vivo. The amyloid-coated fungi bind the damage-associated molecular pattern receptor serum amyloid P component, and there may be a consequential modulation of innate immune responses to the fungi. Structural data now suggest mechanisms for the force-mediated induction of the phase change. We summarize and discuss evidence that the sequences function as triggers for protein aggregation and subsequent cellular aggregation, both in vitro and in vivo.
KEYWORDS: Candida albicans, Saccharomyces cerevisiae, adhesins, amyloids, avidity, beta aggregation, catch bonding, mannoproteins
INTRODUCTION
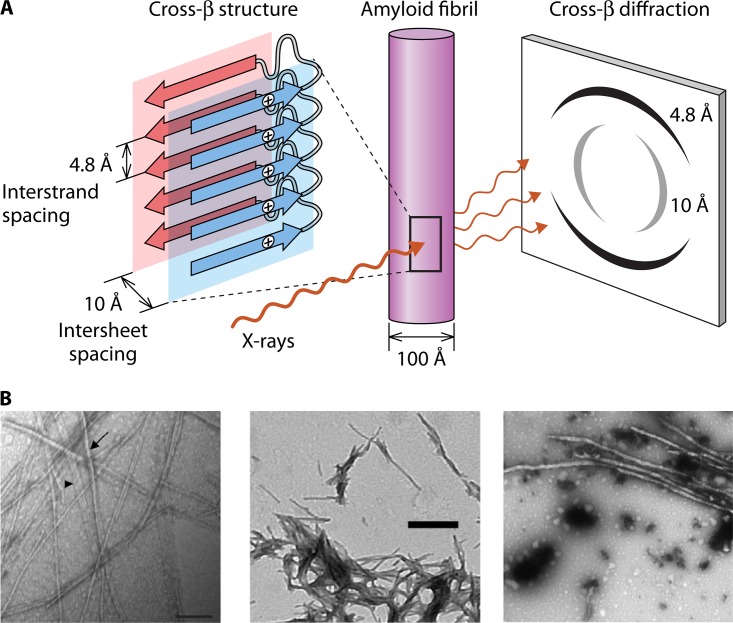
Amyloids are protein aggregates whose basis is regular or periodic interactions of similar or identical amino acid sequences in each protein molecule. These interacting sequences can be extensive, but the formation of many amyloids depends on the presence of specific 4- to 6-residue segments. These amyloid-prone sequences, called “amyloid core sequences,” can be identified in silico based on packing geometry, solubility, and aggregation propensity (1–5). Where the crystalline interactions are due to the formation of β-sheets, they are called β-aggregates. In β-aggregates, β-strands stack laterally to form the β-sheets (Fig. 1). The most common form of amyloids, and a defining characteristic, is the “cross-beta” structure, in which the β-strands run perpendicular to the fiber axis, forming β-sheets along the fiber axis, and pairs of β-sheets tightly mate to form the highly stable amyloid fibers (Fig. 1). The multiple hydrogen bonds between the β-strands stabilize the structure, as do intersheet interdigitations of the amino acid side chains. The anhydrous, close regular fit of the amino acid side chains protruding from the apposed sheets has inspired the name steric zipper for such structures (6, 7). This crystalline structure leads to a gold standard for the identification of amyloids in vitro: a characteristic cross-β-pattern in X-ray diffraction diagrams, with ∼4.8-Å spacing between the β-strands situated along the fiber long axis and 9- to 10-Å orthogonal spacing between the β-sheets. This structure can also be modeled based on solid-state nuclear magnetic resonance (NMR), cryo-electron microscopy, and electron diffraction data (3, 5, 6, 8).
FIG 1.
Amyloid structure. (A) Arrangement of β-strands in cross-β-fibers. Because this model shows sequences “in register,” the same amino acid (e.g., Val326) in each sequence is marked “+.” The figure shows the relative orientations of β-strands, β-sheets, the fiber, and the X-ray diffraction spots. (Modified from reference 5 with permission from Elsevier.) (B) Amyloid fibrils from fungal adhesins visualized by transmission electron microscopy. (Left) Peptide from Flo1p (reprinted from reference 66); (middle) Als5p amyloid peptide (reprinted from reference 67); (right) Als5p20–431 (reprinted from reference 67).
Detection of Amyloids In Vivo
Because X-ray diffraction and solid-state NMR are not practical in vivo, dye binding is commonly used to identify amyloids in cells and tissues. Amyloid fibers bind a characteristic set of amyloidophilic dyes (8–10). Among these, Congo red shows an enhanced and red-shifted absorbance spectrum upon binding to amyloids. Congo red-stained amyloids show an apple-green birefringence when viewed through crossed polaroids. Other dyes show enhanced fluorescence yields in the presence of amyloids; for example, thioflavin T (ThT) is most often used to monitor amyloid formation at a concentration of 10−8 to 10−6 M. At concentrations of ≥10−4 M, these amyloidophilic dyes can disrupt the amyloid structure or inhibit amyloid formation. These characteristics are not shared by amorphous protein aggregates (8, 10).
Amyloid Formation and Phase Transitions
Amyloid formation is a phase transition between monomeric and aggregated states (11–13). This transition is often a result of conformational shifting in amyloid-forming proteins. In some pathogenic amyloids, protein unfolding exposes β-aggregation sequences that were previously buried inside compact protein domains. In other cases, amyloid-forming sequences are in intrinsically disordered regions, and amyloid formation is triggered by intermolecular associations without unfolding. The refolded proteins are likely to be in a nonfunctional state, and the conditions that result are called protein misfolding diseases. For instance, systemic amyloidoses disrupt normal organ anatomy and histology. One form of serum amyloidosis is caused by the dissociation of the native tetrameric form of transthyretin and the subsequent aggregation of the monomers. This condition has been successfully treated with a molecular chaperone, a small-molecule drug that stabilizes the nonaggregative tetrameric conformation of the protein (14, 15).
In other examples, the refolded protein may acquire novel activity. In some fungi, hydrophobins are secreted to the surface, and a soluble-to-solid-phase transition then forms an amyloid-like cell surface coat to make them resistant to environmental insults (16–18). Amyloid formation in the Aβ protein in the central nervous system can trigger amyloid-like aggregation in another protein, Tau; its sequestration and hyperphosphorylation lead to cellular malfunction and neuronal death (19, 20). Another emergent activity in Aβ is the cytotoxicity of folding intermediates that occur during amyloid formation (14, 21). On the other hand, Aβ amyloid formation may limit infections by coating the pathogens (22).
Prions
Amyloid prions are transmissible aggregates and can be beneficial or pathological. In the baker's yeast Saccharomyces cerevisiae, the amyloid-like aggregation of several transcription regulators sequesters these proteins in an inactive aggregate and so epigenetically alters the transcriptional program. Because this happens in only some cells in a population, amyloid formation creates transcriptional variability within the population. The amyloid structures are heritable, so there is an epigenetic heritability of transcriptional variability (23–25). There is debate as to whether such prions are functional, pathological, or both. On the other hand, many prions are solely pathological, such as those associated with bovine spongiform encephalopathy (BSE) (mad cow disease), Kuru in humans, and chronic wasting disease in deer (3, 5, 14, 26, 27). Thus, amyloidogenic refolding of proteins is often accompanied by emergent new activities, which can be beneficial, neutral, or pathological.
Functional Amyloids in Bacterial Biofilms
Many extracellular bacterial proteins form functional amyloids with roles in biofilm formation and maintenance. The commonalities and differences in functional amyloids within the biofilms of Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Streptococcus mutans, and Pseudomonas aeruginosa (28–33) have been highlighted in recent reviews (34–36). These systems are well characterized and serve as models for similar roles of amyloids in biofilms of other Gram-positive and Gram-negative species. In each case, the bacterium secretes and assembles a fibrous structure. In Gram-negative species, the protein curlin assembles into curly fimbriae (“curli”) through β-aggregate-like interactions (Fig. 2). These appendages are natural adhesins and mediate adherence to surfaces and also to other bacteria. Extensive investigations into the genetics, regulation, biogenesis, assembly, and chaperoning of these amyloidogenic proteins make them the best-characterized functional amyloids (33, 37–40). Recent detailed mapping has shown that curli enmesh the more peripheral cells in E. coli biofilms, and curli contribute to biofilm integrity as well (36). Curli are being used to screen for antiamyloid/antibiofilm compounds and as a scaffold for self-assembling biomaterials (38, 41–43). In other bacteria, including Gram-positive strains, amyloid proteins are synthesized, secreted, and assembled not as appendages but as part of the extracellular matrix of the biofilm. In some cases, a nonamyloid form is secreted, and β-aggregated polymers form in response to external conditions such as low pH or the absence of Ca2+ (44). There are also amyloids in archaeal biofilms (45). Thus, emerging knowledge leads to the idea that amyloid proteins are key constituents of biofilms.
FIG 2.
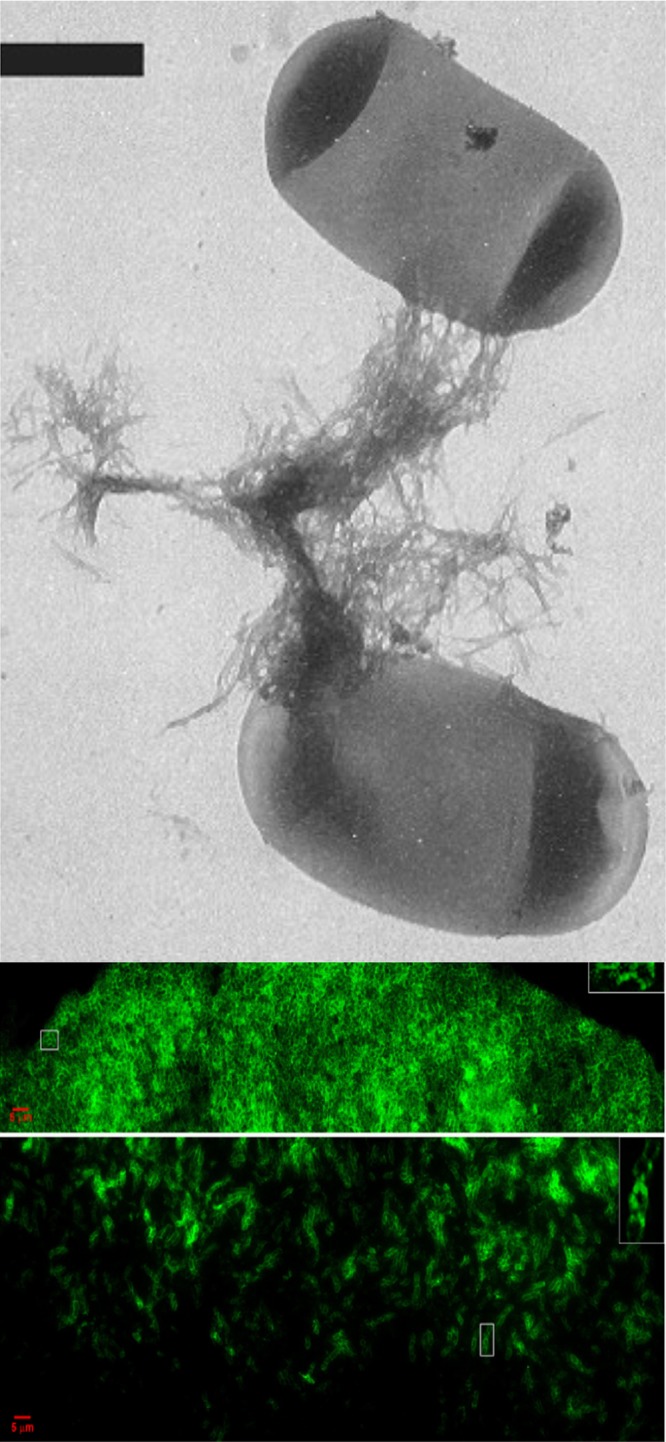
Amyloid curli in bacterial biofilms. (Top) E. coli cells showing curli (scale bar is 500 nm) (reprinted from reference 122 with permission). (Middle and bottom) Fluorescence images of a bacterial colony stained for amyloid with thioflavin T. The middle image shows the top section of the colony, and the bottom image shows a midsection (adapted from reference 35 with permission).
FUNGAL ADHESINS
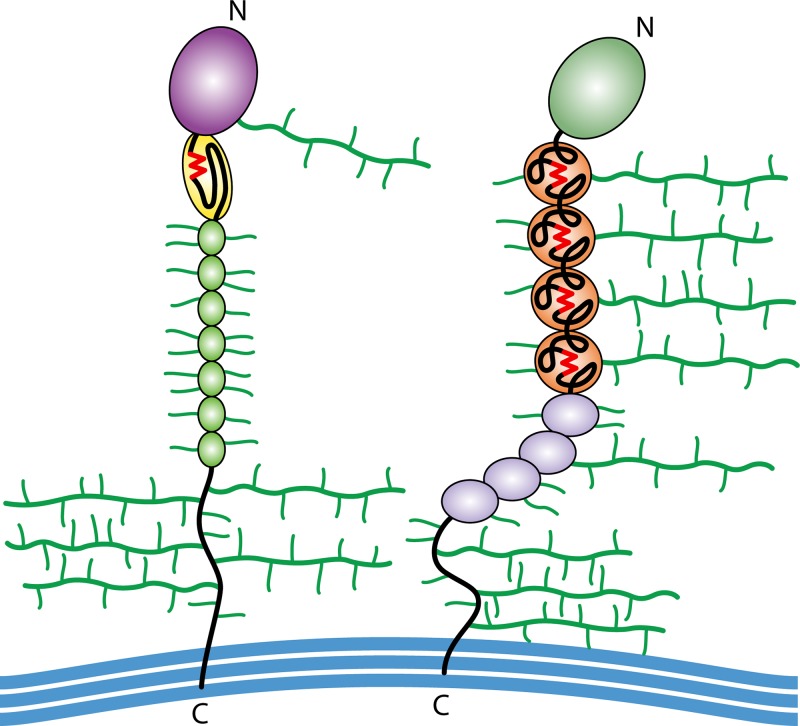
Many fungal adhesins share an architecture reflecting their localization on the surface of the cell wall (Fig. 3) (46, 47). They are β-sheet-rich glycoproteins, composed of 650 to 1,600 amino acids and a similar or larger number of carbohydrate residues, attached as both Asn-linked N-polysaccharides and Ser- and Thr-linked O-oligosaccharides. The fungal adhesins are translated on endoplasmic reticulum (ER)-bound ribosomes, glycosylated in the ER and the Golgi apparatus, and secreted through the conventional vesicular secretion pathway (48, 49). Each well-characterized adhesin has a compact N-terminal ligand-binding domain such as a sugar-binding lectin or an immunoglobulin-like invasin domain (47, 50–52). A variable number of β-sheet-rich tandem repeats is followed by a highly glycosylated unstructured 300- to 800-amino-acid stalk region (47, 53–56). At the C terminus, a glycosylphosphatidylinositol (GPI) membrane anchor is added in the ER. After secretion to the cell surface, the lipid is cleaved off, and the remnant of the C-terminal glycan is covalently linked to cell wall glucan (48, 49, 57, 58).
FIG 3.
Architecture of yeast adhesins. Shown are simplified cartoons of an Als adhesin from C. albicans (left) and Flo1p from S. cerevisiae (right). Each protein has a well-folded β-sheet N-terminal domain (NTD) (purple or green), amyloid-forming sequences (red), tandem repeats (repeated colored ovals), an unstructured stalk region, and a C-terminal cross-link to cell wall β-glucan (blue). N- and O-glycosylations are shown in green.
Fungal adhesins display a recurrent theme of conformational shifting to reinforce cell-cell binding. The Saccharomyces cerevisiae sexual adhesion Sag1p (α-agglutinin) changes conformation upon binding to its ligand Aga2p, with a resulting strengthening of the cell-to-cell bonds (59, 60). In the asexual S. cerevisiae flocculin Flo11p, Ca2+ activates binding but is neither bound to the N-terminal domain (NTD) nor required for the homotypic polymerization of the N-terminal domain. Therefore, Ca2+ likely acts to alter the conformation of a negative regulatory region of the protein (51, 61, 62). This theme is recapitulated in conformational changes leading to the formation of amyloid-like nanodomains on the cell surface. In the C. albicans adhesin Als5p, a conformational shift accompanies the formation of large aggregates of yeast in the presence of ligand-coated beads (63–65). This shift promotes the development of increased adhesiveness over the entire cell surface. This observation fostered the hypothesis that aggregation was an amyloid-like liquid-to-solid-phase or a disordered-to-amyloid-like-phase transition and led to the discovery of fibrous amyloid aggregates of fungal adhesins in vitro (64, 66, 67). Evidence shows that conformational shifting facilitates amyloid-like β-aggregation of adhesin molecules at the cell surface.
AMYLOID-LIKE INTERACTIONS IN Als5p AND OTHER ADHESINS
Cell Surface Conformational Change in Als5p
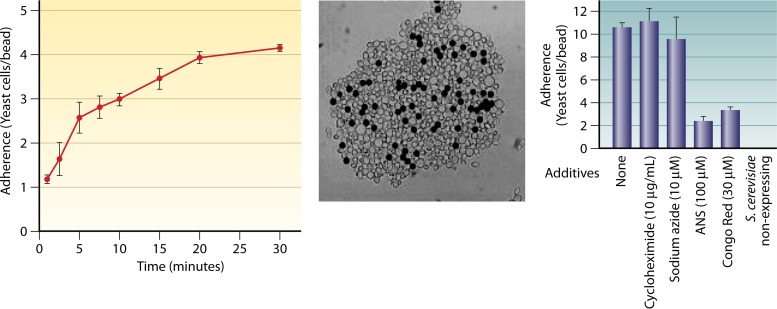
Pioneering work by Gaur et al. showed that the surface display of C. albicans Als5p in laboratory strains of Saccharomyces cerevisiae leads to the cells becoming adherent and aggregative (63, 68). Als5p-expressing yeast cells adhere to beads coated with a protein ligand and then become globally “stickier” and form large aggregates or flocs of cells. Whereas the initial binding to beads happens within about 5 min, the secondary cell-cell aggregation phase takes 15 to 30 min (Fig. 4). We call this secondary phase “aggregation enhancement.” This enhancement is dependent not on new gene expression or protein synthesis during aggregation assays but on a conformational shift in cell surface Als5p (63, 64). Compounds that perturb protein secondary structure changes do not diminish binding to the beads, but they inhibit aggregation enhancement. Therefore, an initial adhesion event triggers a change in the conformation of Als5p on the surface of S. cerevisiae (and in adhesins on C. albicans), leading to the more adhesive enhanced state.
FIG 4.
Als5p shows enhanced cell-to-cell binding in aggregation assays. (Left) Kinetics of adhesion and aggregation. Initial rapid binding (0 to 5 min) is due to cells binding to beads; slower accretion (5 to 30 min) is due to the accumulation of cells binding to cells already bound to beads (adapted from reference 63). (Middle) Large aggregate. Cells are gray, and ligand-coated beads are black. Note that the majority of cells are bound to other cells but not to ligand-coated beads (adapted from reference 64). (Right) Inhibition by 8-anilino-1-naphthalene-sulfonic acid (ANS) and Congo red (adapted from reference 64).
Als5p Is an Amyloid-Forming Protein
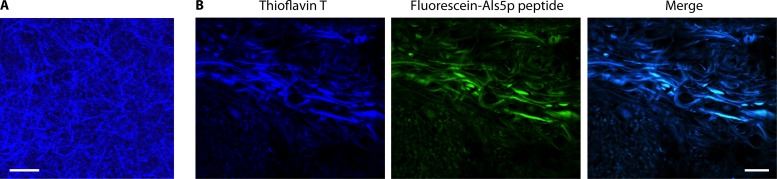
Soluble fragments of Als5p can form amyloids. We identified a 7-amino-acid amyloid core sequence in Als5p with high potential to form β-aggregates, according to the state predictor TANGO (IVIVATT at sequence positions 325 to 331) (2, 67). The modeler ZipperDB also showed this sequence as having high amyloid potential; this algorithm predicts the ability of sequences to form the stable, hydrophobic steric zippers characteristic of amyloids (4). These predictions were experimentally confirmed, because a 13-amino-acid peptide containing this sequence plus flanking residues readily forms amyloid-like fibers (Fig. 1B, middle). The fibers have properties characteristic of amyloids, including gel formation, increased and red-shifted absorbance of Congo red, and enhanced fluorescence of thioflavin T (66, 67). Additionally, the purified soluble protein fragments Als5p20–431, Als5p20–664, and Als5p20–1351 form amyloid-like fibers that bind Congo red and thioflavin T (Fig. 1B, right) (66, 67). Remarkably, the fibers form within a few days at neutral pH and at micromolar protein concentrations. These characteristics demonstrate that Als5p has a high propensity to form amyloids under nondenaturing conditions.
β-Aggregating Sequences in Als Proteins
The amyloid core β-aggregation sequence in Als5p is highly conserved in the ALS gene family. Among the 8 paralogous genes in C. albicans, the sequence 322SNGIVIVATTRTV (the β-aggregation core is underlined) is located in the Thr-rich “T domain,” and its position at residue 325 of the open reading frame is identical in Als5p, Als1p, and many alleles of ALS3 (67). In Als2p, Als4p, Als6p, Als9p, and other alleles of Als3p, a very similar sequence is present at the same position or within a few amino acids. Only Als7p, the most divergent member of the family, has lost the β-aggregation sequence at this position (nevertheless, a similar β-aggregating sequence is present within the T domain at residue 358.) This remarkable sequence conservation argues that the β-aggregating sequences at residue 325 are strongly selected at seven of the eight loci. This observation reinforces the finding that the region containing the sequence is the most highly conserved region in the protein, as measured by the Ka/Ks ratio (the ratio of nonsynonymous to synonymous mutations), an indicator of purifying selection (67, 69, 70). The evidence thus argues for an essential function of the β-aggregating sequence in the T domain in Als adhesins.
β-Aggregating Sequences in Other Fungal Adhesins
The Als adhesin β-aggregation sequences consist mostly of the aliphatic β-branched amino acids Ile, Val, and Thr, which have a high propensity to form β-aggregates and steric zippers. These criteria can be used to identify similar sequences in other fungal adhesins (66, 71). In C. albicans itself, Eap1p, Ece1p, Hwp1p, and Hwp2p are among the surface proteins showing similar sequences. Indeed, TANGO-identified peptides from Eap1p and Hwp2p form amyloid fibers that bind Congo red and thioflavin T (66, 72; S. Singh and P. N. Lipke, unpublished data).
In S. cerevisiae, a number of adhesins and other cell surface proteins also contain sequences predicted to form β-aggregates and steric zippers. Among these proteins, the flocculins Flo1p, Flo5p, and Flo9p show multiple TANGO- and ZipperDB-positive sequences in their tandem repeat domains (62). A peptide containing a sequence from Flo1 forms amyloid fibers at neutral pH (Fig. 1B, left). The nonhomologous adhesin Flo11p has β-aggregating sequences, and a 1,331-residue soluble protein fragment of Flo11p forms amyloid fibers (66). Similar sequences with high β-aggregation potential are present in many fungal cell surface proteins, but amyloid formation has not been tested (66, 71). Overall, there is a recurrent pattern of β-aggregation-prone sequences rich in Ile, Val, and Thr residues present in many fungal adhesins.
Summary of β-Aggregation in Fungal Adhesins
Thus, the evidence that sequences in fungal adhesins can form amyloids and can facilitate amyloid formation by large soluble fragments of the proteins is extensive. These sequences are widespread and highly conserved under purifying selection, a finding that constitutes strong evidence that these sequences confer an advantage to the fungi that express them (23, 25, 73). We argue that the amyloid-like clustering of adhesins enhances aggregation activity and leads to enhanced biofilm formation. Thus, amyloid-like interactions promote the most common lifestyle of fungal growth and survival (74).
AMYLOID-LIKE INTERACTIONS FACILITATE CELL ADHESION
Clustering of Adhesins: Consequences in Cell Adhesion Assays
Aggregation enhancement during adhesion assays is intimately tied to adhesin clustering on the cell surface. The adhesins cluster on cell surfaces during atomic force microscopy (AFM) mapping experiments (Fig. 5A) (71, 75, 76). Confocal microscopy with the amyloidophilic dyes ThT and thioflavin S (ThS) shows the development of fluorescent surface patches during adhesion assays and exposure to shear stress (Fig. 5B) (75, 77, 78). Both genetic and chemical perturbations of amyloid formation inhibit cell surface clustering and also inhibit cellular aggregation. Therefore, surface aggregation of adhesins is necessary and sufficient for robust fungal cell aggregation. This clustering is mediated by the amyloid-like β-aggregation of surface adhesin molecules.
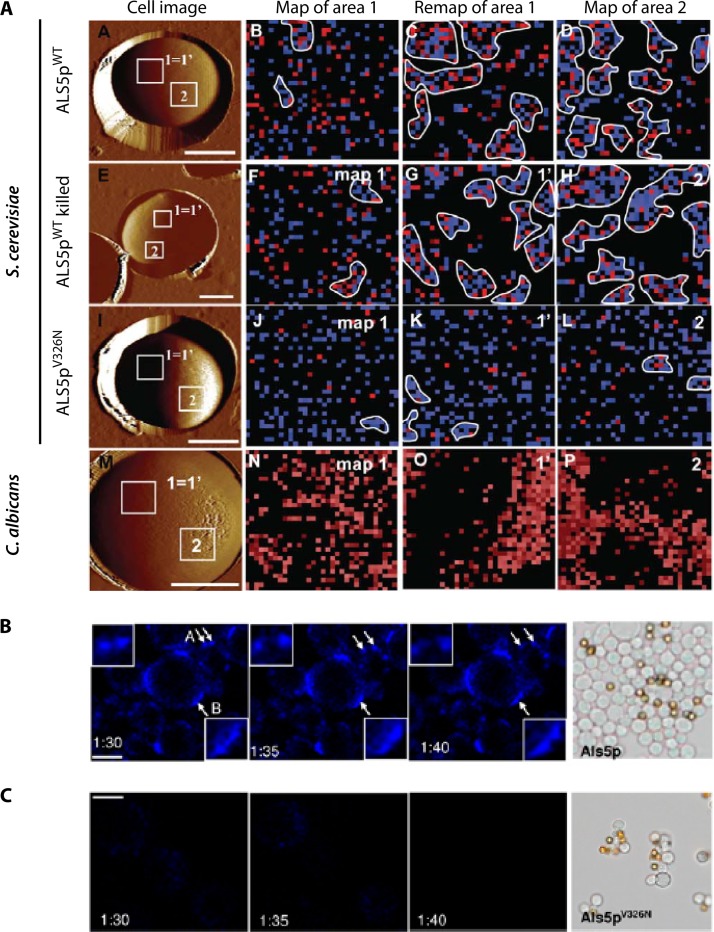
FIG 5.
Surface clustering of Als5p. (A) Atomic force microscopy. The first column (cell image) shows AFM images of live yeast cells entrapped in a microporous membrane on the surface planchet of an atomic force microscope. Column 2 (map of area 1) shows a 32- by 32-pixel map of Als5p in the 1-μm2 area marked 1 = 1′ in the first column. The AFM tip was functionalized with an antibody to an N-terminal epitope tag in Als5p expressed on the surface of S. cerevisiae cells. A pixel is colored if an antigen was detected on the cell surface at that position. The third column (remap of area 1) shows a subsequent map of the same area. The fourth column (map of area 2) shows a map of a remote area marked “2” in column 1, probed after the remap of area 1. (Top row) S. cerevisiae cells expressing Als5pWT; (second row) heat-killed S. cerevisiae Als5pWT-expressing cells; (third row) S. cerevisiae cells expressing the nonamyloid substitution mutant Als5pV326N; (fourth row) mapping of C. albicans surface ligands for Als5p. The AFM tip was functionalized with Als5pWT and used to map the occurrence of ligands for Als5p on the cell surface. (B) Confocal images of ThT-stained surface nanodomains on aggregated S. cerevisiae cells expressing Als5pWT. Arrows show moving domains at 5-min intervals. The right image shows a typical aggregate with gray cells and yellow ligand-coated beads. (Reprinted from reference 75.) (C) ThT-stained Als5pV326N cells at similar intervals and image of an aggregate of these cells, showing reduced cell-cell binding. (Reprinted from reference 75.)
Single-Molecule Imaging Using Atomic Force Microscopy
C. albicans Als5p forms clusters on the surface of S. cerevisiae cells following the extension of single molecules in an atomic force microscope (Fig. 5A) (75, 76, 79). In these experiments, an antibody to an epitope tag in the Als5p N terminus is attached to an AFM tip. The tip is then used to probe the surface of a yeast cell. The force required to move the tip is recorded as the tip is retracted, and wherever the tip binds to the epitope, there is resistance to retraction. In this way, force-distance curves are generated. If the tip is positioned successively at different positions over the cell, we can generate a map of the cell surface, noting whether there is antigen-epitope binding at each position. The resulting map shows Als5p molecules on the surface of expressing cells at a 30-nm resolution (Fig. 5A). At first, Als5p is randomly displayed on the cell surface (Fig. 5A, map of area 1). However, when an area previously mapped is remapped, the number of adhesin-positive 30-nm pixels increases by 50% (remap of area 1). Furthermore, initially, only ∼7% of the detected adhesins are next to pixels with adhesins, but this fraction increases to 65 to 70% on the second map. A map of an area not previously probed also shows similar clustering (map of area 2), implying that clustering propagates all around the cell surface, like a phase transition to the aggregated state. There is similar clustering of Als1p (J. R. Rauceo, D. N. Jackson, P. Herman-Bausier, V. Ho, Y. F. Dufrene, and P. N. Lipke, unpublished data).
The amyloid nature of the clustering has been demonstrated by several approaches (Fig. 5). This clustering occurs similarly in live cells and in heat-killed cells (Fig. 5A, second row), showing that clustering is independent of cellular metabolism and gene expression during mapping. Clustering is also dependent on the high β-aggregation potential of the protein sequence around Val326 in the β-aggregation core (Fig. 5A, first versus third rows). Additionally, ThT staining shows the development of surface patches with enhanced fluorescence during aggregation assays. These patches have a size and mobility similar to those visualized by AFM. Their presence is also dependent on the β-aggregation potential at Val326 in Als1p and Als5p (Fig. 5B and C) (76). Thus, the amyloid-like nature of the interaction leads to a similar phase transition to an aggregated state in dead cells, predictable sequence dependence, and staining with an amyloid-specific dye.
Treatments That Enhance Surface Clustering and Cellular Aggregation
Several treatments can increase adhesin clustering and cellular aggregation. A 13-amino-acid peptide, SNGIVIVATTRTV, contains the β-aggregating sequence from the C. albicans adhesin As1p, Als3p, or Als5p. This peptide (called the “wild-type” [WT] peptide below) readily forms amyloids (Fig. 1B, middle) (67). It also enhances the adhesion and aggregation of Als5p-expressing S. cerevisiae cells and of C. albicans cells (Fig. 6). An alternative treatment is the application of extension forces to Als5p, including pulling by AFM, laminar flow against cells adhered to a substrate, or vortex mixing of the cells. Any of these treatments enhances cellular aggregation and the formation of ThT-fluorescent surface adhesin clusters (71, 75–78, 80). Thus, adhesion surface clustering and enhancement of aggregation are inextricably linked in treatments that enhance amyloid-like interactions.
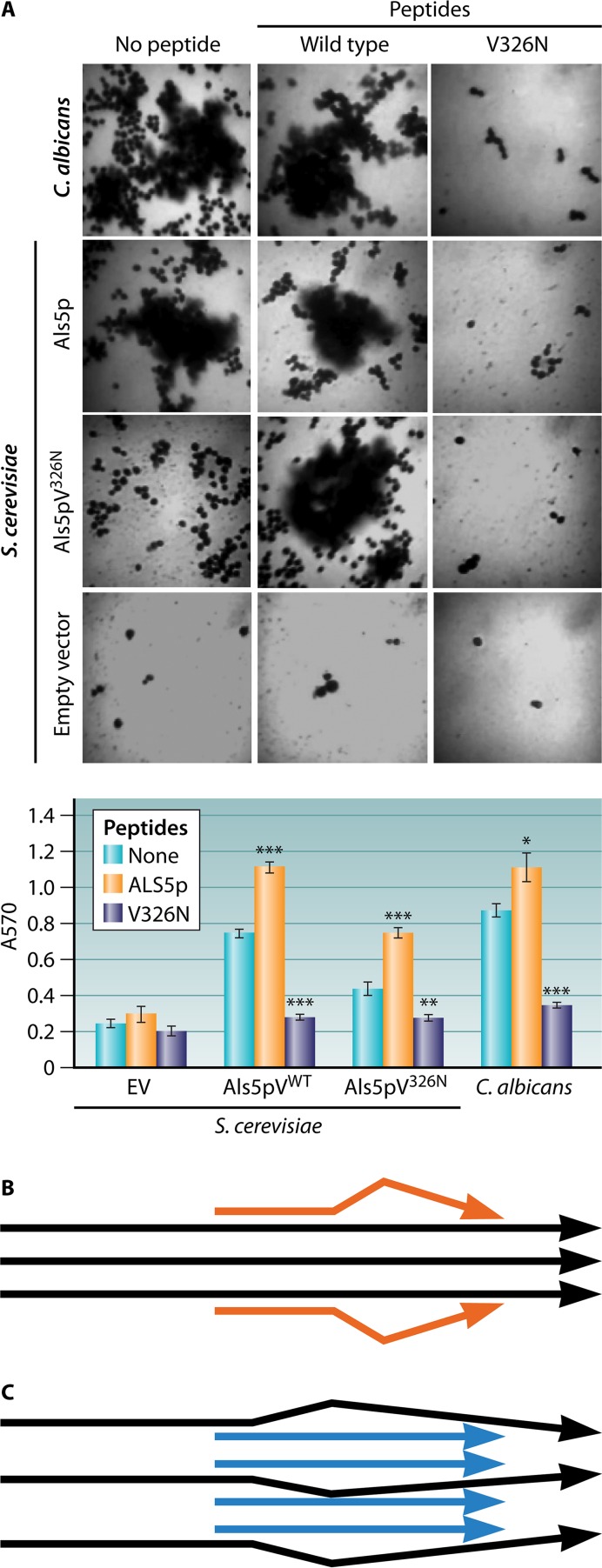
FIG 6.
Effects of WT and V5N peptides on biofilm formation. (A) Twenty-four-hour biofilms of C. albicans or S. cerevisiae cells expressing Als5p, grown for 24 h in the presence or absence of the WT or antiamyloid V5N peptide (labeled “V326N” in the figure) (76). The wild-type peptide enhances the number of cells in the biofilm for C. albicans and Als5p-expressing S. cerevisiae cells and corrects the defect in S. cerevisiae cells expressing the amyloid-impaired Als5pV326N mutant. The antiamyloid peptide inhibits aggregation and biofilm formation. The graph shows crystal violet quantification of cells in biofilms. “EV” represents cells harboring the empty vector. (Reprinted from reference 76.) (B) Model for how the antiamyloid V5N peptide (orange) can “cap” an amyloid-like assembly of proteins (black) and prohibit further growth. (C) Model of WT peptide (blue) templating and assembly of an amyloid-like structure in mutant proteins (black).
Amyloid Perturbants Inhibit Aggregation
Treatments that compromise clustering also inhibit cellular aggregation. Inhibitory treatments include incubation with antiamyloid perturbants, site-specific mutation of the β-aggregation core of Als5p, or incubation with a sequence-specific nonamyloid peptide. Specifically, these treatments have little effect on cell binding to ligand-coated beads or on the affinity for ligands, yet they abrogate the formation of surface nanodomains and also cell-cell aggregation (76). Aniline naphthalene sulfonic acid, which binds to hydrophobic sequences, reduces the cell-to-bead ratio by 76% at a concentration of 100 μM, from 10.6 cells/bead to 2.5 cells/bead. The amyloid indicator and perturbant Congo red at 30 μM reduces cell/bead ratios by 70% (64). Qualitatively, thioflavin S or thioflavin T, used at a 100 to 500 μM concentration (∼103-fold higher than that used to monitor fluorescence), also greatly reduces the cell-to-bead ratio, both for Als5p-expressing S. cerevisiae and for C. albicans cells. In each case, similar numbers of cells are bound directly to ligand-coated beads, but there are very few cells bound to other cells (76). Thus, inhibitors of amyloid formation inhibit cellular aggregation by at least 70% in adhesion/aggregation assays with ligand-coated beads. These results are the ones expected if agents that inhibit adhesin clustering also inhibit cellular aggregation.
In silico mutational studies of the amyloid core sequence of Als5 (325IVIVATT) identified a Val326-to-Asn (V326N) substitution that greatly compromises β-aggregation potential, reducing it from 97% to 3% (71, 76). The corresponding 13-residue peptide (peptide SNGINIVATTRTV, a variant of the native-sequence peptide SNGIVIVATTRTV [substituted residue underlined]) does not form amyloids. This peptide, called “V5N,” can bind to cells that express Als5, Als3, or Als1 and inhibits cellular aggregation and surface clustering of the adhesins (Fig. 6A) (76). Because the peptide is not fluorescent, we can monitor its effects on ThT-bright cell surface nanodomains, and the β-aggregation-impaired V5N peptide extinguishes ThT fluorescence on the cell surface. This result is expected if the peptide binds homotypically to β-aggregating sequences but binds in a different conformation than that with the wild-type β-aggregating sequence. This conformation of V5N would not support the addition of more wild-type peptide to the β-aggregating structure on the cell surface, so this mode of inhibition would be analogous to “capping” inhibitors that stop the growth of amyloid fibers (Fig. 6B). Therefore, the V5N peptide acts as a disruptor of β-aggregation for homologous sequences. As expected, the peptide inhibits the aggregation of cells bearing Als5p or Als1p (76).
Mutation of the β-Aggregation Core Sequence
We constructed Als5pV326N, a single-site mutant of Als5pWT with the same change as that in the V5N peptide. Cells expressing this form of the protein do not cluster Als5p on the surface in response to pulling by AFM and do not form enhanced thioflavin-bright areas during aggregation assays (Fig. 5 and 6). The mutant protein has a similar affinity for its ligand fibronectin and has a similar far-UV circular dichroism (CD) spectrum (76). When displayed on S. cerevisiae, cells expressing this variant of Als5p bind well to ligand-coated beads but form much smaller aggregates than those formed by cells that express Als5pWT, despite more surface expression of the mutant protein. In adhesion assays, antiamyloid concentrations of Congo red, ThT, ThS, or the V5N peptide inhibitor have little effect on Als5pV326N cells (76). This result is expected if the mutant adhesin cannot cluster on the cell surface to enhance cellular aggregation activity, and indeed, clustering is reduced by 90% in AFM maps (Fig. 5A) (76). The mutant's residual activity in adhesion assays reflects the affinity of the unclustered adhesins for ligand proteins present in the assay mixture.
A Semisynthetic β-Aggregating Adhesin
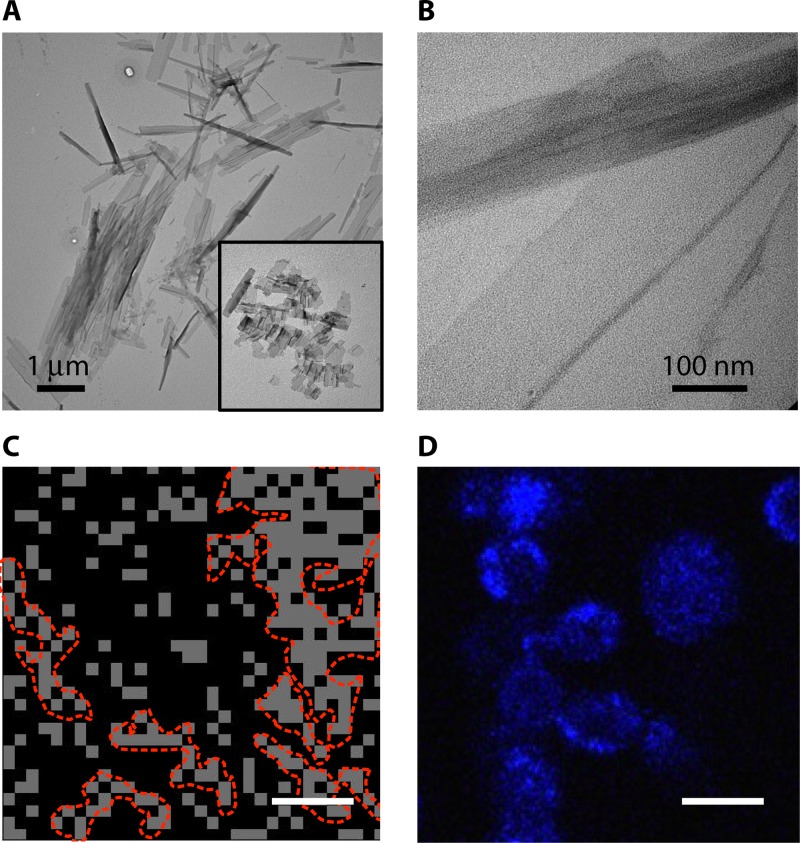
If there are amyloid-like interactions responsible for the formation of surface nanodomains and enhanced aggregation, then a nonnative core sequence should also function in a fungal adhesin. Accordingly, we constructed Als5pLVFFA, with a core amyloid-forming sequence from the human Aβ protein replacing the natural core sequence (81). A tridecapeptide, SNGLVFFATTRTV, analogous to the wild-type proamyloid Als5p peptide, forms amyloid-like ribbons and fibers in vitro (Fig. 7A and B). The chimeric Als5LVFFA protein forms thioflavin-bright adhesin clusters on the cell surface (Fig. 7D), has AFM force-distance profiles similar to those of Als5WT (Fig. 7C), and shows enhanced aggregation that is inhibited by antiamyloid compounds at concentrations of <1 mM. Thus, the substitution of another β-aggregation core sequence into Als5p leads to properties similar to those of the native sequence.
FIG 7.
Amyloid core sequence from Aβ in the context of Als5p. (A and B) Fibers and sheets of peptide SNGLVFFATTRTV (the Aβ sequence is underlined, flanking residues from the sequence of Als5p). (C) AFM mapping of Als5pLVFFA on the surface of S. cerevisiae. Bar, 200 nm. (D) ThT-stained nanodomains. Bar, 2 μm. (Adapted from reference 81.)
HOW β-AGGREGATION INCREASES THE ACTIVITY OF FUNGAL ADHESINS
Surface Clustering and Cellular Aggregation
Amyloid-like clustering is accompanied by increased aggregation activity in each system that we have tested (71, 80, 82). This clustering increases avidity; if an adhesin dissociates from its ligand, the probability of the ligand rebinding is greatly increased because there are so many adhesins nearby, and the effective adhesin concentration is raised manyfold by clustering. This effect is the same as that of the increased avidity of IgM due to the clustering of antigen recognition sites (80). Surface clustering is dependent on the β-aggregation sequence and on the length and flexibility of the adhesins' C-terminal stalks, which extend up to 160 nm away from the cell surface (53, 71).
β-Aggregation between Adhesins on Interacting Cells
There is also evidence that amyloid-like β-aggregates form between aggregating cells. In support of this idea, thioflavin staining shows bright spots wherever cells are in intimate contact (71, 76). Furthermore, treatment of cellular aggregates with 100 to 500 μM thioflavin, Congo red, or the antiamyloid V5N peptide disrupts the preformed aggregates. The most direct evidence comes from AFM. An AFM tip is functionalized with the proamyloid wild-type peptide and used to probe Als5p molecules arrayed on the surface of the AFM planchet (83). The wild-type peptide interacts with the adhesin, and force-distance plots show a signature characteristic of amyloid interactions: the bonds dissociate in small zipper-like steps as the AFM tip is retracted. The wild-type peptide binds to the adhesin molecules four times as frequently as does the V5N mutant peptide, and the zipper-like interactions are never seen when the peptide or the adhesin contains the nonamyloid V→N substitution. Therefore, the peptide-adhesin interaction is dependent on both components being capable of forming amyloids (83). Furthermore, the V5N peptide is an effective disruptor of cell-cell binding in AFM experiments: its addition at a concentration of 140 μM disrupts cell-to-cell binding in AFM single-cell adhesion experiments (Rauceo et al., unpublished).
β-Aggregation of Cell Surface Molecules in C. albicans
C. albicans expresses dozens of surface adhesins (46, 47, 84), so the complexity of cell-cell interactions is greater than that in S. cerevisiae surface display models. Nevertheless, C. albicans shows activities similar to those of Als5p-expressing S. cerevisiae cells. An AFM tip derivatized with Als5p binds to ligands on the C. albicans surface (75, 76). These ligands, which include Als proteins themselves (85, 91), also cluster in response to extension in AFM assays (Fig. 5 and 6). Also, like the model system, C. albicans cells show an aggregative phase with the development of surface birefringence and thioflavin-bright surface domains in adhesion assays (66, 78). Antiamyloid dyes also inhibit aggregation, binding to polystyrene, and the formation of biofilms on plastic, as they do for the S. cerevisiae model surface display system (Fig. 6) (71, 76, 78). It is important to note that these results do not demonstrate that Als5p mediates these activities in C. albicans: similar results would result from adhesion through any adhesins with similar β-aggregation properties.
Other Als-Like Adhesins
The ability of the wild-type peptide SNGIVIVATTRTV to bind similar sequences makes it a specific reagent for Als proteins in C. albicans and non-albicans Candida species (86, 87). The fluorescently labeled peptide binds to Als-expressing cells but not to S. cerevisiae cells lacking an Als protein. Binding is reduced in als1 als3/als1 als3 mutants of C. albicans. (Also, a control peptide of the same composition but with a random sequence does not bind to Als-expressing S. cerevisiae or to C. albicans cells [86].) Therefore, the wild-type peptide has proven useful for the identification of C. albicans in situ. Autopsy sections from candidiasis victims are brightly labeled with the fluorescent wild-type peptide, which colocalizes with the amyloid dyes thioflavin and Congo red as well as with calcofluor white (a fungus-specific dye). Recently, we have found that the Als peptide also binds to some non-albicans Candida species, C. parapsilosis and C. tropicalis, both of which have Als homologs. This peptide does not bind to species that lack ALS homologs. Additionally, the antiamyloid V5N peptide inhibits the adhesion of C. parapsilosis and C. tropicalis to human epithelial cells but does not affect the adhesion of species that lack ALS homologs (M. C. Garcia-Sherman, S. Hamid, D. N. Jackson, J. Thomas, and P. N. Lipke, unpublished data).
β-Aggregation in Fungal Biofilms
Similar β-aggregation interactions reinforce biofilms (76, 86). In Als5p-mediated S. cerevisiae biofilms formed on polystyrene, the biofilm mass is enhanced 67 to 100% for cells expressing Als5pWT relative to cells that express the β-aggregation-impaired mutant form Als5pV326N. The biofilm mass for Als5pWT-mediated biofilms decreases to the level of that of Als5pV326N biofilms in the presence of an antiamyloid peptide or amyloid-inhibitory concentrations of Congo red (500 μM) or ThT (5 μM) (76). Thus, conditions that inhibit amyloid-like β-aggregation simultaneously inhibit the development of robust biofilms.
β-Aggregation of Other Adhesins
The β-aggregating sequences in other fungal adhesins can form amyloid fibers in vitro, including the C. albicans surface proteins Eap1p and Hwp2p as well as the Flo1p and Flo11p adhesins of S. cerevisiae (66). The Flo1p and Flo11p adhesins also show β-aggregation-dependent activity: flocculation is accompanied by the development of cell surface birefringence and an increase in the ThT staining of cell surface nanodomains. The flocculation of cells expressing either Flo1p or Flo11p is strongly inhibited by antiamyloid dyes (300 to 500 μM) at concentrations similar to those that are effective against Als5 and C. albicans aggregation (62, 66, 78, 88). Single-molecule AFM experiments show force-distance curves similar to those for Als5p when the flocculins are extended from the cell surface. Force-distance curves for cell-to-cell adhesion are also similar, and ThS decreases the frequency of cell-cell adhesions by 50 to 90% in single-cell AFM experiments (62). This observation recapitulates the idea that amyloid-like β-aggregation of adhesins occurs between flocculins on different cells in contact to strengthen the cell-to-cell bonds. Therefore, fungal adhesins from three unrelated families (ALS, FLO1, and FLO11/MUC1) show β-aggregated surface nanodomains after extension in AFM experiments, and there is enhanced cell aggregation activity when these nanodomains are present. These observations argue that β-aggregation is a general strategy for increased avidity and/or enhanced strength of cell-cell bonds in fungal adhesins.
Summary of Effects of β-Aggregation on Cell-Cell Adhesion
Interactions between Als5p adhesins cluster them on the surface of the cell wall, with adhesin molecules binding in cis on the cell surface through bonds that have properties of amyloid-like β-aggregation. Clustering into nanodomains increases the avidity of binding, increasing both the size of the aggregates and the number of adhesin-ligand bonds formed between each pair of cells. Clustering is dependent on specific amino acid sequences with extremely high β-aggregation potential, and sequence variants with low β-aggregation potential do not form cell surface clusters. Nanodomain formation can be inhibited by antiamyloid dyes or by a competitive interaction with a similar-sequence peptide with low β-aggregation potential. Furthermore, a peptide containing the high-potential amyloid core sequence binds specifically to the surface of cells with nanodomains, leading to increases in both the formation of surface nanodomains and adhesion. This binding of a homologous sequence is a hallmark of β-aggregates and amyloids. These properties are not limited to Als5p: there are similar interactions between other C. albicans surface adhesins (Fig. 5A, bottom row) and between the S. cerevisiae adhesins Flo1p and Flo11p (62, 66, 78). This clustering is facilitated by the length and flexibility of the unstructured stalk regions of the proteins, which allow neighboring adhesins to interact with each other.
FORCE INITIATES β-AGGREGATION AND AGGREGATION ENHANCEMENT
Force Induces β-Aggregation
β-Aggregation in fungal adhesins can be triggered by the forcible extension of adhesins themselves. The initial observation by Klotz et al. of increased aggregation as the cells and beads are mixed led to the discovery of force-induced surface β-aggregates in AFM experiments (Fig. 4 to 7) (63, 71, 89, 90). Other force-generating regimens also give similar results. C. albicans cells or S. cerevisiae cells expressing Als5p, Flo1p, or Flo11p adhere to a hydrophobic or ligand protein-coated surface in a laminar flow device when a cell suspension flows over the surface (62, 78, 81). As the flow rate increases, the shear stress on the cells increases proportionally, and paradoxically, the cells respond with increased binding to the surface and increased cell-cell aggregation by 25- to 50-fold (Fig. 8) (62, 78, 81). This increased binding under high shear is known as “catch bonding,” a property that allows leukocytes and bacteria to adhere to blood vessel walls under high flow (92, 93). Catch bonding facilitates biofilm formation (94). The catch bonding of C. albicans and Als5p-expressing S. cerevisiae cells is dependent on the β-aggregation sequence, is accompanied by increased surface fluorescence with ThT, and is completely inhibited by antiamyloid compounds (62, 78). Thus, subjecting cells to laminar flow effects both nanodomain formation and increased surface adhesion, and we can add laminar flow-generated shear stress to the list of activating forces. Interestingly, and relevant to the idea of the force sensitivity of these interactions, stirring or shaking of a solution containing an amyloid-forming protein or peptide often promotes amyloid formation (95). Similarly, a non-amyloid-forming mutant of Als3p shows reduced binding to epithelial cells under mixing conditions but is not affected in static assays (96). These results reinforce the idea that in adhesion and biofilm assays, the amount and duration of (physical) agitation significantly affect the results; in our opinion, adhesion and aggregation under flow more accurately reflect conditions in vivo than do assays performed under static conditions.
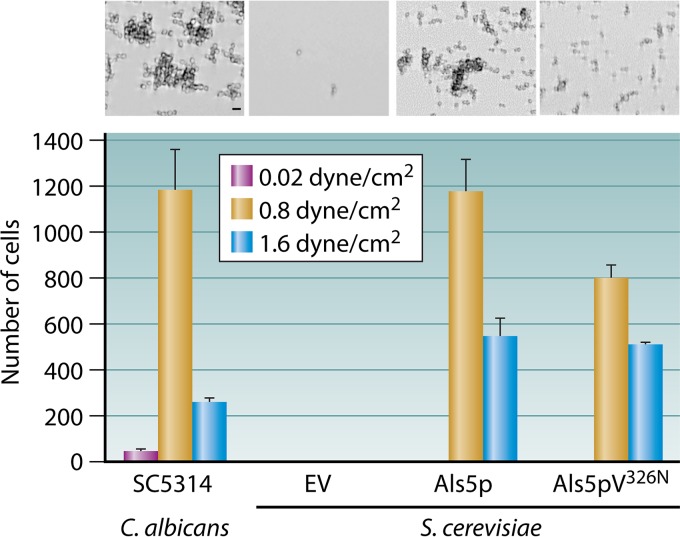
FIG 8.
Catch bonding of yeast to fibronectin under increased flow. (Top) C. albicans SC5314 or S. cerevisiae cells were allowed to adhere to a fibronectin-coated surface at a shear stress of 0.8 dynes/cm2 for 2 h and then imaged. Bar, 5 μm. (Bottom) Adherent cells were quantified with ImageJ. (Adapted from reference 78.)
Centrifugation or vortex mixing can also induce the formation of cell surface adhesin nanodomains. Indeed, centrifugation of S. cerevisiae cells expressing Als5p at 3,200 × g (4,000 rpm in a clinical centrifuge) exerts 16-fold greater frictional force on the cells than does centrifugation at 200 × g (1,000 rpm). The result of the more rapid centrifugation is a 1.5-fold increase in the initial rate of cell adhesion to ligand beads (77). An additional 1.5-fold activation follows vortex mixing. There remains a question as to whether nanodomain formation results from cell-cell collisions or from shear stress due to the laminar flow of the cells through the buffer. Two arguments support the force of the laminar flow argument. First, vortex-induced activation is not dependent on cell density during mixing. This result is expected if the activation event is liquid flow over the cell surface, which is independent of cell density; on the other hand, if the activating force was cell-cell collisions, we would expect much greater activation in denser cell suspensions. The second argument is that centrifugation-induced nanodomain formation is dependent on sedimentation speed. Thus, vortex mixing or centrifugation of cell populations mimics the effects of laminar flow.
Force Activation of Fungal Adhesins in C. albicans
The behavior of C. albicans is highly similar to that shown in the S. cerevisiae surface display model. Force activates β-aggregation and cell adhesins in C. albicans, similarly to its effects on Als5p-expressing S. cerevisiae cells. AFM mapping experiments with an Als5 tip show that >95% of its ligands on the C. albicans cell surface become clustered following force application (Fig. 5A, bottom row) (76). Aggregation and model biofilm formation are enhanced under flow in C. albicans, and these enhancements are exquisitely sensitive to Congo red: 100 nM dye gives 40 to 50% inhibition (see Fig. 3 and S4 in reference 78). Under moderate flow (>0.2 dynes/cm2), C. albicans adhesion to the substrate and aggregation increase 23-fold over those under low-flow conditions, and ThS or Congo red inhibits this increase (78). Thus, C. albicans itself shows force-dependent surface clustering, β-aggregation, and catch bonding, with characteristics similar to those of Als5p-expressing S. cerevisiae (78).
Some of this behavior may result from the activity of Als1p, which can be highly expressed on C. albicans yeast cells and hyphae (97, 98). AFM force-distance profiles of C. albicans show numbers of tandem repeats characteristic of Als1p (99, 100). Als1p expressed on S. cerevisiae also leads to ThT-bright spots during aggregation assays, and aggregation activity is inhibited by antiamyloid treatments (66, 76). Also, antiamyloid treatment inhibits Als1-mediated strong binding to a peptide ligand in AFM experiments with caspofungin-treated C. albicans cells (100). A nonamyloid V326N mutant of Als1p also shows altered adhesion/aggregation activity (our unpublished data). Thus, several properties of Als1p appear to be enhanced by β-aggregation at the cell surface, similarly to Als5p.
Generality of Force Activation of Fungal Adhesins: S. cerevisiae Flocculins
The S. cerevisiae adhesins Flo1p and Flo11p also cluster and increase aggregation activity in response to force. In these proteins, the large number of β-aggregation sequences impedes the identification of specific sequences as being crucial for activity. Like Als1p and Als5p, the flocculins show increased formation of surface ThT-bright nanodomains after laminar flow or vortex mixing, and increased cellular aggregation is also potentiated. Congo red or ThS (<1 mM) inhibits both aggregation activity and the effects of force.
Furthermore, flocculins and Als5p adhesins have similar responses to shear stress when cells are exposed to flowing liquid (62, 77, 78). S. cerevisiae expressing either flocculin adheres to the substrate and aggregates when subjected to shear stress of 0.8 dynes/cm2 but not shear stress of 0.02 dynes/cm2. These aggregates develop into biofilms over 24 h. Thus, flocculin-expressing cells show catch bonding and biofilm formation similar to those of Als5p-expressing cells. With all three adhesins, the shear response reflects characteristics of β-aggregation, i.e., the development of surface birefringence and ThT-bright surface nanodomains, as well as inhibition by antiamyloid dyes. Furthermore, these similarities extend to stimulation by vortex mixing (62, 66, 78). Cells expressing either flocculin aggregate after vortex mixing and simultaneously show the development of surface ThT-bright adhesion nanodomains.
Summary of Force-Induced Nanodomain Formation
The application of any of several extension forces to fungal adhesins increases adhesin clustering through β-aggregation and leads to increased avidities for binding to ligands and for aggregation with other cells (Fig. 9). The ALS, FLO1, and FLO11 adhesins show similar responses to force despite their apparent nonhomology and disparate β-aggregation-prone sequences. This commonality likely reflects the similar overall architectures of the adhesins, the presence of β-aggregation sequences, and their similar roles in mediating the cellular response to shear stress in vivo.
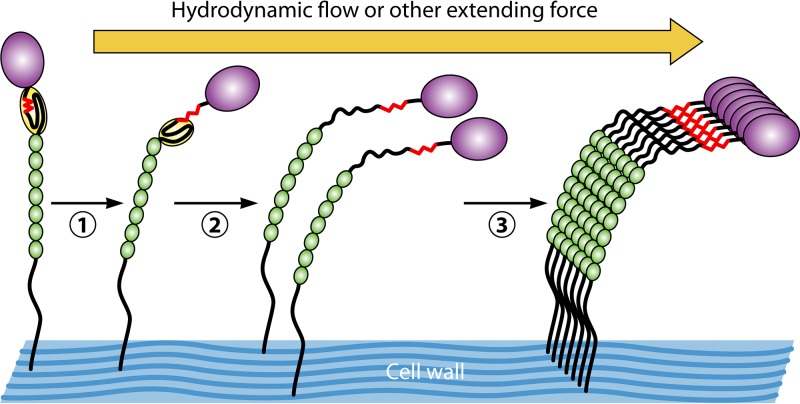
FIG 9.
Model for the effects of hydrodynamic flow on the conformation and aggregation of yeast adhesins. In the initial state, the amyloid core peptide is buried in the interface between the NTD (purple) and the T domain (yellow). In step 1, shear stress unpacks this interface, and in step 2, the T domain unfolds, allowing flexibility to promote interactions among the adhesins to form a nanodomain (step 3).
A MODEL FOR THE RESPONSE TO FORCE
How can a cell adhesion protein respond to shear stress? We have developed a model based on the unfolding of protein domains and subsequent exposure of the β-aggregation sequences. This exposure in turn leads to an association with homologous sequences in other adhesin molecules and transition to the clustered surface structure of adhesin nanodomains (Fig. 9).
Association of the Als5p Amyloid Core with the N-Terminal Domain
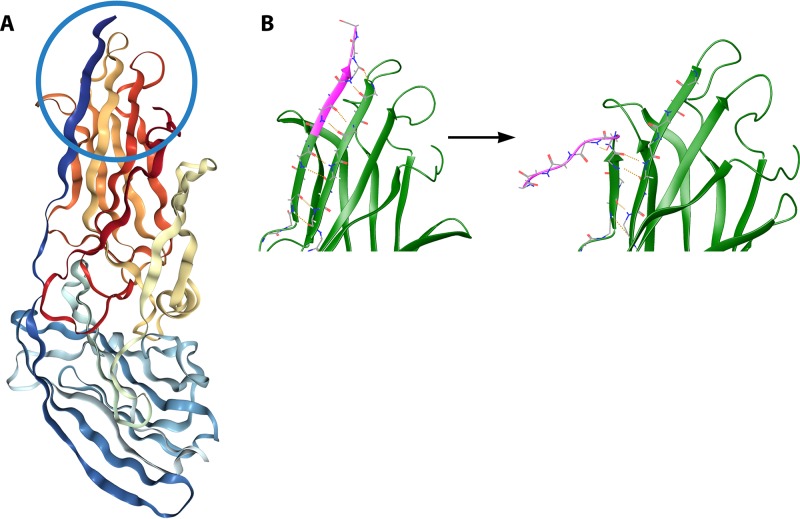
Structures of the Als3p and Als9p invasin-Ig domains (NTDs) have been solved by a combination of X-ray diffraction and NMR spectroscopy (52, 96, 101). Because some of these structures include the β-aggregation sequence at residues 325 to 330, we can use them as a basis for showing how the sequence is buried in the resting state and exposed to the solvent after shear stress. The NTDs are composed of tandem antiparallel β-sheet domains with “Greek key” topology (Fig. 10). There is a deep canyon extending into the domain interface. This canyon binds C-terminal peptides with broad specificity and high affinity, and in some structures, the C-terminal peptide of the crystallized fragment is bound in this canyon. This C-terminal peptide is in fact the β-aggregation core sequence of the adhesins. However, this binding is not geometrically compatible with the native structure of an entire Als protein: there is no C terminus here in the native protein. Indeed, there are 1,000 more amino acid residues after this region, and the C-terminal carboxyl group is bonded to the cell wall polysaccharide through the GPI remnant (Fig. 3 and 9).
FIG 10.
Models of Als protein NTDs. (A) Atomic resolution model of Als3p with the peptide-binding canyon blocked by mutations. The β-aggregating core peptide is dark blue and circled (RCSB PDB accession number 4LEE [96]). (B) Model showing starting points and endpoints of metadynamics simulations of the effect of extension force on the conformation of the amyloid core peptide (pink). The region depicted is the same as the one circled in panel A.
Energetics of the Association of the Amyloid Core Sequence with the NTD
In recognition of this issue, the peptide-binding canyon of Als3p was “blocked off” by site-specific mutations. In this modified protein, the β-aggregating sequence sits in a hydrophobic groove on the side of the NTD with the C-terminal residue available for connection to the rest of the Als3p sequence (Fig. 10A, strand circled in dark blue). We used this structure (RCSB PDB accession number 4LEE) as a basis for calculations of the free energy of dissociation of the β-aggregating sequence from the surface of the NTD in Als5p. Metadynamics simulations calculated the free energy change, the activation energy, and the probability that the peptide would be free from the surface at any specific moment (Fig. 10B) (96, 102). The results showed that the β-aggregating sequence IVIVATT bound tightly to the NTD surface >99.8% of the time, with a dissociation free energy (ΔGdissoc) value of +3.8 kcal/mol and an activation energy value of 6.4 kcal/mol. In contrast, the nonamyloid mutant sequence INIVATT had a 95% probability of being dissociated from the surface, with a ΔGdissoc value of −1.8 kcal/mol. An amyloid-forming sequence from the Alzheimer's disease protein Aβ showed intermediate properties: it was surface bound 95% of the time and had low activation energy for dissociation, with a ΔGdissoc value of +2 kcal/mol.
These calculations can explain the differences in the activities of three forms of Als5p with different β-aggregation sequences: the native sequence, INIVATT (nonamyloid sequence), and LVFFATT (Aβ sequence). For the native protein with the IVIVATT sequence, one side of the β-aggregation sequence is stably docked against the NTD, and the other packs into the all-β-sheet T domain (Fig. 4, 9, and 10). Forces generated by an AFM tip or under laminar flow in blood or saliva are enough to overcome the estimated free energy of dissociation and to “peel” or “lift” the peptide from the surface of the NTD. The exposed sequence then interacts with the same sequence from nearby Als molecules to form surface β-aggregates. In this way, shear forces unfold the T domain, expose the β-aggregating sequence (Fig. 9, steps 1 and 2), and lead to the formation of adhesion nanodomains on the cell surface (Fig. 9, step 3).
In contrast, neither the nonamyloid version of Als5p (INIVATT) nor Aβ-Als5p (LVFFATT) shows force-dependent activation. The nonamyloid sequence is naturally dissociated from the NTD and is therefore constitutively exposed. However, the INIVATT sequence interacts poorly with neighbors and therefore cannot mediate the formation of surface nanodomains of adhesins. For the Aβ sequence, the force needed to expose the sequence and allow β-aggregation to occur is 3- to 4-fold lower and likely to be applied through Brownian motion or incidental handling of the cells. Thus, Aβ-Als5p forms surface nanodomains constitutively, as observed previously (71, 81).
The consequence is that the molecular model explains the cartoon model. The presence of extension or shear force unfolds the T domain in Als proteins and strips the β-aggregation sequence from the surface of the NTD. The exposed sequence then nucleates or adds to an adhesin nanodomain that is held together by amyloid-like interactions (Fig. 9). This model is similar to an emerging theme in catch bonding: shear stress-induced domain unfolding is a key response to force (103, 104). Such unfolding often inactivates inhibitory or regulatory domains (105, 106). The unfolding-associated exposure of a functional amyloid sequences in fungal adhesins appears to be a novel mechanism for catch bonding.
A similar model can apply to Flo1 family proteins, which have an amyloid core sequence in each tandem repeat (Fig. 3) (62). The repeats are unfolded by extension force (88), so the unfolding of any repeat would lead to the exposure of an amyloid core sequence. Amyloid-like interactions can thus form between flocculins on the same cell or different cells. Because amyloid-enhanced aggregation can occur only if both cells express a flocculin with the same amyloid core sequence, aggregation will be stronger between Flo-expressing cells than between an expressing cell and a nonexpressing cell. This difference in adhesion strength can explain the self/nonself discrimination observed in Flo1p-mediated mats of S. cerevisiae and Flo11p-mediated biofilms (74, 107–111). Thus, the activity of S. cerevisiae flocculins is also consistent with the model illustrated in Fig. 9.
Is Force-Dependent β-Aggregation “Real” In Vivo?
We have documented the force-dependent formation of β-aggregated surface adhesion nanodomains primarily in vitro, so a question remains as to whether this phenomenon is applicable in vivo for C. albicans and other fungi. In the laboratory, we can easily visualize amyloids on C. albicans cell surfaces in biofilms (Fig. 11A). Remarkably, there is also compelling evidence for the presence of surface amyloids in abscesses in infected tissue. Autopsy sections from human victims of fungal infections show fungi coated with amyloid and serum amyloid P component (SAP), an amyloid-binding serum protein that is part of the innate immune response, specifically as a part of a damage-associated molecular pattern (DAMP) (9, 112, 113). SAP staining colocalizes with Congo red- and thioflavin-binding amyloid-like material on the cell surface of fungi in aspergillosis, candidiasis, coccidioidomycosis, and Mucorales infections (Fig. 11B) (114). SAP binding may be a cause of the lack of host inflammatory responses to fungal infections (115). Shear force is a plausible factor in biofilm development in deep mycoses, given the presence of significant force within tissues (116). There may also be other mechanisms that trigger unfolding and clustering of adhesins in vivo, but we do not yet know what they are.
FIG 11.
Fungal surface amyloids in situ. (A) C. albicans biofilm stained with ThT. Bar, 25 μm. (Micrograph by Lin Yang.) (B) Abscess from the gastrointestinal tract of a candidiasis victim showing colocalization of ThT and WT peptide binding. This colocalization implies that Als proteins are components of the surface amyloids in the lesion. Bar, 25 μm (86).
These findings demonstrate that amyloid-like β-aggregates are present on infecting fungi and that the inhibition of amyloid-like interactions can prevent fungal adhesion to host cells. In addition, antiamyloid compounds inhibit the natural flocculation of S. cerevisiae, and there is a widespread occurrence of β-aggregating sequences in fungal adhesins. These sequences are strongly conserved, an observation which implies that they contribute to evolutionary fitness, probably by facilitating the formation of biofilms (115). Thus, there is both direct experimental and medical evidence as well as strong evolutionary arguments for an essential role for the β-aggregate-forming sequences in fungal adhesins and extensive demonstrations that they mediate the formation of strongly interacting surface nanodomains in biofilms and host interactions.
OVERALL SUMMARY AND OPEN QUESTIONS
β-Aggregation is based on the formation of β-sheet structures between identical sequence peptides and is the molecular interaction leading to the formation of many amyloid fibers. In many fungal adhesins, there are Ile-Val-Thr-rich peptide sequences that form such β-aggregation-based amyloid fibers. Soluble versions of the adhesins Als5p and Flo11p form amyloids in vitro under nondenaturing conditions, an observation demonstrating that the β-aggregating sequences can be naturally accessible for interactions.
However, adhesin amyloid fibers do not form on fungal cell surfaces, because the geometric constraints of cell surface anchorage prevent the proteins from assembling into fibrillar structures. Instead, adhesins assemble on the cell surface into 2-dimensional nanodomains in which amyloid-like β-aggregation mediates interactions of proteins in cis on the surface of a cell and in trans between cells. The nanodomains are 50 to 500 nm in diameter and bind amyloidophilic dyes at nanomolar concentrations. At 103-fold-higher concentrations, amyloid dyes inhibit interactions, as they do for amyloid fibers themselves. Such β-aggregated nanodomains have been observed in cells expressing four of the best-characterized fungal adhesins from three different gene families: the ALS gene family from C. albicans and the FLO1 family as well as the unrelated adhesin Flo11p in S. cerevisiae. The nanodomains propagate around the entire cell surface, increasing the frequency and avidity of cell-to-cell bonding. At least for the Als proteins, nanodomain formation is dependent on a specific β-aggregating sequence, and indeed, a single residue in this sequence is critical (Val326). There is also evidence for amyloid-like bonds between aggregating cells.
It is the application of extension force that activates β-aggregation of the adhesins and the subsequent enhancement of cellular aggregation. Such force can be applied in an AFM or in vivo through the exposure of cells to laminar flow or through the forces that cells apply to each other during the mixing inherent in aggregation assays. The activating forces are in the range of 10 to 100 pN, sufficient to unfold the protein domains that contain the β-aggregation sequences and to expose the amino acids that participate in amyloid-like interactions. The consequences of the activation of β-aggregation include the formation of large aggregates and biofilms resistant to dislodgement by flow.
Among the most important unanswered questions are the following:
What is the atomic structure of cell surface nanodomain β-aggregates? Although ribbon-like amyloids serve as a first model, solving this problem will require novel biophysical approaches.
Do β-aggregate nanodomains occur in other eukaryotic cells, or are they specific to fungi? Potential β-aggregation sequences are also common in mammalian cell adhesion proteins, but we do not know whether they affect cell-cell adhesion (71). Given the idea that biofilms are organized structures analogous to tissues, are such interactions common in metazoan tissues as well as in biofilms (117, 118)?
In biofilms under flow, are forces transmitted through the nanodomains to the cell interior to alter transcription and metabolism? Are there similar consequences in bacterial and archeal biofilms (119–121)?
Can the amyloid-like interactions be manipulated by pro- or antiamyloid treatments to improve outcomes in fermentations, biofouling prevention, and biomedical treatments (41, 116)?
Thus, we need to investigate the scope of the occurrence and biological consequences of this newly discovered activity in eukaryotic adhesins.
ACKNOWLEDGMENTS
We thank Rajat Kumar Pal and Emilio Gallicchio of the Brooklyn College Chemistry Department for the metadynamics calculations. We also thank Emilio Gallicchio and Meytal Landau for insightful comments.
Work in the authors' laboratories was supported by grants SC1GM 83756 (P.N.L.) and R01GM 098616 (P.N.L.) and by the Fonds de la Recherche Scientifique (FRS)-FNRS (Y.F.D.).
Biographies

Peter N. Lipke is a Professor of Biology at Brooklyn College and the Graduate Center of CUNY. As a Ph.D. student under C. E. Ballou at the University of California—Berkeley, he studied modulation of yeast cell walls during mating. As a postdoc with Jack Lilien at the University of Wisconsin, he worked on roles of cell adhesion proteins in neural pathfinding. His research has focused on mechanisms of adhesin function, and other interests include functional amyloids in cell adhesion and pathogenesis, evolution of fungal cell walls, discovery of wall-targeted antifungal drugs, and biofilm development and pathogenesis. The laboratory's collaboration with the other coauthors is approaching a decade.

Stephen A. Klotz, M.D., is an Infectious Diseases physician with a research interest in fungi, particularly Candida albicans. He has had a rich collaboration with Dr. Lipke for years on how Candida attaches to surfaces, particularly human tissue. Currently, he is working on experiments to demonstrate the role of serum amyloid P component in fungal infections.

Yves F. Dufrene obtained his Bioengineering degree and Ph.D. at the Université Catholique de Louvain (UCL), Belgium. He is now Professor and Research Director at the National Fund for Scientific Research (FNRS). He is interested in the development and use of advanced nanoscale techniques for analyzing the structure and functions of biological systems, with an emphasis on microbial pathogens.

Desmond N. Jackson is a graduate of Stony Brook University, with a Ph.D. from the City University of New York earned in the laboratory of David Foster. His expertise is in cell signaling, pharmacology, and assay design for drug discovery. He is currently Research Associate and Substitute Assistant Professor at Brooklyn College, City University of New York.

Melissa C. Garcia-Sherman received her Ph.D. from the Graduate Center at the City University of New York. As a postdoctoral research fellow at the National Institute of Environmental Health (NIH/NIEHS), in the Cancer Metastasis group directed by Ken Olden and John Roberts, she investigated the role of a Rho small GTPase signaling cascade that regulates cell adhesion in response to inflammatory mediators derived from dietary fatty acids. She joined Peter N. Lipke as a Research associate. She has established the fungus-host interaction facility in his laboratory and is currently the project director of the fungus-host interaction group. She directs projects that seek to elucidate the role of amyloid-forming adhesins and inflammatory mediators in commensal and pathogenic states of Candida-host interactions.
REFERENCES
- 1.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JW, Rousseau F. 2010. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 3.Shewmaker F, McGlinchey RP, Wickner RB. 2011. Structural insights into functional and pathological amyloid. J Biol Chem 286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldschmidt L, Teng PK, Riek R, Eisenberg D. 2010. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A 107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenberg D, Jucker M. 2012. The amyloid state of proteins in human diseases. Cell 148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. 2005. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci U S A 102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. 2007. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson MR. 2004. Techniques to study amyloid fibril formation in vitro. Methods 34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Gilchrist KB, Garcia MC, Sobonya R, Lipke PN, Klotz SA. 2012. New features of invasive candidiasis in humans: amyloid formation by fungi and deposition of serum amyloid P component by the host. J Infect Dis 206:1473–1478. doi: 10.1093/infdis/jis464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisert R, Felau L, Brown LR. 2006. Methods for enhancing the accuracy and reproducibility of Congo red and thioflavin T assays. Anal Biochem 353:144–146. doi: 10.1016/j.ab.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Lee C, Zhang H, Baker AE, Occhipinti P, Borsuk ME, Gladfelter AS. 2013. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell 25:572–584. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. 2015. RNA controls polyQ protein phase transitions. Mol Cell 60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Audas TE, Lee S. 2017. Disentangling a bad reputation: changing perceptions of amyloids. Trends Cell Biol 27:465–467. doi: 10.1016/j.tcb.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ankarcrona M, Winblad B, Monteiro C, Fearns C, Powers ET, Johansson J, Westermark GT, Presto J, Ericzon BG, Kelly JW. 2016. Current and future treatment of amyloid diseases. J Intern Med 280:177–202. doi: 10.1111/joim.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julius RL, Farha OK, Chiang J, Perry LJ, Hawthorne MF. 2007. Synthesis and evaluation of transthyretin amyloidosis inhibitors containing carborane pharmacophores. Proc Natl Acad Sci U S A 104:4808–4813. doi: 10.1073/pnas.0700316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren Q, Kwan AH, Sunde M. 2013. Two forms and two faces, multiple states and multiple uses: properties and applications of the self-assembling fungal hydrophobins. Biopolymers 100:601–612. doi: 10.1002/bip.22259. [DOI] [PubMed] [Google Scholar]
- 17.Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latge JP. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 18.Hakanpaa J, Szilvay GR, Kaljunen H, Maksimainen M, Linder M, Rouvinen J. 2006. Two crystal structures of Trichoderma reesei hydrophobin HFBI—the structure of a protein amphiphile with and without detergent interaction. Protein Sci 15:2129–2140. doi: 10.1110/ps.062326706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dijk G, van Heijningen S, Reijne AC, Nyakas C, van der Zee EA, Eisel UL. 2015. Integrative neurobiology of metabolic diseases, neuroinflammation, and neurodegeneration. Front Neurosci 9:173. doi: 10.3389/fnins.2015.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porzoor A, Macreadie IG. 2013. Application of yeast to study the tau and amyloid-beta abnormalities of Alzheimer's disease. J Alzheimers Dis 35:217–225. doi: 10.3233/JAD-122035. [DOI] [PubMed] [Google Scholar]
- 21.Stroud JC, Liu C, Teng PK, Eisenberg D. 2012. Toxic fibrillar oligomers of amyloid-beta have cross-beta structure. Proc Natl Acad Sci U S A 109:7717–7722. doi: 10.1073/pnas.1203193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar DK, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, Lefkowitz A, McColl G, Goldstein LE, Tanzi RE, Moir RD. 2016. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med 8:340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickner RB, Shewmaker FP, Bateman DA, Edskes HK, Gorkovskiy A, Dayani Y, Bezsonov EE. 2015. Yeast prions: structure, biology, and prion-handling systems. Microbiol Mol Biol Rev 79:1–17. doi: 10.1128/MMBR.00041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes DL, Lancaster AK, Lindquist S, Halfmann R. 2013. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barten DM, Albright CF. 2008. Therapeutic strategies for Alzheimer's disease. Mol Neurobiol 37:171–186. doi: 10.1007/s12035-008-8031-2. [DOI] [PubMed] [Google Scholar]
- 27.Apostol MI, Wiltzius JJ, Sawaya MR, Cascio D, Eisenberg D. 2011. Atomic structures suggest determinants of transmission barriers in mammalian prion disease. Biochemistry 50:2456–2463. doi: 10.1021/bi101803k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besingi RN, Wenderska IB, Senadheera DB, Cvitkovitch DG, Long JR, Wen ZT, Brady LJ. 2017. Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology 163:488–501. doi: 10.1099/mic.0.000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marinelli P, Pallares I, Navarro S, Ventura S. 2016. Dissecting the contribution of Staphylococcus aureus alpha-phenol-soluble modulins to biofilm amyloid structure. Sci Rep 6:34552. doi: 10.1038/srep34552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng G, Vad BS, Dueholm MS, Christiansen G, Nilsson M, Tolker-Nielsen T, Nielsen PH, Meyer RL, Otzen DE. 2015. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front Microbiol 6:1099. doi: 10.3389/fmicb.2015.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. 14 October 2015. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oli MW, Otoo HN, Crowley PJ, Heim KP, Nascimento MM, Ramsook CB, Lipke PN, Brady LJ. 2012. Functional amyloid formation by Streptococcus mutans. Microbiology 158:2903–2916. doi: 10.1099/mic.0.060855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. 2012. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol 20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor JD, Matthews SJ. 2015. New insight into the molecular control of bacterial functional amyloids. Front Cell Infect Microbiol 5:33. doi: 10.3389/fcimb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Gerven N, Klein RD, Hultgren SJ, Remaut H. 2015. Bacterial amyloid formation: structural insights into curli biogensis [sic]. Trends Microbiol 23:693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans ML, Chapman MR. 2014. Curli biogenesis: order out of disorder. Biochim Biophys Acta 1843:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero D, Kolter R. 2011. Will biofilm disassembly agents make it to market? Trends Microbiol 19:304–306. doi: 10.1016/j.tim.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai B, Li D, Xi W, Luo F, Zhang X, Zou M, Cao M, Hu J, Wang W, Wei G, Zhang Y, Liu C. 2015. Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc Natl Acad Sci U S A 112:2996–3001. doi: 10.1073/pnas.1416690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mondal S, Varenik M, Bloch DN, Atsmon-Raz Y, Jacoby G, Adler-Abramovich L, Shimon LJ, Beck R, Miller Y, Regev O, Gazit E. 2017. A minimal length rigid helical peptide motif allows rational design of modular surfactants. Nat Commun 8:14018. doi: 10.1038/ncomms14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero D, Vlamakis H, Losick R, Kolter R. 2014. Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J Bacteriol 196:1505–1513. doi: 10.1128/JB.01363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dueholm MS, Larsen P, Finster K, Stenvang MR, Christiansen G, Vad BS, Boggild A, Otzen DE, Nielsen PH. 24 January 2015. The tubular sheaths encasing Methanosaeta thermophila filaments are functional amyloids. J Biol Chem doi: 10.1074/jbc.M115.654780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dranginis AM, Rauceo JR, Coronado JE, Lipke PN. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev 71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoyer LL, Fundyga R, Hecht JE, Kapteyn JC, Klis FM, Arnold J. 2001. Characterization of agglutinin-like sequence genes from non-albicans Candida and phylogenetic analysis of the ALS family. Genetics 157:1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu CF, Kurjan J, Lipke PN. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol Cell Biol 14:4825–4833. doi: 10.1128/MCB.14.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goossens KV, Ielasi FS, Nookaew I, Stals I, Alonso-Sarduy L, Daenen L, Van Mulders SE, Stassen C, van Eijsden RG, Siewers V, Delvaux FR, Kasas S, Nielsen J, Devreese B, Willaert RG. 2015. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. mBio 6:e00427-15. doi: 10.1128/mBio.00427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraushaar T, Bruckner S, Veelders M, Rhinow D, Schreiner F, Birke R, Pagenstecher A, Mosch HU, Essen LO. 2015. Interactions by the fungal Flo11 adhesin depend on a fibronectin type III-like adhesin domain girdled by aromatic bands. Structure 23:1005–1017. doi: 10.1016/j.str.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 52.Cota E, Hoyer LL. 2015. The Candida albicans agglutinin-like sequence family of adhesins: functional insights gained from structural analysis. Future Microbiol 10:1635–1648. doi: 10.2217/fmb.15.79. [DOI] [PubMed] [Google Scholar]
- 53.Lipke PN, Kurjan J. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol Rev 56:180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen ZM, Wang L, Pike J, Jue CK, Zhao H, de Nobel H, Kurjan J, Lipke PN. 2001. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J Biol Chem 276:15768–15775. doi: 10.1074/jbc.M010421200. [DOI] [PubMed] [Google Scholar]
- 55.Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. 2010. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot Cell 9:405–414. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat Genet 37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu CF, Montijn RC, Brown JL, Klis F, Kurjan J, Bussey H, Lipke PN. 1995. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β1,6-glucan in the S. cerevisiae cell wall. J Cell Biol 128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wojciechowicz D, Lu CF, Kurjan J, Lipke PN. 1993. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol Cell Biol 13:2554–2563. doi: 10.1128/MCB.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H, Chen MH, Shen ZM, Kahn PC, Lipke PN. 2001. Environmentally induced reversible conformational switching in the yeast cell adhesion protein alpha-agglutinin. Protein Sci 10:1113–1123. doi: 10.1110/ps.41701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H, Shen ZM, Kahn PC, Lipke PN. 2001. Interaction of alpha-agglutinin and a-agglutinin, Saccharomyces cerevisiae sexual cell adhesion molecules. J Bacteriol 183:2874–2880. doi: 10.1128/JB.183.9.2874-2880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo WS, Dranginis AM. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J Bacteriol 178:7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan CX, El-Kirat-Chatel S, Joseph IG, Jackson DN, Ramsook CB, Dufrene YF, Lipke PN. 2016. Force sensitivity in Saccharomyces cerevisiae flocculins. mSphere 1:e00128-16. doi: 10.1128/mSphere.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaur NK, Klotz SA, Henderson RL. 1999. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect Immun 67:6040–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, Lipke PN. 2004. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun 72:4948–4955. doi: 10.1128/IAI.72.9.4948-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaur NK, Klotz SA. 2004. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology 150:277–284. doi: 10.1099/mic.0.26738-0. [DOI] [PubMed] [Google Scholar]
- 66.Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O'Meally S, Otoo HN, Khalaf RA, Dranginis AM, Gaur NK, Klotz SA, Rauceo JM, Jue CK, Lipke PN. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell 9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otoo HN, Lee KG, Qiu W, Lipke PN. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell 7:776–782. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaur NK, Klotz SA. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun 65:5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie X, Qiu WG, Lipke PN. 2011. Accelerated and adaptive evolution of yeast sexual adhesins. Mol Biol Evol 28:3127–3137. doi: 10.1093/molbev/msr145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Li WH. 2004. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol 21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- 71.Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrene YF. 2012. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol 20:59–65. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayek P, Dib L, Yazbeck P, Beyrouthy B, Khalaf RA. 2010. Characterization of Hwp2, a Candida albicans putative GPI-anchored cell wall protein necessary for invasive growth. Microbiol Res 165:250–258. doi: 10.1016/j.micres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 73.Halfmann R, Lindquist S. 2010. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science 330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 74.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latge JP, Fink GR, Foster KR, Verstrepen KJ. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci U S A 107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, Lipke PN. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One 6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan CX, Joseph IG, Huang A, Jackson DN, Lipke PN. 2015. Quantitative analyses of force-induced amyloid formation in Candida albicans Als5p: activation by standard laboratory procedures. PLoS One 10:e0129152. doi: 10.1371/journal.pone.0129152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan CX, Lipke PN. 2014. Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot Cell 13:1136–1142. doi: 10.1128/EC.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao J, Dufrene YF. 2016. Optical and force nanoscopy in microbiology. Nat Microbiol 1:16186. doi: 10.1038/nmicrobiol.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lipke PN, Ramsook C, Garcia-Sherman MC, Jackson DN, Chan CX, Bois M, Klotz SA. 2014. Between amyloids and aggregation lies a connection with strength and adhesion. New J Sci 2014:815102. doi: 10.1155/2014/815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rameau RD, Jackson DN, Beaussart A, Dufrene YF, Lipke PN. 2016. The human disease-associated Abeta amyloid core sequence forms functional amyloids in a fungal adhesin. mBio 7:e01815-15. doi: 10.1128/mBio.01815-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia M, Lipke P, Klotz S. 2013. Pathogenic microbial amyloids: their function and the host response. OA Microbiol 1:2. doi: 10.1186/2049-2618-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alsteens D, Ramsook CB, Lipke PN, Dufrene YF. 2012. Unzipping a functional microbial amyloid. ACS Nano 6:7703–7711. doi: 10.1021/nn3025699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. 2013. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell 12:470–481. doi: 10.1128/EC.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE Jr, Lipke PN, El-Azizi M. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol 45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 86.Garcia-Sherman MC, Lysak N, Filonenko A, Richards H, Sobonya RE, Klotz SA, Lipke PN. 2014. Peptide detection of fungal functional amyloids in infected tissue. PLoS One 9:e86067. doi: 10.1371/journal.pone.0086067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Formosa C, Schiavone M, Boisrame A, Richard ML, Duval RE, Dague E. 2015. Multiparametric imaging of adhesive nanodomains at the surface of Candida albicans by atomic force microscopy. Nanomedicine 11:57–65. doi: 10.1016/j.nano.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 88.El-Kirat-Chatel S, Beaussart A, Vincent SP, Abellan Flos M, Hols P, Lipke PN, Dufrene YF. 2015. Forces in yeast flocculation. Nanoscale 7:1760–1767. doi: 10.1039/C4NR06315E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klotz SA. 1992. Fungal adherence to the vascular compartment: a critical step in the pathogenesis of disseminated candidiasis. Clin Infect Dis 14:340–347. doi: 10.1093/clinids/14.1.340. [DOI] [PubMed] [Google Scholar]
- 90.Pendrak ML, Klotz SA. 1995. Adherence of Candida albicans to host cells. FEMS Microbiol Lett 129:103–113. doi: 10.1111/j.1574-6968.1995.tb07566.x. [DOI] [PubMed] [Google Scholar]
- 91.Klotz SA, Gaur NK, Lake DF, Rauceo VCJ, Lipke PN. 2004. Degenerate peptide recognition by Candida albicans adhesins Als5p and Als1p. Infect Immun 72:2029–2034. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Interlandi G, Thomas W. 2010. The catch bond mechanism between von Willebrand factor and platelet surface receptors investigated by molecular dynamics simulations. Proteins 78:2506–2522. doi: 10.1002/prot.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Auton M, Zhu C, Cruz MA. 2010. The mechanism of VWF-mediated platelet GPIbalpha binding. Biophys J 99:1192–1201. doi: 10.1016/j.bpj.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nadell CD, Ricaurte D, Yan J, Drescher K, Bassler BL. 2017. Flow environment and matrix structure interact to determine spatial competition in Pseudomonas aeruginosa biofilms. eLife 6:e21855. doi: 10.7554/eLife.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hill EK, Krebs B, Goodall DG, Howlett GJ, Dunstan DE. 2006. Shear flow induces amyloid fibril formation. Biomacromolecules 7:10–13. doi: 10.1021/bm0505078. [DOI] [PubMed] [Google Scholar]
- 96.Lin J, Oh SH, Jones R, Garnett JA, Salgado PS, Rusnakova S, Matthews SJ, Hoyer LL, Cota E. 2014. The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J Biol Chem 289:18401–18412. doi: 10.1074/jbc.M114.547877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Green CB, Zhao X, Yeater KM, Hoyer LL. 2005. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 151:1051–1060. doi: 10.1099/mic.0.27696-0. [DOI] [PubMed] [Google Scholar]
- 98.Coleman DA, Oh SH, Zhao X, Hoyer LL. 2010. Heterogeneous distribution of Candida albicans cell-surface antigens demonstrated with an Als1-specific monoclonal antibody. Microbiology 156:3645–3659. doi: 10.1099/mic.0.043851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharikova S, Van Dijck P, Dufrene YF. 2012. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.El-Kirat-Chatel S, Beaussart A, Alsteens D, Jackson DN, Lipke PN, Dufrene YF. 2013. Nanoscale analysis of caspofungin-induced cell surface remodelling in Candida albicans. Nanoscale 5:1105–1115. doi: 10.1039/C2NR33215A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. 2011. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 108:15775–15779. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kilburg D, Gallicchio E. 2016. Recent advances in computational models for the study of protein-peptide interactions. Adv Protein Chem Struct Biol 105:27–57. doi: 10.1016/bs.apcsb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 103.Hu X, Margadant FM, Yao M, Sheetz MP. 2017. Molecular stretching modulates mechanosensing pathways. Protein Sci 26:1337–1351. doi: 10.1002/pro.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. 2009. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v) beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci U S A 106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sauer MM, Jakob RP, Eras J, Baday S, Eris D, Navarra G, Berneche S, Ernst B, Maier T, Glockshuber R. 2016. Catch-bond mechanism of the bacterial adhesin FimH. Nat Commun 7:10738. doi: 10.1038/ncomms10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nilsson LM, Thomas WE, Sokurenko EV, Vogel V. 2008. Beyond induced-fit receptor-ligand interactions: structural changes that can significantly extend bond lifetimes. Structure 16:1047–1058. doi: 10.1016/j.str.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 107.Vachova L, Stovicek V, Hlavacek O, Chernyavskiy O, Stepanek L, Kubinova L, Palkova Z. 2011. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J Cell Biol 194:679–687. doi: 10.1083/jcb.201103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lo WS, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by S. cerevisiae. Mol Biol Cell 9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barua S, Li L, Lipke PN, Dranginis AM. 2016. Molecular basis for strain variation in the Saccharomyces cerevisiae adhesin Flo11p. mSphere 1:e00129-16. doi: 10.1128/mSphere.00129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Steinberg MS. 2007. Differential adhesion in morphogenesis: a modern view. Curr Opin Genet Dev 17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 111.Cullen PJ, Sprague GF. 2012. The regulation of filamentous growth in yeast. Genetics 190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Raymond SL, Holden DC, Mira JC, Stortz JA, Loftus TJ, Mohr AM, Moldawer LL, Moore FA, Larson SD, Efron PA. 2017. Microbial recognition and danger signals in sepsis and trauma. Biochim Biophys Acta 1863:2564–2573. doi: 10.1016/j.bbadis.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klotz SA, Sobonya RE, Lipke PN, Garcia-Sherman MC. 2016. Serum amyloid P component and systemic fungal infection: does it protect the host or is it a Trojan Horse? Open Forum Infect Dis 3:ofw166. doi: 10.1093/ofid/ofw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garcia-Sherman MC, Lundgren T, Sobonya R, Lipke PN, Klotz SA. 2015. A unique biofilm in human deep mycoses: fungal amyloid is bound by host serum amyloid P component. NPJ Biofilms Microbiomes 1:15009. doi: 10.1038/npjbiofilms.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pepys MB. 2012. Invasive candidiasis: new insights presaging new therapeutic approaches? J Infect Dis 206:1339–1341. doi: 10.1093/infdis/jis521. [DOI] [PubMed] [Google Scholar]
- 116.Heisenberg CP, Bellaiche Y. 2013. Forces in tissue morphogenesis and patterning. Cell 153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 117.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 118.Watnick P, Kolter R. 2000. Biofilm, city of microbes. J Bacteriol 182:2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harapanahalli AK, Younes JA, Allan E, van der Mei HC, Busscher HJ. 2015. Chemical signals and mechanosensing in bacterial responses to their environment. PLoS Pathog 11:e1005057. doi: 10.1371/journal.ppat.1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Busscher HJ, van der Mei HC. 2012. How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog 8:e1002440. doi: 10.1371/journal.ppat.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Serra DO, Hengge R. 2014. Stress responses go three dimensional—the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ Microbiol 16:1455–1471. doi: 10.1111/1462-2920.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Epstein EA, Reizian MA, Chapman MR. 2009. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol 191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]