SUMMARY
Viruses of the subfamily Orthoretrovirinae are defined by the ability to reverse transcribe an RNA genome into DNA that integrates into the host cell genome during the intracellular virus life cycle. Exogenous retroviruses (XRVs) are horizontally transmitted between host individuals, with disease outcome depending on interactions between the retrovirus and the host organism. When retroviruses infect germ line cells of the host, they may become endogenous retroviruses (ERVs), which are permanent elements in the host germ line that are subject to vertical transmission. These ERVs sometimes remain infectious and can themselves give rise to XRVs. This review integrates recent developments in the phylogenetic classification of retroviruses and the identification of retroviral receptors to elucidate the origins and evolution of XRVs and ERVs. We consider whether ERVs may recurrently pressure XRVs to shift receptor usage to sidestep ERV interference. We discuss how related retroviruses undergo alternative fates in different host lineages after endogenization, with koala retrovirus (KoRV) receiving notable interest as a recent invader of its host germ line. KoRV is heritable but also infectious, which provides insights into the early stages of germ line invasions as well as XRV generation from ERVs. The relationship of KoRV to primate and other retroviruses is placed in the context of host biogeography and the potential role of bats and rodents as vectors for interspecies viral transmission. Combining studies of extant XRVs and “fossil” endogenous retroviruses in koalas and other Australasian species has broadened our understanding of the evolution of retroviruses and host-retrovirus interactions.
KEYWORDS: bioinformatics, endogenous retrovirus, hybrid capture, iatrogenic transmission, orthoretrovirus, retroviral receptor, retroviral transmission, taxonomy
INTRODUCTION
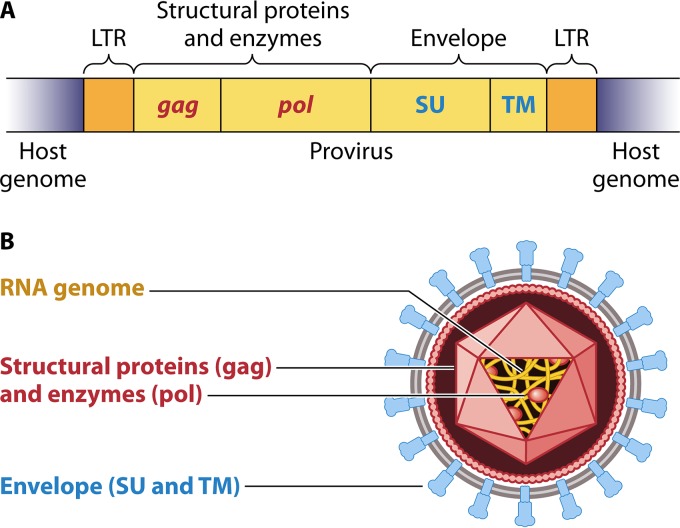
Orthoretroviruses infect host cells by transferring their capsid and viral RNA genomes into the cytoplasm. Subsequently, viral reverse transcriptase (RT) initiates production of a double-stranded DNA copy of the viral genome, which then integrates into the host chromosomal DNA. The viral integrant is termed a provirus. The provirus contains viral regulatory sequences in the long terminal repeats (LTRs) flanking both sides of the provirus (Fig. 1A). The LTRs contain sequences such as the viral promoters, enhancers, transcription initiation and termination sites, and polyadenylation sites. The provirus serves as a template for generating viral genomic RNA and viral mRNA transcripts that are translated by the host cellular machinery into viral proteins. The viral genomic RNA along with host-translated viral proteins self-assemble into particles that are exported from the cell. As shown in Fig. 1B, a simple orthoretrovirus has three basic components found in all infectious orthoretroviruses. First, two copies of the RNA genome are present in the body of the particle (shown in yellow in Fig. 1B). Inside the virion particle are structural core proteins, such as the capsid and matrix proteins, in addition to viral enzymes, such as integrase and RT (depicted in red in Fig. 1B). The env gene encodes a polypeptide that, after translation and furin-mediated proteolytic cleavage, results in two envelope subcomponents: the surface unit (SU), which contains the receptor binding domain, and a transmembrane unit (TM) (shown in blue in Fig. 1B). SU represents the external viral surface envelope components that bind to the receptor on susceptible host cells. The TM unit is the subunit that spans the lipid bilayer of the viral particle.
FIG 1.
(A) An integrated double-stranded DNA provirus (yellow) of a simple orthoretrovirus within the host genome (gray) is shown. The long terminal repeats (LTRs) are at both the 5′ and 3′ ends of the provirus and flank the retroviral gag, pol, and env coding regions. Regions coding for enzymes and other proteins are shown with font colors corresponding to their depiction in panel B. (B) Schematic drawing of a simple orthoretrovirus. All orthoretroviruses have three component parts: (i) the RNA genome, shown in yellow; (ii) internal proteins, shown in red, including internal structural proteins (Gag) as well as the viral enzymes, including the reverse transcriptase (Pol), which makes a DNA copy of the RNA genome which will be integrated into the host cell genome; and (iii) the envelope proteins, shown in blue. Env consists of two components: the TM moiety is embedded in the membrane (depicted in gray and white) of a host cell and is incorporated into the virion during the budding process, and the surface glycoprotein SU forms the knobs and is the part of the virus that binds to receptors on susceptible cells of the host.
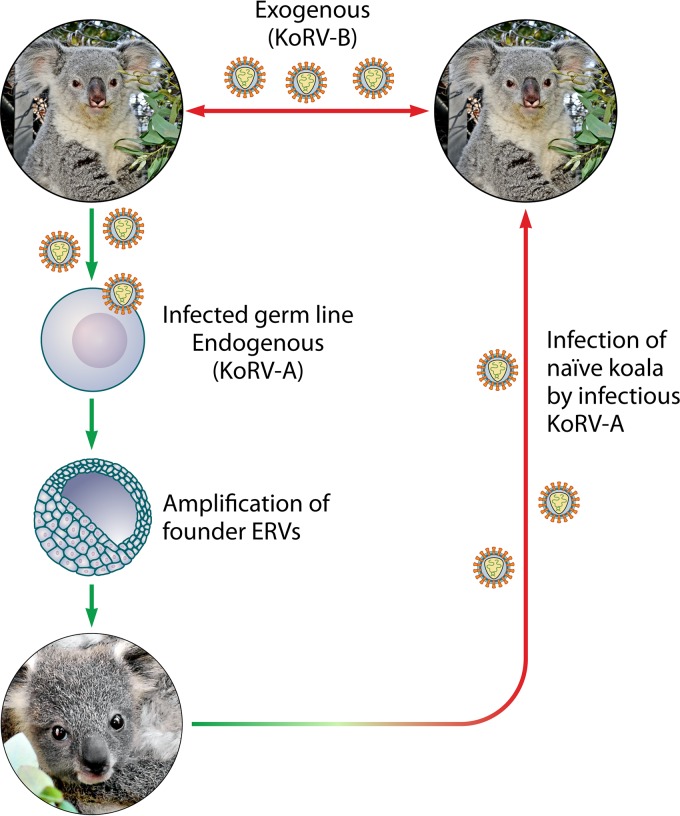
The new generation of virus particles may be competent to infect neighboring cells or other organisms. Retroviruses that maintain a genetic lineage by moving from cell to cell and from organism to organism via horizontal serial infectious events are called exogenous retroviruses (XRVs). In contrast, when a retrovirus infects a germ line cell, the mode of linear transmission and the evolutionary dynamics of the retrovirus can shift profoundly (Fig. 2). The provirus may gain a stable foothold within the germ line of the infected individual and become subject to vertical transmission to the offspring of the host. Vertical transmission of a provirus from parent to progeny within the germ line of a mammalian host species follows the rules of Mendelian segregation. Vertically transmitted proviral genomes within the host germ line are referred to as endogenous retroviruses (ERVs).
FIG 2.
Mechanisms of XRV and ERV transmission. XRVs are transmitted horizontally (red arrows) either in utero or via infected blood, feces, urine, milk, or saliva. After an XRV invades the germ line, ERVs (green arrows) may be amplified by superinfection or retrotransposition in the host germ line. ERVs that remain infectious can potentially infect naive members of the species. (Koala photographs courtesy of Tad Motoyama [Los Angeles Zoo, Los Angeles, CA].)
Following the initial colonization of host germ cells by an ERV, its copy number within the host germ line may be amplified by several mechanisms. First, reinfection across germ cells can occur (1). The process of reinfection of host cells by an infectious ERV, whether they are germ cells or nongerm cells, is restricted by superinfection interference, i.e., the blockade of viral receptors by host ERV envelope proteins preventing subsequent reinfection by viruses with an envelope that targets the same receptor for infection. Thus, this propagation mechanism has an intrinsically low probability. A second mechanism, which overcomes superinfection interference, can occur when the envelope of the infectious ERV mutates or recombines with other viral envelopes, resulting in an XRV bearing an envelope that utilizes a different receptor, thus permitting reinfection of host cells harboring the founder ERV. Other mechanisms include retrotransposition and amplification of the chromosomal fragment in which an ERV provirus is integrated (1). Early after integration, ERVs face several fates. Selection will favor hosts that carry ERVs that have their deleterious effects compromised so as not to endanger host survival. This can involve both mobilization of host restriction mechanisms and degradation of the viral genome (2). Presumably for this reason, many ERVs persist only as host-innocuous genetic remnants of their original XRVs, allowing the XRV-infected host species to survive. ERVs are also subject to genetic drift, the increase or decrease (including loss) of the frequency of an ERV at its chromosomal locus due to stochasticity in host reproductive success.
The processes by which ERVs evolve in the host are often difficult to infer due to the very long periods that may separate current analyses from the initial viral invasion of the germ line. These long periods allow for the accumulation of changes in ERVs that obscure the original invasion events and make it difficult to distinguish critical changes that occurred as the viruses were transitioning from XRVs to ERVs from later changes unrelated to endogenization. Recently, the discovery that the koala retrovirus (KoRV) is a relatively recent integrant in the germ line of its host (3) has provided a means of studying ERV evolution on a prospective basis. KoRV is a member of the genus Gammaretrovirus, one of six genera assigned to the subfamily Orthoretrovirinae, which in turn is one of two subfamilies recognized within the family Retroviridae. Gammaretroviruses include XRVs and ERVs that have been identified in diverse animal host species (Table 1). Endogenous KoRV has been designated “KoRV-A.” While KoRV-A is an established endogenous retrovirus in northern Australian koalas, it is not present in the germ lines of some koalas in southern Australia (4). KoRV-A differs from more ancient ERVs in that it retains its infectivity (3), having colonized the germ line of koalas relatively recently, as established by molecular dating, by the complete absence of KoRV-A in the germ lines of some koalas, and by the presence of KoRV at very different genomic loci across different koalas (i.e., it is insertionally polymorphic) (4–6). This contrasts with ERVs of other species, which are often many millions of years old, are present in all individuals of a species, and are often fixed at the same genomic location across every member of a species. Analysis of the genomes of a sire-dam-joey trio of northern Australian koalas detected no novel KoRV integrants (i.e., there were no KoRVs absent in both parents but present in the progeny), arguing against continuing superinfection by endogenous infectious KoRV (6). Koalas in U.S. and affiliated European zoos originated from northern Australian koalas and are all KoRV-A positive. A second form of KoRV, designated “KoRV-B,” was found in 66% of koalas of a U.S. zoo-based lineage of koalas; KoRV-B infection was strongly associated with T-cell lymphoma, and pedigree analysis within koala families with KoRV-B-positive members established horizontal transmission as the mode of KoRV-B spread (7). KoRV-B was also significantly associated with chlamydial disease in free-ranging koalas in Queensland, Australia (8). KoRV-B most likely arose de novo within KoRV-A animals by mutation or recombination and exists, for now, as an exogenous pathogenic koala retrovirus. The situation of overlapping KoRV endogenization and infection within a single living mammalian species represents a fascinating retrovirological case study in real time and a special challenge for deploying antiretroviral therapy and vaccines in the service of protecting a mammalian species (3).
TABLE 1.
Members of the subfamily Orthroretrovinaea
| Genus | Representative species |
|---|---|
| Alpharetrovirus | Avian leukosis virus, avian carcinoma Mill Hill virus 2, avian myeloblastosis virus, avian myelocytomatosis virus 29, avian sarcoma virus CT10, Rous sarcoma virus, UR2 sarcoma virus, Y73 sarcoma virus |
| Betaretrovirus | Jaagsiekte sheep retrovirus, mouse mammary tumor virus, squirrel monkey retrovirus, Mason-Pfizer monkey virus, Langur virus |
| Deltaretrovirus | Bovine leukemia virus, primate T-lymphotropic virus 1, primate T-lymphotropic virus 2, primate T-lymphotropic virus 3 |
| Epsilonretrovirus | Walleye dermal sarcoma virus, walleye epidermal hyperplasia virus 1, walleye epidermal hyperplasia virus 2 |
| Gammaretrovirus | Reticuloendotheliosis virus, feline leukemia virus, gibbon ape leukemia virus, koala retrovirus, guinea pig type C oncovirus, Moloney murine sarcoma virus, murine leukemia virus, porcine type C oncovirus, Snyder-Theilen feline sarcoma virus, Trager duck spleen necrosis virus, viper retrovirus, woolly monkey sarcoma virus |
| Lentivirus | Human immunodeficiency virus type 1, bovine immunodeficiency virus, caprine arthritis encephalitis virus, equine infectious anemia virus, feline immunodeficiency virus, human immunodeficiency virus type 2, puma lentivirus, simian immunodeficiency virus, visna/maedi virus |
Based on the virus taxonomy 2015 release at http://www.ictvonline.org/virustaxonomy.asp.
In this review, we explore the lessons learned thus far from the study of KoRV. We also examine the relationships among orthoretrovirus receptors, consider how viral interference may provide a barrier to the spread of an infectious endogenous retrovirus in a mammalian species, and how the use of a different receptor can overcome interference by ERVs. The KoRV receptors illustrate a general theme for many mammalian retroviruses, which employ a very limited set of highly related yet critically divergent cell surface solute transporters (the solute carriers [SLCs]). These are expressed with sufficient ubiquity to ensure receptor distribution across divergent cell types, including primary infection portal cells and germ cells.
KoRVs are also paradigmatic for interspecies retrovirus transmission. KoRV-A and KoRV-B are closely related to an XRV isolated from primates, designated gibbon ape leukemia virus (GALV). From a long-term evolutionary perspective, such cross-species transmissions of retroviruses have been common (2). Yet since the geographic range of koalas does not overlap that of gibbons, how did similar viruses come to infect species that do not overlap in range? What were the intermediate vector species? Are there some taxa that serve as reservoirs for interspecies transmission? How do viruses change as they cross the species barrier, and specifically, do retroviral interactions with host receptors change as viruses jump across species? How can highly related gammaretroviruses manifest as an endogenous virus in one species and as an infectious virus in another, what events surround the endogenization of retroviruses into the host germ line, and what are the fates of retroviruses that do integrate into the host germ line? Recent findings address these questions regarding the origins of GALV, KoRV, and other retroviruses. These, together with a more complete knowledge of the receptors used by retroviruses, are profitably synthesized with emerging technologies for XRV and ERV sequencing and genomic and subgenomic bioinformatics to yield a new view of retrovirus transmission and pathogenesis that emphasizes the dynamic coevolution of mammalian retroviral genomes and the cell receptors of their mammalian hosts.
CLASSIFICATION OF ERVs AND XRVs
Morphological Classification
Originally, retroviruses were divided into morphological classes based on the assembly and budding processes of simple retroviruses as visualized by electron microscopy. Retroviruses, originally designated type A to D particles, have now been recategorized into an organized taxonomy. Contemporary alpharetroviruses and gammaretroviruses were originally categorized as having a C-type morphology predicated on their observed ability to form spherical virions with electron-dense cores that assembled at and budded from the plasma membrane. Betaretroviruses were divided into particles having a B- or D-type morphology, with the former forming intracellular preassembled virus-like particles in the cytoplasm. Deltaretroviruses also assemble into intracellular virus-like particles and have a D-type particle morphology. D-type particles exhibit distinct morphological features after budding. Mature B-type particles have an off-center core, whereas D-type particles contain distinctive cylindrical cores (9).
Taxonomic Classification
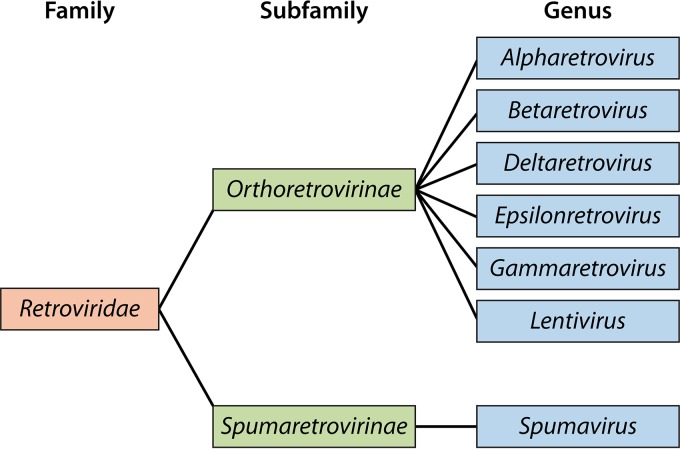
With the discovery of complex retroviruses, the categorization of retroviruses by morphology was replaced with new criteria for defining the Retroviridae. The International Committee on Taxonomy of Viruses (ICTV) currently classifies Retroviridae into two subfamilies: the Orthoretrovirinae and the Spumaretrovirinae (Fig. 3). Taxonomic classification of organisms is based on categorizing organisms in a hierarchical ranking system based on shared genetic and functional features. Features that distinguish Spumaretrovirinae from Orthoretrovirinae are numerous, but most center on how they form infectious particles and subsequently infect cells (10). Reverse transcription appears to occur within the virus particle, and thus spumaviral particles contain double-stranded DNA, whereas an RNA genome is found in orthoretroviruses (11). The Spumaretrovirinae subfamily is not discussed in this review. The retrovirus subfamily Orthoretrovirinae is composed of six genera. These include alpharetroviruses, betaretroviruses, gammaretroviruses, deltaretroviruses, epsilonretroviruses, and lentiviruses. Table 1 lists the six genera and examples of species associated with each. All six of these genera contain not only XRVs but also ERVs. Until recently, epsilon- and deltaretroviruses were also thought to have no ERV correlates. The epsilonretrovirus genus is represented mainly by XRVs in fish; however, Brown et al. recently identified a large number of epsilon-like ERVs in a variety of primate and other mammalian species (12). Similarly, endogenous deltaretroviruses had not been characterized until their recent discovery in long-fingered bats (13).
FIG 3.
Classification of the Retroviridae into subfamilies and genera. The Retroviridae family is not assigned to an order.
Classification Based on Evolutionary Relationships
A different approach to classifying Orthoretrovirinae, based on ERV lineages rather than XRV morphological features, is centered on relationships inferred between ancient endogenous and extant exogenous viruses. These analyses compare homologous sequences, such as gag, pol, or env sequences, across viruses (14–16). Since the pol gene has a relatively low rate of evolutionary change, it is a reliable marker of genetic changes accrued over relatively long periods (17). The inferred phylogeny of ERVs from the six orthoretroviral genera reveals two clades (clusters) or classes, with epsilon- and gammaretroviral ERVs designated class I viruses and betaretroviruses, alpharetroviruses, deltaretroviruses, and lentiviruses and their associated ERVs designated class II viruses (17). Thus, phylogenetic analysis of orthoretroviral ERVs and XRVs based on genetic relationships among pol genes (15, 17, 18) has distributed the six genera into two classes (19).
It is important that subgenomic components of retroviruses may evolve at different rates. The mutation rates for ERVs may be especially low, because ERVs are subject to the DNA repair mechanisms and low evolutionary rates of the genomes of their mammalian hosts. Among XRVs, the surface envelope-, polymerase-, and structural protein-encoding viral regions are under different selective pressures and constraints. The relatively highly conserved transmembrane (TM) domain of the envelope has been found to be useful for classifying retroviruses (19–21), with the exception of epsilonretroviruses, because their envelope structure and processing mechanism have not been evaluated as extensively.
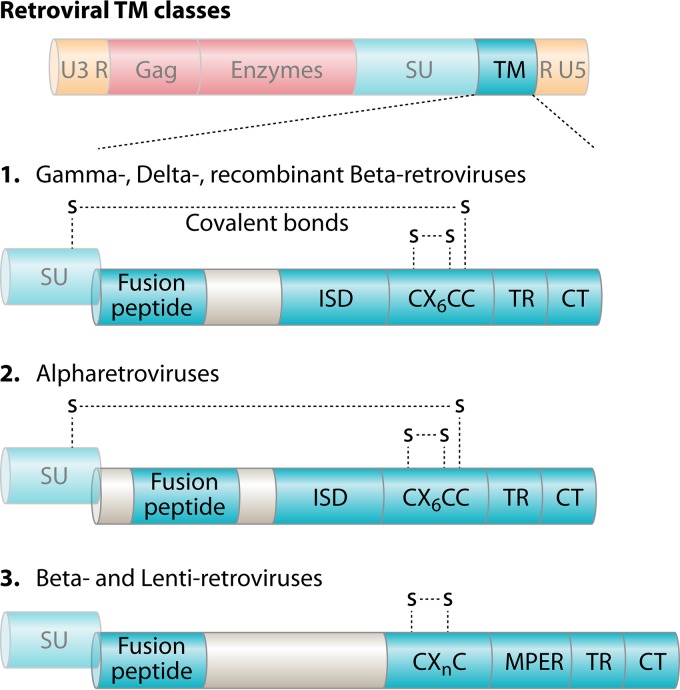
A novel phylogenetic classification of the Orthoretrovirinae was recently proposed based on distinguishing features within the TM domains of ERV and XRV orthoretroviruses (1, 21). The TM subunit is evolutionarily conserved and executes an essential function shared by all XRVs—the fusion of the lipid bilayer of the virus particle with the cell membrane—facilitating viral entry. Unlike SU, the TM subunit is shielded from host immune adaptive pressure and therefore serves as a useful retroviral phylogenetic marker. A fusion peptide is present in all retroviruses, but its position with respect to the N terminus of the TM domain varies. Also conserved among all orthoretroviruses are the transmembrane regions (TRs) and the cytoplasmic tail (CT) (Fig. 4). Features that distinguish the TM domains of orthoretroviruses include the CX6CC motif. This motif consists of three cysteines, the first two of which are separated from the third by six variable residues (represented by “X”). The first two cysteines in the CX6CC motif participate in the formation of an intramolecular loop in the TM ectodomain. The third cysteine is requisite for a covalent bond formed between the TM and SU domains within type D betaretroviruses, gammaretroviruses, and deltaretroviruses (class 1 viral fusion proteins; not to be confused with class I retroviruses), as well as alpharetroviruses (class 2), but not lentiviruses or B-type betaretroviruses (Fig. 4). TM domains of B-type betaretroviruses and lentiviruses (class 3) contain a CXnC motif lacking the third cysteine residue required for covalent association with SU but retaining the ability to form an intramolecular TM covalent loop, with the number of residues separating the two cysteines varying from four to seven (21). Additionally, type D betaretroviruses and all deltaretroviruses, gammaretroviruses, and alpharetroviruses contain an immunosuppressive domain (ISD). The ISD is a region within the TM domain proximal to the CX6CC motif that has been demonstrated to inhibit proliferation and differentiation of cultured T lymphocytes (22) and allows for escape from the innate and adaptive host immune systems (23–26). The ISD is absent in the B-type betaretroviruses and lentiviruses (Fig. 4). Another distinguishing feature of the TM class 3 retroviruses is the membrane-proximal external region (MPER), which extends the TM domains of B-type betaretroviruses and lentiviruses compared to those of alpharetroviruses, type D betaretroviruses, gammaretroviruses, and deltaretroviruses (1, 21). The single feature that distinguishes class 1 retroviruses from class 2 alpharetroviruses is that the fusion peptide is located internally in TM class 1 retroviruses, whereas it is at the immediate site of TM cleavage from SU in alpharetroviruses. Thus, the presence of the CX6CC and ISD motifs along with the absence of an MPER distinguishes class 1 and 2 retroviruses from class 3 retroviruses, and the position of the fusion peptide distinguishes class 1 and 2 retroviruses. The phylogeny of the three TM groups is incongruent with the class I and II groupings of the same viruses based on the pol gene. TM groupings may lay the groundwork for a radical reorganization of orthoretroviruses based on their flexibility to recombine in such a way as to expand their host range. A summary of the various groupings of orthoretroviruses is listed in Table 2. Remarkably, all the diverse receptors that have been identified for class 1 TM orthoretroviruses share an evolutionary feature, namely, the ability to carry solutes across membranes.
FIG 4.
Organization of the three classes of transmembrane (TM) proteins found in orthoretroviruses and the genera of retroviruses in which they are found (21). The immunosuppressive domain (ISD) and the CX6CC cysteine motif with disulfide bonds are depicted for TM classes 1 and 2. In class 3, the alternate cysteine motif CXnC and the membrane-proximal external region (MPER) are present, whereas the transmembrane region (TR), cytoplasmic tail (CT), and fusion peptide are variably positioned within the TM domain but invariably present in all TM proteins.
TABLE 2.
Classifications of ERVs and XRVs
| ERV/XRV genus (no. of species)a | Morphological typeb | TM classc | Pol classd |
|---|---|---|---|
| Alpharetrovirus (9) | C | 2 | II |
| Betaretrovirus (5) | A, B, and D | 1 and 3 | II |
| Deltaretrovirus (4) | C | 1 | II |
| Epsilonretrovirus (3) | Not assigned | Not assigned | I |
| Gammaretrovirus (18) | C | 1 | I |
| Lentivirus (10) | Rod/cone core | 3 | II |
From the list at http://www.ictvonline.org/virustaxonomy.asp.
From reference 304.
From reference 20.
From reference 305.
RETROVIRUS RECEPTORS
Since the topic of retrovirus receptors was last reviewed in this journal (27), a constellation of findings has allowed a deeper understanding of the role of receptors in virus-host coevolution. One of the most noteworthy is that a small group of multipass integral membrane proteins function as solute carrier (SLC) transporters and retroviral receptors. In fact, among the identified receptors for TM group 1 mammalian retroviruses that contain envelopes in which the TM domain is joined to the SU moiety via covalent disulfide linkage, all function as SLC transporters in their mammalian hosts. These include receptors for all gammaretroviruses and deltaretroviruses as well as for all of the recombinant betaretroviruses whose envelopes are of the TM-SU disulfide-linked type.
Retroviruses That Use SLCs as Receptors
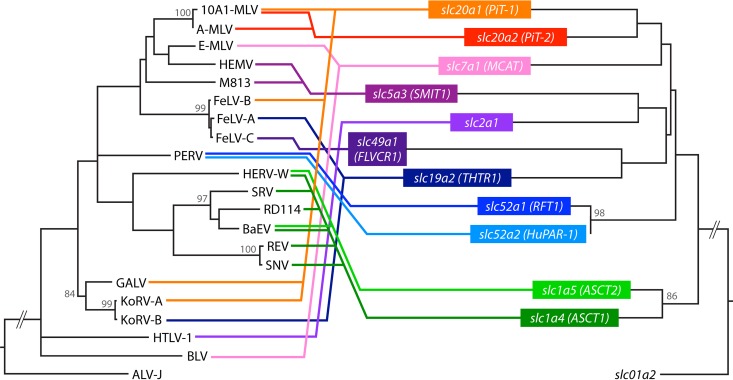
Members of each SLC family share limited sequence similarity with each other, even though they transport a common solute, and even lower sequence identity is found between different SLC families (28, 29). The solute transporters that function as retroviral receptors and the viruses that employ them are listed in Table 3.
TABLE 3.
Retroviruses that employ SLCs as receptorsa
| Virus(es) | Transporter gene name (product) | Transporter function |
|---|---|---|
| KoRV-A, GALV, FeLV-B, 10A1-MLV, FeLV-T, WMV | slc20a1 (previously GLVR1; now PiT1) | Sodium-dependent phosphate transporter |
| KoRV-B, FeLV-A | slc19a2 (THTR1) | Thiamine/folate transporter |
| A-MLV, 10A1-MLV, GALV, FeLV-B | slc20a2 (previously GLVR2; now PiT2) | Sodium-dependent phosphate transporter |
| E-MLV, BLV | slc7a1 (MCAT) | Cationic amino acid transporter |
| M813, HEMV | slc5a3 (Smit1) | Sodium/myoinositol cotransporter |
| RD-114, BaEV, REV, SRVs 1 to 5, HERV-W | slc1a5 (ASCT1) | Glutamate/neutral amino acid transporter |
| BaEV, HERV-W | slc1a4 (ASCT2) | Glutamate/neutral amino acid transporter |
| FeLV-C | slc49a1, slc49a2 | Heme importer and exporter |
| X-MLV, P-MLV | Xpr1 (ND) | Phosphate exporter |
| PERV-A | slc5A1 (huPAR-2) | Riboflavin transporter |
| GLN MLV | slc19A1 | Thiamine/folate transporter |
| ALV-J | slc9A1 (NHE-1) | Sodium/hydrogen exchanger |
| HTLV | slc2A1 (Glut-1) | Glucose transporter |
Abbreviations and nomenclature: KoRV, koala retrovirus; GALV, gibbon ape leukemia virus; FeLV, feline leukemia virus; MLV, murine leukemia virus; WMV, woolly monkey virus; MoMLV, Moloney murine leukemia virus; E-MLV, ecotropic murine leukemia virus; BLV, bovine leukemia virus; M813, an MLV originally isolated from the Asian rodent Mus cervicolor; HEMV, a Mus spicilegus endogenous retrovirus; RD-114, a feline infectious endogenous retrovirus; BaEV, baboon endogenous retrovirus; HERV, human endogenous retrovirus; PERV, porcine endogenous retrovirus; GLN, an infectious endogenous murine retrovirus; ALV-J, a subgroup member of avian leukosis viruses; HTLV, human T-lymphotropic virus.
Phosphate transporters.
The gibbon ape leukemia virus (GALV) and amphotropic murine leukemia virus (A-MLV) receptors were first identified only as retroviral receptors and were designated GLVR1 (30) and -2 (31) and Ram-1 (32), respectively. They were subsequently shown to function as sodium-dependent phosphate symporters (32–34), redesignated type III Na/Pi cotransporters, and assigned the names PIT-1 and PIT-2. PIT-1 and PIT-2 are members of the slc20 family, with the human gene designations slc20a1 and slc20a2, respectively. slc20a1 encodes a highly evolutionarily conserved, ubiquitously expressed sodium-dependent phosphate symporter, PiT1 (33, 35–37). PiT1 functions as a receptor for the primate XRVs woolly monkey virus (WMV) and GALVs. GALVs have been isolated on multiple occasions from captive white-handed gibbons (Hylobates lar). WMV represents a single isolate obtained from a New World woolly monkey (genus Lagothrix) that shared a household with a gibbon, as reviewed previously (38). Thus, WMV and GALV represent a single retroviral clade, and the use of PiT1 as a receptor for both is not unexpected given the close sequence similarity of their SU proteins.
The determination that PiT1 also functions as a receptor for feline XRVs, e.g., FeLV-B (36) and FeLV-T (39), and a murine virus isolate, 10A1-MLV, was somewhat surprising given that GALVs, FeLVs, and 10A1-MLV share limited sequence identity in the receptor binding domains (RBDs) of their envelope genes (40, 41). FeLV-B typically results from in vivo recombination between infectious FeLV-A and endogenous FeLVs present in the genomes of domestic cats (42). 10A1-MLV is a unique murine XRV that also arose by de novo recombination. In the case of 10A1, the recombination was between a murine gammaretrovirus isolated from a feral North American mouse and ERV sequences present in inbred laboratory mice (43). GALV and FeLV-B infect cells from an extremely broad range of mammalian species in vitro, including cells derived from cow, mink, bat, rat, pig, dog, cat, rabbit, and human and nonhuman primates, as well as cells from a variety of birds (but not chickens) (27). Viruses that use PiT1 as a receptor exhibit different host ranges. For example, cells derived from Mus musculus are resistant to FeLV-B, FeLV-T, WMV, and GALV but not 10A1-MLV (27, 38), demonstrating that PiT1, while conserved as a transporter across species, exhibits sharply different viral receptor characteristics depending on the mammalian species in which it is expressed. The koala retrovirus KoRV-A, initially discovered in 2000, was unequivocally identified as an ERV (5) closely related to GALV (44). KoRV-A is a recently endogenized virus that retains its infectious properties and is readily induced from koala peripheral blood mononuclear cells (45). Like GALV, KoRV-A uses the protein encoded by slc20a1 as a receptor (46).
A second sodium-dependent phosphate transporter, PiT2 (encoded by slc20a2), functions as a receptor for A-MLVs and serves as a secondary receptor for 10A1-MLV (31, 32). The slc20a1- and slc20a2-encoded proteins share common transporter features, but studies of knockout mice have shown that slc20a1 is an essential gene; complete deletion of slc20a1 results in lethality at embryonic day 12.5 in mouse knockouts (47). Deletion of slc20a2 is not lethal in mice but results in serious physiological effects, including calcifications in the thalamus and various brain regions (48). A-MLVs, such as 1504A and 4070A, use the transporter as a receptor to infect human and murine cell lines as well as cells derived from a variety of other species (27). FeLV-B can use both slc20a1- and slc20a2-encoded transporters as receptors, but employment of the slc20a2-encoded receptor is restricted to cells expressing the feline ortholog of this receptor/transporter (49).
GALV and KoRV-A, though restricted to using PiT1 to infect human cells and many other cell types, can use proteins encoded by slc20a2 orthologs to infect two types of rodent cells. The PiT2 orthologs expressed in E36 cells derived from the Chinese hamster and cells derived from the feral mouse Mus musculus molossinus function as GALV or KoRV-A receptors (49–52). Some orthologous slc20a2 genes code for proteins that fail to function as GALV receptors, while others code for a functional receptor. Sequence comparisons revealed that the residue present at PiT2 position 522 regulates this aspect of receptor function (34, 53, 54). Furthermore, engineering the appropriate codon into the human slc20A2 cDNA is sufficient to allow the mutated human protein to function as a GALV receptor (54).
Another phosphate transporter, in this case a phosphate exporter (Xpr1), has been proposed to function as a receptor for xenotropic and polytropic gammaretroviruses (X-MLVs and P-MLVs) (55). XPR1 was originally identified as a human cDNA that specifically conferred entry of X-MLVs and P-MLVs into resistant hamster cells (56–58). More recently, it was demonstrated that phosphate efflux is inhibited by the X-MLV envelope binding domain (55), and mutations in Xpr1 are implicated in altering phosphate homeostasis in the neurological disease designated primary familial brain calcification (59). Although it clearly functions as a solute exporter protein, Xpr1 is the only gammaretroviral receptor within the SLC transporter family that has not been assigned SLC nomenclature (Table 3).
Vitamin transporters.
The exogenous koala and feline gammaretroviruses, KoRV-B (7, 60) and FeLV-A (61), respectively, use the thiamine (vitamin B1) transporter THTR1 (corresponding human gene designation slc19a2) as a receptor (Table 3). FeLV-A is an ecotropic virus and is restricted to using the receptor encoded by the feline slc19a2 ortholog to infect feline cells in culture and in vivo. KoRV-B is spread from koala to koala in a manner consistent with the horizontal transmission demonstrated by FeLV-A in cats (7).
The human receptors for one of the three subgroups of porcine endogenous retroviruses, PERV-A, have been identified as being encoded by genes present on human chromosomes 8 and 17. These two human receptors, designated HuPAR-1 and HuPAR-2, share 86.5% amino acid identity (62). One of these receptors, HuPAR-2, is the riboflavin (vitamin B2) transporter RFT1 (corresponding to the human gene slc5a1) (63) (Table 3).
A recently identified gammaretrovirus receptor determined to be a solute transporter is used by the murine infectious GLN retrovirus. The GLN retrovirus is named for the glutamine amino acid (Gln) of the tRNA that is complementary to the primer binding site of the retrovirus. The murine GLN-2 retrovirus is the sole member of the high-copy-number reiterative ERV family present in the C57BL/6 mouse genome and is an infectious retrovirus (64). GLN-2 employs the folate transporter encoded by the murine slc19a1 ortholog to infect mouse cells (Jhen Tsang, David Ribet, Thierry Heidmann, and Marie Dewannieux, submitted for publication).
Amino acid transporters.
Ecotropic murine leukemia viruses (E-MLVs) are a group of related XRVs and ERVs that were isolated from Mus musculus and have a restricted host range, infecting only cells derived from mice or rats. The cDNA of the mouse slc7a1 ortholog is the first gene reported to encode a retroviral receptor whose expression is necessary and sufficient to facilitate the entry of E-MLV into otherwise fully resistant cells (66). Moloney murine leukemia virus (MoMLV) is an archetypal ecotropic MLV and represents the first cloned and fully sequenced retroviral genome (67). The receptor for E-MLVs is a high-affinity, low-capacity transporter of cationic amino acids, e.g., arginine, lysine, and ornithine (68, 69). In addition to murine XRVs, endogenous AKV MLVs can use this transporter as a receptor. However, not all murine orthologs of slc7a1 encode receptors that function for AKV. For example, cells derived from Mus minutoides, an African pygmy mouse, are resistant to AKV MLVs but not to ecotropic murine XRVs (70). Bovine leukemia virus (BLV), the etiological agent of bovine leukemia, is a deltaretrovirus that also employs the cationic amino acid transporter encoded by slc7a1 as a receptor (Svilena Ivanova, Jawida Touhami, Donatella Giovannini, Julien Bellis, Jérôme Feuillard, Lavanya Madakasira, Vincent Petit, Valérie Courgnaud, Marc Sitbon, and Jean-Luc Battini, submitted for publication) to infect its natural host cattle and presumably to horizontally infect zebu, water buffalo, sheep, and capybaras (72).
The human sodium-dependent broad-scope neutral amino acid transporters ASCT1 and ASCT2 (human gene designations slc1a4 and slc1a5) are cell surface receptors used by the largest number of TM group 1 retroviruses (Table 2). The avian XRVs designated reticuloendotheliosis virus (REV) and spleen necrosis virus (SNV) (57, 73, 74), the feline RD-114 isolate (57, 75), the baboon BaEV isolate (57, 76), and human HERV-W (77, 78) all use neutral amino acid transporters for entry. The infectious ERV BaEV was obtained from baboon placental tissue cocultured with various cells (79), and the RD-114 ERV was obtained from human rhabdomyosarcoma cells passaged in fetal cats (80). HERV-W employs both slc1a4- and slc1a5-encoded receptors (77, 78).
slc1a5 encodes a protein that also functions as a receptor for the TM group I squirrel monkey betaretroviruses SRV-1, SRV-2, SRV-3 (Mason-Pfizer monkey virus), SRV-4, and SRV-5 (81, 82). All viruses that use these receptors have broad host ranges that include human cells, but only BaEV can infect mouse cells (27). This suggests that BaEV can employ the murine ortholog-encoded transporter as a receptor. The ability of BaEV (76) and HERV-W (77, 83) to employ slc1a4- in addition to slc1a5-encoded transporters contributes to their expanded host range. Amino acid transporters serve as receptors for the most diverse range of retroviruses, including members of the betaretroviruses, gammaretroviruses, and deltaretroviruses.
Other solute transporters acting as retroviral receptors.
There are four receptor classes for FeLVs. As mentioned above, FeLV-A is a minimally pathogenic ecotropic virus that employs the slc19a2-encoded transporter as a receptor. FeLV-B and -T require the slc20a1-encoded transporter/receptor to infect cells. FeLV-C, a virus associated with aplastic anemia in cats, uses the heme exporters (human gene designations slc49a1 and slc49a2) as receptors (84, 85). FeLV-C is generated by mutation within the FeLV-A envelope variable region (86). Most virus-transporter interactions are not deleterious to the infected cells, as envelope-receptor binding does not compromise transport function; however, this is not the case for FeLV-C. The appropriation of heme exporters as receptors by FeLV-C impairs heme export, resulting in a deficit of erythroid progenitors in the bone marrow, most likely due to heme toxicity. Therefore, the use of this receptor has a direct effect on FeLV-C disease outcomes (84, 87).
In addition to the MLVs discussed previously, other MLVs derived from mice include the XRV M813, isolated from an Asian mouse, Mus cervicolor (88), and an infectious ERV (HEMV) isolated from the Eastern European mouse Mus spicilegus. M813 and HEMV have a different host range in vitro compared to that of the classical E-MLVs mentioned above that use a cationic amino acid transporter as a receptor (encoded by slc7a1). M813 and HEMV infect only cells derived from mice, and unlike classical E-MLVs, they do not infect cells derived from rats. These viruses were independently determined to use a common receptor, Smit1, a sodium-myoinositol cotransporter encoded in humans by the slc5a3 gene (89, 90). Infectious HEMV and M813 were isolated from mice from distinct geographic regions and appear to have evolved independently to use the same receptor.
The human T-lymphotropic viruses (HTLVs) are deltaretroviruses. HTLV-1 was the first human retrovirus identified and the first to be associated with pathogenic outcomes, namely, adult T-cell leukemia/lymphoma (ATL) and tropical spastic paraparesis (TSP) (91). HTLV-1 has a broad host range and employs the glucose transporter Glut-1, encoded by slc2a1, as its receptor (92). Expression of Glut-1 in semiresistant cells confers enhanced susceptibility to both HTLV-1- and HTLV-2-enveloped particles, and Glut-1-expressing cells induce HTLV envelope-host cell fusion (88). Like FeLV-C-mediated interference of heme export by slc49a1- and slc49a2-encoded receptors, Glut-1 protein binding by either HTLV-1 or -2 envelope glycoproteins inhibits glucose transport. This inhibition is hypothesized to contribute to the pathogenesis of HTLV-associated human disease (93).
Avian leukosis virus of subgroup J (ALV-J) is an alpharetrovirus that presumably participated in gammaretrovirus envelope capture, as its SU is significantly different from those for other ALV subgroups, even though the rest of its genome, including the TM-encoding domain, shows little divergence from those of ALV subgroups A to E (94, 95). ALV-J employs a Na+/H+ exchanger transporter (Table 3) encoded by the chNHE1 gene (human gene designation slc9a1) as a receptor in chicken cells. The chicken ortholog of slc9a1 is expressed in all chicken cell lines tested so far and in cell lines derived from jungle fowl and domestic turkey (95). In contrast, the non-SLC receptors for ALV subgroups A to E are not expressed in all chicken cell lines (95, 96). The ALV-J envelope is responsible for inducing myeloid leukemia in chickens, distinguishing ALV-J from ALVs A to E, which primarily cause lymphomas (97).
The discovery that a wide variety of orthoretroviruses use members of the SLC transporter family as receptors indicates that a deeper understanding of viral envelope coevolution with host receptor utilization is needed to determine the deep structure of virus-mammal coevolution and receptor-associated pathogenesis. This requires an understanding of the structural elements within RBDs that allow independent (convergent) evolution of multiple otherwise unrelated RBDs to use the same receptor (e.g., those of FeLV-B, 10A1-MLV, and KoRV-A, which all use PiT1). Likewise, closely related XRVs have diverged sufficiently to use different receptors (e.g., in the cases of FeLV-B, FeLV-A, and FeLV-C; 10A1-MLV, A-MLV, X-MLV, and E-MLV; and KoRVs A and B). Through receptor interference, ERVs may recurrently pressure XRVs to shift receptor usage to sidestep the receptor that may be blocked and the extracellular release of ERV envelope proteins (98).
Diversification of RBD structure leading to differential receptor utilization is driven by evolutionary pressures that are different from those driving other viral genomic regions. Examples of endogenous infectious retroviruses in different host species that employ a common receptor are also instructive in this regard. For example, the RD-114 ERV acquired its envelope as a result of de novo recombination between endogenous feline gag-pol sequences and the envelope of BaEV after an ancestor of the domestic cat and related species was infected by BaEV (99).
Retroviruses That Use Single-Pass or GPI-Anchored Proteins as Receptors
Betaretroviruses such as Jaagsiekte sheep retrovirus (JSRV) and mouse mammary tumor virus (MMTV), as well as many endogenous type B betaretroviruses, lack the third cysteine in the TM ectodomain, consistent with noncovalently associated SU and TM subunits (19) (Fig. 4). These betaretroviruses differ from recombinant D-type betaretroviruses associated with gammaretroviral envelopes in that they do not use SLC membrane proteins as receptors. They instead use single-pass or glycosylphosphatidylinositol (GPI)-anchored proteins as receptors. Hyaluronidase 2 (HYAL2) is the receptor for JRSV (100). HYAL2 is a GPI-anchored cell surface protein with a low hyaluronidase activity (100). The receptor for MMTV is the single-transmembrane-spanning transferrin receptor 1 (Tfr1 or Tfrc), which provides a major means of iron uptake into blood cells (101, 102). Tfr1 is also a cluster of differentiation (CD) cell surface immunomarker (CD71) that serves as a marker of cells of the erythroid lineage.
The receptors for the avian sarcoma and leukosis viruses ALV-A, ALV-B, ALV-C, ALV-D, and ALV-E are encoded by three distinct genes: TVA, TVB, and TVC. TVA encodes a low-density-lipoprotein (LDL)-related protein that serves as a receptor for ALV-A (103, 104) and can be transcribed by alternate splicing into mRNAs encoding two different forms of the receptor, a single-pass membrane form or a GPI-anchored form (105). TVB encodes the single-membrane-spanning receptor for ALV-B, ALV-D, and ALV-E, which is a tumor necrosis factor receptor (TNFR)-related protein (106). TVC encodes a third single-membrane-spanning receptor that is a member of the immunoglobulin superfamily and is used by subgroup C ALVs as a receptor (107). ALVs A to E are thought to exemplify the evolution of avian retroviruses from a common ancestor to use similar paralogs coding for a class of receptors.
Receptors for Lentiviruses
With the observation that AIDS patients manifest CD4+ T-cell decline (108), researchers showed that CD4 serves as a receptor for human immunodeficiency virus type 1 (HIV-1) (109–112), HIV-2 (113), and simian immunodeficiency virus (SIV) (114, 115). CD4 is a cell surface glycoprotein predominantly of immune cells, such as T helper cells, monocytes, macrophages, and dendritic cells. It functions as an associative recognition coreceptor through interactions with major histocompatibility complex (MHC) class II molecules on antigen-presenting cells and facilitates T-cell receptor (TCR)-dependent initiation of the cascade for antigen-specific T-cell activation. However, while in early studies virtually all HIV-1 isolates replicated in vitro in activated CD4+ T cells present in peripheral blood mononuclear cells (PBMC), some replicated in mature macrophage cultures but not in certain established CD4+ T-cell lines (“macrophage tropic”), while some replicated in these established CD4+ T-cell lines but not in mature macrophage cultures (“T cell tropic”) (116–118). This property was later explained by the alternate use of one of two coreceptors in addition to CD4. Viruses historically designated macrophage (or M) tropic utilize the chemokine receptor CCR5, which is not expressed on the established CD4+ T-cell lines used in early studies, and are now designated R5 tropic (119–123), while viruses originally identified as T-cell line (or T) tropic use a different chemokine receptor, CXCR4, and are now described as X4 tropic (124). Both CCR5 and CXCR4 are G-protein-coupled receptors and function as chemokine receptors. The ligands for CCR5 include β chemokines, particularly MIP-1β, while the ligand of CXCR4 is SDF-1. These chemokines are secreted by many cell types and serve as chemoattractants to draw lymphocytes to sites of inflammation. HIV-2 and SIV also use these coreceptors. Transmitter/founder (T/F) viruses are almost exclusively R5 viruses, with X4 or dual-tropic R5X4 viruses developing later in infection in some infected individuals, although many T/F isolates of HIV-1 do not productively infect primary macrophage cultures, indicating the importance of factors other than CCR5 expression. This coreceptor dependence of HIV-1 explains the apparent resistance of CCR5Δ32 homozygous individuals to HIV infection (125–127). This mutation results in the retention of CCR5 in the endoplasmic reticulum (128); therefore, those homozygous for the allele have no surface expression of CCR5, which is required to render cells permissive for infection with R5-tropic strains of HIV-1 (126). In addition, CCR5 has been exploited as a drug target (129) and figures in current gene therapy approaches to treat HIV infection (130), including in the one individual, to date, who has been cured (131, 132). Individuals homozygous for CCR5Δ32 have become infected with HIV-1 and progressed to AIDS, but the T/F viruses in these cases were either X4 tropic or R5X4 tropic (133–139).
Like HIV, feline immunodeficiency virus (FIV) induces a disease state characterized by a progressive depletion of CD4+ T lymphocytes and an AIDS-like condition (140, 141). FIV employs CD134 (OX40) as its main receptor (142, 143) and CXCR4 as the coreceptor (144, 145). CD134 is a member of the tumor necrosis factor-nerve growth factor receptor family and is associated with enhanced cellular survival of T cells and prolonged activation. While it is abundant on CD4+ T cells, CD134 is also found in a subpopulation of CD8+ T cells, CD45R+ B cells, and monocyte-derived macrophages, consistent with FIV's cellular tropism (146). FIV binds the first cysteine-rich domain (CRD1) of CD134 (147), with some strains additionally requiring the second cysteine-rich domain (CRD2) (148, 149). CRD2-dependent strains appear to be transmitted preferentially (150), while CRD2-independent strains arise during chronic infection (151), analogous to the coreceptor dependence of HIV and SIV.
The presence of functional receptors is a critical element for determining the host range of an infectious retrovirus. However, many other factors are involved in the transmission of infectious retroviruses among and between species.
XRV TRANSMISSION
Zoonotic Cross-Species Transmission
The lentivirus HIV-1 group M, the main form of the virus responsible for the global AIDS epidemic, originated in the region of Kinshasa (formerly Leopoldville) in what is now the Democratic Republic of Congo around 1920 (152–154). Related lentiviruses, the SIVs, are naturally endemic to sub-Saharan Africa, with approximately 40 different primate species harboring strains. In the naturally occurring hosts that have been characterized, infections appear to be nonpathogenic (155). A virus closely related to HIV-1 was discovered in chimpanzees (SIVcpz) (156, 157), with only the subspecies Pan troglodytes troglodytes serving as a natural reservoir. The subspecies P. t. schweinfurthii also harbors SIVcpz, but it is divergent from HIV-1 and that of P. t. troglodytes (158). Unlike other nonhuman primate hosts in the region, infected chimpanzees can manifest an AIDS-like illness (159). SIVcpz resulted from the recombination of two SIVs: one related to SIVrcm of red-capped mangabeys (160) and one infecting several Cercopithecus species (Cercopithecus nictitans, C. cephus, and C. mona) (161–163). Both sets of monkeys overlap in geographic range with P. t. troglodytes, which likely acquired the relevant SIVs in the context of predation (155).
Phylogenetic analysis revealed that chimpanzees in the far southeastern region of Cameroon are the probable source of HIV-1 group M (159). Chimpanzee-to-human cross-species transmission likely occurred through exposure to infected blood and bodily fluids during the butchering of bushmeat (164). Water traffic may have conveyed infected animals from Cameroon to Kinshasa, a port and former capital of the Belgian Congo. In the early 20th century, this area of Africa was experiencing rapid sociocultural changes, population growth, and medical intervention (153, 165, 166) that may have allowed the virus time to adapt to a human host (167, 168). The chimpanzee-to-human cross-species transmission did not occur once but many times, yielding the additional HIV groups N and O. Additionally, HIV-2 emerged as a cross-species transmission from sooty mangabeys (Cercocebus atys) (169), with evidence of eight distinct events (170). SIVsmm has also inadvertently been transmitted multiple times to various macaque species in laboratory settings, as distinct events (171) giving rise to AIDS-like illness. Evidence also exists for chimpanzee-to-gorilla cross-species transmission (172), estimated to have taken place at least 100 to 200 years ago (173) and culminating in SIVgor. HIV-1 group P likely originates from SIVgor (174). Cross-species transmissions of lentiviruses between nonhuman primates and humans have likely occurred for millennia without reaching epidemic/pandemic scales until modern times (175). The current evidence suggests that this is not due to a change in the rate of interspecies transmission but has more to do with sociodemographic changes, such as massive-scale urbanization in Africa, an increased reliance on bushmeat, and increasing global travel, all of which are highly conducive to the spread of infectious agents (153, 176, 177).
The deltaretrovirus HTLV-1 was the first infectious, oncogenic human retrovirus discovered (178). It is the etiological factor for adult T-cell leukemia (ATL) and tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM); however, disease manifestation is rare among infected individuals (179). HTLV-1 is present globally but not homogenously and is endemic in isolated regions, i.e., the southwestern part of Japan, sub-Saharan Africa, the Caribbean Basin, South America, and foci in the Middle East and Australo-Melanesia (180). While the prevalence in some areas likely arose from mass migration of an infected population (181), that in other areas likely stems from frequent interspecies transmissions (182).
Studies in Africa have found individuals with strains more closely related to those in sympatric nonhuman primates (sooty mangabeys, chimpanzees, colobus monkeys, mandrills, crested mona monkeys, and gorillas) than to those in other local human individuals (183, 184). HTLV-1 and simian T-lymphotropic virus type 1 (STLV-1), the related simian counterpart, do not separate into distinct phylogenetic lineages. Furthermore, strains cluster together by geographic origin of their hosts rather than by host species, which is indicative of local cross-species transmissions. Each of the seven subtypes (A to G) is believed to have originated from a distinct cross-species transmission between a nonhuman primate and a human (182, 185–187).
An outbreak of the FeLV gammaretrovirus among Florida panthers (Puma concolor coryi, a subspecies of puma) was reported in 2008, which resulted from a cross-species transmission event between a domestic cat and a Florida panther (188). This cross-species transfer was most likely the result of a single transmission of FeLV-A from a domestic cat resulting in rapid, widespread transmission among panthers beginning in 2001, at which time 33% of panthers were seropositive for FeLV antibodies, with five FeLV-associated deaths reported. This was a surprising finding given that, as recently as 1991, no evidence of exposure to FeLV-A was reported for the 38 Florida panthers surveyed (188). In addition to FeLV-A, feline lentiviruses, particularly an isolate of puma lentivirus clade A (PLVA), originally recovered from bobcats (Lynx rufus), have gone on to infect pumas in Florida and California (189, 190).
Intraspecies XRV Transmission
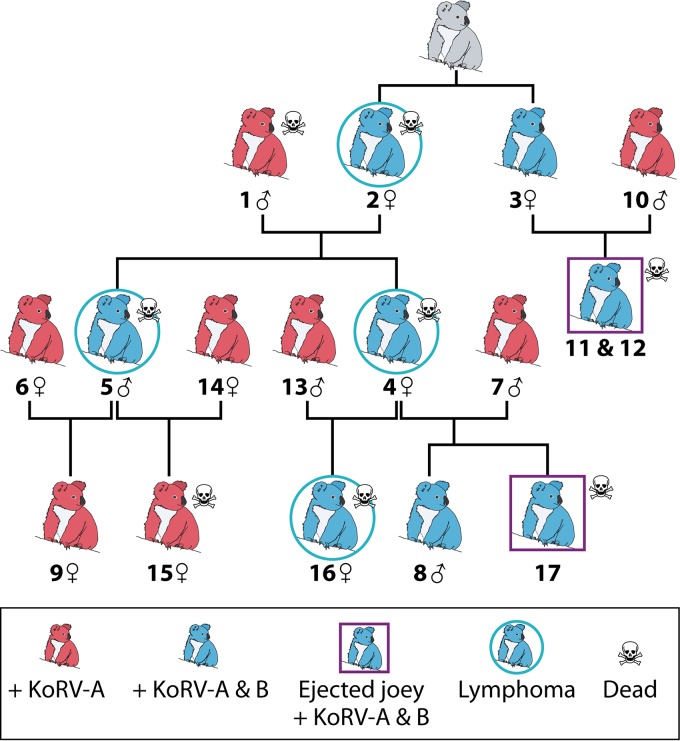
The original KoRV-A ERV genomic sequence was obtained from DNA extracted from the blood and tissues of an Australian koala in 2000. An XRV KoRV, KoRV-B, was later obtained from biological materials obtained from Los Angeles Zoo (LAZ) koalas. KoRV-B most likely arose de novo in KoRV-A-infected koalas as a result of mutations introduced during error-prone reverse transcription during KoRV-A replication or by recombination. The 17 related koalas at LAZ all contained the KoRV-A ERV, and nine koalas, including three joeys that died and were ejected from the pouch and six adult koalas, were both KoRV-A and KoRV-B positive (Fig. 5). Four of the six KoRV-B-positive adult koalas died of lymphoid neoplasias, establishing a strong etiological link between KoRV-B and malignant neoplasias (3). KoRV-B is not vertically transferred in the germ line, as the KoRV-B-positive sire did not transmit KoRV-B to any of their offspring. In contrast, all KoRV-B-positive dams transmitted the virus to offspring (Fig. 5), suggesting that the virus may be transmitted in utero and/or in milk. Because KoRV-B uses a receptor distinct from that employed by KoRV-A, it can infect KoRV-A-positive koalas and even rescue and transmit the KoRV-A genome.
FIG 5.
Summary of koala retrovirus (KoRV) statuses within a pedigree of 17 northern Australian koalas kept at the Los Angeles Zoo. Red koala symbols denote individuals positive for KoRV-A (which is ubiquitous in U.S. zoos), whereas the blue koala symbols denote individuals positive for KoRV-B (which is not ubiquitous) and KoRV-A. Dead joeys ejected from the maternal pouch are depicted within squares; each was positive for KoRV-B. Note that the pattern is consistent with maternal transmission of KoRV-B.
The feline leukemia viruses also participate in interspecies XRV transmission. Curiously, FeLV-A and FeLV-B employ the same receptors as KoRV-B and KoRV-A, which use the slc19a2 and slc20a1 transporters, respectively, as receptors. Additional KoRV XRV envelope variants have now been discovered that apparently arise as a result of evolutionary pressures that favor both host resistance and virus escape mutants (191).
Iatrogenic Transmission of GALV, REV, and RD-114
The first GALV isolates were partially sequenced in the early 1970s. Surprisingly, given their use as vectors in cancer therapy, the complete sequences of five isolates were not reported until 2016 (192). These are four isolates of GALV isolated from captive gibbons, i.e., GALV SEATO (193, 194), GALV-H (195), GALV-Br (196), and GALV-SF (197), and one GALV obtained as a contaminant in a human cell line (GALV-X) (198). Substantial variation has been observed among the different GALVs, with two isolates, GALV-X and GALV SEATO, sharing only 87.1% nucleotide identity across their genomes (44). Other genomic regions exhibit even lower identity; for example, the long terminal repeats (LTRs) of GALV-X and GALV SEATO share only 69.6% identity (44). A closely related gammaretrovirus, woolly monkey virus (WMV), is the apparent result of cross-species transmission from a pet gibbon infected with GALV to a woolly monkey (a New World species) kept in the same household (3). WMV, though referenced by ICTV as a separate species owing to its retention of an oncogene, is also considered a strain of GALV. Despite the high level of divergence between GALV-X and GALV SEATO, WMV represents the most basal GALV and exhibits several strain-specific insertions in its LTRs (192). Because gibbons, in contrast to koalas (see above), have not been reported to be unusually susceptible to leukemias/lymphomas and GALV has not been isolated from wild gibbons or gibbons in zoos (199), it is possible that GALV represents an inadvertently acquired pathogen that has run its course among captive gibbons.
GALV most probably originated when a gammaretrovirus was accidentally introduced into captive gibbons in the SEATO colony in Thailand during experimental studies of a malaria parasite in the latter half of the 20th century. In the early 1960s, juvenile white-handed gibbons (Hylobates lar) were wild caught and then maintained in the SEATO Medical Research Laboratory in Bangkok, Thailand, for research purposes (200, 201). Reports on the number of animals in the SEATO colony suggest that somewhere between 120 (202) and 195 (203) were present in the colony at any time. An unprecedented incidence of chronic granulocytic leukemia (CGL) was reported for the SEATO colony, occurring in 12 animals in total (201–203). The incidence of leukemia was confined to the 92 experimental animals involved in malaria studies (13%), compared to 0% of animals in the control group of gibbons (203).
In 1965, Southeast Asian gibbons were afforded legal protection, and the SEATO laboratory was closed. However, laboratories in the United States imported captive laboratory gibbons from Thailand for research purposes, through irregular channels (200, 204). GALV isolates, including GALV SEATO, were subsequently obtained. GALV SEATO was isolated from a gibbon in a U.S. research facility that had been inoculated with bone marrow homogenate from one of the gibbons (designated S-74) that succumbed to CGL after receiving a malaria inoculation at the Bangkok SEATO research facility (194). Additional GALV isolates were obtained from healthy gibbons and from the offspring of GALV-infected mothers, suggesting that GALV, once introduced into gibbons, was efficiently horizontally transmitted (205). It is interesting that gibbons that developed CGL were those receiving viral or malaria injections (194, 203), whereas those that acquired GALV via horizontal transmission developed T-cell lymphomas (205, 206), the disease found in cats infected with FeLV-B (42) and koalas infected with KoRV-B (3). The route of infection is also relevant for HTLV-2, with a profound increase in viral evolutionary rate observed in intravenous drug users relative to that for sexually transmitted viruses (207, 208). This likely reflects a larger and more diverse infectious dose of virus transmitted by needle than by sexual transmission, with the latter often representing a severe bottleneck on retroviral diversity.
Interestingly, a similar laboratory origin has been proposed for another gammaretrovirus transferred across species to an avian host. Although many endogenous gammaretroviruses in birds and reptiles have phylogenetic patterns indicative of cospeciation with their host (209), the reticuloendotheliosis viruses (REVs) are a group of closely related viruses isolated from gallinaceous birds that are similar to ERV sequences present in two species of mongoose (210) and to a full-length REV present in short-beaked echidnas (211). Their presence across these taxa strongly suggests the interclass transmission of a mammalian ancestral retrovirus (209). REVs such as spleen necrosis virus (SNV) were very likely derived iatrogenically from a mammalian gammaretrovirus during a malaria study involving avian host animals in the late 1930s (210). It is presently unknown if the retrovirus ancestral to avian REVs was present in the source animal from which the malarial agent was originally obtained or represents a contaminant introduced during serial passage. Whatever the case, animals that harbor REV-related ERVs do not overlap in biogeographic regions or overlap geographically with the avian species that harbor REV XRVs, suggesting that a series of intermediate vectors, such as bats (212–215), may have been involved in the dissemination of the ancestral virus, but this remains speculative (210). REVs can also be found as homologs inserted in the viral genomes of fowlpox virus and gammaherpesvirus-2 (210). The fowlpox virus containing REV is particularly relevant because it is found in both captive and wild fowl populations. Hitchhiking of REVs is not the only example of an ERV integrating into DNA viruses. Cocultivation of REVs and ALVs with turkey herpesvirus and Marek's disease virus, respectively, resulted in retroviral integration into the herpesvirus, in both cases, after several passages (216). Similarly, field and vaccine strains of fowlpox virus have been demonstrated to carry REV integrations (217). The demonstrated interactions between retroviruses and unrelated DNA viruses suggest that recombination is a critical means by which retroviruses may transport themselves from one host to another via unrelated DNA viruses and independent of retroviral replication.
Live attenuated feline and canine vaccines have been demonstrated to harbor RD-114, an infectious feline ERV (45). Contamination of vaccines, including the canine parvovirus type 2 vaccine, is apparently the result of the manufacture of these vaccines by use of cell lines, such as Crandell-Rees feline kidney cells or human rhabdomyosarcoma TE671 cells, that are persistently infected with RD-114. RD-114 replicates efficiently in both canine primary cells and established cell lines. The likelihood of iatrogenic transmission of retroviruses is probably underestimated and perhaps undetected, with the exception of the cases, such as those described above, that become prevalent.
ERV ESTABLISHMENT AND MOBILIZATION
KoRV as an Example of a Retroviral Germ Line Invasion Leading to Endogenization
Rodent source animals for GALV and KoRV.
Most cross-species transmissions are restricted to XRV somatic cell infections without invasion of the host germ line. However, retrovirus-like elements in most mammalian genomes suggest that cross-species transmissions can also result in germ cell infection, leading to the establishment of ERVs. The host-virus evolutionary dynamics resulting in exogenous virus invasion of the host germ line and ERV establishment within the host species are often obscured by the millions of years that have elapsed since ERV colonization of the host species germ line (218). An exception is the relatively recent endogenization of the koala ERV, KoRV-A. KoRV-A is believed to have invaded the germ line of the koala recently, within the last 50,000 years (6). Thus, the earliest stages of host germ line colonization, i.e., conversion from an XRV to an ERV, can be studied in the koala.
In 2000, Hanger et al. obtained a full-length KoRV-A genome that, based on its genetic structure and functional motifs, was determined to be a gammaretrovirus (44). Southern blot analysis suggested that KoRV-A integration patterns varied greatly among unrelated koalas (44), as one might expect for an XRV that had integrated at random chromosomal locations in each individual of the species. However, the KoRV chromosomal integration patterns across the various tissues (including germ cells) of a given individual koala were identical, which is characteristic of an ERV provirus (44). This paradoxical picture suggested that KoRV was insertionally polymorphic, with integrants present at different chromosomal locations in different individuals, rather than being fixed across the species. This was also supported by pedigree analysis of a dam-sire-joey triad study that examined 39 KoRV-A integration sites across the triad (6). The joey carried only KoRV integrants present in either the sire or dam (showing that each KoRV-A was transmitted by inheritance, not by infection). Yet the unrelated sire and dam had only one integration site in common, supporting a high degree of insertional polymorphisms across unrelated koala individuals, with KoRV-A at any one genomic locus being shared by only a small proportion of koalas. Additional evidence also supports a recent invasion of the koala germ line by KoRV-A. In another experiment testing seven unrelated koalas, 429 5′ and 331 3′ distinct sequences were found to flank KoRV-A integration sites across the koalas, and 93% of these were unique to single individuals (219). A subsequent study examined KoRV-A flanking sequences across 10 museum koala specimens, identifying hundreds of integration sites (214). Again, in that study, more than 90% of these sites were unique to individuals, i.e., unshared by any of the other sampled members of the species (219, 220). It is not clear whether the small percentage of integrations shared across koalas may represent older integration events. The insertional polymorphisms among KoRV-A proviruses are especially consistent with a recent retroviral endogenization because no proviral integrations have yet become fixed in the koala. Had sufficient time passed since endogenization, the insertionally polymorphic KoRV-A loci would have either become fixed or been lost entirely from the population. For very ancient ERVs, the genomic locations of ERVs are fixed across all individuals in a lineage. Insertional polymorphisms in ERVs are thus an indication of the recency of ERV endogenization. There also appears to be a trend that sharing of integration sites among koalas is increasing with time, as more recently collected museum samples share more integration sites than previously collected samples (4). This is consistent with the expectation of the gradual loss or fixation of ERVs at different integration sites due to random genetic drift. Additionally, within each provirus there is an absence of mutational divergence between LTRs. Since the 5′ and 3′ LTRs are generated by duplication, they are identical within a provirus at the time of proviral integration. Divergence via mutation of the two LTRs of an integrated provirus can be used as a molecular clock to date the time since integration. In one study, KoRV-A sequences were determined for the 5′ and 3′ LTRs of the proviral integrants at 10 separate koala genomic loci. Within each integrant, the 5′ and 3′ LTRs were identical, indicating that these integrations occurred less than 22,200 to 49,900 years ago (6).
A final line of evidence that KoRV-A integration into the germ line is a recent event is provided by the prevalence and copy number of KoRV among koalas, which vary greatly with geographic origin (3). In northern Australia (Queensland and parts of New South Wales), KoRV prevalence is 100%, with ca. 165 copies of KoRV detected per cell (4). However, this may represent an overestimation, as the copy number of the recent koala genome assembly had only 58 full-length KoRVs and an unrelated koala comprehensively examined for integration sites had 66 (Rebecca N. Johnson, Denis O'Meally, Zhiliang Chen, Graham J. Etherington, Simon Y. W. Ho, Will J. Nash, Catherine E. Grueber, Yuanyuan Cheng, Camilla M. Whittington, Siobhan Dennison, Emma Peel, Wilfried Haerty, Rachel J. O'Neill, Don Colgan, Tonia L. Russell, David E. Alquezar-Planas, Val Attenbrow, Jason G. Bragg, Parice A. Brandies, Amanda Yoon-Yee Chong, Janine E. Deakin, Federica Di Palma, Zachary Duda, Mark D. B. Eldridge, Kyle M. Ewart, Carolyn J. Hogg, Greta J. Frankham, Arthur Georges, Amber K. Gillett, Merran Govendir, Alex D. Greenwood, Takashi Hayakawa, Kristofer M. Helgen, Matthew Hobbs, Clare Holleley, Thomas N. Heider, Elizabeth A. Jones, Andrew King, Danielle Madden, Jennifer A. Marshall Graves, Katrina M. Morris, Linda E. Neaves, Hardip R. Patel, Adam Polkinghorne, Marilyn B. Renfree, Charles Robin, Ryan Salinas, Kyriakos Tsangaras, Paul D. Waters, Shafagh A. Waters, Belinda Wright, Marc R. Wilkins, Peter Timms, and Katherine Belov, unpublished data; Ulrike Löber, Matthew Hobbs, Kyriakos Tsangaras, Kiersten Jones, David E. Alquezar-Planas, Yasuko Ishida, Joanne Meers, Jens Mayer, Anisha Dayaram, Claudia Quedenau, Wei Chen, Rebecca N. Johnson, Peter Timms, Paul Young, Alfred L. Roca, and Alex D. Greenwood, unpublished data). Some of the differences may be accounted for by the large number of structurally rearranged KoRVs that may still be detected as KoRV by quantitative PCR (qPCR) (306). In contrast, in southern Australia, and particularly on southern Australian islands, such as Kangaroo Island, the prevalence of KoRV is low (0 to 14%), and the detected copy number is also low (less than 1 copy, on average, per cell) (4). Such a result, particularly for an ERV, makes sense if KoRV-A first invaded the germ line of koalas in northern Australia, with only gradual gene flow to (or infection of) southern Australian koala populations. One would expect that, over time, the distribution of KoRV-A among koalas across Australia will gradually come to resemble that seen for species with ERVs that are more ancient. Genetic drift would be expected to remove some KoRV-A loci while fixing others. Gene flow across geographic regions would be expected to increase KoRV-A prevalence to the point that all Australian koala populations would become 100% positive for endogenous KoRVs. However, the number of eventual fixed KoRV loci in koalas would be gradually equalized to an intermediate level between the low number in southern Australia and the high number in northern Australia. Of course, this proposed scenario presumes the persistence of koalas and of gene flow across the koala range, which may be questionable.
The spread and evolution of endogenous KoRV have been slow processes, as established from historical DNA samples. Koalas were extensively hunted in the late 19th century and early 20th century, causing a large reduction in population that was particularly pronounced in southern Australia. Repopulation of southern Australia was heavily influenced by reintroduction of koalas to the mainland from populations established on Phillip, French, Raymond, and Kangaroo Islands (4), which have low KoRV prevalences. As these largely KoRV-free populations were established about a century ago, it was postulated that KoRV entered the koala population in the last 100 to 200 years. However, when 28 koala skin museum samples were assessed for the presence or absence of KoRV-A, the provirus was found to have already been ubiquitous in northern Australian koalas in the late 1800s, directly excluding a very recent origin (i.e., a few hundred years) for KoRV-A in koalas (221). The full envelope gene was sequenced from five museum samples of koalas that ranged in year of collection from 1870 to 1980. Surprisingly little variation (20 polymorphic sites) was identified among the KoRV-A sequences, regardless of the year of collection, suggesting that KoRV-A had changed little across more than a century. This is consistent with KoRV-A being endogenous and therefore subject to the low rate of mutation of the mammalian host genome and inconsistent with the high rate of mutation that would be expected for an exogenous gammaretrovirus due to the much higher effective population size and lack of DNA repair mechanisms of an XRV. To provide a more comprehensive analysis, full KoRV-A genomes were later sequenced from six historical koala specimens and one modern zoo koala, spanning 130 years (219). This identified 138 polymorphisms, among which 72 were detected in more than one of the koalas. There was evidence of purifying selection (removal of deleterious genetic material) acting on the envelope gene, although this likely reflects evolution of the virus before endogenization.
For retroviruses to manifest as they have in gibbons and koalas (i.e., as XRVs), they must emerge from another host source (i.e., a host vector). Determining this preceding host or reservoir species is a daunting task. No obvious characteristics of host ERVs (or, for that matter, XRVs) mark them as particularly likely to engage in cross-species transmission (211). However, genomic screening of 42 Australian vertebrate species identified partial retroviral gene sequences with homology to GALV in DNA from the rodent Melomys burtoni, a species endemic to Australia (222). However, as the full-length genome and integration sites for Melomys burtoni ERV (MbERV) have not yet been reported, it is unclear whether MbERV has exogenous potential or can be transmitted to other species. Phylogenetically, MbERV is sister to the woolly monkey virus (WMV), a basal member of the GALV clade. Recently, Alfano et al. screened 49 rodent samples belonging to 26 species from Southeast Asia, including Indonesia and Papua New Guinea, by using target enrichment hybridization capture and high-throughput sequencing (50). A subspecies of Melomys burtoni from one of the Maluku Islands (Moluccas) of Indonesia contained genomic sequences with 98% identity to WMV (a strain similar to but basal to GALVs). The virus, designated MelWMV, was fully sequenced, with a single integration site identified in 6 different individuals of the same species. The sequence contained a premature stop codon in the gag gene and a large deletion within pol. Thus, MelWMV is a GALV ERV in a subspecies of Melomys burtoni on an Indonesian island. The LTRs were identical, suggesting that it is also a relatively recent integrant (not older than 291,000 years) into the Melomys germ line.
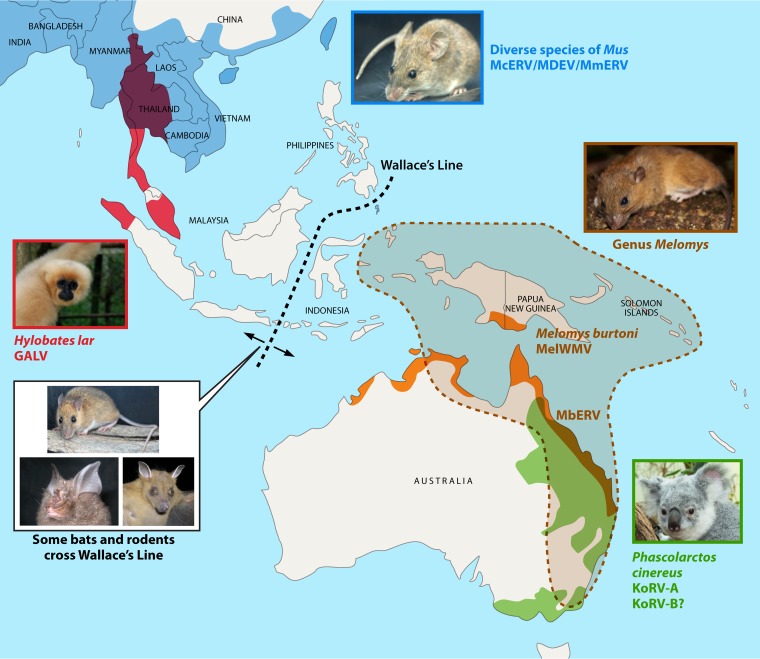
The two melomys GALVs identified suggest a complex picture for the origins of GALV and KoRV. Curiously, the habitats of gibbons and Melomys burtoni do not overlap. MbERV is in a species that is biogeographically separated from gibbons. Melomys species are not present west of Wallace's Line (a faunal boundary maintained by deep oceanic separation of islands) (223), while gibbons are not present east of Wallace's Line (Fig. 6). This appears to rule out a direct transmission from the Maluku melomys to gibbons, as the Indonesian subspecies of Melomys burtoni is not sympatric with gibbons. Regardless, the detection of GALVs in wild rodents suggests that they may have played a role in the transmission of GALV- and KoRV-like viruses. A tentative scenario based on current findings is that one or more melomys or related species, possibly no longer detectably extant, may have crossed Wallace's Line while carrying a virus similar to GALV. At some point, the virus infected gibbons, e.g., at the SEATO facility in Thailand. Alternatively, a GALV-infected rodent (or bat) from the Australo-Papuan region was imported into the SEATO facility. As more species are sampled, additional GALV- or KoRV-like viruses may be detected among the Southeast Asian or Australo-Papuan fauna. There are several dozen untested species of Melomys in this region, in addition to many other widely dispersed rodent and bat lineages that have crossed Wallace's Line.
FIG 6.
Geographic distributions of species harboring KoRV, GALV, and related retroviruses. Red shading indicates the historical distribution of the white-handed gibbon (Hylobates lar); five GALV strains have been isolated from this species. The range of several species of Mus (Mus cervicolor, Mus dunni, and Mus musculus), carrying McERV (296), MDEV (297), and MmERV (298), respectively, is indicated in blue; Mus was originally proposed as the source of GALV. The green region approximates the range of koalas (Phascolarctos cinereus) in eastern Australia, many of which carry KoRV-A. The distribution of KoRV-B among wild koalas has yet to be determined. Orange shading indicates the range of Melomys burtoni and the two GALVs identified in this species: MbERV and MelERV. The broken brown line and shading indicate the overall distribution of the genus Melomys. Wallace's Line (dashed black line) is a biogeographic (deep water) barrier separating the terrestrial fauna of Southeast Asia, including western Indonesia, from that of eastern Indonesia, New Guinea, and Australia. (Photographs courtesy of Serge Morand and CeroPath [rodents], Tilo Nadler [gibbon], Daniel Zupanc [koala], and Neil Furey [bats].)
Rodents and bats as potential recurrent vectors for retroviruses transmitted across species.
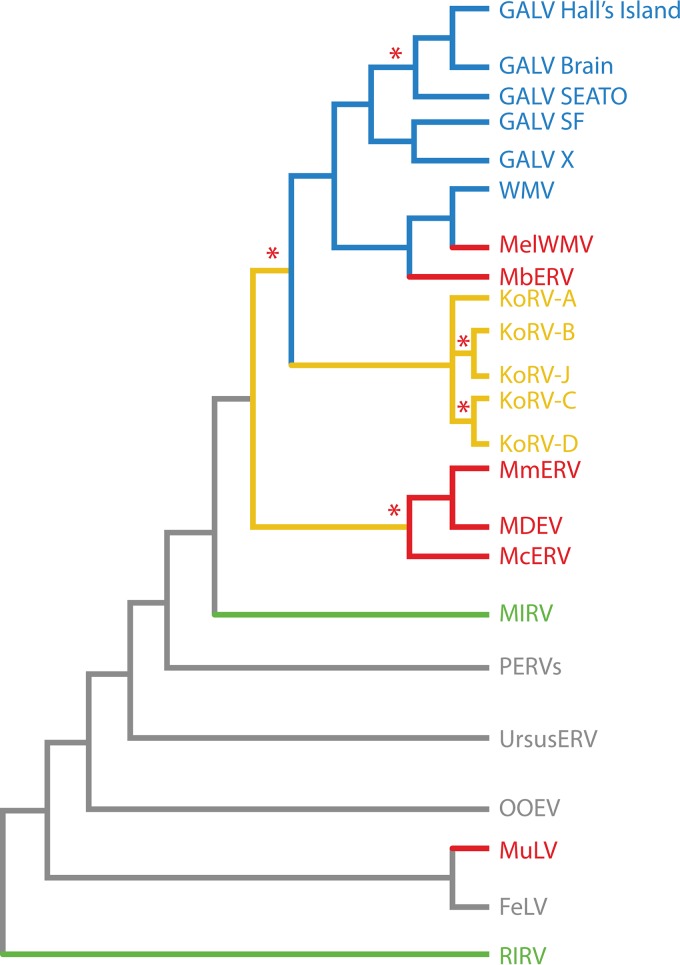
KoRV, GALV, and the ERVs found in Melomys form part of a clade that includes related ERV and XRV isolates from swine, bats, bears, orcas, felids, and humans, which is indicative of cross-species transmissions that occurred during the evolution and spread of gammaretroviruses among mammalian species (Fig. 7). There is evidence that the evolution of this viral genus has involved many bat lineages, as several gammaretroviral ERVs have been identified in bats. The bat viruses occupy basal positions in the phylogeny (Fig. 7), suggesting that some gammaretroviruses in other species may have originated in bats. However, a recent analysis of gammaretroviral diversity in a larger number of rodents and bats suggests that for ERVs in the bat Rhinolophus ferrumequinum, related gammaretroviruses in shrews were basal to those in bats (214). The study also suggests that bats and rodents generally harbor more retroviral diversity than other mammals and that rodents may have been the source of several retroviral lineages that infect bats. A series of studies found that, in general, rodents are the originators of class I retroviruses, and specifically, 9 of 14 class II retroviral clades identified previously (201) had rodent viruses in a basal position (213, 215, 224, 225). However, bats clearly have played a role in retroviral cross-species transmission, with phylogenomic analysis of ERV diversity across mammals placing bat as well as rodent viruses at basal positions in major retroviral clades (214, 225).
FIG 7.
Summarized phylogenetic relationships of KoRVs, GALVs, and related gammaretroviruses. Rodent retroviruses are shown in red, bat retroviruses are shown in green, retroviruses within the GALV clade are shown in blue, KoRVs are shown in yellow, and related gammaretroviruses from other taxa are shown in gray. Lineages under episodic diversifying selection within the GALV-like viruses are marked with red asterisks.
One hypothesis is that smaller mammals can tolerate higher levels of retroviral replication than those tolerated by larger mammals, which would explain why rodent and bat viruses occupy basal positions in so many retroviral clades (226). As retrotransposition is intrinsically mutagenic, there is an associated risk of oncogenesis with active retroviral replication. Smaller mammals tend to reproduce early and frequently and to produce large litters. In contrast, large mammals reproduce at older ages and are therefore under greater selective pressure against the development of cancers early in life. Such selective pressure would tend to make them relatively intolerant of retrotransposition. This could account for a relatively high diversity of ERVs in rodent and some bat species, since many species in these orders mature rapidly and reproduce at a relatively young age.
Given that gibbons are not likely to be the original source of GALVs, the natural strain diversity of GALV is probably undersampled. An increasing number of retroviruses with various degrees of similarity to GALV have been described for diverse mammalian taxa. For bats, two types of retroviruses homologous to GALV have been described. Rousettus leschenaultia retrovirus (RlRV) has been detected in the megabat Rousettus leschenaultia, which ranges across South Asia, southern China, and Southeast Asia (213). On phylogenetic trees, RlRV is present at a basal position (i.e., at the root or ancestral split) or groups with the porcine endogenous retrovirus (PERV) retroviral clade. RlRV and PERV are basal to the diverse clade that is formed by Mus caroli endogenous retrovirus (McERV), Mus musculus endogenous retrovirus (MmERV), Mus dunni endogenous retrovirus (MDEV), GALV, and KoRV (Fig. 7). Megaderma lyra retrovirus (MlRV) has been detected in the microbat Megaderma lyra, which is distributed across South Asia, southern and central China, and the Southeast Asian mainland. In phylogenies, MlRV either clusters with MDEV or is basal to MDEV and the other retroviruses in the diverse clade. The grouping of these gammaretroviruses coheres to some degree with biogeography, because the two bat species are found in Southeast Asia, and the region is also a center for swine biodiversity. Another mammalian lineage harboring homologous retroviruses is comprised of the ursine bears, including black (Ursus americanus), brown (Ursus arctos), and polar (Ursus maritimus) bears (227). UrsusERV is found in all ursine bears but not in pandas or tremarctine (spectacled) bears. Some copies of UrsusERV in polar bears appear to have intact open reading frames (ORFs) and are insertionally polymorphic within this species, suggesting a relatively recent origin for polar bear UrsusERVs. Based on gag and env sequences, UrsusERVs are basal to PERVs (Fig. 7). Placement on phylogenetic trees may be affected by recombination; for example, the pol gene sequences of McERV and MDEV place them in positions different from those seen with the gag and env sequences.
Our understanding of the evolutionary history of gammaretroviruses is affected by the availability of host genome sequences. For example, although the extant ursids are not a particularly species-rich group, full genomic screens for ERVs are possible for each of these species. In contrast, rodents and bats are the most species-rich mammalian orders (at least 2,268 and 1,133 species, respectively), but only a very small number of their species relative to the total have been screened for viruses. It seems very likely that as genomic sequences are generated for larger numbers of mammalian species, many additional gammaretroviruses (perhaps including GALV relatives) will be identified.
Identification of reservoir or intermediate host species is challenging, and in cases of ancient cross-species transmissions, it may be impossible if the original virus or viral host is extinct. This limitation to a comprehensive knowledge of mammalian-retroviral coevolution must be acknowledged, especially because frequent cross-species transmissions of ERVs (211, 228) and XRVs have occurred across a broad range of mammalian taxa. Insights into how retroviruses have spread may be provided by examining the diverse fates of retroviral invasions of the germ line in diverse taxa.
Activation of Established ERVs
One retrovirus, three fates.
Once a retrovirus becomes established in the germ line of a species, multiple fates await. A rigorous study of a single retroviral group (Myotis lucifugus ERV [MlERV] and related viruses) in three different taxa—bats, pangolins, and felids—demonstrated three distinct evolutionary trajectories (226). In the pangolin lineage, shortly after endogenization, the MlERV-like Manis pentadactyla ERV1 (MPERV) failed to proliferate substantially, generating only two ERV copies. In contrast, the feline MlERV-like virus FcERV_γ6 generated several hundred ERV copies that fell within a single viral clade and could be identified in domestic cats (Felis catus) and tigers (Panthera tigris) but not in nonfelid carnivores. Among bats, MlERVs are found in four vesper bat genera. The proliferation and diversification of MlERVs were more profound in bats than in the other lineages, with three distinct MlERV lineages identified and with hundreds of proviruses per clade. Few of the integration sites were shared among bat species, suggesting that MlERVs have repeatedly been infecting and/or intracellularly proliferating within the vesper bat lineage over millions of years, a process that may still be ongoing. Therefore, one retroviral group produced three different outcomes in mammals: limited proliferation in pangolins, clade-specific proliferation of a single viral type within felids, and repeated infection and proliferation among vesper bats.
env changes mediating ERV success.
It has been suggested that following endogenization, the ability of retroviruses to proliferate in the host germ line is enhanced by degradation of the env gene (229). Proviruses that maintain coding potential in all open reading frames tend to have lower copy numbers in the host germ line than those of proviruses that have degraded genomes. In particular, proviruses that lose most or all of the env region appear to become genomic superspreaders (229). This holds true for a variety of retroviral groups and follows the expected trajectory for an infection whereby 20% of the viral lineages account for 80% of the viral loci (Pareto effect). What is not clear is whether this process occurs early in endogenization or over time. Because most ERVs are quite ancient, the events that enabled and followed endogenization are difficult to infer (see above). However, the findings are consistent with the critical role that Env proteins play in retroviral function. The complete abrogation of Env function in many ERVs suggests that it is important to virus and host that the ability of ERVs to interact with the viral receptor be restricted.
In the case of intracisternal-A-particle-related envelope-encoding (IAPE) elements, proviral elements of betaretroviral origin, a mechanistic explanation has been proposed to account for the transition from an ERV with exogenous potential to an ERV that can proliferate endogenously (230). IAP elements, in contrast to IAPE elements, are a group of ERVs that typically contain LTRs and gag and pol genes but lack env (231). The IAPE elements are IAP-related elements that carry the env gene (232). A single endogenous “living fossil” copy of IAPE behaves as if it were an exogenous retrovirus in that it produces particles that can bud from the extracellular membrane. In contrast, other IAPE elements are present at high copy number but lack the ability to generate viral particles. Sequence comparisons between the two types of elements revealed that IAPE elements that are unable to bud from the cell membrane have acquired a sequence that targets them to the endoplasmic reticulum, which prevents them from budding at the cellular membrane and producing exogenous virus. Additionally, laboratory-generated IAPE mutants missing all or most of env would undergo retrotransposition, whereas the intact “living fossil” virus could not. Since only minor changes within the sequences coding for the matrix protein led to the dramatic switch to exclusive within-cell retrotransposition, this could occur quite rapidly from an evolutionary perspective. While IAPE elements are not gammaretroviruses, the rapid transition from conditions favoring exogenous states to those favoring endogenous states, mediated by relatively few genomic changes, may be generally applicable to retroviral endogenization. However, IAPE elements are millions of years old, like many known ERVs, and thus it is unclear which mutations occurred at which point during the endogenization process or when such dramatic switches occurred within different host and viral lineages. Such an analysis requires examination of more recently endogenized or currently endogenizing retroviruses (3).
Recombination and successful ERV invasion.
Postinvasion, there can be exchanges of envelope sequences among retroviruses (233). Surprisingly, such exchanges can involve retroviruses that affect taxonomically distant hosts. For example, the alpharetrovirus Tg-ERV-F, an ERV of songbirds, was found to have a mammalian gammaretrovirus-derived env gene (20). How this recombination occurred is not clear, but the acquisition of a gammaretroviral envelope may have aided in the spread of this alpharetrovirus among songbirds over the last 4 million years. Envelope swapping is likely common, as mammalian IAP element invasions in shrews and guinea pigs were preceded by an envelope switch that exchanged the IAP envelope with a betaretroviral env sequence (229). Another potential example is the similarity observed between MLV and HTLV Env SU domains (233). The structural and sequence identities of the SU domain between MLV and HTLV and the dissimilarity of all other retroviral sequences may be the result of an MLV env capture by HTLV. In the case of feline leukemia viruses (FeLVs), recombination between FeLV-A and endogenous FeLVs leads to the generation of FeLV-B retroviruses with altered cellular tropism (234). In the case of MLVs, endogenous ecotropic MLVs can be remobilized as exogenous MLVs by recombination with polytropic MLVs (235). The use of ERVs as reservoirs for acquisition of new env sequences by XRVs or the use of XRVs to remobilize ERVs can have a significant impact on the evolution of the recombinant exogenous retroviruses.
ERVs can be coopted by their hosts.
Another fate of ERVs is to be coopted or “domesticated” by the host for a variety of biological processes. For example, the LTRs of ERVs may alter gene expression of neighboring genes in ways useful to the host, as is the case for the human salivary amylase gene (236). Using transgenic mice, it was shown that the integrated ERVA1C element upstream of the AMY1C gene is sufficient to promote tissue-specific expression, suggesting that the ERV was coopted into modifying AMY1C expression during primate evolution. Many additional examples of such retroviral control of genes have been documented (237) and appear to be a general evolutionary benefit of retroviral colonization of germ lines.
However, coopting of LTR sequences to alter host gene expression is not the only phenomenon observed. Protein coding sequences of ERVs have also been coopted in mammalian evolution to perform crucial biological roles. One of the most intensively investigated domestication events involves syncytin genes. Initially, a HERV-W ERV env gene was identified as a gene encoding a crucial fusogenic protein involved in human placentation, and it is now designated syncytin 1 (238). HERV-FRD, a second domesticated human ERV with a similar function, has been designated syncytin 2. Domesticated ERVs with similar biological roles have also been identified in mice, rabbits, cattle, and sheep (239, 240). A surprising finding is that the coopted ERVs do not have a common origin, and thus endogenous retroviruses that integrated independently of each other in different mammalian lineages have been coopted for similar roles. Another recent example of ERV domestication involves a HERV-T Env protein that has been uniquely preserved during evolution. The Env protein can block a receptor (monocarboxylate transporter-1 [MCT-1]) that was used by the ancestor of HERV-T and prevent infection with an ancient HERV-T-pseudotyped virus by blocking the receptor on the cell surface (241). Thus, ERV proteins likely play a role in host defenses against retroviral infection.
Postendogenization silencing of ERVs.
Uncontrolled proliferation of ERVs in the germ line by retrotransposition must be blocked if the host lineage is to survive. Expression of ERVs is required for their retrotransposition, and therefore epigenetic modification of ERVs has been found to be important in controlling them. Studies of primate ERV expression, including expression of HERVs, indicate a complex and tissue-specific expression of ERVs (242, 243). However, not all ERVs are expressed, and many are silenced by CpG methylation. A study of porcine endogenous retroviruses (PERV-A and PERV-B) reported that the vast majority of proviruses in 15 different tissues were heavily methylated, resulting in reduced expression (244). PERV expression derives from only a few proviral insertions that are hypomethylated. The expression of these PERVs indicates that methylation control of ERVs is not absolute but that, nonetheless, methylation can substantially reduce the number of potentially active ERVs.
Methylation-based control is also determined by developmental stage. Some HERV-K elements (those of the HML-2 group) are insertionally polymorphic in humans (245). Some of the youngest of these elements can escape suppression by methylation during early embryogenesis and are expressed starting at the eight-cell stage (246). Expression of intracellular viral particles can be detected and HERV-K expression can inhibit viral infection in experimental systems. Therefore, some of the limited permissiveness of ERV expression may be tolerated because of benefits to the host, much like coopting gene products of ERVs that can serve host functional roles.
Repression of IAP elements involves both positive and negative epigenetic mechanisms whereby methylation of cytosines enhances and hinders, respectively, activation of a given IAP element (247). The role of methylation in silencing ERVs is not always a direct mechanistic way of preventing transcription factor access to viral LTRs or prevention of transcription of viral protein coding sequences. A recent study of DNA (cytosine-5)-methyltransferase 1 (Dnmt1) knockout embryonic stem cell lines (ESCs) suggested that additional factors are required to repress IAP element expression (247). Although hypomethylation of DNA occurs at IAP ERVs in Dnmt1 knockout mice, IAP elements are not strongly derepressed (247). The nuclear protein Np95 (Uhrf1) recruits Dnmt1 to hemimethylated DNA. In knockouts of Np95, hypomethylation occurs as in the Dnmt1 knockouts. Aberrant IAP element expression is observed; however, this is associated with accumulation of hemimethylated DNA. In a double knockout of Dnmt1 and Np95, IAP elements were not repressed, suggesting that Np95 is necessary for IAP element expression in hypomethylated DNA. In vivo studies of mouse embryos recapitulated this result, showing that Dnmt1 knockout mice had derepressed IAP elements, whereas Np95 or double knockout mice did not show derepressed IAP elements to the same degree. The hemimethylated DNA binding protein Setdb1 (SET domain, bifurcated 1) is able to suppress ERV expression itself, even in hypomethylated DNA. It appears that Np95 binding to IAP element DNA disrupts Setdb1 binding, leading to derepression of the IAP elements. The degree of disruption is largely IAP element specific, which can be explained by the unusually high CpG content of IAP elements relative to those of other ERVs. Moreover, there is evidence for a role of these proteins in repression and derepression of other ERVs. The example of IAP elements serves to show that methylation-based ERV suppression is intricate and complex.
Retroviral restriction genes.
Retroviruses are ancient and are estimated to be almost half a billion years old in vertebrates (248). This suggests that conflicts between retroviruses and their hosts have been ongoing throughout the evolution of vertebrate lineages. A result of this host-pathogen conflict has been the evolution of a suite of genes that hamper the replication of retroviruses. Many were discovered through the study of genetic variants in HIV-1 cohorts. However, HIV infections of humans are evolutionarily very recent (153), and antiretroviral genes evolved in response to repeated infections by XRVs and ERVs (249). The best-characterized antiretroviral genes include the apolipoprotein B mRNA editing enzyme catalytic polypeptide-like (APOBEC), tripartite motif containing 5α (TRIM5α), tetherin (BST2), and sterile alpha motif histidine-aspartic domain-containing protein 1 (SAMHD1) genes.
The APOBEC gene family varies among vertebrates. While primates have 11 APOBECs, more primitive vertebrates have fewer, and invertebrates have only one (250). Three of the APOBEC proteins within the APOBEC 3 group have retroviral restriction ability (APOBEC3G, -F, and -H). Of the three, APOBEC3G has the strongest restriction activity. APOBECs are copackaged in viral particles, bind to the polymerase during reverse transcription, and cause the deamination of cytosine, causing guanosine-to-adenosine hypermutation in the resulting viral DNA genomes that are integrated into the host genome. The same pattern is observed in a variety of endogenous retroviruses and other retroelements, such as LINES and SINES, that are also susceptible to restriction factors (249). This is evident as an excess of G-A transitions in ERV genomes. Of particular relevance to KoRV and koalas, the APOBEC 3 group originated after the marsupial and eutherian lineages diverged from a common ancestor, leaving marsupials without this retroviral restriction enzyme (251). This may partly explain the invasion of the koala genome by KoRV.
TRIM5α is also a post-viral-entry retroviral restriction protein. Although it is not entirely clear how TRIM5α inhibits retroviruses, it is known to bind the viral capsid postentry and results in the failure of cDNA production as a result of this binding (252). TRIM5α proteins of a given species are relatively poor at restricting viruses isolated from the host, most likely because the viruses that are isolated will be those most resistant to the host protein. However, TRIM5α proteins are likely to be formidable barriers to some retroviruses that attempt to cross species barriers. For example, although human TRIM5α is largely ineffective at restricting HIV-1, it is effective at inhibiting a revived chimpanzee ERV (PtERV1) (253).
Tetherin or BST2 restricts retroviruses postreplication by binding the cellular membrane and the lipid envelope of budding viral particles, preventing their release (252). This results in an accumulation of budding viral particles and subsequent sequestration in endosomes. While it is known as a retroviral restriction factor, BST2 can restrict other enveloped viruses as well.
A more recent discovery is SAMHD1, which can inhibit cDNA synthesis of HIV-1 when viral protein X (Vpx) is inhibited (254). Less is known about its ability to restrict the replication of endogenous retroviruses; however, it is likely that gammaretroviruses, such as MLV, are also blocked by SAMHD1 (255). More retroviral restriction genes are likely to be discovered, potentially including genes that inhibit the invasion of the germ line by novel ERVs. The long evolutionary conflict between retroviruses and vertebrates has led to the evolution in vertebrates of an effective set of countermeasures to retroviral infection and ERV proliferation.
CONCLUSIONS: RETROVIRUS-HOST COEVOLUTION
For the Orthoretrovirinae, the major determinant of transmission and, in many cases, of pathogenesis is the viral envelope. Envelope complementation and recombination events that occur among gammaretroviruses or between gammaretroviruses and alpharetroviruses, betaretroviruses, or deltaretroviruses have led to an assortment of retroviruses with altered receptor usage. Remarkably, this multiplicity of recombinant and nonrecombinant TM class 1 orthoretroviruses as well as the singular class 2 recombinant TgERV-F (20) (Fig. 4) all rely on a small number of receptors that share the ability to carry solutes across membranes (Fig. 8).
FIG 8.
Phylogeny of retroviral envelope sequences (left) and gene sequences (right) of the host receptors used by viruses. Phylogenies were inferred using the neighbor-joining method implemented in MEGA v7.0.18 (299). Human gene sequences were used to generate the tree of receptor gene sequences, with slc01a2 (28) used as the outgroup. ALV-J was used as the outgroup for the virus phylogeny, which includes the viruses listed in Table 2. Bootstrap values of ≥80% for 10,000 replicates are shown. Colored lines connect each virus to the gene for the receptor that it uses. GenBank accession numbers for gammaretroviruses are as follows: 10A1-MLV, M33470; A-MLV, AF411814; X-MLV, m59793; P-MLV, KJ668271; E-MLV, KJ668270; M813, AF327437; HEMV, AY818896; FeLV-B, K01209; FeLV-A, AF052723; FeLV-C, U58951; GALV, KT724048; KoRV-A, AF151794; KoRV-B, NC_021704; HERV-W, AH013146; REV, KU204702; RD-114, NC_009889; BaEV-M7, NC_022517; SRV, U85505; ALV-J, KP284572; BLV, M35242; and HTLV-I, AB600229. GenBank accession numbers for receptor genes (human gene product) are as follows: XPR1 (ND), NM_004736; slc20a1 (PiT-1), NM_005415; slc20a2 (PiT-2), NM_001257180; slc19a2 (THTR1), NM_006996; slc49a1-4 (FLVCR1), NM_014053; slc5a3 (SMIT1), NM_006933; slc7a1 (MCAT), NM_003045; slc1a5 (ASCT1), NM_005628; slc1a4 (ASCT2), NM_003038; slc01a2, NM_134431; and slc2a1, NM_006516.
Some 362 proteins (in humans) are encoded by the solute carrier (SLC) gene superfamily; these are organized into 55 families based on the substrates that they transport, e.g., fatty acids and lipids, amino acids, sugars, inorganic anions and organic anions, oligopeptides, essential metals, urea, vitamins, bile salts, and others (28). Yet among this vast array of transporters, retroviruses generally employ only those encoded by 11 genes (Fig. 8; Table 3) across species. For example, the bovine leukemia deltaretrovirus (BLV) (Ivanova et al., submitted) and the ecotropic murine retrovirus (E-MLV) (66) are in distinct retroviral genera, yet both use the cationic amino acid transporter as a receptor (Fig. 8; Table 3). Likewise, the SLC used by the recently endogenized KoRV-A, recombinant FeLV-B, and exogenous GALV and 10A1 retroviruses is encoded by slc20a1 (human designation). Thus, the SLC transporters shown in Fig. 8 have repeatedly been used by TM class 1 orthoretroviruses (Fig. 4).
The interplay between XRV, host, and ERV provides opportunities to examine very long-term and recurrent interactions among pathogens and their hosts. Viral envelope coevolution with a host receptor may occur in the absence or presence of a related ERV. Because this process occurs in many different lineages of virus and host yet typically involves the same small group of SLC receptors, a comparative approach holds promise for the study of virus-host coevolution and receptor-associated pathogenesis, especially if analogous changes are recurrent across lineages. The structural elements within the receptor binding domains of Env that allow independent (convergent or parallel) evolution may be identified by comparing otherwise unrelated viruses that use the same receptor; for example, FeLV-B, GALV, 10A1-MLV, and KoRV-A have all evolved to use PiT1 (27, 46). Likewise, receptors may show common elements; for example, the identified receptors for mammalian retroviruses that contain envelopes in which the TM domain is joined to the SU moiety via covalent disulfide linkage (Fig. 4) function as SLC transporters in their mammalian hosts (Table 3; Fig. 8).
Cross-species infections by retroviruses provide an additional level of complexity, because the viral envelope must adapt to the immune system of a novel host that may also have SLC transporters divergent in structure from those of the original host. A number of studies have successfully explored the temporal, geographic, and genetic requirements for expansion and restriction of viral host ranges and the balance between viral pathogenesis and propagation (17, 224, 257). The lentiviruses that gave rise to HIV recurrently crossed the species barrier from other species before one of the interspecies transfers gave rise to a pandemic in humans (164), and the GALV-like viruses only recently infected a number of gibbons and a woolly monkey. Signatures of episodic diversifying selection that may reflect adaptation to the receptors and immune systems of novel hosts have been detected in GALVs. There is now evidence that GALV is likely to have had an iatrogenic origin (204). Since iatrogenic transmission is also supported for REV (210), and potentially for RD-114 (45), the risk of such interspecies transfers should not be underestimated.
The dynamics of virus-host coevolution in some cases extend across vast periods. Genomic studies can identify endogenous retroviruses, which have been referred to as molecular fossils, since they record events that occurred in the distant past. Like fossils, ERVs provide only a very limited record of the history of retroviruses. Only a minuscule proportion of exogenous retroviruses would have invaded host germ lines, while the genomic record of invasions is constantly being eroded by mutations or recombination in host genomes. Nonetheless, information about past retrovirus-host interactions can be obtained by using currently available genomic methods. The cataloging of retroviruses into taxonomic categories should also be useful in providing a broad systematic overview of retroviral relationships and evolution (Table 1). Phylogenetic studies have identified a strong role for recombination in retroviral evolution (218), with chimeric subgenomic components showing differences in evolutionary rates (20). Rates of mutation are also much lower among ERVs than among XRVs, since the former are subject to the lower evolutionary rates of host genomes.
Most retroviral studies have been conducted on humans, model organisms, and agricultural animals. Additional insights into the biogeography and interspecies transfer of retroviruses can be provided by comparative studies involving species not typically utilized for biomedical research. The clade containing GALVs, KoRV, and related retroviruses now comprises an important system for examining the evolution and biogeography of retroviral interspecies transmission (Fig. 7). Retroviruses in this clade have been detected among koalas (KoRV) and rodents of the genus Melomys (MelWMV and MbERV) in the Australo-Papuan biogeographic region (Fig. 6). Related viruses have been found in Southeast Asian taxa (Fig. 6), including gibbons (GALV), mice of the genus Mus (MmERV, MDEV, and McERV), bats (MIRV), and pigs (PERVs). The phylogeography of this clade indicates that interspecies transfers of retroviruses have occurred within each biogeographic region as well as between the two biogeographic regions. As genomic sequences are generated for larger numbers of host species, many additional gammaretroviruses will be identified, perhaps including additional members of this clade. Greater insights into viral transmission across species within these biogeographic regions or between the two biogeographic regions may result. The degree to which retroviral clades are biogeographically constrained or widespread would thus be further elucidated. Phylogenetic analyses of currently identified retroviruses have often placed rodents or bats in basal positions (214, 225). Additional genome sequences would help to establish whether bat and rodent retroviruses play an outsized role in the dynamics of retroviral transmission across species within and (especially for bats) across biogeographic regions.
Following endogenization, ERVs are known to face a variety of fates in their host germ lines. They may lose infectivity through disruptive mutations, recombination (often within the same ERV provirus), methylation, and other host defenses (258). Some integrants have the potential to be disruptive and detrimental to the host (259). LTRs may function as promoter elements that can affect nearby host genes (228). ERVs may affect exogenous retroviruses through receptor interference and through recombination (2). Some ERVs have been coopted or “domesticated” for a functional role benefiting the host lineage (260). Thus, ERVs affect host evolution while also shaping future interactions between the host and XRVs. Genomic approaches comparing retroviruses across taxa provide additional insights. For example, examination of MlERV-like ERVs across mammals revealed different outcomes in each of three taxa: limited proliferation in pangolins, proliferation of a single viral type within felids, and repeated infection and proliferation among vesper bats (226). Likewise, examination of ERVs across 38 taxa revealed that the loss of the env gene was associated with a greater proliferation of ERVs in host germ lines (229). The characterization of syncytin genes in various mammals has shown that these domesticated ERVs do not have a common origin, and thus endogenous retroviruses representing independent integrations in different mammalian lineages have been coopted for similar functions (261–263). Identification of syncytins across a broader array of mammalian species may identify how different ERVs affect placental formation and chronicle the appearance and turnover of domesticated ERVs. Other host functions performed by coopted ERVs may also be revealed through genomic screening.
Comparative studies are vital for understanding ERV-like elements, which comprise 8% of the human genome, representing many distinct retroviral invasions of the germ line (260). KoRV is especially noteworthy due to the relatively recent invasion of the koala germ line by this ERV (264). Koalas and KoRV provide a system for examining ERV-host interactions at the earliest stages of endogenization, and in particular the roles of population genetic processes, such as selection, drift, mutation, population structure, gene flow, and recombination. By use of museum samples, KoRV proviral genomes have been shown to remain conserved for many generations (219). The relatively low evolutionary rates of the host may hinder mutualistic accommodation between KoRVs and koalas, causing the detrimental effects of ERVs on hosts to persist for extended periods. KoRVs in the koala germ line are found at a very large number of genomic loci, each at a very low frequency, likely producing a high mutational load in host populations (6). Thus, the many retroviral germ line invasions detected in the human lineage may each have produced deleterious effects for long periods. As genomes are sequenced for additional species, additional ERV invasions may be identified that are occurring in “real time,” allowing for determination and comparisons across species of the earliest events that follow retroviral endogenization.
ACKNOWLEDGMENTS
A.D.G., Y.I., and A.L.R. were supported by grant R01GM092706 from the National Institute of General Medical Sciences (NIGMS). S.P.O. was supported by federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E, and M.V.E. was supported by the NIMH intramural program.
The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
APPENDIX
Methods for Genome-Wide Detection of ERVs
This appendix describes methods that can be used to obtain information on ERVs across the genomes of host species. There are several challenges associated with characterizing ERVs genome-wide, notably the high sequence similarity of proviruses at different genomic locations, mutations and divergence of individual proviral sequences, and the high copy numbers of many sequences. Approaches to these challenges depend on the question being addressed, such as the following. Is a specific ERV present in a given species? What is the full-length proviral sequence(s)? Where and how often is each ERV integrated into the genome? Each of these questions can be addressed by relying on different methodologies (Fig. A1).
FIG A1.
Methods used to determine whether a given sequence is present, to characterize full genomes by hybridization capture (HC), and to characterize ERV integration sites by inverse or linker-based PCR approaches. (A) Southern blotting and pan- or broad-specificity PCRs are shown as examples of low-throughput methods. Southern blotting will provide information on whether homologous sequences are present in a given DNA extract but not whether the sequences are closely related, divergent, or a mixture of both. PCR-based approaches using conserved retroviral gene regions can produce limited sequence information from single or low-copy-number ERVs. However, if a high-copy-number ERV that has diversified is being amplified, amplification may fail for some loci due to primer mismatch, while the amplification of many loci at once will result in many polymorphisms at different positions in the sequence. Cloning and sequencing of the PCR product would be needed to sequence distinct ERVs. To determine sequence diversity comprehensively, high-throughput sequencing (HTS) methods must be used. (B) Hybridization capture. HTS libraries are built from fragmented genomic DNA or cDNA. The sequencing platform employed is largely irrelevant if the correct sequencing platform adaptors are used in building the libraries (and correct blocking oligonucleotides are used during capture). Baits for enriching the target sequence of interest can be generated by PCR, generally involving large amplicons (depending on the target) and fragmenting them to avoid having baits that are much larger than target library molecules (300). Alternatively, the baits can be synthesized much like the synthesis of PCR primers and can be made of DNA or RNA. Recently, entire genomes have been used to enrich highly contaminated samples for genomic DNA, for example, using elephant DNA to enrich woolly mammoth DNA from fossils (301). In some cases, microarrays, such as virus microarrays, may be used as baits (302, 303). In the figure, solution-based hybridization capture is shown. Regardless of strategy, the resulting captured library is massively enriched for target sequences of interest, while nontarget sequences are washed away prior to HTS. In addition to target sequence enrichment, the sequences flanking the target will also be captured as library sequences extending just beyond the end of the bait or even further are also captured by the annealing of overlapping sequences, or “CapFlank” (219). (C) Additional methods for enriching ERV integration sites. By fragmenting genomic DNA, circularizing the fragments in a ligation step, and then applying inverse PCR to the resulting circular DNA, one can extend known sequences into flanking unknown sequences based on PCR primers anchored entirely within the known sequence (inverse PCR). Linker-based approaches use a somewhat similar strategy; however, instead of using two primers based on a known sequence, a known linker sequence is ligated to the ends of fragmented DNA, and one PCR primer is located on the linker sequence, extending across the unknown sequence toward the other primer, which is based on the known sequence. Sequencing of the amplicons can then be performed by low- or high-throughput methods.
PCR and low-throughput methods for ERV detection.
PCR based on conserved retroviral domains has been successful in identifying many viruses, but it has drawbacks. Only fragmentary information is generated, and the products may represent a mixture of ERVs that does not cover the complete, diverse population of high-copy-number viruses when cloned and subsequently sequenced. This is not a trivial issue, as it may be difficult or impossible to extend such short sequences by PCR to recover full-length proviruses. This is illustrated by the Melomys burtoni ERV, MbERV, which is represented by 2 PCR products. Twenty-two primers were developed to amplify the provirus, among which 18 failed to yield a product, and thus the full genome could not be completed (222). While the pol gene is relatively useful for primer design, primers designed to amplify conserved regions may also amplify a wide variety of related retroviruses, while the extent of sequence divergence in other parts of the retroviral genome precludes the design of comprehensive pan-retroviral PCR primers.
An additional drawback to PCR-based approaches for identifying retroviruses is the sensitivity of PCR to reagent contamination, particularly in the case of poorly characterized species. The xenotropic murine retrovirus-like retrovirus (XMRV) was first identified in human prostate cancer patients and subsequently found at a high prevalence in human patients suffering from chronic fatigue syndrome (265, 266). In both instances, it was postulated that rodent-to-human cross-species transmission of XMRV was the underlying causal factor leading to clinical symptoms. However, these findings were later found to be spurious and due in part to contamination by a recombinant retrovirus derived from mouse cell lines used for culture of human prostate tumors (267) or due to the high frequency of low levels of contaminating rodent DNA, including rodent ERVs, in commercially available kits for reverse transcription, DNA and RNA extraction, and Taq DNA polymerase (268–272). Xenotransplantation that includes grafting of human and murine tissues can also lead to the acquisition and propagation of MLVs (273). The origins of the various MLV contaminations are not entirely clear but strongly suggest that when employing conserved PCR primers to identify novel retroviruses, one must be extremely cautious in interpreting the origins of the amplified fragments.
Proviral genome characterization.
Full-genome proviral characterization is more difficult than determining a fragment of a novel provirus, particularly if it is present at a high copy number. The simplest method, yet the rarest employed because of cost and difficulty, is high-coverage sequencing of a given genome in large fragments, for example, by using PacBio long sequence reads. Each integrant may be sequenced independently and the unique integration site linked to a specific provirus. If sequenced to a deep enough depth, rarer XRV somatic integrations will also be detected. Although sequencing costs continue to rapidly decline and bioinformatics pipelines have been developed to assist in assembling genomes, the cost and analytical effort are still beyond the ability of most individual laboratories. Thus, most of such available data has been the by-product of sequencing projects, such as the Human Genome Project or the Genome 10K Project (224).
High-throughput sequencing (HTS) demonstrated a great deal of KoRV variation and variants that were not detectable using low-throughput approaches. For example, Ávila-Arcos et al. (221) used 30 primer pairs spanning the KoRV envelope gene to amplify the entire gene from 5 koala museum skins in a multiplex PCR by using a protocol applied to sequencing of woolly mammoth mitochondrial DNA genomes (274, 275). The 30 primers were pooled into “odd” and “even” groups which did not overlap, multiplex PCR was performed for 25 cycles, and then the individual PCR products were amplified from the initial multiplex reaction mixture. The products were sequenced on a Roche 454 platform. This resulted in full coverage of the envelope gene and the identification of several novel KoRV polymorphisms spanning a 120-year span. Although a multiplex approach allows for the determination of more sequence from a limited amount of material, it suffers from the same drawbacks as those of individual pan-retroviral PCRs, including the inability to place polymorphisms in phase if they do not occur within the single sequenced PCR amplicon. Nonetheless, the results did indicate that while KoRV is not unusually diverse among individual proviral sequences, at least for full-length KoRV-A envelope-containing sequences, there is variability not observed in the original PCR-based approaches for determining the genome sequence.
Target enrichment by HC.
Both the host mitochondrial and non-ERV nuclear genome sequences vastly outnumber any given ERV sequence. The use and drawbacks of PCR to overcome this are discussed above. Recently, another approach has been successful for post-DNA-extraction enrichment of viral sequences in a variety of contexts. Targeted-enrichment hybridization capture (HC) followed by HTS was developed to isolate and characterize specific genetic regions from genomic DNA (276–278). It was quickly modified for use in fields as diverse as biomedicine and ancient DNA (279). The first application of HC to virology was the examination of integration sites of the Merkel cell polyomavirus from formalin-fixed tissues (280), which provided an unprecedented level of sequence coverage for the 5.3-kb genome of this virus and identified most, but not all, of the 5′ and 3′ viral integration sites. Using similar methods on DNA extracts from museum skins (219) and a modern zoo koala DNA extract, the full proviral genomes of KoRV for 7 koalas were determined. Coverage depth for each koala sample was high, revealing substantial KoRV-A diversity not detected using the multiplex PCR approach employed previously. In addition to the full KoRV genomes, 429 5′ integration sites and 331 3′ integration sites were also captured and sequenced. This occurred even though the flanking regions were not part of the bait used to target KoRV. The explanation turned out to be a phenomenon, designated “capturing flanking sequences” (CapFlank), by which target library molecules bound to immobilized baits draw additional molecules to the bound bait-target molecule and generate concatemers (219). Recently, HC was applied to determine the full-length GALV genomes for the original GALV isolates (X, SEATO, Brain, SF, and WMV) (192). Similarly, a novel GALV was identified by HC for a subspecies of Melomys burtoni from Indonesia and was termed MelWMV (50). The virus was recalcitrant to PCR amplification but was easily assembled by HC, using GALV and KoRV sequences as baits. As for KoRV subjected to HC (219), the integration sites of MelWMV were also identified despite not being part of the bait sequence (50). The general benefits of HC are as follows: entire viral genomes can be determined in a single experiment; baits are inexpensive and easy to make, particularly if they are PCR products; it is scalable such that, if libraries are barcoded, dozens can be processed in an individual HC experiment; and HC tolerates substantial mismatches between bait and target. Mismatches can be substantial and still lead to successful capture. For example, in the work of Alfano et al. (50), species of Mus negative for KoRV and GALV yielded coverage of viruses belonging to the sister clade of this viral group, including MDEV, McERV, and MmERV, which are divergent from KoRVs and GALVs. Therefore, little prior knowledge of the target viral sequence is required to characterize novel sequences, but the specificity is high enough not to overwhelm the output sequences with distantly related viruses; for example, MLVs from Mus were not captured at high frequencies with the GALV and KoRV baits (50).
The utility of HC applies to characterization of integration sites as well. Integration site analysis presents different challenges because the target is unknown. Thus, other than certain types of sequencing, e.g., sequencing of bacterial artificial chromosome (BAC) clones or PacBio sequencing of long fragments, methods must rely on anchoring strategies based on the viral sequence, notably the LTRs. Inverse PCR is a common strategy for identifying integration sites and is particularly suitable in cases where the DNA quality is high and the ERV copy number is low (281). Other methods that have been applied to ERV integration site characterization have been rapid amplification of cDNA ends (RACE), ligation-mediated PCR, linker selection-mediated PCR, linear amplification-mediated PCR, and genome walking (282–286). Each of these methods depends on using a known sequence, such as the LTR, to prime an extension or a PCR into the unknown flanking sequence and adding a reverse sequence, such as ligation of a linker sequence, which makes it possible to amplify across the unknown sequence by using the known anchor sequence. Recently, three methods for identifying KoRV integration sites from museum samples were compared that could, in principle, be applied to modern DNA as well. HC, primer extension capture (PEC), and single primer extension (SPEX) were compared based on their performances in identifying KoRV integration sites (220). HC methods performed best in retrieving integration sites. However, despite the better performance of the capture-based approaches, none comprehensively retrieved all integration sites, as suggested by the uneven number of total 5′ and 3′ flanking sites retrieved, which should, in principle, be the same if all KoRVs had been identified. The samples tested were museum specimens, and it is possible that modern DNA may perform better in terms of comprehensive analysis using capture-based approaches.
Bioinformatics methodology.
Sifting through complex HTS data also presents challenges. Mapping of HTS reads to a reference genome is reasonably straightforward. However, identification of polymorphisms can be challenging if DNA damage or sequencing errors are present (287). However, polymorphisms have been confirmed by examining historical DNA-specific nucleotide misincorporation patterns (287), and statistical modeling of the patterns has been applied by using bioinformatics tools to compute and identify the misincorporation patterns from HTS reads (288, 289). In some instances of using a bait to capture divergent ERVs, the presence of nonhomologous regions may yield a sequence with substantial numbers of gaps. One approach that has been applied successfully is to use an iterative baiting and mapping approach called Mitobim (290). In effect, if a sequence read maps to a given reference, the algorithm identifies sequences that partially overlap the mapped sequence and extend it in both directions. The more extended the reads, the longer is the consensus sequence that is built. While originally applied to mapping mitochondrial DNA genomes, the method was shown to be useful for extending divergent foamy virus-related ERVs from modern and extinct sloths (291). The resulting extended consensus sequences allowed for the reconstruction of divergent mitochondrial and ERV sequences from samples of highly variable quality and average library sequence length, with the ancient DNA samples having very short library inserts.
Integration site analysis is also quite challenging. This is facilitated if a reference genome has an ERV on one of the chromosomes but retains the proviral sequence in the other chromosome, in which the 5′ and 3′ integration sites will be adjacent to one another. A variety of bioinformatics packages exist for mapping retroviral integrations if a reference sequence is available, using different algorithms. For example, SLOPE, VirusFinder, and VirusSeq have all been applied successfully (280, 292, 293). Lack of a quality reference genome sequence is a serious impediment to identifying integration sites. For example, koala sequences queried against wallaby sequences often show a very poor match (219). Cui et al. and Ishida et al. (220, 294) attempted to overcome these problems by using a sequence clustering approach for defining integration sites independent of a reference sequence (220). Sequences were first identified that contained both the host genome flanking the ERV and the viral LTR. Sequences were then trimmed to remove the LTR to avoid clustering based on the identity of the LTR. The LTR-less host genome sequences were clustered, and consensus sequences were determined separately for the 5′ and 3′ integrations. By mapping to the wallaby or koala genome, the matching 5′ and 3′ flanks at a single proviral locus were detected in many cases. Similar methods have been applied to viral integrations in carnivores (12, 295).
REFERENCES
- 1.Johnson WE. 2015. Endogenous retroviruses in the genomics era. Annu Rev Virol 2:135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- 2.Weiss RA. 2013. On the concept and elucidation of endogenous retroviruses. Philos Trans R Soc Lond B Biol Sci 368:20120494. doi: 10.1098/rstb.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu W, Eiden MV. 2015. Koala retroviruses: evolution and disease dynamics. Annu Rev Virol 2:119–134. doi: 10.1146/annurev-virology-100114-055056. [DOI] [PubMed] [Google Scholar]
- 4.Simmons GS, Young PR, Hanger JJ, Jones K, Clarke D, McKee JJ, Meers J. 2012. Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust Vet J 90:404–409. doi: 10.1111/j.1751-0813.2012.00964.x. [DOI] [PubMed] [Google Scholar]
- 5.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 6.Ishida Y, Zhao K, Greenwood AD, Roca AL. 2015. Proliferation of endogenous retroviruses in the early stages of a host germ line invasion. Mol Biol Evol 32:109–120. doi: 10.1093/molbev/msu275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Stadler CK, Gorman K, Jensen N, Kim D, Zheng H, Tang S, Switzer WM, Pye GW, Eiden MV. 2013. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A 110:11547–11552. doi: 10.1073/pnas.1304704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waugh CA, Hanger J, Loader J, King A, Hobbs M, Johnson R, Timms P. 2017. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci Rep 7:134. doi: 10.1038/s41598-017-00137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bannert N, Fiebig U, Hohn O. 2010. Retroviral particles, proteins and genomes, p 71–106. In Kurth R, Bannert N (ed), Retroviruses. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 10.Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 11.Yu SF, Sullivan MD, Linial ML. 1999. Evidence that the human foamy virus genome is DNA. J Virol 73:1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown K, Emes RD, Tarlinton RE. 2014. Multiple groups of endogenous epsilon-like retroviruses conserved across primates. J Virol 88:12464–12471. doi: 10.1128/JVI.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkasova H, Hron T, Paces J, Hulva P, Benda P, Gifford RJ, Elleder D. 2017. Discovery of an endogenous deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae). Proc Natl Acad Sci U S A 114:3145–3150. doi: 10.1073/pnas.1621224114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClure MA, Johnson MS, Feng DF, Doolittle RF. 1988. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci U S A 85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jern P, Sperber GO, Blomberg J. 2005. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology 2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benit L, Dessen P, Heidmann T. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J Virol 75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 18.Bolisetty M, Blomberg J, Benachenhou F, Sperber G, Beemon K. 2012. Unexpected diversity and expression of avian endogenous retroviruses. mBio 3:e00344-12. doi: 10.1128/mBio.00344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henzy JE, Coffin JM. 2013. Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class. J Virol 87:1937–1946. doi: 10.1128/JVI.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henzy JE, Gifford RJ, Johnson WE, Coffin JM. 2014. A novel recombinant retrovirus in the genomes of modern birds combines features of avian and mammalian retroviruses. J Virol 88:2398–2405. doi: 10.1128/JVI.02863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henzy JE, Johnson WE. 2013. Pushing the endogenous envelope. Philos Trans R Soc Lond B Biol Sci 368:20120506. doi: 10.1098/rstb.2012.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cianciolo GJ, Bogerd HP, Kipnis RJ, Copeland TD, Oroszlan S, Snyderman R. 1985. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to envelope proteins of human and animal retroviruses. Trans Assoc Am Physicians 98:30–41. [PubMed] [Google Scholar]
- 23.Blaise S, Mangeney M, Heidmann T. 2001. The envelope of Mason-Pfizer monkey virus has immunosuppressive properties. J Gen Virol 82:1597–1600. doi: 10.1099/0022-1317-82-7-1597. [DOI] [PubMed] [Google Scholar]
- 24.Mangeney M, de Parseval N, Thomas G, Heidmann T. 2001. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J Gen Virol 82:2515–2518. doi: 10.1099/0022-1317-82-10-2515. [DOI] [PubMed] [Google Scholar]
- 25.Mangeney M, Heidmann T. 1998. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci U S A 95:14920–14925. doi: 10.1073/pnas.95.25.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlecht-Louf G, Renard M, Mangeney M, Letzelter C, Richaud A, Ducos B, Bouallaga I, Heidmann T. 2010. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc Natl Acad Sci U S A 107:3782–3787. doi: 10.1073/pnas.0913122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overbaugh J, Miller AD, Eiden MV. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol Mol Biol Rev 65:371–389. doi: 10.1128/MMBR.65.3.371-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Vasiliou K, Nebert DW. 2009. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics 3:195–206. doi: 10.1186/1479-7364-3-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins JF, Bai L, Ghishan FK. 2004. The SLC20 family of proteins: dual functions as sodium-phosphate cotransporters and viral receptors. Eur J Physiol 447:647–652. doi: 10.1007/s00424-003-1088-x. [DOI] [PubMed] [Google Scholar]
- 30.O'Hara B, Johann SV, Klinger HP, Blair DG, Rubinson H, Dunn KJ, Sass P, Vitek SM, Robins T. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ 1:119–127. [PubMed] [Google Scholar]
- 31.van Zeijl M, Johann SV, Closs E, Cunningham J, Eddy R, Shows TB, O'Hara B. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci U S A 91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kavanaugh MP, Miller DG, Zhang W, Law W, Kozak SL, Kabat D, Miller AD. 1994. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci U S A 91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olah Z, Lehel C, Anderson WB, Eiden MV, Wilson CA. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem 269:25426–25431. [PubMed] [Google Scholar]
- 34.Wilson CA, Eiden MV, Anderson WB, Lehel C, Olah Z. 1995. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol 69:534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johann SV, Gibbons JJ, O'Hara B. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol 66:1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller AD. 1996. Cell-surface receptors for retroviruses and implications for gene transfer. Proc Natl Acad Sci U S A 93:11407–11413. doi: 10.1073/pnas.93.21.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eiden MV, Taliaferro DL. 2011. Emerging retroviruses and cancer, p 307–333. In Dudley J. (ed), Retroviruses and insights into cancer. Springer, New York, NY. [Google Scholar]
- 39.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 40.Wilson CA, Farrell KB, Eiden MV. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol 68:7697–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller DG, Miller AD. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol 68:8270–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolin LL, Levy LS. 2011. Viral determinants of FeLV infection and pathogenesis: lessons learned from analysis of a natural cohort. Viruses 3:1681–1698. doi: 10.3390/v3091681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasheed S, Pal BK, Gardner MB. 1982. Characterization of a highly oncogenic murine leukemia virus from wild mice. Int J Cancer 29:345–350. doi: 10.1002/ijc.2910290319. [DOI] [PubMed] [Google Scholar]
- 44.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to gibbon ape leukemia virus. J Virol 74:4264–4272. doi: 10.1128/JVI.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazawa T, Yoshikawa R, Golder M, Okada M, Stewart H, Palmarini M. 2010. Isolation of an infectious endogenous retrovirus in a proportion of live attenuated vaccines for pets. J Virol 84:3690–3694. doi: 10.1128/JVI.02715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J Virol 80:3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck L, Leroy C, Beck-Cormier S, Forand A, Salaun C, Paris N, Bernier A, Urena-Torres P, Prie D, Ollero M, Coulombel L, Friedlander G. 2010. The phosphate transporter PiT1 (Slc20a1) revealed as a new essential gene for mouse liver development. PLoS One 5:e9148. doi: 10.1371/journal.pone.0009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen N, Schroder HD, Hejbol EK, Fuchtbauer EM, de Oliveira JR, Pedersen L. 2013. Loss of function of Slc20a2 associated with familial idiopathic basal ganglia calcification in humans causes brain calcifications in mice. J Mol Neurosci 51:994–999. doi: 10.1007/s12031-013-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alfano N, Michaux J, Morand S, Aplin K, Tsangaras K, Lober U, Fabre PH, Fitriana Y, Semiadi G, Ishida Y, Helgen KM, Roca AL, Eiden MV, Greenwood AD. 2016. Endogenous gibbon ape leukemia virus identified in a rodent (Melomys burtoni subsp.) from Wallacea (Indonesia). J Virol 90:8169–8180. doi: 10.1128/JVI.00723-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eglitis MA, Eiden MV, Wilson CA. 1993. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J Virol 67:5472–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneiderman RD, Farrell KB, Wilson CA, Eiden MV. 1996. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol 70:6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaudry GJ, Farrell KB, Ting YT, Schmitz C, Lie SY, Petropoulos CJ, Eiden MV. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J Virol 73:2916–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eiden MV, Farrell KB, Wilson CA. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol 70:1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giovannini D, Touhami J, Charnet P, Sitbon M, Battini JL. 2013. Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep 3:1866–1873. doi: 10.1016/j.celrep.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 56.Battini JL, Rasko JE, Miller AD. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci U S A 96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci U S A 96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang YL, Guo L, Xu S, Holland CA, Kitamura T, Hunter K, Cunningham JM. 1999. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat Genet 21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- 59.Legati A, Giovannini D, Nicolas G, Lopez-Sanchez U, Quintans B, Oliveira JR, Sears RL, Ramos EM, Spiteri E, Sobrido MJ, Carracedo A, Castro-Fernandez C, Cubizolle S, Fogel BL, Goizet C, Jen JC, Kirdlarp S, Lang AE, Miedzybrodzka Z, Mitarnun W, Paucar M, Paulson H, Pariente J, Richard AC, Salins NS, Simpson SA, Striano P, Svenningsson P, Tison F, Unni VK, Vanakker O, Wessels MW, Wetchaphanphesat S, Yang M, Boller F, Campion D, Hannequin D, Sitbon M, Geschwind DH, Battini JL, Coppola G. 2015. Mutations in XPR1 cause primary familial brain calcification associated with altered phosphate export. Nat Genet 47:579–581. doi: 10.1038/ng.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shojima T, Yoshikawa R, Hoshino S, Shimode S, Nakagawa S, Ohata T, Nakaoka R, Miyazawa T. 2013. Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J Virol 87:9943–9948. doi: 10.1128/JVI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ericsson TA, Takeuchi Y, Templin C, Quinn G, Farhadian SF, Wood JC, Oldmixon BA, Suling KM, Ishii JK, Kitagawa Y, Miyazawa T, Salomon DR, Weiss RA, Patience C. 2003. Identification of receptors for pig endogenous retrovirus. Proc Natl Acad Sci U S A 100:6759–6764. doi: 10.1073/pnas.1138025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yonezawa A, Masuda S, Katsura T, Inui K. 2008. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am J Physiol Cell Physiol 295:C632–C641. doi: 10.1152/ajpcell.00019.2008. [DOI] [PubMed] [Google Scholar]
- 64.Ribet D, Harper F, Esnault C, Pierron G, Heidmann T. 2008. The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J Virol 82:4413–4419. doi: 10.1128/JVI.02141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reference deleted.
- 66.Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 67.Sutcliffe JG, Shinnick TM, Verma IM, Lerner RA. 1980. Nucleotide sequence of Moloney leukemia virus: 3′ end reveals details of replication, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A 77:3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim JW, Closs EI, Albritton LM, Cunningham JM. 1991. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 69.Yoshimoto T, Yoshimoto E, Meruelo D. 1991. Molecular cloning and characterization of a novel human gene homologous to the murine ecotropic retroviral receptor. Virology 185:10–17. doi: 10.1016/0042-6822(91)90748-Z. [DOI] [PubMed] [Google Scholar]
- 70.Yan Y, Kozak CA. 2008. Novel postentry resistance to AKV ecotropic mouse gammaretroviruses in the African pygmy mouse, Mus minutoides. J Virol 82:6120–6129. doi: 10.1128/JVI.00202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reference deleted.
- 72.Eiden MV, Radke K, Rovnak J, Quackenbush SL. 2010. Non-primate mammalian and fish retroviruses, p 371–416. In Kurth R, Bannert N (ed), Retroviruses: molecular biology, genomics and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 73.Kewalramani VN, Panganiban AT, Emerman M. 1992. Spleen necrosis virus, an avian immunosuppressive retrovirus, shares a receptor with the type D simian retroviruses. J Virol 66:3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koo H-M, Gu J, Varela-Echavarria A, Ron Y, Dougherty JP. 1992. Reticuloendotheliosis type C and primate type D oncoretroviruses are members of the same receptor interference group. J Virol 66:3448–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rasko JE, Battini JL, Gottschalk RJ, Mazo I, Miller AD. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci U S A 96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marin M, Tailor CS, Nouri A, Kabat D. 2000. Sodium-dependent neutral amino acid transporter type 1 is an auxiliary receptor for baboon endogenous retrovirus. J Virol 74:8085–8093. doi: 10.1128/JVI.74.17.8085-8093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ponferrada VG, Mauck BS, Wooley DP. 2003. The envelope glycoprotein of human endogenous retrovirus HERV-W induces cellular resistance to spleen necrosis virus. Arch Virol 148:659–675. doi: 10.1007/s00705-002-0960-x. [DOI] [PubMed] [Google Scholar]
- 79.Benveniste RE, Lieber MM, Livingston DM, Sherr CJ, Todaro GJ, Kalter SS. 1974. Infectious C-type virus isolated from a baboon placenta. Nature 248:17–20. doi: 10.1038/248017a0. [DOI] [PubMed] [Google Scholar]
- 80.McAllister RM, Nicolson M, Gardner MB, Rongey RW, Rasheed S, Sarma PS, Huebner RJ, Hatanaka M, Oroszlan S, Gilden RV, Kabigting A, Vernon L. 1972. C-type virus released from cultured human rhabdomyosarcoma cells. Nat New Biol 235:3–6. [DOI] [PubMed] [Google Scholar]
- 81.Sommerfelt MA. 1999. Retrovirus receptors. J Gen Virol 80:3049–3064. doi: 10.1099/0022-1317-80-12-3049. [DOI] [PubMed] [Google Scholar]
- 82.Tailor CS, Nouri A, Zhao Y, Takeuchi Y, Kabat D. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol 73:4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lavillette D, Marin M, Ruggieri A, Mallet F, Cosset F-L, Kabat D. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J Virol 76:6442–6452. doi: 10.1128/JVI.76.13.6442-6452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khan AA, Quigley JG. 2013. Heme and FLVCR-related transporter families SLC48 and SLC49. Mol Aspects Med 34:669–682. doi: 10.1016/j.mam.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duffy SP, Shing J, Saraon P, Berger LC, Eiden MV, Wilde A, Tailor CS. 2010. The Fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol Cell Biol 30:5318–5324. doi: 10.1128/MCB.00690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rigby MA, Rojko JL, Stewart MA, Kociba GJ, Cheney CM, Rezanka LJ, Mathes LE, Hartke JR, Jarrett O, Neil JC. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol 73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 87.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. [PubMed] [Google Scholar]
- 88.Prassolov V, Hein S, Ziegler M, Ivanov D, Munk C, Lohler J, Stocking C. 2001. Mus cervicolor murine leukemia virus isolate M813 belongs to a unique receptor interference group. J Virol 75:4490–4498. doi: 10.1128/JVI.75.10.4490-4498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hein S, Prassolov V, Zhang Y, Ivanov D, Lohler J, Ross SR, Stocking C. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J Virol 77:5926–5932. doi: 10.1128/JVI.77.10.5926-5932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tipper CH, Cingoz O, Coffin JM. 2012. Mus spicilegus endogenous retrovirus HEMV uses murine sodium-dependent myo-inositol transporter 1 as a receptor. J Virol 86:6341–6344. doi: 10.1128/JVI.00083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reitz MS, Gallo RC. 2010. HTLV and HIV, p 445–477. In Kurth R, Bannert N (ed), Retroviruses: molecular biology, genomics and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 92.Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115:449–459. doi: 10.1016/S0092-8674(03)00881-X. [DOI] [PubMed] [Google Scholar]
- 93.Manel N, Taylor N, Kinet S, Kim FJ, Swainson L, Lavanya M, Battini JL, Sitbon M. 2004. HTLV envelopes and their receptor GLUT1, the ubiquitous glucose transporter: a new vision on HTLV infection? Front Biosci 9:3218–3241. doi: 10.2741/1474. [DOI] [PubMed] [Google Scholar]
- 94.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 95.Payne LN, Howes K, Gillespie AM, Smith LM. 1992. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol 73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 96.Payne LN, Gillespie AM, Howes K. 1992. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia 6:1167–1176. [PubMed] [Google Scholar]
- 97.Chesters PM, Howes K, Petherbridge L, Evans S, Payne LN, Venugopal K. 2002. The viral envelope is a major determinant for the induction of lymphoid and myeloid tumours by avian leukosis virus subgroups A and J, respectively. J Gen Virol 83:2553–2561. doi: 10.1099/0022-1317-83-10-2553. [DOI] [PubMed] [Google Scholar]
- 98.McDougall AS, Terry A, Tzavaras T, Cheney C, Rojko J, Neil JC. 1994. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol 68:2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van der Kuyl AC, Dekker JT, Goudsmit J. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEV(Gag-Pol)/baboon endogenous virus BaEV(Env) recombinant. J Virol 73:7994–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miller AD. 2003. Identification of Hyal2 as the cell-surface receptor for Jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr Top Microbiol Immunol 275:179–199. [DOI] [PubMed] [Google Scholar]
- 101.Golovkina TV, Dzuris J, van den Hoogen B, Jaffe AB, Wright PC, Cofer SM, Ross SR. 1998. A novel membrane protein is a mouse mammary tumor virus receptor. J Virol 72:3066–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang E, Obeng-Adjei N, Ying Q, Meertens L, Dragic T, Davey RA, Ross SR. 2008. Mouse mammary tumor virus uses mouse but not human transferrin receptor 1 to reach a low pH compartment and infect cells. Virology 381:230–240. doi: 10.1016/j.virol.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bates P, Young JAT, Varmus HE. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 104.Young JT, Bates P, Varmus HE. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol 67:1811–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barnard RJ, Young JA. 2003. Alpharetrovirus envelope-receptor interactions. Curr Top Microbiol Immunol 281:107–136. [DOI] [PubMed] [Google Scholar]
- 106.Adkins HB, Blacklow SC, Young JAT. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J Virol 75:3520–3526. doi: 10.1128/JVI.75.8.3520-3526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elleder D, Stepanets V, Melder DC, Senigl F, Geryk J, Pajer P, Plachy J, Hejnar J, Svoboda J, Federspiel MJ. 2005. The receptor for the subgroup C avian sarcoma and leukosis viruses, Tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J Virol 79:10408–10419. doi: 10.1128/JVI.79.16.10408-10419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gottlieb MS, Schroff R, Schanker HM, Weisman JD, Fan PT, Wolf RA, Saxon A. 1981. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med 305:1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 109.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 110.Klatzmann D, Barre-Sinoussi F, Nugeyre MT, Danquet C, Vilmer E, Griscelli C, Brun-Veziret F, Rouzioux C, Gluckman JC, Chermann JC, Montagnier L. 1984. Selective tropism of lymphadenopathy associated virus (LAV) for helper-inducer T lymphocytes. Science 225:59–63. doi: 10.1126/science.6328660. [DOI] [PubMed] [Google Scholar]
- 111.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 112.McDougal JS, Kennedy MS, Sligh JM, Cort SP, Mawle A, Nicholson JK. 1986. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 113.McClure MO, Sattentau QJ, Beverley PC, Hearn JP, Fitzgerald AK, Zuckerman AJ, Weiss RA. 1987. HIV infection of primate lymphocytes and conservation of the CD4 receptor. Nature 330:487–489. doi: 10.1038/330487a0. [DOI] [PubMed] [Google Scholar]
- 114.Hoxie JA, Haggarty BS, Bonser SE, Rackowski JL, Shan H, Kanki PJ. 1988. Biological characterization of a simian immunodeficiency virus-like retrovirus (HTLV-IV): evidence for CD4-associated molecules required for infection. J Virol 62:2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kornfeld H, Riedel N, Viglianti GA, Hirsch V, Mullins JI. 1987. Cloning of HTLV-4 and its relation to simian and human immunodeficiency viruses. Nature 326:610–613. doi: 10.1038/326610a0. [DOI] [PubMed] [Google Scholar]
- 116.Fisher AG, Ensoli B, Looney D, Rose A, Gallo RC, Saag MS, Shaw GM, Hahn BH, Wong-Staal F. 1988. Biologically diverse molecular variants within a single HIV-1 isolate. Nature 334:444–447. doi: 10.1038/334444a0. [DOI] [PubMed] [Google Scholar]
- 117.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 118.Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 119.Alkhatib G, Broder CC, Berger EA. 1996. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol 70:5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 121.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 122.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. doi: 10.1016/S0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 123.Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 124.Feng Y, Broader C, Kennedy P, Berger E. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 125.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien SJ. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856–1862. [DOI] [PubMed] [Google Scholar]
- 126.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 127.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 128.Benkirane M, Jin DY, Chun RF, Koup RA, Jeang KT. 1997. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J Biol Chem 272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 129.Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, Montana JB, McHale M, Sullivan J, Ridgway C, Felstead S, Dunne MW, van der Ryst E, Mayer H, MOTIVATE Study Teams. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 132.Symons J, Vandekerckhove L, Hutter G, Wensing AM, van Ham PM, Deeks SG, Nijhuis M. 2014. Dependence on the CCR5 coreceptor for viral replication explains the lack of rebound of CXCR4-predicted HIV variants in the Berlin patient. Clin Infect Dis 59:596–600. doi: 10.1093/cid/ciu284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Balotta C, Bagnarelli P, Violin M, Ridolfo AL, Zhou D, Berlusconi A, Corvasce S, Corbellino M, Clementi M, Clerici M, Moroni M, Galli M. 1997. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS 11:F67–F71. doi: 10.1097/00002030-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 134.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med 3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 135.Gorry PR, Zhang C, Wu S, Kunstman K, Trachtenberg E, Phair J, Wolinsky S, Gabuzda D. 2002. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5 Delta 32 allele. Lancet 359:1832–1834. doi: 10.1016/S0140-6736(02)08681-6. [DOI] [PubMed] [Google Scholar]
- 136.Michael NL, Nelson JA, KewalRamani VN, Chang G, O'Brien SJ, Mascola JR, Volsky B, Louder M, White GC II, Littman DR, Swanstrom R, O'Brien TR. 1998. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 delta32. J Virol 72:6040–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Naif HM, Cunningham AL, Alali M, Li S, Nasr N, Buhler MM, Schols D, de Clercq E, Stewart G. 2002. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Delta32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J Virol 76:3114–3124. doi: 10.1128/JVI.76.7.3114-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.O'Brien TR, Winkler C, Dean M, Nelson JA, Carrington M, Michael NL, White GC II. 1997. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet 349:1219. [DOI] [PubMed] [Google Scholar]
- 139.Sheppard HW, Celum C, Michael NL, O'Brien S, Dean M, Carrington M, Dondero D, Buchbinder SP. 2002. HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion. J Acquir Immune Defic Syndr 29:307–313. doi: 10.1097/00126334-200203010-00013. [DOI] [PubMed] [Google Scholar]
- 140.Ackley CD, Yamamoto JK, Levy N, Pedersen NC, Cooper MD. 1990. Immunologic abnormalities in pathogen-free cats experimentally infected with feline immunodeficiency virus. J Virol 64:5652–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Novotney C, English RV, Housman J, Davidson MG, Nasisse MP, Jeng CR, Davis WC, Tompkins MB. 1990. Lymphocyte population changes in cats naturally infected with feline immunodeficiency virus. AIDS 4:1213–1218. doi: 10.1097/00002030-199012000-00005. [DOI] [PubMed] [Google Scholar]
- 142.de Parseval A, Chatterji U, Sun P, Elder JH. 2004. Feline immunodeficiency virus targets activated CD4+ T cells by using CD134 as a binding receptor. Proc Natl Acad Sci U S A 101:13044–13049. doi: 10.1073/pnas.0404006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shimojima M, Miyazawa T, Ikeda Y, McMonagle EL, Haining H, Akashi H, Takeuchi Y, Hosie MJ, Willett BJ. 2004. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 303:1192–1195. doi: 10.1126/science.1092124. [DOI] [PubMed] [Google Scholar]
- 144.Willett BJ, Hosie MJ, Neil JC, Turner JD, Hoxie JA. 1997. Common mechanism of infection by lentiviruses. Nature 385:587. [DOI] [PubMed] [Google Scholar]
- 145.Willett BJ, Picard L, Hosie MJ, Turner JD, Adema K, Clapham PR. 1997. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol 71:6407–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Willett BJ, McMonagle EL, Logan N, Spiller OB, Schneider P, Hosie MJ. 2007. Probing the interaction between feline immunodeficiency virus and CD134 by using the novel monoclonal antibody 7D6 and the CD134 (Ox40) ligand. J Virol 81:9665–9679. doi: 10.1128/JVI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.de Parseval A, Chatterji U, Morris G, Sun P, Olson AJ, Elder JH. 2005. Structural mapping of CD134 residues critical for interaction with feline immunodeficiency virus. Nat Struct Mol Biol 12:60–66. doi: 10.1038/nsmb872. [DOI] [PubMed] [Google Scholar]
- 148.Willett BJ, McMonagle EL, Bonci F, Pistello M, Hosie MJ. 2006. Mapping the domains of CD134 as a functional receptor for feline immunodeficiency virus. J Virol 80:7744–7747. doi: 10.1128/JVI.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Willett BJ, McMonagle EL, Ridha S, Hosie MJ. 2006. Differential utilization of CD134 as a functional receptor by diverse strains of feline immunodeficiency virus. J Virol 80:3386–3394. doi: 10.1128/JVI.80.7.3386-3394.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Willett BJ, Kraase M, Logan N, McMonagle E, Varela M, Hosie MJ. 2013. Selective expansion of viral variants following experimental transmission of a reconstituted feline immunodeficiency virus quasispecies. PLoS One 8:e54871. doi: 10.1371/journal.pone.0054871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Willett BJ, Kraase M, Logan N, McMonagle EL, Samman A, Hosie MJ. 2010. Modulation of the virus-receptor interaction by mutations in the V5 loop of feline immunodeficiency virus (FIV) following in vivo escape from neutralising antibody. Retrovirology 7:38. doi: 10.1186/1742-4690-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Negre D, Mangeot P-E, Duisit G, Blanchard S, Vidalain P-O, Leissner P, Winter A-J, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset F-L. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Therapy 7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 153.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 155.Sharp PM, Hahn BH. 2010. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci 365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Peeters M, Honore C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper RW, Delaporte E. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 157.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 158.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 159.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Beer BE, Foley BT, Kuiken CL, Tooze Z, Goeken RM, Brown CR, Hu J, St Claire M, Korber BT, Hirsch VM. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J Virol 75:12014–12027. doi: 10.1128/JVI.75.24.12014-12027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 162.Barlow KL, Ajao AO, Clewley JP. 2003. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona). J Virol 77:6879–6888. doi: 10.1128/JVI.77.12.6879-6888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Courgnaud V, Salemi M, Pourrut X, Mpoudi-Ngole E, Abela B, Auzel P, Bibollet-Ruche F, Hahn B, Vandamme AM, Delaporte E, Peeters M. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J Virol 76:8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 165.Chitnis A, Rawls D, Moore J. 2000. Origin of HIV type 1 in colonial French Equatorial Africa? AIDS Res Hum Retroviruses 16:5–8. doi: 10.1089/088922200309548. [DOI] [PubMed] [Google Scholar]
- 166.Pepin J, Labbe AC. 2008. Noble goals, unforeseen consequences: control of tropical diseases in colonial Central Africa and the iatrogenic transmission of blood-borne viruses. Trop Med Int Health 13:744–753. doi: 10.1111/j.1365-3156.2008.02060.x. [DOI] [PubMed] [Google Scholar]
- 167.Wain LV, Bailes E, Bibollet-Ruche F, Decker JM, Keele BF, Van Heuverswyn F, Li Y, Takehisa J, Ngole EM, Shaw GM, Peeters M, Hahn BH, Sharp PM. 2007. Adaptation of HIV-1 to its human host. Mol Biol Evol 24:1853–1860. doi: 10.1093/molbev/msm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 170.Damond F, Worobey M, Campa P, Farfara I, Colin G, Matheron S, Brun-Vezinet F, Robertson DL, Simon F. 2004. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res Hum Retroviruses 20:666–672. doi: 10.1089/0889222041217392. [DOI] [PubMed] [Google Scholar]
- 171.Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol 79:8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 173.Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, Ngole EM, Shaw GM, Peeters M, Sharp PM, Hahn BH. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol 83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. 2009. A new human immunodeficiency virus derived from gorillas. Nat Med 15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 175.Zhao K, Ishida Y, Oleksyk TK, Winkler CA, Roca AL. 2012. Evidence for selection at HIV host susceptibility genes in a West Central African human population. BMC Evol Biol 12:237. doi: 10.1186/1471-2148-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. 2007. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A 104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Worobey M, Watts TD, McKay RA, Suchard MA, Granade T, Teuwen DE, Koblin BA, Heneine W, Lemey P, Jaffe HW. 2016. 1970s and ‘patient 0′ HIV-1 genomes illuminate early HIV/AIDS history in North America. Nature 539:98–101. doi: 10.1038/nature19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A 77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Murphy EL, Hanchard B, Figueroa JP, Gibbs WN, Lofters WS, Campbell M, Goedert JJ, Blattner WA. 1989. Modelling the risk of adult T-cell leukemia/lymphoma in persons infected with human T-lymphotropic virus type I. Int J Cancer 43:250–253. doi: 10.1002/ijc.2910430214. [DOI] [PubMed] [Google Scholar]
- 180.Gessain A, Cassar O. 2012. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol 3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gessain A, Gallo RC, Franchini G. 1992. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol 66:2288–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Van Dooren S, Salemi M, Vandamme AM. 2001. Dating the origin of the African human T-cell lymphotropic virus type-I (HTLV-I) subtypes. Mol Biol Evol 18:661–671. doi: 10.1093/oxfordjournals.molbev.a003846. [DOI] [PubMed] [Google Scholar]
- 183.Calvignac-Spencer S, Adjogoua EV, Akoua-Koffi C, Hedemann C, Schubert G, Ellerbrok H, Leendertz SA, Pauli G, Leendertz FH. 2012. Origin of human T-lymphotropic virus type 1 in rural Cote d'Ivoire. Emerg Infect Dis 18:830–833. doi: 10.3201/eid1805.111663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, Torimiro JN, Prosser AT, Lebreton M, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Switzer WM. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A 102:7994–7999. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koralnik IJ, Boeri E, Saxinger WC, Monico AL, Fullen J, Gessain A, Guo HG, Gallo RC, Markham P, Kalyanaraman V, Hirsch V, Allan J, Murthy K, Alford P, Slattery JP, O'Brien SJ, Franchini G. 1994. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J Virol 68:2693–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu HF, Goubau P, Van Brussel M, Van Laethem K, Chen YC, Desmyter J, Vandamme AM. 1996. The three human T-lymphotropic virus type I subtypes arose from three geographically distinct simian reservoirs. J Gen Virol 77:359–368. doi: 10.1099/0022-1317-77-2-359. [DOI] [PubMed] [Google Scholar]
- 187.Salemi M, Vandamme AM, Desmyter J, Casoli C, Bertazzoni U. 1999. The origin and evolution of human T-cell lymphotropic virus type II (HTLV-II) and the relationship with its replication strategy. Gene 234:11–21. doi: 10.1016/S0378-1119(99)00169-9. [DOI] [PubMed] [Google Scholar]
- 188.Brown MA, Cunningham MW, Roca AL, Troyer JL, Johnson WE, O'Brien SJ. 2008. Genetic characterization of feline leukemia virus from Florida panthers. Emerg Infect Dis 14:252–259. doi: 10.3201/eid1402.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee J, Malmberg JL, Wood BA, Hladky S, Troyer R, Roelke M, Cunningham M, McBride R, Vickers W, Boyce W, Boydston E, Serieys L, Riley S, Crooks K, VandeWoude S. 2017. Feline immunodeficiency virus cross-species transmission: implications for emergence of new lentiviral infections. J Virol 91:e02134-16. doi: 10.1128/JVI.02134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee JS, Bevins SN, Serieys LE, Vickers W, Logan KA, Aldredge M, Boydston EE, Lyren LM, McBride R, Roelke-Parker M, Pecon-Slattery J, Troyer JL, Riley SP, Boyce WM, Crooks KR, VandeWoude S. 2014. Evolution of puma lentivirus in bobcats (Lynx rufus) and mountain lions (Puma concolor) in North America. J Virol 88:7727–7737. doi: 10.1128/JVI.00473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xu W, Gorman K, Santiago JC, Kluska K, Eiden MV. 2015. Genetic diversity of koala retroviral envelopes. Viruses 7:1258–1270. doi: 10.3390/v7031258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Alfano N, Kolokotronis SO, Tsangaras K, Roca AL, Xu W, Eiden MV, Greenwood AD. 2015. Episodic diversifying selection shaped the genomes of gibbon ape leukemia virus and related gammaretroviruses. J Virol 90:1757–1772. doi: 10.1128/JVI.02745-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kawakami TG, Buckley PM. 1974. Antigenic studies on gibbon type-C viruses. Transplant Proc 6:193–196. [PubMed] [Google Scholar]
- 194.Kawakami TG, Kollias GV Jr, Holmberg C. 1980. Oncogenicity of gibbon type-C myelogenous leukemia virus. Int J Cancer 25:641–646. doi: 10.1002/ijc.2910250514. [DOI] [PubMed] [Google Scholar]
- 195.Gallo RC, Gallagher RE, Wong-Staal F, Aoki T, Markham PD, Schetters H, Ruscetti F, Valerio M, Walling MJ, O'Keefe RT, Saxinger WC, Smith RG, Gillespie DH, Reitz MS. 1978. Isolation and tissue distribution of type-C virus and viral components from a gibbon ape (Hylobates lar) with lymphocytic leukemia. Virology 84:359–373. doi: 10.1016/0042-6822(78)90255-6. [DOI] [PubMed] [Google Scholar]
- 196.Todaro GJ, Lieber MM, Benveniste RE, Sherr CJ. 1975. Infectious primate type C viruses: three isolates belonging to a new subgroup from the brains of normal gibbons. Virology 67:335–343. doi: 10.1016/0042-6822(75)90435-3. [DOI] [PubMed] [Google Scholar]
- 197.Snyder SP, Dungworth DL, Kawakami TG, Callaway E, Lau DT-L. 1973. Lymphosarcoma in two gibbons (Hylobates lar) with associated C-type virus. J Natl Cancer Inst 51:89–95. doi: 10.1093/jnci/51.1.89. [DOI] [PubMed] [Google Scholar]
- 198.Parent I, Qin Y, Vandenbroucke AT, Walon C, Delferriere N, Godfroid E, Burtonboy G. 1998. Characterization of a C-type retrovirus isolated from an HIV infected cell line: complete nucleotide sequence. Arch Virol 143:1077–1092. doi: 10.1007/s007050050357. [DOI] [PubMed] [Google Scholar]
- 199.Siegal-Willott JL, Jensen N, Kimi D, Taliaferro D, Blankenship T, Malinsky B, Murray S, Eiden MV, Xu W. 2015. Evaluation of captive gibbons (Hylobates spp., Nomascus spp., Symphalangus spp.) in North American zoological institutions for gibbon ape leukemia virus (GALV). J Zoo Wildl Med 46:27–33. doi: 10.1638/2014-0034R.1. [DOI] [PubMed] [Google Scholar]
- 200.Brockelman WY, Ross BA, Pantuwatana S. 1973. Social correlates of reproductive success in the gibbon colony on Ko Klet Kaeo, Thailand. Am J Phys Anthropol 38:637–640. doi: 10.1002/ajpa.1330380280. [DOI] [PubMed] [Google Scholar]
- 201.Whitehead RH, Chaicumpa V, Olson LC, Russell PK. 1970. Sequential dengue virus infections in the white-handed gibbon (Hylobates lar). Am J Trop Med Hyg 19:94–102. doi: 10.4269/ajtmh.1970.19.94. [DOI] [PubMed] [Google Scholar]
- 202.Johnsen DO, Wooding WL, Tanticharoenyos P, Bourgeois CH Jr. 1971. Malignant lymphoma in the gibbon. J Am Vet Med Assoc 159:563–566. [PubMed] [Google Scholar]
- 203.De Paoli A, Johnson DO, Noll WW. 1973. Granulocytic leukemia in whitehanded gibbons. J Am Vet Med Assoc 163:624–628. [PubMed] [Google Scholar]
- 204.Brown K, Tarlinton RE. 2016. Is gibbon ape leukaemia virus still a threat. Mamm Rev 47:53–61. doi: 10.1111/mam.12079. [DOI] [Google Scholar]
- 205.Kawakami TG, Sun L, McDowell TS. 1978. Natural transmission of gibbon leukemia virus. J Natl Cancer Inst 61:1113–1115. [PubMed] [Google Scholar]
- 206.Kawakami TG, Huff SD, Buckley PM, Dungworth DL, Synder SP, Gilden RV. 1972. C-type virus associated with gibbon lymphosarcoma. Nat New Biol 235:170–171. [DOI] [PubMed] [Google Scholar]
- 207.Salemi M, Lewis M, Egan JF, Hall WW, Desmyter J, Vandamme AM. 1999. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc Natl Acad Sci U S A 96:13253–13258. doi: 10.1073/pnas.96.23.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Vandamme AM, Bertazzoni U, Salemi M. 2000. Evolutionary strategies of human T-cell lymphotropic virus type II. Gene 261:171–180. doi: 10.1016/S0378-1119(00)00473-X. [DOI] [PubMed] [Google Scholar]
- 209.Martin J, Kabat P, Tristem M. 2002. Tangled trees: phylogeny, co-speciation and coevolution. University of Chicago Press, Chicago, IL. [Google Scholar]
- 210.Niewiadomska AM, Gifford RJ. 2013. The extraordinary evolutionary history of the reticuloendotheliosis viruses. PLoS Biol 11:e1001642. doi: 10.1371/journal.pbio.1001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Martin J, Herniou E, Cook J, O'Neill RW, Tristem M. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J Virol 73:2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Breed AC, Field HE, Smith CS, Edmonston J, Meers J. 2010. Bats without borders: long-distance movements and implications for disease risk management. Ecohealth 7:204–212. doi: 10.1007/s10393-010-0332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cui J, Tachedjian G, Tachedjian M, Holmes EC, Zhang S, Wang LF. 2012. Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J Gen Virol 93:2037–2045. doi: 10.1099/vir.0.043760-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cui J, Tachedjian G, Wang LF. 2015. Bats and rodents shape mammalian retroviral phylogeny. Sci Rep 5:16561. doi: 10.1038/srep16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cui J, Tachedjian M, Wang L, Tachedjian G, Wang LF, Zhang S. 2012. Discovery of retroviral homologs in bats: implications for the origin of mammalian gammaretroviruses. J Virol 86:4288–4293. doi: 10.1128/JVI.06624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Isfort RJ, Qian Z, Jones D, Silva RF, Witter R, Kung HJ. 1994. Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology 203:125–133. doi: 10.1006/viro.1994.1462. [DOI] [PubMed] [Google Scholar]
- 217.Hertig C, Coupar BE, Gould AR, Boyle DB. 1997. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology 235:367–376. doi: 10.1006/viro.1997.8691. [DOI] [PubMed] [Google Scholar]
- 218.Weiss RA. 2015. What's the host and what's the microbe? The Marjory Stephenson Prize Lecture 2015. J Gen Virol 96:2501–2510. doi: 10.1099/jgv.0.000220. [DOI] [PubMed] [Google Scholar]
- 219.Tsangaras K, Siracusa MC, Nikolaidis N, Ishida Y, Cui P, Vielgrader H, Helgen KM, Roca AL, Greenwood AD. 2014. Hybridization capture reveals evolution and conservation across the entire koala retrovirus genome. PLoS One 9:e95633. doi: 10.1371/journal.pone.0095633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cui P, Lober U, Alquezar-Planas DE, Ishida Y, Courtiol A, Timms P, Johnson RN, Lenz D, Helgen KM, Roca AL, Hartman S, Greenwood AD. 2016. Comprehensive profiling of retroviral integration sites using target enrichment methods from historical koala samples without an assembled reference genome. PeerJ 4:e1847. doi: 10.7717/peerj.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ávila-Arcos MC, Ho SY, Ishida Y, Nikolaidis N, Tsangaras K, Honig K, Medina R, Rasmussen M, Fordyce SL, Calvignac-Spencer S, Willerslev E, Gilbert MT, Helgen KM, Roca AL, Greenwood AD. 2013. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol Biol Evol 30:299–304. doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Simmons G, Clarke D, McKee J, Young P, Meers J. 2014. Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): a putative link between gibbon ape leukemia virus and koala retrovirus. PLoS One 9:e106954. doi: 10.1371/journal.pone.0106954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wallace AR. 1876. The geographic distribution of animals. With a study of the relations of living and extinct faunas as elucidating the past changes of the earth's surface, vol 2 Macmillan and Co, London, United Kingdom. [Google Scholar]
- 224.Hayward A, Cornwallis CK, Jern P. 2015. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci U S A 112:464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hayward A, Grabherr M, Jern P. 2013. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci U S A 110:20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhuo X, Feschotte C. 2015. Cross-species transmission and differential fate of an endogenous retrovirus in three mammal lineages. PLoS Pathog 11:e1005279. doi: 10.1371/journal.ppat.1005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Tsangaras K, Mayer J, Alquezar-Planas DE, Greenwood AD. 2015. An evolutionarily young polar bear (Ursus maritimus) endogenous retrovirus identified from next generation sequence data. Viruses 7:6089–6107. doi: 10.3390/v7112927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bromham LD. 2002. The human zoo: endogenous retroviruses in the human genome. Trends Ecol Evol 17:91–97. doi: 10.1016/S0169-5347(01)02394-1. [DOI] [Google Scholar]
- 229.Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. 2012. Env-less endogenous retroviruses are genomic superspreaders. Proc Natl Acad Sci U S A 109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. 2008. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res 18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mietz JA, Grossman Z, Lueders KK, Kuff EL. 1987. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J Virol 61:3020–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Reuss FU, Schaller HC. 1991. cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J Virol 65:5702–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kim FJ, Battini JL, Manel N, Sitbon M. 2004. Emergence of vertebrate retroviruses and envelope capture. Virology 318:183–191. doi: 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 234.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Evans LH, Alamgir AS, Owens N, Weber N, Virtaneva K, Barbian K, Babar A, Malik F, Rosenke K. 2009. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J Virol 83:2429–2435. doi: 10.1128/JVI.01926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. 1992. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev 6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- 237.Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 238.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 239.Gong R, Huang L, Shi J, Luo K, Qiu G, Feng H, Tien P, Xiao G. 2007. Syncytin-A mediates the formation of syncytiotrophoblast involved in mouse placental development. Cell Physiol Biochem 20:517–526. doi: 10.1159/000107535. [DOI] [PubMed] [Google Scholar]
- 240.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci U S A 103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Blanco-Melo D, Gifford RJ, Bieniasz PD. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. doi: 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol 79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stengel A, Roos C, Hunsmann G, Seifarth W, Leib-Mosch C, Greenwood AD. 2006. Expression profiles of endogenous retroviruses in Old World monkeys. J Virol 80:4415–4421. doi: 10.1128/JVI.80.9.4415-4421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Matouskova M, Vesely P, Daniel P, Mattiuzzo G, Hector RD, Scobie L, Takeuchi Y, Hejnar J. 2013. Role of DNA methylation in expression and transmission of porcine endogenous retroviruses. J Virol 87:12110–12120. doi: 10.1128/JVI.03262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A 113:E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RA, Wysocka J. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind'Amour J, Sun MA, Sun Z, Ishikura T, Mizutani-Koseki Y, Ohara O, Shinkai Y, Nakanishi M, Xie H, Lorincz MC, Koseki H. 2016. Activation of endogenous retroviruses in Dnmt1(−/−) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell 19:81–94. doi: 10.1016/j.stem.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 248.Aiewsakun P, Katzourakis A. 2017. Marine origin of retroviruses in the early Palaeozoic Era. Nat Commun 8:13954. doi: 10.1038/ncomms13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 250.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Conticello SG. 2008. The AID/APOBEC family of nucleic acid mutators. Genome Biol 9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kaiser SM, Malik HS, Emerman M. 2007. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 254.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wittmann S, Behrendt R, Eissmann K, Volkmann B, Thomas D, Ebert T, Cribier A, Benkirane M, Hornung V, Bouzas NF, Gramberg T. 2015. Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology 12:103. doi: 10.1186/s12977-015-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Reference deleted.
- 257.Gifford RJ. 2006. Evolution at the host-retrovirus interface. Bioessays 28:1153–1156. doi: 10.1002/bies.20504. [DOI] [PubMed] [Google Scholar]
- 258.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 259.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, Stadhouders R, Johrens K, Wurster KD, Callen DF, Harte MF, Giefing M, Barlow R, Stein H, Anagnostopoulos I, Janz M, Cockerill PN, Siebert R, Dorken B, Bonifer C, Mathas S. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 16:571–579. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 260.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 261.Dupressoir A, Lavialle C, Heidmann T. 2012. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 262.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of ‘syncytins,’ retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, Blanchard-Gutton N, Charrin S, Tiret L, Dumonceaux J, Dupressoir A, Heidmann T. 2016. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet 12:e1006289. doi: 10.1371/journal.pgen.1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Stoye JP. 2006. Koala retrovirus: a genome invasion in real time. Genome Biol 7:241. doi: 10.1186/gb-2006-7-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog 2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 266.Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 267.Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ Jr, Coffin JM, Pathak VK. 2011. Recombinant origin of the retrovirus XMRV. Science 333:97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Oakes B, Tai AK, Cingoz O, Henefield MH, Levine S, Coffin JM, Huber BT. 2010. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology 7:109. doi: 10.1186/1742-4690-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tuke PW, Tettmar KI, Tamuri A, Stoye JP, Tedder RS. 2011. PCR master mixes harbour murine DNA sequences. Caveat emptor! PLoS One 6:e19953. doi: 10.1371/journal.pone.0019953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zheng H, Jia H, Shankar A, Heneine W, Switzer WM. 2011. Detection of murine leukemia virus or mouse DNA in commercial RT-PCR reagents and human DNAs. PLoS One 6:e29050. doi: 10.1371/journal.pone.0029050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Erlwein O, Robinson MJ, Dustan S, Weber J, Kaye S, McClure MO. 2011. DNA extraction columns contaminated with murine sequences. PLoS One 6:e23484. doi: 10.1371/journal.pone.0023484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wolff D, Gerritzen A. 2011. Presence of murine leukemia virus (MLV)-related virus gene sequences in a commercial RT-PCR reagent. Clin Lab 57:631–634. [PubMed] [Google Scholar]
- 273.Naseer A, Terry A, Gilroy K, Kilbey A, Watts C, Mackay N, Bell M, Mason S, Blyth K, Cameron E, Neil JC. 2015. Frequent infection of human cancer xenografts with murine endogenous retroviruses in vivo. Viruses 7:2014–2029. doi: 10.3390/v7042014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Paabo S, Hofreiter M. 2006. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature 439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 275.Rompler H, Dear PH, Krause J, Meyer M, Rohland N, Schoneberg T, Spriggs H, Stiller M, Hofreiter M. 2006. Multiplex amplification of ancient DNA. Nat Protoc 1:720–728. doi: 10.1038/nprot.2006.84. [DOI] [PubMed] [Google Scholar]
- 276.Devault AM, McLoughlin K, Jaing C, Gardner S, Porter TM, Enk JM, Thissen J, Allen J, Borucki M, DeWitte SN, Dhody AN, Poinar HN. 2014. Ancient pathogen DNA in archaeological samples detected with a microbial detection array. Sci Rep 4:4245. doi: 10.1038/srep04245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hofreiter M, Paijmans JL, Goodchild H, Speller CF, Barlow A, Fortes GG, Thomas JA, Ludwig A, Collins MJ. 2015. The future of ancient DNA: technical advances and conceptual shifts. Bioessays 37:284–293. doi: 10.1002/bies.201400160. [DOI] [PubMed] [Google Scholar]
- 278.Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, NISC Comparative Sequencing Program, Margulies EH, Green ED, Collins FS, Mullikin JC, Biesecker LG. 2010. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res 20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Duggan AT, Perdomo MF, Piombino-Mascali D, Marciniak S, Poinar D, Emery MV, Buchmann JP, Duchene S, Jankauskas R, Humphreys M, Golding GB, Southon J, Devault A, Rouillard JM, Sahl JW, Dutour O, Hedman K, Sajantila A, Smith GL, Holmes EC, Poinar HN. 2016. 17th century variola virus reveals the recent history of smallpox. Curr Biol 26:3407–3412. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Duncavage EJ, Magrini V, Becker N, Armstrong JR, Demeter RT, Wylie T, Abel HJ, Pfeifer JD. 2011. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J Mol Diagn 13:325–333. doi: 10.1016/j.jmoldx.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, von Kalle C, Rethwilm A. 2006. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol 87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- 282.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. 2005. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 283.Ciuffi A, Barr SD. 2011. Identification of HIV integration sites in infected host genomic DNA. Methods 53:39–46. doi: 10.1016/j.ymeth.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 284.Kustikova OS, Modlich U, Fehse B. 2009. Retroviral insertion site analysis in dominant haematopoietic clones. Methods Mol Biol 506:373–390. doi: 10.1007/978-1-59745-409-4_25. [DOI] [PubMed] [Google Scholar]
- 285.Moalic Y, Blanchard Y, Felix H, Jestin A. 2006. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J Virol 80:10980–10988. doi: 10.1128/JVI.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, Braun S, Glimm H, von Kalle C. 2007. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat Methods 4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 287.Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prufer K, Meyer M, Krause J, Ronan MT, Lachmann M, Paabo S. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci U S A 104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ginolhac A, Rasmussen M, Gilbert MT, Willerslev E, Orlando L. 2011. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics 27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 289.Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res 41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Slater GJ, Cui P, Forasiepi AM, Lenz D, Tsangaras K, Voirin B, de Moraes-Barros N, MacPhee RD, Greenwood AD. 2016. Evolutionary relationships among extinct and extant sloths: the evidence of mitogenomes and retroviruses. Genome Biol Evol 8:607–621. doi: 10.1093/gbe/evw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Chen Y, Yao H, Thompson EJ, Tannir NM, Weinstein JN, Su X. 2013. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics 29:266–267. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang Q, Jia P, Zhao Z. 2013. VirusFinder: software for efficient and accurate detection of viruses and their integration sites in host genomes through next generation sequencing data. PLoS One 8:e64465. doi: 10.1371/journal.pone.0064465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ishida Y, McCallister C, Nikolaidis N, Tsangaras K, Helgen KM, Greenwood AD, Roca AL. 2015. Sequence variation of koala retrovirus transmembrane protein p15E among koalas from different geographic regions. Virology 475:28–36. doi: 10.1016/j.virol.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Tarlinton RE, Barfoot HK, Allen CE, Brown K, Gifford RJ, Emes RD. 2013. Characterisation of a group of endogenous gammaretroviruses in the canine genome. Vet J 196:28–33. doi: 10.1016/j.tvjl.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 296.Prassolov V, Ivanov D, Hein S, Rutter G, Munk C, Lohler J, Stocking C. 2001. The Mus cervicolor MuLV isolate M813 is highly fusogenic and induces a T-cell lymphoma associated with large multinucleated cells. Virology 290:39–49. doi: 10.1006/viro.2001.1145. [DOI] [PubMed] [Google Scholar]
- 297.Wolgamot G, Bonham L, Miller AD. 1998. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol 72:7459–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Bromham L, Clark F, McKee JJ. 2001. Discovery of a novel murine type C retrovirus by data mining. J Virol 75:3053–3057. doi: 10.1128/JVI.75.6.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Maricic T, Whitten M, Paabo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Enk JM, Devault AM, Kuch M, Murgha YE, Rouillard JM, Poinar HN. 2014. Ancient whole genome enrichment using baits built from modern DNA. Mol Biol Evol 31:1292–1294. doi: 10.1093/molbev/msu074. [DOI] [PubMed] [Google Scholar]
- 302.Wylie TN, Wylie KM, Herter BN, Storch GA. 2015. Enhanced virome sequencing using targeted sequence capture. Genome Res 25:1910–1920. doi: 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, Lipkin WI. 2015. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio 6:e01491-15. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Goff SP. 2007. Retroviridae: the retroviruses and their replication, p 1999–2069. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 305.Tristem M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol 74:3715–3730. doi: 10.1128/JVI.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hobbs M, King A, Salinas R, Chen Z, Tsangaras K, Greenwood AD, Johnson RN, Belov K, Wilkins MR, Timms P. 2017. Long-read genome sequence assembly provides insight into ongoing retroviral invasion of the koala germline. Sci Rep 7:15838. doi: 10.1038/s41598-017-16171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]