Abstract
Insulin-degrading enzyme (IDE) is an atypical zinc-metalloendopeptidase that hydrolyzes insulin and other intermediate-sized peptide hormones, many of which are implicated in skin health and wound healing. Pharmacological inhibitors of IDE administered internally have been shown to slow the breakdown of insulin and thereby potentiate insulin action. Given the importance of insulin and other IDE substrates for a variety of dermatological processes, pharmacological inhibitors of IDE suitable for topical applications would be expected to hold significant therapeutic and cosmetic potential. Existing IDE inhibitors, however, are prohibitively expensive, difficult to synthesize and of undetermined toxicity. Here we used phage display to discover novel peptidic inhibitors of IDE, which were subsequently characterized in vitro and in cell culture assays. Among several peptide sequences tested, a cyclic dodecapeptide dubbed P12-3A was found to potently inhibit the degradation of insulin (Ki = 2.5 ± 0.31 μM) and other substrates by IDE, while also being resistant to degradation, stable in biological milieu, and highly selective for IDE. In cell culture, P12-3A was shown to potentiate several insulin-induced processes, including the transcription, translation and secretion of alpha-1 type I collagen in primary murine skin fibroblasts, and the migration of keratinocytes in a scratch wound migration assay. By virtue of its potency, stability, specificity for IDE, low cost of synthesis, and demonstrated ability to potentiate insulin-induced processes involved in wound healing and skin health, P12-3A holds significant therapeutic and cosmetic potential for topical applications.
Introduction
Insulin is a pleiotropic peptide hormone that, although best known for its role in blood sugar regulation, is implicated in a wide array of physiological processes relevant to skin health and wound repair [1]. Insulin stimulates the proliferation [2, 3], differentiation [4] and migration [5, 6] of skin fibroblasts and keratinocytes, as well as the production and secretion of extracellular matrix (ECM) proteins, particularly collagen [7–13]. Conversely, all of these processes are impaired in the skin of mice with genetic deletion of the insulin receptor [14]. Moreover, impairments in wound healing and other skin disorders are common among patients with diabetes [15], a disease characterized by defects in insulin production or action.
Given the importance of insulin signaling to wound healing, topical insulin has been investigated in numerous studies in animals [6, 16–20] and humans [21], including several clinical trials [22–24]. However, the routine clinical use of topical insulin for wound management is not generally accepted as a first-line treatment, and significant adverse effects—including life-threatening hypoglycemia—have been reported [25].
Our group has been exploring an alternative approach to boosting insulin signaling that obviates the risk of hypoglycemia: namely, pharmacological inhibition of insulin-degrading enzyme (IDE) [26], the principal protease implicated in the catabolism and inactivation of insulin [27]. IDE inhibitors have been shown to potentiate insulin action in cultured cells [28] and in vivo [29–31]. Recently developed, highly selective IDE inhibitors exhibited potent antidiabetic properties [29], effects that were attributable to reduced catabolism of insulin. Importantly, mice with genetic deletion of IDE are viable [32–34]; thus—unlike insulin—IDE inhibitors possess no intrinsic risk of triggering life-threatening hypoglycemia. IDE is expressed to high levels in skin [35, 36] and—notably—is especially abundant in wound fluid [37, 38] where it degrades insulin [37, 38]. Thus, topical application of IDE inhibitors is strongly predicted to enhance insulin signaling in skin.
Although a number of IDE inhibitors have been developed [28, 29, 39–43], existing compounds are not ideal for topical applications due to their high cost of synthesis and undetermined toxicity. To overcome these limitations, we sought here to develop peptidic inhibitors of IDE that, by their intrinsic nature, would be inexpensive to manufacture and unlikely to be toxic. To that end, we used phage display to discover cyclic and linear peptide sequences that bind with high affinity to IDE. Among the sequences analyzed, a dodecameric, cyclic peptide dubbed P12-3A, proved to be a potent inhibitor of IDE that was stable in biologic milieu and highly selective for IDE. P12-3A was found to potentiate a number of insulin-stimulated processes in cultured skin cells, including collagen production in fibroblasts and migration of keratinocytes in a scratch wound assay. Given its high potency, selectivity for IDE, minimal potential for toxicity, and its low cost of manufacture, P12-3A possesses the characteristics needed to further explore the therapeutic and cosmetic potential of topical IDE inhibition.
Results
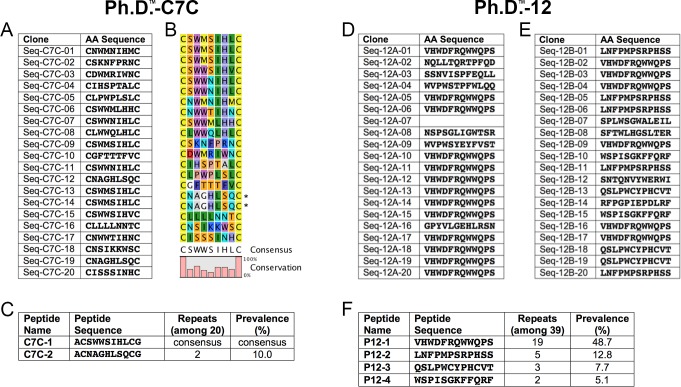
To identify novel peptidic inhibitors of IDE, we utilized phage display technology [44] to search for sequences that bind with high affinity to immobilized recombinant human IDE. Reasoning that IDE possesses an intrinsic affinity for cyclic peptides, we screened a library of cyclic peptides (Ph.D.TM-C7C, New England Biolabs) comprised of essentially all combinations of seven natural amino acids flanked by two cysteines (ACXXXXXXXCGGG…, where X represents any amino acid). The two cysteines form a disulfide bond that cyclizes each peptide, the alanine serves to protect the N-terminal cysteine from off-target interactions, and the three glycines form a flexible linker with the bacteriophage coat protein. Three rounds of panning were conducted using immobilized recombinant human IDE, with elution performed by addition of excess insulin in order to enrich for sequences that bind to the internal chamber of IDE. Twenty clones were selected for DNA sequencing, yielding sixteen unique amino acid sequences, some of which appeared more than once (Fig 1A). From analysis of all sequences (Fig 1B), a clear consensus sequence emerged (ACSWWSIHLCGGG…). This sequence, dubbed C7C-1 (Fig 1C, Panel A in S1 Fig), was selected for subsequent synthesis and testing, together with another that appeared two times (ACNAGHLSQCGGG…), dubbed C7C-2 (Fig 1C, Panel B in S1 Fig).
Fig 1. Peptides derived by phage display.
A, Peptide sequences deduced from DNA sequencing of 20 clones from the Ph.D.™-C7C library. B, Consensus sequence derived from analysis of all data. C, Parent peptides selected for synthesis and testing. C,D, Peptide sequences deduced from DNA sequencing of 39 clones from the Ph.D.™-12 library, conducted as two independent runs (D and E). Note that Seq-12A-07 did not yield a decipherable sequence. F, Parent peptides selected for subsequent synthesis and testing based on prevalence.
A second library of dodecapeptides (Ph.D.TM-12, New England Biolabs), consisting of ~109 possible combinations of twelve amino acids, was also screened, in this case two separate times, yielding the sequences in Fig 1D and 1E. In this case, four sequences appeared multiple times (dubbed 12–1, 12–2, 12–3 and 12–4), and these dodecapeptides were selected for subsequent synthesis and testing (Fig 1F, Panels C-F in S1 Fig).
The six selected peptide sequences (C7C-1, C7C-2, 12–1, 12–2, 12–3 and 12–4) were synthesized, together with a variety of modifications, yielding a total of 25 peptides (Table 1). For the 12-mer peptides, we synthesized variants of the parent peptides truncated at one or both termini, to test whether the entire sequence was necessary or not, in the hope that shorter, and thus less expensive, sequences might be effective. Other modifications included N-terminal acetylation and C-terminal amidation, included to mimic the charge state of the peptides when incorporated in the phage coat protein (Table 1). To assess the extent to which these peptide sequences inhibit IDE, we quantified their potency in protease activity assays using recombinant human IDE and two different substrates: FRET1, a fluorogenic peptide [45], and recombinant human insulin. Peptides were initially tested at a few concentrations (10 μM, 100 μM and/or 500 μM) then, for those peptides showing good potency, dose-response relationships were obtained to quantify IC50 values, which were subsequently converted to inhibitory constants (Ki values) with the Cheng-Prusoff equation [46].
Table 1. Peptide sequences synthesized and their potency in activity assays with FRET1 and insulin.
| Name | N-term | Sequence | C-term | Ki FRET1 (μM) | Ki Insulin (μM) |
|---|---|---|---|---|---|
| C7C-1 | NH3+ | ACSWWSIHLCG | COO- | 112 ± 6 | 3.7 ± 0.7 |
| C7C-1A | NH3+ | ACSWWSIHLCG | amide | >100 | >10 |
| C7C-1B | NH3+ | ACSWWSIHLCGGG | COO- | >100 | >10 |
| C7C-2 | NH3+ | ACNAGHLSQCG | COO- | >500 | >10 |
| C7C-2A | NH3+ | ACNAGHLSQCG | amide | >500 | >10 |
| P12-1 | NH3+ | VHWDFRQWWQPS | COO- | 7.7 ± 0.7 | 0.8 ± 0.04 |
| P12-1A | NH3+ | VHWDFRQWWQPS | amide | 5.0 ± 0.3 | 1.3 ± 0.2 |
| P12-1B | NH3+ | VHWDFRQW | amide | 41 ± 6.4 | 7.0 ± 1.7 |
| P12-1C | acetyl | FRQWWQPS | COO- | 139 ± 21 | >10 |
| P12-1D | acetyl | WDFRQWWQ | amide | 140 ± 28 | >10 |
| P12-2 | NH3+ | LNFPMPSRPHSS | COO- | >100 | >10 |
| P12-2A | NH3+ | LNFPMPSRPHSS | amide | >100 | >10 |
| P12-2B | NH3+ | LNFPMPSR | amide | >500 | >10 |
| P12-2C | acetyl | MPSRPHSS | COO- | >500 | >10 |
| P12-2D | acetyl | FPMPSRPH | amide | >500 | >10 |
| P12-3 | NH3+ | QSLPWCYPHCVT | COO- | 8.9 ± 0.3 | 3.9 ± 1.6 |
| P12-3A | NH3+ | QSLPWCYPHCVT | amide | 10 ± 0.4 | 4.1 ± 0.3 |
| P12-3B | NH3+ | QSLPWCYP | amide | 39 ± 20 | >10 |
| P12-3C | acetyl | WCYPHCVT | COO- | 35 ± 4.7 | >10 |
| P12-3D | acetyl | LPWCTPHC | amide | 102 ± 14 | >10 |
| P12-4 | NH3+ | WSPISGKFFQRF | COO- | 3.9 ± 0.5 | 1.5 ± 0.3 |
| P12-4A | NH3+ | WSPISGKFFQRF | amide | 4.2 ± 1.3 | 2.6 ± 0.7 |
| P12-4B | NH3+ | WSPISGKF | amide | >500 | >10 |
| P12-4C | acetyl | SGKFFQRF | COO- | >500 | >10 |
| P12-4D | acetyl | PISGKFFQ | amide | >500 | >10 |
Among the cyclic peptides tested, C7C-1 exhibited a modest Ki value of 112 ± 6 μM against FRET1 but yielded considerably improved potency against insulin (Ki = 3.7 ± 0.7 μM) (Table 1). None of the other C7C-1 derivatives exhibited Ki values below 100 μM for FRET1 or 10 μM for insulin, nor did C7C-2 or its amidated derivative (Table 1).
Relative to the cyclic peptides C7C-1 and C7C-2 and their derivatives, the unmodified linear peptide P12-1 exhibited significantly lower Ki values against FRET1 (7.7 ± 0.7 μM) as well as insulin (0.8 ± 0.04 μM) (Table 1). The C-terminally amidated version of P12-1, P12-1A, exhibited comparable Ki values against FRET1 and insulin (5.0 ± 0.3 μM and 1.3 ± 0.2 μM, respectively) (Table 1). Among three different 8-amino acid truncated versions of P12-1, the C-terminally truncated variant (P12-1B) exhibited slightly higher Ki values (41 ± 6.4 μM and 7.0 ± 1.7 μM for FRET1 and insulin, respectively), while the other N-terminally truncated (P12-1C) and N- and C-terminally truncated (P12-1D) variants exhibited relatively poor potency, with Ki values >100 μM for FRET1 and >10 μM for insulin (Table 1). These results suggest the N-terminal residues of P12-1 (VHWD…) are the most critical determinants of its potency.
The dodecapeptide P12-2 contained 3 proline residues (Table 1), which constrain the flexibility of the peptide backbone (Panel D in S1 Fig) and, in general, tend to render peptides less vulnerable to proteolytic degradation. However, P12-2 and all its variants exhibited Ki values >100 μM for FRET1 and >10 μM for insulin (Table 1) and were not characterized further.
Peptide P12-3 is noteworthy for containing two cysteine residues (Table 1), which are predicted to form a disulfide bond that cyclizes the peptide, together with 2 proline residues (Panel E in S1 Fig). The unmodified peptide, P12-3, and its C-terminally amidated variant, P12-3A, both potently inhibited the degradation of both FRET1 (Ki = 8.9 ± 0.3 μM and 10 ± 0.4 μM, respectively) and insulin (Ki = 3.9 ± 1.6 μM and 4.1 ± 0.3 μM, respectively) (Table 1). For the FRET1 substrate, the 8-amino acid truncated variants exhibited poorer potency, with the C-terminally (P12-3B) and N-terminally (P12-3C) truncated variants exhibiting of Ki values of 39 ± 20 μM and 35 ± 4.7 μM, respectively, and the dual N- and C-terminally truncated variant (P12-3D) exhibiting even higher Ki values of 102 ± 14 μM (Table 1). When insulin was used as a substrate, none of the truncated variants of P12-3 exhibited Ki values <10 μM.
For the final peptide series, P12-4 and its derivatives (Table 1, Panel F in S1 Fig), the full-length unmodified (P12-4) and amidated (P12-4A) versions showed good potency against FRET1 (Ki = 3.9 ± 0.5 μM and 4.2 ± 1.3 μM, respectively) and insulin (Ki = 1.5 ± 0.3 μM and 2.6 ± 0.7 μM, respectively), while none of the truncated variants (P12-4B, P12-4C and P12-4D) exhibited Ki values below 500 μM or 10 μM for FRET1 or insulin, respectively (Table 1).
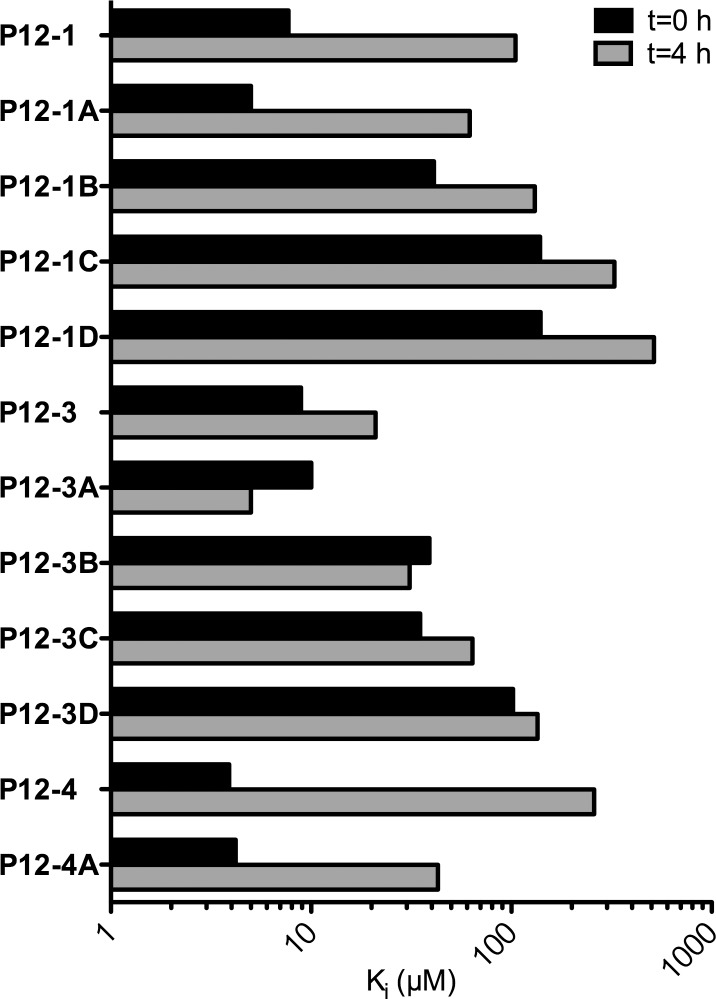
The sequences exhibiting high potency could be used either as conventional peptides—a preferred outcome due to their low cost of synthesis and low intrinsic toxicity—or, instead, as the starting point for the development of derivatives containing modifications that confer resistance to degradation (e.g., D-amino acids, beta-amino acids, etc.). To determine which peptides were susceptible to degradation by IDE, we incubated twelve peptides showing quantifiable inhibitory potency together with IDE for an extended period (4 h), then the potency of each was determined and compared to the potency immediately after addition of the enzyme (0 h). As shown in Fig 2, most peptides exhibited significant reductions in potency (i.e., increases in Ki values) after a 4-h incubation with IDE, reflecting proteolytic degradation by IDE. Notable exceptions to this trend included P12-3A and P12-3B, particularly P12-3A (QSLPWCYPHCVT-amide).
Fig 2. Vulnerability of peptides to degradation by IDE assessed by activity assays.
IC50 values obtained for selected peptides pre-incubated with IDE 0 or 4 h before testing with the FRET1 assay. Note that all peptides except P12-3A and P12-3B showed reductions in apparent potency after prolonged incubation with IDE, reflecting degradation. Data are the average of duplicate assays that did not differ by more than 5%.
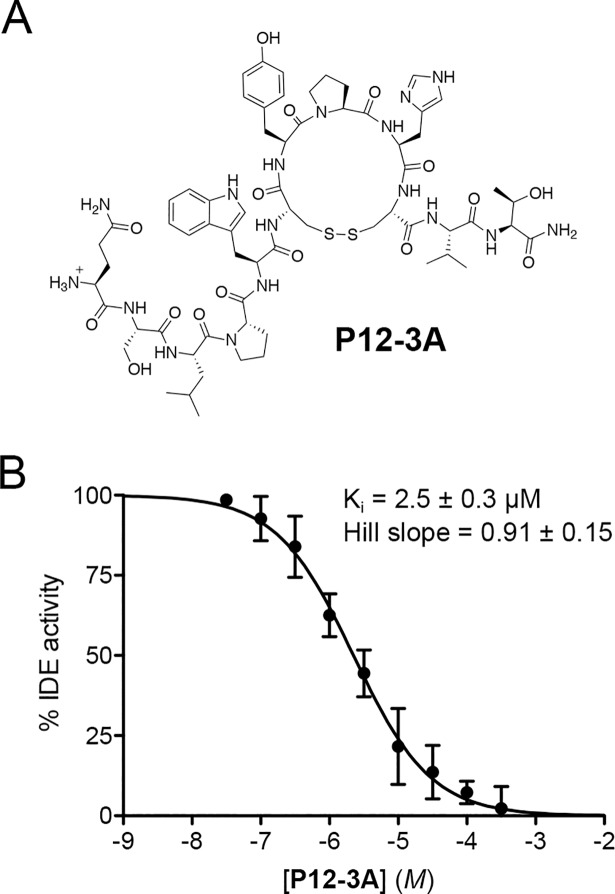
Taken together with the potency of all peptides against insulin (Table 1), we concluded that P12-3A represented the best inhibitor for further studies, and a highly purified, deliberately cyclized version (Fig 3A) was synthesized. This cosmetic-grade version of P12-3A was found to be soluble up to ~500 μM in assay medium (PBS/0.05%BSA) and up to ~100 μM in cell culture medium, and was confirmed to exhibit similar potency against insulin degradation (Ki = 2.5 ± 0.31 μM, n = 5) (Fig 3B), and highly consistent inhibition constants were also observed for the degradation of two other substrates, FRET1 (Ki = 2.7 ± 0.50 μM, n = 6) and amyloid ß-protein (Aß) (Ki = 2.1 ± 0.34 μM, n = 6). Notably, P12-3A exhibited essentially no inhibition against 15 different proteases tested (S4 Fig), suggesting it is highly selective for IDE. Based on its potency, stability and selectivity for IDE, P12-3A was selected for use in downstream assays.
Fig 3. Structure and activity of P12-3A.
A, Structure of cyclized P3-12A. B, Dose-response of P12-3A against insulin degradation by IDE. Data are mean ± SEM of 5 independent experiments.
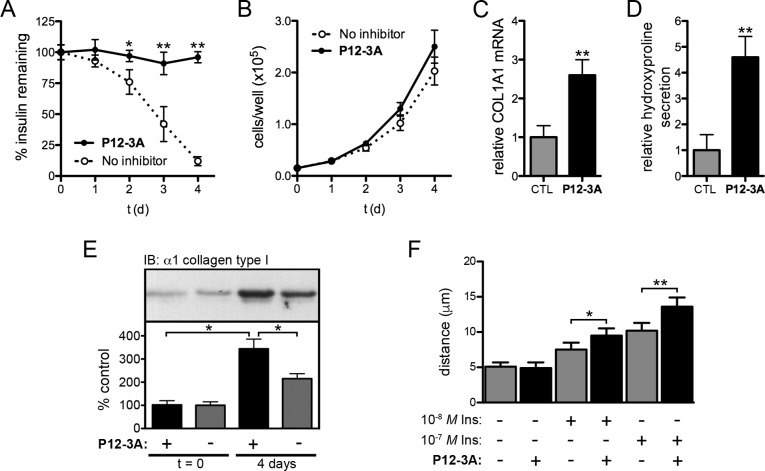
Insulin promotes wound healing and overall skin health by affecting a number of processes, including cell proliferation [2, 3], cell migration [5, 6], and the production and secretion of ECM components, particularly type I collagen [7–13]. We therefore assessed the ability of P12-3A to influence these processes in cultured skin fibroblasts and keratinocytes. To that end, primary mouse skin fibroblasts were grown for 4 d in multiple concentrations of insulin in the presence or absence of P12-3A, and insulin concentration over time was monitored by ELISA. In untreated cells, 10 nM insulin was degraded approximately 90% by day 4 in logarithmically growing cells, whereas insulin levels remained constant in cells treated with either 100 μM P12-3A (Fig 4A). These results demonstrate that P12-3A is effective at inhibiting insulin degradation by skin fibroblasts and, moreover, confirm that the compound is stable in biological milieu. Relative to control cells, cell proliferation in the presence of 10 nM insulin was found to be modestly increased in the presence of 100 μM P12-3A (Fig 4B), but this did not achieve statistical significance. Finally, the effects of P12-3A on collagen production in were assessed at both the transcriptional and the posttranslational level in confluent monolayers of fibroblasts. After 4 d of treatment with P12-3A (100 μM), mRNA levels for the major form of collagen, alpha-1 type I collagen (COL1A1), were quantified by RT-PCR and found to be increased to ~2.6 times the levels of untreated cells (Fig 4C). To assess overall levels of collagen production and secretion, we quantified levels of hydroxyproline, a modified amino acid present almost exclusively in collagen proteins [47]. In the presence of P12-3A, hydroxyproline levels secreted into the medium were found to be increased to levels ~4.6 times that secreted by untreated cells (Fig 4D). Western blotting also confirmed that mature, cell-associated collagen levels were increased in the presence of P12-3A (Fig 4E). Finally, using an in vitro scratch wound assay, P12-3A (100 μM) was found to result in statistically significant increases in the migration of keratinocytes in the presence of different concentrations of insulin (Fig 4F).
Fig 4. Effects of P12-3A on cultured skin cells.
A, Insulin concentrations as a function of time in logarithmically growing primary murine skin fibroblasts in the absence or presence of P12-3A (100 μM). Note that insulin levels remain constant in the presence of P12-3A, reflecting both the effectiveness of the peptide inhibiting insulin degradation and also the stability of the peptide in biological milieu. Data are mean ± SEM of 4 independent replications. *P<0.05, **P<0.01. B, Proliferation of cells in the absence or presence of P12-3A (100 μM). Data are mean ± SEM of 6 independent replications. No significant differences were observed. C,D,E, P12-3A (100 μM) potentiates insulin-induced collagen production in skin fibroblasts. Collagen production was assessed by COL1A1 mRNA levels (C), levels of hydroxyproline secreted into the medium (D), and cell-associated mature alpha-1 type I collagen levels detected by Western blotting (E). Data are mean ± SD of 4 independent replications. *P<0.05, **P<0.01. F, P12-3A (100 μM) potentiates the migration of keratinocytes in a scratch wound assay. Migration of HaCaT cells 48 h after induction of a scratch wound in the presence of the indicated quantities of insulin and/or P12-3A (100 μM). Data are mean ± SEM of 6 independent replications. *P<0.05, **P<0.01.
Discussion
Although a variety of potent and selective IDE inhibitors have been developed [28, 29, 39–43], current inhibitors are difficult to synthesize, expensive to generate and/or contain chemical moieties or constituents with established or undetermined potential for toxicity. Due to these and other considerations, existing IDE inhibitors are poorly suited for topical applications. To overcome these limitations, in the present study we aimed to develop peptidic inhibitors of IDE suitable for use in wound healing or cosmetic applications. Peptides are easy to manufacture and therefore inexpensive to scale up and, being composed solely of all-natural amino acids, are unlikely to possess any degree of toxicity. To that end, we used phage display technology to select for a range of peptides that bind to IDE with strong affinity, which were then screened for resistance to degradation by IDE. One cyclic dodecapeptide in particular, P12-3A, proved to be a potent and stable inhibitor of IDE that showed excellent selectivity and also showed no evidence of toxicity in cell culture experiments. Critically, P12-3A was found to potentiate a number of insulin-stimulated processes relevant to wound healing and skin health, including collagen production by fibroblasts and migration of keratinocytes in response to scratch wounds.
Phage display proved to be a highly effective approach for developing peptidic inhibitors. From among six parent peptides selected for further testing, four exhibited low-micromolar Ki values against insulin, and one (P12-1) exhibited sub-micromolar potency (Ki = 0.8 ± 0.04 μM). Among the modified versions of these parent peptides, an additional five exhibited Ki values <10 μM; thus nine of the twenty-five peptides tested (36%) showed good activity. These hit rates are markedly higher than those obtained through high-throughput compound screening [41, 45, 48] or other approaches [29]. Notably, the potency of P12-1 (Ki = 800 nM) compares favorably to that of the highly optimized and extensively characterized macrocyclic IDE inhibitor, 6bK, which shows a IC50 value of ~100 nM against insulin [29]. Given that 6bK underwent considerable optimization [29], the potency of P12-1, a simple, unmodified dodecapeptide, is notable.
Although the majority of peptides showed good affinity for IDE, most were also degraded by it. Due to the peculiarities of its structure [26, 49–51], IDE is a pure peptidase that cannot degrade proteins; thus it is unsurprising that phage display would reveal sequences that bind strongly while attached to the bacteriophage coat protein but are nevertheless degraded when synthesized as a short peptide. The particular stability of P12-3A (and related peptides) likely derives from the fact that it contains two cysteines and can therefore form a cyclic peptide. Of note, it is unusual for cyclic peptides to emerge from a library of linear peptides, because cyclization tends to slow the maturation of the bacteriophage, leaving the phage expressing them at a competitive disadvantage when grown in parallel with phage expressing linear peptides. This suggests this peptide sequence was strongly favored during the selection process.
Based on the ability of P12-3A to potentiate insulin-stimulated collagen production and cell migration, topical IDE inhibition would appear to hold significant therapeutic potential in wound healing, particularly for diabetic patients [1]. Given the accruing evidence that insulin signaling pathways are critical for wound healing [1, 22] and, given that IDE is abundant in wound fluid [37, 38], where it actively degrades insulin, there is a strong prediction that pharmacological inhibition of IDE will promote wound healing [28]. This prediction is strongly supported by the finding that IDE inhibitors potentiate insulin action in vivo in part by preserving endogenous insulin [32, 33] and possibly via actions downstream of insulin receptor binding [28]. Importantly, by contrast to direct topical administration insulin, which can cause life-threatening hypoglycemia [25], pharmacological inhibition of IDE possesses no intrinsic risk of triggering hypoglycemia [32–34].
One of the most immediately implementable potential cosmetic applications for P12-3A may be as an adjuvant for microneedling procedures [52]. Also known as percutaneous collagen induction [53, 54], microneedling is a minimally invasive, widely used technique by which production of ECM proteins in the dermis can be stimulated by introducing uniform, sterile wounds in a controlled manner [52]. Although originally developed for skin rejuvenation, this technique is now being used as a novel treatment for a wide range of cosmetic and medical conditions, including acne, alopecia, stretch marks, hyperhidrosis and scarring of multiple types [52, 53]. Topical application of P12-3A prior to microneedling would permit the delivery of the peptide subcutaneously [52], thus maximizing its impact on the processes involved in wound repair.
In sum, using phage display technology, we have generated novel peptidic inhibitors of IDE that, by virtue of their low cost of synthesis and minimal risk of toxicity, have the properties needed to explore the therapeutic and cosmetic potential of topical IDE inhibition. Given the importance of insulin in wound healing and normal skin health, these novel inhibitors, as well as future derivatives thereof, will be useful for exploring the involvement of IDE in these processes, and may also hold significant value as adjuvants for medicinal and cosmetic treatments.
Materials and methods
Materials
Anti-alpha-1 type I collagen antibody (Cat. No. AB765P) and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (Cat. No. A0545) were from Sigma-Aldrich (St. Louis, MO, USA). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Cat. No. AF5718) antibody was from R&D Systems (Minneapolis, MN, USA). HRP-conjugated anti-goat IgG antibody (Cat. No. sc-2354) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Materials for Western blotting and cell culture were from Thermo Fisher Scientific (Waltham, MA, USA). Insulin ELISAs (Cat. No. 90082) were from Crystal Chem (Downers Grove, IL, USA). Primary murine skin fibroblasts were a generous gift from Dr. Jorge Busciglio (UC Irvine). HaCaT cells and optimized growth medium were purchased from AddexBio Technologies (San Diego, CA, USA). Unless specified, all other reagents were from Sigma-Aldrich (St. Louis, MO, USA).
Phage display
The selection of IDE-binding peptide sequences was conducted by phage display using the Ph.D.™-C7C and Ph.D.™-12 Phage Display Library Kits from New England Biolabs (Ipswich, MA, USA) according to manufacturer’s recommendations. Briefly, purified, glycerol-free, recombinant human IDE (100 μg/mL) [35] was immobilized onto Corning® High Bind, 96-well, round-bottom plates (Cat. No. CLS3366). After washing and prior to addition of bacteriophage, activity assays with FRET1 (see below) were used to confirm the presence of proteolytically active IDE in wells coated in parallel with those used for panning. Three rounds of panning were conducted, with 2 x 1011 phage/well added at each step. After incubation at room temperature for 60 min, bound phage were eluted by addition of excess recombinant human insulin (100 μg/mL) and amplified for the subsequent round of panning. After the third round of panning, the eluate was titered and individual clones were selected for DNA sequencing. Peptide sequences were decoded and consensus sequences searched for using CLC Sequence Viewer (Version 7.5).
Peptide synthesis
Peptides were synthesized by automated solid-phase peptide synthesis by Sigma-Aldrich, with the exception of C7C-1, which was synthesized in-house essentially as described [28] and analyzed by electrospray-ionization mass spectrometry (ESI-MS; S2 Fig) and HPLC (S3 Fig). Cosmetic-grade, gram-scale quantities of P12-3A were synthesized by GenScript Biotechnology Corp. (Piscataway Township, NJ, USA).
Degradation assays
IDE activity was quantified by monitoring the degradation of Mca-GGFLRKVGQK(Dnp) (FRET1, 5 μM) [49, 55], fluoresceinated and biotinylated amyloid ß-protein (FAßB; 500 nM) or recombinant human insulin (50 nM) in PBS supplemented with 0.05% BSA. FRET1 degradation was measured by changes in fluorescence (λex = 340 nm, λem = 420 nm); FAßB degradation was monitored by fluorescence polarization (λex = 488 nm, λem = 525 nm), as described [56]; and insulin degradation was quantified by ELISA. In vitro activity assays incorporated recombinant human IDE (1 nM) purified from bacteria [35]. For quantitation of insulin degradation in primary murine skin fibroblasts, cells (1 x 105/well) were plated in 96-well plates and maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 2mM glutamine, penicillin and streptomycin. After addition of insulin (10 nM or 100 nM), samples of conditioned medium were removed daily and quickly frozen, then insulin levels were quantified in parallel by ELISA according to manufacturer’s recommendations (Crystal Chem, Downers Grove, IL, USA). Assessment of the activity of P12-3A against a variety of matrix-metalloproteases was conducted using the Matrix Metalloproteinase (MMP) Inhibitor Profiling Kit, Fluorometric RED (Enzo Life Sciences, Inc., Farmingdale, NY, USA) according to manufacturer’s recommendations using the broad-spectrum MMP inhibitor, NNGH, as a positive control. Activity assays on additional proteases were conducted using the FAßB degradation assay, using a custom protease inhibitor cocktail (PIC) comprised of cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail supplemented with 1,10-phenanthroline (2 mM) and pepstatin A (5 μM).
Cell proliferation
Primary murine skin fibroblasts cells (1 x 105/well) were plated in 96-well plates, using separate plates for each timepoint and endpoint. Cell proliferation was quantified using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega Corp., Madison, WI, USA) according to manufacturer’s recommendations.
RNA quantification
RNA was extracted from freshly lysed cells, reverse transcribed and amplified using the Ambion® Fast SYBR® Green Cells-to-CT™ Kit according to manufacturer’s recommendations (Thermo Fisher Scientific, Waltham, MA, USA). The quantitative real-time PCR reaction was conducted using a 7500 real-time PCR system and analyzed using System SDS software v2.0.5 (Applied Biosystems). Murine COL1A1 mRNA was detected using the following primers (forward: 5’-ACCTAAGGGTACCGCTGGA and reverse: 5’ TCCAGCTTCTCCATCTTTGC). Fold change differences between samples were determined using the comparative Ct (ΔΔCt) method, normalized to internal standards detected with the SYBR® Green Cells-to-CT™ Control Kit according to manufacturer’s recommendations (Thermo Fisher Scientific, Waltham, MA, USA) calculated by 2-ΔΔCt.
Hydroxyproline quantification
For quantitation of hydroxyproline secretion by primary murine skin fibroblasts, cells (1 x 105/well) were plated in 24-well plates DMEM supplemented with 10% FBS, 2mM glutamine, penicillin, streptomycin and 100 nM insulin, in the absence or presence of 100 μM P12-3A. After incubation for 4 days, the conditioned medium was removed, centrifuged at 1000 x g for 10 min to remove cellular debris, and hydroxyproline levels were quantified using the Hydroxyproline Assay Kit according to manufacturer’s recommendations (Sigma-Aldrich, St. Louis, MO, USA).
Western blotting
Protein was collected using the M-Per Mammalian Extraction Reagent and the concentration was quantified using the Pierce™BCA Protein Assay Kit according to manufacturer’s recommendations (Thermo Fisher Scientific, Waltham, MA, USA). Protein (30 μg/well) was separated SDS-PAGE under reducing conditions using Novex™ 10% polyacrylamide tris-glycine mini gels and transferred to nitrocellulose membranes as described [57]. Briefly, membranes were blocked in 5% non-fat milk in tris-buffered saline supplemented with 0.2% Tween-20 (TBST), cut into segments and incubated for 1 h at room temperature with anti-alpha-1 type I collagen (1:5000) and anti-GAPDH (1:10,000) antibodies, washed extensively in TBST, then probed with anti-rabbit (1:20,000) or anti-goat (1:50,000) secondary antibodies, respectively, and detected by enhanced chemoluminescence using SuperSignal West Pico Substrate. Protein expression, normalized to GAPDH levels, was quantified using the band analysis tools of ImageLab software, version 4.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
In vitro scratch wound assay
Cell migration in HaCaT cells after induction of scratch wounds was quantified essentially as described [58]. Briefly, HaCaT cells maintained in optimized DMEM (AddexBio Technologies, San Diego, CA, USA) supplemented with 10% FBS, penicillin and streptomycin, were grown to confluency in 24-well tissue culture plates, and scratch wounds were induced with a 200-μL pipette tip. After growth for 48 h in the absence or presence of insulin (10 nM or 100 nM) and/or P12-3A (100 μM), cell migration distance was quantified by two independent, blinded observers using a Nikon TMS inverted light microscope (Nikon Corp., Melville, NY, USA) fitted with a ruler reticle.
Statistical analyses
Tests for statistical significance were performed by using the two-tailed Student's t test with various levels of significance (P = 0.05, 0.01). For comparisons with unequal numbers of replications per group, Hartley's Fmax was calculated to check for homogeneity of variance. All calculations and curve fitting were performed in Prism for Mac OS X, version 5.0b (GraphPad Software, Inc., La Jolla, CA, USA).
Supporting information
Note that P12-3, although derived from a library of primarily linear peptides, is predicted to be a cyclic peptide.
(TIF)
The entire spectrum as well as expanded regions are shown.
(TIF)
Note that the purity is ~95%.
(TIF)
Activity of P12-3A (100 μM) against (A) multiple matrix-metalloproteases (MMPs) and (B) multiple peptidases of different protease classes. Note that significant inhibition was observed exclusively for IDE, with modest inhibition (~18%) observed for just one of 15 other proteases tested (MMP-7). Data are mean ± SD, n = 8–16 per group. P<0.05 by 2-tailed Student’s t-test. Note that all positive controls (NNGH or protease inhibitor cocktail (PIC)) exhibited significant inhibition (P< 0.01). See Materials and Methods for details. NEP, neprilysin; mCatD, murine cathepsin D; hCatD, human cathepsin D.
(TIF)
(ZIP)
(XLSX)
Acknowledgments
We thank Drs. Jorge Busciglio and Pinar Coskun for providing the primary mouse fibroblasts and Dr. Richard Chamberlin for access to his automated peptide synthesizer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was generously supported by grants from the National Institutes of Health (R01-GM115617; www.nih.gov), the American Diabetes Association (Career Development Award 7-11-CD-13; www.diabetes.org), and the UCI Applied Innovation Technology Development Innovation Fund (www.innovation.uci.edu) to MAL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hrynyk M, Neufeld RJ. Insulin and wound healing. Burns. 2014;40(8):1433–46. doi: 10.1016/j.burns.2014.03.020 [DOI] [PubMed] [Google Scholar]
- 2.Aaronson SA, Rubin JS, Finch PW, Wong J, Marchese C, Falco J, et al. Growth factor-regulated pathways in epithelial cell proliferation. Am Rev Respir Dis. 1990;142(6 Pt 2):S7–10. [DOI] [PubMed] [Google Scholar]
- 3.Monaco S, Illario M, Rusciano MR, Gragnaniello G, Di Spigna G, Leggiero E, et al. Insulin stimulates fibroblast proliferation through calcium-calmodulin-dependent kinase II. Cell Cycle. 2009;8(13):2024–30. doi: 10.4161/cc.8.13.8813 [DOI] [PubMed] [Google Scholar]
- 4.Wertheimer E, Trebicz M, Eldar T, Gartsbein M, Nofeh-Moses S, Tennenbaum T. Differential roles of insulin receptor and insulin-like growth factor-1 receptor in differentiation of murine skin keratinocytes. J Invest Dermatol. 2000;115(1):24–9. doi: 10.1046/j.1523-1747.2000.00008.x [DOI] [PubMed] [Google Scholar]
- 5.Benoliel AM, Kahn-Perles B, Imbert J, Verrando P. Insulin stimulates haptotactic migration of human epidermal keratinocytes through activation of NF-kappa B transcription factor. J Cell Sci. 1997;110 (Pt 17):2089–97. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Petreaca M, Yao M, Martins-Green M. Cell and molecular mechanisms of keratinocyte function stimulated by insulin during wound healing. BMC Cell Biol. 2009;10:1 doi: 10.1186/1471-2121-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villee DB, Powers ML. Effect of glucose and insulin on collagen secretion by human skin fibroblasts in vitro. Nature. 1977;268(5616):156–8. [DOI] [PubMed] [Google Scholar]
- 8.Kjellstrom T, Malmquist J. Insulin effects on collagen and protein production in cultured human skin fibroblasts from diabetic and non-diabetic subjects. Horm Metab Res. 1984;16(4):168–71. doi: 10.1055/s-2007-1014734 [DOI] [PubMed] [Google Scholar]
- 9.Goldstein RH, Poliks CF, Pilch PF, Smith BD, Fine A. Stimulation of collagen formation by insulin and insulin-like growth factor I in cultures of human lung fibroblasts. Endocrinology. 1989;124(2):964–70. doi: 10.1210/endo-124-2-964 [DOI] [PubMed] [Google Scholar]
- 10.Krupsky M, Fine A, Kuang PP, Berk JL, Goldstein RH. Regulation of type I collagen production by insulin and transforming growth factor-beta in human lung fibroblasts. Connect Tissue Res. 1996;34(1):53–62. [DOI] [PubMed] [Google Scholar]
- 11.Trevisan R, Yip J, Sarika L, Li LK, Viberti G. Enhanced collagen synthesis in cultured skin fibroblasts from insulin-dependent diabetic patients with nephropathy. J Am Soc Nephrol. 1997;8(7):1133–9. [DOI] [PubMed] [Google Scholar]
- 12.Gore-Hyer E, Pannu J, Smith EA, Grotendorst G, Trojanowska M. Selective stimulation of collagen synthesis in the presence of costimulatory insulin signaling by connective tissue growth factor in scleroderma fibroblasts. Arthritis Rheum. 2003;48(3):798–806. doi: 10.1002/art.10953 [DOI] [PubMed] [Google Scholar]
- 13.Musselmann K, Kane B, Alexandrou B, Hassell JR. Stimulation of collagen synthesis by insulin and proteoglycan accumulation by ascorbate in bovine keratocytes in vitro. Invest Ophthalmol Vis Sci. 2006;47(12):5260–6. doi: 10.1167/iovs.06-0612 [DOI] [PubMed] [Google Scholar]
- 14.Wertheimer E, Spravchikov N, Trebicz M, Gartsbein M, Accili D, Avinoah I, et al. The regulation of skin proliferation and differentiation in the IR null mouse: implications for skin complications of diabetes. Endocrinology. 2001;142(3):1234–41. doi: 10.1210/endo.142.3.7988 [DOI] [PubMed] [Google Scholar]
- 15.Baltzis D, Eleftheriadou I, Veves A. Pathogenesis and treatment of impaired wound healing in diabetes mellitus: new insights. Adv Ther. 2014;31(8):817–36. doi: 10.1007/s12325-014-0140-x [DOI] [PubMed] [Google Scholar]
- 16.Hanam SR, Singleton CE, Rudek W. The effect of topical insulin on infected cutaneous ulcerations in diabetic and nondiabetic mice. J Foot Surg. 1983;22(4):298–301. [PubMed] [Google Scholar]
- 17.Belfield WO, Golinsky S, Compton MD. The use of insulin in open-wound healing. Vet Med Small Anim Clin. 1970;65(5):455–60. [PubMed] [Google Scholar]
- 18.Weringer EJ, Kelso JM, Tamai IY, Arquilla ER. Effects of insulin on wound healing in diabetic mice. Acta Endocrinol (Copenh). 1982;99(1):101–8. [DOI] [PubMed] [Google Scholar]
- 19.Madibally SV, Solomon V, Mitchell RN, Van De Water L, Yarmush ML, Toner M. Influence of insulin therapy on burn wound healing in rats. J Surg Res. 2003;109(2):92–100. [DOI] [PubMed] [Google Scholar]
- 20.Apikoglu-Rabus S, Izzettin FV, Turan P, Ercan F. Effect of topical insulin on cutaneous wound healing in rats with or without acute diabetes. Clin Exp Dermatol. 2010;35(2):180–5. doi: 10.1111/j.1365-2230.2009.03419.x [DOI] [PubMed] [Google Scholar]
- 21.Wilson JM, Baines R, Babu ED, Kelley CJ. A role for topical insulin in the management problematic surgical wounds. Ann R Coll Surg Engl. 2008;90(2):160 doi: 10.1308/003588408X261816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima MH, Caricilli AM, de Abreu LL, Araujo EP, Pelegrinelli FF, Thirone AC, et al. Topical insulin accelerates wound healing in diabetes by enhancing the AKT and ERK pathways: a double-blind placebo-controlled clinical trial. PLoS One. 2012;7(5):e36974 doi: 10.1371/journal.pone.0036974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenway SE, Filler LE, Greenway FL. Topical insulin in wound healing: a randomised, double-blind, placebo-controlled trial. J Wound Care. 1999;8(10):526–8. doi: 10.12968/jowc.1999.8.10.26217 [DOI] [PubMed] [Google Scholar]
- 24.Rezvani O, Shabbak E, Aslani A, Bidar R, Jafari M, Safarnezhad S. A randomized, double-blind, placebo-controlled trial to determine the effects of topical insulin on wound healing. Ostomy Wound Manage. 2009;55(8):22–8. [PubMed] [Google Scholar]
- 25.Coid DR. Hypoglycaemia during treatment of decubitus ulcer with topical insulin. Br Med J. 1977;2(6094):1063–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang WJ. Targeting Insulin-Degrading Enzyme to Treat Type 2 Diabetes Mellitus. Trends Endocrinol Metab. 2016;27(1):24–34. doi: 10.1016/j.tem.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leal MC, Morelli L. Insulysin In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. 1 3rd ed: Academic Press; 2013. p. 1415–20. [Google Scholar]
- 28.Leissring MA, Malito E, Hedouin S, Reinstatler L, Sahara T, Abdul-Hay SO, et al. Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin. PLoS One. 2010;5(5):e10504 doi: 10.1371/journal.pone.0010504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maianti JP, McFedries A, Foda ZH, Kleiner RE, Du XQ, Leissring MA, et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature. 2014;511(7507):94–8. doi: 10.1038/nature13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirsky IA, Perisutti G. Effect of insulinase-inhibitor on hypoglycemic action of insulin. Science. 1955;122(3169):559–60. [PubMed] [Google Scholar]
- 31.Mirsky IA, Perisutti G, Diengott D. Effect of insulinase-inhibitor on destruction of insulin by intact mouse. Proc Soc Exp Biol Med. 1955;88(1):76–8. [DOI] [PubMed] [Google Scholar]
- 32.Abdul-Hay SO, Kang D, McBride M, Li L, Zhao J, Leissring MA. Deletion of insulin-degrading enzyme elicits antipodal, age-dependent effects on glucose and insulin tolerance. PLoS One. 2011;6(6):e20818 doi: 10.1371/journal.pone.0020818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):4162–7. doi: 10.1073/pnas.0230450100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, et al. Amyloid-beta peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):6221–6. doi: 10.1073/pnas.1031520100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farris W, Leissring MA, Hemming ML, Chang AY, Selkoe DJ. Alternative splicing of human insulin-degrading enzyme yields a novel isoform with a decreased ability to degrade insulin and amyloid beta-protein. Biochemistry. 2005;44(17):6513–25. doi: 10.1021/bi0476578 [DOI] [PubMed] [Google Scholar]
- 36.Kuo WL, Montag AG, Rosner MR. Insulin-degrading enzyme is differentially expressed and developmentally regulated in various rat tissues. Endocrinology. 1993;132(2):604–11. doi: 10.1210/endo.132.2.7678795 [DOI] [PubMed] [Google Scholar]
- 37.Shearer JD, Coulter CF, Engeland WC, Roth RA, Caldwell MD. Insulin is degraded extracellularly in wounds by insulin-degrading enzyme (EC 3.4.24.56). Am J Physiol. 1997;273(4 Pt 1):E657–64. [DOI] [PubMed] [Google Scholar]
- 38.Duckworth WC, Fawcett J, Reddy S, Page JC. Insulin-degrading activity in wound fluid. J Clin Endocrinol Metab. 2004;89(2):847–51. doi: 10.1210/jc.2003-031371 [DOI] [PubMed] [Google Scholar]
- 39.Durham TB, Toth JL, Klimkowski VJ, Cao JX, Siesky AM, Alexander-Chacko J, et al. Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo. J Biol Chem. 2015;290(33):20044–59. doi: 10.1074/jbc.M115.638205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charton J, Gauriot M, Totobenazara J, Hennuyer N, Dumont J, Bosc D, et al. Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme. European journal of medicinal chemistry. 2015;90:547–67. doi: 10.1016/j.ejmech.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdul-Hay SO, Bannister TD, Wang H, Cameron MD, Caulfield TR, Masson A, et al. Selective targeting of extracellular insulin-degrading enzyme by quasi-irreversible thiol-modifying inhibitors. ACS Chem Biol. 2015;10(12):2716–24. doi: 10.1021/acschembio.5b00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charton J, Gauriot M, Guo Q, Hennuyer N, Marechal X, Dumont J, et al. Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis. European journal of medicinal chemistry. 2014;79:184–93. doi: 10.1016/j.ejmech.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdul-Hay SO, Lane AL, Caulfield TR, Claussin C, Bertrand J, Masson A, et al. Optimization of peptide hydroxamate inhibitors of insulin-degrading enzyme reveals marked substrate-selectivity. J Med Chem. 2013;56(6):2246–55. doi: 10.1021/jm301280p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–7. [DOI] [PubMed] [Google Scholar]
- 45.Cabrol C, Huzarska MA, Dinolfo C, Rodriguez MC, Reinstatler L, Ni J, et al. Small-molecule activators of insulin-degrading enzyme discovered through high-throughput compound screening. PLoS One. 2009;4(4):e5274 doi: 10.1371/journal.pone.0005274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–108. [DOI] [PubMed] [Google Scholar]
- 47.Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339(1):247–57. doi: 10.1007/s00441-009-0844-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bannister TD, Wang H, Abdul-Hay SO, Masson A, Madoux F, Ferguson J, et al. ML345, A Small-Molecule Inhibitor of the Insulin-Degrading Enzyme (IDE). Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD)2010. [PubMed] [Google Scholar]
- 49.Neant-Fery M, Garcia-Ordonez RD, Logan TP, Selkoe DJ, Li L, Reinstatler L, et al. Molecular basis for the thiol sensitivity of insulin-degrading enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(28):9582–7. doi: 10.1073/pnas.0801261105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443(7113):870–4. doi: 10.1038/nature05143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leissring MA, Selkoe DJ. Structural biology: enzyme target to latch on to. Nature. 2006;443(7113):761–2. doi: 10.1038/nature05210 [DOI] [PubMed] [Google Scholar]
- 52.Singh A, Yadav S. Microneedling: Advances and widening horizons. Indian dermatology online journal. 2016;7(4):244–54. doi: 10.4103/2229-5178.185468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aust MC, Fernandes D, Kolokythas P, Kaplan HM, Vogt PM. Percutaneous collagen induction therapy: an alternative treatment for scars, wrinkles, and skin laxity. Plastic and reconstructive surgery. 2008;121(4):1421–9. doi: 10.1097/01.prs.0000304612.72899.02 [DOI] [PubMed] [Google Scholar]
- 54.Fernandes D. Minimally invasive percutaneous collagen induction. Oral and maxillofacial surgery clinics of North America. 2005;17(1):51–63, vi. doi: 10.1016/j.coms.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 55.Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, et al. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007;282(35):25453–63. doi: 10.1074/jbc.M701590200 [DOI] [PubMed] [Google Scholar]
- 56.Leissring MA, Lu A, Condron MM, Teplow DB, Stein RL, Farris W, et al. Kinetics of amyloid beta-protein degradation determined by novel fluorescence- and fluorescence polarization-based assays. J Biol Chem. 2003;278(39):37314–20. doi: 10.1074/jbc.M305627200 [DOI] [PubMed] [Google Scholar]
- 57.Delledonne A, Kouri N, Reinstatler L, Sahara T, Li L, Zhao J, et al. Development of monoclonal antibodies and quantitative ELISAs targeting insulin-degrading enzyme. Molecular neurodegeneration. 2009;4:39 doi: 10.1186/1750-1326-4-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nature protocols. 2007;2(2):329–33. doi: 10.1038/nprot.2007.30 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note that P12-3, although derived from a library of primarily linear peptides, is predicted to be a cyclic peptide.
(TIF)
The entire spectrum as well as expanded regions are shown.
(TIF)
Note that the purity is ~95%.
(TIF)
Activity of P12-3A (100 μM) against (A) multiple matrix-metalloproteases (MMPs) and (B) multiple peptidases of different protease classes. Note that significant inhibition was observed exclusively for IDE, with modest inhibition (~18%) observed for just one of 15 other proteases tested (MMP-7). Data are mean ± SD, n = 8–16 per group. P<0.05 by 2-tailed Student’s t-test. Note that all positive controls (NNGH or protease inhibitor cocktail (PIC)) exhibited significant inhibition (P< 0.01). See Materials and Methods for details. NEP, neprilysin; mCatD, murine cathepsin D; hCatD, human cathepsin D.
(TIF)
(ZIP)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.