Abstract
The small G-protein Ras is a conserved regulator of cell and tissue growth. These effects of Ras are mediated largely through activation of a canonical RAF-MEK-ERK kinase cascade. An important challenge is to identify how this Ras/ERK pathway alters cellular metabolism to drive growth. Here we report on stimulation of RNA polymerase III (Pol III)-mediated tRNA synthesis as a growth effector of Ras/ERK signalling in Drosophila. We find that activation of Ras/ERK signalling promotes tRNA synthesis both in vivo and in cultured Drosophila S2 cells. We also show that Pol III function is required for Ras/ERK signalling to drive proliferation in both epithelial and stem cells in Drosophila tissues. We find that the transcription factor Myc is required but not sufficient for Ras-mediated stimulation of tRNA synthesis. Instead we show that Ras signalling promotes Pol III function and tRNA synthesis by phosphorylating, and inhibiting the nuclear localization and function of the Pol III repressor Maf1. We propose that inhibition of Maf1 and stimulation of tRNA synthesis is one way by which Ras signalling enhances protein synthesis to promote cell and tissue growth.
Author summary
The Ras oncogene is one of the primary drivers of cell and tissue growth in both normal development and in diseases such as cancer. In this report, we identify the stimulation of tRNA synthesis as an important mechanism by which Ras functions. Using fruit fly genetics, we show that Ras promotes tRNA synthesis by inhibiting Maf1, a protein that normally inhibits RNA polymerase III, the enzyme complex that stimulates tRNA synthesis. We further show that stimulation of tRNA synthesis is required for Ras to promote growth in two important cell types—stem cells and epithelial cells. This work provides new insight into mechanisms that are important for growth and that may contribute to cancer.
Introduction
The Ras small G-protein is one of the key conserved regulators of cell growth and proliferation. Over three decades of research have defined the textbook model of how Ras is activated by growth factors to stimulate a core RAF kinase, MEK (Mitogen-activated protein kinase kinase) and ERK (Extracellular signal–regulated kinase) signalling cascade. Work in model organisms such as Drosophila, C elegans and mouse has shown how this Ras/ERK pathway coordinates tissue growth and patterning to control organ size during development and homeostatic growth in adults.
Given its central role in development it is not surprising that defects in Ras signalling contribute to disease. Most notably, activating mutations in Ras and RAF occur in a large percentage of cancers, and lead to hyper-activation of ERK, which drives tumour formation in both epithelial and stem cells [1]. Ras pathway mutations are also seen in several genetic developmental disorders–described collectively as RASopathies–often characterized by abnormal growth[2]. Understanding how Ras promotes cell proliferation and tissue growth is therefore an important concern in biology.
Drosophila has been a powerful model system to understand the biological roles of Ras signalling. In flies, Ras functions downstream of epidermal growth factor (EGF) and activation of its tyrosine kinase receptor (the EGFR). A series of genetic studies initiated over 25 years ago were pivotal in defining the canonical EGFR/Ras/ERK pathway in Drosophila (for reviews of this early work see:[3,4]). Extensive studies since then have established when, where and how the pathway is activated during the fly life cycle to control development. This work has emphasized the importance of Ras signalling in the control of cell growth and proliferation (e.g. [5–9]. Notably, during larval development Ras/ERK promotes EGFR-mediated cell proliferation and tissue growth in epithelial organs such as the imaginal discs, which eventually give rise to adult structures such as the legs, wings and eyes [10–14]. In addition, in the adult the EGFR/Ras/ERK signalling controls proliferation of stem cell populations to maintain homeostasis and promote regenerative growth [15–19].
How does Ras mediate these effects on cell and tissue growth? Most work on this area has focused on transcriptional effects of Ras signalling. Work in Drosophila has identified several transcription factors that are targeted by ERK such as fos, capicua, and pointed, and that regulate growth [19–22]. Ras signalling has also been shown to crosstalk with other transcriptional regulators of growth such as the hippo/yorkie pathway and dMyc [10,23–27]. These transcriptional effects control expression of metabolic and cell cycle genes important for growth[20,21]. Less is known, however, about how Ras/ERK may regulate mRNA translation to drive growth. The prevailing view, arising mostly from mammalian tissue culture experiments, is that ERK controls protein synthesis by stimulating the activity of translation initiation factors[28]. In particular, these effects are mediated via two ERK effector families—the MNK (MAP kinase-interacting serine/threonine-protein kinase) and RSK (ribosomal s6 kinase) kinases [28–30]. These kinases are important for cellular transformation and tumour growth in mammalian cells [31–34]. However, MNK and RSK mutants in mice and Drosophila have little growth or developmental phenotypes, and mouse MNK mutant cells show no alterations in protein synthesis [33–37]. These findings suggest Ras uses additional mechanisms to control translation and growth in vivo during animal development.
In this paper, we report that the Ras/ERK pathway can stimulate RNA polymerase III-dependent tRNA synthesis. We find that these effects are required for Ras to drive proliferation in both epithelial and stem cells. Finally, we show that ERK promotes tRNA synthesis by inhibiting the Pol III repressor Maf1. These findings suggest that stimulation of tRNA synthesis may be one way that Ras promotes mRNA translation to drive cell and tissue growth.
Results
Activation of Ras/ERK signalling leads to increased protein synthesis
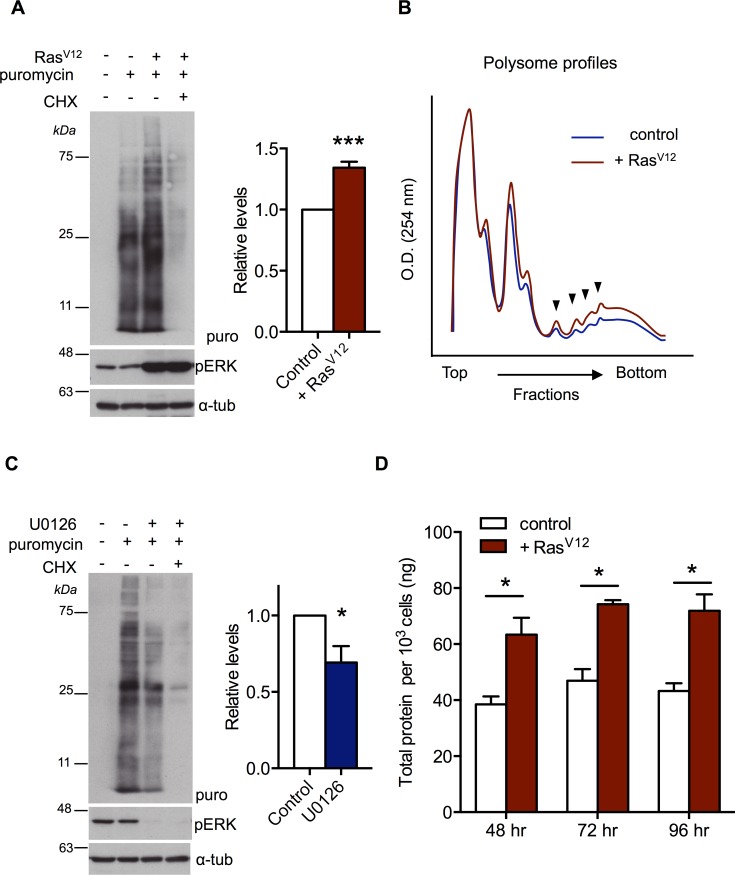
We first examined whether Ras signalling regulates protein synthesis in Drosophila S2 cells using a puromycin-labelling assay [38]. When a constitutively active Ras mutant (RasV12) was expressed in Drosophila S2 cells using an inducible expression vector, we found an increase in protein synthesis, which was blocked by treatment of cells with cycloheximide (CHX), an inhibitor of mRNA translation (Fig 1A). Also, using polysome profiling to measure mRNA translation, we saw an increase in polysome levels in RasV12 overexpressing cells when compared with control cells (Fig 1B). Conversely when we blocked Ras/ERK signalling by treating cells with the MEK inhibitor, U0126, protein synthesis was decreased (Fig 1C). Finally, we found that total protein content/cell increased after RasV12 was overexpressed in S2 cells (Fig 1D). Our findings suggest that one way that the Ras/ERK signalling pathway may drive growth in Drosophila is by promoting protein synthesis.
Fig 1. Activated Ras/ERK signalling pathway stimulates protein synthesis.
(A) Left: RasV12 expression was induced in cultured Drosophila S2 cells for 24 hrs. Cells were then incubated in puromycin for 30 min. Protein extracts were separated by SDS-PAGE and analyzed by western blot with an antibody to puromycin to measure the levels of puromycin-labelled peptides. Cycloheximide treatment was for 15 mins prior to addition of puromycin. A phospho-ERK immunoblot is shown as an indication of Ras/ERK signalling pathway activation. An alpha-tubulin immunoblot is shown as a loading control. Right: Experiments were performed in at least three biological replicates and western blots were quantified using NIH Image J software. Data are represented as relative levels (mean +/- SEM) compared to control (B) A Representative polysome profiles from control S2 cells (blue) and S2 cells with induced RasV12 expression (red). Polysome peaks (arrowheads) in RasV12 expressing cells were higher compared to controls, suggesting translation was increased. (C) Left: Drosophila S2 cells were treated with in the presence or absence of 10 μM U0126 for 2 hours at 25°C. Cells were then incubated in puromycin for 30 min. Protein extracts were separated by SDS-PAGE and analyzed by western blot with an antibody to puromycin to measure the levels of puromycin-labelled peptides. A phospho-ERK immunoblot is shown as an indication of Ras/ERK signalling pathway activation. An alpha-tubulin immunoblot is shown as a loading control. Right: Experiments were performed in at least three biological replicates and western blots were quantified using NIH Image J software. Data are represented as relative levels (mean +/- SEM) compared to control. (D) RasV12 expression was induced in cultured Drosophila S2 cells for 48–96 hrs. Total protein content per 103 cells was calculated using a Bradford assay. Data represent mean +/- SEM for at least three independent replicates per time point.
Ras/ERK signalling promotes tRNA synthesis
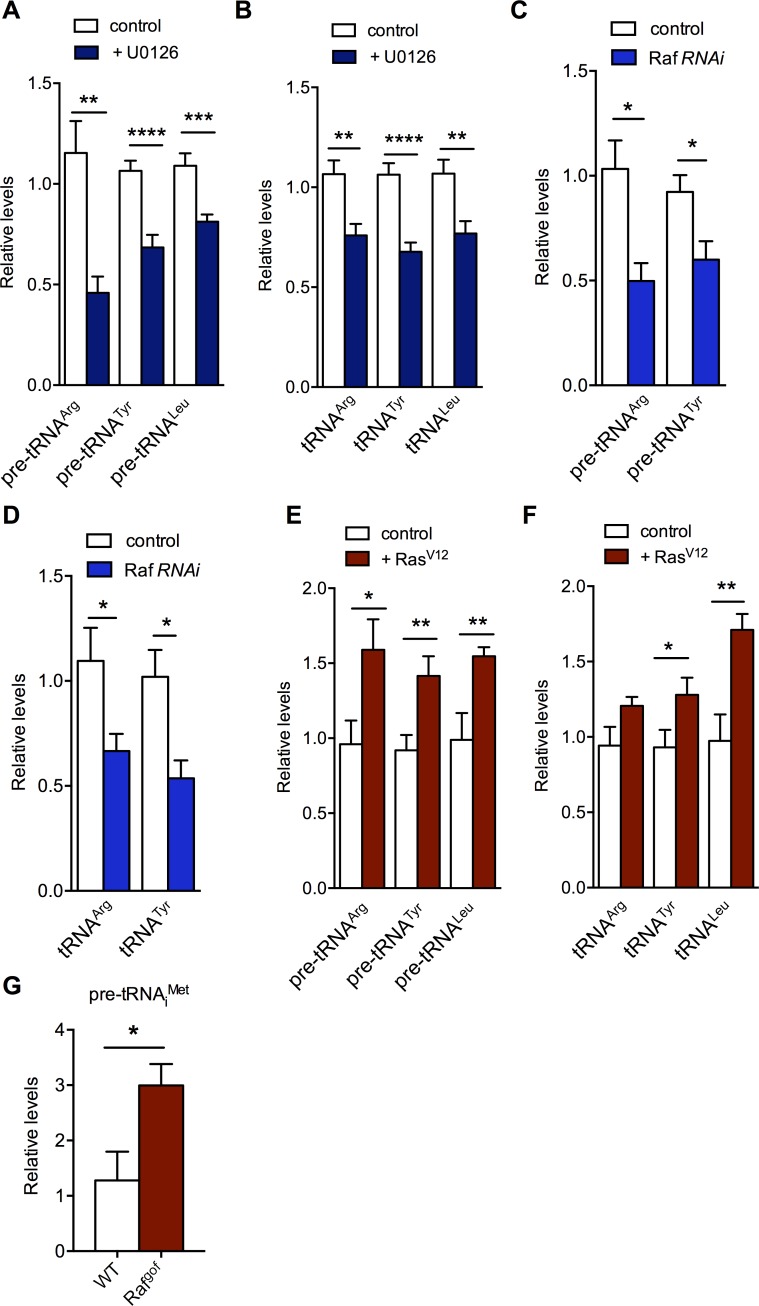
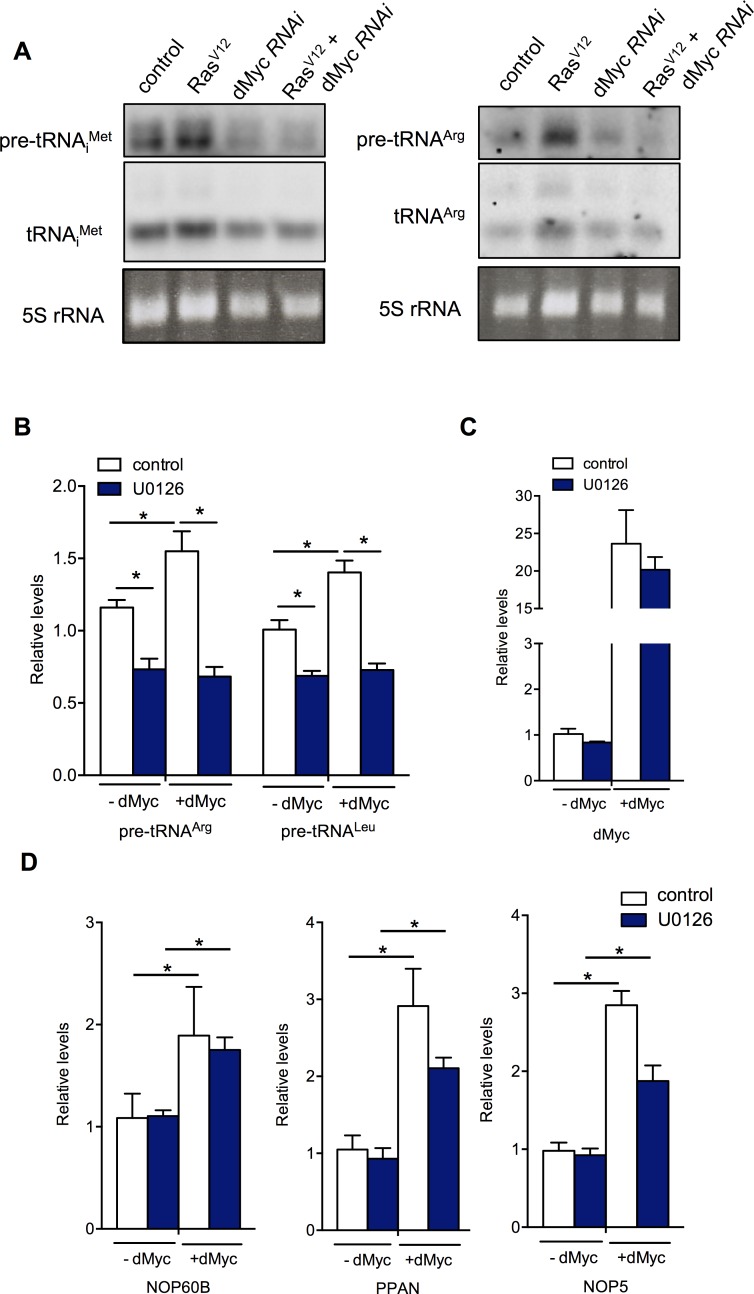
We previously identified the regulation of RNA Polymerase III and tRNA synthesis as a mechanism for controlling protein synthesis in Drosophila larvae [39,40]. We showed that these processes were regulated by TORC1 kinase signalling, and that they were important for driving tissue and body growth[39,40]. We were therefore interested in examining Ras signalling could also promote tRNA synthesis. We first used qRT-PCR to examine both pre-tRNA and total tRNA levels in S2 cells. We began by using the pharmacological MEK inhibitor U0126 to examine the effects of blocking Ras signalling. We found that treatment of S2 cells with UO126 lead to a decrease in levels of both pre-tRNAs and total tRNAs (Fig 2A and 2B). Also, using Northern blots, we saw that treatment with UO126 lead to reduced pre-tRNA and tRNA levels in S2 cells (S1A Fig). We also examined the effects of Ras pathway inhibition by using RNAi to knockdown Raf. We found that treatment of cells with dsRNA to Raf lead to reduced levels of both pre-tRNA and total tRNAs (Fig 2C and 2D). In contrast to Ras pathway inhibition, we found that RasV12 (constitutively active Ras) overexpression lead to an increase in both pre-tRNA and mature tRNA levels as measured by both qRT-PCR (Fig 2E and 2F) and Northern blot (S1B Fig), indicating enhanced tRNA synthesis. In contrast, to these effects on tRNA levels, we found no effect of inhibiting Ras signalling on expression levels of TBP (which was previously reported[41]) or on levels of Brf1 or Trf1 –both of which are components of TFIIIB complex, which is required for Pol III recruitment to tRNA genes (S1C Fig). We also found that altering Ras signalling (either by MEK inhibition or overexpression of RasV12) had no effect on levels of 5S or 7SL RNA in S2 cells (S1C and S1D Fig). Both of these genes transcribed by RNA polymerase III, but they are different Pol III gene types (type I and III respectively) that use a different set of core promoter factors compared to tRNA genes (which are type II Pol III genes). These finding suggest that the changes in tRNA synthesis we observed upon altering Ras signalling are not due to alterations in the levels of the basal transcriptional machinery required for tRNA transcription. These data also suggest Ras signalling may predominantly affect type II RNA pol III genes.
Fig 2. The Ras/ERK signalling pathway stimulates tRNA synthesis.
(A, B) Drosophila S2 cells were treated with 10 μM U0126 for 2 hours. Total RNA was isolated and levels of either pre-tRNAs (A), or total tRNAs (B) measured by qRT-PCR. N = 15 independent samples per condition. (C, D) Raf was knocked down in Drosophila S2 cells by incubating cells with dsRNAs against Raf. Control cells were treated with dsRNA to GFP. Total RNA was isolated and levels of either pre-tRNAs (C), or total tRNAs (D) measured by qRT-PCR. N = 4 independent samples per condition. (E, F) RasV12 expression was induced in Drosophila S2 cells for 24 hours. Total RNA was isolated and levels of either pre-tRNAs (E), or total tRNAs (F) measured by qRT-PCR. N = 9 independent samples per condition. (G) UAS-Rafgof was expressed in imaginal tissues using the esg-GAL4ts system. Control flies were esg-GAL4ts flies crossed to w1118. Transgenes were induced by shifting larvae to 29°C at 48hrs of larval development, and then discs were dissected from wandering L3 stage larvae. Total RNA was isolated and levels of pre-tRNAs measured by qRT-PCR. N = 4 independent samples per condition. Data are presented as mean +/- SEM.
We also examined the effects of Ras signalling on tRNA levels in the developing wing imaginal discs. We used the temperature-sensitive escargot-Gal4 (esg-Gal4ts) system, which allows for inducible transgenes expression in all imaginal tissues. When we overexpressed UAS-Rafgof using this system, we found a marked increase in pre-tRNA levels in wing discs as measured by qRT-PCR on dissected wing discs (Fig 2G).
Brf1 is required for Ras-induced tRNA synthesis and cell proliferation in wing discs
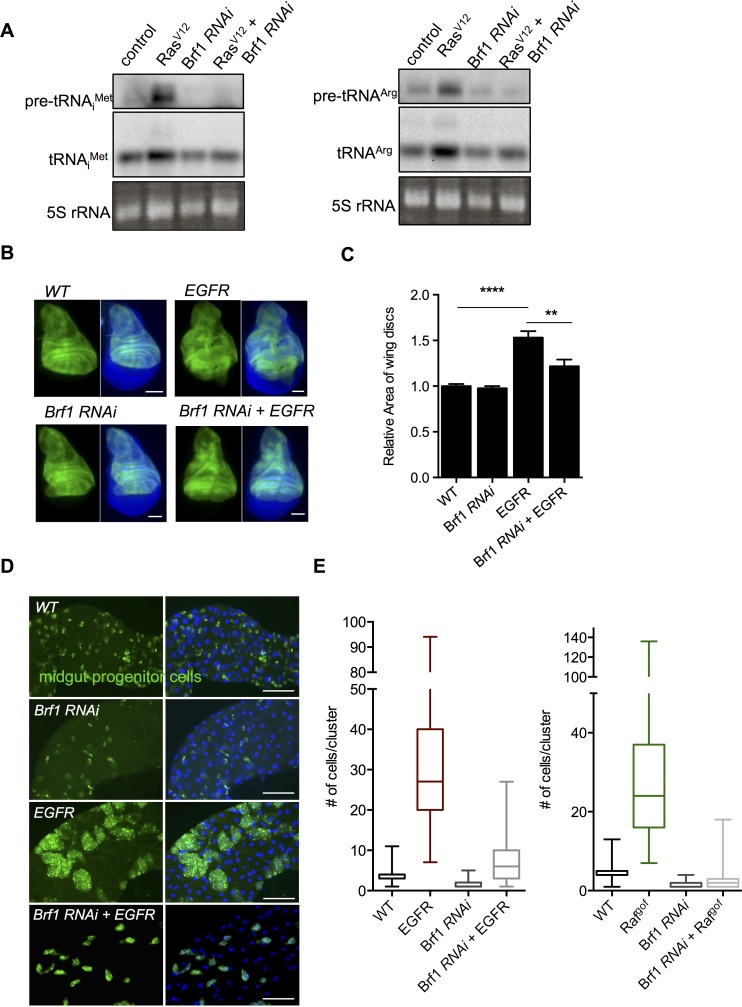
Brf1 is a conserved component of TFIIIB complex, which is required for Pol III recruitment to tRNA genes [42]. We previously showed that Brf1 is involved in controlling Pol III-dependent transcription, and tissue and body growth in Drosophila larvae [39]. Here we examined whether Brf1 is required for Ras-induced tRNA synthesis. We found that knocking down Brf1 using dsRNA in S2 cells (S2A and S2B Fig) suppressed the RasV12 induced increase in tRNA levels (Fig 3A) without altering the strong induction of ERK phosphorylation seen with RasV12 expression (S2B Fig). Hence, our data here suggest that the elevation of tRNA levels upon Ras activation is due to increased Pol III transcription. We then examined whether Brf1 is required for Ras-induced growth in Drosophila wing discs. We expressed UAS-driven transgenes in the dorsal compartment of the wing imaginal disc (using an apterous-Gal4 driver, ap-GAL4) and then, in each case, measured tissue size in wandering stage third instar larvae, Overexpression of UAS-EGFR (UAS-λtop) in the dorsal compartment of the wing imaginal disc stimulates Ras/ERK signalling and leads to tissue growth (Fig 3B and 3C). We found that RNAi-mediated knockdown of Brf1 by expression of a UAS-Brf1 inverted repeat line (UAS-Brf1 RNAi) in the dorsal compartment had little effect on tissue growth. However, expression of UAS-Brf1 RNAi blocked the overgrowth seen with UAS-EGFR expression. Expression of UAS-Brf1 RNAi with ap-GAL4 had little effect on tissue growth, suggesting we are not knocking down Brf1 to a level that cannot support any growth. We previously showed that Brf1 knockdown had no effect on ribosome synthesis, suggesting that its predominant effect was to block Pol III function [39]. Hence, these data indicate that Brf1 and Pol III transcription is required for EGFR/Ras/ERK-mediated increases in epithelial tissue growth in Drosophila.
Fig 3. Brf1 is required for Ras-induced tRNA synthesis and growth in both wing imaginal discs and adult midgut progenitor cells (AMPs).
(A). RasV12 expression was induced in Drosophila S2 cells for 24 hours in either control cells or Brf1 knockdown cells, Brf1 was knocked down by incubating cells with dsRNA against Brf1. Control cells were treated with dsRNA to GFP. Total RNA was isolated with Trizol and analyzed by northern blotting using DIG-labelled antisense probes to tRNAiMet or tRNAArg. Ethidium bromide stained 5S rRNA band was used as a loading control. (B, C) UAS-EGFR and UAS-Brf1 RNAi were expressed, either alone or together, in the dorsal compartment of larval wing imaginal discs using an ap-Gal4 driver. Control discs were from ap-Gal4 crossed to w1118. Wing discs were dissected at the wandering L3 larval stage and the area of the GFP-marked dorsal compartment quantified using NIH imaging software (n > 50 wings per genotype, data presented as mean +/- SEM). Representative images are shown in (B), quantification of tissue area is shown in (C). (D) UAS-EGFR and UAS-Brf1 RNAi were expressed, either alone or together, in the Drosophila larval AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24 hrs of development to induce transgene expression and dissected as L3 larvae. AMPs are marked by UAS-GFP expression. DNA is stained with Hoechst dye (blue). (E) The number of cells in each AMP cluster was quantified for each of the genotypes in D (left), and an additional similar experiment in which the Ras pathway was activated by expression of a UAS-Rafgof transgene (right). Data are presented as box plots (25%, median and 75% values) with error bars indicating the min and max values.
Brf1 is required for Ras/ERK-induced proliferation in adult mid-gut progenitor cells (AMPs) and adult intestinal stem cells (ISCs)
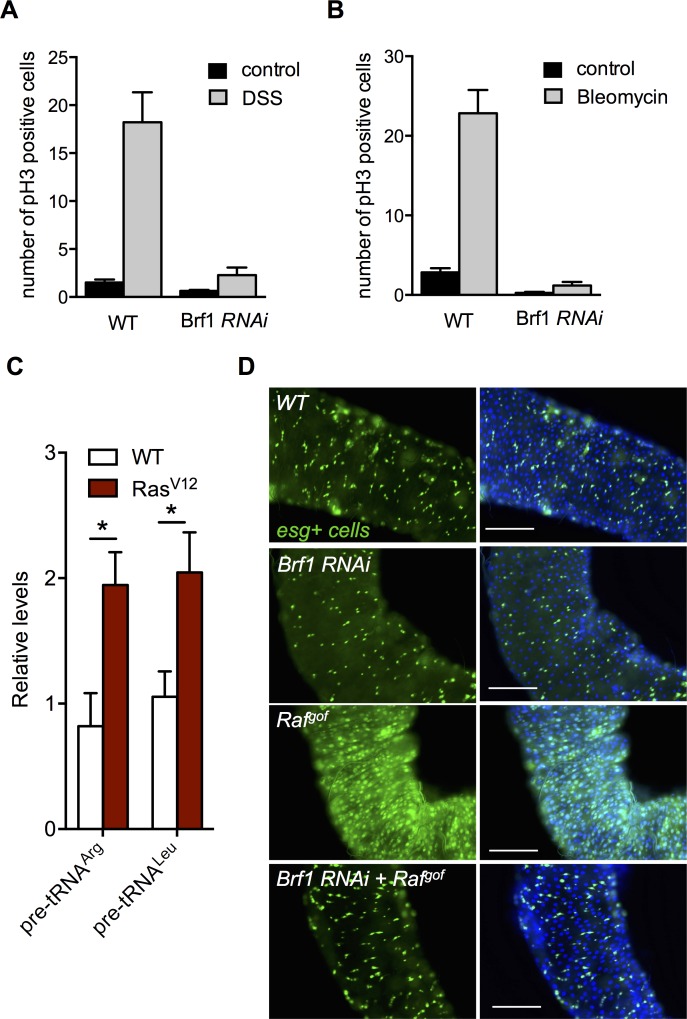
A major role for the EGFR/Ras/ERK pathway is in the growth and maintenance of the Drosophila intestine. In larvae, activation of the pathway plays a central role in controlling the proliferation of adult midgut progenitor cells (AMPs), which eventually give rise to the adult intestine [9]. In the adult the EGFR/Ras/ERK pathway is required to promote stem cell proliferation and tissue regeneration [15–17,19,43]. We therefore examined whether Brf1-mediated Pol III transcription was required for these proliferative effects of Ras/ERK signalling. We first examined the larval intestine. During the larval period, AMPs proliferate and give rise to clusters of ~5–10 cells scattered throughout the larval intestine. These cell clusters eventually proliferate and fuse during metamorphosis to give rise to the adult intestinal epithelium. The EGFR/Ras/ERK pathway controls the proliferation of AMPS [9]. Overexpression of either UAS-EGFR or UAS-Rafgof in the AMPs using the temperature-sensitive escargot-Gal4 (esg-Gal4ts) system lead to a massive increase AMP proliferation and an increase in the numbers of AMP cells per cluster as previously reported. We found that expression of UAS-Brf1 RNAi (Fig 3D and 3E and S2C Fig) lead to a small reduction in the number of cells per cluster. However, we found that when co-expressed along with UAS-EGFR or UAS-Rafgof, UAS-Brf1 RNAi blocked the increase in AMP cell numbers. These data indicate Brf1 is required for EGFR/Ras/ERK mediated cell proliferation.
We next examined Brf1 function in homeostatic growth in the adult intestine. Damage to intestinal epithelial cells leads to an increase in expression and release of EGF ligands from both intestinal cells and underlying visceral muscle [15]. These EGF ligands then act on the intestinal stem cells (ISCs) to stimulate the Ras/ERK pathway, which triggers stem cell growth and division, and promotes regeneration of the intestinal epithelium. This damage-induced increase in ISC proliferation is dependent on EGFR/Ras/ERK signalling and can be mimicked by genetically activating the pathway specifically in the stem cells [15–17,19]. We tested a requirement for Brf1 in this Ras-mediated homeostatic growth response. We first examined the effects of intestinal damage. As previously reported [19], we found that feeding flies either DSS or bleomycin–two different gut stressors–leads to an increase in ISC proliferation. However, we found that this effect was inhibited when we knocked down Brf1 (using UAS-Brf1 RNAi expression) specifically in the ISCs and their transient daughter cells, the enteroblasts (EBs), using the inducible esg-Gal4ts system (Fig 4A and 4B). We next examined the effects of activation of the Ras/ERK pathway. We first overexpressed UAS-RasV12 in the adult intestine using the inducible esg-Gal4ts system, and observed an increase in pre-tRNA levels (Fig 4C). As previously reported, when we overactivated the pathway in stem cells by expressing UAS-Rafgof using esg-Gal4ts, we saw an increase cell proliferation as indicated by a marked increase in GFP labelled ISCs and EBs (Fig 4D). Expression of a UAS-Brf1 RNAi had little effect on GFP labelled cells, but when co-expressed with UAS-Rafgof it blocked the increase in cell proliferation. These results suggest that Brf1 and Pol III-dependent transcription is required for stem cell proliferation in the adult intestine.
Fig 4. Brf1 is required for intestinal stem cells (ISCs) homeostasis and for Ras-induced cell proliferation.
(A, B) UAS-Brf1 RNAi was expressed adult ISCs and EBs using the esg-GAL4ts system. Control flies were esg-GAL4ts flies crossed to w1118. Flies were then fed with sucrose or sucrose plus DSS (A) or Bleomycin (B) for 2 days. Intestines were then dissected and stained for phospho-histone H3 positive cells. Data represent the mean number of phospho-histone H3 cells per intestine +/ SEM. N >15 intestines per condition. (C) A UAS-RasV12 transgene was expressed in adult intestines using the esg-GAL4ts driver. Control samples (WT) expressed UAS-GFP alone. Total RNA was isolated and levels of pre-tRNAs measured by qRT-PCR. N = 4 independent samples per condition. Data are presented as mean +/- SEM. (D) UAS-Rafgof and UAS-Brf1 RNAi were expressed, either alone or together, in the adult ISCs and EBs using the esg-Gal4ts system. esg positive cells are marked with GFP and DNA is stained with Hoechst dye. Knockdown of Brf 1(UAS-Brf RNAi) suppresses the increased proliferation seen with UAS-Rafgof expression.
dMyc is required but not sufficient for Ras-induced tRNA synthesis
We next wanted to examine how Ras signalling stimulates Pol III-dependent tRNA transcription. One candidate regulator we tested was dMyc. In both mammalian cells and Drosophila, Myc can interact with Brf1 and stimulate Pol III-dependent transcription [39,44,45]. Moreover, studies in both mammalian cells and Drosophila suggest Ras signalling can regulate dMyc levels and that Myc is required for Ras-induced growth[10,23,24,46,47]. Indeed, we found that the UAS-EGFR- and UAS-RasV12S35-induced proliferation of larval AMPs was blocked when we knocked down dMyc by expression of a UAS-dMyc RNAi construct (S3A–S3C Fig). We therefore examined whether dMyc functions downstream of Ras in the control of Pol III. Using S2 cells we found that the increase in tRNA levels seen following RasV12 expression was blocked when cells were treated with dsRNA to knockdown dMyc (Fig 5A). In contrast, we found that overexpression of dMyc in S2 cells was not able to induce tRNA synthesis when the Ras pathway was inhibited by treatment with the MEK inhibitor UO126 (Fig 5B). Under these conditions of Ras pathway inhibition, however, dMyc mRNA levels were not affected (Fig 5C) and overexpressed dMyc was still able to significantly stimulate expression of Nop60B, PPAN and NOP5—three dMyc Pol II target genes (Fig 5D)–although the effect on PPAN and NOP5 was somewhat reduced. Nevertheless, these data suggest that U0126 does not simply abrogate dMyc’s ability to stimulate transcription of its target genes, and that dMyc is required, but not sufficient, to mediate the effects of Ras signalling on tRNA synthesis. These data suggest that Ras/ERK signalling can use an additional mechanism to control Pol III transcription.
Fig 5. dMyc is required but not sufficient for Ras-induced tRNA synthesis.
(A) RasV12 expression was induced in Drosophila S2 cells for 24 hours in either control cells or dMyc knockdown cells. dMyc was knocked down by incubating cells with dsRNA against dMyc. Control cells were treated with dsRNA to GFP. Total RNA was isolated with Trizol and analyzed by northern blotting using DIG-labelled antisense probes to tRNAiMet or tRNAArg. Ethidium bromide stained 5S rRNA band was used as a loading control. (B, C and D) dMyc expression was induced in S2 cells for 24hrs, and then cells were treated with 10 μM U0126 or DMSO for 2 hours. Total RNA was isolated with Trizol and analyzed by qRT-PCR to measure levels of (B) pre-tRNAs, (C) dMyc mRNA, or (D) mRNA levels of three dMyc target genes—NOP60B, PPAN and NOP5. N = 4 independent samples per condition. Data are presented as mean +/-SEM.
Ras signalling promotes tRNA synthesis by inhibiting the RNA pol III repressor dMaf1
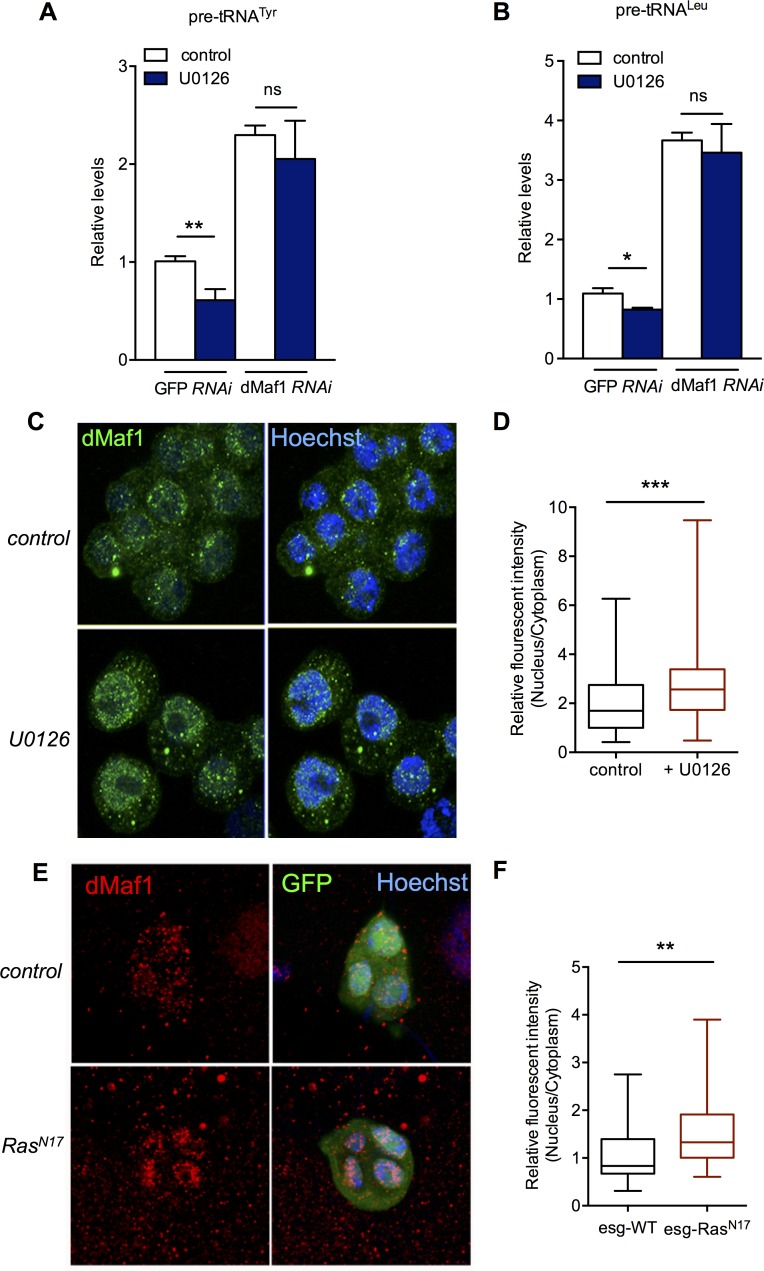
Another candidate that we considered as a mediator of Ras-induced tRNA synthesis was the conserved Pol III repressor, Maf1. Studies in yeast, Drosophila and mammalian cells have shown that inhibition of Maf1 is the main way that the nutrient-dependent TORC1 kinase pathway stimulates Pol III and tRNA synthesis [39,48–51]. Knockdown of Drosophila Maf1 (dMaf1) has been shown to promote tRNA synthesis, and to enhance tissue and body growth [40]. Here, we found that when we expressed UAS-dMaf1 RNAi in the Ras-responsive AMP cells during larval development using esg-GAL4ts, we observed a modest, but significant increase in the number of AMP cells per cluster (S4A Fig). Although considerably weaker than the effect of Ras pathway activation (e.g. see comparison with effect of UAS-EGFR, S4B Fig) this effect of dMaf1 knockdown was similar to the increase in AMP numbers seen with overexpression of dMyc, another stimulator of tRNA synthesis and mRNA translation (S4C Fig). We therefore next examined whether the Ras/ERK pathway functions to promote tRNA synthesis by inhibiting dMaf1. We examined pre-tRNA levels using qRT-PCR in S2 cells, and, as described above, we saw that treatment of cells with the MEK inhibitor UO126 led to reduced tRNA synthesis (Fig 6A and 6B). However, we found that this decrease in tRNA synthesis was reversed when cells were treated with dsRNA to knockdown dMaf1 levels (Fig 6A and 6B). We observed similar effects when we used Northern blotting to measure pre-tRNA and tRNA levels (S4D Fig). We also used treatment of cell with dsRNA to Ras to block Ras signalling, and saw a decrease in tRNA synthesis (S4E Fig). However, as with UO126 treatment, we found that this decrease in tRNA synthesis caused by dsRNA to Ras was reversed by co-treatment of cells with dsRNA to dMaf1. These data suggest that one main way that Ras/Erk signalling functions to promote tRNA synthesis is by inhibiting the Pol III repressor function of dMaf1.
Fig 6. Ras induces tRNA synthesis by via inhibition of the Pol III repressor, dMaf1.
(A, B) dMaf1 was knocked down in Drosophila S2 cells by incubating cells with dsRNAs against dMaf1. Control cells were treated with dsRNA to GFP. Cells were then treated with DMSO (control) or 10 μM U0126 for 2 hrs. Total RNA was isolated and levels of pre-tRNATyr (A) and pre-tRNALeu measured by qRT-PCR. N = 4 independent samples per condition. Data are presented as mean +/-SEM. (C) dMaf1 subcellular localization was assessed by immunostaining with an anti-dMaf1 antibody in both control and 10 μM U0126 treated S2 cells. Green: dMaf1 staining; blue: Hoechst-stained nuclei. (D) The differential localization of dMaf1 in S2 cells from the experiment shown in (C) was quantified by measuring the ratio of nucleus: cytoplasmic intensity quantified using NIH Image J. Data are plotted in the graph as relative nuclear: cytoplasmic staining intensity for both experimental conditions. Data are presented as box plots (25%, median and 75% values) with error bars indicating the min and max values. N>90 cells per condition fom three independent experiments. (E) A UAS-RasN17 transgene was expressed in GFP-marked adult midgut progenitor cells using the esg-GAL4ts driver. Control samples (WT) expressed UAS-GFP alone. dMaf1 subcellular localization was then assessed in third instar larvae by immunostaining with an anti-dMaf1 antibody. (F) The differential localization of dMaf1 in the AMPs from the experiment shown in (E) was quantified by measuring the ratio of nucleus: cytoplasmic intensity quantified using NIH Image J. Data are plotted in the graph as relative nuclear: cytoplasmic staining intensity for both experimental conditions. Data are presented as box plots (25%, median and 75% values) with error bars indicating the min and max values. N>30 cells per condition fom three independent experiments.
Studies in both yeast and mammals indicate that Maf1can be regulated by controlling its nuclear localization (e.g [50,52]). We first tested this in S2 cells using an antibody to endogenous dMaf1. Under our normal media culture conditions, we observed that dMaf1 was localized throughout the cell (Fig 6C). When we carried out antibody staining in dMaf1 dsRNA-treated cells (which leads to a strong knockdown of both dMaf1 mRNA, S5A Fig, and dMaf1 protein, S5B Fig) we saw minimal background staining, suggesting that the antibody is specific for dMaf1 (S5C Fig). We found that treatment of cells with the MEK inhibitor U0126 lead to a significant increase in nuclear localization of dMaf1 (Fig 6C and 6D), without having any effect on overall dMaf1 protein levels (S5D Fig). We also found that genetic inhibition of Ras signalling in AMPs, by overexpression of dominant-negative Ras (UAS-RasN17), lead to an increase in nuclear localization of dMaf1 (Fig 6E and 6F). Similar results were seen when we used expression of either UAS-EGFR or UAS-Ras RNAi to block Ras signalling in AMPs (Supplemental Fig 6A). Thus, Ras/ERK signalling functions to prevent nuclear accumulation of dMaf1, hence blocking its Pol III repressor activity and promoting tRNA synthesis.
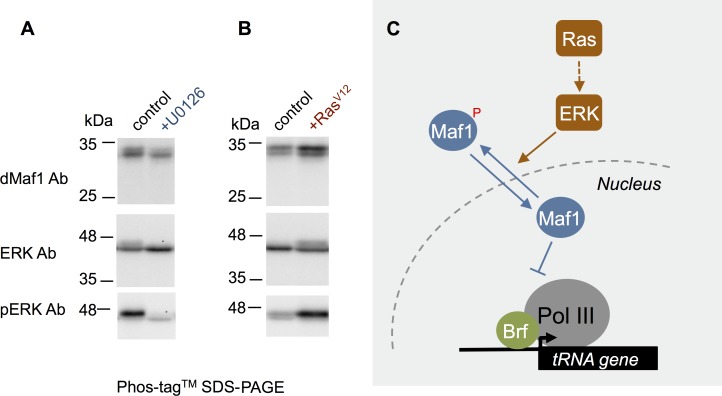
Previous studies showed that the TORC1 pathway can regulate Maf1 nuclear localization and repressor function via phosphorylation [48–51,53]. We therefore explored whether the Ras pathway could also control the phosphorylation status of dMaf1 in S2 cells. We used the phos-tag reagent, which slows the migration of phosphorylated proteins in SDS-PAGE gels, and hence helps resolve phosphorylated vs. non-phosphorylated versions of a protein on a western blot. For example, when we examined total ERK levels by western blotting following SDS-PAGE with Phos-tag, we observed two ERK bands. The relative levels of the upper band were reduced when we treated cells with the MEK inhibitor (Fig 7A), while levels of the upper band were increased in cells overexpressing RasV12 (Fig 7B), thus indicating this method can detect protein phosphorylation changes. We then examined dMaf1 protein levels in western blots following SDS-PAGE with Phos-tag. As with ERK, we observed two dMaf1 bands, and the relative levels of the upper band were reduced when we treated the sample with phosphatase prior to SDS-PAGE (S6B Fig), suggesting this upper band is a phosphorylated version of dMaf1. Also, like ERK, we found that relative levels of the upper band were reduced when we treated cells with the MEK inhibitor (Fig 7A), while levels of the upper band were increased in cells overexpressing RasV12 (Fig 7B). Together these data suggest that Ras signalling may regulate dMaf1 phosphorylation, and based on previous work with TORC1 signalling, this may be one way that Ras regulates dMaf1 nuclear vs. cytoplasmic localization (Fig 7C).
Fig 7. Ras signalling regulates dMaf1 phosphorylation.
(A) Drosophila S2 cells were treated with 10 μM U0126 for 2 hours. Cells were then lysed and processed for SDS-PAGE and western blotting using the phos-tag reagent, as described in the Methods. The blots were then probed with an anti-dMaf1 antibody (top panel), an anti-total ERK antibody (middle panel) or an anti-phospho ERK antibody (lower panel) (B) RasV12 expression was induced in Drosophila S2 cells for 24 hours. Cells were then lysed and processed for SDS-PAGE and western blotting using the phos-tag reagent, as described in the Methods. The blots were then probed with an anti-dMaf1 antibody (top panel), an anti-total ERK antibody (middle panel) or an anti-phospho ERK antibody (lower panel). (C) A model for how Ras signalling may regulate Pol III and tRNA synthesis.
Discussion
We propose that stimulation of RNA polymerase III and tRNA synthesis contributes to the ability of the conserved Ras/ERK pathway to promotes mRNA translation and growth. Our data indicate that Ras can control Pol III by inhibiting the Maf1 repressor, in part by preventing its nuclear accumulation. Maf1 is a phospho protein and studies in yeast and mammalian cells have described how phosphorylation can regulate Maf1 nuclear localization. For example, both TORC1 and PKA can phosphorylate Maf1 on several conserved residues[48–53]. This phosphorylation prevents Maf1 nuclear accumulation and allows both kinases to stimulate Pol III. In contrast, dephosphorylation of Maf1 by both PP2A and PP4 protein phosphatases leads to nuclear accumulation of Maf1 and Pol III repression [54–56]. Thus, it is possible that ERK may function by promoting Maf1 phosphorylation–either directly or indirectly–to prevent its function. Other mechanisms may also be important for Ras to simulate tRNA synthesis. For example, one study in mammalian cells showed that ERK could phosphorylate and regulate Brf1 function [57]. Also, Ras was shown to upregulate TBP, which can increase transcription by all three RNA polymerases [41], although we did not see a similar effect. Interestingly, we found that the decrease in tRNA synthesis caused by inhibiting Ras signalling could be completely reversed by dMaf1 knockdown. This result suggests that while Ras signalling may exert multiple effects to control Pol III transcription, inhibition of dMaf1 seems to be an important effector of Ras in the control of tRNA synthesis. Maf1 function is conserved suggesting that the Ras/ERK-dependent regulation of Maf1 and tRNA synthesis that we describe in Drosophila may operate in other organisms, particularly human cells.
Our data using the phos-tag reagent suggest that one way that Ras/ERK signalling may control dMaf1 is via phosphorylation. Previous studies in both yeast and mammalian cells have shown that the TORC1 pathway can control the nuclear localization and repressor function of dMaf1 via phosphorylation of several conserved residues [48–50,53]. One can therefore speculate that Ras signalling may work in a similar manner. Although further studies are required to identify if ERK directly phosphorylates dMaf1 and to identify the phosphorylated residues, it is interesting to note that two of the conserved TORC1 phosphorylation sites on dMaf1 are serine residues followed by proline, which are sites that are often phosphorylated by ERK, a proline-directed kinase.
We also show that the transcription factor dMyc is required for the effects of Ras on Pol III and tRNA synthesis. Previous work from both mammalian cells and Drosophila has shown that in some cells Ras can promote Myc levels and that Ras-mediated growth requires Myc function[10,23,24,46,47]. We previously showed that Drosophila Myc could stimulate expression of the Pol III transcription factor, Brf1, and also other Pol III subunits [39]. In addition, Myc can directly interact with Brf1 and localize at Pol III to directly stimulate tRNA transcription in Drosophila and mammalian cells [39,44,45]. We suggest that both these effects are under the upstream control of Ras/ERK signalling and may, in part, explain the requirements for Myc in Ras-induced growth in both animal development and cancer.
Given our findings with dMaf1 and dMyc, we attempted to address which of the two mechanisms—inhibition of dMaf1 or activation of Myc—might explain the main effects of Ras/ERK signalling on tRNA synthesis. To do this, we inhibited Ras/ERK signalling in S2 cells and then asked whether knockdown of dMaf1 or overexpression of dMyc could maintain tRNA synthesis. We found that, of these two manipulations, only dMaf1 inhibition could restore tRNA synthesis when ERK signalling was inhibited. We interpret these findings to suggest that, while dMyc is required for tRNA synthesis, it is the inhibition of dMaf1 that explains a substantial part of the mechanism of action of Ras/ERK signalling in the regulation of Pol III and tRNA synthesis. We previously showed that dMaf1 knockdown does not alter expression of dMyc target genes [39], suggesting that enhancement of dMyc function doesn’t explain why Maf1 knockdown can maintain tRNA synthesis in cells in which Ras/ERK signalling is inhibited.
Previous studies in mammalian cells have shown that Ras/ERK signalling can promote protein synthesis by stimulating translation initiation factor function. We suggest that inhibition of Maf1 represents another target of Ras/ERK signalling, and that the subsequent increase in tRNA levels may cooperate with enhanced translation initiation factor activity to promote maximal stimulation of mRNA translation. Most of the work on Ras-mediated gene expression has focused on the effect of several Pol II transcription factors identified downstream of Ras in Drosophila such as fos, pointed, and capicua [19–22]. Stimulation of Pol III transcription to enhance tRNA levels and mRNA translation may provide another layer of control on overall gene expression by Ras signalling. For example, translational control of cell cycle genes has been proposed as one way to couple growth signalling pathways to cellular proliferation [58,59]. Furthermore, selective translational regulation of certain mRNAs has been shown to regulate growth and metastatic behaviour of tumour cells [60–62]. It important to note though that we find that simply knocking down dMaf1 alone has only a modest effect on cell proliferation AMPs, compared to the strong hyperproliferative effect of overactivation of Ras signalling. This is likely because increasing Pol III is only one downstream effect of Ras signalling and that the full Ras effect on cell proliferation requires the coordinated increase in the expression of many genes. Indeed, it is likely that Ras stimulates the activity of all three RNA polymerases to drive cell growth and proliferation.
Ras is one of the most often overactivated or mutated pathways in cancer, hence our findings may also have implications for processes that contribute to tumour growth and metastasis. Indeed, there is increasing appreciation for potential roles for alterations in tRNA biology in cancer cells [63]. For example, tRNA expression profiling has revealed that levels of many tRNAs are elevated in different cancer types [64,65]. Interestingly, these changes in tRNA levels have been shown to correlate with codon usage in mRNAs whose expression also changes in cancer cells [66]. Several studies have reported that increasing the levels of specific tRNAs can promote tumour growth and metastatic behavior [67–70]. Previous work also showed that increasing tRNA levels alone is sufficient to drive growth in Drosophila [40,71]. Hence, an increase in tRNA levels caused by oncogenic Ras signalling may be a driver of tumour growth and progression, rather than simply a consequence of increased growth. Ras also controls other process such as cell fate specification, differentiation and cell survival. Many of these effects are mediated through translation and so may also rely on the effects of Ras on tRNA synthesis.
Materials and methods
Drosophila stocks
Flies were raised on standard medium (150 g agar, 1600 g cornmeal, 770 g Torula yeast, 675 g sucrose, 2340 g D-glucose, 240 ml acid mixture (propionic acid/phosphoric acid) per 34 L water) and maintained at 25°C, unless otherwise indicated. The following fly stocks were used:
w1118,
yw,
UAS-RasV12, [24]
UAS-RasN17, [24]
UAS-RasV12S35, [24]
UAS-EGFR (UAS-λTOP), [24]
UAS-Rafgof, [24]
UAS-Brf RNAi (NIG, Japan),
UAS-dMyc [72]
UAS-Maf1 RNAi [40],
esg-gal4, tub-GAL80ts, UAS-GFP, [15]
ap-gal4/CyO,
dpp-gal4/TM6B.
For all GAL4/UAS experiments, GAL4 lines were crossed to the relevant UAS line(s) and the larval or adult progeny were analyzed. Control animals were obtained by crossing the relevant GAL4 line to either w1118 or yw depending on the genetic background of the particular experimental UAS transgene line. For the esg-gal4ts system, larvae and were flies were initially raised at 18°C and then for each experiment they were shifted to 29°C to inactivate the temperature sensitive GAL80 and to allow GAL4-mediated transgene expression.
Cell culture and transfection
Drosophila Schneider S2 cells were grown at 25°C in Schneider’s medium (Gibco; 11720–034) supplemented with 10% fetal bovine serum (Gibco; 10082–139), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco; 15140). Stably transfected inducible RasV12 cells were a gift from the lab of Marc Therrien [73]. Stably transfected inducible dMyc cells were a gift from the lab of Paula Bellosta [74]. Both RasV12 and dMyc expression are under the control of a metallothionein promoter. For all experiments RasV12 or dMyc were induced by addition of copper sulphate to the culture media.
dsRNA Treatment of S2 cells: dsRNAs were synthesized with RiboMAX large-scale RNA production system (Promega) using PCR products from either cDNAs or genomic DNA (primer sequences in S2 Table). Cells were pretreated with 15 μg of dsRNAs in the absence of serum for 30 mins and then 2 mls of media plus serum was added, and cells were then incubated for 96 to 120 hrs. Control cells were treated with ds RNA to Green Fluorescent Protein (GFP). Cells were harvested by centrifugation at 4 °C and washed with cold PBS and frozen for RNA isolation or protein extraction.
MEK inhibitor (U0126) treatment of Drosophila S2 cells: S2 cells were cultured at 25°C in Schneider’s medium (Gibco; 11720–034) supplemented with 10% fetal bovine serum (Gibco; 10082–139), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco; 15140). Cells were treated with either 10 μM U0126 (Promega Cat. No. V1121) or DMSO (Sigma; D2650) for 2 hours. Then cells were washed twice with ice-cold PBS. Cells were then used to isolate RNA or make protein extracts as described below.
Preparation of protein extracts
Drosophila S2 cells were lysed with a buffer containing 20 mM Tris-HCl (pH 8.0), 137 mM NaCl, 1 mM EDTA, 25% glycerol, 1% NP-40 and with following inhibitors 50 mM NaF, 1 mM PMSF, 1 mM DTT, 5 mM sodium ortho vanadate (Na3VO4) and Protease Inhibitor cocktail (Roche Cat. No. 04693124001) and Phosphatase inhibitor (Roche Cat. No. 04906845001) according to the manufacturer’s instruction.
Phos-tag SDS-PAGE
Drosophila S2 cells were lysed with a buffer containing 20 mM Tris-HCl (pH 8.0), 137 mM NaCl, 25% glycerol, 1% NP-40 and with following inhibitors 1 mM PMSF, 1 mM DTT and Protease Inhibitor cocktail (Roche Cat. No. 04693124001) and Phosphatase inhibitor without EDTA. Phos-tag SDS-PAGE was prepared according to the manufacturer’s instruction (Wako Chemicals USA, Inc). Cell lysates were separated on 12.5% SDS-polyacrylamide gel containing 20 uM Phos-tag acrylamide (AAL-107 Wako Chemicals USA, Inc), and transferred onto PVDF membranes (Bio Rad).
Western blot and antibodies
Protein concentrations were measured using the Bio-Rad Dc Protein Assay kit II (5000112). Protein lysates (15 μg to 30μg) were resolved by SDS–PAGE and electrotransferred to a nitrocellulose membrane, subjected to Western blot analysis with specific antibodies, and visualized by chemiluminescence (enhanced ECL solution (Perkin Elmer). Brf primary antibodies were against a C-terminal fragment of Drosophila Brf, alpha-tubulin (E7, Drosophila Studies Hybridoma Bank), dMyc [24], phospho-ERK (Cell Signalling Technology 4370) and ERK (Cell Signalling Technology 4695). Peptide antiserum against Drosophila Maf1 was raised by immunizing rabbits with synthetic peptide LADFSPNFRC corresponding to residues 65–74 (GL Biochem (Shanghai) Ltd).
Puromycin-labelling protein synthesis assay
10 μM puromycin was added to Drosophila S2 cell culture media and the cells were incubated with puromycin for 30 min at 25 °C. Cells were harvested by centrifugation at 4°C and washed with cold PBS. Cells were frozen on dry ice and then lysed according to the Western blot protocol described above and analyzed by SDS-PAGE and western blotting using an anti-puromycin antibody (3RH11) (Kerafast, Catalog No.EQ0001) at 1:2000 dilution.
Northern blot analysis
Total RNA was extracted from Drosophila S2 cells using TRIzol. 5 μg total RNA was separated on a 5% denaturing polyacrylamide/urea gel and northern blotting was carried using alkaline transfer. Hybridization of tRNA probes were carried out as described in Roche DIG Easy Hyb (Cat. No.11603558001). Digoxigenin-labelled probes were made by in vitro transcription using either full-length cDNAs or PCR fragments as templates. Primers used for PCR are included in S1 Table.
Immunostaining
Drosophila S2 cells were fixed in 4% paraformaldehyde at room temperature for 20 mins on cover slips. Cells were then washed with 1x PBS and permeabilized with 0.1% Triton X in PBS by washing 2x for 5 mins. Cells were blocked with 5% FBS, 0.1% Triton X in PBS for 2 hours. Primary dMaf1 antibody was diluted in 5% BSA in PBS at 1:500 dilution and incubated overnight at 4°C. Then washed 3x with 0.1% Triton X in PBS for 5 min each and Alexa 568 (Molecular probes) goat-anti rabbit secondary antibody was diluted at 1:400 in 5% BSA in PBS for 2 hours at room temperature. Then, cells were washed 3x with 0.1% Triton X in PBS for 5 min each and mounted using VectaShield mounting medium.
Drosophila larvae were inverted and fixed in 8% paraformaldehyde/PBS at room temperature for 45 mins. After blocking for 2hrs in 1%BSA in PBS/0.1% Triton-X 100, larval carcasses were incubated overnight in anti-dMaf1 antibody (1:1000). Primary antibody staining was detected using Alexa 488 (Molecular probes) goat-anti rabbit secondary antibodies.
For experiments looking at dMaf1 subcellular localization, we used Image J to measure dMaf1 staining intensity. Nuclear localization was measured was calculated by measuring the total intensity of signal in the nucleus and dividing this by the total intensity in the cytoplasm (calculated as total overall cellular signal intensity minus total nuclear signal intensity).
Real-time quantitative PCR
Total RNA was extracted using TRIzol according to manufacturer’s instructions (Invitrogen; 15596–018). RNA samples were DNase treated according to manufacturer’s instructions (Ambion; 2238G) and reverse transcribed using Superscript II (Invitrogen; 100004925). The generated cDNA was used as a template to perform qRT–PCRs (ABI 7500 real time PCR system using SyBr Green PCR mix) using specific primer pairs (sequences available upon request). PCR data were normalized to either actin or Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Each experiment was independently repeated a minimum of three times. All primer sequences are in S3 Table.
Polysome gradient centrifugation
Polysome gradient centrifugation was performed as described [40]. 100 million Drosophila S2 cells were lysed in 1 ml of lysis buffer (25 mM Tris pH 7.4, 10 mM MgCl2, 250 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.5 mM DTT, 100 mg/ml cycloheximide, 1 mg/ml heparin, Complete mini Roche protease inhibitor (Roche), 2.5 mM PMSF, 5 mM sodium fluoride, 1 mM sodium orthovanadate and 200 U/ml ribolock RNAse inhibitor (Fermentas) using a Dounce homogenizer. The lysates were centrifuged at 15,000 rpm for 20 minutes and the supernatant was removed carefully. 150 to 250 g μg RNA was layered gently on top of a 15–45% w/w sucrose gradient (made using 25 mM Tris pH 7.4, 10 mM MgCl2, 250 mM NaCl, 1 mg/ml heparin, 100 mg/ml cycloheximide in 12 ml polyallomer tube) and centrifuged at 37,000 rpm for 150 minutes in a Beckmann Coulter Optima L-90K ultracentrifuge using a SW-41 rotor. Polysome profiles were obtained by pushing the gradient using 70% w/v Sucrose pumped at 1.5 ml/min into a continuous OD254 nm reader (ISCO UA6 UV detector) showing the OD corresponding to the RNA present from the top to the bottom of the gradient.
Statistical analysis
All qRT-PCR data and quantification of immunostaining data were analyzed by Students t-test, or two-way ANOVA followed by post-hoc students t-test where appropriate. All statistical analysis and data plots were performed using Prism software. In all figures, statistically significant differences are presented as: * p<0.05, ** p<0.005, *** p<0.0005, **** p<0.0001.
Supporting information
(A) Left: S2 cells were treated with (10μM) U0126 for 2 hrs. Total RNA was isolated and analyzed by Northern blot. Levels of tRNAArg were detected using DIG-labelled tRNA probes. Ethidium bromide stained 5S rRNA bands were used as loading controls, since total 5S rRNA levels are unchanged by manipulations in Ras/Erk signalling (see C). (B) RasV12 expression was induced in S2 cells for 24hrs. Total RNA was isolated and analyzed by northern blot. Levels of tRNAArg were detected using DIG-labelled tRNA probes. Ethidium bromide stained 5S rRNA bands were used as loading controls, since total 5S rRNA levels are unchanged by manipulations in Ras/Erk signalling (see C) (C) qRT-PCR measurement of Brf1 mRNA, Trf1 mRNA, TBP mRNA, 5S rRNA and 7SL RNA levels in U0126 treated S2 cells. (D) qRT-PCR measurement of 5S rRNA levels in RasV12 overexpressing S2 cells.
(TIFF)
(A) Brf1 mRNA levels were measured by qRT-PCR in Drosophila S2 cells treated with dsRNA against Brf1 or GFP (control). Control cells were treated with GFP dsRNA (B) Brf1, phospho-ERK levels and alpha-tubulin protein levels were measured by western blot in Drosophila S2 cells treated with dsRNA against Brf1 and overexpressing RasV12, both alone and together. (C) UAS-Rafgof and UAS-Brf1 RNAi were expressed, either alone or together, in the Drosophila larval AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24 hrs of development to induce transgene expression and dissected as L3 larvae. AMPs are marked by UAS-GFP expression. DNA is stained with Hoechst dye (blue) Representative images are shown for each genotype.
(TIFF)
UAS-EGFR (A) or UAS-RasV12S35 (C) were expressed, either alone or together with UAS-dMyc RNAi in the Drosophila larval AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24 hrs of development to induce transgene expression and dissected as L3 larvae. AMPs are marked by UAS-GFP expression. DNA is stained with Hoechst dye (blue) (B) (related to experiment in A) Numbers of cells in each AMP cluster were counted and expressed as box plots.
(TIFF)
(A, B) UAS-dMaf1 RNAi (A), UAS-EGFR (B), or UAS-dMyc (C) were expressed in AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24hrs of development and dissected at wandering stage. The numbers of cells in each AMP cluster were counted and expressed in box plots. (D) dMaf1 and Ras were knocked down, both alone and together, in Drosophila S2 cells by incubating cells with dsRNAs against dMaf1 and Ras. Control cells were treated with dsRNA to GFP. Total RNA was isolated with Trizol and analyzed by Northern blot using DIG-labelled tRNAArg probe. Ethidium bromide stained 5S rRNA band was used as a loading control.
(TIFF)
(A, B) dMaf1 mRNA levels (A) or protein levels (B) were measured by qRT-PCR or Western blot respectively in cells treated with dsRNA to GFP (control) or dMaf1 (dMaf1 RNAi). dsRNA treatment produced a strong knockdown of dMaf1 levels. (C) Control and dMaf1 dsRNA-treated S2 cells were stained with an anti-dMaf1 antibody (green) and Hoechst dye (blue). (D) dMaf1and Brf1 protein levels were analyzed with western blotting after treatment with U0126 for 2 hours. Decreased phospho-ERK levels served as a positive control for UO126-mediated MEK inhibition. Tubulin levels served as a loading control.
(TIFF)
WT, UAS-EGFR RNAi, UAS-Ras RNAi were expressed in AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24hrs of development and dissected at wandering stage and stained with dMaf1 antibody. (B) Drosophila S2 cell lysates (left, control samples; right, RasV12 induced samples) were treated with either Alkaline phosphatase or λ-phosphatase for 1 hr at 37°C and samples were analysed by phos-tag SDS-PAGE and western blotting using an anti-dMaf1 antibody.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Acknowledgments
We thank Dr. Marc Therrien and Dr. Paola Bellosta for Drosophila S2 cells stably transfected with RasV12 and Myc cell lines. Stocks obtained from the NIG-Fly Stock Centre, Kyoto, Japan and from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We also thank Dr. Shinako Takada for providing the Brf1 antibody
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
THis works was supported by CIHR (grants #MOP-86622 and #PJT-152892) to SSG, and an AIHS fellowship to SSP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D (2011) RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11: 761–774. doi: 10.1038/nrc3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rauen KA (2013) The RASopathies. Annu Rev Genomics Hum Genet 14: 355–369. doi: 10.1146/annurev-genom-091212-153523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin GM, Chang HC, Karim F, Laverty T, Michaud NR, et al. (1997) Signal transduction downstream from Ras in Drosophila. Cold Spring Harb Symp Quant Biol 62: 347–352. [PubMed] [Google Scholar]
- 4.Wassarman DA, Therrien M, Rubin GM (1995) The Ras signalling pathway in Drosophila. Curr Opin Genet Dev 5: 44–50. [DOI] [PubMed] [Google Scholar]
- 5.Ninov N, Manjon C, Martin-Blanco E (2009) Dynamic control of cell cycle and growth coupling by ecdysone, EGFR, and PI3K signalling in Drosophila histoblasts. PLoS Biol 7: e1000079 doi: 10.1371/journal.pbio.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asha H, Nagy I, Kovacs G, Stetson D, Ando I, et al. (2003) Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Read RD, Cavenee WK, Furnari FB, Thomas JB (2009) A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet 5: e1000374 doi: 10.1371/journal.pgen.1000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker J (2006) Control of compartment size by an EGF ligand from neighboring cells. Curr Biol 16: 2058–2065. doi: 10.1016/j.cub.2006.08.092 [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Edgar BA (2009) EGFR signalling regulates the proliferation of Drosophila adult midgut progenitors. Development 136: 483–493. doi: 10.1242/dev.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prober DA, Edgar BA (2000) Ras1 promotes cellular growth in the Drosophila wing. Cell 100: 435–446. [DOI] [PubMed] [Google Scholar]
- 11.Karim FD, Rubin GM (1998) Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125: 1–9. [DOI] [PubMed] [Google Scholar]
- 12.Zecca M, Struhl G (2002) Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signalling. Development 129: 1369–1376. [DOI] [PubMed] [Google Scholar]
- 13.Nagaraj R, Pickup AT, Howes R, Moses K, Freeman M, et al. (1999) Role of the EGF receptor pathway in growth and patterning of the Drosophila wing through the regulation of vestigial. Development 126: 975–985. [DOI] [PubMed] [Google Scholar]
- 14.Halfar K, Rommel C, Stocker H, Hafen E (2001) Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 128: 1687–1696. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA (2011) EGFR/Ras/MAPK signalling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8: 84–95. doi: 10.1016/j.stem.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu N, Wang SQ, Tan D, Gao Y, Lin G, et al. (2011) EGFR, Wingless and JAK/STAT signalling cooperatively maintain Drosophila intestinal stem cells. Dev Biol 354: 31–43. doi: 10.1016/j.ydbio.2011.03.018 [DOI] [PubMed] [Google Scholar]
- 17.Buchon N, Broderick NA, Kuraishi T, Lemaitre B (2010) Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol 8: 152 doi: 10.1186/1741-7007-8-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castanieto A, Johnston MJ, Nystul TG (2014) EGFR signalling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biteau B, Jasper H (2011) EGF signalling regulates the proliferation of intestinal stem cells in Drosophila. Development 138: 1045–1055. doi: 10.1242/dev.056671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, Ha N, Fores M, Xiang J, Glasser C, et al. (2015) EGFR/Ras Signalling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet 11: e1005634 doi: 10.1371/journal.pgen.1005634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baonza A, Murawsky CM, Travers AA, Freeman M (2002) Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol 4: 976–980. doi: 10.1038/ncb887 [DOI] [PubMed] [Google Scholar]
- 22.Tseng AS, Tapon N, Kanda H, Cigizoglu S, Edelmann L, et al. (2007) Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signalling pathway. Curr Biol 17: 728–733. doi: 10.1016/j.cub.2007.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren F, Shi Q, Chen Y, Jiang A, Ip YT, et al. (2013) Drosophila Myc integrates multiple signalling pathways to regulate intestinal stem cell proliferation during midgut regeneration. Cell Res 23: 1133–1146. doi: 10.1038/cr.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prober DA, Edgar BA (2002) Interactions between Ras1, dMyc, and dPI3K signalling in the developing Drosophila wing. Genes Dev 16: 2286–2299. doi: 10.1101/gad.991102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herranz H, Hong X, Cohen SM (2012) Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr Biol 22: 651–657. doi: 10.1016/j.cub.2012.02.050 [DOI] [PubMed] [Google Scholar]
- 26.Herranz H, Hong X, Hung NT, Voorhoeve PM, Cohen SM (2012) Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev 26: 1602–1611. doi: 10.1101/gad.192021.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy BV, Irvine KD (2013) Regulation of Hippo signalling by EGFR-MAPK signalling through Ajuba family proteins. Dev Cell 24: 459–471. doi: 10.1016/j.devcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux PP, Topisirovic I (2012) Regulation of mRNA translation by signalling pathways. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, et al. (1999) Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 19: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, et al. (2007) RAS/ERK signalling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem 282: 14056–14064. doi: 10.1074/jbc.M700906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romeo Y, Moreau J, Zindy PJ, Saba-El-Leil M, Lavoie G, et al. (2013) RSK regulates activated BRAF signalling to mTORC1 and promotes melanoma growth. Oncogene 32: 2917–2926. doi: 10.1038/onc.2012.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo Y, Roux PP (2011) Paving the way for targeting RSK in cancer. Expert Opin Ther Targets 15: 5–9. doi: 10.1517/14728222.2010.531014 [DOI] [PubMed] [Google Scholar]
- 33.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R (2004) Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, et al. (2010) Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci U S A 107: 13984–13990. doi: 10.1073/pnas.1008136107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumont J, Umbhauer M, Rassinier P, Hanauer A, Verlhac MH (2005) p90Rsk is not involved in cytostatic factor arrest in mouse oocytes. J Cell Biol 169: 227–231. doi: 10.1083/jcb.200501027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arquier N, Bourouis M, Colombani J, Leopold P (2005) Drosophila Lk6 kinase controls phosphorylation of eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr Biol 15: 19–23. doi: 10.1016/j.cub.2004.12.037 [DOI] [PubMed] [Google Scholar]
- 37.Reiling JH, Doepfner KT, Hafen E, Stocker H (2005) Diet-dependent effects of the Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr Biol 15: 24–30. doi: 10.1016/j.cub.2004.12.034 [DOI] [PubMed] [Google Scholar]
- 38.Deliu LP, Ghosh A, Grewal SS (2017) Investigation of protein synthesis in Drosophila larvae using puromycin labelling. Biol Open 6: 1229–1234. doi: 10.1242/bio.026294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall L, Rideout EJ, Grewal SS (2012) Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J 31: 1916–1930. doi: 10.1038/emboj.2012.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rideout EJ, Marshall L, Grewal SS (2012) Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signalling. Proc Natl Acad Sci U S A 109: 1139–1144. doi: 10.1073/pnas.1113311109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong S, Zhang C, Johnson DL (2004) Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol Cell Biol 24: 5119–5129. doi: 10.1128/MCB.24.12.5119-5129.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geiduschek EP, Kassavetis GA (2001) The RNA polymerase III transcription apparatus. J Mol Biol 310: 1–26. doi: 10.1006/jmbi.2001.4732 [DOI] [PubMed] [Google Scholar]
- 43.Cordero JB, Stefanatos RK, Myant K, Vidal M, Sansom OJ (2012) Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development 139: 4524–4535. doi: 10.1242/dev.078261 [DOI] [PubMed] [Google Scholar]
- 44.Steiger D, Furrer M, Schwinkendorf D, Gallant P (2008) Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40: 1084–1091. doi: 10.1038/ng.178 [DOI] [PubMed] [Google Scholar]
- 45.Gomez-Roman N, Grandori C, Eisenman RN, White RJ (2003) Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294. doi: 10.1038/nature01327 [DOI] [PubMed] [Google Scholar]
- 46.Sears R, Leone G, DeGregori J, Nevins JR (1999) Ras enhances Myc protein stability. Mol Cell 3: 169–179. [DOI] [PubMed] [Google Scholar]
- 47.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, et al. (2013) Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27: 504–513. doi: 10.1101/gad.205542.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ (2010) mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A 107: 11823–11828. doi: 10.1073/pnas.1005188107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, et al. (2010) mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 30: 3749–3757. doi: 10.1128/MCB.00319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, et al. (2010) Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem 285: 15380–15392. doi: 10.1074/jbc.M109.071639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei Y, Tsang CK, Zheng XF (2009) Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J 28: 2220–2230. doi: 10.1038/emboj.2009.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, et al. (2006) Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci U S A 103: 15044–15049. doi: 10.1073/pnas.0607129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, et al. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943. doi: 10.1101/gad.532109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, et al. (2006) General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell 22: 623–632. doi: 10.1016/j.molcel.2006.04.008 [DOI] [PubMed] [Google Scholar]
- 55.Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR (2006) Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell 22: 633–644. doi: 10.1016/j.molcel.2006.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oler AJ, Cairns BR (2012) PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J 31: 1440–1452. doi: 10.1038/emboj.2011.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, et al. (2003) Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2: 181–184. [PubMed] [Google Scholar]
- 58.Polymenis M, Schmidt EV (1997) Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev 11: 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, et al. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176. doi: 10.1126/science.1187532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, et al. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61. doi: 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, et al. (2013) mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711. doi: 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 62.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, et al. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. doi: 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grewal SS (2015) Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta 1849: 898–907. doi: 10.1016/j.bbagrm.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T (2009) High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun 385: 160–164. doi: 10.1016/j.bbrc.2009.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, et al. (2009) tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280. doi: 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, et al. (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. doi: 10.1016/j.cell.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 67.Goodarzi H, Nguyen HC, Zhang S, Dill BD, Molina H, et al. (2016) Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 165: 1416–1427. doi: 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarke CJ, Berg TJ, Birch J, Ennis D, Mitchell L, et al. (2016) The Initiator Methionine tRNA Drives Secretion of Type II Collagen from Stromal Fibroblasts to Promote Tumor Growth and Angiogenesis. Curr Biol 26: 755–765. doi: 10.1016/j.cub.2016.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birch J, Clarke CJ, Campbell AD, Campbell K, Mitchell L, et al. (2016) The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol Open 5: 1371–1379. doi: 10.1242/bio.019075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pavon-Eternod M, Gomes S, Rosner MR, Pan T (2013) Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19: 461–466. doi: 10.1261/rna.037507.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rojas-Benitez D, Thiaville PC, de Crecy-Lagard V, Glavic A (2015) The Levels of a Universally Conserved tRNA Modification Regulate Cell Growth. J Biol Chem 290: 18699–18707. doi: 10.1074/jbc.M115.665406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA (2005) Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302. doi: 10.1038/ncb1223 [DOI] [PubMed] [Google Scholar]
- 73.Ashton-Beaucage D, Udell CM, Gendron P, Sahmi M, Lefrancois M, et al. (2014) A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signalling in Drosophila. PLoS Biol 12: e1001809 doi: 10.1371/journal.pbio.1001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bellosta P, Hulf T, Balla Diop S, Usseglio F, Pradel J, et al. (2005) Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci U S A 102: 11799–11804. doi: 10.1073/pnas.0408945102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Left: S2 cells were treated with (10μM) U0126 for 2 hrs. Total RNA was isolated and analyzed by Northern blot. Levels of tRNAArg were detected using DIG-labelled tRNA probes. Ethidium bromide stained 5S rRNA bands were used as loading controls, since total 5S rRNA levels are unchanged by manipulations in Ras/Erk signalling (see C). (B) RasV12 expression was induced in S2 cells for 24hrs. Total RNA was isolated and analyzed by northern blot. Levels of tRNAArg were detected using DIG-labelled tRNA probes. Ethidium bromide stained 5S rRNA bands were used as loading controls, since total 5S rRNA levels are unchanged by manipulations in Ras/Erk signalling (see C) (C) qRT-PCR measurement of Brf1 mRNA, Trf1 mRNA, TBP mRNA, 5S rRNA and 7SL RNA levels in U0126 treated S2 cells. (D) qRT-PCR measurement of 5S rRNA levels in RasV12 overexpressing S2 cells.
(TIFF)
(A) Brf1 mRNA levels were measured by qRT-PCR in Drosophila S2 cells treated with dsRNA against Brf1 or GFP (control). Control cells were treated with GFP dsRNA (B) Brf1, phospho-ERK levels and alpha-tubulin protein levels were measured by western blot in Drosophila S2 cells treated with dsRNA against Brf1 and overexpressing RasV12, both alone and together. (C) UAS-Rafgof and UAS-Brf1 RNAi were expressed, either alone or together, in the Drosophila larval AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24 hrs of development to induce transgene expression and dissected as L3 larvae. AMPs are marked by UAS-GFP expression. DNA is stained with Hoechst dye (blue) Representative images are shown for each genotype.
(TIFF)
UAS-EGFR (A) or UAS-RasV12S35 (C) were expressed, either alone or together with UAS-dMyc RNAi in the Drosophila larval AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24 hrs of development to induce transgene expression and dissected as L3 larvae. AMPs are marked by UAS-GFP expression. DNA is stained with Hoechst dye (blue) (B) (related to experiment in A) Numbers of cells in each AMP cluster were counted and expressed as box plots.
(TIFF)
(A, B) UAS-dMaf1 RNAi (A), UAS-EGFR (B), or UAS-dMyc (C) were expressed in AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24hrs of development and dissected at wandering stage. The numbers of cells in each AMP cluster were counted and expressed in box plots. (D) dMaf1 and Ras were knocked down, both alone and together, in Drosophila S2 cells by incubating cells with dsRNAs against dMaf1 and Ras. Control cells were treated with dsRNA to GFP. Total RNA was isolated with Trizol and analyzed by Northern blot using DIG-labelled tRNAArg probe. Ethidium bromide stained 5S rRNA band was used as a loading control.
(TIFF)
(A, B) dMaf1 mRNA levels (A) or protein levels (B) were measured by qRT-PCR or Western blot respectively in cells treated with dsRNA to GFP (control) or dMaf1 (dMaf1 RNAi). dsRNA treatment produced a strong knockdown of dMaf1 levels. (C) Control and dMaf1 dsRNA-treated S2 cells were stained with an anti-dMaf1 antibody (green) and Hoechst dye (blue). (D) dMaf1and Brf1 protein levels were analyzed with western blotting after treatment with U0126 for 2 hours. Decreased phospho-ERK levels served as a positive control for UO126-mediated MEK inhibition. Tubulin levels served as a loading control.
(TIFF)
WT, UAS-EGFR RNAi, UAS-Ras RNAi were expressed in AMPs using the esg-Gal4ts system. Larvae were shifted to 29°C at 24hrs of development and dissected at wandering stage and stained with dMaf1 antibody. (B) Drosophila S2 cell lysates (left, control samples; right, RasV12 induced samples) were treated with either Alkaline phosphatase or λ-phosphatase for 1 hr at 37°C and samples were analysed by phos-tag SDS-PAGE and western blotting using an anti-dMaf1 antibody.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.