Abstract
Human skin contains an abundant and diverse population of microbial organisms. Many of these microbes inhabit follicular structures of the skin. Furthermore, numerous studies have shown that the interaction of some members of the skin microbiome with host cells will result in changes in cell function. However, estimates of the potential for the microbiome to influence human health through skin have ignored the inner follicular surface, and therefore vastly underestimated the potential of the skin microbiome to have a systemic effect on the human body. By calculating the surface area of follicular and the interfollicular epithelial surface it is shown that skin provides a vast interface for interactions with the microbiome.
Studies of the function of the microbiome depend on understanding how microbes that live outside of the body can influence systemic behaviors. The location of this interaction is at epithelial surfaces, specially designed interfaces between the external environment and the delicate internal cellular networks necessary for life. Classical physiology assigned skin, lung, and intestine with their respective functions of protection and temperature regulation, gas exchange, and nutrient absorption. Human skin, lung, and gut are generally considered to have the largest organs, but based on the classical perspective of their functions, skin has been considered to have the smallest surface area. Estimates of the surface areas that depend on height, weight, and other assumptions have led to widely accepted surface area values of 2 m2 for the skin (Mosteller, 1987), 30 m2 for the gut (Helander and Fandriks, 2014), and 50 m2 for the lung (Hasleton, 1972).
We have taken these surface area calculations at face value for decades, with little reason to question their accuracy. This essay seeks to clarify the estimate of the surface area of human skin to demonstrate the potential impact of the skin microbiome on human health. Although the hair follicle has been recognized as a potential reservoir for topical delivery systems (Blume-Peytavi and Vogt, 2011), most of the medical and scientific community assumes that the skin surface area is only 2 m2. This assumption is misleading if the functions of the skin microbiome are to be incorporated into modern models of cutaneous biology.
Abundant evidence now exists that supports the hypothesis that microbial communities play important roles in human health (Sanford and Gallo, 2013; Schroeder and Backhed, 2016). The best studied of these systems has been the gut where a villous epithelium provides a large surface area for both nutrient absorption and microbe-host interactions. Using stool samples as distant surrogates, gut microbiome research has proposed a wide range of functions for these microbes that include food processing, control of allergy, influences on autoimmunity, neural and psychological functions, weight control, and others. In contrast, despite the ease of direct sampling of the skin microbiome, the skin has been less extensively studied. Data from our lab and others have shown that functions of the skin microbiome can include direct host defense against pathogens, control of inflammation, and education of adaptive immune pathways (Cogen et al., 2008; Lai et al., 2009; Naik et al., 2012).
A careful study of the skin using laser-capture microdissection and in situ hybridization has revealed that microbes reside not only on the external interfollicular epithelial surface but also on the entire skin appendage surface and even below the basement membrane (Nakatsuji et al., 2013, 2016). Although these studies do not prove that the bacteria are alive below the dermis, observations of the presence of bacterial DNA, RNA, and antigens are consistent with much prior work that has shown that an abundant amount of bacteria reside within the hair follicle (Lange-Asschenfeldt et al., 2011). Importantly, although much of the epithelial lining of hair follicles, eccrine ducts, apocrine ducts, and sebaceous glands are not directly exposed to the general external environment, these epithelia are in contact with microbes. Therefore, it should be recognized that the epithelial surfaces of skin appendages are relevant interfaces for communication between microbes and host.
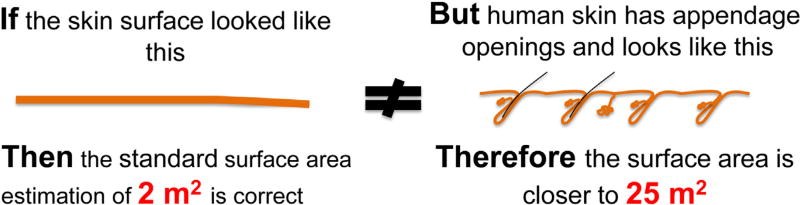
Figure 1 illustrates the simple argument for revising the value of the skin surface area that is important for communication with the microbiome. If one estimates the depth of an average human follicle to be 3 mm and the diameter of that tube is approximately 0.5 mm, then the surface area of a hair follicle is 3.14 × 0.5 × 3 = 4.71 mm2 or 4.71 × 10 −6 m2. The human body is estimated to have 5 × 106 follicles. Therefore, the surface area of the follicular surface could be approximated as 4.71 × 10ȡ6 × 5 × 106 = 24 m2. Added to the 2 m2 surface area estimate based on the exposed interfollicular epithelium, and accounting that other appendage structures like sweat and sebaceous glands also provide epithelial surfaces for microbes, the total skin surface area is at least 30 m2. This is more than 10 times greater than the surface area commonly reported for the skin!
Figure 1. The surface area of skin has been miscalculated.
Human skin is not a flat surface. The presence of approximately 5 million appendages such as hair follicles and sweat ducts greatly increases the epithelial surface area that is uniquely accessible to the microbiome.
This estimate of the skin surface area shows how the skin has a large potential interface with the microbiome, and thus the microbial community that exists on the skin has the greatest potential to influence human health. This interface is special, as the majority of the epithelial surface is uniquely protected below the follicular opening, and not easily accessed by topical products. Therefore, the old estimate of 2 m2 is more appropriate for calculations of heat and water loss, and for topical drug delivery of agents that cannot penetrate into the follicle. However, for the world of microbiome research, it is time to set the record straight.
Footnotes
CONFLICT OF INTEREST
Consultant and equity interest in Matrisys and Sente.
References
- Blume-Peytavi U, Vogt A. Human hair follicle: reservoir function and selective targeting. Br J Dermatol. 2011;165(Suppl. 2):13–7. doi: 10.1111/j.1365-2133.2011.10572.x. [DOI] [PubMed] [Google Scholar]
- Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008;158:442–55. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasleton PS. The internal surface area of the adult human lung. J Anat. 1972;112(Pt 3):391–400. [PMC free article] [PubMed] [Google Scholar]
- Helander HF, Fandriks L. Surface area of the digestive tract—revisited. Scand J Gastroenterol. 2014;49:681–9. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–82. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange-Asschenfeldt B, Marenbach D, Lang C, Patzelt A, Ulrich M, Maltusch A, et al. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Skin Pharmacol Physiol. 2011;24:305–11. doi: 10.1159/000328728. [DOI] [PubMed] [Google Scholar]
- Mosteller RD. Simplified calculation of body-surface area. N Engl J Med. 1987;317:1098. doi: 10.1056/NEJM198710223171717. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol. 2016;136:2192–200. doi: 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25:370–7. doi: 10.1016/j.smim.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–89. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]