Abstract
Although cellular therapies represent a promising strategy for a number of conditions, current approaches face major translational hurdles, including limited cell sources and the need for cumbersome pre-processing steps (for example, isolation, induced pluripotency)1–6. In vivo cell reprogramming has the potential to enable more-effective cell-based therapies by using readily available cell sources (for example, fibroblasts) and circumventing the need for ex vivo pre-processing7,8. Existing reprogramming methodologies, however, are fraught with caveats, including a heavy reliance on viral transfection9,10. Moreover, capsid size constraints and/or the stochastic nature of status quo approaches (viral and non-viral) pose additional limitations, thus highlighting the need for safer and more deterministic in vivo reprogramming methods11,12. Here, we report a novel yet simple-to-implement non-viral approach to topically reprogram tissues through a nanochannelled device validated with well-established and newly developed reprogramming models of induced neurons and endothelium, respectively. We demonstrate the simplicity and utility of this approach by rescuing necrotizing tissues and whole limbs using two murine models of injury-induced ischaemia.
Recent advances in nuclear reprogramming in vivo have opened up the possibility for the development of ‘on-site’, patient-specific, cell-based therapies. We have developed a novel yet simple to implement non-viral approach to topically and controllably deliver reprogramming factors to tissues through a nanochannelled device (Fig. 1). This tissue nano-transfection (TNT) approach allows direct cytosolic delivery of reprogramming factors by applying a highly intense and focused electric field through arrayed nanochannels11,13, which benignly nanoporates the juxtaposing tissue cell membranes and electrophoretically drives reprogramming factors into the cells (Fig. 1a–d). Detailed information regarding the TNT system fabrication process and simulation results is provided in Supplementary Figs 1 and 2. In contrast to current in vivo transfection technologies (for example, viruses and conventional tissue bulk electroporation (BEP)), in which gene delivery is highly stochastic in nature and could lead to adverse side effects (such as inflammatory response and cell death)14, nanochannel-based transfection enables more focused (Fig. 1b,c) and ample (Fig. 1d) reprogramming factor delivery at the single-cell level, thus making this a powerful tool for deterministic in vivo gene transfection and reprogramming11,13.
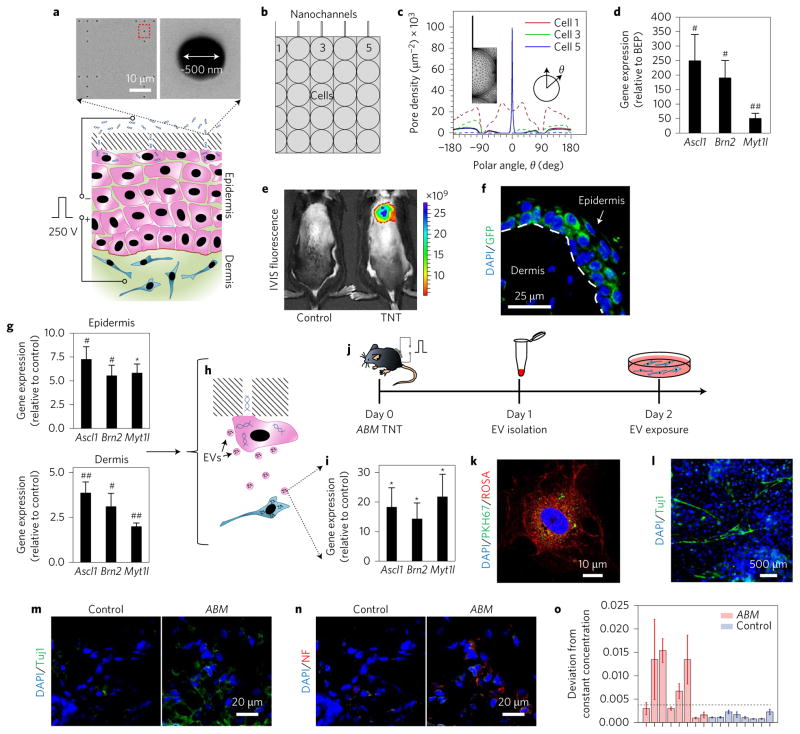
Figure 1. TNT mediates enhanced reprogramming factor delivery and propagation beyond the transfection boundary.
a, Schematic diagram of the TNT process on exfoliated skin tissue. Exfoliation is required to remove dead cells from the skin surface. The positive electrode is inserted intradermally, and the negative electrode is put in contact with the cargo solution. A pulsed electric field (250 V, 10 ms pulses, 10 pulses) is then applied across the electrodes to nanoporate exposed cell membranes and inject the cargo directly into the cytosol. Scanning electron micrographs (top) of the TNT platform surface show the nanopore array. b, Schematic diagram showing the boundary conditions for simulation purposes. Nanochannels are in direct contact with the outermost cell layer. c, Simulation of the poration profile for different cells (cells 1, 3 and 5 from b) undergoing TNT (solid lines) versus BEP (dashed lines). The plot shows that TNT leads to focused poration, and BEP results in widespread poration. d, ABM expression results for TNT versus BEP 24 h after transfection (n = 5). TNT resulted in superior ABM expression. BEP was conducted via intradermal injection of the ABM plasmids followed by a pulsed electric field. Controls for BEP experiments involved intradermal injections of ABM plasmids with no electric field implementation. e,f, Representative in vivo imaging system (IVIS) fluorescence (e) and confocal microscopy (f) images of mouse skin after TNT treatment with labelled DNA and the ABM factors, respectively. GFP is the reporter gene in the Ascl1 plasmid. g, Laser capture microdissection (LCM) and qRT–PCR results of gene expression in the epidermis and dermis (t = 24 h) showing that gene expression propagated beyond the epidermal transfection boundary (n = 5–6). h, Schematic diagram illustrating the concept of EV-mediated transfection propagation from epidermis to dermis. i, qRT–PCR analysis of the EV cargo showing significant loading of ABM cDNAs/mRNAs (n = 6–8). j, Experimental design to confirm whether EVs are a viable vehicle for propagating transfection and reprogramming. k, Confocal micrograph showing a mouse embryonic fibroblast (red) that has spontaneously internalized the EVs (green) isolated from TNT-treated skin. l, Mouse embryonic fibroblast cultures showing iNs at day 7 after a 24 h exposure to ABM-laden EVs isolated from ABM-transfected skin. m,n, Immunostaining results (week 4) showing increased Tuj1 (m) and neurofilament (NF) (n) expression in the skin after ABM transfection. o, Electrophysiological activity shown as a statistically representative bar plot indicating changes in ionic concentration (quantified as average standard deviations from the norm per insertion site) of the extracellular niche as a result of neuronal cell cluster excitability (n = 8, P < 0.05, Fisher’s exact test). This average was calculated for 5–10 trials (with 100 sequential discrete measurements per trial) for each ABM or control mouse. Activity was defined as changes in ionic concentration in excess of the baseline (dashed line: experimental noise measured in physiological saline solution). Each bar shows the results collected in individual mice. *P < 0.01 (Dunn’s), #P < 0.01 (Tukey test), ##P < 0.05 (Holm–Sidak method).
Experiments with fluorescein amidite (FAM)-labelled DNA on C57BL/6 mice established that TNT can deliver cargo into the skin in a rapid (<1 s) and non-invasive/topical manner (Fig. 1e). We next tested whether TNT-based topical delivery of reprogramming factors could lead to successful skin reprogramming using a robust model where overexpression of Ascl1/Brn2/Myt1l (ABM) is known to directly reprogram fibroblasts into induced neurons (iNs) in vitro11,15. Our findings showed that TNT can not only be used for topical delivery of reprogramming factors (Fig. 1f), but it can also orchestrate a coordinated response that results in reprogramming stimuli propagation (that is, epidermis to dermis) beyond the initial transfection boundary (the epidermis) (Fig. 1g–i), possibly via dispatch of extracellular vesicles (EVs) rich in target gene cDNAs/mRNAs (Fig. 1h,i)16, among other plausible mechanisms17. Exposing naive cells to ABM-loaded EVs isolated from TNT-treated skin (Fig. 1j–l) established that these EVs can be spontaneously internalized by remote cells and trigger reprogramming (Fig. 1k,l and Supplementary Fig. 3). Moreover, gene expression analysis indicated that intradermal ABM EV injection triggered changes in the skin consistent with neuronal induction (Supplementary Fig. 4), as evidenced by increased Tuj1 expression. The neurotrophic effect of skin-derived ABM-loaded EVs was further confirmed in a middle cerebral artery occlusion stroke mouse model (Supplementary Fig. 5)18.
Successful skin cell reprogramming was verified by immunofluorescence, which showed increased Tuj1 and neurofilament expression over time (Fig. 1m,n). Further characterization was conducted via genome-wide transcriptome array analysis comparing in vitro and in vivo derived iNs (Supplementary Fig. 6). Electrophysiological activity, indicative of neuronal excitability, was successfully detected and monitored (in situ) in ~50% of the ABM-transfected mice (Fig. 1o) through a novel polypyrrole (PPy)-based biosensing platform (Supplementary Fig. 7)19,20. No such activity was detected in any of the control mice. Lineage tracing experiments with a K14-Cre reporter mouse model established that the newly induced neurons partly originated from K14+ skin cells (Supplementary Fig. 8). Hair follicles also consistently showed marked Tuj1 immunoreactivity, suggesting that follicular cells could participate in the reprogramming process21,22. Additional experiments with a Col1A1-enhanced green fluorescent protein (eGFP) mouse model (Supplementary Fig. 8), where cells with an active Col1A1 promoter (for example, dermal fibroblasts) express eGFP, showed a number of collagen/eGFP+ cells in the dermis in a transition phase to Tuj1+, thus suggesting a fibroblastic origin for some of the reprogrammed cells in the skin.
Having validated the TNT platform for successful in vivo reprogramming using iNs as a case study, we then set out to develop a robust and simple non-viral methodology that would be capable of reprogramming skin cells into induced endothelial cells (iECs). To this end, we first identified and validated (in vitro) a set of reprogramming factors—Etv2, Foxc2 and Fli1 (EFF)—to promote more rapid and effective reprogramming of somatic cells into iECs (Supplementary Figs 9 and 10) compared to previous reports10. In vitro non-viral transfection and reprogramming experiments11 showed that EFF could reprogram human and mouse primary fibroblasts into iECs rapidly (<1 week) and efficiently (Supplementary Fig. 9).
Once the efficacy of EFF to induce direct endothelial cell reprogramming was established in vitro, we then proceeded to test this paradigm in vivo. Co-transfection of these three genes into the dorsal skin of C57BL/6 mice resulted in marked stroma reprogramming within a week, as evidenced by a significant increase in Pecam-1 and vWF expression compared to control skin (Fig. 2a–c), in addition to enhanced proliferative activity (Supplementary Fig. 11). Experiments with K14-Cre reporter and Col1A1-eGFP mouse models demonstrated that the reprogrammed cell population had, for the most part, a dermal origin (Supplementary Fig. 12). High-resolution laser speckle (HRLS) imaging of dorsal skin showed that TNT-based delivery of EFF enhanced blood flow to the treated area within 3 days (Fig. 2d,e). Ultrasound imaging detected unexpected pulsatile blood flow only 3 mm away from the surface of the skin (Fig. 2f, right), demonstrating successful anastomosis of the newly formed blood vessels with local functional cutaneous arteries. Note that, in control mice, blood vessels were not typically detected near the skin surface (Fig. 2f, left).
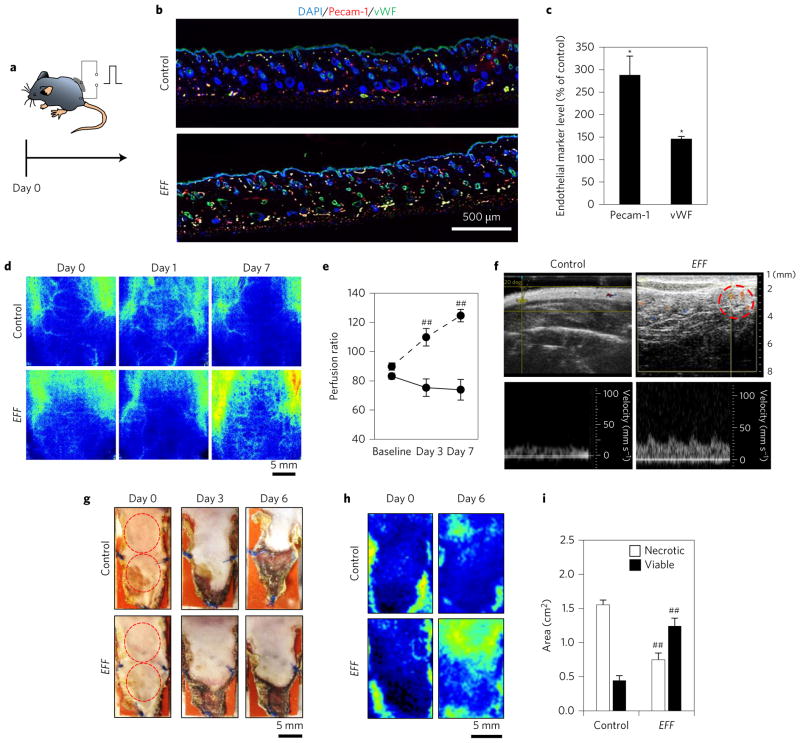
Figure 2. EFF TNT leads to increased vascularization and rescue of skin tissue under ischaemic conditions.
a–c, A one-time treatment of dorsal skin lasting only a few seconds led to increased angiogenesis (Pecam-1, vWF) of skin tissue (day 7) (n = 3). d,e, High-resolution laser speckle imaging shows enhanced perfusion to the EFF-treated area over time (n = 5). f, Ultrasound imaging of EFF-treated skin confirmed the presence of superficial blood vessels (dashed circle) with pulsatile behaviour, which suggests successful anastomosis with the parent circulatory system. g, Monopedicle flap experiment showing increased flap necrosis for controls compared to EFF-treated skin. h, Laser speckle imaging showing increased blood flow to the flapped tissue treated with EFF TNT. i, Quantification of flap necrosis (n = 6). *P < 0.05 (t-test), ##P < 0.05 (Holm–Sidak method).
Once the robustness of the EFF cocktail to induce vascular endothelium was demonstrated both in vitro and in vivo, we studied whether EFF TNT-mediated topical skin reprogramming could lead to functional reperfusion of ischaemic tissues. We first tested this concept with a full-thickness 2 × 1 cm2 monopedicle dorsal skin flap in C57BL/6 mice, whereby blood supply to the flapped tissue only came from the cephalad attachment (Fig. 2g). Laser speckle monitoring after EFF treatment showed higher blood perfusion compared to control flaps (Fig. 2h). As expected, control flaps showed significant signs of tissue necrosis (Fig. 2g,top,i). This tissue damage was significantly limited in response to EFF transfection. Thus, TNT-mediated EFF delivery and subsequent stroma reprogramming effectively counteracted tissue necrosis under ischaemic conditions.
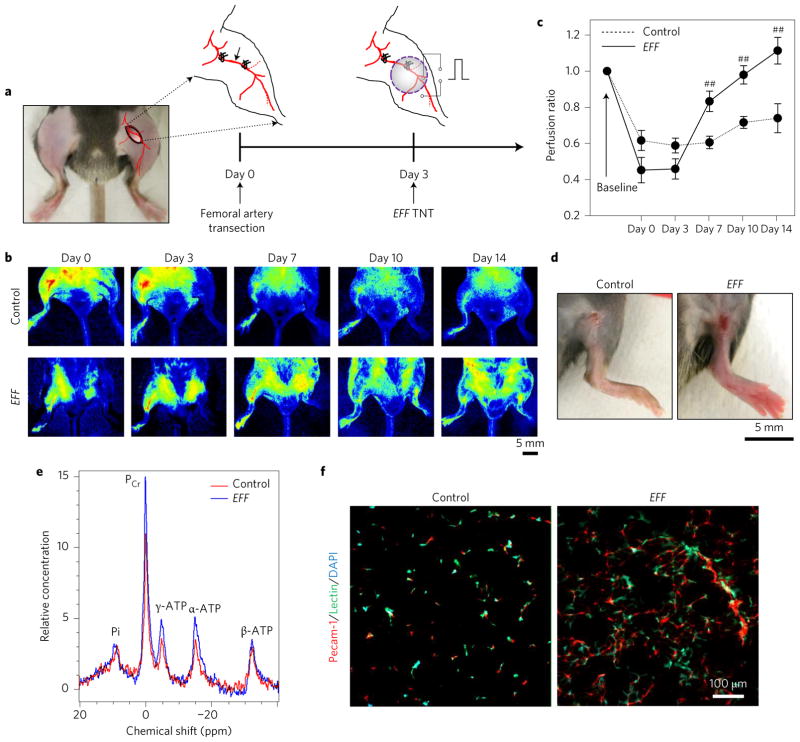
Finally, to verify whether TNT-based delivery of EFF could lead to whole limb rescue, we tested TNT in a hindlimb ischaemia C57BL/6 mouse model (Fig. 3a). EFF TNT was conducted on the inner thigh skin three days after transection of the femoral artery. Laser speckle monitoring recorded a significant reduction in blood flow to the limb immediately after surgery (Fig. 3b). Compared to control ischaemic limbs, EFF-treated limbs showed improved perfusion as early as day 7 post-TNT (Fig. 3b,c). HRLS imaging demonstrated an increased incidence of small collaterals in the EFF-treated limbs compared to controls (Supplementary Fig. 13). Macroscopic analysis showed more pronounced signs of tissue necrosis in the control limbs compared to the EFF-treated ones (Fig. 3d). Additional experiments in BALB/c mice, which have a tendency to experience more deleterious side effects from injury-induced limb ischaemia23,24, showed that EFF transfection also led to successful limb perfusion and minimized the incidence of necrosis and auto-amputation (Supplementary Fig. 14). Muscle energetics testing by NMR imaging showed increased levels of adenosine triphosphate (ATP) and phosphocreatine (PCr) in EFF-treated limbs compared to controls (Fig. 3e). Immunofluorescence analysis revealed marked revascularization far beyond the treatment area. Angiogenesis was also induced in more distal locations within the limb, such as the gastrocnemius muscle (Fig. 3f and Supplementary Fig. 15). Although the underlying mechanisms for this response need to be elucidated further, we found that autologous EVs, isolated from EFF-treated dorsal skin, have the potential to induce blood vessel formation when injected directly into the gastrocnemius muscle in a hindlimb ischaemia mouse model (Supplementary Fig. 16). Parallel in vitro experiments demonstrated that these EVs can induce reprogramming in naive cells (Supplementary Fig. 17). We thus propose that EVs dispatched from EFF-treated tissue serve as a mediator of the propagation of pro-iEC reprogramming signals. PCR analysis revealed that in addition to the transduced EFF cDNAs/mRNAs, the EVs also appeared to be preloaded with pro-angiogenic vascular endothelial growth factor and basic fibroblast growth factor mRNAs (Supplementary Fig. 16). This suggests that EVs derived from EFF-treated skin not only represent a viable mechanism for propagating EFF reprogramming signals throughout the target tissue, but may also play a role in niche preconditioning by spreading pro-angiogenic signals within the first hours after transfection.
Figure 3. EFF TNT rescues whole limbs from necrotizing ischaemia.
a–c, A one-time treatment of thigh skin lasting only a few seconds led to increased limb reperfusion following transection of the femoral artery. Perfusion was calculated based on the ratio of the ischaemic versus normal/contralateral limb (n = 5–7). d, Control limbs showing more pronounced signs of tissue necrosis compared to EFF-treated limbs (day 14). e, NMR-based measurements of muscle energetics confirmed increased ATP and PCr levels for EFF-treated limbs compared to controls. f, Immunofluorescence analysis of the gastrocnemius muscle showing enhanced angiogenesis. ##P < 0.05 (Holm–Sidak method).
Here, we show for the first time that TNT can be used to deliver reprogramming factors into the skin in a rapid, highly effective and non-invasive manner. Such TNT delivery leads to tailored skin tissue reprogramming, as demonstrated with well-established and newly developed reprogramming models of iNs and iECs, respectively. TNT-induced skin-derived iECs rapidly formed blood vessel networks that successfully anastomosed with the parent circulatory system and restored tissue and limb perfusion in two murine models of injury-induced ischaemia. TNT-based tissue reprogramming has the potential to ultimately enable the use of a patient’s own tissue as a prolific immunosurveilled bioreactor to produce autologous cells that can resolve conditions locally/on site or distally upon harvesting. This simple to implement TNT approach, which elicits and propagates powerfully favourable biological responses through a topical one-time treatment that only lasts seconds, could also find applications beyond plasmid DNA-based reprogramming strategies, including oligo RNA (for example, miRs and siRNAs)-mediated reprogramming (Supplementary Fig. 18)25, gene modulation, editing and so on.
Methods
TNT platform fabrication
TNT devices were fabricated from thinned (~200 μm) double-side-polished (100) silicon wafers (Supplementary Fig. 1). Briefly, ~1.5-μm-thick layers of AZ5214E photoresist were first spin-coated on the silicon wafers at ~3,000 r.p.m. Nanoscale openings were subsequently patterned on the photoresist using a GCA 6100C stepper. Up to 16 dies of nanoscale opening arrays were patterned on each 100 mm wafer. These openings were then used as etch masks to drill ~10-μm-deep nanochannels on the silicon surface using deep reactive ion etching (DRIE) (Oxford Plasma Lab 100 system). Optimized etching conditions were as follows: SF6 gas, 13 s/100 s.c.c.m. gas flow/700 W inductance-coupled plasma (ICP) power/40 W RF power/30 mT automatic pressure controller (APC) pressure; C4F8 gas, 7 s/100 s.c.c.m. gas flow/700 W ICP power/10 W RF power/30 mT APC pressure. Microscale reservoirs were then patterned on the back side of the wafers by means of contact photolithography and DRIE. Finally, a ~50-nm-thick insulating/protective layer of silicon nitride was deposited on the TNT platform surface.
Animal husbandry
C57BL/6 mice were obtained from Harlan Laboratory. B6.129 (Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J mice obtained from Jackson Laboratories were bred with K14cre to produce K14cre/Gt(ROSA)26Sortm4 (ACTB-tdTomato-EGFP)Luo/J mice. pOBCol3.6GFPtpz mice were gifts from T. Wilgus (The Ohio State University). repTOP mitoIRE mice were obtained from Charles River Laboratories. Fsp1-Cre mice were donated by A. Deb (University of California, Los Angeles). Fsp1-Cre mice were crossed with the B6.Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze/J mice (Jackson Laboratories) to generate mice with tdTomato expression specific to fibroblasts. All mice were male and 8–12 weeks old at the time of the study. Genotyping PCR for ROSAmT/mG mice was conducted using primers oIMR7318-CTC TGC TGC CTC CTG GCT TCT, oIMR7319-CGA GGC GGA TCA CAA GCA ATA and oIMR7320-TCA ATG GGC GGG GGT CGT T, and the K-14 Cre transgene was confirmed using primers oIMR1084-GCG GTC TGG CAG TAA AAA CTA TC and oIMR1085-GTG AAA CAG CAT TGC TGT CAC TT. Genotyping PCR for Fsp1-Cre mice was conducted using the following primers: forward-CTAGGCCACAGAATTGAAAGATCT and reverse-GTAGGTGGAAATTCTAGCATCATCC (for wild type, product length = 324 bp) and forward-GCGGTCTGGCAGTAAAAACTATC and reverse-GTGAAACAGCATTGCATTGCTGTCACTT (for Cre transgene, product length = 100 bp). td tomato was confirmed using the following primers: forward-AAGGGAGCTGCAGTGGAGTA and reverse-CCGAAAATCTGTGGGAAGTC (for wild type, product length = 196 bp) and forward-GGCATTAAAGCAGCGTATCC and reverse-CTGTTCCTGTACGGCATGG (mutant type, product length = 297 bp). All animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of The Ohio State University. No statistical method was used to predetermine the sample size. Power analysis was not necessary for this study. The animals were tagged and grouped randomly using a computer-based algorithm (www.random.org).
Mammalian cell culture and in vitro reprogramming
Primary human adult dermal fibroblasts (ATCC PCS-201-012) were purchased, mycoplasma-free and certified, directly from ATCC. No further cell line authentication/testing was conducted. The cells were expanded in fibroblast basal medium supplemented with fibroblast growth kit–serum-free (ATCC PCS 201–040) and penicillin/streptomycin. E12.5–E14 mouse embryonic fibroblasts (MEFs) were cultured in DMEM/F12 supplemented with 10% fetal bovine serum. Non-viral cell transfection and reprogramming experiments were conducted via three-dimensional nanochannel electroporation (NEP), as described previously11. Briefly, the cells were first grown to full confluency overnight on the 3D NEP device. Subsequently, a pulsed electric field was used to deliver a cocktail of plasmids (0.05 μg μl−1) into the cells consisting of a 1:1:1 mixture of Fli1:Etv2:Foxc2. The cells were then collected 24 h after plasmid delivery, placed in EBM-2 basal medium (CC-3156, Lonza) supplemented with EGM-2 MV SingleQuot kit (CC-4147, Lonza) and further processed for additional experiments/measurements. Etv2 and Fli1 plasmids were donated by A. Ferdous (Department of Internal Medicine, UT Southwestern Medical Center, Texas). Foxc2 plasmids were donated by T. Kume (Department of Medicine–Cardiology and Pharmacology, Northwestern University–FCVRI, Chicago).
In vivo reprogramming
The areas to be treated were first depilated 24–48 h before TNT. The skin was then exfoliated to eliminate the dead/keratin cell layer and expose nucleated cells in the epidermis. The TNT devices were placed directly over the exfoliated skin surface. ABM or EFF plasmid cocktails were loaded in the reservoir at a concentration of 0.05–0.1 μg μl−1. A gold-coated electrode (the cathode) was immersed in the plasmid solution, and a 24G needle counterelectrode (the anode) was inserted intradermally, juxtaposed to the TNT platform surface. Pulsed electrical stimulation (10 pulses, 250 V in amplitude, duration of 10 ms per pulse) was then applied across the electrodes to nanoporate the exposed cell membranes and drive the plasmid cargo into the cells through the nanochannels. ABM plasmids were mixed at a 2:1:1 molar ratio as described previously11. Unless otherwise specified, control specimens involved TNT treatments with a blank, phosphate buffer saline (PBS)/mock plasmid solution (Supplementary Fig. 19).
Electrophysiological activity measurements
The general principle of extracellular recordings was used to detect electrophysiological activity in the skin. Chronoamperometric measurements were conducted using PPy-based probes to detect neuronal excitability through two small incisions on the skin of sedated mice. For details see Supplementary Fig. 7.
Middle cerebral artery occlusion stroke surgery and analysis
Transient focal cerebral ischaemia was induced in mice by middle cerebral artery occlusion (MCAO), achieved using the intraluminal filament insertion technique as previously described18. Magnetic resonance imaging (MRI) images were used to determine infarct size as a percentage of the contralateral hemisphere after correcting for oedema.
Ischaemic skin flaps
Monopedicle (that is, random-pattern) ischaemic flaps measuring 20 mm × 10 mm were created on the dorsal skin of C57BL/6 mice. Briefly, eight- to ten-week-old mice were anaesthetized with 1–3% isoflurane. The dorsa were depilated, cleaned and sterilized with betadine. A monopedicle flap was created on the dorsal skin of the mice by making 20-mm-long full-thickness parallel incisions 10 mm apart. The bottom part of the skin was cut to create a free hanging flap. Flap edges were cauterized. A 0.5 mm silicon sheet was placed under the flap and then sutured to the adjacent skin with 5–0 ethicon silk suture. Finally, a single dose of buprenorphine was administered subcutaneously to control pain. Laser speckle imaging (Perimed) was conducted 2 h post-surgery to confirm successful blood flow occlusion. TNT-based transfections were conducted 24 h before skin flapping.
Hindlimb ischaemia surgery
Unilateral hind-limb ischaemia was induced via occlusion and subsequent transection of the femoral artery26. Briefly, eight- to ten-week-old week mice were anaesthetized with 1–3% isoflurane, and placed supine under a stereomicroscope (Zeiss OPMI) on a heated pad. The femoral artery was exposed and separated from the femoral vein via a ~1 cm incision. Proximal and distal end occlusions were induced with 7–0 silk suture, which was then followed by complete transection of the artery. Finally, a single dose of buprenorphine was administered subcutaneously to control pain. Laser speckle imaging (PeriCam PSI High Resolution, PeriMed) was conducted 2 h post-surgery to confirm successful blood flow occlusion.
Isolation of EVs
EVs were isolated from 12-mm-diameter skin biopsies, which were collected in optimal cutting temperature compound (OCT) blocks and stored frozen for later use. Briefly, the blocks were thawed and washed with PBS to eliminate the OCT. Following removal of the fat tissue with a scalpel, the skin tissue was minced into ~1 mm pieces and homogenized with a microgrinder in PBS. After centrifugation at 3,000g, an Exoquick kit was used at a 1:5 ratio (Exoquick: supernatant) to isolate EVs from the supernatant for 12 h at 4 °C. EVs were precipitated via centrifugation at 1,500g for 30 min. Total RNA was then extracted from the pellets using a Mirvana kit (Life Technologies) following the recommendations of the manufacturer.
DNA plasmid preparation
Plasmids were prepared using a plasmid DNA purification kit (Qiagen Maxi-prep, cat. no. 12161 and Clontech Nucleobond cat. no. 740410). DNA concentrations were obtained from a Nanodrop 2000c Spectrophotemeter (Thermoscientific). For a list of plasmid DNA constructs and their original sources see Supplementary Table 1.
LCM and quantitative real-time PCR
LCM was performed using a laser microdissection system from PALM Technologies (Zeiss). Specific regions of tissue sections, identified based on morphology and/or immunostaining, were cut and captured under a ×20 ocular lens. The samples were catapulted into 25 μl of cell direct lysis extraction buffer (Invitrogen). Approximately 1,000,000 μm2 of tissue area was captured into each cap, and the lysate was then stored at −80 °C for further processing. Quantitative real-time (qRT)–PCR of the LCM samples was performed from cell direct lysis buffer following the manufacturer’s instructions. For a list of primers see Supplementary Table 2.
Immunohistochemistry and confocal microscopy
Tissue immunostaining was carried out using specific antibodies and standard procedures. Briefly, OCT-embedded tissue was cryosectioned at 10 μm thick, fixed with cold acetone, blocked with 10% normal goat serum and incubated with specific antibodies (Supplementary Table 3). The signal was visualized by subsequent incubation with appropriate fluorescence-tagged secondary antibodies (Alexa 488-tagged α-guinea pig, 1:200; Alexa 488-tagged α-rabbit, 1:200; Alexa 568-tagged α-rabbit, 1:200) and counter-stained with DAPI. Lectin-based visualization of blood vessels was conducted via tail vein injection of FITC-labelled lectin 30 min before tissue collection. Images were captured by a laser scanning confocal microscope (Olympus FV 1000 filter/spectral).
IVIS imaging
The animals were imaged under anaesthesia using an IVIS Lumina II optical imaging system. repTOP mitoIRE mice were pre-injected with substrate luciferin (potassium salt of beetle luciferin, Promega) at a dose of 100 mg kg−1, 5–10 min before imaging. Overlay images with luminescence images were made using Living Image software.
MRI of stroke brains
Magnetic resonance angiography was used to validate our MCAO model in mice and to optimize the occluder size and the internal carotid artery insertion distance for effective MCAO. T2-weighted MRI was performed on anaesthetized mice 48 h after MCA–reperfusion using a 9.4 T MRI (Bruker Corporation, Bruker BioSpin). Images were acquired using a Rapid Acquisition with Relaxation Enhancement (RARE) sequence using the following parameters: field of view (FOV) 30 × 30 mm2, acquisition matrix 256 × 256, TR 3,500 ms, TE 46.92 ms, slice gap 1.0 mm, rare factor 8, number of averages 3. The resolution was 8.5 pixels per mm. Raw MR images were converted to the standard DICOM format and processed. After appropriate software contrast enhancement of images using Osirix v.3.4, digital planimetry was performed by a masked observer to delineate the infarct area in each coronal brain slice. Infarct areas from brain slices were summed, multiplied by slice thickness, and corrected for oedema-induced swelling as previously described to determine the infarct volume18.
Analysis of muscle energetics
Muscle energetics was evaluated by NMR spectroscopy measurements on a 9.4 T scanner (Bruker BioSpec) using a volume coil for RF transmission and a 31P coil for reception27. In vivo imaging was conducted in a custom-made 1H/31P transceiver coil array. Data were acquired using a single pulse sequence. The raw data were windowed for noise reduction and Fourier-transformed to the spectral domain.
Ultrasound-based imaging and characterization of blood vessels
Blood vessel formation was parallel monitored via ultrasound imaging. Briefly, a Vevo 2100 system (Visual Sonics) was used to obtain ultrasound images on the B-mode with an MS 250 linear array probe28. Doppler colour flow imaging was implemented to monitor and quantify the blood flow characteristics under systole and diastole.
GeneChip probe array and ingenuity pathway (IPA) analyses
LCM was used to prepare tissue isolates enriched for in vivo-derived iNs from ABM-transfected mouse skin29,30. Tissue isolates were processed in lysis buffer using a PicoPure RNA Isolation Kit (ThermoFisher). RNA extraction, target labelling, GeneChip and data analysis were performed as described previously29–31. The samples were hybridized to Affymetrix Mouse transcriptome Array 1.0 (MTA1.0). The arrays were washed and scanned with the GeneArray scanner (Affymetrix) at The Ohio State University facilities as described already29,31. Expression data have been submitted to the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) under series accession no. GSE92413. Raw data were normalized using RMA16 and analysed using Genespring GX (Agilent). Additional data processing was performed using dChip software (Harvard University)29,31. Functional annotation of the similar genes across groups was performed using IPA analysis.
Statistical analysis
Samples were coded, and data collection was performed in a blinded fashion. Data are reported as mean ± standard error of 3–8 biological replicates. Unsuccessful transfections (for example, due to poor contact between the skin and the nanochannels, or nanochannel clogging) were excluded from the analysis. Experiments were replicated at least twice to confirm reproducibility. Comparisons between groups were made by analysis of variance (ANOVA). Statistical differences were determined using parametric/non-parametric tests, as appropriate, with SigmaPlot version 13.0.
Data availability
GeneChip expression data can be accessed through the Gene Expression Omnibus. Additional data are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
Funding for C.K.S. was partly provided by NIGMS/NINR (R01GM07718507, R01GM10801402, R01NR01567601, R01NR01389804 and R01NS42617) and a philanthropic gift from Leslie and Abigail Wexner. Funding for L.J.L. was partly provided by NIBIB (R21EB017539), NSF (NSEC EEC-0914790) and the National Center for the Advancing Translational Sciences (UL1TR001070). Funding for D.G.-P., S.K. and C.R. was partly provided by NINDS (R21NS099869). Additional funding for D.G.-P. and S.K. was provided in part by the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org), grant DK076169 (U24DK076169). J.J.O., V.B.S., S.K., C.R. and S.R. acknowledge financial support from NIH (R01HL132355, R21EB017539, R01NS099869 and R01DK076566) and NSF (1325114), respectively. The authors thank T. Wilgus and B. Wulff (Department of Pathology, The Ohio State University) for providing the Col1A1 mice used in this study. Fsp1-Cre mice were a gift from A. Deb (University of California, Los Angeles). Etv2 and Fli1 plasmids were donated by A. Ferdous (Department of Internal Medicine, UT Southwestern). Foxc2 plasmid was donated by T. Kume (Department of Medicine-Cardiology and Pharmacology, Northwestern University-FCVRI, Chicago). X. Wang (The Ohio State University) provided support with plasmid design and preparation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences, National Science Foundation or the National Institutes of Health. This work was sponsored by and represents activity of The Ohio State University Center for Regenerative Medicine and Cell Based Therapies (regenerativemedicine.osu.edu) and Nanoscale Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices.
Footnotes
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
Author contributions
TNT platform design, implementation and optimization (for different applications) was performed by D.G.-P., L.J.L., D.P., S.G. and C.K.S. TNT chip fabrication, transductions, cell/tissue imaging, transgenic mouse experiments and histology were performed by D.G.-P., D.P., S.G., N.H.-C., V.M., S.G., L.C., M.S., E.S., A.S., J.M., P.B., W.L., J.J.O., L.J.L. and C.K.S. Electrophysiological activity measurements were conducted by T.Z., R.G.N., V.B.S. and J.J.O., with support from S.G. and N.H.-C. Global gene expression analyses were conducted by S.R., S.K., K.S. and C.K.S. TNT chip simulations were conducted by W.-C.L., J.S., L.C., D.G.-P. and L.J.L. S.R. oversaw and participated in the LCM work. Stroke recovery experiments were conducted by D.G.-P., V.M., A.S., R.S., M.H., S.K., C.R. and C.K.S. The manuscript was written by D.G.-P., L.J.L., D.P., S.G. and C.K.S. C.K.S. and L.J.L. jointly supervised this work.
References
- 1.Rosova I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26:2173–2182. doi: 10.1634/stemcells.2007-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinoshita M, et al. Long-term clinical outcome after intramuscular transplantation of granulocyte colony stimulating factor-mobilized CD34 positive cells in patients with critical limb ischemia. Atherosclerosis. 2012;224:440–445. doi: 10.1016/j.atherosclerosis.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 3.Losordo DW, Dimmeler S. Therapeutic angiogenesis and vasculogenesis for ischemic disease. Part II: cell-based therapies. Circulation. 2004;109:2692–2697. doi: 10.1161/01.CIR.0000128596.49339.05. [DOI] [PubMed] [Google Scholar]
- 4.Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham JJ, Ulbright TM, Pera MF, Looijenga LH. Lessons from human teratomas to guide development of safe stem cell therapies. Nat Biotechnol. 2012;30:849–857. doi: 10.1038/nbt.2329. [DOI] [PubMed] [Google Scholar]
- 6.Leduc PR, et al. Towards an in vivo biologically inspired nanofactory. Nat Nanotech. 2007;2:3–7. doi: 10.1038/nnano.2006.180. [DOI] [PubMed] [Google Scholar]
- 7.Heinrich C, Spagnoli FM, Berninger B. In vivo reprogramming for tissue repair. Nat Cell Biol. 2015;17:204–211. doi: 10.1038/ncb3108. [DOI] [PubMed] [Google Scholar]
- 8.Karagiannis P, Yamanaka S. The fate of cell reprogramming. Nat Methods. 2014;11:1006–1008. doi: 10.1038/nmeth.3109. [DOI] [PubMed] [Google Scholar]
- 9.Grande A, et al. Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat Commun. 2013;4:2373. doi: 10.1038/ncomms3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita R, et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc Natl Acad Sci USA. 2015;112:160–165. doi: 10.1073/pnas.1413234112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallego-Perez D, et al. Deterministic transfection drives efficient nonviral reprogramming and uncovers reprogramming barriers. Nanomedicine. 2016;12:399–409. doi: 10.1016/j.nano.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marx V. Cell biology: delivering tough cargo into cells. Nat Methods. 2016;13:37–40. doi: 10.1038/nmeth.3693. [DOI] [PubMed] [Google Scholar]
- 13.Boukany PE, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotech. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 14.Sen CK, Ghatak S. miRNA control of tissue repair and regeneration. Am J Pathol. 2015;185:2629–2640. doi: 10.1016/j.ajpath.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 17.Davis DM, Sowinski S. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol. 2008;9:431–436. doi: 10.1038/nrm2399. [DOI] [PubMed] [Google Scholar]
- 18.Khanna S, et al. Loss of miR-29b following acute ischemic stroke contributes to neural cell death and infarct size. J Cereb Blood Flow Metab. 2013;33:1197–1206. doi: 10.1038/jcbfm.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venugopal V, Sundaresan VB. Polypyrrole-based amperometric cation sensor with tunable sensitivity. J Intel Mat Syst Str. 2016;27:1702–1709. [Google Scholar]
- 20.Venugopal V, Venkatesh V, Northcutt RG, Maddox J, Sundaresan VB. Nanoscale polypyrrole sensors for near-field electrochemical measurements. Sensors Actuat B Chem. 2017;242:1193–1200. [Google Scholar]
- 21.Hunt DP, et al. A highly enriched niche of precursor cells with neuronal and glial potential within the hair follicle dermal papilla of adult skin. Stem Cells. 2008;26:163–172. doi: 10.1634/stemcells.2007-0281. [DOI] [PubMed] [Google Scholar]
- 22.Higgins CA, et al. Reprogramming of human hair follicle dermal papilla cells into induced pluripotent stem cells. J Invest Dermatol. 2012;132:1725–1727. doi: 10.1038/jid.2012.12. [DOI] [PubMed] [Google Scholar]
- 23.Helisch A, et al. Impact of mouse strain differences in innate hindlimb collateral vasculature. Arterioscler Thromb Vasc Biol. 2006;26:520–526. doi: 10.1161/01.ATV.0000202677.55012.a0. [DOI] [PubMed] [Google Scholar]
- 24.Chalothorn D, Clayton JA, Zhang H, Pomp D, Faber JE. Collateral density, remodeling, and VEGF-A expression differ widely between mouse strains. Physiol Genomics. 2007;30:179–191. doi: 10.1152/physiolgenomics.00047.2007. [DOI] [PubMed] [Google Scholar]
- 25.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limbourg A, et al. Evaluation of postnatal arteriogenesis and angiogenesis in a mouse model of hind-limb ischemia. Nat Protoc. 2009;4:1737–1746. doi: 10.1038/nprot.2009.185. [DOI] [PubMed] [Google Scholar]
- 27.Fiedler GB, et al. Localized semi-LASER dynamic 31P magnetic resonance spectroscopy of the soleus during and following exercise at 7 T. MAGMA. 2015;28:493–501. doi: 10.1007/s10334-015-0484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnyawali SC, et al. High-frequency high-resolution echocardiography: first evidence on non-invasive repeated measure of myocardial strain, contractility, and mitral regurgitation in the ischemia-reperfused murine heart. J Vis Exp. 2010;2010:e1781. doi: 10.3791/1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy S, et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rink C, et al. Oxygen-sensitive outcomes and gene expression in acute ischemic stroke. J Cereb Blood Flow Metab. 2010;30:1275–1287. doi: 10.1038/jcbfm.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy S, Khanna S, Rink C, Biswas S, Sen CK. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics. 2008;34:162–184. doi: 10.1152/physiolgenomics.00045.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GeneChip expression data can be accessed through the Gene Expression Omnibus. Additional data are available from the corresponding authors upon reasonable request.