Abstract
Skin colonization by Staphylococcus aureus is associated with severity of atopic dermatitis (AD). Two papers in this issue of Cell Host & Microbe by Nakagawa et al. (2017) and Liu et al. (2017) define a pathway by which epicutaneous Staphylococcus aureus promotes skin inflammation and may contribute to AD.
With the recent technological advancements to determine the identity of microbes that are present on or in humans, it appears that most microbial communities enter a state of dysbiosis, or a change from their normal composition, when the host is diseased. This is perhaps best understood in the context of atopic dermatitis (AD), a form of the common skin disease eczema. Staphylococcus aureus (S. aureus) is frequently cultured from AD patients, and its abundance can be more than 100 times that on normal skin. This large increase in S. aureus is observed often despite the absence of clinical signs of skin infection. However, a key question has remained controversial: does S. aureus cause AD, or is the increased survival of this organism on the skin simply a consequence of the disease?
An answer to this important question is beginning to emerge by considering the results of next-generation sequencing together with recent experimental evidence from human and animal model systems. A prospective observational study of culturable skin bacteria of children from birth to age 2 has shown that S. aureus colonization precedes the onset of the disease (Meylan et al., 2017), while another recent bacterial metagenomic study before onset of AD has shown the bacterial community can be protective (Kennedy et al., 2017). Additionally, whole-genome sequencing of skin swabs from AD subjects has shown again that S. aureus is increased and clonal on AD skin and suggested these clones promote changes in the skin of mice (Byrd et al., 2017). These observations support the hypothesis that S. aureus can cause the disease to occur in susceptible individuals. To advance the plausibility of this causal hypothesis, it is necessary to provide a rational mechanism for how S. aureus could drive this disease.
To advance our mechanistic understanding of how skin surface microbes could promote an inflammatory skin disease, recent work has focused on how individual virulence factors from S. aureus can influence host defense in the mouse. An area of interest has been to study the role of bacterially expressed phenolsoluble modulins (PSMs), a group of peptides split into several groups including PSMα1–4, δ-toxin, PSMβ1–2, and occasionally PSM-mec. In particular, PSMα was demonstrated to induce cytokine expression in both human keratinocytes and in tape-stripped murine skin epicutaneous models (Syed et al., 2015). S. aureus δ-toxin has also been observed to induce mast cell degranulation, increase IgE levels, and promote skin inflammation (Nakamura et al., 2013).
In this issue of Cell Host & Microbe, two papers by Nakagawa et al. (2017) and Liu et al. (2017) have further outlined pathways through which the epicutaneous application of S. aureus can induce an AD-like phenotype on murine skin (Nakagawa et al., 2017; Liu et al., 2017). The Nakagawa et al. (2017) study shows that PSMα from S. aureus activates keratinocyte production of IL-1α and IL-36α and this in turn stimulates γδ T cells and innate lymphoid cell type 3 (ILC3)-mediated IL-17 release as well as neutrophil recruitment. Similarly, the Liu et al. (2017) study shows that PSMα can induce keratinocyte IL-36α to stimulate IL-17-producing γδ T cells and CD4+ T cells. Interestingly, only application of S. aureus on the skin surface produced this response while subcutaneous injections of S. aureus promoted a distinct IL-1β response. Both studies show that S. aureus PSMα can stimulate keratinocytes to produce cytokines that promote skin damage and inflammation resembling AD.
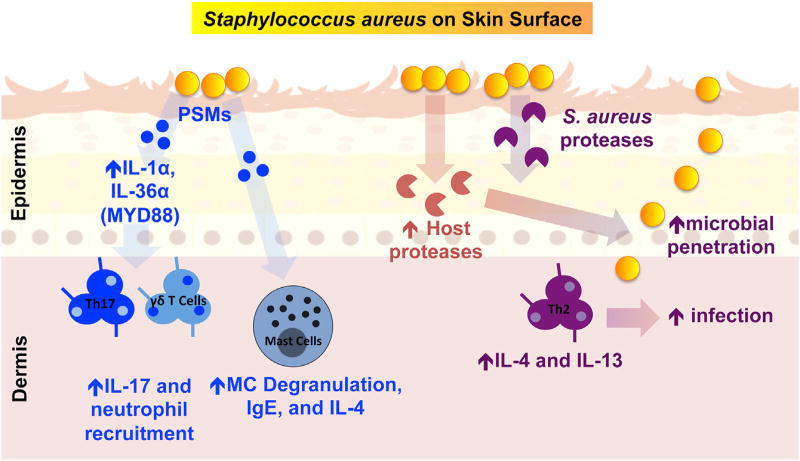
Other important virulence factors from S. aureus have also been shown to affect inflammation and damage the skin. Our group has observed that an S. aureus protease null strain failed to penetrate into the deeper layers of the skin and also failed to induce TH2 cytokine production in mice (Nakatsuji et al., 2016). Interestingly, this penetration of S. aureus into the skin may be linked to the differences Liu et al. (2017) observed in subcutaneous versus epicutaenous S. aureus infections. Furthermore, we have shown that a secreted S. aureus factor(s) can stimulate human keratinocytes to increase expression of epithelial serine proteases that further damage the skin barrier (Williams et al., 2017). These S. aureus-induced serine proteases have been shown to stimulate the protease-activated receptor 2 (PAR-2) in keratinocytes to induce IL-1-type cytokines. These findings link those observed by Nakagawa et al. (2017) and Liu et al. (2017). Overall, there is mounting evidence that S. aureus-secreted proteases as well as PSMs combine with induced keratinocyte proteases to disrupt the skin barrier, activate the local immune response, and initiate an AD-like phenotype. An overview of the pathways involved in the process of driving AD is illustrated in Figure 1.
Figure 1. S. aureus Promotes Skin Inflammation through Multiple Pathways.
Increased colonization of the skin surface by S. aureus leads to secretion of virulence factors including phenol-soluble modulins (PSMs) and proteases. PSMα can stimulate epidermal keratinocytes to produce IL-1-type cytokines (e.g., IL-36α and IL-1α) that further induce γδ T cells, lymphoid cells type 3 (ILC3), and neutrophil recruitment. Another PSM, σ-toxin, can stimulate mast cell degranulation, IL-4 release, increased IgE levels, and increased skin inflammation. In addition to PSMs, S. aureus-secreted proteases combine with induced keratinocyte serine proteases to drive skin barrier damage. This damage promotes bacterial penetration, further drives a Th2 response, and promotes infection.
Therapeutic strategies in AD have sought to target S. aureus by using broad-spectrum oral antibiotics and/or topical antibiotics and antiseptics. These strategies have rarely prevented S. aureus colonization and lacked specificity for targeting S. aureus. This lack of antimicrobial specificity may have an unintended detrimental effect by suppressing the positive impact of other bacteria on the skin. Commensal bacteria such as coagulase-negative Staphylococci (CoNS) co-exist with S. aureus on AD subjects. Many of the strains of CoNS inhabiting the skin of healthy subjects produce lantibiotics or bacteriocin-type antimicrobial peptides that kill S. aureus, thus providing the host with an additional layer of immune defense. Interestingly, these strains of CoNS were infrequently cultured from AD subjects. When antimicrobial CoNS strains were applied to AD lesional skin, S. aureus colonization was significantly reduced (Nakatsuji et al., 2017). Thus, consistent with other observations such as the Kennedy et al. (2017) study, strain-specific functions of the skin bacterial community protect against S. aureus and subsequent disease.
The role of increased S. aureus colonization in skin disease is an important area of research that is distinct from understanding bacterial virulence factors that promote invasive superficial and deep infections. The recent work by Nakagawa et al. (2017) and Liu et al. (2017) has provided important new information about how S. aureus can manipulate the epidermal environment to promote AD-like inflammation. Shaping the host environment to promote the AD immunologic response suppresses the capacity of the host to inhibit microbial infections (Ong et al., 2002), and is therefore desirable for survival of S. aureus. Understanding these mechanisms and complex community interactions will provide new therapeutic strategies to target S. aureus and inhibit its capacity to promote skin disease.
Footnotes
CONFLICTS OF INTEREST
R.L.G. is a consultant for and has equity interest in Sente and MatriSys.
References
- Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Belkaid Y, Segre JA, Kong HH NISC Comparative Sequencing Program. Sci. Transl. Med. 2017;9:eaal4651. doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WH, Murray D, Jo JH, Segre JA, Kong HH, Irvine AD. J. Allergy Clin. Immunol. 2017;139:166–172. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, Ortines RV, Kao T, Lee SK, Cai SS, Miller RJ, et al. Cell Host Microbe. 2017;22(this issue):653–666. doi: 10.1016/j.chom.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, Vial Y, Prod’hom G, Greub G, Kypriotou M, Christen-Zaech S. J. Invest. Dermatol. 2017 doi: 10.1016/j.jid.2017.07.834. http://dx.doi.org/10.1016/j.jid.2017.07.834. [DOI] [PubMed]
- Nakagawa S, Matsumoto M, Katayama Y, Oguma R, Wakabayashi S, Nygaard T, Saijo S, Inohara N, Otto M, Matsue H, et al. Cell Host Microbe. 2017;22(this issue):667–677. doi: 10.1016/j.chom.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GY, McGavin MJ, Travers JB, et al. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, Hata TR, Gallo RL. J. Invest. Dermatol. 2016;136:2192–2200. doi: 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, et al. Sci. Transl. Med. 2017;9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DY. N. Engl. J. Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- Syed AK, Reed TJ, Clark KL, Boles BR, Kahlenberg JM. Infect. Immun. 2015;83:3428–3437. doi: 10.1128/IAI.00401-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL. J. Invest. Dermatol. 2017;137:377–384. doi: 10.1016/j.jid.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]