Abstract
For many bacterial infections, drug resistant mutants are likely present by the time antibiotic treatment starts. Nevertheless, such infections are often successfully cleared. It is commonly assumed that this is due to the combined action of drug and immune response, the latter facilitating clearance of the resistant population. However, most studies of drug resistance emergence during antibiotic treatment focus almost exclusively on the dynamics of bacteria and the drug and neglect the contribution of immune defenses. Here, we develop and analyze several mathematical models that explicitly include an immune response. We consider different types of immune responses and investigate how each impacts the emergence of resistance. We show that an immune response that retains its strength despite a strong drug-induced decline of bacteria numbers considerably reduces the emergence of resistance, narrows the mutant selection window, and mitigates the effects of non-adherence to treatment. Additionally, we show that compared to an immune response that kills bacteria at a constant rate, one that trades reduced killing at high bacterial load for increased killing at low bacterial load is sometimes preferable. We discuss the predictions and hypotheses derived from this study and how they can be tested experimentally.
Keywords: Bacterial infection dynamics, Immune response, Drug resistance, Antibiotic treatment, Mutant selection window
1. Introduction
By the time bacterial infections cause symptoms and thereby call for antibiotic treatment, the bacterial population is often so large that it likely includes mutants that are resistant to the treating antibiotic (Drusano, 2004; Drlica, 2003). One might therefore expect treatment with single antimicrobial agents to fail. One reason why this is frequently not the case is the host's immune defense, which contributes to bacteria clearance (Pamer, 2007; Happel et al., 2004). Despite the general recognition of the important role of the host's defenses, most studies of the within-host dynamics of bacteria and antibiotics focus almost exclusively on the pharmacokinetics (PK) and the pharmacodynamics (PD) of the drug and bacteria, without explicitly considering the immune response (Lipsitch and Levin, 1997; Mueller et al., 2004; Mager et al., 2003; Drusano, 2004; Schentag et al., 2001; DeRyke et al., 2006; Mouton et al., 1999; Craig, 1998; Andes and Craig, 2002; Ambrose et al., 2007).
Such omission of the immune response also applies to models that are specifically intended to develop treatment protocols to prevent the ascent of resistant mutants, like those based on the mutant selection window (MSW) theory (Zhao and Drlica, 2001, 2008; Jumbe et al., 2003; Drlica and Zhao, 2004). The MSW is defined as the range of drug concentrations for which the drug is strong enough to remove the sensitive population, but not strong enough to remove the (partially) resistant population. In the absence of an immune response, this is expected to lead to selection of the resistant mutant, which can ascend to high levels (Negri et al., 2000; Drlica and Zhao, 2007). As we show here, the presence of an immune response can alter the MSW. Several theoretical and experimental studies have addressed the role of the immune response during antimicrobial treatment, and the issue has also been studied in the context of viral infections (Dalhoff and Shalit, 2003; Labro, 2000; Tsai and Standiford, 2004; Dalhoff, 2005; Imran and Smith, 2007; Austin et al., 1998; Curlin et al., 2007). But, to our knowledge, the interactions between antibiotics and the host's immune response as it affects the emergence of resistance during bacteria infections has not been addressed.
While the lack of data precludes the development of detailed and quantitatively accurate models of the contribution of the immune response for the antibiotic treatment of specific bacterial infections, simple mathematical models can provide a way to generate hypotheses for the design and interpretation of future experiments. We develop such simple mathematical models and use them to analyze the effects of different types of immune responses on the ascent of resistance during antibiotic treatment. We show that an immune response that retains its strength despite a strong drug-induced decline of bacteria numbers considerably reduces the emergence of resistance, narrows the MSW, and mitigates the effects of non-adherence to treatment. Additionally, we show that compared to an immune response that kills bacteria at a constant rate, one that trades reduced killing at high bacterial load for increased killing at low bacterial load is sometimes preferable. We discuss the implications of these theoretical results to antibiotic treatment and how the hypotheses generated from our analysis can be tested experimentally.
2. The mathematical models
2.1. Bacteria and drug dynamics
To describe bacterial growth, PK and PD, we use a model that has previously been shown to successfully fit data from in vitro experiments on the emergence of fluoroquinolone (ciprofloxacin) resistance in Staphylococcus aureus (Campion et al., 2005; Chung et al., 2006). We consider two populations of bacteria, one susceptible to the antibiotic (Bs) and the other resistant (Br). The susceptible and resistant bacteria grow at rates gs and gr. Growth slows down as the total number of bacteria approaches a maximum population size, N0. During growth, susceptible bacteria generate resistant mutants at a rate μ back mutations to the sensitive genotype are ignored. The antibiotic is administered at a dose C0 every T hours. It decays according to a standard first-order PK function with an exponential decline in concentration at rate d. The antibiotic kills bacteria according to a hyperbolic (Monod like) Emax function with maximum kill rates ks and kr and half-maximum antibiotic concentrations and , for the sensitive and resistant bacteria, respectively. (This corresponds to a Hill function with Hill-coefficient of 1: Regoes et al., 2004). Note that the resistant bacteria are not fully resistant to the drug, instead they require higher drug concentration for clearance, which reflected in the values of the kill and half maximum kill concentrations, i.e. kr<ks and . This is the type of resistance usually found in experimental or clinical situations. The model is expressed by the set of coupled differential equations (a dot denotes differentiation as a function of time)
| (1) |
| (2) |
| (3) |
While the model was shown to describe in vitro data, it is not clear how it applies to an in vivo situation. However, since not enough is known about the in vivo dynamics of bacteria and drugs during the process of resistance generation, we decided to choose the present model because of the availability of experimentally measured values for the model parameters (Table 1).
Table 1.
Variables and parameters for the part of the model describing bacteria and drug dynamics.
| Symbol | Meaning | Values | |
|---|---|---|---|
| Bs(0) | Susceptible bacteria inoculum | 103 | |
| Br(0) | Resistant bacteria inoculum | 0 | |
| N | Total bacteria | Bs + Br | |
| N0 | Carrying capacity | 109 | |
| gs | Maximum sensitive growth | 1 h−1 | |
| gr | Maximum resistant growth | 0.65 h−1 | |
| ks | Maximum kill rate of sensitives | 1.5 h−1 | |
| kr | Maximum kill rate of resistant | 1.1 h−1 | |
|
|
Half-maximum kill rate of sensitives | 0.25 μg/ml | |
|
|
Half-maximum kill rate of resistant | 5 μg/ml | |
| μ | Mutation rate | 10−8 | |
| d | Drug decay rate | 0.15h−1 | |
| T | Times at which drug is administered | Varied | |
| C0 | Drug dose administered at times T | Varied |
Parameter values are chosen in accordance with Campion et al. (2005) and Chung et al. (2006).
2.2. Immune responses
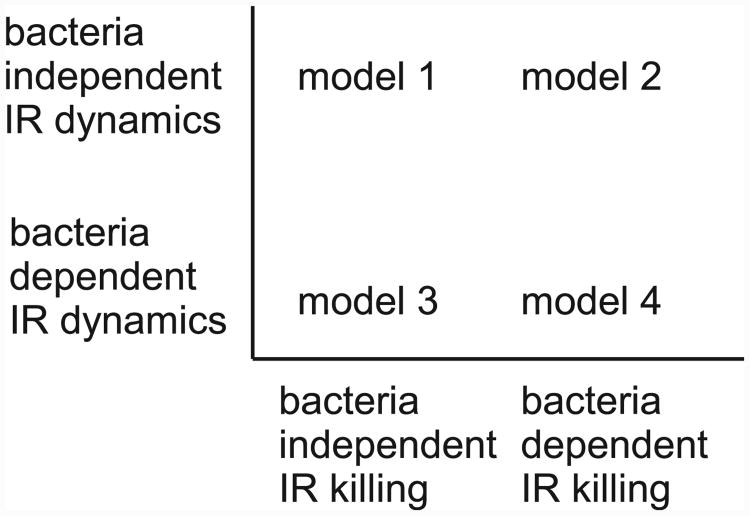
The important and novel aspect of our study is that we explicitly model the dynamics of an immune response. The immune response is immensely complex; many interdependent players, such as different cell types and cytokines, participate at varying degrees. This complexity, combined with the experimental difficulty in accurately measuring all the different immune response components, leads to a lack of detailed, quantitative data. Because of this lack of data, we do not try to create a detailed model of the immune response for a specific infection, such as S. aureus. Instead, we employ several simple, heuristic models that are meant to capture known aspects of immune response dynamics, while at the same time realizing that these are simplified caricatures and the obtained insights are therefore conceptual. We consider the following four models:
Immune response model 1
For the first model, we consider an immune response, I, that is triggered upon onset of infection and increases exponentially at rate gi (clonal expansion: De Boer et al., 2003; Antia et al., 2003; De Boer et al., 2001), independent of the bacteria. Pathogen independent aspects of the expansion dynamics have recently been found for CD4 and CD8 T-cells (Kaech and Ahmed, 2001; Mercado et al., 2000; van Stipdonk et al., 2001; Bajenoff et al., 2002; Lee et al., 2002), which are known to play important roles against obligate or facultative intracellular bacteria, such as Listeria monocytogenes and S. aureus (Pamer, 2004; McLoughlin et al., 2006). The immune response saturates once it reaches some maximum strength. Such dynamics seems to hold for CD4 and CD8 T-cells in the lung after tuberculosis infection (Kamath et al., 2004, 2006). Since I is given in arbitrary units, we choose the maximum strength to be Imax = 1. This leads to a term for the immune dynamics given by İ = giI(1 −I). We set the immune response at the beginning of the infection to I0 = 10−6 and a rate of expansion of gi = 1 h−1. These values was chosen based on data from CD4 and CD8 T-cells (De Boer et al., 2003). However, the exact choices are not important, the results presented below also hold for different values. Killing of bacteria by the immune system is assumed to occur at a rate directly proportional to the strength of the immune response, with a killing rate constant b. In all our immune response models, killing of sensitive and resistant bacteria occurs in the same manner. This leads to the terms −bIBs and −bIBr added to the equations for sensitive and resistant bacteria, respectively. This mass-action type killing term is consistent with observations from CD8 T-cells and neutrophils (Li et al., 2002; Regoes et al., 2007; Yates et al., 2007).
Immune response model 2
For the second model, we consider the same dynamics of the immune response as in model 1, but now the rate at which bacteria are killed saturates at some maximum level as the bacterial load increases. We implement this by scaling the killing term with the total number of bacteria present, so that these terms become −bIBs/(N + s) and −bIBr/(N + s), where N = Bs + Br and s is a saturation constant for the killing rate. This captures the observation that immune cells need time to kill and under these conditions, the mass-action formulation of model 1 breaks down at high pathogen load (Pilyugin and Antia, 2000; Mempel et al., 2004). This has been observed experimentally for neutrophils and is also likely to apply to CD8 T-cell mediated killing at high infected cell densities (Leijh et al., 1979; Li et al., 2004; Regoes et al., 2007; Yates et al., 2007).
Immune response model 3
For the third model, we change the dynamics of the immune response. We assume that the immune response grows proportional to the bacterial load and decays at a fixed rate. This leads to the term İ = giN − diI. If the decay of the immune response is reasonably fast—and we will focus on that situation in the following—this model approximates an immune response that closely tracks the bacterial load, i.e. I ≈ gi/diN. Such dynamics might apply to cytokines or highly activated immune cells (Fritz et al., 1999; Hayden et al., 1998; Green et al., 2003). For the killing, we again assume a mass-action term, as described for model 1.
Immune response model 4
The fourth model uses the same assumption for the dynamics of the immune response as model 3, and combines it with the saturated killing term of model 2.
We want to again stress that these four models we use here are simple and heuristic, meant to capture aspects of the spectrum of possible immune response dynamics. Fig. 1 shows graphically the different conditions for the models just described, Table 2 summarizes the model equations. All models were implemented in Matlab R2007a (The Mathworks), the code is available from the authors upon request.
Fig. 1.
Graphical representation of the differences between the four models. IR: immune response.
Table 2.
Summary and parameters of the immune response (IR) models.
| IR model | IR term | Killing term | |
|---|---|---|---|
| Model 1 | I = gi I(1 − I) | −bIBn | |
| Model 2 | I = giI(1 − I) |
|
|
| Model 3 | I = giN − diI | −bIBn | |
| Model 4 | I = giN − diI |
|
Subscript i denotes terms for the immune response, the subscript n is a placeholder for either s or r, indicating susceptible and resistant bacteria, respectively. Values for the parameters are given in the results section.
3. Results
3.1. Resistance emergence in the absence of an immune response
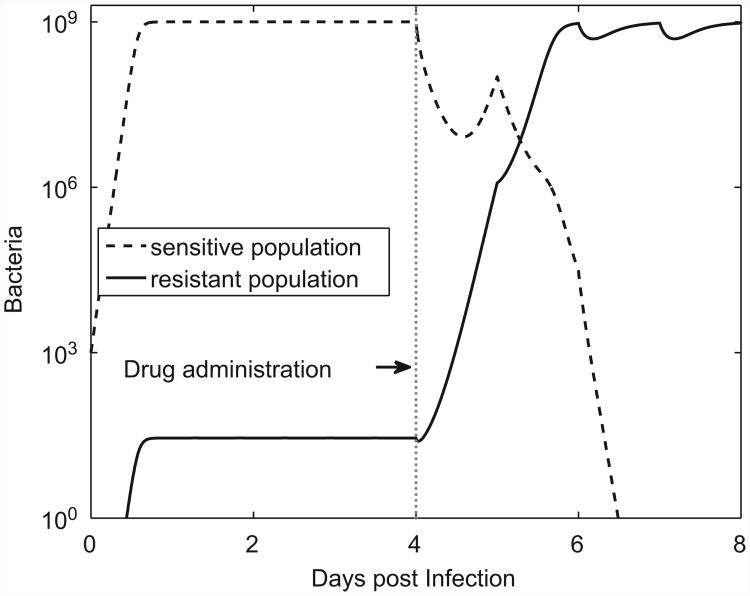
We start by considering the dynamics of susceptible and resistant bacteria in a situation where there is no immune response. The infection starts with a few drug sensitive bacteria. As the bacteria grow, resistant mutants are generated and because they also replicate (at a lower rate compared to the drug sensitive population), they too increase. Both the resistant and susceptible populations level off when the total bacterial load reaches the saturation level. In the absence of drug treatment, the less-fit resistant population constitutes a small fraction of the total population. Once antimicrobial treatment is started, the sensitive population is cleared by the drug and the total bacterial load falls below the saturation level. This allows the resistant population to increase until its net growth rate equals the rate of killing by the antibiotic (Fig. 2). (Recall that resistance is not complete, the drug can kill the resistant population, albeit at a very low rate.)
Fig. 2.
Dynamics of infection in the absence of an immune response. Antimicrobial treatment is started at day 4. Every T = 24 h, a dose of C0 = 4μg/ml is administered. Parameters as given in Table 1.
3.2. Resistance emergence in the presence of immune responses
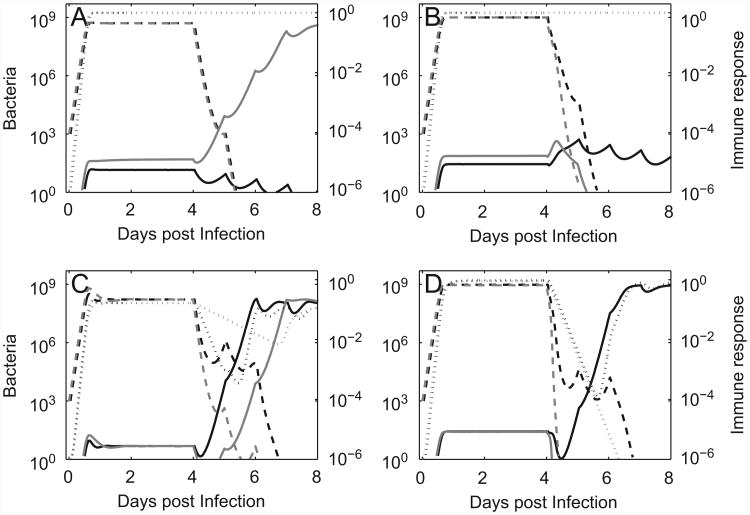
In the worst case, even the combined effect of antimicrobial drug and immune response cannot eradicate the susceptible population. While this will likely prevent the ascent of the resistant population, this scenario represents treatment failure not due to resistance but simply due to an ineffective drug. We do not consider this situation further. If the immune response alone is not strong enough to clear the susceptible population, but a combination of antimicrobial treatment and immune response can do so, two possibilities exist for the resistant subpopulation: Either the immune response (together with the weak effect of the antibiotic on the resistant mutants) can prevent the emergence of the resistant population. This will likely occur if the cost of resistance is non-negligible (Fig. 3A, black lines). Or, alternatively, the immune response cannot prevent the emergence of resistance. This might be the case if the fitness of the resistant strain is close to that of the sensitive strain (Fig. 3A, gray lines), or if the immune response is less potent (e.g. a value for b half that shown for the first scenario in Fig. 3A leads to resistance emergence. Graph not shown).
Fig. 3.
Dynamics of infection in the presence of immune responses. Dotted lines show immune response (with axis label on the right side), solid and dashed lines show the drug resistant and drug sensitive bacteria. Drug dosing as described in legend of Fig. 2. The parameters b, gr, di, gi are all given in units of h−1. All parameter values are as given in Table 1 unless otherwise stated. (A) Model 1. Black: low-fitness (growth rate) resistant strain (gr = 0.65), prevention of resistance. Gray: high-fitness resistant strain (gr = 0.9), resistance emerges (killing rate b = 0.5). (B) Model 2. Black: maximum killing at low bacterial load is the same as in model 1, killing rate declines once the bacteria increase beyond 1% of the carrying capacity and is about one-hundredth that of model 1 for N → N0 (s = N0/100, b = 0.5s, gr = 0.65). Gray: maximum killing at low bacterial load is twice that of model 1 but still only one-fiftieth that of model 1 for N → N0 (s = N0/100, b = s, gr = 0.9). (C) Model 3. Black: the immune response changes rapidly as bacterial load changes (b = 5, di = 0.25, gi = di/N0). Gray: the immune response changes less rapidly as bacterial load changes (b = 5, di = 0.05, gi = di/N0). (D) Model 4. Black: maximum killing at low bacterial is the same as in model 3, killing rate declines once the bacteria increase beyond 1% of the maximum carrying capacity and is about one-hundredth that of model 3 for N → N0 (s = N0/100, b = 5s, di = 0.25, gi = di/N0). Gray: maximum killing at low bacterial load is twice that of model 1 but still only one-fiftieth that of model 1 for N → N0 (s = N0/100, b = 10s, di = 0.25, gi = di/N0).
The type of killing can also impact the outcome. If the killing rate saturates for high bacterial load (model 2) and the rate is the same as for the non-saturating model at low bacterial load, the result is a weaker immune response and less good control of the resistant population (compare Figs. 3A and B, black lines). In contrast, if the killing at low bacterial load is more effective, it can lead to better clearance of the resistant subpopulation once the drug has reduced the sensitive subpopulation, even if the killing strength at high bacterial load is much weaker compared to the non-saturating, mass-action killing (compare Fig. 3A and B gray lines).
For the two models considered so far, the immune response quickly reaches a constant level and remains at this level, independent of the bacteria dynamics. We now turn to models 3 and 4 to investigate how an immune response that depends on bacteria numbers affects resistance emergence. If the immune response decays quickly (di large), a decline in bacteria due to drug treatment will result in a rapid decline of the immune response and subsequent emergence of resistance is likely, even if the immune response is very potent. The black lines in Fig. 3C show this for a scenario where resistance emerges, even though the killing rate, b, is 10 times larger compared to model 1. Note how closely the immune response (dotted line) tracks the bacterial load. If the decay of the immune response is slower, it will lead to less immediate tracking of the bacteria decline and improved ability to control the resistant population. the gray lines in Fig. 3C show a situation where resistance emergence is almost prevented. A further reduction in the decay rate leads to a situation that approaches the pathogen-independent immune response shown in panels A + B, where resistance emergence is prevented (not shown).
As seen for models 1 and 2, a saturating immune response that kills less efficiently at high bacterial load but more efficiently at low bacteria numbers can help to prevent resistance emergence. This is also true if the immune response declines almost immediately as the bacteria decline. The higher killing efficiency at low bacteria numbers can make the difference between resistance emergence or clearance (compare Fig. 3C and D, gray lines).
3.3. Immune responses can reduce the mutant selection window
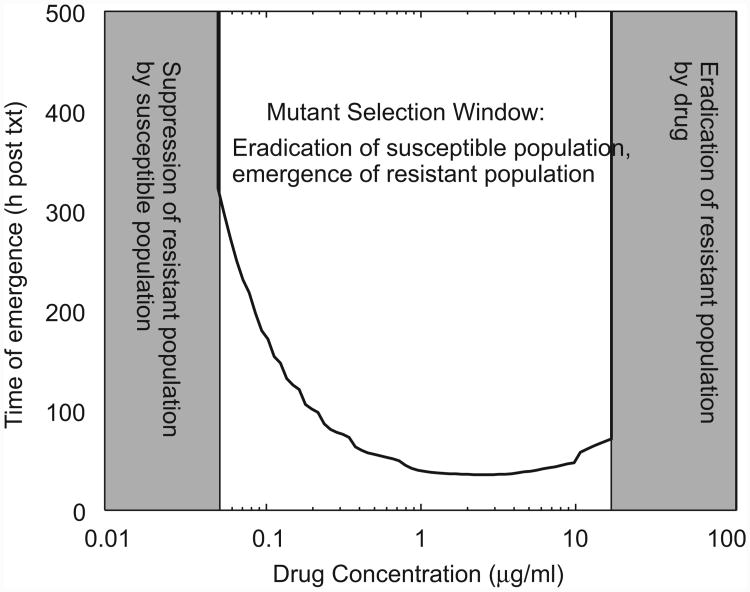
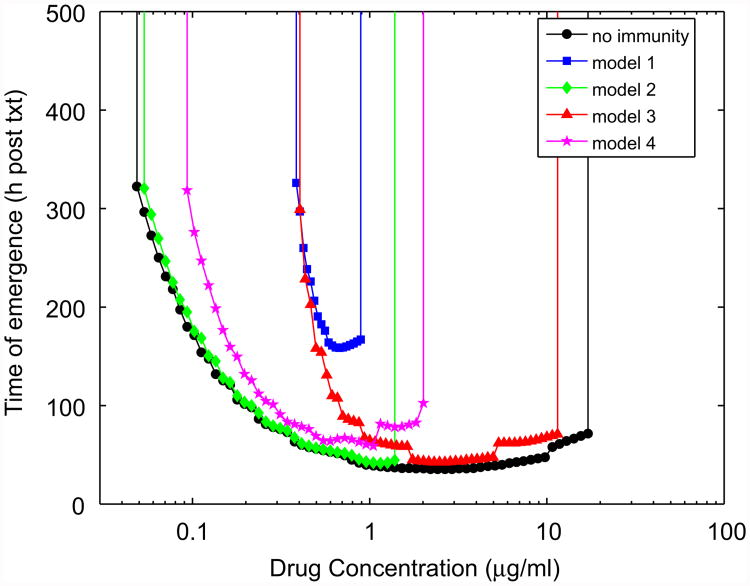
So far, we have shown illustrative examples how different types of immune responses can affect the emergence of resistance. We now investigate the impact of the different immune responses in more detail. We begin by exploring how the immune response can change the size of the MSW. The MSW is defined as the range of drug concentrations for which the drug is strong enough to remove the sensitive population, but not strong enough to remove the resistant population. In the absence of an immune response, this is expected to lead to selection of the resistant mutant, which can ascend to high levels (Negri et al., 2000; Drlica and Zhao, 2007). Fig. 4 illustrates the MSW idea.
Fig. 4.
The mutant selection window (MSW) in the absence of an immune response. Shown is the time of resistance emergence following treatment (txt), as a function of drug concentration (C0). Emergence is defined as the resistant population reaching 10% of the carrying capacity. The simulation is run until 14 days (txt) post-treatment start. If the resistant population has not reached 10% by day 14, the time of emergence is set to infinity. At low drug concentrations, the drug sensitive population is not removed and the resistant population cannot emerge. Very high drug doses kill both sensitive and resistant populations. Intermediate drug doses clear the sensitive population only, and thereby allow the resistant population to reach high levels.
The presence of an immune response is expected to change the MSW. One would expect to see the MSW shrink at the right side, for high drug concentrations, simply because in the presence of additional killing by an immune response, a lower drug dose is required to remove both the susceptible and the (partially) resistant subpopulations. This is indeed what one finds (Fig. 5). Also, as expected, the immune responses that do not tightly follow the decline of bacteria (models 1 and 2) are able to shrink this part of the window by a larger amount, though model 4, which declines in strength as bacterial load declines, but at the same time has an increasing killing rate, is able to perform almost as good as models 1 and 2.
Fig. 5.
The MSW in the presence of the different immune responses. Time to emergence is defined as described in the caption for Fig. 4. Immune responses are chosen as in Fig. 2 with gr = 0.65, b = 0.5 (model 1), s = N0/100 and b = s (model 2), b = 5, di = 0.25, gi = di/N0 (model 3), and s = N0/100, b = 10s, di = 0.25, gi = di/N0 (model 4).
Less intuitively obvious is why the MSW shrinks at the left side, for low drug concentrations. Since the immune response acts together with the drug, the sensitive population is cleared at lower drug concentrations. One might therefore also expect resistance to emerge at lower, not higher, drug concentrations compared to the situation without an immune response. This would indeed be the case if the immune response were to act only on the sensitive population. However, it affects both populations equally. In the absence of an immune response, any drug-induced decline of the sensitive population allows the resistant population to quickly increase in numbers. In contrast, if an immune response is present, it can prevent the resistant population from growing, even if some of the competitive pressure is removed by a reduction in the sensitive population. It therefore takes higher drug doses, corresponding to a stronger selective pressure in favor of the resistant population, before resistance can emerge. Since at high bacteria loads, the saturating immune responses (models 2 and 4) are rather weak, they are not very good in preventing resistance to emerge at low drug concentrations (Fig. 5, green diamonds and magenta stars). In contrast, the mass-action models retain a potent response at high bacteria numbers, therefore preventing the emergence of resistance (Fig. 5, blue squares and red triangles) and significantly shrinking the MSW at the left side.
3.4. Immune responses can dampen the effect of imperfect adherence to treatment
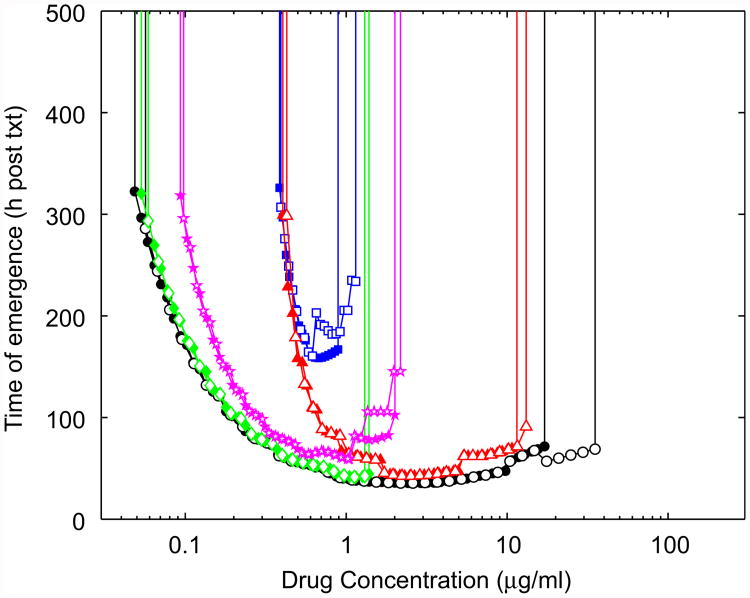
The emergence of resistance is often due to the failure of patients to follow a prescribed antibiotic treatment protocol. A prominent example is treatment failure of long-term infections like tuberculosis (Lipsitch and Levin, 1998; Gillespie, 2002; Gomez and McKinney, 2004). It is therefore of interest to understand how the immune response might modulate the effect of imperfect adherence to an antibiotic treatment regime. As our model for non-adherence we use a scheme similar to one of those considered previously (Lipsitch and Levin, 1997). In this scenario, patients are less likely to take the prescribed drug dose as symptoms reduce. We assume that symptoms are proportional to bacterial load. This is implemented by assuming that a dose is taken with probability p = 0.25 + 0.75 log10(N)/log10(N0)). This means that for maximum bacterial burden (N = N0), adherence is perfect. As bacteria load declines, so does the probability that the patient takes the drug. At very low bacteria load, the probability that the patient takes the antibiotic is reduced to ≈ 25%. We simulate 5000 infections using a Monte Carlo routine where at each scheduled dosing interval the drug will be taken with probability p. Fig. 6 shows the MSW for perfect and imperfect adherence in the absence or presence of immune responses. We plot the median value for the time to resistance emergence, defined as previously. As expected, in the absence of an immune response, non-adherence increases the MSW (Fig. 6, open and filled circles). The presence of an immune response dampens the impact of non-adherence. The immune response models with a bacteria load-dependent killing (models 2 and 4) seem to perform slightly better, compared with the bacteria load-independent killing models (models 1 and 3). This is presumably because killing in these models is improved at low bacteria numbers, which is exactly the regime where the probability of taking the prescribed drug dose is the lowest.
Fig. 6.
The mutant selection window for imperfect adherence (empty markers). For comparison, results for complete adherence are replotted from Fig. 5 (solid markers). Everything else as described for Fig. 5.
3.5. Immune responses can change optimal dosing strategies
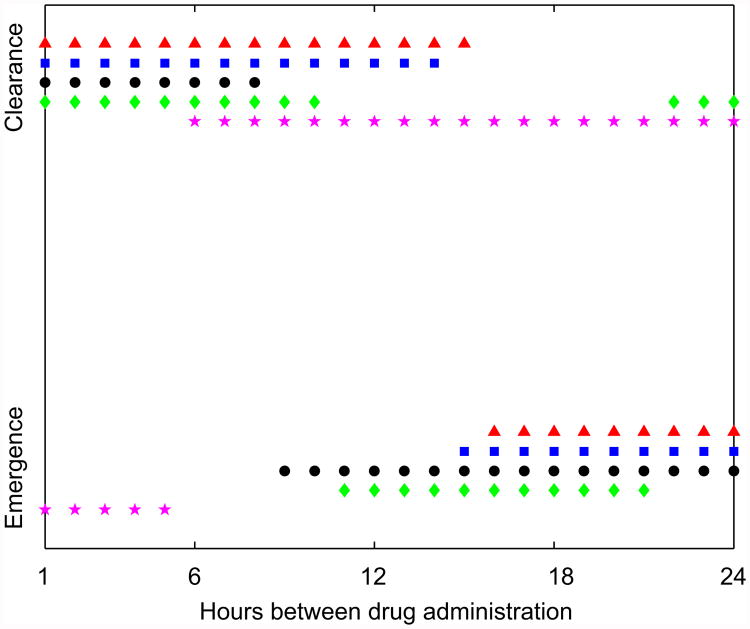
The goal of an optimal treatment strategy is to achieve all specified goals (e.g. clearance, prevention of resistance emergence), while at the same time ensuring that the smallest possible amount of drug is used (to reduce toxicity and financial costs). Additionally, one might want to reduce the frequency at which the drug is administered. If the killing action of the drug (the PD) is well described by an Emax model, such as the one we use here, overall killing of bacteria is larger if a given amount of drug is administered in frequent small doses instead of few large ones (Lipsitch and Levin, 1997). Because the Emax model applies to both the sensitive and resistant populations, we expect that in the absence of an immune response, more frequent drug doses are better at killing the resistant strain and clearing the infection. This is indeed the case (Fig. 7, black circles). We also find that this holds for the immune response models 1 and 3, which kill at a rate independent of bacterial load (blue squares and red triangles). Interestingly, the bacterial load-dependent killing models, model 2 and 4, behave differently. For model 2 (green diamonds), small frequent doses can prevent resistance emergence, but so can few large ones. The reason for this is that a high drug dose strongly reduces the sensitive population, which leads to improved per-capita killing by the immune response and subsequent clearance of the resistant population. For model 4, this latter mechanism works as well, while the improved overall killing by the drug due to the Emax PD has less impact.
Fig. 7.
Bacteria clearance or resistance emergence as a function of dosing regime. Drug is administered at the indicated time intervals, in doses such that the total amount of drug administered over one day, Ĉ, remains fixed. Ĉ for the situation without immunity and the four immune response models are 10, 0.75, 1.5, 8 and 2.5 μg/ml (see text). Everything else as described for Fig. 5.
Note that we chose the total drug dose administered over the 24 h interval (Ĉ) for the different models such that a change in dosing frequency lead to a switch between resistance emergence and resistance prevention. There are of course a wide range of values for the drug concentration that lead to less interesting results, namely resistance emergence or bacteria clearance, no matter how the dosing schedule over a 24 h period is chosen. While it is not clear if for a realistic situation, any of the results shown in Fig. 7 might occur, it is nevertheless important to illustrate what could happen, and how the complicated dynamical interactions between the antibiotic, bacteria and immune response can lead to unexpected outcomes, which depend on the details of the immune response.
4. Discussion
Help from the immune response is often necessary to clear bacterial infections, even in the presence of antibiotic treatment. An example is the success of drugs that—at least in vitro—are only bacteriostatic (Pankey and Sabath, 2004). Here we developed mathematical models that combine the dynamics of bacteria and drugs with different models for the immune response. We used these models to analyze the emergence of resistance during the course of treatment.
4.1. Caveats and limitations
As with all mathematical (as well as verbal) models of biological systems, the models employed in this study represent strong simplifications of the complex interactions between bacteria, antibiotics, and the host's immune response. Although the equations for bacteria and drug dynamics provided a reasonable fit to data for S. aureus and ciprofloxacin generated in vitro (Campion et al., 2005; Chung et al., 2006), it is not clear how well such models apply in vivo. In fact, S. aureus readily evolves resistance to ciprofloxacin (Dalhoff and Schmitz, 2003) which makes this fluoroquinolone a less than optimal drug for treating staphylococcal infections.
However, the focus of this study was not on prediction of resistance emergence for a specific infection but rather to generate a conceptual framework for addressing questions about the contribution of the immune response to preventing the evolution of antibiotic resistance during the course of therapy. With minor modifications, this same theory could apply to many different antibiotics and bacteria for which resistance can be generated by mutation. Of particular relevance in this regard is multi-drug therapy in situations where mutants resistant to single drugs are almost always present (Lipsitch and Levin, 1998; Sacchettini et al., 2008). Although our model of the immune response is a simplistic caricature of the plethora of host responses to a bacterial infection, it is based on biologically realistic assumptions and captures aspects of the anticipated dynamics of the host response. More specifically, we chose scenarios were the strength of the immune response is independent of or strongly dependent upon the bacterial load, and where the rate of immune-mediated killing does and does not saturate at high bacterial load. We believe these models capture aspects of the dynamics of the induction, buildup and waning of the immune defenses and immune-mediated killing of bacteria.
Our models focused on resistance generated by genetic mutation through a one-step process. We did not formally consider non-inherited resistance due to physiological processes, biofilm formation, or persistence, which can prolong therapy and promote the generation and ascent of inherited resistance (Hogan and Kolter, 2002; Gomez and McKinney, 2004; Levin and Rozen, 2006; Dhar and McKinney, 2007; Lewis, 2007). We also assumed that a resistant population already existed prior to treatment start. As mentioned in the introduction, we chose this setup because it is likely to occur for many clinical infections, where the bacteria have reached such high numbers by the time symptoms occur and treatment starts that the existence of a resistant subpopulation is likely. For a situation where resistance had not yet emerged at the beginning of the treatment period, an immune response that helps to eradicate the sensitive population as quickly as possible—and thereby minimize the chance that a resistant mutant is created—is expected to perform best.
4.2. Predictions and hypotheses
Our results suggest that the presence of an immune response narrows the MSW, helps to mitigate the negative effects of non-adherence, and influences the optimal dosing strategy. We find that if antibiotic drug concentrations can be maintained at relatively high levels (the right border of the MSW), the synergism between immune response and drug in reducing resistance emergence is best for immune response components that are largely independent of the dynamics of the pathogen (Fig. 5, models 1 and 2). Such dynamics applies probably most strongly to parts of the adaptive immune responses. For an immune response that is tightly linked to pathogen load (cytokines, parts of the innate immune response, highly activated CD8 T-cells), trading reduced killing at high bacterial load for increased killing at low bacterial load can be better at preventing resistance emergence (Fig. 5, model 4). In contrast, if antibiotic drug concentrations cannot reach levels that are high enough to be above the MSW (for instance due to toxicity), immune responses that do not saturate in their strength of killing at high bacterial load will perform best (Fig. 5, models 1 and 3). Saturated killing might be unavoidable due to biological constraints (e.g. time it takes to kill Pilyugin and Antia, 2000), one can speculate that reduced killing at high pathogen load might be a “choice” made by the immune response in certain situations to prevent excessive immuno-pathology. It is worth pointing out that the type of killing function depends on the exact numbers of immune players and bacteria and the killing mechanism, and can switch from non-saturated to a saturated regime in some but not other situations. In general, a higher number of immune players (e.g. a certain type of immune cell) and a faster killing process will reduce the potential for killing saturation at high bacterial load.
The predictions from our models can be tested experimentally in laboratory animals. One can use a “resistance competition assay” (Negri et al., 2000; Bull et al., 2002), whereby animals are inoculated with low numbers of bacteria resistant to the treating antibiotic and high numbers of the susceptible population, and the changes in frequency of resistance during the course of antibiotic treatment are followed. In addition to measuring the bacterial load and concentrations of antibiotics in these experiments, different components of the immune responses should also be quantified. Applying this protocol to laboratory animals with normal immune systems and animals with specific components of the immune system impaired can provide information into the role different components of the immune response play in affecting the MSW, dosing strategies, etc. (Drlica, 2003). We would expect that animals with impaired immune response components which are largely independent of bacterial load, both in their dynamics and their killing behavior, will be most susceptible to resistance emergence, compared with healthy animals or animals that have bacteria load-dependent immune dynamics or killing. Of course, the main problem with such experimental tests (and the direct applicability of our results) is the fact that immune response components tightly interact, therefore it might be difficult to knock out certain components without affecting the performance of others.
To summarize, we have shown that the host's immune defenses can play an important role in the emergence or prevention of drug resistance. This highlights the need for further studies that consider the joint impact of the immune response and antimicrobial drug treatment on the emergence of resistance. Eventually, such a combination of experimental and theoretical studies should allow us to design treatment protocols that prevent resistance emergence and lead to complete bacteria clearance, while also optimizing drug dose, robustness against non-adherence and treatment schedule.
Acknowledgments
This project was supported by grants from the US National Institutes of Health AI40662 (BRL) and an NIH Training Grant (EM). Pfizer Inc. also generously provided financial support for this project without imposing review or any other constraints that could possibly be construed as a conflict of interest. We thank members of the Levin and Antia labs for stimulating discussions during this project and the reviewers for feedback that helped to substantially improve the paper.
References
- Ambrose PG, Bhavnani SM, Rubino CM, Louie A, Gumbo T, Forrest A, Drusano GL. Pharmacokinetics–pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin Infect Dis. 2007;44(1):79–86. doi: 10.1086/510079. URL 〈 http://dx.doi.org/10.1086/510079〉. [DOI] [PubMed] [Google Scholar]
- Andes D, Craig WA. Animal model pharmacokinetics and pharmacody-namics: a critical review. Int J Antimicrob Agents. 2002;19(4):261–268. doi: 10.1016/s0924-8579(02)00022-5. [DOI] [PubMed] [Google Scholar]
- Antia R, Bergstrom CT, Pilyugin SS, Kaech SM, Ahmed R. Models of CD8+ responses: 1. What is the antigen-independent proliferation program. J Theor Biol. 2003;221(4):585–598. doi: 10.1006/jtbi.2003.3208. [DOI] [PubMed] [Google Scholar]
- Austin DJ, White NJ, Anderson RM. The dynamics of drug action on the within-host population growth of infectious agents: melding pharmacokinetics with pathogen population dynamics. J Theor Biol. 1998;194(3):313–339. doi: 10.1006/jtbi.1997.0438. URL 〈 http://dx.doi.org/10.1006/jtbi.1997.0438〉. [DOI] [PubMed] [Google Scholar]
- Bajenoff M, Wurtz O, Guerder S. Repeated antigen exposure is necessary for the differentiation, but not the initial proliferation, of naive CD4(+) T cells. J Immunol. 2002;168(4):1723–1729. doi: 10.4049/jimmunol.168.4.1723. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Levin BR, DeRouin T, Walker N, Bloch CA. Dynamics of success and failure in phage and antibiotic therapy in experimental infections. BMC Microbiol. 2002;2:35. doi: 10.1186/1471-2180-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion JJ, McNamara PJ, Evans ME. Pharmacodynamic modeling of ciprofloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49(1):209–219. doi: 10.1128/AAC.49.1.209-219.2005. URL 〈 http://dx.doi.org/10.1128/AAC.49.1.209-219.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P, McNamara PJ, Campion JJ, Evans ME. Mechanism-based pharmacodynamic models of fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50(9):2957–2965. doi: 10.1128/AAC.00736-05. URL 〈 http://dx.doi.org/10.1128/AAC.00736-05〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis. 1998;26(1):1–10. doi: 10.1086/516284. quiz 11-2. [DOI] [PubMed] [Google Scholar]
- Curlin ME, Iyer S, Mittler JE. Optimal timing and duration of induction therapy for hiv-1 infection. PLoS Comput Biol. 2007;3(7):e133. doi: 10.1371/journal.pcbi.0030133. URL 〈 http://dx.doi.org/10.1371/journal.pcbi.0030133〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhoff A. Contribution of immunocompetence to the antibacterial activities of ciprofloxacin and moxifloxacin in an in vitro pharmacodynamic model. Infection. 2005;33(Suppl. 2):44–49. doi: 10.1007/s15010-005-8207-x. URL 〈 http://dx.doi.org/10.1007/s15010-005-8207-x〉. [DOI] [PubMed] [Google Scholar]
- Dalhoff A, Schmitz FJ. In vitro antibacterial activity and pharmacodynamics of new quinolones. Eur J Clin Microbiol Infect Dis. 2003;22(4):203–221. doi: 10.1007/s10096-003-0907-5. URL 〈 http://dx.doi.org/10.1007/s10096-003-0907-5〉. [DOI] [PubMed] [Google Scholar]
- Dalhoff A, Shalit I. Immunomodulatory effects of quinolones. Lancet Infect Dis. 2003;3(6):359–371. doi: 10.1016/s1473-3099(03)00658-3. [DOI] [PubMed] [Google Scholar]
- De Boer R, Oprea M, Antia R, Murali-Krishna K, Ahmed R, Perelson A. Recruitment times, proliferation, and apoptosis rates during the CD8(+) T-cell response to lymphocytic choriomeningitis virus. J Virol. 2001;75(22):10663–10669. doi: 10.1128/JVI.75.22.10663-10669.2001. URL 〈 http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=11602708〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer RJ, Homann D, Perelson AS. Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J Immunol. 2003;171(8):3928–3935. doi: 10.4049/jimmunol.171.8.3928. [DOI] [PubMed] [Google Scholar]
- DeRyke CA, Lee SY, Kuti JL, Nicolau DP. Optimising dosing strategies of antibacterials utilising pharmacodynamic principles: impact on the development of resistance. Drugs. 2006;66(1):1–14. doi: 10.2165/00003495-200666010-00001. [DOI] [PubMed] [Google Scholar]
- Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10(1):30–38. doi: 10.1016/j.mib.2006.12.007. URL 〈 http://dx.doi.org/10.1016/j.mib.2006.12.007〉. [DOI] [PubMed] [Google Scholar]
- Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. 2003;52(1):11–17. doi: 10.1093/jac/dkg269. URL 〈 http://dx.doi.org/10.1093/jac/dkg269〉. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao X. Mutant selection window hypothesis updated. Clin Infect Dis. 2007;44(5):681–688. doi: 10.1086/511642. URL 〈 http://dx.doi.org/10.1086/511642〉. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao XL. Is ‘dosing-to-cure’ appropriate in the face of antimicrobial resistance? Rev Med Microbiol. 2004;15(2):73–80. [Google Scholar]
- Drusano GL. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug’. Nat Rev Microbiol. 2004;2(4):289–300. doi: 10.1038/nrmicro862. URL 〈 http://dx.doi.org/10.1038/nrmicro862〉. [DOI] [PubMed] [Google Scholar]
- Fritz RS, Hayden FG, Calfee DP, Cass LM, Peng AW, Alvord WG, Strober W, Straus SE. Nasal cytokine and chemokine responses in experimental influenza A virus infection: results of a placebo-controlled trial of intravenous zanamivir treatment. J Infect Dis. 1999;180(3):586–593. doi: 10.1086/314938. [DOI] [PubMed] [Google Scholar]
- Gillespie SH. Evolution of drug resistance in mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother. 2002;46(2):267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004;84(1–2):29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Green DR, Droin N, Pinkoski M. Activation-induced cell death in t cells. Immunol Rev. 2003;193:70–81. doi: 10.1034/j.1600-065x.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- Happel KI, Bagby GJ, Nelson S. Host defense and bacterial pneumonia. Semin Respir Crit Care Med. 2004;25(1):43–52. doi: 10.1055/s-2004-822304. URL 〈 http://dx.doi.org/10.1055/s-2004-822304〉. [DOI] [PubMed] [Google Scholar]
- Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection, Relation to symptom formation and host defense. J Clin Invest. 1998;101(3):643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D, Kolter R. Why are bacteria refractory to antimicrobials? Curr Opin Microbiol. 2002;5(5):472–477. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- Imran M, Smith HL. The dynamics of bacterial infection, innate immune response, and antibiotic treatment. Discrete Continuous Dynamical Syst Ser B. 2007;8(1):127–143. [Google Scholar]
- Jumbe N, Louie A, Leary R, Liu W, Deziel MR, Tam VH, Bachhawat R, Freeman C, Kahn JB, Bush K, Dudley MN, Miller MH, Drusano GL. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J Clin Invest. 2003;112(2):275–285. doi: 10.1172/JCI16814. URL 〈 http://dx.doi.org/10.1172/JCI200316814〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath A, Woodworth JSM, Behar SM. Antigen-specific cd8+ t cells and the development of central memory during mycobacterium tuberculosis infection. J Immunol. 2006;177(9):6361–6369. doi: 10.4049/jimmunol.177.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath AB, Woodworth J, Xiong X, Taylor C, Weng Y, Behar SM. Cytolytic cd8+ t cells recognizing cfp10 are recruited to the lung after mycobacterium tuberculosis infection. J Exp Med. 2004;200(11):1479–1489. doi: 10.1084/jem.20041690. URL 〈 http://dx.doi.org/10.1084/jem.20041690〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labro MT. Interference of antibacterial agents with phagocyte functions: immunomodulation or “immuno-fairy tales”? Clin Microbiol Rev. 2000;13(4):615–650. doi: 10.1128/cmr.13.4.615-650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WT, Pasos G, Cecchini L, Mittler JN. Continued antigen stimulation is not required during cd4(+) t cell clonal expansion. J Immunol. 2002;168(4):1682–1689. doi: 10.4049/jimmunol.168.4.1682. [DOI] [PubMed] [Google Scholar]
- Leijh PC, van den Barselaar MT, van Zwet TL, Dubbeldeman-Rempt I, van Furth R. Kinetics of phagocytosis of Staphylococcus aureus and Escherichia coli by human granulocytes. Immunology. 1979;37(2):453–465. [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4(7):556–562. doi: 10.1038/nrmicro1445. URL 〈 http://dx.doi.org/10.1038/nrmicro1445〉. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. URL 〈 http://dx.doi.org/10.1038/nrmicro1557〉. [DOI] [PubMed] [Google Scholar]
- Li Y, Karlin A, Loike JD, Silverstein SC. A critical concentration of neutrophils is required for effective bacterial killing in suspension. Proc Natl Acad Sci USA. 2002;99(12):8289–8294. doi: 10.1073/pnas.122244799. URL 〈 http://dx.doi.org/10.1073/pnas.122244799〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Karlin A, Loike JD, Silverstein SC. Determination of the critical concentration of neutrophils required to block bacterial growth in tissues. J Exp Med. 2004;200(5):613–622. doi: 10.1084/jem.20040725. URL 〈 http://dx.doi.org/10.1084/jem.20040725〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Levin BR. The population dynamics of antimicrobial chemotherapy. Antimicrob Agents Chemother. 1997;41(2):363–373. doi: 10.1128/aac.41.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Levin BR. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int J Tuberc Lung Dis. 1998;2(3):187–199. [PubMed] [Google Scholar]
- Mager DE, Wyska E, Jusko WJ. Diversity of mechanism-based pharmacodynamic models. Drug Metab Dispos. 2003;31(5):510–518. doi: 10.1124/dmd.31.5.510. [DOI] [PubMed] [Google Scholar]
- McLoughlin RM, Solinga RM, Rich J, Zaleski KJ, Cocchiaro JL, Risley A, Tzianabos AO, Lee JC. Cd4+ t cells and cxc chemokines modulate the pathogenesis of Staphylococcus aureus wound infections. Proc Natl Acad Sci USA. 2006;103(27):10408–10413. doi: 10.1073/pnas.0508961103. URL 〈 http://dx.doi.org/10.1073/pnas.0508961103〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Andrian UHV. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427(6970):154–159. doi: 10.1038/nature02238. URL 〈 http://dx.doi.org/10.1038/nature02238〉. [DOI] [PubMed] [Google Scholar]
- Mercado R, Vijh S, Allen SE, Kerksiek K, Pilip IM, Pamer EG. Early programming of T cell populations responding to bacterial infection. J Immunol. 2000;165(12):6833–6839. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- Mouton JW, van Ogtrop ML, Andes D, Craig WA. Use of pharmacodynamic indices to predict efficacy of combination therapy in vivo. Antimicrob Agents Chemother. 1999;43(10):2473–2478. doi: 10.1128/aac.43.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M, de la Pena A, Derendorf H. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: kill curves versus MIC. Anti-microb Agents Chemother. 2004;48(2):369–377. doi: 10.1128/AAC.48.2.369-377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri MC, Lipsitch M, Blazquez J, Levin BR, Baquero F. Concentration-dependent selection of small phenotypic differences in TEM beta-lactamase-mediated antibiotic resistance. Antimicrob Agents Chemother. 2000;44(9):2485–2491. doi: 10.1128/aac.44.9.2485-2491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4(10):812–823. doi: 10.1038/nri1461. URL 〈 http://dx.doi.org/10.1038/nri1461〉. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8(11):1173–1178. doi: 10.1038/ni1526. URL 〈 http://dx.doi.org/10.1038/ni1526〉. [DOI] [PubMed] [Google Scholar]
- Pankey GA, Sabath LD. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin Infect Dis. 2004;38(6):864–870. doi: 10.1086/381972. URL 〈 http://dx.doi.org/10.1086/381972〉. [DOI] [PubMed] [Google Scholar]
- Pilyugin S, Antia R. Modeling immune responses with handling time. Bull Math Biol. 2000;62(5):869–890. doi: 10.1006/bulm.2000.0181. URL 〈 http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=11016088〉. [DOI] [PubMed] [Google Scholar]
- Regoes RR, Wiuff C, Zappala RM, Garner KN, Baquero F, Levin BR. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob Agents Chemother. 2004;48(10):3670–3676. doi: 10.1128/AAC.48.10.3670-3676.2004. URL 〈 http://dx.doi.org/10.1128/AAC.48.10.3670-3676.2004〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes RR, Barber DL, Ahmed R, Antia R. Estimation of the rate of killing by cytotoxic T lymphocytes in vivo. Proc Natl Acad Sci USA. 2007;104(5):1599–1603. doi: 10.1073/pnas.0508830104. URL 〈 http://dx.doi.org/10.1073/pnas.0508830104〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6(1):41–52. doi: 10.1038/nrmicro1816. URL 〈 http://dx.doi.org/10.1038/nrmicro1816〉. [DOI] [PubMed] [Google Scholar]
- Schentag JJ, Gilliland KK, Paladino JA. What have we learned from pharmacokinetic and pharmacodynamic theories? Clin Infect Dis. 2001;32(Suppl. 1):S39–S46. doi: 10.1086/319375. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Standiford TJ. Immunomodulatory effects of macrolides in the lung: lessons from in-vitro and in-vivo models. Curr Pharm Des. 2004;10(25):3081–3093. doi: 10.2174/1381612043383430. [DOI] [PubMed] [Google Scholar]
- van Stipdonk MJB, Lemmens EE, Schoenberger S. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2:415–422. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- Yates A, Graw F, Barber DL, Ahmed R, Regoes RR, Antia R. Revisiting estimates of CTL killing rates in vivo. PLoS ONE. 2007;2(12):e1301. doi: 10.1371/journal.pone.0001301. URL 〈 http://dx.doi.org/10.1371/journal.pone.0001301〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis. 2001;33(Suppl. 3):S147–S156. doi: 10.1086/321841. [DOI] [PubMed] [Google Scholar]
- Zhao X, Drlica K. A unified anti-mutant dosing strategy. J Antimicrob Chemother. 2008 doi: 10.1093/jac/dkn229. URL 〈 http://dx.doi.org/10.1093/jac/dkn229〉. [DOI] [PMC free article] [PubMed]