Abstract
Even with the advances in molecular or automated methods for detection of red blood cells of interest (such as reticulocytes or parasitized cells), light microscopy continues to be the gold standard especially in laboratories with limited resources. The conventional method for determination of parasitemia and reticulocytemia uses a Miller reticle, a grid with squares of different sizes. However, this method is prone to errors if not used correctly and counts become inaccurate and highly time-consuming at low frequencies of target cells. In this report, we outline the correct guidelines to follow when using a reticle for counting, and present a new counting protocol that is a modified version of the conventional method for increased accuracy in the counting of low parasitemias and reticulocytemias.
Keywords: Light microscopy, parasitemia, reticulocytemia, Miller reticle, malaria
Introduction
Visual examination of patient blood samples under a microscope provides important information. For example, quantification of reticulocytes – the most immature red blood cells (RBCs) found in circulation in peripheral blood – is a commonly used diagnostic, prognostic and monitoring parameter as it helps determine the health and activity of the patient's bone marrow [1]. Blood smears from suspected malaria patients are also used to determine Plasmodium species and parasite burden, identify developmental stages of the parasite, and assess response of the patient to treatments [2].
Both parameters become relevant in cases of Plasmodium vivax malaria patients, as it is thought that P. vivax preferentially, if not exclusively, invades reticulocytes [3-5]. In addition, reticulocytosis is indicative of anemia, a common symptom in malaria patients [1]. While the exact determination of parasitemia and reticulocytemia does not impact immediate treatment of P.vivax malaria, records of these parameters are vital in scientific studies.
In recent years, several automated or semi-automated methods using flow cytometry [1,6-9] or image analysis of slides, especially for parasitized cells [10-12], have been developed to reduce the tediousness of the manual counting process and increase accuracy. However, these methods remain expensive and inaccessible in many resource-poor field settings.
Examination of a patient's blood sample in field studies thus remains largely reliant on manual counts of thin smears through a light microscope, consisting of enumerating and either categorizing all RBCs in a visual field as infected or uninfected or classifying them as normocytes or reticulocytes. A practical way used by both parasitologists and hematologists is to use a Miller reticle (Figure 1A) [13]. This method uses the ratio of two squares of different sizes to effectively count a large number of cells [14]. However, the reticle can be easily misused without proper guidelines and is also prone to several unconscious biases from the technician, especially when the frequency of the cell types of interest is low [15,16]. For example, fields containing more target cells than the average can be preferentially chosen by the technician, thus overestimating the frequency. On the other hand, one may also underestimate the frequency if not enough fields are observed with a target cell falling in the reticle by chance. This method thus becomes less reliable for reticulocyte and P. vivax counts, as both are found at relatively low frequencies in circulation.
Figure 1. Measuring the accuracy of microscope reticles.
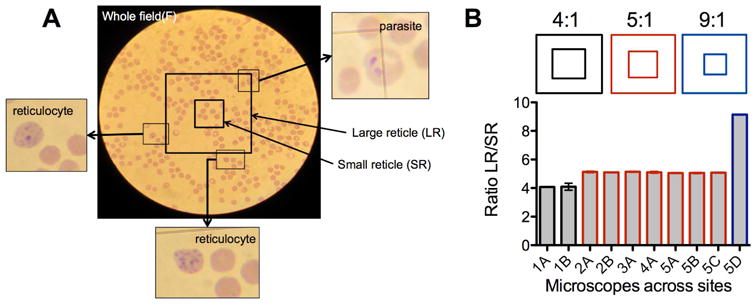
(A) Typical image of a microscopic field with a Miller reticle. The reticle is placed at the middle of a randomly chosen field. Parasitized cells and reticulocytes can be observed in and around the reticle. (B) Ratio of the areas of the small (SR) and large (LR) squares of reticles in microscopes at different study sites. While not evident by eye as shown in different reticle schematics, the different sites from one research group use three different reticles, a factor to consider when standardizing cell counts in the lab. Measurements were done 2-5 times for each microscope. Error bars indicate 95% Confidence Interval (CI).
Here, we describe a simple modified version of the reticle counting, called New Whole Field counting (NWF). This method takes advantage of the partitioned reticle for accurate counting of a large number of total RBCs (denominator) while using the whole visual field to increase the number of cells of interest encountered (numerator).
Methods
Sample collection and slide preparation
The Malaria Evolution in South Asia International Center of Excellence in Malaria Research (MESA-ICEMR) and the University of Washington (Seattle, WA, USA; Site 1) established several field sites across India, including at Goa Medical College and Hospital (Bambolim, Goa), Shalini Memorial Hospital (Ranchi, Jharkand), Acharya Vinoba Bhave Rural Hospital (Wardha, Maharashtra) and the Regional Medical Research Center-NE (Dibrugarh, Assam) (Sites 2-5).
The human subjects protocol for this study was approved by the institutional ethics boards at Goa Medical College and Hospital, the University of Washington, and the Division of Microbiology and Infectious Diseases of the US National Institutes of Health as well as by the the Government of India Health Ministry Screening Committee (HMSC).
Patients presenting to Goa Medical College and Hospital with malaria symptoms and subsequently diagnosed as only P.vivax positive by Rapid Diagnostic Test (RDT; FalciVax, Zephyr Biomedicals) were included in the study. For each patient, thin smears were prepared, fixed with 100% methanol and stained with filtered 100% Giemsa (Sigma Aldrich) for 20 minutes or with Hemacolor® Rapid staining, as per manufacturer's directions (EMD Millipore). A second set of thin smears was also stained for both reticulocytes and parasites. Approximately 2-5 uL of packed RBCs were incubated with an equal amount of Reticulocyte Stain (Sigma Aldrich) at room temperature for 20 minutes. A thin smear was made from 1.5 uL of the mixture, fixed with 100% methanol, and then stained with Giemsa (Sigma Aldrich) for at least 20 minutes.
Reticle size and cell size measurements
Pictures of the whole visual field of a microscope slide were taken through the eyepiece using compact digital cameras (Panasonic Lumix DMC-FH22, Canon SD870 and Canon Powershot A4000). The images were processed and analyzed using ImageJ image analysis software [17] to calculate the ratio of the different areas (large and small squares of Miller reticles, circle of whole field) and the sizes of the cells in the field.
Statistics
All statistical analyses were done using GraphPad Software v5 (GraphPad®).
Results
Improper use of Miller reticle method can introduce significant error
The standard equation for parasitemia or reticulocytemia using the Miller reticle is as follows:
| (1) |
where LR is the large square of the reticle and SR the small square of the reticle. The Reticle Factor (RF) is derived from the ratio of areas of SR to LR (100/ratio) (Fig 1A). However, the correct ratio of the reticle is not always evident and it is especially difficult to distinguish between similar ratios (such as 1:4/1:5 or 1:9/1:10) (Fig 1B) [1]. This can be due to miscataloging, or miscommunication, especially when setting up new laboratories in field sites and can introduce a significant error in cell counts (for example, using a 1:5 reticle while applying a 1:4 RF introduces a 25% overestimation). A simple way of verifying the ratio of the reticle used is to take a picture of the field through the eyepiece with an appropriate compact digital camera and use image analysis software to calculate the ratio of the area of the two squares. To verify the ratio of all reticles in microscopes deployed in different field sites of our research group, we analyzed several images taken through the eyepiece (Fig 1B). It was evident that the reticles that had been assumed identical at different sites were in fact different, with most sites in India (Site 2 – 5) using the 1:5 reticle (except one at 1:9), while the Seattle laboratory (Site 1) used a 1:4 reticle.
Another aspect of reticle use that can lead to inaccurate estimation of counts is the edge effect [18]. Cells falling on the corner and edges of both squares of the reticle must be counted following the edge rule as not to introduce errors. The different scenarios and the correct way of considering the edge effect are explained in Figure 2.
Figure 2. Proper use of reticle accounting for the edge effect.
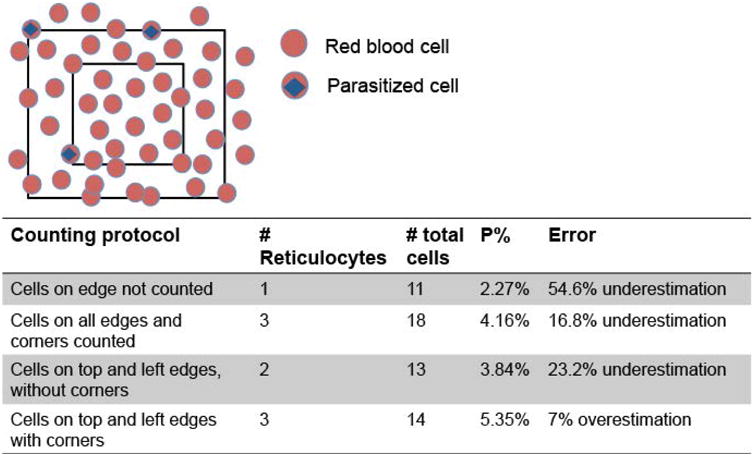
In this simple scenario of a field containing 5% parasitemia, we estimated the parasitemia using the counting protocols described in the table below with the equation (1). The table shows how improper consideration of the edge effect greatly increases error. This also applies when the cells of interest are reticulocytes.
Whole field and reticle counting can be combined to increase accuracy of low-frequency cell counts
The accuracy of a frequency count increases with increased denominator, but also with higher numerator (see equation 1). A major improvement of the introduction of the Miller reticle was the efficient way of counting a larger number of total cells (denominator). However, it further limits the number of cells of interest (numerator) the technician can consider as it restricts the visual field to the reticle. To overcome this limitation, we established a new whole field counting method (NWF method), with the equation for frequency calculation as follows:
| (2) |
The effect of considering the whole field is demonstrated in Figure 3.
Figure 3. Counting cells of interests using the New Whole Field method.
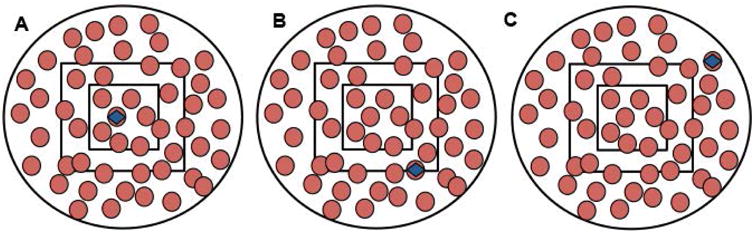
These schematic diagrams show the whole field of a microscope with RBCs (red circles), parasitized RBCs (red circles with blue square) and Miller reticle in the center, depicting three different scenarios for counting the parasitized cells. For parasitemia determination using the reticle method, only parasitized RBCs in (a) and (b) will be considered, whereas the whole field counting method would include parasitized RBCs in all three scenarios. One can approximate the area of half of the whole field and include parasitized cells falling at the edge of that half.
To get an accurate value of the RF, we measured the ratio of the area of the whole field to that of the small square of the reticle (Fig. 4A). With this method, the numerator is significantly higher than when using the conventional reticle method. This is especially true for counts of low frequency that would have been easily missed, even when counting a total of 1000 cells (Fig. 4B).
Figure 4. An approach for calculating cells of low frequency with increased accuracy.
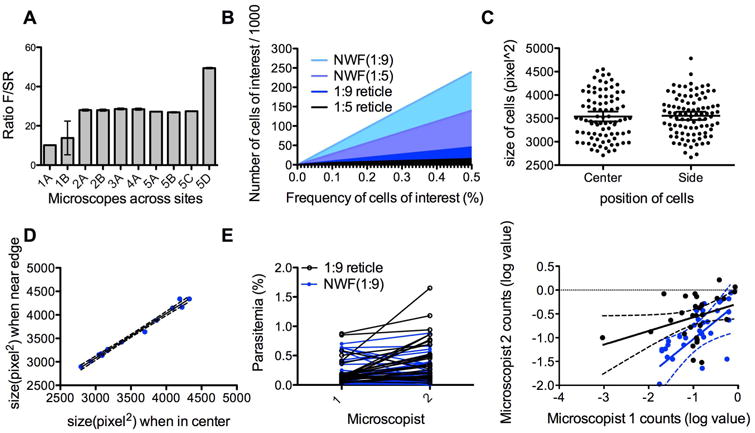
(A) Ratio of the areas of the small square (SR) of the reticle and the whole field circle (F) for microscopes at different study sites. This ratio can be used to determine the correct reticle factor for each microscope-reticle combination. Measurements were done 2-5 times for each microscope. Error bars indicate 95% Confidence Interval. (B) Estimated number of target cells observed when counting a total of 1000 cells using different methods (reticle method with a 1:5 or 1:9 reticle, and NWF method using a 1:5 or 1:9 reticle). (C) To test whether there is distortion at the edge of the pictures taken for analysis, sizes of the cells at the center (in reticle) of the field and at the edge of the field were measured. Measurements were made in at least three separate fields, using three different microscopes. The graph is from one representative microscope. (Median and standard deviation values for cells in the center versus side :3269.5± 541.5 vs 3341.5 ±517.654) Error bars indicate 95% Confidence Interval. (D) Pictures were taken of the same cells at different positions (in center of field or at edge) and the sizes of the cells analyzed. Dotted lines represent the 90% range. (Median and standard deviation values for cells in the center versus side : 3468 ± 491.2 vs 3584 ± 428.3 (E) Counts of two microscopists (1 and 2) were compared when using the 1:9 reticle (black) or the NWF method (blue). The left graph is a direct comparison of individual values using the two methods. The right graph shows the linearity between the counts. The parasitemia values were log-transformed for better comparison. The correlation between the two microscopists' counts is greater using the NWF method (Pearson r: 0.6654, p < 0.0001 for NWF, r:0.3898, p=0.03 for 1:9 reticle). Dotted lines represent the 95% Confidence Interval.
The RF is a critical value for this new method. To ensure that distortions due to photography did not influence the calculated ratio and thus the RF, we measured the size of the cells at the center of the square and compared to those at the edge in one slide (Fig. 4C). We also measured the size of the same cell when it is at the center versus when put at the edge (Fig. 4D). These cell sizes showed no significant difference, indicating that the edges of the whole field were not subject to distortions that would influence its area calculation. This was also consistent across three different microscopes.
To compare accuracy of counts between technicians, we chose slides of P. vivax patients with a range of parasitemia. Two experienced microscopists counted the slides using the Miller reticle method or the NWF method (Fig. 4E). There was significantly less discrepancy between the microscopists when using the NWF method. The NWF method also detected much lower ranges of parasitemia than the reticle counts.
Discussion
By combining counts of the small area of the reticle and observation of the large whole field, our method retains the advantages of scanning greater areas of a slide, while introducing significant advantages in detecting a much greater number of cells of interests. Compared to the traditional method of counting all cells in a given field or the conventional method of counting the cells just in a reticle, the effective number of cells counted by the NWF method is 5 to 50 times greater. Accordingly, the number of cells needed to be counted to attain the same level of accuracy would thus be much lower using our new method, considerably reducing the time commitment and tediousness of manual counts. Finally, at low frequency the error is minimized as the number of target cells observed increases. Using our method, the number of target cells one would encounter when counting the same number of denominator cells can reach a significantly higher number, giving more power to the final count.
With proper use of the reticle and taking advantage of the whole visual field of a light microscope, the NWF method provides an improvement to light-microscopy based counting, which remains the most cost-effective and reliable way of evaluating blood samples from patients in under-resourced settings.[19]. In addition to accurately determining P. vivax parasitemia and reticulocytemia, which are frequently low, this may also be useful in eradication studies of P. falciparum, where analysis of low parasitemia samples is crucial. Experiments that require accurate quantification of parasitized RBCs, such as parasite invasion assays, can also benefit from this new method. Finally, this method is widely applicable to any microscope and reticles of any ratio, as long as the RF is accurately calculated.
Acknowledgments
The authors thank the study participants and research support staff at the Goa Medical College and Hospital. The authors also thank Dr. Kristen Skillman for critical reading of the manuscript. The Program Project on ‘Malaria Evolution in South Asia’ is an International Center of Excellence for Malaria Research (ICEMR) supported by the US NIH/NIAID agreement U19 AI089688 (Program Director, PKR). This study was also supported by the Bill and Melinda Gates Foundation (OPP1023594).
References
- 1.Riley RS, Ben-Ezra JM, Goel R, Tidwell A. Reticulocytes and reticulocyte enumeration. J Clin Lab Anal. 2001;15(5):267–294. doi: 10.1002/jcla.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Routine examination of blood film for malaria parasites. WHO; 2009. pp. 69–76. [Google Scholar]
- 3.Kitchen SF. The infection of reticulocytes by Plasmodium vivax. 1938;18:347–353. [Google Scholar]
- 4.Mons B, Collins WE, Skinner JC, et al. Plasmodium vivax: in vitro growth and reinvasion in red blood cells of Aotus nancymai. Exp parasitol. 1988;66(2):183–188. doi: 10.1016/0014-4894(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 5.Malleret Benoit, Li Ang, Zhang Rou, et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood. 2015;125(8):1314–1324. doi: 10.1182/blood-2014-08-596015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roobsoong Wanlapa, Maher Steven P, Rachaphaew Nattawan, et al. A rapid sensitive, flow cytometry-based method for the detection of Plasmodium vivax-infected blood cells. Malar J. 2014;13(1):55. doi: 10.1186/1475-2875-13-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guy Rebecca, Liu Paul, Pennefather Peter, Crandall Ian. The use of fluorescence enhancement to improve the microscopic diagnosis of falciparum malaria. Malar J. 2007;6(1):89. doi: 10.1186/1475-2875-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BH, Bigelow NC, Koepke JA, et al. Flow cytometric reticulocyte analysis. Multiinstitutional interlaboratory correlation study. Am J Clin Pathol. 1994;102(4):468–477. doi: 10.1093/ajcp/102.4.468. [DOI] [PubMed] [Google Scholar]
- 9.Yu PH, So CC, Wong KF, et al. Automated reticulocyte counting--an evaluation of GEN-S, Cell-Dyn 3500 and Cell-Dyn 4000. Clin Lab Haematol. 1999;21(2):145–147. doi: 10.1046/j.1365-2257.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- 10.Le Minh-Tam, Bretschneider Timo R, Kuss Claudia, Preiser Peter R. A novel semi-automatic image processing approach to determine Plasmodium falciparum parasitemia in Giemsa-stained thin blood smears. BMC Cell Biol. 2008;9(1):15–12. doi: 10.1186/1471-2121-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savkare SS, Narote SP. Automatic System for Classification of Erythrocytes Infected with Malaria and Identification of Parasite's Life Stage. Procedia Technology. 2013;6:405–410. [Google Scholar]
- 12.Díaz Gloria, González Fabio A, Romero Eduardo. A semi-automatic method for quantification and classification of erythrocytes infected with malaria parasites in microscopic images. J Biomed Inform. 2009;42(2):296–307. doi: 10.1016/j.jbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Moll Kirsten, Kaneko Akira, Scherf Arthur, Wahlgren Mats., editors. Methods in malaria research. 2013. pp. 1–499. [Google Scholar]
- 14.Brecher G, Schneiderman M. A time-saving device for the counting of reticulocytes. Am J Clin Pathol. 1950;20(11):1079–1083. doi: 10.1093/ajcp/20.11_ts.1079. [DOI] [PubMed] [Google Scholar]
- 15.Tatsumi N, Tsuda I, Yokomatsu Y, et al. Inaccuracy and imprecision of reticulocyte counting. Osaka City Med J. 1989;35(1):39–47. [PubMed] [Google Scholar]
- 16.Peebles DA, Hochberg A, Clarke TD. Analysis of manual reticulocyte counting. Am J Clin Pathol. 1981;76(5):713–717. doi: 10.1093/ajcp/76.5.713. [DOI] [PubMed] [Google Scholar]
- 17.Schneider Caroline A, Rasband Wayne S, Eliceiri Kevin W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowgill Elizabeth S, Nee Jennifer A, Grindem, Carol B. Clinical application of reticulocyte counts in dogs and cats. Vet Clin N Am-Small. 2003;33(6):1223–1244. doi: 10.1016/s0195-5616(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 19.Ali Afsheen Farzand, Moiz Bushra, Omer Sadia. Is manual reticulocyte count a reliable option for under resourced countries? J Pak Med Assoc. 2010;60(11):892–896. [PubMed] [Google Scholar]


