Abstract
Wheat germ cell-free translation is shown to be an effective method to produce integral membrane proteins in the presence of unilamelar liposomes. In this chapter, we describe the expression vectors, preparation of mRNA, two types of cell-free translation reactions performed in the presence of liposomes, a simple and highly efficient purification of intact proteoliposomes using density gradient ultracentrifugation, and some of the types of characterization studies that are facilitated by this facile preparative approach. The in vitro transfer of newly translated, membrane proteins into liposomes compatible with direct measurements of their catalytic function is contrasted with existing approaches to extract membrane proteins from biological membranes using detergents and subsequently transfer them back to liposomes for functional studies.
1. Introduction
Membrane proteins provide the molecular mechanisms through which useful molecules gain controlled entry into a cell, and likewise provide the portals through which cellular products are exported from the cell. Membrane enzymes synthesize the molecules that make up cellular membranes (e.g., saturated and unsaturated fatty acids, phospholipids, glycerolipids, sphingolipids, sterols, polyisoprenes). Membrane enzyme complexes are responsible for electron transport and generate electrochemical gradients, and an exquisite membrane motor ATPase uses these gradients to generate ATP. They harvest light, provide allosteric receptors that trans-duce the information from binding of external molecules into cellular responses via signaling cascades, provide essential surface contacts as differentiating embryonic stem cells begin to assemble into more complex tissues, and help to elicit the antigenic responses observed in response to pathogen infection. These few examples do not do justice to the incredible diversity of functions and the essential roles that membrane proteins and enzymes contribute to cellular function.
Achieving control of integral membrane protein expression, transfer into the lipid bilayer, and cofactor incorporation are significant experimental challenges, and the ability to manipulate these events would be of great scientific utility. Furthermore, identification of novel ways to address increasing structural complexity, leading to the expression and facile purification of fully folded, functional membrane proteins or complexes embedded in easily handled and manipulated lipid environments or in functionally compatible detergent mixtures would also be of great utility.
Recent studies arising from structural genomics efforts suggest that cell-free translation may have unique possibilities for addressing these challenges. Although known for a long time, cell-free protein translation is undergoing a renaissance (Endo and Sawasaki, 2006; Klammt et al., 2004; Schwarz et al., 2007; Spirin and Swartz, 2008; Vinarov et al., 2006; Yokoyama, 2003). This approach to protein synthesis, using extracts from either prokaryotic (Boyer et al., 2008; Kigawa et al., 2004; Klammt et al., 2006; Schwarz et al., 2007; Wuu and Swartz, 2008; Yokoyama, 2003) or eukaryotic (Endo and Sawasaki, 2006; Madin et al., 2000) sources, offers an alternative to E. coli or other living cell-based platforms that are the mainstay of most recombinant protein expression efforts. Because it decouples the production of proteins and enzymes from cellular homeostasis, cell-free translation removes variability associated with the use of living expression hosts (Hall et al., 2005). Cell-free systems permit the production of proteins that form complexes in the cell (Matsumoto et al., 2008), inclusion bodies (Klammt et al., 2004; Schwarz et al., 2007), that undergo proteolysis in cells (Goren and Fox, 2008; Yokoyama, 2003), or that are toxic to cells (Klammt et al., 2004; Madin et al., 2000; Schwarz et al., 2007). Cell-free translation can offer dramatic increases in the speed of evaluation of different conditions for expression, considerable simplification of the steps needed to purify a recombinant protein, and direct possibilities for automation of all steps after sequence-verified cloning to catalytic assay of an expressed and purified protein.
Because it is an open system, cell-free translation allows the addition of many types of reagents to the protein synthesis reaction in order to address biological complexity. Some examples of added reagents include detergents to solubilize proteins (Klammt et al., 2007), liposomes to capture functional forms of membrane proteins (Goren and Fox, 2008; Nozawa et al., 2007), cofactors, coenzymes, and metal ions to reconstitute catalytic function (Abe et al., 2004; Boyer et al., 2008; Goren and Fox, 2008), multiple mRNAs to yield cotranslation of heteromeric proteins and protein–protein complexes (Matsumoto et al., 2008), or other enzymes to effect posttranslational modifications (Goerke and Swartz, 2008; Kanno et al., 2007). Moreover, many different types of amino acid (AA) labeling strategies can be accomplished in cell-free translation by a simple substitution of unlabeled AAs with AAs containing isotopic substitutions such as 2H, 13C, or 15N for NMR or selenomethionine (SeMet) for determination of X-ray diffraction phasing (Klammt et al., 2004; Matsuda et al., 2007; Vinarov and Markley, 2005). Residue type selective labeling can be carried out without the need for auxotrophs or specialized feeding, and unnatural AAs can be incorporated in a straightforward manner (Kiga et al., 2002). Another advantage is that wheat germ cell-free translation has been successfully automated (Vinarov and Markley, 2005; Vinarov et al., 2006) in 24-, 96-, and 384-well formats for screening in volumes as low as 50 μL, and 6- and 24-well formats for protein production in volumes up to 6 mL.
2. Overview of Cell-Free Translation
This chapter describes the use of wheat germ extract, a highly stable and productive eukaryotic system for cell-free translation (Kanno et al., 2007; Madin et al., 2000). Since wheat germ extract is prepared from a eukaryote, it provides differences in the mechanisms for translation and folding of nascent proteins as compared to bacterial translation (Endo and Sawasaki, 2006). Moreover, the eukaryotic system may provide unique chaperones and trans-location factors that may aid in the expression of some proteins (Goren and Fox, 2008), possibly including the examples described here.
Figure 37.1 provides a schematic summary of the steps used for cell-free translation and shows time-course photographs of an expression of green fluorescent protein (GFP, photo images provided courtesy of our collaborator and mentor, Prof. Yaeta Endo, Cell-Free Science and Technology Research Center, Ehime University, Japan). For this work, the gene of interest is cloned into a specialized expression vector that adds 5′-translation enhancer and 3′-UTR sequences to the transcribed mRNA. Genes are transcribed using SP6 RNA polymerase, and the highly purified mRNA is used in the translation reaction. The quality and quantity of the expression plasmid and mRNA preparations are critical, and often underappreciated inputs to the success of high-yield cell-free translation (Tyler et al., 2005; Vinarov et al., 2006). Along with the mRNA template, the cell-free translation utilizes exogenously added AAs as substrates for the protein synthesis.
Figure 37.1.
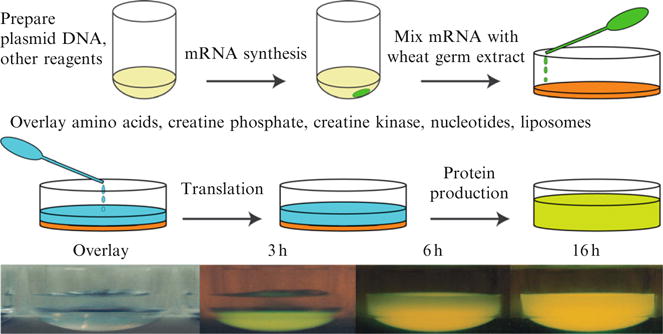
A schematic representation of the steps used for cell-free translation in a bilayer mode reaction. A customized plasmid is used to prepare reagent mRNA, which is added to the translation mixture, overlaid with amino acids, other substrates, and desired additives, and then the reaction can begin. The photos show an experimental demonstration of GFP translation over a 16 h period.
3. Expression Vectors
Table 37.1 summarizes specialized vectors used for this work. pEU, the original cell-free translation vector optimized for wheat germ cell-free translation (Madin et al., 2000), was modified for high-throughput Flexi Vector cloning (Promega) to contain 5′-SgfI and 3′-PmeI restriction sites and a toxic selection cassette in the multicloning site (Blommel et al., 2006). The vector named pEU-FV produces a protein with no purification tag, while pEU-His-FV and pEU-HSBC produces a protein with an N-terminal His6 purification tag. Vectors pEU-NGFP and pEU-GFPC produce fusions of GFP to either the N- or C-termini of the target protein. These vectors are useful for control studies and as vehicles for simplified detection of the translated fusion proteins during purification (Drew et al., 2001). With proper design of the primer sequences, the GFP tag can be removed by treatment with tobacco etch virus (TEV) protease (Blommel and Fox, 2007; Sobrado et al., 2008). The vectors pEU-Nb5R and pEU-Cb5 create fusion proteins with the membrane anchor signals from human cytochrome b5 reductase and human cytochrome b5, respectively. Fusion proteins containing these tags spontaneously associate with liposomes upon translation (Nomura et al., 2008), and thus become amenable to purification by density gradient ultracentrifugation (Goren and Fox, 2008). Other posttranslational modifications may also be used to target proteins to liposomes (Nosjean and Roux, 2003). The vectors described herein are available from the NIH Protein Structure Initiative Material Repository (http://psimr.asu.edu).
Table 37.1.
Flexi Vector compatible vectors for cell-free translation
| Name | Producta | Utility |
|---|---|---|
| pEU | Target | Expression screening, native protein |
| pEU-FV | Target | Expression screening |
| pEU-His-FV | His6-Target | Expression screening; His6 purification |
| pEU-HSBC | His6-Target | Expression screening; His6 purification |
| pEU-NGFP | GFP-target | Expression screening; detection |
| pEU-GFPC | Target-GFP | Expression screening; detection |
| pEU-Nb5R | Target-(cyt b5 reductase N-terminal anchor peptide)b | Spontaneous association of N-terminal anchor peptide to liposomes |
| pEU-Cb5 | Target-(cyt b5 C-terminal anchor peptide)c | Spontaneous association of C-terminal anchor peptide to liposomes |
The sequence of targets, domains, or tags found in the translated protein. For example, pEU-GFPC produces a target protein with GFP fused to the C-terminus, that is, target-GFP.
The N-terminal anchor peptide sequence is GAQLSTLGHMVLFPVWFLYSLLM.
The C-terminal anchor peptide sequence is TLITTIDSSSSWWTNWVIPAISAVAVALMYRLYMAED.
4. Gene Cloning
This approach requires a two-step PCR procedure (Blommel et al., 2006; Thao et al., 2004). Figure 37.2 provides a vector map for pEU-HSCB and an example of primers designed for cloning of His6-bacteriorhodopsin (Blommel and Fox, 2007). By using this vector and primer design, the sequence MGHHHHHHAIAENLYFQ- can be liberated from the translated protein upon treatment with TEV protease to leave Ser as the first residue of the mature protein. The tobacco mosaic virus omega sequence is a translation enhancement sequence (Sawasaki et al., 2000). Incorporation of the SgfI and PmeI restriction sites in the indicated positions in the respective primers allows the cloned gene to be transferred by Flexi Vector cloning (Blommel et al., 2006) into any (or all) of the vectors described in Table 37.1, along with a number of other commercially available vectors (see http:// www.promega.com for other examples). The sacB-CAT cassette shown in Fig. 37.2A provides toxic selection when transformants containing this construct are grown on plates containing 5% sucrose. In this manner, positive selection for cloning the desired gene can be enforced. The insert also confers resistance to chloramphenicol. The pF1K homology region enhances the efficiency of transfer of cloned genes between different Flexi Vectors (Blommel et al., 2006).
Figure 37.2.
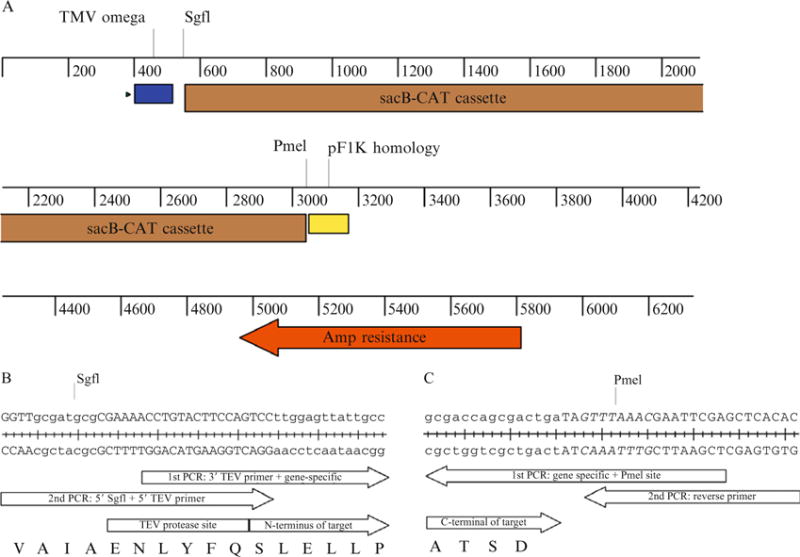
(A) Linear map of pEU-HSCB showing important features of the vector construction. The TMV omega sequence (blue box) enhances translation efficiency. The pF1K homology sequence (yellow box) prevents ligation of two plasmid backbones. (B) Example of the 5′ primer designed for the cloning of Halobacterium salinarium bacteriorhodopsin (GenBank M11720.1). The first-step PCR forward primer is 5′-ACCTGTACTTCCAGTCCttggagttattgcc, with the upper case nucleotides corresponding to the 3′ TEV primer and the lower case nucleotides corresponding to the gene-specific sequence. The second-step universal forward primer is 5′-GGTTgcgatcgcCGAAAACCTGTACTTCCAGTCC, with the SgfI site in lower case. There is an 18 bp overlap between the first-step forward primer and universal forward primer. The TEV protease recognition sequence is ENLYFQ/S, with proteolysis between Q and S. (C) Example of the 3′ primer designed for the cloning of Halobacterium salinarium bacteriorhodopsin (GenBank M11720.1). The first-step PCR reverse complement primer is 5′-GCTCGAATTCGTTTAAACTAtcagtcgctggtcgc, with the upper case, italic nucleotides corresponding to the PmeI site and the lower case nucleotides corresponding to the gene-specific sequence. The second-step universal reverse primer is 5′-GTGTGAGCTCGAATTCGTTTAAAC. There is an 18 bp overlap between the first-step reverse primer and universal reverse primer.
In the example shown in Fig. 37.2B, the first-step PCR uses a 5′ forward primer containing gene-specific nucleotides. An invariant sequence is added to the 5′ end of the first-step PCR forward primer to encode a portion of the TEV protease site. The first-step PCR also uses a 3′ reverse complement primer as shown in Fig. 37.2C, which contains gene-specific nucleotides and the PmeI site. Other genes may be cloned by substitution of the gene-specific sequence in the designated primer sequences.
For the second-step PCR, a universal forward primer (Fig. 37.2B) is used to add the nucleotides required to complete the TEV site and the SgfI site. A universal reverse PCR primer is used to duplicate the PmeI site and add additional nucleotides (Fig. 37.2C). For the second-step PCR, a portion of the first-step PCR reaction is added into a new PCR reaction containing the universal forward primer and reverse primers to obtain the PCR product properly functionalized for Flexi Vector cloning.
4.1. Materials and reagents
Gene-specific primers (25 nmol synthesis with standard desalting) can be obtained from IDT (Coralville, IA).
The dNTP mix (10 mM of each nucleotide) is from Promega (Madison, WI).
Pfu Ultra II fusion hotstart DNA polymerase is from Stratagene (La Jolla, CA).
A 10× Flexi Enzyme Blend containing restriction endonucleases SgfI and PmeI is from Promega.
PCR plates (T-3069-B) are from ISC Bioexpress (Kaysville, UT).
Adhesive covers for PCR plates (4306311) are from Applied Biosystems (Foster City, CA).
PCR plates are centrifuged in an Allegra 6R centrifuge with a GH3.8 rotor (Beckman Coulter, Fullerton, CA).
An MJ DNA Engine, DYAD, Peltier Thermal Cycler (MJ Research, Waltham, MA) can be used for thermocycling reactions.
4.1.1. Protocol
The following steps are used to PCR amplify the desired genes and append the sequences required for cloning. A sequence-verified gene in a plasmid is used for the PCR template. These may be obtained from a variety of other sources or research projects. This PCR protocol can also be used on genomic DNA, if the gene of interest has no introns, or on reverse transcribed mRNA (Thao et al., 2004).
If the plasmid being used for the template and the Flexi Vector that the PCR product will be cloned into have the same antibiotic resistance, DpnI can be added to the PCR reaction followed by incubation for 1 h at 37°C. The DpnI will digest the template plasmid while leaving the PCR product fully intact.
Create a PCR Primers plate by combining forward and reverse primers for each target gene to 10 μM each in a well of an ISC PCR plate.
Create a PCR Master Mix consisting of 2.195 mL of water, 250 μL of 10 × Pfu Ultra II Buffer, 25 μL of dNTPs (10 μM each), and 50 μL of Pfu Ultra II Hotstart DNA polymerase This Master Mix will provide up to 96 reactions, and can be scaled as appropriate. Discard the unused mix.
Aliquot 23.0 μL of PCR Master Mix to each well of an ISC PCR plate that will be used. This is the PCR Reaction plate.
Add 1 μL of the mixture from each used well of the PCR primers plate to each used well of the PCR Reaction plate.
Add 1 μL (~100 ng) of purified plasmid DNA for each gene to be cloned to a separate well of the PCR Reaction plate.
Centrifuge the PCR Reaction plate briefly in an Allegra 6R centrifuge and 6H3.B rotor to get liquid to the bottom of the wells and then cover the plate with sealing tape.
Put the PCR Reaction plate in the thermocycler and cycle using the following parameters: (1) 95 °C for 2.00 min; (2) 94 °C for 20 s; (3) 50 °C for 20 s; (4) 72 °C, 15 s/kb; (5) repeat steps 1–4 for 19 more times.
For the second-step PCR, 20% of the volume of the first-step reaction is added into a new PCR reaction along with 0.2 μM of each of the universal forward and reverse primers. The second-step reaction is then completed using the following cycling parameters: (1) 95 °C for 2.00 min; (2) 94 °C for 20 s; (3) 50 °C for 20 s; (4) 72 °C, 15 s/kb; (5) repeat steps 2–4 for four more times; (6) 94 °C for 20 s; (7) 55 °C for 20 s; (8) 72 °C for 15 s/kb; (9) repeat steps 6–8 for 24 more times; (10) 72 °C for 3.00 min; and (11) 4 °C and hold.
Analyze the completed PCR reactions for the appropriately sized products using agarose gel electrophoresis.
5. PCR Product Cleanup
The PCR products are purified prior to SgfI/PmeI digestion.
5.1. Materials and reagents
Quickstep 2 PCR purification kit (EdgeBio, Gaithersburg, MD), containing SOPE resin, Secure Seal Sterile tape, Performa Ultra 96-well Plate and V-Bottom 96-well Plate.
5.1.1. Protocol
Add 4 μL of well-mixed SOPE resin to 20 μL of PCR product from the FLEXI-TEV PCR protocol.
Cover the plate with Secure Seal Sterile tape and vortex. Let the suspension stand at room temperature while preparing the Performa Ultra 96-Well Plate.
Remove adhesive plate sealers from the top and bottom of the Performa Ultra 96-Well Plate. Cover with a lid.
Stack the Performa Ultra 96-Well Plate on top of the 96-well flat-bottom microplate.
Place the assembly in a cushioned centrifuge plate carrier.
Centrifuge for 5 min at 850 × g.
Briefly spin the SOPE/PCR mix to the bottom of the wells. Transfer the SOPE/PCR reaction mixture by slowly pipetting directly into the wells of the Performa Ultra 96-Well Plate. Be sure the fluid runs into the gel matrix. Cover with a lid.
Stack the Performa Ultra 96-Well Plate on top of the 96-well V-bottom microplate.
Place the assembly in the centrifuge plate carrier and centrifuge for 5 min at 850 × g. Retain the eluates, which contain the purified PCR products. PCR products can be stored at −20 °C until ready for use.
6. Flexi Vector and PCR Product Digestion Reaction
The following steps are used to digest the pEU vector variant and the purified PCR products with SgfI and PmeI prior to the ligation. Additional descriptions of cloning using the Flexi Vector system are presented elsewhere (Blommel and Fox, 2007; Blommel et al., 2006). For success in Flexi Vector cloning, it is important to avoid overdigesting either the PCR product or the vector. SgfI has star activity so overdigestion removes the overhanging nucleotide sequence leaving bunt ends on the digested vector that efficiently ligate and yield a high background of clones with no insert after transformation.
6.1. Reagents
5 × Flexi Digest Buffer (Promega)
10 × SgfI/PmeI Enzyme Blend (Promega)
pEU vector variant (see Table 37.1)
Purified PCR products from PCR Product Cleanup protocol, step 9
6.1.1. Protocol
Create a pEU Vector Digest Master Mix consisting of 158.3 μL of sterile, deionized water, 44.0 μL of 5 × Flexi Digest Buffer, 2.20 μL of 10 × SgfI/PmeI Enzyme Blend, and 13.5 μL of the pEU vector variant desired (e.g., purified pEU-His-FV at a concentration of 150 ng/μL). The enzyme blend is dense and tends to settle, so must be mixed thoroughly into the remainder of the mix.
Place the pEU Vector Digest Master Mix in the thermocycler and cycle as follows: (1) 37 °C for 40.00 min; (2) 65 °C for 20.00 min; and (3) hold at 4 °C until needed.
Create the PCR Product Digest Master Mix consisting of 638 μL of sterile, deionized water, 220 μL 5 × of Flexi Digest Buffer, and 22 μL of 10 × SgfI/PmeI Enzyme Blend. This Master Mix will provide up to 96 reactions, and can be scaled as appropriate. Discard the unused mix.
Add 8.0 μL of the PCR Product Digest Master Mix to each well of an ISC PCR plate. This is the PCR Digest plate.
Add 2.0 μL of the purified PCR products in the PCR Reaction plate from Gene Cloning protocol, step 8 to each used well of the PCR Digest plate.
Place the PCR Digest plate in the thermocycler and cycle as follows: (1) 37 °C for 40.00 min; (2) 65 °C for 20.00 min; and (3) hold at 4 °C until needed. If transformation yields no colonies, or colonies containing vector without the desired insert, decrease the 37 °C incubation time to minimize star activity.
7. Ligation Reaction
Digested and purified PCR products and pEU vector variants are ligated in this step. For the best efficiency in this ligation reaction, it is important to use a high concentration of ligase.
7.1. Reagents
10 × T4 DNA ligase buffer (Promega)
High concentration T4 DNA ligase (Promega)
pEU vector variant from Flexi Vector Digestion Reaction, step 2
Purified PCR product in PCR Cleanup plate from PCR Product Digestion Reaction Cleanup, step 6
7.1.1. Protocol
Create a Ligation Master Mix containing 225 μL of sterile, deionized water, 110 μL of 10 × T4 ligase buffer, and 50 μL of high-concentration T4 DNA ligase.
Add 5.0 μL of purified PCR product from the PCR Cleanup plate, 2.0 μL of the pEU vector digest, and 3.5 μL of Ligation Master Mix to each well of a new PCR plate. This is the Ligation plate.
Incubate the Ligation plate in a thermocycler at 25 °C for 3 h.
Immediately proceed to the transformation step or store the Ligation plate overnight at 4 °C.
8. Transformation Reaction
The material from the ligation reaction is used to transform competent cells by the following steps. Competent cells from Invitrogen and Promega have also been used successfully by following the manufacturers protocols.
8.1. Materials and reagents
10G cells, chemically competent for transformation (Lucigen, Middleton, WI)
10G cells recovery medium (Lucigen).
Ligation Plate from Ligation Reaction, step 4.
Fisherbrand sterile disposable Petri dishes (60 × 15 mm) (Fisher Scientific, Pittsburgh, PA).
YT agar plates containing 0.5% (w/v) glucose, 50 μg/mL of kanamycin, and 5% sucrose for selection with pEU-HSCB vector (Fig. 37.2).
ColiRoller Plating Beads (Novagen, Gibbstown, NJ).
8.1.1. Protocol
Remove 10G chemically competent cells from −80 °C freezer and thaw on ice.
Aliquot 10 μL of cells into a PCR plate or strip tubes that have been chilled on ice.
Add 1.0 μL of the ligation reaction and stir with the pipet tip.
Incubate on ice for at least 5 min. Set a thermocycler block to 34 °C.
Heat shock at 34 °C for 30 s (per the manufacturer’s directions for small reaction volumes).
Return the transformation reactions to ice and incubate for 2 min.
Remove from ice and add 80 μL of room temp recovery medium.
Incubate at 37 °C for 1 h without shaking.
While transformations are incubating, label the bottoms of 96 YT plates, containing the appropriate antibiotic, A1–H12 and add 5–10 sterile ColiRoller glass beads to each plate.
Apply the entire transformation reaction on each of the correspondingly labeled plates. Shake the plates in a circular motion to spread the culture. Carefully remove ColiRoller beads by tipping the plate over an appropriate waste container, or, if they will be reused, 100% ethanol.
Incubate the plates overnight at 37 °C.
9. Purification of Plasmid DNA
All reagents used for in vitro transcription and translation must be RNase-free. Therefore, all glassware used to prepare reagents for cell-free translation reactions must be baked for 3 h at 180 °C to eliminate contaminating RNases. Furthermore, wear gloves, keep one’s mouth closed, and avoid sneezing while handling reagents in order to prevent RNase contamination from hands or saliva. All buffers must be sterilized by passage through a 0.2-μm filter, and stored at −20 °C unless otherwise indicated.
Also, unless otherwise stated, 18 MΩ water (Milli-Q water, Millipore, Billerica, MA) is used to prepare all reagents. Diethylpyrocarbonate-treated water should not be used in these protocols as the degradation products of DEPC can inhibit in vitro transcription and translation reactions.
9.1. Reagents
The 10 × buffer used with proteinase K is 100 mM Tris–HCl, pH 8.0, containing 50 mM EDTA and 1% (w/v) SDS.
Proteinase K is from Sigma/Aldrich (St. Louis, MO). Prepare a 10 × proteinase K solution by addition of 0.5 mg of proteinase K to 1.0 mL of 10 × proteinase K buffer. Aliquot the proteinase K solution into 10 μL samples and store them at −80 °C.
9.1.1. Protocol
Inoculate 150 mL of 2 × YT medium with a single colony from a pEU-His-FV transformation and grow the culture in a shaking incubator overnight at 37 °C. Recover the cells by centrifugation.
Purify the plasmid using a Marligen high-purity plasmid Maxiprep kit (Marligen Biosciences, Ijamsville, MD) and the manufacturer’s instructions. Resuspend each separate DNA pellet in 500 μL of Milli-Q water and measure the absorbance at 260 nm to determine the plasmid DNA concentration. A typical yield is 600–900 μg of plasmid DNA.
Plasmid DNA prepared by commercially available kits often contains a trace contamination of RNase. This contaminant must be removed for successful transcription and translation. In order to remove trace RNase contamination, treat the purified plasmid with 50 ng/μL of proteinase K for at least 60 min at 37 °C in 1 × proteinase K buffer.
Extract the remaining protein by adding an equal volume of a 1:1 phenol/chloroform solution to the plasmid preparation, vortex it vigorously, and then centrifuge the preparation at 14,000 rpm (18,000 × g) and 4 °C in an F2402H rotor and Allegra 21R centrifuge or comparable instrument. Remove the upper aqueous phase to a separate new tube and repeat the extraction, centrifugation of the phenol/chloroform solution, and transfer of the aqueous phase to a new tube.
Add 0.1 volume of 3 M sodium acetate, pH 5.2, and 2.5 volumes of ethanol to the aqueous phase obtained from step 4 and mix well. Chill each tube at −20 °C for 10 min to facilitate precipitation of the plasmid DNA.
Centrifuge each tube at 14,000 rpm (18,000 × g) and 4 °C in an F2402H rotor and Allegra 21R centrifuge. Wash each pellet with 500 μL of ice-cold 70% ethanol.
Centrifuge each tube again for 5 min as above, discard the supernatant, and thoroughly air-dry the pellet, which contains the desired plasmid DNA.
Dissolve the plasmid DNA pellets in 400 μL of Milli-Q water and measure the absorbance of each at 260 nm to determine the concentration of each separate plasmid DNA preparation. Adjust the volume of each preparation with Milli-Q water to obtain a DNA concentration for the solution of 1 μg/μL based on the fact that 1 μg/μL of DNA gives an A260 = 20 (40-fold dilution = 0.5).
10. Preparation of mRNA
In cell-free translation, mRNA is an essential reagent for the protein synthesis reaction. In order to obtain maximal translation efficiency, the mRNA preparations must be added in sufficient quantity to saturate ribosomes present in the translation reaction.
10.1. Reagents
Transcription Buffer plus Mg (5×) is 400 mM HEPES-KOH, pH 7.8, containing 100 mM magnesium acetate, 10 mM spermidine hydrochloride, and 50 mM DTT. Store this buffer at −20 °C.
An NTP solution containing 25 mM each of ATP, GTP, CTP, and UTP is prepared from 0.2 μm filter-sterilized, 100 mM solutions of each NTP prepared in Milli-Q water. NTP solutions are stored at −80 °C.
SP6 RNA polymerase and RNase inhibitor (RNasin) are from Promega.
10.1.1. Protocol
Immediately before use, prepare a transcription mixture containing 2 × Transcription Buffer plus Mg, 8 mM NTPs, 3.2 unit/μL of SP6 RNA polymerase, and 1.6 unit/μL of RNasin.
Dispense 2.5 μL of each separate plasmid DNA from Purification of Plasmid DNA, step 8, to a separate well of a PCR plate. To the PCR plate, dispense 2.5 μL of the transcription mixture into each well and mix. This is called the Transcription plate.
Tightly cap the wells of the Transcription plate to avoid concentrating the samples by evaporation. Incubate the Transcription plate at 37 °C for 4 h. If the transcription reaction is proceeding correctly, a white precipitate of magnesium pyrophosphate will form and make the transcription solution turbid.
Remove the white precipitate from the transcription reaction by centrifugation in the C0650 rotor and Allegra X-22R centrifuge for 5 min at 6230 rpm (4000 × g) and 26 °C. To avoid coprecipitation of mRNA, the reaction should not be chilled. Transfer the supernatant to a new tube. This clarified solution will be used in the translation reactions as the mRNA solution.
11. Preparation of Liposomes
Unilamelar liposomes are added to the cell-free translation reaction to capture membrane proteins as they are translated. This step provides an alternative to solubilization of membrane proteins by detergents, which often does not allow direct determination of function.
11.1. Materials and reagents
Soybean total extract (20% lecithin) is from Avanti Polar Lipids (Alabaster, AL).
The lipid rehydration buffer is 25 mM HEPES, pH 7.5, containing 100 mM NaCl.
Track-etch polycarbonate membranes, 0.4 and 0.1 μm, are from Nucleopore (Pleasanton, CA).
A mini-extruder for preparation of unilamelar liposomes is from Avanti Polar Lipids.
11.1.1. Protocol
Dissolve 1 g of soybean total extract (20% lecithin) in 3 mL of chloroform.
Flush the lipid solution with a stream of N2 gas in order to remove the bulk of the organic solvent. Dry the remaining lipid further under vacuum for 30 min.
Resuspend the dried lipid in 67 mL of lipid rehydration buffer (final concentration of 15 mg/mL). Vortex the lipid solution until homogeneous. Hydration of the lipids can be aided by 3–5 freeze/thaw cycles.
Form unilamelar liposomes by extrusion through a mini-extruder. Pass the liposome solution 11 times through a 0.4-μm track-etch polycarbonate membrane and then 11 times through a 0.1-μm membrane.
Aliquot the liposomes and flash-freeze them. Liposomes prepared in this way can be stored at −80 °C.
12. Wheat Germ Translation Reaction
Figure 37.3 compares the set up of the bilayer reaction with that of a dialysis reaction. The bilayer reaction separates the extract and mRNA from other reagents by the difference in density of the extract and upper buffer solutions. In this case, diffusion of substrates and products occurs through the entire buffer interface. Because of this simplicity in set up, the bilayer cell-free translation reaction is amenable to automation (Sawasaki et al., 2002a; Tyler et al., 2005; Vinarov et al., 2004, 2006). In our hands, the bilayer reaction has yielded ~ 0.2 mg/mL of various membrane proteins. However, as diffusion dilutes the reaction, the yield is limited.
Figure 37.3.
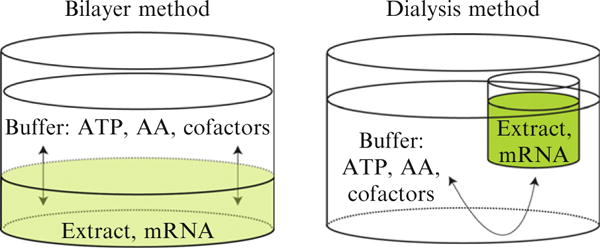
Schematic representation of two methods of cell-free translation. In the bilayer method, the extract and mRNA are separated from ATP, amino acids (AA), cofactors, and other buffer additives by the difference in density of the two solutions (also see Fig. 37.1). In the dialysis method, the extract and mRNA are contained within a dialysis membrane. In both cases, diffusion gives transfer of substrates, products, and additives between the extract and buffer.
The dialysis reaction is another method for performing cell-free translation (Fig. 37.3). A 12-kDa cutoff membrane at the bottom of the reactant cup retains the concentrated wheat germ extract, mRNA, and expressed protein. Diffusion replenishes ATP, AAs, cofactors, and other additives required for continued translation and removes inhibitory products. The dialysis method of cell-free translation can yield a 5- to 10-fold higher level of expression than the bilayer method because the constituents of the extract are not diluted by diffusion, which occurs during the course of the translation in the bilayer reaction. Thus the dialysis method can yield purified membrane proteins in the range of 0.2 mg/mL to greater than 2 mg/mL in the standard reactions (Goren and Fox, 2008). Although not as easily adaptable to robotics, the dialysis reaction can be performed on the benchtop and is scalable from 50 μL to 10 mL or greater with no overall changes in volumetric productivity. In the smaller volumes, this method has utility for simple screening of a few proteins, while in the larger volumes it also has utility for scale-up of the production of proteins whose properties have been investigated at small scale and found to be favorable for more extensive studies. Protocols for cell-free translation in automated batch, bilayer, and dialysis reactions have been published (Endo and Sawasaki, 2006; Sawasaki et al., 2002a; Vinarov et al., 2006). For translation of integral membrane proteins, we have found that translation reactions are productively modified by the addition of unilamellar liposomes (Goren and Fox, 2008; Nozawa et al., 2007).
12.1. Materials and reagents
WEPRO2240 wheat germ extract is from CellFree Sciences, Ltd. (Yokohama, Japan). This preparation has ~ 240 OD260/mL and does not contain AAs. Store the extract at −80 °C, thaw on the bench, and flash-freeze the unused lysate before storing. For addition to translation reactions, the extract is diluted from the concentrated commercial preparation to a final OD260 of 60 with 1 × Reaction Buffer.
Unlabeled AAs are from Advanced ChemTech (Louisville, KY). A mixture of the 20 unlabeled AAs is prepared in Milli-Q water with each AA present at 2 mM. Do not filter these preparations because some AAs will not dissolve in these solutions.
5 × Reaction Buffer is prepared from 150 mM HEPES–KOH, pH 7.8, and contains 500 mM potassium acetate, 12.5 mM magnesium acetate, 2 mM spermidine hydrochloride, 20 mM DTT, 6 mM ATP, 1.25 mM GTP, 80 mM creatine phosphate, and 0.025% (w/v) sodium azide. Store this buffer at −80 °C.
1 × Overlay Buffer is prepared by fivefold dilution of 5 × Reaction Buffer. The overlay buffer is then amended with AAs solution to give a final concentration of 0.3 mM for each AA.
Creatine kinase is from Roche Applied Sciences (Indianapolis, IN). Dissolve in Milli-Q water to make a 50 mg/mL solution and store at −80 °C. Dilute the stock solution to 1 mg/mL prior to use. Avoid multiple cycles of freezing and thawing of the concentrated solution. Discard the diluted solution.
12 kDa molecular weight cutoff (MWCO) dialysis cups are from Cosmo Bio (Tokyo, Japan). Prior to use, check the integrity of the membrane by adding 500 μL of Milli-Q water and monitor for any leakage. If there is no leakage, shake out the water prior to use.
The purified mRNA preparation is from Preparation of mRNA, step 4.
The liposome solution from Preparation of Liposomes, step 6.
24-Deep well pyramid-bottom plate, maximum volume of 10 mL (Artic-white, Bethlehem, PA).
96-Well U-Bottom Plate (Greiner Bio-One, Monroe, NC).
12.1.1. Protocol for a bilayer reaction
Prepare a 20 μL translation mixture by mixing 2 μL of a 15 mg/mL liposome solution, 4.25 μL of Milli-Q water, 2.75 μL of 5 × Reaction Buffer, 3.75 μL of 2 mM unlabeled AAs, 1 μL of 1 mg/mL creatine kinase, and 6.25 μL of WEPRO2240 wheat germ extract. Depending on the number of separate reactions desired, scale these volumes and include ~ 10% extra volume to account for handling losses.
Transfer 5 μL of an mRNA preparation to an individual well of a 96-well U-bottom plate.
Add 20 μL of translation mixture to the mRNA sample in each well, and mix.
Form a bilayer by carefully adding 125 μL of 1 × Overlay Buffer. Be careful not to disrupt the layers as that can dilute the extract and reduce protein production.
Incubate the reaction for 20 h at 26 °C. Do not disturb the bilayer during this time.
Protein translation levels can be determined by denaturing electrophoresis SDS–PAGE with creatine kinase serving as an internal intensity standard.
12.1.2. Protocol for a dialysis reaction
Dissolve the purified mRNA pellet in 50 μL of the translation mixture prepared as described in Protocol for Bilayer Reaction, step 1.
Place the translation mixture into a 12-kDa MWCO dialysis cup.
Prepare the reservoir dialysis buffer by mixing 6.5 mL of Milli-Q water, 2.0 mL of 5 × Reaction Buffer, and 1.5 mL of 2 mM unlabeled AAs. Sonicate the mixture for 5 min, and then pass the solution through a 0.2-μm filter. Add 2.5 mL of reservoir dialysis buffer to each well of the 24-deep well plate.
Suspend the dialysis cup in a reservoir dialysis buffer. Be careful not to trap air bubbles underneath the dialysis cup, which will disrupt replenishment of additives.
Cover the 24-deep well plate with Saran Wrap to prevent evaporation of the reservoir dialysis buffer. Incubate the translation reaction at 26 °C for 16 h.
Protein levels are determined by denaturing electrophoresis with creatine kinase serving as an internal intensity standard.
13. Purification by Density Gradient Ultracentrifugation
Figure 37.4 shows a schematic representation of the purification of proteoliposomes containing membrane proteins produced by wheat germ cell-free translation. After assembly of the density gradient, the proteoliposomes are separated from the cell-free extract proteins by a centrifugation step. In most cases, the proteoliposomes are sharply concentrated at the interface between the 30% Accudenz solution and the upper buffer.
Figure 37.4.
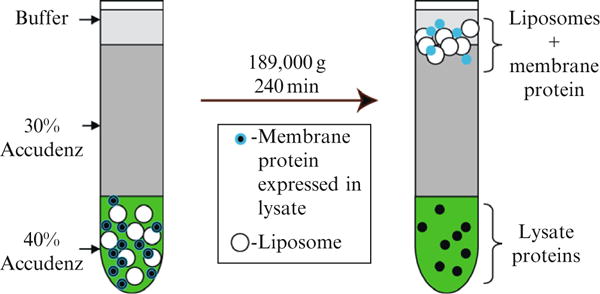
Schematic representation of the purification of proteoliposomes containing membrane proteins produced by cell-free translation.
Figure 37.5 shows a denaturing PAGE analysis of the separation of proteoliposomes obtained by cotranslation of human stearoyl-CoA desaturase (hSCD1) and cytochrome b5 (cytb5) in the wheat germ extract. hSCD1 accounted for ~ 4% of the total protein present after translation in the presence of liposomes. The density gradient purification yielded 24 μg of hSCD1 from a 50-μL translation reaction with greater than 80% purity. The density gradient purification also gave a 25-fold increase in the specific activity of the enzyme, and provided near complete recovery of the enzyme activity from the extract.
Figure 37.5.
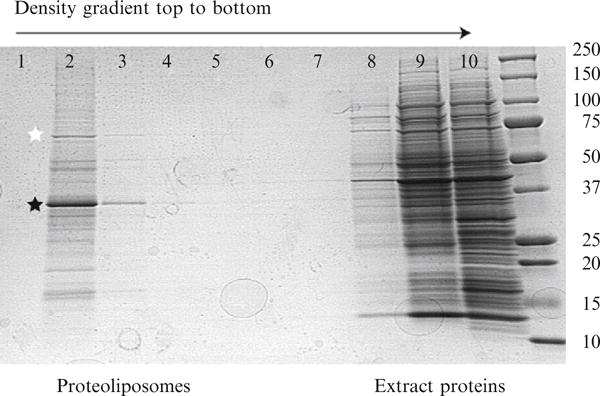
Denaturing PAGE analysis of the density gradient obtained after cell-free translation of hSCD1. The order of fractions from the density gradient is indicated. hSCD1 (black star) and a plant Hsp70-like protein (white star) are noted in lane 2, which represents the interface between the upper buffer (lane 1) and the remainder of the 30% Accudenz layer. The majority of wheat germ extract proteins remain in the 40% Accudenz layer. The rightmost lane contains molecular weight markers (kDa, noted alongside image). Adapted from Goren and Fox (2008).
13.1. Materials and reagents
Accudenz is from Accurate Chemical and Scientific (Westbury, NY). Prepare 80% (w/v) and 30% (w/v) solutions in 25 mM HEPES, pH 7.5, containing 100 mM NaCl and 10% (w/v) glycerol. Store these solutions at room temperature.
Ultraclear centrifuge tubes (5 mm id × 41 mm h) are from Beckman Coulter (Fullerton, CA).
The lipid rehydration buffer is 25 mM HEPES, pH 7.5, containing 100 mM NaCl.
13.1.1. Protocol
Carefully mix 75 μL of 80% (w/v) Accudenz solution with up to 75 μL of the translation reaction, and place the mixed sample in the bottom of an ultraclear centrifuge tube. This creates the 40% Accudenz layer.
Carefully layer 350 μL of 30% (w/v) Accudenz solution on top of the mixture in the centrifuge tube. Minimize mixing of the gradient by using a gel loading pipet tip to add the successive buffer layers.
Carefully layer 100 μL of lipid rehydration buffer on top of two other layers in the centrifuge tube. This is the density gradient tube.
Spin the density gradient tube in an SW 50.1 rotor and L-60 ultracentrifuge for at least 4 h at 45,000 rpm (189,000 × g) and 4 °C. The time required for floatation is dependent on the properties of the proteoliposomes created, so optimization of density gradient and the centrifugation time may be required to obtain the best separation for other proteins.
Carefully remove 60 μL fractions from the top to the bottom of the gradient in order to fractionate the density gradient. Store the individual fractions in separate 1.5-mL microfuge tubes. Typically, proteoliposomes will migrate to the interface between the 30% (w/v) Accudenz solution and the liposome rehydration buffer.
Use denaturing SDS–PAGE to analyze the individual fractions for protein content.
14. Characterization of Proteoliposomes
Another chapter has elegantly described the process of transfer of solubilized membrane proteins from detergent into liposomes for functional assays (Rigaud and Levy, 2003). In this chapter, the liposome is an integral part of the enzyme production and purification process, so functional assays are immediately feasible without further manipulations of the preparation. The facile recovery of proteoliposomes from the cell-free translation reaction by density gradient centrifugation permits functional assays on highly enriched preparations with a minimal investment of time and effort. This provides some obvious advantages in the discovery and characterization of the membrane proteome (Sawasaki et al., 2002b; Wu et al., 2003), a large and still poorly understood fraction of all proteins.
Figure 37.6 shows results of assays for conversion of 14C-labeled stearoyl-CoA (18:0) to oleoyl-CoA (18:1) obtained from different combinations of proteoliposomes of hSCD1 and cyt5b produced by cotranslation or independently with wheat germ extract (Goren and Fox, 2008). There is no desaturase activity in the cell-free extract only control (lane 9). In order to obtain hSCD activity, both cytb5 and Fe2+ must be added (lanes 1–3 versus lanes 4–8). Furthermore, iron and hemin must be optimized to obtain maximal activity (lanes 4–8). Furthermore, in this case, cotranslation of hSCD1 and cytb5 versus separate translation and subsequent mixing of the proteoliposomes gave equivalent catalytic activity (lane 6 versus lane 8).
Figure 37.6.
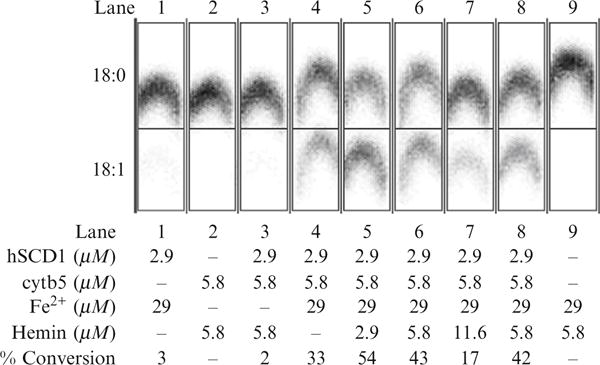
Catalytic activity observed from various reconstitutions of the complex of human stearoyl-CoA desaturase and human cytochrome b5 produced by cell-free translation and purified by density gradient centrifugation. The soluble domain of human cytochrome b5 reductase was expressed in E. coli and added to these assays. The influence of adding Fe2+ (required for hSCD1 activity) and hemin (required for cytb5 activity) is also demonstrated. Lane 1, hSCD1 plus iron. Lane 2, cytb5 plus hemin. Lane 3, cotranslation of hSCD1 and cytb5 supplemented with hemin. Lane 4, cotranslation of hSCD1 and cytb5 supplemented with Fe2+. Lane 5, cotranslation of hSCD1 and cytb5 supplemented with Fe2+ and 0.5 equiv. of hemin. Lane 6, cotranslation of hSCD1 and cytb5 supplemented with Fe2+ and 1 equiv. of hemin. Lane 7, cotranslation of hSCD1 and cytb5 supplemented with Fe2+ and 2 equiv. of hemin. Lane 8, mixture of separately translated and purified hSCD1 with separately translated and purified cytb5 supplemented with Fe2+ and 1 equiv. of hemin. Lane 5, cotranslation of hSCD1 and cytb5 supplemented with Fe2+ and 0.5 equiv. of hemin. Adapted from Goren and Fox (2008).
In many cases, the cell-free extract will not exhibit competing enzymatic reactions that are present in whole cells, microsomes, and other impure natural membrane preparations. This possibility can be experimentally verified by use of the cell-free extract as a control. If verified, the low background reactivity of the cell-free extract can have important implications for discovery of the function of unknown membrane proteins and complexes. Moreover, the absence of competing background reactions may facilitate additional characterizations using site-directed mutagenesis, inhibitors, and other catalytic methods that have been more routinely carried out with purified soluble enzymes.
The availability of functional proteoliposomes also facilitates investigations of the topology of cell-free translated membrane proteins by the combination of proteolytic digestion and mass spectrometry (Speers and Wu, 2007; Wu et al., 2003). In some cases, the ability to perform protein synthesis using radiolabeled AAs, unnatural AAs, or to specifically target one or more AAs with diagnostic mass tags can advance these studies.
Further purification likely requires that the proteoliposomes be dissolved with detergents, much as would be required for purification of a membrane protein starting with a microsomal preparation from a living tissue. However, the combination of cell-free translation, liposomes, and density gradient centrifugation allows these additional purification efforts to start at a much higher level of purity. Monitoring the decrease in light scattering from the proteoliposome as detergent is added provides a simple diagnostic for detergent optimization (Seddon et al., 2004; Womack et al., 1983). New approaches using NMR, analytical ultracentrifugation, and gel filtration can provide insight into the ability of a given detergent to yield monodisperse protein-detergent micelles (Maslennikov et al., 2007; Slotboom et al., 2008), which is generally considered to be an indicator of the compatibility of protein and detergent. It is also possible to perform cell-free translation of membrane proteins in the presence of detergents (Klammt et al., 2007; Nozawa et al., 2007), with the constraints that the detergents used are compatible with the cell-free translation reaction and with solubilization of the nascent membrane protein.
15. Considerations for Scale-Up
With demonstration of suitable behavior using the bilayer or small-volume dialysis reactions, scale-up of protein production is most effectively carried out by use of the dialysis approach. Methods for performing the dialysis reaction in large scale have been published (Madin et al., 2000; Spirin and Swartz, 2008). More recently, a robotic dialysis device has become available to address the need to produce large quantities of proteins using cell-free translation. Wheat germ cell-free translation has been used to solve the NMR structures of numerous soluble proteins (Phillips et al., 2007; Tyler et al., 2005), and more recently an X-ray structure has been solved (Makino et al., 2009). Correspondingly, E. coli cell-free translation has been effectively used to produce membrane proteins for refolding and, in the case of EmrE, preparation of a SeMet-labeled sample for structure determination (Klammt et al., 2007; Chen et al., 2007). To prepare sufficient mRNA for a large-scale protein production, carry out a transcription reaction in a 50 mL conical tube with a total volume of 4 mL of 1 × Transcription Buffer plus Mg containing 4 mM NTPs, 0.05 mg/mL of plasmid DNA, 0.5 unit/μL of SP6 RNA polymerase, and 0.25 unit/μL of RNasin. Incubate the reaction at 37 °C for 3–5 h.
16. Isotopic Labeling for Structural Studies
Because AAs are added to the wheat germ extract as substrates for the protein synthesis, SeMet, 2H-, 13C-, 15N- or other isotopically labeled AAs can be easily substituted. There is no significant metabolism of AAs in wheat germ extract except for alanine transaminase (beta-chloro-L-alanine), aspartate transaminase (aminooxyacetate), and glutamine synthetase (L-methionine sulfoximine), which can be inhibited by the compounds indicated (Endo and Sawasaki, 2006; Morita et al., 2004). Otherwise, the level of isotopic enrichment in translated proteins consistently matches that of the AA precursors used in the translation reactions. 15N and 13C, 15N-labeled AAs are from Cambridge Isotope Laboratories (Andover, MA) or other sources. For NMR studies, the 15N-labeled AAs are prepared at 8 mM, and the 13C, 15N-labeled AAs are prepared at 5 mM, also in Milli-Q water. These preparations are substituted into the standard protocols to give the same final concentrations as described above. Likewise SeMet (Acros Organics, Morris Plains, NJ) is prepared as a 2-mM stock solution and substituted for methionine as described earlier.
17. Conclusions
The process simplifications afforded by the cell-free translation approaches described herein provide significant labor and time advantages, and this conclusion is emphasized when cell-free translation can produce integral membrane proteins and enzymes that cannot be reasonably obtained in a catalytically active state by any other method. By comparison, other methods for the preparation of membrane proteins beginning with expression in living hosts are time-, labor-, and material-intensive. Through application of these methods, uniform samples of membrane proteins can be easily obtained as the starting point for optimization of catalytic assays, purification procedures, antibody production, structure determination, and many other types of studies. This work provides an attractive alternative to the sequence of expressing in a living host, extracting membrane proteins with detergents, and then transferring them back into liposomes or other lipid bilayers for functional studies. It is reasonable to assume that continued application of this approach could open many new avenues to investigations of membrane protein structure and function.
Acknowledgments
This work was supported by NIGMS grant GM50853 to BGF, NIGMS Protein Structure Initiative grant 1U54 GM074901 (J. L. Markley, PI, G. N. Phillips, Jr., and B. G. Fox, Co-Investigators) and NSF East Asia and Pacific Summer Institutes for U.S. Graduate Students to MAG.
References
- Abe M, Hori H, Nakanishi T, Arisaka F, Ogasawara T, Sawasaki T, Kitamura M, Endo Y. Application of cell-free translation systems to studies of cofactor binding proteins. Nucleic Acids Symp Ser (Oxf) 2004;48:143–144. doi: 10.1093/nass/48.1.143. [DOI] [PubMed] [Google Scholar]
- Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High efficiency single step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Expr Purif. 2006;47:562–570. doi: 10.1016/j.pep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Boyer ME, Stapleton JA, Kuchenreuther JM, Wang CW, Swartz JR. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol Bioeng. 2008;99:59–67. doi: 10.1002/bit.21511. [DOI] [PubMed] [Google Scholar]
- CellFree Sciences, Ltd.; Yokohama, Japan: http://www.cfsciences.com. [Google Scholar]
- Chen YJ, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17:373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Goerke AR, Swartz JR. Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol Bioeng. 2008;99:351–367. doi: 10.1002/bit.21567. [DOI] [PubMed] [Google Scholar]
- Goren MA, Fox BG. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr Purif. 2008;62:171–178. doi: 10.1016/j.pep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JF, Ellis MJ, Kigawa T, Yabuki T, Matsuda T, Seki E, Hasnain SS, Yokoyama S. Towards the high-throughput expression of metalloproteins from the Mycobacterium tuberculosis genome. J Synchrotron Radiat. 2005;12:4–7. doi: 10.1107/S0909049504027864. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kitano M, Kato R, Omori A, Endo Y, Tozawa Y. Sequence specificity and efficiency of protein N-terminal methionine elimination in wheat-embryo cell-free system. Protein Expr Purif. 2007;52:59–65. doi: 10.1016/j.pep.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, Hasegawa Y, Endo Y, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc Natl Acad Sci USA. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- Klammt C, Lohr F, Schafer B, Haase W, Dotsch V, Ruterjans H, Glaubitz C, Bernhard F. High level cell-free expression and specific labeling of integral membrane proteins. Eur J Biochem. 2004;271:568–580. doi: 10.1111/j.1432-1033.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- Klammt C, Schwarz D, Lohr F, Schneider B, Dotsch V, Bernhard F. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273:4141–4153. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- Klammt C, Schwarz D, Dotsch V, Bernhard F. Cell-free production of integral membrane proteins on a preparative scale. Methods Mol Biol. 2007;375:57–78. doi: 10.1007/978-1-59745-388-2_3. [DOI] [PubMed] [Google Scholar]
- Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc Natl Acad Sci USA. 2000;97:559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino SI, Bingman CA, Berge S, Larkin A, Fox BG, Phillips GN, Jr, Markley JL. Crystal structure of agmatine iminohydrolase produced by wheat germ cell-free translation. 2009 in preparation. [Google Scholar]
- Maslennikov I, Kefala G, Johnson C, Riek R, Choe S, Kwiatkowski W. NMR spectroscopic and analytical ultracentrifuge analysis of membrane protein detergent complexes. BMC Struct Biol. 2007;7:74. doi: 10.1186/1472-6807-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J Biomol NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Tomikawa C, Toyooka T, Ochi A, Takano Y, Takayanagi N, Abe M, Endo Y, Hori H. Production of yeast tRNA (m(7)G46) methyl-transferase (Trm8-Trm82 complex) in a wheat germ cell-free translation system. J Biotechnol. 2008;133:453–460. doi: 10.1016/j.jbiotec.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Morita EH, Shimizu M, Ogasawara T, Endo Y, Tanaka R, Kohno T. A novel way of amino acid-specific assignment in 1H–15N HSQC spectra with a wheat germ cell-free protein synthesis system. J Biomol NMR. 2004;30:37–45. doi: 10.1023/B:JNMR.0000042956.65678.b8. [DOI] [PubMed] [Google Scholar]
- Nomura SM, Kondoh S, Asayama W, Asada A, Nishikawa S, Akiyoshi K. Direct preparation of giant proteo-liposomes by in vitro membrane protein synthesis. J Biotechnol. 2008;133:190–195. doi: 10.1016/j.jbiotec.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Nosjean O, Roux B. Anchoring of glycosylphosphatidylinositol-proteins to liposomes. Methods Enzymol. 2003;372:216–232. doi: 10.1016/S0076-6879(03)72012-6. [DOI] [PubMed] [Google Scholar]
- Nozawa A, Nanamiya H, Miyata T, Linka N, Endo Y, Weber AP, Tozawa Y. A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 2007;48:1815–1820. doi: 10.1093/pcp/pcm150. [DOI] [PubMed] [Google Scholar]
- Phillips GN, Jr, Fox BG, Markley JL, Volkman BF, Bae E, Bitto E, Bingman CA, Frederick RO, McCoy JG, Lytle BL, Pierce BS, Song J, et al. Structures of proteins of biomedical interest from the Center for Eukaryotic Structural Genomics. J Struct Funct Genomics. 2007;8:73–84. doi: 10.1007/s10969-007-9023-6. [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Hasegawa Y, Tsuchimochi M, Kasahara Y, Endo Y. Construction of an efficient expression vector for coupled transcription/translation in a wheat germ cell-free system. Nucleic Acids Symp Ser. 2000;44:9–10. doi: 10.1093/nass/44.1.9. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Hasegawa Y, Tsuchimochi M, Kamura N, Ogasawara T, Kuroita T, Endo Y. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 2002a;514:102–105. doi: 10.1016/s0014-5793(02)02329-3. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Ogasawara T, Morishita R, Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci USA. 2002b;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Klammt C, Koglin A, Lohr F, Schneider B, Dotsch V, Bernhard F. Preparative scale cell-free expression systems: New tools for the large scale preparation of integral membrane proteins for functional and structural studies. Methods. 2007;41:355–369. doi: 10.1016/j.ymeth.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Sobrado P, Goren MA, James D, Amundson CK, Fox BG. A protein structure initiative approach to expression, purification, and in situ delivery of human cytochrome b5 to membrane vesicles. Protein Expr Purif. 2008;58:229–241. doi: 10.1016/j.pep.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, Wu CC. Proteomics of integral membrane proteins—Theory and application. Chem Rev. 2007;107:3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- Spirin AS, Swartz JR, editors. Cell-Free Protein Synthesis-Methods and Protocols. WILEY-VCH Verlag Gmbh & Co.; Weinheim: 2008. [Google Scholar]
- Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5:267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- Tyler RC, Aceti DJ, Bingman CA, Cornilescu CC, Fox BG, Frederick RO, Jeon WB, Lee MS, Newman CS, Peterson FC, Phillips GN, Jr, Shahan MN, et al. Comparison of cell-based and cell-free protocols for producing target proteins from the Arabidopsis thaliana genome for structural studies. Proteins. 2005;59:633–643. doi: 10.1002/prot.20436. [DOI] [PubMed] [Google Scholar]
- Vinarov DA, Markley JL. High-throughput automated platform for nuclear magnetic resonance-based structural proteomics. Expert Rev Proteomics. 2005;2:49–55. doi: 10.1586/14789450.2.1.49. [DOI] [PubMed] [Google Scholar]
- Vinarov DA, Lytle BL, Peterson FC, Tyler EM, Volkman BF, Markley JL. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat Methods. 2004;1:149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- Vinarov DA, Loushin Newman CL, Markley JL. Wheat germ cell-free platform for eukaryotic protein production. FEBS J. 2006;273:4160–4169. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- Womack MD, Kendall DA, Macdonald RC. Detergent effects on enzyme activity and solubilization of lipid bilayer membranes. Biochim Biophys Acta. 1983;733:210–215. doi: 10.1016/0005-2736(83)90524-2. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- Wuu JJ, Swartz JR. High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim Biophys Acta. 2008;1778:1237–1250. doi: 10.1016/j.bbamem.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Protein expression systems for structural genomics and proteomics. Curr Opin Chem Biol. 2003;7:39–43. doi: 10.1016/s1367-5931(02)00019-4. [DOI] [PubMed] [Google Scholar]


