Abstract
Survival rates for osteosarcoma, the most common primary bone cancer, have changed little over the past three decades and are particularly low for patients with metastatic disease. We conducted a multi-institutional genome-wide association study (GWAS) to identify germline genetic variants associated with overall survival in 632 patients with osteosarcoma including 523 patients of European ancestry and 109 from Brazil. We conducted a time-to-event analysis and estimated hazard ratios (HR) and 95% confidence intervals (CI) using Cox proportional hazards models, with and without adjustment for metastatic disease. The results were combined across the European and Brazilian case sets using a random-effects meta-analysis. The strongest association after meta-analysis, was for rs3765555 at 9p24.1, which was inversely associated with overall survival (HR=1.76; 95% CI 1.41-2.18, P = 4.84×10−7). After imputation across this region, the combined analysis identified two SNPs that reached genome-wide significance. The strongest single association was with rs55933544 (HR=1.9; 95% CI 1.5-2.4; P=1.3×10−8), which localizes to the GLDC gene, adjacent to the IL33 gene and was consistent across both the European and Brazilian case sets. Using publicly available data, the risk allele was associated with lower expression of IL33 and low expression of IL33 was associated with poor survival in an independent set of patients with osteosarcoma. In conclusion, we have identified the GLDC/IL33 locus on chromosome 9p24.1 as associated with overall survival in patients with osteosarcoma. Further studies are needed to confirm this association and shed light on the biological underpinnings of this susceptibility locus.
Keywords: Osteosarcoma, overall survival, genome-wide association study, osteosarcoma specific survival
INTRODUCTION
Osteosarcoma is the most common primary bone cancer in children and adolescents.1–4 The introduction of effective neo-adjuvant and adjuvant chemotherapy in the 1980s resulted in improved long-term survival for non-metastatic osteosarcoma patients, increasing survival from 20%–30% to more than 70%.4–7 However, over the past three decades, there has been little improvement in survival rates for patients with metastatic disease at diagnosis8 with 5-year overall survival rates remaining at 25-30%.9, 10 Several factors have been suggested to influence survival of patients with osteosarcoma11, including age at diagnosis, metastatic disease at presentation, tumor histology, blood alkaline phosphatase levels, tumor size and location, and response to chemotherapy (estimated by the percentage of tumor necrosis after chemotherapy).12–14 Recently, we reported that germline genetic variants in the NFIB gene locus (9p23-9p22.3) are associated with metastatic disease at osteosarcoma diagnosis, suggesting that genetic susceptibility could contribute to survival.15
There is growing interest in whether germline genetic variants could influence outcomes of patients with cancer. A population-based family study showed that cancer-specific survival can be partly related to inherited factors within families, suggesting there are germline genetic determinants of survival.16 While earlier candidate gene studies have identified variants associated with prognosis that have not been confirmed, more recent large genome-wide association studies (GWAS) have identified associations between common SNPs and survival in adult cancers of the pancreas,17 breast,18–23 ovary,24 and in a rare pediatric cancer, neuroblastoma.25–27 There have also been exploratory pharmacogenomic studies of pediatric Ewing sarcoma and osteosarcoma that have identified SNPs associated with response to treatment and survival,28–31 although most await further validation. We performed a GWAS in order to explore whether germline genetics may contribute to survival in patients with osteosarcoma.
METHODS
Study populations
A summary of the participating studies is provided in Supplemental Table 1. Previously, we reported 689 histologically confirmed osteosarcoma patients of >80% European ancestry based on a STRUCTURE analysis32 employing principal components analyses of a set of 12,000 unlinked markers (pairwise r2 < 0.004).15, 33 A subset of 523 European ancestry osteosarcoma patients had available data on mortality (European set) for a survival analysis. After performing the GWAS for survival in this data set, we evaluated the most promising SNPs from the European set (P < 10−4) in 109 Brazilian osteosarcoma patients from the Instituto de Oncologia Pediátrica GRAACC/UNIFESP and Universidade Federal de Sao Paulo, Brazil (Brazilian set; Supplemental Table 1). Participating subjects provided informed consent under the auspices of local Institutional Review Boards.
Genotyping and quality control
All subjects were previously genotyped as part of our osteosarcoma susceptibility GWAS (dbGaP Study Accession: phs000734.v1.p1).33 In brief, genotyping was conducted on the Illumina OmniExpress BeadChip, SNPs included in the analyses were autosomal with a minor allele frequency (MAF) of 5% or more; had a 90% or more completion rate; and no evidence of deviation from Hardy-Weinberg equilibrium (P > 10−7). SNPs were also excluded if they had abnormal heterozygosity values. After quality control assessment, 510,856 SNPs were advanced in our survival analysis.
Statistical analyses
We conducted a time-to-event analysis to investigate the effect of genetic variation on overall survival. The outcome variable of interest was time until the event of death. The overall survival time was calculated as the time from the date of osteosarcoma diagnosis until the date of death for those deceased or the last date known to be alive; patients were censored at the last date known to be alive or when lost to follow-up. All events were identified and verified through medical record review and/or death certificates at each participating study center. We modeled each genome-wide association between one or more SNPs and survival using Cox proportional hazards regression and estimated hazard ratios (HR) and 95% confidence intervals (CI) per copy of the minor allele (log-additive genetic model).34 We tested the hazards proportionality assumption (i.e., the hazard ratio is constant over time)35 of the Cox model using Schoenfeld’s residuals;36, 37 we did not detect nominally significant violations of the proportional hazards assumption.
Cox models were adjusted for age at diagnosis, gender, significant principal components, and study/center (except for the Brazilian study, since all samples were from the same hospital). We did not adjust for metastatic disease agnostically at the genome-wide level, because some SNPs may be associated with metastatic disease and survival, as we have shown previously,15 and thus adjusting for potential intermediate factors that lie on the causal pathway could introduce bias.38–40 However, since metastatic disease is a prognostic factor for overall survival,12 we performed sensitivity analyses for the top signals by also adjusting for metastatic disease .41 We constructed Kaplan-Meier survival curves42 for the top SNPs under dominant models and compared their statistical equivalence for each model with the log-rank test.
SNPs that reached P < 10−4 in the European set were followed-up in the Brazilian set. We used a random-effects meta-analysis with inverse-variance weighting to estimate the summary effect across the sets.43 We evaluated between-sets heterogeneity using Cochran’s Q chi-squared statistic and quantified heterogeneity with the I2 metric.44 SNPs that demonstrated significant between-set heterogeneity (P < 0.05) were excluded.
Based on our GWAS results, we conducted region-specific imputation analysis of flanking SNPS, namely, 1 Mb region on either side of the strongest SNP using the IMPUTE2 software and the reference data from the 1000 Genomes project (Phase 3 genotype data).45
To investigate whether signals in the same genomic region represent independent associations, we conducted conditional analyses by adjusting the Cox models for the top SNP in each region as applicable.
Statistical analyses were performed in Stata version 13 and R 3.0.2. All P-values are two-sided.
eQTL and survival-expression analyses
We performed expression quantitative trait locus (eQTL) based analyses using publicly available genotype and expression data from 29 osteosarcoma tumors (GSE33383)46 and separately in osteosarcoma tumors from 89 patients included in the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) osteosarcoma dataset (http://ocg.cancer.gov/programs/target).47 The data used for TARGET are available at dbGaP accession phs000218, accession phs000468, in which we conducted survival-expression analysis.47 For the 127 patients from the combined dataset (Kuijjer et al46 and Buddingh et al48) survival-expression was analyzed using the R2: Genomics analysis and visualization platform (http://r2.amc.nl/).49
Bioinformatic analyses
Linkage disequilibrium (LD) was evaluated with r2 based on the 1000 Genomes Project (Phase 3 genotype data)45 using LDlink (http://analysistools.nci.nih.gov/LDlink/).23 Chromosome location for human genome assembly hg19 was retrieved from the National Center for Biotechnology Information’s (NCBI) Gene database (http://www.ncbi.nlm.nih.gov/gene/). Genomic annotation of SNP markers was conducted using the Encyclopedia of DNA Elements (ENCODE)50 tool HaploReg51 and RegulomeDB52 for all cell lines. Surrogate SNPs were identified using the 1000 Genomes data for individuals of European ancestry with an r2 > 0.4 and within ±500kb. LocusZoom53 was used to plot regional associations.
RESULTS
Patient characteristics
Table 1 shows the characteristics of the patients included in this study. There were 632 total osteosarcoma patients included in the overall survival analyses. In the European set, there were 170 (33%) mortality events, and 37 (34%) in the Brazilian set. Age (P < 0.001) and presence of metastases at diagnosis (P < 0.001) were associated with patient survival (Table 1, Supplemental Figure 1).
Table 1.
Patient characteristics in the European and Brazilian set
| Endpoint | European* | Brazil | Combined Analysis | ||
|---|---|---|---|---|---|
|
| |||||
| Overall Survival | N=523 | N=109 | N=632 | 75% ST† (years) | P-value†† |
| Vital status, N (%) | |||||
| Dead | 170 (33) | 37 (34) | 207 (33) | 2.8 | |
| Alive | 353 (67) | 72 (66) | 425 (67) | ||
| Age (years) | <0.001 | ||||
| <25 | 432 (83) | 93 (85) | 525 (83) | 3.3 | |
| ≥25 to<60 | 69 (13) | 12 (11) | 81 (13) | 3.4 | |
| ≥60 | 21 (4) | 0 (0) | 21 (3) | 4.3 | |
| Missing | 1 (0) | 4 (4) | 5 (1) | ||
| Gender, N (%) | 0.737 | ||||
| Males | 295 (56) | 58 (53) | 353 (56) | 2.9 | |
| Females | 228 (44) | 51 (47) | 279 (44) | 2.7 | |
| Metastasis at diagnosis, N (%) | <0.001 | ||||
| Yes | 131 (25) | 40 (37) | 171 (27) | 4.2 | |
| No | 392 (75) | 69 (63) | 455 (72) | 1.5 | |
All subjects included in the European set were of >80% European ancestry.
Shows the time at which 75% of patients had not experienced the event of interest (i.e. death or progression).
P-values are from log-rank test.
ST: survival time; NA: non-applicable
SNPs associated with overall survival
In a case-case analysis, 81 SNPs were associated with overall survival at P < 10−4 in the European set (Supplemental Table 2) and were followed-up in the independent set of 109 osteosarcoma cases from Brazil. The strongest association with overall survival in the European set is at chromosome 5q11.1 tagged by rs1030228 (located 14kb 5' of NDUFS4) with a HR for mortality of 1.71 (95% CI 1.39-2.12, P = 7.10×10−7; Supplemental Figure 2, Supplemental Table 2). However, this SNP is not associated with survival in the Brazilian study (P=0.80). There is a large degree of between-set heterogeneity (Phet=0.052, I2=73.5%) for this variant (Supplemental Table 2).
In the combined analysis, the strongest association was SNP rs3765555, which is inversely associated with overall survival (HR=1.76 per copy of the A allele, 95% CI 1.41-2.18, P=4.84×10−7; I2=0%; Table 2, Figure 1A). This SNP is located in intron 23 of the glycine dehydrogenase (decarboxylating) gene (GLDC) on chromosome 9p24.1. The MAF for rs3765555 in the European (MAF=0.26) and Brazilian (MAF=0.21) populations are similar and we did not observe significant between-set heterogeneity (Phet=0.34; Table 2, Supplemental Table 2).
Table 2.
SNPs associated with overall survival.
| SNP* | Method | Gene Locus** | Position† | Set | MAF | Unadjusted for metastatic disease | Adjusted for metastatic disease | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||||||
| rs3765555-C|A | Genotyped | GLDC | Chr9: 6535956 | European | 0.259 | 1.67 (1.32–2.13) | 2.70×10−5 | 1.71 (1.34–2.18) | 1.60×10−5 |
| Brazil | 0.205 | 2.23 (1.31–3.81) | 3.29×10−3 | 2.12 (1.27–3.53) | 3.87×10−3 | ||||
| Combined | 1.76 (1.41–2.18) | 4.84×10−7 | 1.78 (1.43–2.21) | 2.74×10−7 | |||||
| rs55933544-C|T | Imputed | GLDC | Chr9: 6534080 | European | 0.231 | 1.85 (1.44–2.37) | 1.58×10−6 | 1.91 (1.49–2.45) | 3.20×10−7 |
| Brazil | 0.231 | 2.11 (1.21–3.69) | 8.44×10−3 | 1.98 (1.16–3.38) | 0.012 | ||||
| Combined | 1.89 (1.50–2.37) | 4.81×10−8 | 1.92 (1.53–2.41) | 1.34×10−8 | |||||
Alleles are shown as major/minor.
Gene locus information is based on the GENCODE data from HaploReg v2.
Position is based on hg19.
Hazard ratios are shown per copy of the minor allele in the discovery stage.
MAF: minor allele frequency; HR: hazard ratio; CI: confidence interval
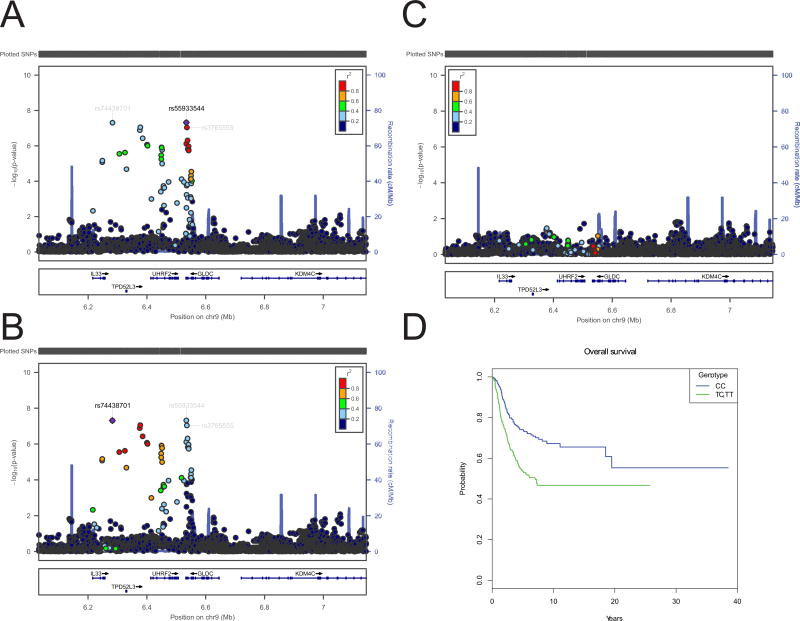
Figure 1.
Regional plots of the combined association results, recombination hotspots and linkage disequilibrium (LD) for the 9p24.1 region that harbors rs55933544 and rs74438701 that are associated with overall survival. Results are shown for unconditional (A, B) and conditional (C) analyses. Also shown is the Kaplan-Meier curve (D) for overall survival for the strongest SNP (rs55933544) under a dominant model in the combined European and Brazilian sets. In panels A-C, Y-axes represent the statistical significance (−log10 transformed P values) of SNP association results from a trend test (left) and the recombination rate (right). SNPs are color-coded based on pairwise linkage disequilibrium (r2) with the most statistically significant SNP. The most statistically significant SNP is labeled and shown in purple. Allelic P values are the P-values from Cox models. Physical locations of the SNPs are based on NCBI human genome build 36, and gene annotation was based on the NCBI RefSeq genes from the UCSC Genome Browser.
In a further exploration of the promising regions, we imputed SNPs across a 1 Mb region centered on rs3765555 to further evaluate this locus. After a random-effects meta-analysis for the imputed SNPs, we identified rs55933544 (Table 2, Figure 1A) as the SNP most strongly associated with overall survival (HR=1.89 per copy of the T allele, 95% CI 1.50-2.37; P=4.81×10−8; I2=0%), which is in strong LD with rs3765555 (r2=0.86 in Europeans and r2=0.94 in admixed Americans). The results remained the same after adjustment for metastatic disease at diagnosis (Table 2), suggesting that the 9p24.1 locus marked by rs55933544 affects overall survival independent of metastatic disease at genome-wide significance (HR=1.92 per copy of the T allele, 95% CI 1.53-2.41; P=1.34×10−8; I2=0%). rs55933544 (chr9:6534080) was not correlated with SNP rs7034162 at 9p23-9p22.3 (chr9:14190287) that we previously identified as associated with metastatic disease at osteosarcoma diagnosis (r2=0.0008, 1,000 Genomes Project CEU data).15
A second SNP at 9p24.1, rs74438701 located approximately 25kb downstream of the interleukin 33 (IL33) gene, is also inversely associated with overall survival in the combined analysis (HR=2.00 per copy of the C allele, 95% CI 1.56-2.57, P=4.90×10−8; I2=0%; Figure 1B). However, the conditional analysis showed that rs74438701 is not an independent signal (Figure 1C).
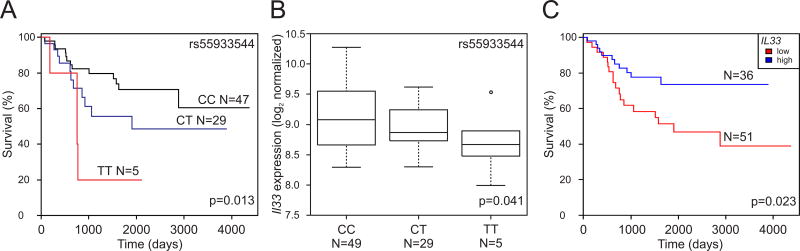
Kaplan-Meier curve analysis indicate a statistically significant difference between survival rates over time (log-rank test P < 0.001) for both the dominant (Figure 1D) and a multiplicative model (data not shown) for rs55933544. In addition, we confirmed this association in an independent dataset of 89 cases (TARGET47; log-rank P=0.013; Figure 2A).
Figure 2.
IL33 expression levels associated with survival in osteosarcoma patients. (A) Kaplan-Meier curve for overall survival by rs55933544 genotype; (B) eQTL of rs55933544 and IL33; and, (C) patient survival by IL33 low and high expression levels (independent of genotype), all using TARGET data.
We examined the set of highly correlated surrogate SNPs (n=31) across the 9p24.1 region (based on an r2>0.4, 1,000 Genomes Project CEU data) to identify putative regulatory elements using the ENCODE data resource50 tools HaploReg51 and RegulomeDB52 (Supplemental Table 3). A subset of the surrogate SNPs (n=29) are located in predicted promoter and/or enhancer histone marks, DNAse sensitivity regions, and/or transcription factor binding sites in a variety of different cell types and may have a functional impact (Supplemental Table 3).
IL33 expression levels associated with survival
We performed expression quantitative trait locus (eQTL) analyses to evaluate whether rs55933544 was associated with expression of GLDC, IL33 or other neighboring protein-encoding genes, using publicly available expression and genotyping data on osteosarcoma tumors. Interestingly, a previous eQTL was observed between rs55933544 and IL33 expression in human skin54 and human brain tissue55 (Supplemental Table 3). We found that the risk allele (T) of rs55933544 was significantly associated with a decrease in IL33 expression in both osteosarcoma tumor data sets from TARGET47 (N=83, P=0.041) and Kuijjer et al.46 (N=29, P=0.020) (Figure 2B and Supplemental Figure 3). In addition, lower expression of IL33 in osteosarcoma tissue was independently associated with worse osteosarcoma patient survival in TARGET47 (log-rank test P=0.023; Figure 2C) and Kuijjer et al.46 (raw P=7.9x10−3; Supplemental Figure 3). There was no association between rs55933544 genotypes and expression of GLDC or other nearby protein-encoding genes (TPD52L3, UHRF2 and KDM4C; data not shown).
DISCUSSION
We conducted a genome-wide association study for overall survival in osteosarcoma cases using data from a multi-stage, international collaborative effort.33 One locus, GLDC/IL33 at 9p24.1, was associated with overall survival of patients with osteosarcoma, which suggests that germline genetics can influence osteosarcoma outcomes, independent of metastatic disease. Here we observed moderate to large effect sizes for a SNP associated with overall survival, a finding similar to that observed in our GWAS of metastatic disease at osteosarcoma diagnosis.15 The observed effect sizes are also comparable to GWAS of other pediatric and young adulthood cancers,25, 56–58 and higher than those observed in adult GWAS of common cancer susceptibility.59–61
The most promising signal for overall survival in patients with osteosarcoma localizes to the 9p24.1 region, downstream and independent of the NFIB gene locus (9p23-9p22.3) previously reported for metastatic disease.15 The SNP marker, rs55933544, in the GLDC gene region is associated with decreased survival. High expression of GDLC has been associated with poor survival in other cancers,62, 63 however, we did not detect an eQTL for rs55933544 and GDLC in osteosarcoma cells. Interestingly, rs55933544 alleles have also been associated with expression of the nearby gene, IL3354, 55 and we detected an eQTL with IL33 in osteosarcoma cells. In addition, lower expression of IL33 was associated with poor survival in patients with osteosarcoma. IL-33 is an inhibitor of bone reabsorption, blocking osteoclastic activity,64 which may be important in osteosarcoma. Lower levels of IL-33 have also been associated with worse prognosis or more advanced disease in several other tumor types,65–67 consistent with our data.
This exploratory study requires further follow up and has limitations. Treatment likely varied among patients in our studies, but we could not control for this because treatment information was not available. However, it is unlikely that individual treatment modalities varied systematically by germline genotype. We also performed a combined analysis with osteosarcoma patients from Brazil, with a relatively small sample size. LD patterns differ in many parts of the genome between admixed Brazil populations and Europeans, and this could lead to false negative results in our analysis. This could explain the lack of replication in Brazilian patients of some of the top SNPs in the European osteosarcoma patients. However, an important strength, is that this two-stage design also reduces the likelihood of false positive results.
In conclusion, we provide evidence that germline genetic variants are associated with overall survival in osteosarcoma patients. These findings warrant follow-up in additional populations and functional characterization to investigate the biologic mechanisms by which polymorphisms at this locus impact survival.
Supplementary Material
Novelty and Impact.
To date, few prognostic factors have been identified associated with survival in patients with osteosarcoma. The authors conducted a genome-wide association study (GWAS) of overall survival in two sets of patients with osteosarcoma. They identified a common single nucleotide polymorphism (SNP), rs55933544, located in the GLDC gene on chromosome 9, associated with poor survival. The rs55933544 risk allele was associated with lower expression of the nearby gene, IL33. These findings, if replicated in additional populations, form the foundation for future studies of the molecular basis of the association of the GLDC/IL33 (rs55933544) variant with survival in osteosarcoma.
Acknowledgments
Funding Support
This study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
This work was supported by the Bone Cancer Research Trust UK to A.M.F.
Research is supported by the Chair's Grant U10 CA98543 and Human Specimen Banking Grant U24 CA114766 of the Children's Oncology Group from the National Cancer Institute, National Institutes of Health, Bethesda, MD, USA. Additional support for research is provided by a grant from the WWWW (QuadW) Foundation, Inc. to the Children's Oncology Group.
This work was supported by grants to I.L.A. and J.S.W. from the Ontario Research Fund, and Canadian Foundation for Innovation.
This study was also supported by biobank grants from the Regione Emilia-Romagna, by the infrastructure and personnel of the Royal National Orthopaedic Hospital Musculoskeletal Research Programme and Biobank. Support was also provided to A.M.F. (UCL) by the National Institute for Health Research UCLH Biomedical Research Centre, and UCL Experimental Cancer Centre, funding from PI13/01476, FIS, ISCIII and La Fundación Bancaria "La Caixa”, Fundación Caja Navarra to AP-G, and AECC project to F.L.
The International Sarcoma Kindred Study was supported by the Rainbows for Kate Foundation, the Liddy Shriver Sarcoma Initiative, the Victorian Cancer Agency, the Australian National Health and Medical Research Council (APP1004017) and Cancer Australia (APP1067094).
Abbreviations used
- GWAS
genome-wide association study
- HR
hazard ratio
- CI
confidence intervals
- MAF
minor allele frequency
- LD
linkage disequilibrium
- ENCODE
Encyclopedia of DNA Elements
- NCBI
National Center for Biotechnology Information
- eQTL
expression quantitative trait loci
- SNP
single nucleotide polymorphisms
- GLDC
glycine dehydrogenase (decarboxylating) gene
- IL33
Interleukin 33 gene
- NFIB
Nuclear Factor I B gene
- TPD52L3
Tumor Protein D52 Like 3 gene;
- KDM4C
Lysine Demethylase 4C gene
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Stiller CA, Bielack SS, Jundt G, Steliarova-Foucher E. Bone tumours in European children and adolescents, 1978–1997. Report from the Automated Childhood Cancer Information System project. European journal of cancer. 2006;42:2124–35. doi: 10.1016/j.ejca.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary C, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al. SEER Cancer Statistics Review, 1975–2011, National Cancer Institute. Bethesda, MD: 2014. [Google Scholar]
- 3.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–34. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill J, Ahluwalia MK, Geller D, Gorlick R. New targets and approaches in osteosarcoma. Pharmacology & therapeutics. 2013;137:89–99. doi: 10.1016/j.pharmthera.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer treatment reviews. 2014;40:523–32. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015;39:593–9. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–91. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 9.Bielack S, Carrle D, Casali PG, Group EGW. Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2009;20(Suppl 4):137–9. doi: 10.1093/annonc/mdp154. [DOI] [PubMed] [Google Scholar]
- 10.Kempf-Bielack B, Bielack SS, Jurgens H, Branscheid D, Berdel WE, Exner GU, Gobel U, Helmke K, Jundt G, Kabisch H, Kevric M, Klingebiel T, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS) Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:559–68. doi: 10.1200/JCO.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 11.Bramer JA, van Linge JH, Grimer RJ, Scholten RJ. Prognostic factors in localized extremity osteosarcoma: a systematic review. European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2009;35:1030–6. doi: 10.1016/j.ejso.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Pakos EE, Nearchou AD, Grimer RJ, Koumoullis HD, Abudu A, Bramer JA, Jeys LM, Franchi A, Scoccianti G, Campanacci D, Capanna R, Aparicio J, et al. Prognostic factors and outcomes for osteosarcoma: an international collaboration. European journal of cancer. 2009;45:2367–75. doi: 10.1016/j.ejca.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Bernthal NM, Federman N, Eilber FR, Nelson SD, Eckardt JJ, Eilber FC, Tap WD. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118:5888–93. doi: 10.1002/cncr.27651. [DOI] [PubMed] [Google Scholar]
- 14.Pakos EE, Ioannidis JP. The association of P-glycoprotein with response to chemotherapy and clinical outcome in patients with osteosarcoma. A meta-analysis. Cancer. 2003;98:581–9. doi: 10.1002/cncr.11546. [DOI] [PubMed] [Google Scholar]
- 15.Mirabello L, Koster R, Moriarity BS, Spector LG, Meltzer PS, Gary J, Machiela MJ, Pankratz N, Panagiotou OA, Largaespada D, Wang Z, Gastier-Foster JM, et al. A Genome-Wide Scan Identifies Variants in NFIB Associated with Metastasis in Patients with Osteosarcoma. Cancer discovery. 2015;5:920–31. doi: 10.1158/2159-8290.CD-15-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstrom LS, Hall P, Hartman M, Wiklund F, Gronberg H, Czene K. Familial concordance in cancer survival: a Swedish population-based study. The Lancet Oncology. 2007;8:1001–6. doi: 10.1016/S1470-2045(07)70282-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Kraft P, Stolzenberg-Solomon R, Steplowski E, Brotzman M, Xu M, Mudgal P, Amundadottir L, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2014;63:152–60. doi: 10.1136/gutjnl-2012-303477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, Bojesen SE, Nordestgaard BG, Flyger H, Milne RL, Arias JI, Menendez P, et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. Journal of the National Cancer Institute. 2010;102:650–62. doi: 10.1093/jnci/djq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Q, Schmidt MK, Kraft P, Canisius S, Chen C, Khan S, Tyrer J, Bolla MK, Wang Q, Dennis J, Michailidou K, Lush M, et al. Identification of novel genetic markers of breast cancer survival. Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/djv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fagerholm R, Schmidt MK, Khan S, Rafiq S, Tapper W, Aittomaki K, Greco D, Heikkinen T, Muranen TA, Fasching PA, Janni W, Weinshilboum R, et al. The SNP rs6500843 in 16p13.3 is associated with survival specifically among chemotherapy-treated breast cancer patients. Oncotarget. 2015;6:7390–407. doi: 10.18632/oncotarget.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu XO, Long J, Lu W, Li C, Chen WY, Delahanty R, Cheng J, Cai H, Zheng Y, Shi J, Gu K, Wang WJ, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 2012;72:1182–9. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrdahl M, Canzian F, Lindstrom S, Shui I, Black A, Hoover RN, Ziegler RG, Buring JE, Chanock SJ, Diver WR, Gapstur SM, Gaudet MM, et al. Association of breast cancer risk loci with breast cancer survival. Int J Cancer. 2015;137:2837–45. doi: 10.1002/ijc.29446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirie A, Guo Q, Kraft P, Canisius S, Eccles DM, Rahman N, Nevanlinna H, Chen C, Khan S, Tyrer J, Bolla MK, Wang Q, et al. Common germline polymorphisms associated with breast cancer-specific survival. Breast Cancer Res. 2015;17:58. doi: 10.1186/s13058-015-0570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun R, Finney R, Yan C, Chen QR, Hu Y, Edmonson M, Meerzaman D, Buetow K. Discovery analysis of TCGA data reveals association between germline genotype and survival in ovarian cancer patients. PloS one. 2013;8:e55037. doi: 10.1371/journal.pone.0055037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, Diamond M, Carpenter EL, Winter C, Lee H, Jagannathan J, Latorre V, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–30. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, Glessner J, Kim C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, McDaniel LD, Diamond M, Hart LS, Zhu S, Durbin AD, Abraham BJ, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–21. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hattinger CM, Serra M. Role of pharmacogenetics of drug-metabolizing enzymes in treating osteosarcoma. Expert Opin Drug Metab Toxicol. 2015;11:1449–63. doi: 10.1517/17425255.2015.1060220. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Pinto S, Pita G, Patino-Garcia A, Garcia-Miguel P, Alonso J, Perez-Martinez A, Sastre A, Gomez-Mariano G, Lissat A, Scotlandi K, Serra M, Ladenstein R, et al. Identification of genetic variants in pharmacokinetic genes associated with Ewing Sarcoma treatment outcome. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2016;27:1788–93. doi: 10.1093/annonc/mdw234. [DOI] [PubMed] [Google Scholar]
- 30.Vos HI, Coenen MJ, Guchelaar HJ, Te Loo DM. The role of pharmacogenetics in the treatment of osteosarcoma. Drug Discov Today. 2016;21:1775–86. doi: 10.1016/j.drudis.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 31.Serra M, Hattinger CM. The pharmacogenomics of osteosarcoma. Pharmacogenomics J. 2017;17:11–20. doi: 10.1038/tpj.2016.45. [DOI] [PubMed] [Google Scholar]
- 32.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savage SA, Mirabello L, Wang Z, Gastier-Foster JM, Gorlick R, Khanna C, Flanagan AM, Tirabosco R, Andrulis IL, Wunder JS, Gokgoz N, Patino-Garcia A, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45:799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox DR. Regression Models and Life-Tables. J R Stat Soc B. 1972;34 187-+ [Google Scholar]
- 35.Kleinbaum DG, Klein M. Survival analysis : a self-learning text. 3. xv. New York: Springer; 2012. p. 700. [Google Scholar]
- 36.Schoenfeld D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–41. [Google Scholar]
- 37.Harrell FE, Lee KL. On Choosing Logistic and Cox Regression-Models and Verifying Their Assumptions. Am J Epidemiol. 1986;124:543. [Google Scholar]
- 38.VanderWeele TJ, Tchetgen Tchetgen EJ, Halloran ME. Interference and Sensitivity Analysis. Statistical science : a review journal of the Institute of Mathematical Statistics. 2014;29:687–706. doi: 10.1214/14-STS479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schisterman EF, Cole SR, Platt RW. Overadjustment bias and unnecessary adjustment in epidemiologic studies. Epidemiology. 2009;20:488–95. doi: 10.1097/EDE.0b013e3181a819a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenland S. Quantifying biases in causal models: classical confounding vs collider-stratification bias. Epidemiology. 2003;14:300–6. [PubMed] [Google Scholar]
- 41.VanderWeele TJ. Explanation in causal inference : methods for mediation and interaction. xvi. New York: Oxford University Press; 2015. p. 706. [Google Scholar]
- 42.Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. J Am Stat Assoc. 1958;53:457. [Google Scholar]
- 43.Panagiotou OA, Willer CJ, Hirschhorn JN, Ioannidis JP. The power of meta-analysis in genome-wide association studies. Annual review of genomics and human genetics. 2013;14:441–65. doi: 10.1146/annurev-genom-091212-153520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuijjer ML, Peterse EF, van den Akker BE, Briaire-de Bruijn IH, Serra M, Meza-Zepeda LA, Myklebost O, Hassan AB, Hogendoorn PC, Cleton-Jansen AM. IR/IGF1R signaling as potential target for treatment of high-grade osteosarcoma. BMC Cancer. 2013;13:245. doi: 10.1186/1471-2407-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.TARGET. Therapeutically Applicable Research to Generate Effective Treatments. [Google Scholar]
- 48.Buddingh EP, Kuijjer ML, Duim RA, Burger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC, Lankester AC, Cleton-Jansen AM. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–9. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 49.R2: Genomics Analysis and Visualization Platform. http://r2.amc.nl.
- 50.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, Consortium UKBE, North American Brain Expression C. Coin L, de Silva R, Cookson MR, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung CC, Kanetsky PA, Wang Z, Hildebrandt MA, Koster R, Skotheim RI, Kratz CP, Turnbull C, Cortessis VK, Bakken AC, Bishop DT, Cook MB, et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet. 2013;45:680–5. doi: 10.1038/ng.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, Oberlin O, Lapouble E, Ballet S, Lucchesi C, Kontny U, Gonzalez-Neira A, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 58.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, Allan JM, Tomlinson IP, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–10. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koster R, Chanock SJ. Hard Work Ahead: Fine Mapping and Functional Follow-up of Susceptibility Alleles in Cancer GWAS. Current Epidemiology Reports. 2015;2:205–17. [Google Scholar]
- 61.Panagiotou OA, Evangelou E, Ioannidis JP. Genome-wide significant associations for variants with minor allele frequency of 5% or less--an overview: A HuGE review. Am J Epidemiol. 2010;172:869–89. doi: 10.1093/aje/kwq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 63.Kwon JE, Kim DH, Jung WH, Koo JS. Expression of serine and glycine-related enzymes in phyllodes tumor. Neoplasma. 2014;61:566–78. doi: 10.4149/neo_2014_069. [DOI] [PubMed] [Google Scholar]
- 64.Schulze J, Bickert T, Beil FT, Zaiss MM, Albers J, Wintges K, Streichert T, Klaetschke K, Keller J, Hissnauer TN, Spiro AS, Gessner A, et al. Interleukin-33 is expressed in differentiated osteoblasts and blocks osteoclast formation from bone marrow precursor cells. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:704–17. doi: 10.1002/jbmr.269. [DOI] [PubMed] [Google Scholar]
- 65.Hu W, Wu C, Li X, Zheng Z, Xie Q, Deng X, Jiang J, Wu C. Serum IL-33 level is a predictor of progression-free survival after chemotherapy. Oncotarget. 2017;8:35116–23. doi: 10.18632/oncotarget.16627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Musolino C, Allegra A, Profita M, Alonci A, Saitta S, Russo S, Bonanno A, Innao V, Gangemi S. Reduced IL-33 plasma levels in multiple myeloma patients are associated with more advanced stage of disease. Br J Haematol. 2013;160:709–10. doi: 10.1111/bjh.12146. [DOI] [PubMed] [Google Scholar]
- 67.Rossle M, Cathomas G, Bonapace L, Sachs M, Dehler S, Storz M, Huber G, Moch H, Junt T, Mertz KD. Interleukin-33 Expression Indicates a Favorable Prognosis in Malignant Salivary Gland Tumors. Int J Surg Pathol. 2016;24:394–400. doi: 10.1177/1066896916633856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.