Abstract
In current era, majority of microRNA (miRNA) are being discovered through computational approaches which are more confined towards model plants. Here, for the first time, we have described the identification and characterization of novel miRNA in a non-model plant, Persicaria minor (P. minor) using computational approach. Unannotated sequences from deep sequencing were analyzed based on previous well-established parameters. Around 24 putative novel miRNAs were identified from 6,417,780 reads of the unannotated sequence which represented 11 unique putative miRNA sequences. PsRobot target prediction tool was deployed to identify the target transcripts of putative novel miRNAs. Most of the predicted target transcripts (mRNAs) were known to be involved in plant development and stress responses. Gene ontology showed that majority of the putative novel miRNA targets involved in cellular component (69.07%), followed by molecular function (30.08%) and biological process (0.85%). Out of 11 unique putative miRNAs, 7 miRNAs were validated through semi-quantitative PCR. These novel miRNAs discoveries in P. minor may develop and update the current public miRNA database.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1164-8) contains supplementary material, which is available to authorized users.
Keywords: Deep sequencing, In silico, Novel miRNA, Transcriptomic library, Persicaria minor
Introduction
Persicaria minor (formerly known as Polygonum minus Huds.) locally known as ‘kesum’ in Malay is a significant foodstuff in many Malaysian dishes. It also has been used in folk medicine to treat dandruff and digestive problems (Qader et al. 2011; Vikram et al. 2014). Besides, this plant has nice and pleasant aroma, thus has big potential in perfume industry (Vikram et al. 2014; Christapher et al. 2015). Persicaria minor is commonly distributed in Europe and Southeast Asia such as Malaysia, Indonesia, Thailand, and Vietnam. It can grow well in damp areas, river banks and lakes and also survives well on cool and hilly areas (Christapher et al. 2015). P. minor have demonstrated several pharmacological activities such as antibacterial, antifungal, antiviral, antiulcer, antioxidant and cytoprotective activities either in vivo or in vitro (Vikram et al. 2014; Christapher et al. 2015). Interestingly, recent study revealed that P. minor extract could improve cognitive function in middle-aged women in terms of mood, short-term memory and intelligence quotient (IQ) as well (Shahar et al. 2015).
MiRNAs are among the majority class of newly identified non-coding small RNAs (~ 20 nucleotides (nt)) that act as negative regulator of gene expression at the post-transcriptional level by targeting messenger RNA (mRNAs) for cleavage or inhibiting the translation process (Ha and Kim 2014; Rogers and Chen 2013; Samad et al. 2017a). The first miRNA (lin-4) was discovered by Ambros, and is now recognized as the pioneer of the large miRNA family (Esquela-Kerscher 2014). In a couple of decades, enormous progress has been achieved in this field. MiRNAs have been acknowledged to play crucial roles in many physiological and biological processes, including developmental pattern, cell fate, apoptosis, and response to biotic and abiotic stresses (Sunkar et al. 2012; Wang and Schiefelbein 2014). To date, abundances of miRNAs have been characterized either from plant or animal and well-documented in miRNA databases (Kozomara and Griffiths-Jones 2014). In the beginning, miRNA was discovered through forward genetic approaches. However, later studies revealed that miRNA can be identified and characterized using computational approaches (Gomes et al. 2013). An important characteristic possessed by miRNA is the stem-loop structure of precursor miRNA (pre-miRNA) which can be predicted using RNA folding software such as UNAfold and psRobot (Wu et al. 2012; Markham and Zuker 2008). However, this characteristic does not only belong to precursor miRNA since another RNA species can form the stem-loop structure such as rRNA and tRNA (Mathews et al. 2010). Additionally, the stem-loop structure formed by pre-miRNA is usually more complex than another RNA species (Okamura et al. 2013).
During the last decade, Zhang and his colleagues computationally analyzed the unique characteristic of miRNA secondary structure which leads to the value of Minimum Folding Energy Index (MFEI) (Zhang et al. 2006). Here, we have reported our discovery of novel miRNA in P. minor using the parameters established and this finding may improve the available dataset in public miRNA database.
Materials and methods
Persicaria minor culture and RNA extraction
P. minor plants were cultured in Murashige and Skoog (MS) media in a controlled chamber room, Universiti Kebangsaan Malaysia approximately for 6 weeks. Two replicate of P. minor were prepared for this experiment. Healthy leaves were harvested and quickly stored in liquid nitrogen. Approximately 0.1 g of the harvested leaves was grinded for total RNA extraction using PureLink® Plant RNA Reagent (Thermos Fisher Scientific, USA) (Samad et al. 2016). The purity of total RNA was determined by NanoDrop ND-1000 spectrophotometer (NanoDrop, USA) at 260/280 nm (ratio ~ 2.0) and 260/230 nm (ratio ~ 2.2). Total RNA concentration and integrity were determined using a Qubit Fluorometer and 1% gel electrophoresis, respectively. The integrity of total RNA was confirmed by Agilent Bioanalyzer 2100 and RNA 6000 NanoLab Chip Kit (Agilent Technologies, USA) with RIN number at least 7.0.
Small RNA library preparation
In an attempt to construct the small RNA libraries, NEBNext Small RNA Preparation kit (New England Biolabs, USA) was used according to manufacturer’s protocol. Briefly, RNA was ligated with 3’ adapter. Before 5’ adapter was ligated to the RNA, primer hybridization was carried out to minimize the formation of primer dimer. After adapter ligation, first-strand cDNA synthesis was carried out followed by PCR amplification. The PCR product was subjected to 6% polyacrylamide gel for size selection. The samples were sequenced using 2500 HiSeq Illumina platform.
Bioinformatic analysis
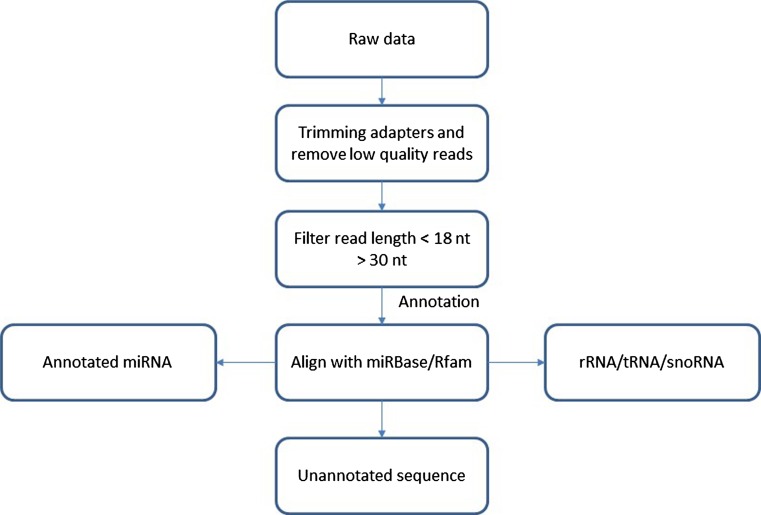
After the raw data was retrieved, further analysis was carried out using CLC workbench version 8 (https://www.qiagenbioinformatics.com/). The details of raw data analysis were carried out as shown in previous report (Samad et al. 2017b). Briefly, begin with trimming of the adapter sequence, low-quality sequences were discarded and the sequences were filtered to remove any reads less than 18 nt and more than 30 nt. Then, the filtered sequences were annotated using miRBase with two mismatches allowed to obtain conserve miRNA (version 21; http://www.mirbase.org) (Kozomara and Griffiths-Jones 2014) and Rfam (version 12; (http://www.sanger.ac.uk/Software/Rfam/) to annotate another non-coding RNA (Nawrocki et al. 2015). The filtering and annotation pipeline was summarized in Fig. 1. The rest of the sequences (unannotated sequences) were used for novel miRNA prediction.
Fig. 1.
Pipeline of data processing from raw data until annotation using CLC workbench
Secondary structure and target prediction for novel miRNA
Since no available genome for P. minor, transcriptomic data were used for this prediction. Transcriptomic data of P. minor were accessed through GenBank under accession number SRX669305 (leaf) to map against unannotated sequences of small RNA (Loke et al. 2016). The result of this mapping was used to form the secondary structure using UNAfold software with default parameter (http://unafold.rna.albany.edu/) (Markham and Zuker 2008). Minimal Folding Free Energy (MFE, ΔG in kcal/mol) was recorded for further analysis. After that, parameters for pre-miRNA prediction from previous report were applied (Alptekin and Budak 2017; Zhang et al. 2006). First, candidate of pre-miRNA sequence must contain approximately 22 nt within one arm in the loop which consists of 30–70% of adenine and uracil base. Second, mature miRNA must not have more than six mismatches against the opposite arm. Third, pre-miRNA sequence must have maximum 3-nt bulge and no loops or breaks are allowed. The list of secondary structures was included in Supplementary file_pre-miRNA. Finally, pre-miRNA sequence must have MFEI value at least 0.85. Before MFEI is calculated, Adjusted Minimal Folding Free Energy (AMFE) was obtained. The equations below were used to calculate AMFE and MFEI:
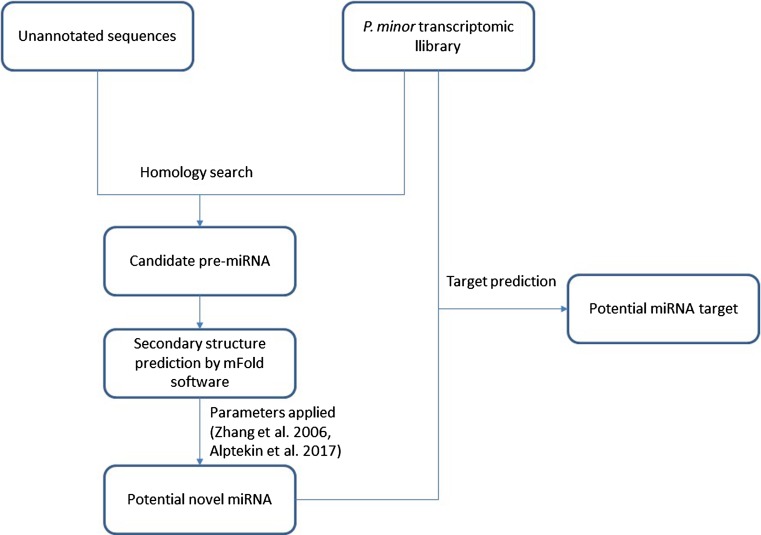
Any secondary structures that did not match the above criteria were discarded. Annotated transcriptomic data were used for miRNA target prediction (Loke et al. 2016; Rahnamaie-Tajadod et al. 2017). We utilized psRobot software for target prediction developed by Wu et al. (2012) (http://omicslab.genetics.ac.cn/psRobot/) with score 2.5 which indicates 90% accuracy and reduced false positive prediction. The work flow is summarized in Fig. 2.
Fig. 2.
Schematic illustration of novel miRNA search and target prediction in P. minor
Validation of secondary structure
cDNA was synthesized using RevertAid Reverse Transcriptase (Thermofisher) according to manufacturer protocols. Briefly, 1 µg of RNA was mixed with 1 µL of oligo(dT). Then, 4 µL of 5× Reaction Buffer, 1 µL of RiboLock RNAse Inhibitor (20 U/µL), 2 µL of 10 mM dNTP Mix and 1 µL of RevertAid RT (200 U/µL) was added to the mixture. Sample was incubated in thermocycler for 1 h at 42 °C. The reaction was terminated by heating the sample at 70 °C for 5 min. For PCR amplification, forward and reverse primers were designed using Integrated DNA Technologies (https://sg.idtdna.com/) (Supplementary file_primer). The PCR product was subjected to 4% agarose gel electrophoresis.
Results and discussion
Sequencing data
Illumina sequencing of P. minor library generated a total of 12,409,685 ± 7.0 million reads. After filtering, 7,724,932 ± 4.7 million clean reads in the library which have length from 18 to 30 nt were retrieved, representing 1,451,635 ± 0.8 million unique sequences as shown in Table 1. Data mapping to miRBase and Rfam database yield 111,235 ± 0.08 million and 1,195,917 ± 0.6 million sequences which represent 2610 ± 0.001 million (0.18%) and 100,356 ± 0.04 m million (6.91%) of unique sequences, respectively. The annotation result against miRBase identified 101 conserved miRNA. Unannotated sequence dominates the statistics of small RNA library where they contribute 6,417,780 ± 3.9 million (83.07%) of total reads, represent 1,348,660 ± 0.8 million (92.91%) of unique reads. The raw sequencing reads of small RNA were deposited in NCBI under two accession numbers, SRX2645686 and SRX2645687.
Table 1.
Statistic of small RNA sequences from P. minor library
| Total reads (million; m) | Percent (%) | Unique | Percent (%) | |
|---|---|---|---|---|
| Raw reads | 12,409,685 ± 7.0 m | |||
| Clean reads | 7,724,932 ± 4.7 m | 100.00 | 1,451,635 ± 0.8 m | 100.00 |
| miRNA | 111,235 ± 0.08 m | 1.44 | 2,610 ± 0.001 m | 0.18 |
| Rfam | 1,195,917 ± 0.6 m | 15.48 | 100,356 ± 0.04 m | 6.91 |
| Unannotated | 6,417,780 ± 3.9 m | 83.07 | 1,348,660 ± 0.8 m | 92.91 |
Novel miRNA detection
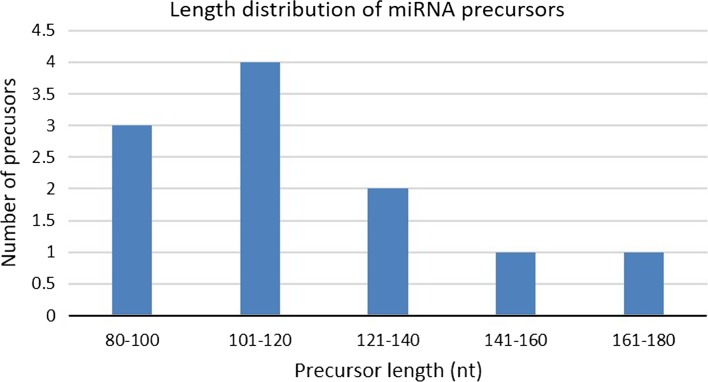
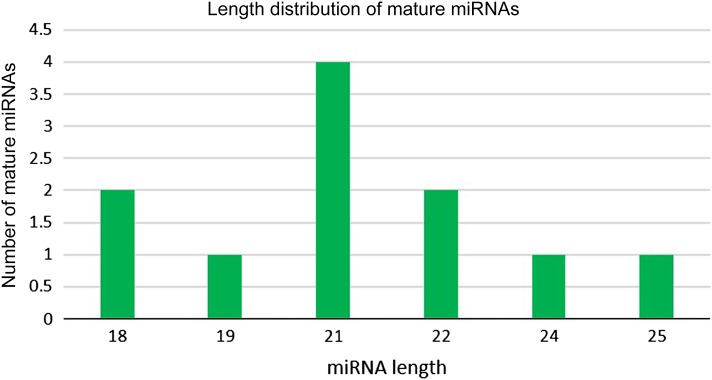
The analysis had revealed that 24 putative novel miRNAs were discovered from 6,417,780 reads of the unannotated sequences which represent 11 unique sequences of putative novel miRNAs. The precursor length of the putative miRNA detected varied between 73 nt to 163 nt and the length of mature miRNA retrieved are in range from 18 nt to 25 nt. The length distribution of pre-miRNA showed majority of pre-miRNA have length between 101–120 nt followed by 80–100 nt, 121–140 nt and finally 141–160 nt and 161–180 nt (Fig. 3). Pre-miRNA and mature miRNA should contain more A and U bases than G and C bases. A high content of A and U possibly makes the secondary structure of pre-miRNA less stable, therefore, becomes more accessible to be processed by the RNA Induced Silencing Complex (RISC) complex into mature miRNA (Zhang et al. 2006; Ni et al. 2010). An important aspect of the secondary structure of DNA and RNA is MFE. The lower value of MFE indicates that the sequence has the higher stability to form secondary structure (Prabu and Mandal 2010). In this study, the lowest value of MFE is − 61.60 kcal/mol belong to Pmi-nov_9 while the highest is − 13.80 kcal/mol belong to Pmi-nov_8. The MFEI for the putatives miRNA varies from 0.86 to 1.96 (Table 2). Analysis of mature miRNA length in Fig. 4 showed that most of the miRNA sequences have 21 nt while only single miRNA has length 19, 24, and 25 nt, respectively.
Fig. 3.
The putative pre-miRNA with different length
Table 2.
List of novel miRNAs discoveries and their information
| ID | miRNA | Length of Precursor | ID transcript | % A and U | ΔG | AMFE | MFEI |
|---|---|---|---|---|---|---|---|
| Pmi-nov_1 | ATGGATATGCAAGCATGATA | 133 | comp66662_c8_seq2 | 66.92 | − 37.1 | 27.89 | 1.19 |
| Pmi-nov_2 | CGGGGAAGAGGCTGAGCAAGGA | 103 | comp64725_c0_seq1 | 50.49 | − 57.5 | 55.83 | 0.89 |
| Pmi-nov_3 | CTGTTAGCAGTAGTACAATTA | 133 | comp60703_c0_seq2 | 59.40 | − 55.1 | 41.43 | 0.98 |
| Pmi-nov_4 | CTTGATTGAAATGGTTCTCGACGA | 83 | comp52499_c0_seq1 | 60.24 | − 24.6 | 29.64 | 1.34 |
| Pmi-nov_5 | GGAGCGACCTTAGACCACATGA | 163 | comp6546_c0_seq1 | 51.53 | − 59.0 | 36.20 | 1.34 |
| Pmi-nov_6 | GTGCTCTCTCTCATTGTCATA | 113 | comp51053_c0_seq1 | 55.75 | − 58.0 | 51.33 | 0.86 |
| Pmi-nov_7 | TCCCACTCTCAACACCAA | 83 | comp62823_c0_seq1 | 44.58 | − 28.6 | 34.46 | 1.61 |
| Pmi-nov_8 | AATGTATTTTTGGACGGAGGTAGTA | 82 | comp66383_c1_seq1 | 67.07 | − 13.8 | 16.83 | 1.96 |
| Pmi-nov_9 | GTGCTCTCTCTCGTTGTCA | 143 | comp7042_c0_seq1 | 59.44 | − 61.6 | 43.07 | 0.94 |
| Pmi-nov_10 | ACGGCTGCTCGATGACCA | 113 | comp51416_c0_seq1 | 48.67 | − 42.7 | 37.79 | 1.36 |
| Pmi-nov_11 | AGGTCACAAATGGACGGTTGA | 113 | comp59558_c0_seq1 | 48.67 | − 57.9 | 51.24 | 1.00 |
From left, miRNA candidate, novel mature miRNA sequence, length of secondary structure, ID transcript that used for secondary structure prediction, percentage (%) of A and U base, MFE (ΔG), AMFE and MFEI value
Fig. 4.
The putative novel mature miRNA with different length
Novel miRNA target prediction
As a single miRNA can target multiple mRNAs, and on the other hand numerous miRNAs can regulate a single mRNA (Thomson et al. 2011; Samad et al. 2017a). Target prediction resulted with 10 putative novel miRNAs, targeting 18 mRNA transcripts involved in plant growth, development and signal transduction as well as stress responses (Table 3). For instance, Pmi-nov_6 targets three mRNAs encoding F-box protein, histidinol dehydrogenase and WEB family protein families. These three genes actively participated in plant development where F-box involved in protein degradation, histidinol dehydrogenase involved in histidine biosynthesis and WEB family protein involved in chloroplast movement (Stefanowicz et al. 2015; Petersen et al. 2010; Kodama et al. 2010). Additionally, F-box proteins were known as multifunctional proteins since they were involved in defense response and hormone signaling as well (Piisilä et al. 2015; Kelley and Estelle 2012; Stefanowicz et al. 2015). F-box protein in Arabidopsis, MORE AXILLARY GROWTH2 (MAX2) contributed in plant defense system against P. syringae and P. carotovorum, since max2 mutant showed susceptibility to both bacteria (Piisilä et al. 2015). Besides, F-box proteins were able to regulate different types of hormone such as auxin, jasmonate, gibberillic acid, ethylene, and salicylic acid which were crucial in plant defense and development (Kelley and Estelle 2012).
Table 3.
List of target genes for putative novel miRNAs in P. minor
| miRNA | Score | Target ID | Target annotation |
|---|---|---|---|
| Pmi-nov_1 | 2.5 2.0 2.2 |
comp66946_c3_seq5 comp60103_c0_seq2 comp65298_c0_seq4 |
F-box protein Histidinol dehydrogenase WEB family protein |
| Pmi-nov_2 | 2.0 2.5 |
comp65018_c0_seq3 comp52866_c1_seq1 |
1-aminocyclopropane-1-carboxylate oxidase Homeobox-leucine zipper protein |
| Pmi-nov_3 | 2.5 | comp68390_c0_seq6 | Putative disease-resistance protein |
| Pmi-nov_4 | 0 | comp52499_c0_seq1 | Cytochrome c biogenesis protein |
| Pmi-nov_5 | N/A | N/A | N/A |
| Pmi-nov_6 | 2.5 2.5 2.5 |
comp64912_c2_seq1 comp63854_c3_seq4 comp53735_c0_seq3 |
Serine/threonine protein kinase Transcription factor TCP4 UDP-glycosyltransferase |
| Pmi-nov_7 | 2.5 1.2 1.2 |
comp60227_c1_seq1 comp67662_c1_seq5 comp58603_c0_seq4 |
WRKY transcription factor Homeobox-leucine zipper protein Protease-Do like |
| Pmi-nov_8 | 2.5 2.0 |
comp67443_c0_seq9 comp65934_c1_seq7 |
Sucrose-phosphate synthase Cysteine-rich receptor-like protein kinase |
| Pmi-nov_9 | 2.2 | comp60840_c0_seq9 | Serine/threonine protein kinase |
| Pmi-nov_10 | 2.5 | comp26923_c0_seq1 | Peroxidase |
| Pmi-nov_11 | 2.0 | comp39280_c0_seq1 | U-box domain-containing protein |
From left, miRNA candidate, alignment score, target ID and target annotation. Non-applicable (N/A) represents the putative novel miRNA does not hit any target
Pmi-nov_8 targeted cysteine-rich receptor-like protein kinase which also participated actively in signal transduction (Burdiak et al. 2015). In addition to that, Pmi-nov_5 and Pmi-nov_9 were also involved in signal transduction via targeting same target that plays role in signal transduction, serine/threonine protein kinase (Kulik et al. 2011). In Arabidopsis, 44 members of cysteine-rich receptor-like protein kinase were discovered (Burdiak et al. 2015). Overexpression of this protein had increased the plant sensitivity to abscisic acid, resulting in enhanced drought tolerance (Lu et al. 2016). Serine/threonine protein kinase worked as negative regulator to immune system. In Arabidopsis, the regulation by serine/threonine protein kinase prevents improper activation of plant defense activity in the absence of plant pathogen (Lin et al. 2015). By contrast, in tomato, serine/threonine protein kinase was encoded by Pto gene that contributed resistance against Pseudomonas syringae pv. tomato that produced AvrPto (Piquerez et al. 2014).
On the other hand, Pmi-nov_2 and Pmi-nov_10 target putative disease-resistance protein and peroxidase that contribute in regulating plant defense system, respectively. Disease-resistant proteins in plant serve as pathogen recognition and responsible for triggering innate immunity system to halt pathogen proliferation (Wen et al. 2015; Wang et al. 2016). In Arabidopsis, transgenic plant overexpressing disease-resistant gene showed resistance to powdery mildew caused by Golovinomyces cichoracearum, and also to a virulent bacteria, Pseudomonas syringae pv. tomato DC3000 (Wen et al. 2015). Pmi-nov_10 target, peroxidase, was well known as antioxidant defense that detoxify reactive oxygen species (ROS) (Ozyigit et al. 2016).
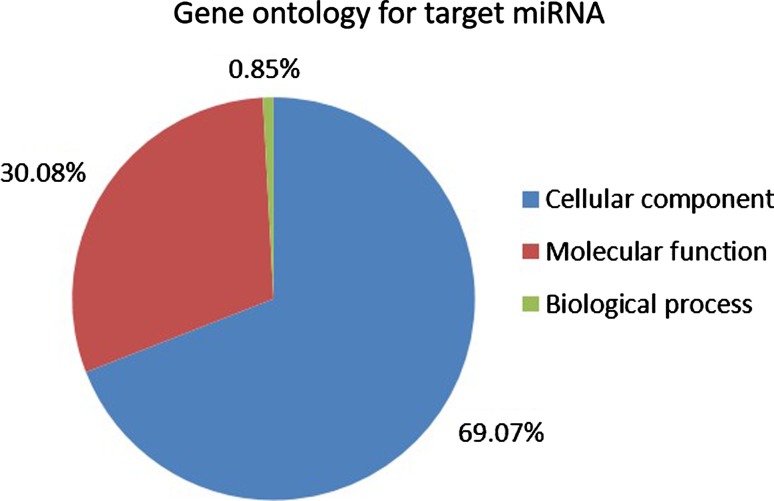
From all of the miRNA candidates, Pmi-nov_5 has no hit to any target. This might be because of the limitation of genome sequences in P. minor. Overall, gene ontology analysis for target enrichment (Fig. 5) showed that most of the target gene belongs to cellular component (69.07%) followed by molecular function (30.08%) and biological process (0.85%).
Fig. 5.
Pie chart showing the distribution of GO annotations of the 10 novel miRNA targets which belong to three major classes
Experimental validation
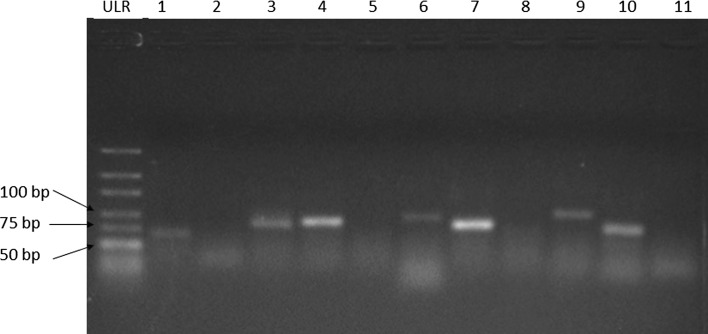
Out of 11 miRNA precursors, 7 were successfully validated through experimental method. The size range of the miRNA precursors varies between ~ 70 and ~ 100 nt (Fig. 6). The validated precursors belonged to Pre-nov_1 (~ 70 nt), Pre-nov_3 (~ 80 nt), Pre-nov_4 (~ 80 nt), Pre-nov_6 (~ 90 nt), Pre-nov_7 (~ 80 nt), Pre-nov_9 (~ 100 nt) and Pre-nov_10 (~ 80 nt). The size of precursors detected was smaller than predicted because the primers were designed to avoid the bulges region to prevent inefficient primer annealing (Stadhouders et al. 2010). However, no miRNA precursors were detected for Pre-nov_2, Pre-nov_5, Pre-nov_8, and Pre-nov_11. Troubleshooting steps had been taken into account for the amplification of these precursor sequences such as temperature adjustment during PCR and primers substitution, but none of them worked. These problems may arise due to false positive prediction from bioinformatics tools (Alptekin et al. 2017).
Fig. 6.
Gel picture showing miRNA precursors. From left Ultra Low Range DNA ladder (ULR), Pre-nov_1 (1), Pre-nov_2 (2), Pre-nov_3 (3), Pre-nov_4 (4), Pre-nov_5 (5), Pre-nov_6 (6), Pre-nov_7 (7), Pre-nov_8 (8), Pre-nov_9 (9), Pre-nov_10 (10) and Pre-nov_11 (11)
Conclusion
Despite the fact that this type of miRNA discovery technique is one of the common research in miRNA field, but miRNA research on P. minor is still quite behind from other model plant species. In this work, we have successfully characterized potential novel miRNA in P. minor since, it may be developed as a genetic tool to enhance the production of valuable compound which can be beneficial in pharmaceutical industry. We have identified 11 potential miRNAs through computational approach, together with their target genes except for one miRNA. Further analysis lead to the validation of 7 miRNAs. In our study, we have used a well-developed transcriptomic library for miRNA homology search method. Our finding may improve the current public miRNA database by depositing this miRNAs sequence from non-model plant species where the genome is still unknown.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The author, Abdul Fatah A. Samad is sponsored by MyBrain 15 under MyPhD scholarship from Ministry of Higher Education (Malaysia). This research was supported by Dana Impak Perdana (DIP-2015-018).
Compliance with ethical standards
Conflict of interest
There are no conflicts to declare by authors.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1164-8) contains supplementary material, which is available to authorized users.
References
- Alptekin B, Budak H. Wheat miRNA ancestors: evident by transcriptome analysis of A, B, and D genome donors. Funct Integr Genom. 2017;17(2):171–187. doi: 10.1007/s10142-016-0487-y. [DOI] [PubMed] [Google Scholar]
- Alptekin B, Akpinar BA, Budak H. A Comprehensive prescription for plant miRNA identification. Front Plant Sci. 2017;7:2058. doi: 10.3389/fpls.2016.02058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdiak P, Rusaczonek A, Witon D, Glow D, Karpinski S. Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J Exp Bot. 2015;66(11):3325–3337. doi: 10.1093/jxb/erv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christapher PV, Parasuraman S, Christina JM, Asmawi MZ, Vikneswaran M. Review on Polygonum minus. Huds, a commonly used food additive in Southeast Asia. Pharmacogn Res. 2015;7(1):1–6. doi: 10.4103/0974-8490.147125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A. The lin-4 microRNA: the ultimate micromanager. Cell Cycle. 2014;13(7):1060–1061. doi: 10.4161/cc.28384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C, Cho J-H, Hood L, Franco O, Pereira R, Wang K. A review of computational tools in microRNA discovery. Front Genet. 2013;4:81. doi: 10.3389/fgene.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Estelle M. Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 2012;160(1):47–55. doi: 10.1104/pp.112.200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Suetsugu N, Kong S-G, Wada M. Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc Natl Acad Sci. 2010;107(45):19591–19596. doi: 10.1073/pnas.1007836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS. 2011;15(12):859–872. doi: 10.1089/omi.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z-JD, Liebrand TWH, Yadeta KA, Coaker GL. PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 2015 doi: 10.1104/pp.15.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke K-K, Rahnamaie-Tajadod R, Yeoh C-C, Goh H-H, Mohamed-Hussein Z-A, Mohd Noor N, Zainal Z, Ismail I. RNA-seq analysis for secondary metabolite pathway gene discovery in Polygonum minus. Genom Data. 2016;7:12–13. doi: 10.1016/j.gdata.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Liang S, Wu Z, Bi C, Yu YT, Wang XF, Zhang DP. Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J Exp Bot. 2016;67(17):5009–5027. doi: 10.1093/jxb/erw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol (Clifton, NJ) 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol. 2010;2(12):a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43(D1):D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Shu W, Bo X, Wang S, Li S. Correlation between sequence conservation and structural thermodynamics of microRNA precursors from human, mouse, and chicken genomes. BMC Evol Biol. 2010;10(1):329. doi: 10.1186/1471-2148-10-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27(7):778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyigit II, Filiz E, Vatansever R, Kurtoglu KY, Koc I, Öztürk MX, Anjum NA. Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci. 2016;7:301. doi: 10.3389/fpls.2016.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LN, Marineo S, Mandalà S, Davids F, Sewell BT, Ingle RA. The missing link in plant histidine biosynthesis: Arabidopsis myoinositol monophosphatase-like2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol. 2010;152(3):1186–1196. doi: 10.1104/pp.109.150805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piisilä M, Keceli MA, Brader G, Jakobson L, Jõesaar I, Sipari N, Kollist H, Palva ET, Kariola T. The F-box protein MAX2 contributes to resistance to bacterial phytopathogens in Arabidopsis thaliana. BMC Plant Biol. 2015;15(1):53. doi: 10.1186/s12870-015-0434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquerez SJM, Harvey SE, Beynon JL, Ntoukakis V. Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front Plant Sci. 2014;5:671. doi: 10.3389/fpls.2014.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabu GR, Mandal AK. Computational identification of miRNAs and their target genes from expressed sequence tags of tea (Camellia sinensis) Genom Proteom Bioinform. 2010;8(2):113–121. doi: 10.1016/S1672-0229(10)60012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qader SW, Abdulla MA, Chua LS, Najim N, Zain MM, Hamdan S. Antioxidant, total phenolic content and cytotoxicity evaluation of selected Malaysian plants. Molecules. 2011;16(4):3433–3443. doi: 10.3390/molecules16043433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamaie-Tajadod R, Loke K-K, Goh H-H, Mohd Noor N. Differential gene expression analysis in Polygonum minus leaf upon 24 h of Methyl Jasmonate Elicitation. Front Plant Sci. 2017;8:109. doi: 10.3389/fpls.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell Online. 2013;25(7):2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad AFA, Ali NM, Ismail I, Murad AMA. Analysis of miRNAs targeting transcription factors in Persicaria minor induced by Fusarium oxysporum. AIP Conf Proc. 2016;1784(1):020009. doi: 10.1063/1.4966719. [DOI] [Google Scholar]
- Samad AFA, Sajad M, Nazaruddin N, Fauzi IA, Murad AMA, Zainal Z, Ismail I. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad AFA, Nazaruddin N, Sajad M, Jani J, Murad AMA, Zainal Z, Ismail I. Small RNA sequencing for secondary metabolite analysis in Persicaria minor. Genom Data. 2017;13:3–4. doi: 10.1016/j.gdata.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahar S, Aziz AF, Ismail SNA, Yahya HM, Din NC, Manaf ZA, Badrasawi MM. The effect of Polygonum minus extract on cognitive and psychosocial parameters according to mood status among middle-aged women: a randomized, double-blind, placebo-controlled study. Clin Interv Aging. 2015;10:1505–1520. doi: 10.2147/CIA.S86411. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stadhouders R, Pas SD, Anber J, Voermans J, Mes THM, Schutten M. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Mol Diagn. 2010;12(1):109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz K, Lannoo N, Van Damme EJM. Plant F-box proteins—judges between life and death. Crit Rev Plant Sci. 2015;34(6):523–552. doi: 10.1080/07352689.2015.1024566. [DOI] [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39(16):6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram P, Chiruvella KK, Ripain IHA, Arifullah M. A recent review on phytochemical constituents and medicinal properties of kesum (Polygonum minus Huds.) Asian Pac J Trop Biomed. 2014;4(6):430–435. doi: 10.12980/APJTB.4.2014C1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Schiefelbein J. Regulation of cell fate determination in plants. Front Plant Sci. 2014;5:368. doi: 10.3389/fpls.2014.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-M, Liu P-Q, Xu Y-J, Xiao S. Protein trafficking during plant innate immunity. J Integr Plant Biol. 2016;58(4):284–298. doi: 10.1111/jipb.12426. [DOI] [PubMed] [Google Scholar]
- Wen Z, Yao L, Wan R, Li Z, Liu C, Wang X. Ectopic expression in Arabidopsis thaliana of an NB-ARC encoding putative disease resistance gene from wild Chinese Vitis pseudoreticulata enhances resistance to phytopathogenic fungi and bacteria. Front Plant Sci. 2015;6:1087. doi: 10.3389/fpls.2015.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012;40:W22–W28. doi: 10.1093/nar/gks554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006;63(2):246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.