Abstract
The major causes of congenital heart diseases (CHDs) are the interactions of genetic and environmental factors. We conducted a case–control study in 357 mothers of CHDs fetuses and 270 control mothers to investigate the association of maternal PAHs exposure, AHR, CYP1A1, CYP1A2, CYP1B1 and CYP2E polymorphisms, the interaction between PAHs exposure and genetic variants with the risk of CHDs. The higher level PAHs exposure was associated with the risk of CHDs (aOR = 2.029, 95% CI: 1.266, 3.251) or subtypes. The haplotypes of AHR or CYP1A2 were associated with the risk of CHDs: AHR: C-G-A-C: aOR = 0.765; T-A-G-A: aOR = 1.33; CYP1A2: A-T:aOR = 1.75; C-C: aOR = 0.706. When exposed to higher level PAHs, the risk of CHDs among the mothers carrying rs2158041 “C/T or T/T” genotype or rs7811989 “G/A or A/A” genotype in AHR was 1.724 (χ2 = 7.209, P = 0.007) or 1.735 (χ2 = 7.364, P = 0.007) times greater than the aOR in the mothers carrying wild genotype. The multiplicative-scale interactions between PAHs exposure and polymorphisms of CYP1A2 rs4646425 (P = 0.03) or CYP2E1 rs915908 (P = 0.0238) on the risk of CHDs were observed. Our study suggests that maternal AHR polymorphisms may modify the association of PAHs exposure with CHDs, CYP1A2 or CYP2E1 polymorphisms significantly interact with PAHs exposure on CHDs.
Introduction
Congenital heart diseases (CHDs) are the most common type of birth defect, accounting for one-third of all major congenital anomalies1. Approximately, 4~10 of live births are affected by CHDs2,3. In China, the average total prevalence of CHDs was 40.95 per 10,000 live births in 2011, and 130 thousand infants are born with CHDs each year4. Consequently, CHDs cause considerable suffering to patients and their families, and CHDs have become a sizable public health concern.
Although advances in the understanding of the genetic risk factors and environmental risk factors affecting the development of CHDs have been made, the aetiology of the majority of CHDs remains unknown5–7. Now, it is widely believed that most CHDs arise from a complex and ill-defined combination of genetic and environmental factors8,9.
Polycyclic aromatic hydrocarbons (PAHs) are widespread environmental pollutants formed by the incomplete burning of coal, tobacco, or other organic substances10. The general population is unavoidably exposed to PAHs through the inhalation of tobacco smoke, smoke from other sources of combustion, and ambient air and through the consumption of PAHs, particularly PAHs in charbroiled foods11. PAHs have been found in placental tissues12 and umbilical cord blood12, which suggests that transplacental transfer of these chemicals to the foetus can have a significant impact on foetal development, including increased risk of neural tube defects, cleft lip with or without cleft palate, gastroschisis, low birth weight, preterm birth, and intrauterine growth restriction13–18. Although studies of experimental model systems have suggested that prenatal exposure to PAHs is associated with CHDs19,20, the results of a large, population-based study did not support the association between potential maternal occupational exposure to PAHs and CHDs21. Owing to the limitation of using expert industrial hygienists’ assessments of exposure to PAHs, further evidence and quantitative data are needed to illustrate the association between prenatal exposure to PAHs and CHDs.
The cytochrome P450 (CYP) enzymes CYP1A1, CYP1A2, and CYP1B1 have been shown to play important roles in the metabolic activation of PAHs22. CYP2E1 is expressed at higher levels in Asians than in Caucasians; therefore, it is thought to be responsible for the metabolism of pyrene in Asian people23. The aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor, mediates the toxic effects of a variety of environmental chemicals, including PAHs24, as well as the induction of three members of the CYP1 family, CYP1A1, CYP1A2 and CYP1B125. Common genetic polymorphisms in these genes could affect individual susceptibility to adverse effects of exposure to PAHs. Maternal genotypes, such as those involving the gene CYP1B1, have been shown to enhance the association between maternal exposure to PAHs and neural tube defects26. However, few studies have investigated maternal genetic susceptibility to CHDs related to PAHs or have explored possible gene-environment interactions.
1-Hydroxypyrene-glucuronide (1-OHPG) is a stable PAH metabolite that is excreted in the urine and is an index biomarker that reflects recent exposure to mixed PAHs27. In the present study, we first analysed the association between maternal exposure to PAHs by measuring urine 1-OHPG concentration during pregnancy and the risk of foetal CHDs. Then, we investigated the association between maternal genetic polymorphisms and the risk of foetal CHDs. Finally, we evaluated the potential interaction between maternal genetic variants and exposure to PAHs on the risk of foetal CHDs.
Results
Characteristics of the study participants
In this study, a total of 627 participants were analysed (357 cases and 270 controls). The baseline characteristics of the participants are presented in Table 1. There were significant differences between the two groups with respect to gestational week, cooking at home, maternal alcohol consumption, and folic acid supplements.
Table 1.
Characteristics of the case and control participants.
| Variables/Characteristic | Cases | Controls | χ 2 | P-values |
|---|---|---|---|---|
| No.(%) | No.(%) | |||
| Maternal age (yrs)a | 4.096 | 0.129 | ||
| <25 | 99(27.7) | 58(21.5) | ||
| 25–34 | 211(59.1) | 166(61.5) | ||
| ≥35 | 47(13.2) | 46(17.0) | ||
| Gestational weeka | 103.038 | <0.001 | ||
| <19 | 13(3.6) | 88(32.6) | ||
| 20–25 | 194(54.3) | 126(46.7) | ||
| 26–31 | 109(30.5) | 42(15.6) | ||
| ≥32 | 41(11.5) | 14(5.2) | ||
| Housing renovationb | 0.766 | 0.381 | ||
| Yes | 79(22.1) | 52(19.3) | ||
| No | 278(77.9) | 218(80.7) | ||
| Factory or landfill nearbyb | 0.460 | 0.498 | ||
| Yes | 56(15.7) | 37(13.7) | ||
| No | 298(83.5) | 230(85.2) | ||
| Cooking at homeb | 13.021 | 0.001 | ||
| Often | 203(56.9) | 122(45.2) | ||
| Never | 76(21.3) | 91(33.7) | ||
| Occasional | 75(21.0) | 54(20.0) | ||
| Parental smoking or ETS exposureb | 1.758 | 0.185 | ||
| Yes | 223(62.5) | 155(57.4) | ||
| No | 133(37.3) | 115(42.6) | ||
| Maternal alcohol consumptionb | 10.671 | 0.005 | ||
| Often | 6(1.7) | 8(3.0) | ||
| Occasional | 55(15.4) | 68(25.2) | ||
| Never | 293(82.1) | 194(71.9) | ||
| Folic acid supplementsb | 4.057 | 0.044 | ||
| Yes | 297(83.2) | 240(88.9) | ||
| No | 60(16.8) | 30(11.1) |
aUsing base data in following multivariate analysis as continuous variables.
bThe exposure was defined from the 3 months before pregnancy to the first trimester.
Association between maternal exposure to PAHs and the risk of CHDs
Table 2 displays the relation between maternal exposure to PAHs and foetal CHDs. Significant positive associations were observed between maternal exposure to PAHs and various CHDs phenotypes when comparing high exposure to low exposure after adjusting for potential confounders: all CHDs (aOR = 2.029; 95% CI: 1.266, 3.251), septal defects (aOR = 2.373, 95% CI: 1.376, 4.093), conotruncal heart defects (aOR = 2.349, 95% CI: 1.250, 4.416), right-sided obstructive malformations (aOR = 2.423, 95% CI: 1.190, 4.933), left-sided obstructive malformations (aOR = 2.662, 95% CI: 1.085, 6.529), anomalous pulmonary venous return (aOR = 2.962, 95% CI: 1.068, 8.212), and other heart abnormalities (aOR = 2.327, 95% CI: 1.129, 4.795).
Table 2.
Association between maternal PAHs exposure and the risk of CHDs.
| PAHs exposure | Cases | Controls | cOR(95%CI) | aORb(95%CI) |
|---|---|---|---|---|
| No.(%) | No.(%) | |||
| Any CHDs | ||||
| low | 43(12.0) | 62(23.0) | Ref. | Ref. |
| high | 314(88.0) | 208(77.0) | 2.177(1.421,3.335) | 2.029(1.266,3.251)* |
| septal defects | ||||
| low | 27(11.5) | 62(23.0) | Ref. | Ref. |
| high | 208(88.5) | 208(77.0) | 2.296(1.405,3.752) | 2.373(1.376,4.093)* |
| conotruncal heart defects | ||||
| low | 16(10.0) | 62(23.0) | Ref. | Ref. |
| high | 144(90.0) | 208(77.0) | 2.683(1.488,4.836) | 2.349(1.250,4.416)* |
| right-sided obstructive malformations | ||||
| low | 11(9.6) | 62(23.0) | Ref. | Ref. |
| high | 103(90.4) | 208(77.0) | 2.791(1.409,5.528) | 2.423(1.190,4.933)* |
| left-sided obstructive malformations | ||||
| low | 7(9.7) | 62(23.0) | Ref. | Ref. |
| high | 65(90.3) | 208(77.0) | 2.768(1.207,6.345) | 2.662(1.085,6.529)* |
| anomalous pulmonary venous return | ||||
| low | 5(7.8) | 62(23.0) | Ref. | Ref. |
| high | 59(92.2) | 208(77.0) | 3.517(1.352,9.149) | 2.962(1.068,8.212)* |
| other cardiac structural abnormalities | ||||
| low | 11(10.8) | 62(23.0) | Ref. | Ref. |
| high | 91(89.2) | 208(77.0) | 2.466(1.241,4.906) | 2.327(1.129,4.795)* |
cOR: crude odds ration; aOR: adjusted odds ration, adjusted for maternal age, gestational week, housing renovation, factory or landfill nearby, cooking at home, parental smoking or ETS exposure, maternal alcohol consumption, folic acid supplements.
Association between maternal gene polymorphisms and the risk of CHDs
The genotype frequencies for polymorphisms of AHR, CYP1A1, CYP1A2, CYP1B1 and CYP2E1 in the controls were in Hardy-Weinberg equilibrium (see Supplementary Appendix A, Table S1).
Table 3 shows the association between single gene loci polymorphisms and the risk of CHDs, assuming various genetic models. In the AHR gene, the SNPs rs2158041 and rs7811989 were associated with an increased risk of CHDs under the dominant model (aOR = 1.454, 95% CI: 1.024, 2.065; aOR = 1.46, 95% CI: 1.027, 2.075), and the SNPs rs2066853 and rs2040623 were associated with a decreased risk of CHDs under the additive model (aOR = 0.7648, 95% CI: 0.5859, 0.9983; aOR = 0.761, 95% CI: 0.5867, 0.9872). In the CYP1A2 gene, the SNP rs762551 was associated with a decreased risk of CHDs (under the dominant model: aOR = 0.6529, 95% CI: 0.4608, 0.9252; under the additive model: aOR = 0.7062, 95% CI: 0.5444, 0.916) and the SNP rs4646425 was associated with an increased risk of CHDs (under the dominant model: aOR = 1.723, 95% CI: 1.048, 2.833; under the additive model: aOR = 1.748, 95% CI: 1.077, 2.839). The SNPs rs2158041, rs7811989, rs762551 and rs4646425 were associated with some subtypes of CHDs; the data are shown in Supplementary Appendix B, Table S2. However, the associations were not statistically significant after the false discovery rate (FDR) correction. No significant association was found between any of the remaining 16 selected loci and the risk of CHDs or any CHDs subtype before or after the FDRcorrection.
Table 3.
Association between maternal genotypes and the risk of CHDs.
| dbSNP_ID | Model | Genotype | Controls | Cases | aOR(95%CI) | P-value | FDR-BH P value |
|---|---|---|---|---|---|---|---|
| N(%) | N(%) | ||||||
| rs2158041 | Dominant | C/C | 168 (62.2) | 194 (54.3) | 1 | 0.03656 | 0.1828 |
| C/T- T/T | 102 (37.8) | 163 (45.7) | 1.454 (1.024, 2.065)* | ||||
| Recessive | C/C- C/T | 253 (93.7) | 331 (92.7) | 1 | 0.6361 | 0.8959 | |
| T/T | 17 (6.3) | 26 (7.3) | 1.179 (0.596, 2.332) | ||||
| Log-additive | — | — | — | 1.303 (0.9841, 1.725) | 0.06457 | 0.2544 | |
| rs7811989 | Dominant | G/G | 169 (62.6) | 194 (54.3) | 1 | 0.03491 | 0.1828 |
| G/A-A/A | 101 (37.4) | 163 (45.7) | 1.46 (1.027, 2.075)* | ||||
| Recessive | G/G-G/A | 252 (93.3) | 331 (92.7) | 1 | 0.7877 | 0.8959 | |
| A/A | 18 (6.7) | 26 (7.3) | 1.097 (0.5606, 2.145) | ||||
| Log-additive | — | — | — | 1.287 (0.9736, 1.702) | 0.07631 | 0.2544 | |
| rs2066853 | Dominant | G/G | 111 (41.1) | 163 (45.7) | 1 | 0.1473 | 0.453 |
| G/A-A/A | 159 (58.9) | 194 (54.3) | 0.7704 (0.5414,1.096) | ||||
| Recessive | G/G-G/A | 237 (87.8) | 326 (91.3) | 1 | 0.0622 | 0.4806 | |
| A/A | 33 (12.2) | 31 (8.7) | 0.5861 (0.3342,1.028) | ||||
| Log-additive | — | — | — | 0.7648 (0.5859,0.9983)* | 0.04855 | 0.2428 | |
| rs2040623 | Dominant | A/A | 96 (35.6) | 141 (39.5) | 1 | 0.1137 | 0.453 |
| A/C-C/C | 174 (64.4) | 216 (60.5) | 0.7453 (0.5178,1.073) | ||||
| Recessive | A/A-A/C | 228 (84.4) | 314 (88.0) | 1 | 0.07209 | 0.4806 | |
| CC | 42 (15.6) | 43 (12.0) | 0.6322 (0.3836,1.042) | ||||
| Log-additive | — | — | — | 0.761 (0.5867,0.9872)* | 0.03969 | 0.2428 | |
| rs1048943 | Dominant | T/T | 146 (54.1) | 194 (54.3) | 1 | 0.9835 | 0.9835 |
| T/C- C/C | 124 (45.9) | 163 (45.7) | 1.004 (0.7102, 1.418) | ||||
| Recessive | T/T- T/C | 254 (94.1) | 330 (92.4) | 1 | 0.3767 | 0.8126 | |
| C/C | 16 (5.9) | 27 (7.6) | 1.375 (0.6786, 2.786) | ||||
| Log-additive | — | — | — | 1.055 (0.7976, 1.395) | 0.708 | 0.885 | |
| rs4646422 | Dominant | C/C | 201 (74.4) | 284 (79.6) | 1 | 0.1585 | 0.453 |
| C/T-T/T | 69 (25.6) | 73 (20.4) | 0.7448 (0.4944, 1.122) | ||||
| Recessive | C/C-C/T | 268 (99.3) | 351 (98.3) | 1 | 0.439 | 0.8126 | |
| T/T | 2 (0.7) | 6 (1.7) | 1.934 (0.364, 10.27) | ||||
| Log-additive | — | — | — | 0.8108 (0.5575, 1.179) | 0.2725 | 0.545 | |
| rs4642421 | Dominant | G/G | 84 (31.1) | 108 (30.3) | 1 | 0.6549 | 0.9835 |
| G/A-A/A | 186 (68.9) | 249 (69.7) | 1.089 (0.7487,1.585) | ||||
| Recessive | G/G-G/A | 225 (83.3) | 290 (81.2) | 1 | 0.546 | 0.8401 | |
| A/A | 45 (16.7) | 67 (18.8) | 1.151 (0.7289,1.818) | ||||
| Log-additive | — | — | — | 1.087 (0.8425,1.402) | 0.5219 | 0.8718 | |
| rs762551 | Dominant | A/A | 119 (44.1) | 187 (52.4) | 1 | 0.01651 | 0.1828 |
| A/C- C/C | 151 (55.9) | 170 (47.6) | 0.6529 (0.4608, 0.9252)* | ||||
| Recessive | A/A- A/C | 235 (87.0) | 328 (91.9) | 1 | 0.0703 | 0.4806 | |
| C/C | 35 (13.0) | 29 (8.1) | 0.5969 (0.3414, 1.044) | ||||
| Log-additive | — | — | — | 0.7062 (0.5444, 0.916)* | 0.008771 | 0.1754 | |
| rs4646425 | Dominant | C/C | 238 (88.1) | 291 (81.5) | 1 | 0.03186 | 0.1828 |
| C/T-T/T | 32 (11.9) | 66 (18.5) | 1.723 (1.048, 2.833)* | ||||
| Recessive | C/C-C/T | 270 (100.0) | 354 (99.2) | 1 | 0.9988 | 0.9988 | |
| T/T | 0 (0.0) | 3 (0.8) | — | ||||
| Log-additive | — | — | — | 1.748 (1.077, 2.839)* | 0.02394 | 0.2394 | |
| rs2472304 | Dominant | G/G | 189 (70.0) | 244 (68.3) | 1 | 0.206 | 0.4578 |
| G/A-A/A | 81 (30.0) | 113 (31.7) | 1.278 (0.874,1.867) | ||||
| Recessive | G/G-G/A | 265 (98.1) | 349 (97.8) | 1 | 0.7375 | 0.8959 | |
| A/A | 5 (1.9) | 8 (2.2) | 1.231 (0.3656,4.143) | ||||
| Log-additive | — | — | — | 1.241 (0.8806,1.748) | 0.2175 | 0.5438 | |
| rs2470890 | Dominant | G/G | 189 (70.0) | 244 (68.3) | 1 | 0.1935 | 0.4578 |
| G/A-A/A | 81 (30.0) | 113 (31.7) | 1.287 (0.88,1.881) | ||||
| Recessive | G/G-G/A | 265 (98.1) | 349 (97.8) | 1 | 0.7375 | 0.8959 | |
| A/A | 5 (1.9) | 8 (2.2) | 1.231 (0.3656,4.143) | ||||
| Log-additive | — | — | — | 1.248 (0.8855,1.758) | 0.2058 | 0.5438 | |
| rs2855658 | Dominant | C/C | 217 (80.4) | 283 (79.3) | 1 | 0.9736 | 0.9835 |
| C/T- T/T | 53 (19.6) | 74 (20.7) | 1.007 (0.6578, 1.542) | ||||
| Recessive | C/C - C/T | 265 (98.1) | 353 (98.9) | 1 | 0.9292 | 0.9781 | |
| T/T | 5 (1.9) | 4 (1.1) | 0.9364 (0.2199, 3.987) | ||||
| Log-additive | — | — | — | 1.001 (0.6818, 1.47) | 0.995 | 0.995 | |
| rs1056837 | Dominant | G/G | 217 (80.4) | 282 (79.0) | 1 | 0.8692 | 0.9835 |
| G/A-A/A | 53 (19.6) | 75 (21.0) | 1.036 (0.6772, 1.586) | ||||
| Recessive | G/G-G/A | 265 (98.1) | 353 (98.9) | 1 | 0.4875 | 0.8126 | |
| A/A | 6 (1.9) | 4 (1.1) | 0.6042 (0.1457, 2.506) | ||||
| Log-additive | — | — | — | 0.9918 (0.6774, 1.452) | 0.9663 | 0.995 | |
| rs1056836 | Dominant | G/G | 216 (80.0) | 280 (78.4) | 1 | 0.8848 | 0.9835 |
| G/C- C/C | 54 (20.0) | 77 (21.6) | 1.032 (0.6764, 1.573) | ||||
| Recessive | G/G - G/C | 264 (97.8) | 353 (98.9) | 1 | 0.4875 | 0.8126 | |
| C/C | 6 (2.2) | 4 (1.1) | 0.6042 (0.1457, 2.506) | ||||
| Log-additive | — | — | — | 0.9887 (0.6769, 1.444) | 0.9532 | 0.995 | |
| rs1056827 | Dominant | C/C | 171 (63.3) | 228 (63.9) | 1 | 0.7812 | 0.9835 |
| C/A- A/A | 99 (36.7) | 129 (36.1) | 1.052 (0.7341, 1.508) | ||||
| Recessive | C/C - C/A | 260 (96.3) | 343 (96.1) | 1 | 0.3418 | 0.8126 | |
| A/A | 10 (3.7) | 14 (3.9) | 1.554 (0.6262, 3.856) | ||||
| Log-additive | — | — | — | 1.092 (0.8036, 1.485) | 0.5729 | 0.8718 | |
| rs10012 | Dominant | G/G | 171 (63.3) | 230 (64.4) | 1 | 0.9318 | 0.9835 |
| G/C-C/C | 99 (36.7) | 127 (35.6) | 1.016 (0.708, 1.458) | ||||
| Recessive | G/G-G/C | 260 (96.3) | 342 (95.8) | 1 | 0.2845 | 0.8126 | |
| C/C | 10 (3.7) | 15 (4.2) | 1.631 (0.6658, 3.995) | ||||
| Log-additive | — | — | — | 1.072 (0.7899, 1.456) | 0.6543 | 0.8724 | |
| rs3813867 | Dominant | G/G | 171 (63.3) | 213 (59.7) | 1 | 0.3651 | 0.6637 |
| G/C- C/C | 99 (36.7) | 144 (40.3) | 1.178 (0.8266, 1.678) | ||||
| Recessive | G/G- G/C | 258 (95.6) | 344 (96.4) | 1 | 0.4491 | 0.8126 | |
| C/C | 12 (4.4) | 13 (3.6) | 0.7175 (0.3037, 1.695) | ||||
| Log-additive | — | — | — | 1.082 (0.7998, 1.463) | 0.6103 | 0.8718 | |
| rs2031920 | Dominant | C/C | 171 (63.3) | 213 (59.7) | 1 | 0.3651 | 0.6637 |
| C/T-T/T | 99 (36.7) | 144 (40.3) | 1.178 (0.8266, 1.678) | ||||
| Recessive | C/C-C/T | 258 (95.6) | 344 (96.4) | 1 | 0.4491 | 0.8126 | |
| T/T | 12 (4.4) | 13 (3.6) | 0.7175 (0.3037, 1.695) | ||||
| Log-additive | — | — | — | 1.082 (0.7998, 1.463) | 0.6103 | 0.8718 | |
| rs915908 | Dominant | G/G | 190 (70.4) | 252 (70.6) | 1 | 0.7128 | 0.9835 |
| G/A-A/A | 80 (29.6) | 105 (29.4) | 1.074 (0.7346, 1.57) | ||||
| Recessive | G/G-G/A | 258 (95.6) | 345 (96.6) | 1 | 0.8063 | 0.8959 | |
| A/A | 12 (4.4) | 12 (3.4) | 0.8921 (0.3581, 2.222) | ||||
| Log-additive | — | — | — | 1.037 (0.7549, 1.424) | 0.8237 | 0.9691 | |
| rs6413432 | Dominant | T/T | 145 (53.7) | 198 (55.5) | 1 | 0.5408 | 0.9014 |
| T/A-A/A | 125 (46.3) | 159 (44.5) | 0.8977 (0.6352, 1.269) | ||||
| Recessive | T/T-T/A | 248 (91.9) | 338 (94.7) | 1 | 0.1336 | 0.6682 | |
| A/A | 22 (8.1) | 19 (5.3) | 0.5923 (0.2988, 1.174) | ||||
| Log-additive | — | — | — | 0.8556 (0.6482, 1.129) | 0.2707 | 0.545 |
aOR: adjusted odds ration, adjusted for maternal age, gestational week, housing renovation, factory or landfill nearby, cooking at home, parental smoking or ETS exposure, maternal alcohol consumption, folic acid supplements.
Table 4 displays the association between maternal haplotypes and the risk of CHDs. In the AHR gene, the haplotype C-G-A-C was associated with a decreased risk of CHDs (aOR = 0.765, P = 0.0486), and the haplotype T-A-G-A was associated with an increased risk of CHDs (aOR = 1.33, P = 0.0447). In the CYP1A2 gene, one haplotype block defined by 2 SNPs (rs762551 and rs4646425) showed a significant association with the risk of CHDs (the haplotype A-T: aOR = 1.75, P = 0.0239; the haplotype C-C: aOR = 0.706, P = 0.00877). No significant association was found between any of other haplotypes and the risk of CHDs. Linkage disequilibrium analysis is shown in Supplementary Appendix C, Figure S1.
Table 4.
Association between maternal haplotypes and the risk of CHDs
| Gene | Haplotype | Frequency | P-value | aOR | ||
|---|---|---|---|---|---|---|
| All | Controls | Cases | ||||
| AHR | ||||||
| Block1 | 4-SNPs: rs2158041- rs7811989 - rs2066853- rs2040623 | |||||
| C-G-A-C | 0.333 | 0.3556 | 0.3137 | 0.0486 | 0.765* | |
| C-G-G-C | 0.0434 | 0.04019 | 0.04622 | 0.866 | 0.948 | |
| T-A-G-A | 0.241 | 0.2124 | 0.2633 | 0.0447 | 1.33* | |
| C-G-G-A | 0.375 | 0.3783 | 0.3739 | 0.655 | 1.06 | |
| CYP1A1 | ||||||
| Block1 | 3-SNPs: rs1048943- rs4646422 – rs4646421 | |||||
| C-C-A | 0.259 | 0.2548 | 0.2627 | 0.695 | 1.06 | |
| T-C-A | 0.177 | 0.173 | 0.1799 | 0.704 | 1.07 | |
| T-T-G | 0.12 | 0.1302 | 0.1106 | 0.273 | 0.811 | |
| T-C-G | 0.44 | 0.4375 | 0.4434 | 0.91 | 1.01 | |
| CYP1A2 | ||||||
| Block1 | 2-SNPs: rs762551- rs4646425 | |||||
| A-T | 0.0805 | 0.05926 | 0.09664 | 0.0239 | 1.75* | |
| C-C | 0.307 | 0.3444 | 0.2787 | 0.00877 | 0.706* | |
| A-C | 0.612 | 0.5963 | 0.6246 | 0.182 | 1.19 | |
| Block2 | 2-SNPs: rs2472304- rs2470890 | |||||
| A-T | 0.164 | 0.1593 | 0.1681 | 0.24 | 1.23 | |
| G-C | 0.834 | 0.8407 | 0.8291 | 0.186 | 0.793 | |
| CYP1B1 | ||||||
| Block1 | 3-SNPs: rs2855658- rs1056837- rs1056836 | |||||
| T-A-C | 0.107 | 0.1056 | 0.1078 | 0.923 | 1.02 | |
| C-G-G | 0.888 | 0.8889 | 0.8866 | 0.953 | 1.01 | |
| Block2 | 2-SNPs: rs1056827- rs10012 | |||||
| A-C | 0.199 | 0.2019 | 0.1961 | 0.732 | 1.06 | |
| C-G | 0.797 | 0.7981 | 0.7969 | 0.504 | 0.901 | |
| CYP2E1 | ||||||
| Block1 | 3-SNPs: rs3813867- rs2031920- rs915908 | |||||
| G-C-A | 0.165 | 0.1704 | 0.1603 | 0.864 | 1.03 | |
| C-T-G | 0.212 | 0.2056 | 0.2163 | 0.643 | 1.07 | |
| G-C-G | 0.621 | 0.6241 | 0.6199 | 0.574 | 0.93 | |
aOR: adjusted odds ration, adjusted for maternal age, gestational week, housing renovation, factory or landfill nearby, cooking at home, parental smoking or ETS exposure, maternal alcohol consumption, folic acid supplements.
Interaction between maternal genotypes and exposure to PAHs on the risk of CHDs
Assuming a dominant genetic model (minor allele considered to be the risk allele) and a 1 df association test, Table 5 shows the interaction between maternal genotypes and exposure to PAHs and the risk of CHDs. When exposed to the higher level of PAHs, the risk of CHDs for the children of mothers carrying the C/T or T/T genotypes of SNP rs2158041 in the AHR gene was 1.724 (χ2 = 7.209, P = 0.007) times greater than the aOR for the children of the mothers carrying the C/C genotype. The risk of CHDs for the children of mothers carrying the G/A or A/A genotypes of SNP rs7811989 was 1.735 (χ2 = 7.364, P = 0.007) times greater than the aOR of the children of mothers carrying the G/G genotype. Multiplicative-scale interactions between maternal exposure to PAHs and the SNP rs4646425 in the CYP1A2 gene (P = 0.03) and the SNP rs915908 in the CYP2E1 gene (P = 0.0238) and the risk of CHDs were observed. No multiplicative-scale interactions were observed between maternal exposure to PAHs or the other SNPs and the risk of CHDs.
Table 5.
Interaction between maternal PAHs exposure and genotypes on the risk of CHDs.
| Genotype | PAHs exposure | Count (%) | aOR (95% CI) | Interaction Test | ||
|---|---|---|---|---|---|---|
| Controls | Cases | G2 | P | |||
| rs2158041 | 3.044 | 0.081 | ||||
| C/C | low | 33 (12.2) | 25 (7.0) | Ref. | ||
| C/C | high | 135 (50.0) | 169 (47.3) | 1.421 (0.762,2.648) | ||
| C/T or T/T | low | 29 (10.7) | 18 (5.0) | 0.739 (0.310,1.762) | ||
| C/T or T/T | high | 73 (27) | 145 (40.6) | 2.449 (1.281, 4.682) | ||
| rs7811989 | 3.605 | 0.0576 | ||||
| G/G | low | 34 (12.6) | 26 (7.3) | Ref. | ||
| G/G | high | 135 (50.0) | 168 (47.1) | 1.383 (0.749,2.544) | ||
| G/A or A/A | low | 28 (10.4) | 17 (4.8) | 0.687 (0.286,1.648) | ||
| G/A or A/A | high | 73 (27.0) | 146 (40.9) | 2.399 (1.267,4.544) | ||
| rs2066853 | 0.139 | 0.7093 | ||||
| G/G | low | 23 (8.5) | 22 (6.2) | Ref. | ||
| G/G | high | 88 (32.6) | 141 (39.5) | 1.815 (0.871,3.781) | ||
| G/A or A/A | low | 39 (14.4) | 21 (5.9) | 0.652 (0.270,1.574) | ||
| G/A or A/A | high | 120 (44.4) | 173 (48.5) | 1.422 (0.693,2.916) | ||
| rs2040623 | 0.09 | 0.7642 | ||||
| A/A | low | 19 (7.0) | 18 (5.0) | Ref. | ||
| A/A | high | 77 (28.5) | 123 (34.5) | 1.830 (0.829,4.042) | ||
| A/C or C/C | low | 43 (15.9) | 25 (7.0) | 0.678 (0.274,1.673) | ||
| A/C or C/C | high | 131 (48.5) | 191 (53.5) | 1.443 (0.671,3.105) | ||
| rs1048943 | 0.002 | 0.9643 | ||||
| T/T | low | 34 (12.6) | 26 (7.3) | Ref. | ||
| T/T | high | 112 (41.5) | 168 (47.1) | 2.048 (1.098,3.819) | ||
| T/C or C/C | low | 28 (10.4) | 17 (4.8) | 1.021 (0.427,2.439) | ||
| T/C or C/C | high | 96 (35.6) | 146 (40.9) | 2.045 (1.088, 3.844) | ||
| rs4646422 | 0.605 | 0.4367 | ||||
| C/C | low | 49 (18.1) | 36 (10.1) | Ref. | ||
| C/C | high | 152 (56.3) | 248 (69.5) | 1.895 (1.117,3.217) | ||
| C/T or T/T | low | 13 (4.8) | 7 (2.0) | 0.487 (0.159,1.490) | ||
| C/T or T/T | high | 56 (20.7) | 66 (18.5) | 1.431 (0.775,2.644) | ||
| rs4646421 | 0.096 | 0.7567 | ||||
| G/G | low | 18 (6.7) | 13 (3.6) | Ref. | ||
| G/G | high | 66 (24.4) | 95 (26.6) | 2.275 (0.963,5.372) | ||
| G/A or A/A | low | 44 (16.3) | 30 (8.4) | 1.267 (0.496,3.238) | ||
| G/A or A/A | high | 142 (52.6) | 173 (61.3) | 2.449 (1.079,5.561) | ||
| rs762551 | 1.232 | 0.267 | ||||
| A/A | low | 26 (9.6) | 23 (6.4) | Ref. | ||
| A/A | high | 93 (34.4) | 164 (45.9) | 1.519 (0.765,3.018) | ||
| A/C or C/C | low | 36 (13.3) | 20 (5.6) | 0.431 (0.180,1.028) | ||
| A/C or C/C | high | 115 (42.6) | 450 (42.0) | 1.120 (0.567, 2.212) | ||
| rs4646425 | 4.736 | 0.03 | ||||
| C/C | low | 58 (21.5) | 32 (9.0) | Ref. | ||
| C/C | high | 180 (66.7) | 259 (72.5) | 2.50 (1.484,4.213) | ||
| C/T or T/T | low | 4 (1.5) | 11 (3.1) | 5.988 (1.588,22.57) | ||
| C/T or T/T | high | 28 (10.4) | 55 (15.4) | 3.441 (1.728,6.853) | ||
| rs2472304 | 0.564 | 0.4527 | ||||
| G/G | low | 41 (15.2) | 31 (8.7) | Ref. | ||
| G/G | high | 148 (54.8) | 213 (59.7) | 1.803 (1.036,3.140) | ||
| G/A or A/A | low | 21 (7.8) | 12 (3.4) | 0.852 (0.321,2.258) | ||
| G/A or A/A | high | 60 (22.2) | 101 (28.3) | 2.301 (1.241,4.264) | ||
| rs2740890 | 0.591 | 0.442 | ||||
| C/C | low | 41 (15.2) | 31 (8.7) | Ref. | ||
| C/C | high | 148 (54.8) | 213 (59.7) | 1.79 (1.033,3.131) | ||
| C/T or T/T | low | 21 (7.8) | 12 (3.4) | 0.852 (0.322,2.259) | ||
| C/T or T/T | high | 60 (22.2) | 101 (28.3) | 2.317 (1.250,4.295) | ||
| rs2855658 | 0.001 | 0.9748 | ||||
| C/C | low | 49 (18.1) | 33 (9.2) | Ref. | ||
| C/C | high | 168 (62.2) | 250 (70.0) | 2.022 (1.184,3.450) | ||
| C/T or T/T | low | 13 (4.8) | 10 (2.8) | 1.067 (0.382,2.979) | ||
| C/T or T/T | high | 40 (14.8) | 64 (17.9) | 2.210 (1.152, 4.240) | ||
| rs1056837 | 0.051 | 0.8213 | ||||
| G/G | low | 48 (17.8) | 33 (9.2) | Ref. | ||
| G/G | high | 169 (62.6) | 249 (69.7) | 1.977 (1.157,3.380) | ||
| G/A or A/A | low | 14 (5.2) | 10 (2.8) | 1.015 (0.367,2.805) | ||
| G/A or A/A | high | 39 (14.4) | 65 (18.2) | 2.286 (1.188,4.399) | ||
| rs1056836 | 0.041 | 0.8395 | ||||
| G/G | low | 48 (17.8) | 33 (9.2) | Ref. | ||
| G/G | high | 168 (62.2) | 247 (69.2) | 1.981 (1.158,3.387) | ||
| G/C or C/C | low | 14 (5.2) | 10 (2.8) | 1.015 (0.367,2.805) | ||
| G/C or C/C | high | 40 (14.8) | 67 (18.8) | 2.259 (1.179,4.329) | ||
| rs1056827 | 0.683 | 0.4086 | ||||
| C/C | low | 42 (15.6) | 25 (7.0) | Ref. | ||
| C/C | high | 129 (47.8) | 203 (56.9) | 2.359 (1.300,4.281) | ||
| C/A or A/A | low | 20 (7.4) | 18 (5.0) | 1.475 (0.605,3.596) | ||
| C/A or A/A | high | 79 (29.3) | 111 (31.1) | 2.304 (1.227,4.326) | ||
| rs10012 | 0.836 | 0.3605 | ||||
| G/G | low | 42 (15.6) | 25 (7.0) | Ref. | ||
| G/G | high | 129 (47.8) | 205 (57.4) | 2.398 (1.321,4.350) | ||
| G/C or C/C | low | 20 (7.4) | 18 (5.0) | 1.475 (0.605,3.596) | ||
| G/C or C/C | high | 79 (29.3) | 109 (30.5) | 2.240 (1.192,4.209) | ||
| rs3813867 | 0.477 | 0.4898 | ||||
| G/G | low | 36 (13.3) | 26 (7.3) | Ref. | ||
| G/G | high | 135 (50) | 187 (52.4) | 1.780 (0.968,3.273) | ||
| G/C or C/C | low | 26 (9.6) | 17 (4.8) | 0.915 (0.382,2.195) | ||
| G/C or C/C | high | 73 (27.0) | 127 (35.6) | 2.286 (1.204, 4.342) | ||
| rs2031920 | 0.477 | 0.4898 | ||||
| C/C | low | 36 (13.3) | 26 (7.3) | Ref. | ||
| C/C | high | 135 (50) | 187 (52.4) | 1.780 (0.968,3.273) | ||
| C/T or T/T | low | 26 (9.6) | 17 (4.8) | 0.915 (0.382,2.195) | ||
| C/T or T/T | high | 73 (27.0) | 127 (35.6) | 2.286 (1.204, 4.342) | ||
| rs915908 | 5.108 | 0.0238 | ||||
| G/G | low | 41 (15.2) | 34 (9.5) | Ref. | ||
| G/G | high | 149 (55.2) | 218 (61.1) | 1.448 (0.832,2.520) | ||
| G/A or A/A | low | 21 (7.8) | 9 (2.5) | 0.343 (0.124,0.953) | ||
| G/A or A/A | high | 59 (21.9) | 96 (26.9) | 1.728 (0.931,3.208) | ||
| rs6413432 | 0.003 | 0.9563 | ||||
| T/T | low | 32 (11.9) | 23 (6.4) | Ref. | ||
| T/T | high | 113 (41.9) | 175 (49.0) | 2.003 (1.054,3.806) | ||
| T/A or A/A | low | 30 (11.1) | 20 (5.6) | 0.869 (0.367,2.056) | ||
| T/A or A/A | high | 95 (35.2) | 139 (38.9) | 1.783 (0.928,3.427) | ||
aOR: adjusted odds ration, adjusted for maternal age, gestational week, housing renovation, factory or landfill nearby, cooking at home, parental smoking or ETS exposure, maternal alcohol consumption, folic acid supplements.
Discussion
In this case-control study, we evaluated the association between maternal exposure to PAHs and the risk of CHDs and the association between maternal genetic variants and the risk of CHDs. We further explored possible interactions between the risk of CHDs and maternal exposure to PAHs and genetic variants.
PAHs are lipophilic, which means that they can pass through the placenta. The estimated transplacental dose of PAHs is about ten times lower than the dose in maternal tissues12,28. The developing embryos may be as much as 10 times more susceptible than the mother to PAH-induced DNA damage12. Suggested mechanisms of the teratogenicity of PAHs include oxidative stress29; changes in signal transduction pathways30,31; the formation of bulky PAH-DNA adducts that result in a spectrum of cellular mutations that may be teratogenic12,32; or epigenetic changes, including DNA methylation33.
Previous studies in experimental model systems have suggested that prenatal exposure to PAHs is associated with CHDs19,20. PAHs are important components of PM2.5 and PM1034. One human study found that PM10 exposure during weeks 3–8 of pregnancy is associated with isolated atrial septal defects (OR = 2.27, 95% CI: 1.43, 3.60)35. Another study suggested that PM10 is associated with increased odds of pulmonary valve stenosis (aORFourth Quartile = 2.6, 95% CI: 1.2, 5.7) and perimembranous ventricular septal defects (aORThird Quartile = 2.1, 95% CI: 1.1, 3.9)36. Consistent with these results, our study found that higher maternal level of exposure to PAHs during pregnancy is associated with an increased risk of CHDs or CHDs phenotypic subtypes after adjusting for a series of potential confounders. However, one published US study did not support the association between potential maternal occupational exposure to PAHs and various CHDs subtypes21. Because there have been only a limited number of human studies regarding the relationship between maternal exposure to PAHs and CHDs, and the results were inconsistent, more research is necessary to elucidate the potential relations between maternal exposure to PAHs and CHDs.
Genetic polymorphisms in AhR lead to substantial differences in sensitivity to the biochemical and toxic effects of chemical compounds in laboratory animals37. There have been a few reports on SNPs in the maternal AHR gene, environmental exposure to PAHs during pregnancy and the impacts on the foetus, but the results have been inconsistent. Two previous studies showed that infants born to continuously smoking mothers with the AhR rs2066853 wild-type genotype had significantly lower estimated birth weights and lengths compared with the offspring of non-smokers38,39. One previous study did not observe an association between SNPs (including rs2158041, rs7811989, and rs2040623) of the maternal AHR gene and NTD or between NTD and the interaction between maternal genes and indoor air pollution26. Another later study did not find that the AhR rs2066853 polymorphism modified the effect of prenatal dioxin levels on birth size40. The polymorphism of the AHR gene affected the expression of the protein41. Our study showed that SNPs (including rs2158041, rs7811989, rs2066853, and rs2040623) in the maternal AHR gene were associated with CHDs or CHDs subtypes, but they were not observed after the FDR correction. The haplotypes of the AHR gene were associated with the risk of CHDs. The SNPs rs2158041 and rs7811989 may modify the association of maternal exposure to PAHs with foetal CHDs. The molecular mechanism remains unclear. Perhaps these two intronic polymorphisms influence the alternative spicing of the gene products42, or they might be in LD with other causal loci or genes, further affecting the metabolism of PAHs.
Four members of the CYP family, CYP1A1, CYP1A2, CYP1B1 and CYP2E1, are the main Phase I enzymes mediating the toxicity of xenobiotic chemicals, including PAHs. Their polymorphisms are associated with increased risk of many types of cancer, such as renal cell carcinoma, lung cancer, colorectal cancer, and breast cancer43–46. Several animal studies have demonstrated that knocking out CYP1A1, CYP1A2, or CYP1B1 expression had no deleterious effects when the mice were raised in a clean environment; however, severe adverse effects appeared in the knockout mice when they were challenged with toxins and carcinogens47,48. In human studies, the results regarding the association between maternal gene polymorphisms or gene-environmental interactions and the offspring’s health effects have been inconsistent. One previous study did not find an association between the maternal CYP1A1 rs1048943 polymorphism and the risk of non-syndromic oral cleft49. Another study did not find an association between the maternal CYP1B1 rs1056836 polymorphism and childhood medulloblastoma50. One study in 2006 showed that the maternal CYP1B1 rs1048943 polymorphism was associated with first-trimester miscarriage and that it might also modify the risk of first-trimester miscarriage among coffee drinkers51. One later study in 2013 did not find an association between the maternal CYP1A1 rs1048943 polymorphism and the risk of preterm delivery, but it showed an interaction between a high organochlorine pesticide level and the CYP1A1 rs1048943 polymorphism that might magnify the risk of PTD52. One report did not find an association between maternal CYP1A1, CYP1A2, CYP1B1 polymorphisms and neural tube defects, but it found that the maternal CYP1B1 rs2855658 polymorphism modified the effect of indoor air pollution on the risk of neural tube defects26. Another study did not find an association between the maternal CYP1A2 rs762551 polymorphism and the risk of gastroschisis or an association between the risk of gastroschisis and gene-maternal smoking interaction53. Our study observed that the maternal CYP1A2 rs762551 and rs4646425 polymorphisms are associated with CHDs or CHDs subtypes but that those associations were not observed after the FDR correction. The haplotypes of the CYP1A2 gene were associated with the risk of CHDs. Multiplicative-scale interactions were observed between maternal exposure to PAHs, the SNP rs4646425 in CYP1A2 and the SNP rs915908 in CYP2E, and the risk of CHDs. The molecular mechanism underlying these interactions remains to be determined.
This study has several strengths. First, to our knowledge, this is the first study to evaluate the effect of the interaction between maternal exposure to PAHs during pregnancy and maternal genotypes on the risk of CHDs. Second, we used a urinary biomarker-based approach to examine the relationship between maternal exposure to PAHs and the risk of CHDs, offering an objective measure of exposure to PAHs, in contrast to the previous exposure assessment that relied solely on expert industrial hygienist consensus or the self-report of pregnant women, whose knowledge of their exposure to PAHs is likely to be limited54. Finally, the subjects of our study were pregnant women non-occupationally exposed to PAHs; thus, the results can be generalized to all women because the environmental factors can be assessed at the individual level.
Our study is subject to certain limitations. First, given the relatively moderate number of subjects, future studies with larger sample sizes are warranted to confirm or refute our findings. Second, only maternal exposure to PAHs and genetic susceptibilities were considered; however, the genetics of cardiac development is likely to be a complex interplay between both maternal and foetal genetic susceptibilities. Therefore, future studies are needed to investigate the effects of foetal exposure, foetal genotypes, and the interaction between them on the risk of CHDs.
In summary, our findings indicate that higher maternal levels of exposure to PAHs during pregnancy might be associated with increased risk of foetal CHDs and CHDs subtypes. The haplotypes of the AHR or CYP1A2 genes were associated with the risk of CHDs, and the AHR gene rs2158041 and rs7811989 polymorphisms may modify the association of maternal exposure to PAHs with foetal CHDs. The maternal SNPs rs4646425 in CYP1A2 and rs915908 in CYP2E1 significantly interacted with the effect of maternal exposure to PAHs on CHDs.
Materials and Methods
Study population
This case–control study was performed from February 2010 to July 2015. The study subjects were recruited from six tertiary maternal and child health hospitals with expertise in foetal echocardiography to screen for foetal CHDs.
The main inclusion criteria were as follows: pregnant women with foetuses diagnosed with CHDs and without any extracardiac abnormalities determined by echocardiography and with gestational age more than 12 weeks. CHDs was confirmed in the foetuses by appointed senior ultrasonic doctors. For the CHDs-affected foetuses that were aborted, CHDs was further confirmed by humanitarian examination of the pathological anatomy. For the CHDs-affected foetuses that were born, a further ultrasound examination was performed within 30 postnatal days. Furthermore, a telephone follow-up was performed within 60 days. The exclusion criteria were as follows: (1) foetuses with syndromic diseases and chromosomal aberrations and (2) woman with multiple pregnancies.
The controls were pregnant women with foetuses with no major congenital malformations diagnosed by echocardiography in the same hospital and gestational age more than 12 weeks. A further ultrasound examination was performed within 30 postnatal days, and a telephone follow-up was performed within 60 days.
Based on the anatomic lesions, CHDs cases were categorized into six subgroups: (1) septal defects, (2) conotruncal heart defects, (3) right-sided obstructive malformations, (4) left-sided obstructive malformations, (5) anomalous pulmonary venous return, and (6) other heart abnormalities55,56.
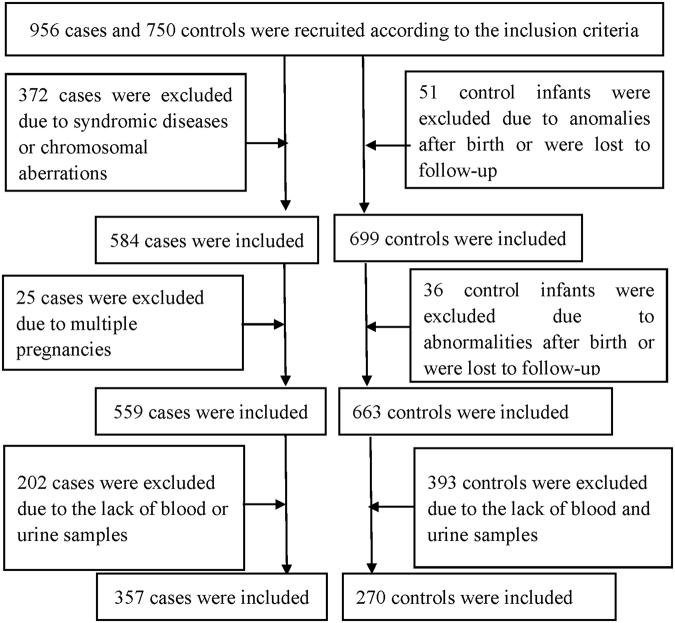
During the study period, 956 cases and 750 controls were initially recruited. According to the exclusion criteria, 397 cases and 87 controls were excluded from the study. Additionally, 202 cases and 393 controls were excluded due to a lack of maternal blood or urine samples. Therefore, 357 cases and 270 controls were included. The flowchart of case and control inclusion and exclusion is shown in Fig. 1.
Figure 1.
Flowchart presenting the inclusion and exclusion of case and control participants.
All the participants signed an informed consent form. This research was approved by the Ethics Committee of Sichuan University (No. 2010004) and followed the tenets of the Declaration of Helsinki.
Data and biological sample collection
Each participant participated in a face-to-face interview when they were recruited during the antenatal examination. The questionnaire comprised eight parts, including parental social demographics, living environment, living habits, working environment, maternal reproductive history, maternal illness and drug use history, maternal diet and nutrition, and maternal life events and mental state. Information on potential confounders was obtained for inclusion as covariate factors.
Referring to the literature, potential confounders are those factors that correlate with both the main determinant and CHDs. These potential confounders included maternal age (at the time of the last menstrual period), gestational week, home or workplace renovation (yes or no), exposure to a factory or landfill nearby (<1000 metres, yes or no), cooking at home (often: ≥4 times/week, never, or occasional: 1~4 times/week), parental smoking or environmental tobacco smoke (ETS) exposure (yes or no), maternal alcohol consumption (often: ≥1 time(s)/week, occasional: <1 time/week, or never), and use of folic acid supplements (yes or no). The age and the gestational week were used as continuous variables in the multivariate analysis.
Ten millilitres of urine was collected from each participant in the morning and stored at −70°C until analysis. Four millilitres of blood was collected in EDTA from each participant by venepuncture and stored at −70°C until genotyping.
Assessment of exposure to PAHs
1-OHPG concentration was measured in 1 ml urine specimens at the West China School of Public Health, Sichuan University, using ultra-high-performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS).
The analysis was performed on a Nexera X2 LC-30AD Liquid Chromatograph System (Shimadzu, Japan) coupled with a TSQ Vantage tandem mass spectrometer (Thermo Fisher Scientific, USA) with an electrospray ionization (ESI) interface operated in a negative ion mode. The limit of detection was 0.015 ng 1-OHPG/ml urine. The details regarding the preparation and analysis of the samples are available in Supplementary appendix D.
Some environmental factors are risk factors for disease, but the association is not simply linear57–59. In previous studies that conducted an association analysis between environmental exposures and birth defects, researchers generally did not directly incorporate the concentrations of environmental exposures into the models but, rather, grouped environmental exposures according to certain criteria (such as the general population reference value or the normal range) to obtain the odds ratios under different environmental exposures13,60,61. In this study, the subjects were non-occupationally exposed pregnant women, who are representative of the general population. We searched many studies but were unable to obtain a reference value for exposure to polycyclic aromatic hydrocarbons or the dose-effect threshold related to CHDs in the general population. Therefore, the Youden Index (sensitivity + specificity − 1) within the receiver operating characteristic (ROC) curve was used to identify the optimal cut-off point used to define the binary exposure to PAHs. The receiver operating characteristic curve and area are shown in Supplementary Appendix E, Figure S2 and Table S5. When the Youden Index reached the maximum value of 0.10918, the concentration of 1-OHPG was 0.03186 µg/g Cr. Therefore, maternal exposure to PAHs was categorized into two groups: “high” if the 1-OHPG concentration was above 0.03186 µg/g Cr and “low” if the 1-OHPG concentration was less than or equal to 0.03186 µg/g Cr.
DNA extraction and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Blood Mini Kit (Qiagen, Cat. No. 51106, Germany) according to the recommended protocol.
SNPs in the AHR, CYP1A1, CYP1A2, CYP1B1, and CYP2E1 genes were selected based on the following principal criteria: (1) an association with diseases in previous studies or the metabolic level of PAHs26,43–46,62–70 and (2) a minor allele frequency >0.05 in Han Chinese. In total, 20 SNPs were selected. The SNPs were genotyped using an improved multiplex ligation detection reaction (iMLDR) technique that was newly developed by Genesky Biotechnologies Inc. (Shanghai, China). More detailed information about the studied genetic variants and genotyping is presented in Supplementary Appendix F, Table S5.
For quality-control assessment, genotyping was repeated in 10% of samples, and the consistency rate was 100%.
Statistical Analysis
Chi-square statistics tested the differences in covariates between the cases and controls. Unconditional logistic regression analysis was performed to investigate the association between maternal exposure to PAHs and foetal CHDs using Statistical Package for Social Sciences (SPSS) version 16.0 software (SPSS Inc., IBM, Chicago, USA).
Hardy–Weinberg equilibrium was assessed in the controls using Plink software (http://pngu.mgh.harvard.edu/~purcell/plink/). The pairwise linkage disequilibrium (LD) patterns and haplotype structures of CYP1A1, CYP1A2, CYP1B1, CYP2E1 and AHR genes were analysed using Haploview 4.2 software. Unconditional logistic regression analysis was performed to investigate the association between individual genetic polymorphisms and CHDs using Plink software.
The effects of the gene-exposure interactions on CHDs occurrence were evaluated by logistic models using SPSS version 16.0 software (SPSS Inc., IBM, Chicago, USA).
All analyses were adjusted for covariates/potential confounders. False discovery rate (FDR) correction of multiple hypothesis testing was performed. Two-sided P < 0.05 was considered statistically significant.
Electronic supplementary material
Acknowledgements
We are indebted to the paediatric cardiologists, geneticists, and epidemiologists who collaborated in this programme and made the study possible. We thank the obstetricians, paediatricians, pathologists, experimental technicians and other participants involved in the project for recruiting the case and control participants and collecting the data. We thank all participating families for their cooperation and for providing personal information. We also thank the reviewers for their helpful comments. This study is funded by the Natural Science Foundation of China (ID: 81573165, 81602865), the National “Twelfth Five-Year” Plan for Science & Technology Support of China (grant ID: 2014BAI06B01), the National Science & Technology basic work Project of the Ministry of Science and Technology of China (grant ID: 2014FY110700), and the Program for Changjiang Scholars and Innovative Research Team in University (ID: IRT0935).
Author Contributions
J.Z. and Y.P.W. developed the study design. N.N.L. and Y.M. conducted the experiment and drafted the manuscript. Z.L. and Y.D. assisted in analyzing the data. Y.X.G. and X.J.Z. assisted in organizing and collecting the samples. X.H.L. and P.Y. participated in reviewing, editing, and revising the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Nana Li and Yi Mu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21380-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yanping Wang, Email: wyxyanping@163.com.
Jun Zhu, Email: zhujun028@163.com.
References
- 1.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr. 2008;153:807–813. doi: 10.1016/j.jpeds.2008.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christianson, A., Howson, C. P. & Modell, B. March of Dimes Global Report on Birth Defects: The Hidden Toll of Dying and Disabled Children. White Plains, New York (2006).
- 4.National Health and Family Planning Commission of PRC National Stocktaking Report on Birth defect Prevention (2012).
- 5.Gelb BD, Chung WK. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb Perspect Med. 2014;4:a013953. doi: 10.1101/cshperspect.a013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SS, Burns TL. Nongenetic risk factors and congenital heart defects. Pediatr Cardiol. 2013;34:1535–1555. doi: 10.1007/s00246-013-0775-4. [DOI] [PubMed] [Google Scholar]
- 7.van der Bom T, et al. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 8.Tiret L. Gene-environment interaction: a central concept in multifactorial diseases. Proc Nutr Soc. 2002;61:457–463. doi: 10.1079/PNS2002178. [DOI] [PubMed] [Google Scholar]
- 9.Krauss RS, Hong M. Gene-Environment Interactions and the Etiology of Birth Defects. Curr Top Dev Biol. 2016;116:569–580. doi: 10.1016/bs.ctdb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Tao S. Global atmospheric emission inventory of polycyclic aromatic hydrocarbons (PAHs) for 2004. Atmos Environ 2009. 2009;43:812–819. [Google Scholar]
- 11.Arey, J. & Atkinson, R. Photochemical reactions of PAH in the atmosphere. In: Douben PET, editor. PAHs: An ecotoxicological perspective. New York: John Wiley and Sons Ltd, 47–63 (2003).
- 12.Perera F, Tang D, Whyatt R, Lederman SA, Jedrychowski W. DNA damage from polycyclic aromatic hydrocarbons measured by benzo[a]pyrene-DNA adducts in mothers and newborns from Northern Manhattan, the World Trade Center Area, Poland, and China. Cancer Epidemiol Biomark Prev. 2005;14:709–714. doi: 10.1158/1055-9965.EPI-04-0457. [DOI] [PubMed] [Google Scholar]
- 13.Ren A, et al. Association of selected persistent organic pollutants in the placenta with the risk of neural tube defects. Proc Natl Acad Sci USA. 2011;108:12770–12775. doi: 10.1073/pnas.1105209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langlois PH, et al. Maternal occupational exposure to polycyclic aromatic hydrocarbons and risk of oral cleft-affected pregnancies. Cleft Palate Craniofac J. 2013;50:337–346. doi: 10.1597/12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupo PJ, et al. Maternal occupational exposure to polycyclic aromatic hydrocarbons: effects on gastroschisis among offspring in the National Birth Defects Prevention Study. Environ Health Perspect. 2012;120:910–915. doi: 10.1289/ehp.1104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhane DK, et al. Impact of prenatal exposure to polycyclic aromatic hydrocarbons from maternal diet on birth outcomes: a birth cohort study in Korea. Public Health Nutr. 2016;19:2562–2571. doi: 10.1017/S1368980016000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padula AM, et al. Exposure to airborne polycyclic aromatic hydrocarbons during pregnancy and risk of preterm birth. Environ Res. 2014;135:221–226. doi: 10.1016/j.envres.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi H, Rauh V, Garfinkel R, Tu Y, Perera FP. Prenatal exposure to airborne polycyclic aromatic hydrocarbons and risk of intrauterine growth restriction. Environ Health Perspect. 2008;116:658–665. doi: 10.1289/ehp.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Incardona JP, Collier TK, Scholz NL. Defects in cardiac function precede morphological abnormalities in fish embryos exposed to polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol. 2004;196:191–205. doi: 10.1016/j.taap.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Farwell A, Nero V, Croft M, Bal P, Dixon DG. Modified Japanese medaka embryo-larval bioassay for rapid determination of developmental abnormalities. Arch Environ Contam Toxicol. 2006;51:600–607. doi: 10.1007/s00244-005-0319-x. [DOI] [PubMed] [Google Scholar]
- 21.Lupo PJ, et al. Maternal occupational exposure to polycyclic aromatic hydrocarbons and congenital heart defects among offspring in the national birth defects prevention study. Birth Defects Res A Clin Mol Teratol. 2012;94:875–881. doi: 10.1002/bdra.23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nan HM, et al. Effects of occupation, lifestyle and genetic polymorphisms of CYP1A1, CYP2E1, GSTM1 and GSTT1 on urinary 1-hydroxypyrene and 2-naphthol concentrations. Carcinogenesis. 2001;22:787–793. doi: 10.1093/carcin/22.5.787. [DOI] [PubMed] [Google Scholar]
- 24.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, et al. Indoor air pollution and neural tube defects: effect modification by maternal genes. Epidemiology. 2014;25:658–665. doi: 10.1097/EDE.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 27.Strickland MJ, et al. High urine 1-hydroxypyrene glucuronide concentrations in Linxian, China, an area of high risk for squamous oesophageal cancer. Biomarkers. 2001;6:381–386. doi: 10.1080/13547500110044780. [DOI] [PubMed] [Google Scholar]
- 28.Gladen BC, et al. Polycyclic aromatic hydrocarbons in placenta. Hum Exp Toxicol. 2000;19:597–603. doi: 10.1191/096032700671433928. [DOI] [PubMed] [Google Scholar]
- 29.Klaunig JE, Wang Z, Pu X, Zhou S. Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicol Appl Pharmacol. 2011;254:86–99. doi: 10.1016/j.taap.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, et al. Benzo(a)pyrene-induced mitochondrial dysfunction and cell death in p53-null Hep3B cells. Mutat Res. 2011;726:75–83. doi: 10.1016/j.mrgentox.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Donauer J, Schreck I, Liebel U, Weiss C. Role and interaction of p53, BAX and the stress-activated protein kinases p38 and JNK in benzo(a)pyrene-diolepoxide induced apoptosis in human colon carcinoma cells. Arch Toxicol. 2012;86:329–337. doi: 10.1007/s00204-011-0757-3. [DOI] [PubMed] [Google Scholar]
- 32.Wells PG, McCallum GP, Lam KC, Henderson JT, Ondovcik SL. Oxidative DNA damage and repair in teratogenesis and neurodevelopmental deficits. Birth Defects Res C Embryo Today. 2010;90:103–109. doi: 10.1002/bdrc.20177. [DOI] [PubMed] [Google Scholar]
- 33.Herbstman JB, et al. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect. 2012;120:733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bostrom CE, et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.02110s3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilboa SM, et al. Relation between ambient air quality and selected birth defects, seven county study, Texas, 1997-2000. Am J Epidemiol. 2005;162:238–252. doi: 10.1093/aje/kwi189. [DOI] [PubMed] [Google Scholar]
- 36.Padula AM, et al. Ambient air pollution and traffic exposures and congenital heart defects in the San Joaquin Valley of California. Paediatr Perinat Epidemiol. 2013;27:329–339. doi: 10.1111/ppe.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu Y, et al. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2000;97:779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki S, et al. Maternal smoking during pregnancy and genetic polymorphisms in the Ah receptor, CYP1A1 and GSTM1 affect infant birth size in Japanese subjects. Mol Hum Reprod. 2006;12:77–83. doi: 10.1093/molehr/gal013. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi S, et al. Combined effects of AHR, CYP1A1, and XRCC1 genotypes and prenatal maternal smoking on infant birth size: Biomarker assessment in the Hokkaido Study. Reprod Toxicol. 2016;65:295–306. doi: 10.1016/j.reprotox.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi S, et al. Dioxin-metabolizing genes in relation to effects of prenatal dioxin levels and reduced birth size: The Hokkaido study. Reprod Toxicol. 2017;67:111–116. doi: 10.1016/j.reprotox.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Harper PA, Wong J, Lam MS, Okey AB. Polymorphisms in the human AH receptor. Chem Biol Interact. 2002;141:161–187. doi: 10.1016/S0009-2797(02)00071-6. [DOI] [PubMed] [Google Scholar]
- 42.Hirose Y, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Am J Hum Genet. 2008;82:1122–1129. doi: 10.1016/j.ajhg.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang I, et al. Cytochrome P450 1B1 polymorphisms and risk of renal cell carcinoma in men. Tumour Biol. 2014;35:10223–10230. doi: 10.1007/s13277-014-2292-3. [DOI] [PubMed] [Google Scholar]
- 44.Ye XH, et al. Association between the CYP2E1 polymorphisms and lung cancer risk: a meta-analysis. Mol Genet Genomics. 2015;290:545–558. doi: 10.1007/s00438-014-0941-2. [DOI] [PubMed] [Google Scholar]
- 45.Zhu X, et al. Associations between CYP1A1 rs1048943 A > G and rs4646903 T > C genetic variations and colorectal cancer risk: Proof from 26 case-control studies. Oncotarget. 2016;7:51365–51374. doi: 10.18632/oncotarget.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, et al. Cytochrome P450 1A1 (CYP1A1) Gene Polymorphisms and Susceptibility to Breast Cancer: a Meta-Analysis in the Chinese Population. Clin Lab. 2017;63:67–72. doi: 10.7754/Clin.Lab.2016.160535. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez FJ. The use of gene knockout mice to unravel the mechanisms of toxicity and chemical carcinogenesis. Toxicol Lett. 2001;120:199–208. doi: 10.1016/S0378-4274(01)00296-X. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez FJ, Kimura S. Study of P450 function using gene knockout and transgenic mice. Arch Biochem Biophys. 2003;409:153–158. doi: 10.1016/S0003-9861(02)00364-8. [DOI] [PubMed] [Google Scholar]
- 49.Chevrier C, et al. Genetic susceptibilities in the association between maternal exposure to tobacco smoke and the risk of nonsyndromic oral cleft. Am J Med Genet A. 2008;146A:2396–2406. doi: 10.1002/ajmg.a.32505. [DOI] [PubMed] [Google Scholar]
- 50.Lupo PJ, Nousome D, Okcu MF, Chintagumpala M, Scheurer ME. Maternal variation in EPHX1, a xenobiotic metabolism gene, is associated with childhood medulloblastoma: an exploratory case-parent triad study. Pediatr Hematol Oncol. 2012;29:679–685. doi: 10.3109/08880018.2012.722747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karypidis AH, et al. Association of cytochrome P450 1B1 polymorphism with first-trimester miscarriage. Fertil Steril. 2006;86:1498–1503. doi: 10.1016/j.fertnstert.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 52.Mustafa MD, Banerjee BD, Ahmed RS, Tripathi AK, Guleria K. Gene-environment interaction in preterm delivery with special reference to organochlorine pesticides. Mol Hum Reprod. 2013;19:35–42. doi: 10.1093/molehr/gas039. [DOI] [PubMed] [Google Scholar]
- 53.Jenkins MM, et al. Maternal smoking, xenobiotic metabolizing enzyme gene variants, and gastroschisis risk. Am J Med Genet A. 2014;164A:1454–1463. doi: 10.1002/ajmg.a.36478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsson AC, et al. Occupational exposure to polycyclic aromatic hydrocarbons and lung cancer risk: a multicenter study in Europe. Occup Environ Med. 2010;67:98–103. doi: 10.1136/oem.2009.046680. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, et al. Association between maternal exposure to housing renovation and offspring with congenital heart disease: a multi-hospital case-control study. Environ Health. 2013;12:25. doi: 10.1186/1476-069X-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng K, et al. Periconceptional paternal smoking and the risk of congenital heart defects: a case-control study. Birth Defects Res A Clin Mol Teratol. 2013;97:210–216. doi: 10.1002/bdra.23128. [DOI] [PubMed] [Google Scholar]
- 57.Tovey, E. R., Almqvist, C., Li, Q., Crisafulli, D. & Marks, G. B. Nonlinear relationship of mite allergen exposure to mite sensitization and asthma in a birth cohort. J Allergy Clin Immunol122, 114–118, 118 e111–115, 10.1016/j.jaci.2008.05.010 (2008). [DOI] [PubMed]
- 58.Wu, D. M. et al. Relationship Between Neonatal Vitamin D at Birth and Risk of Autism Spectrum Disorders: the NBSIB Study. J Bone Miner Res, 10.1002/jbmr.3326 (2017). [DOI] [PubMed]
- 59.Xu M, et al. Non-Linear Association between Exposure to Ambient Temperature and Children's Hand-Foot-and-Mouth Disease in Beijing, China. PLoS One. 2015;10:e0126171. doi: 10.1371/journal.pone.0126171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, et al. Modification of the association between maternal smoke exposure and congenital heart defects by polymorphisms in glutathione S-transferase genes. Sci Rep. 2015;5:14915. doi: 10.1038/srep14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ou Y, et al. Associations between toxic and essential trace elements in maternal blood and fetal congenital heart defects. Environ Int. 2017;106:127–134. doi: 10.1016/j.envint.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Chen D, et al. Association of human aryl hydrocarbon receptor gene polymorphisms with risk of lung cancer among cigarette smokers in a Chinese population. Pharmacogenet Genomics. 2009;19:25–34. doi: 10.1097/FPC.0b013e328316d8d8. [DOI] [PubMed] [Google Scholar]
- 63.Gu A, et al. Contributions of aryl hydrocarbon receptor genetic variants to the risk of glioma and PAH-DNA adducts. Toxicol Sci. 2012;128:357–364. doi: 10.1093/toxsci/kfs158. [DOI] [PubMed] [Google Scholar]
- 64.Choi YH, Kim JH, Hong YC. CYP1A1 genetic polymorphism and polycyclic aromatic hydrocarbons on pulmonary function in the elderly: haplotype-based approach for gene-environment interaction. Toxicol Lett. 2013;221:185–190. doi: 10.1016/j.toxlet.2013.06.229. [DOI] [PubMed] [Google Scholar]
- 65.Stasiukonyte, N., Liutkeviciene, R., Vilkeviciute, A., Banevicius, M. & Kriauciuniene, L. Associations between Rs4244285 and Rs762551 gene polymorphisms and age-related macular degeneration. Ophthalmic Genet 1–8, 10.1080/13816810.2016.1242018 (2017). [DOI] [PubMed]
- 66.Lin KM, et al. CYP1A2 genetic polymorphisms are associated with treatment response to the antidepressant paroxetine. Pharmacogenomics. 2010;11:1535–1543. doi: 10.2217/pgs.10.128. [DOI] [PubMed] [Google Scholar]
- 67.Abnet CC, et al. The influence of genetic polymorphisms in Ahr, CYP1A1, CYP1A2, CYP1B1, GST M1, GST T1 and UGT1A1 on urine 1-hydroxypyrene glucuronide concentrations in healthy subjects from Rio Grande do Sul, Brazil. Carcinogenesis. 2007;28:112–117. doi: 10.1093/carcin/bgl131. [DOI] [PubMed] [Google Scholar]
- 68.Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- 69.Ye LL, et al. Are polymorphisms in metabolism protective or a risk for reduced white blood cell counts in a Chinese population with low occupational benzene exposures? Int J Occup Environ Health. 2015;21:232–240. doi: 10.1179/2049396714Y.0000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang S, et al. Cytochrome P450 2E1 gene polymorphisms/haplotypes and anti-tuberculosis drug-induced hepatitis in a Chinese cohort. PLoS One. 2013;8:e57526. doi: 10.1371/journal.pone.0057526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.