Abstract
Ubiquitin-specific protease 2 (USP2) belongs to the family of deubiquitinases that can rescue protein targets from proteasomal degradation by reversing their ubiquitination. In various cancers, including prostate cancer and ovarian carcinoma, upregulation of USP2 leads to an increase in the levels of deubiquitinated substrates such as fatty acid synthase, MDM2, cyclin D1 and Aurora-A. USP2 thus plays a critical role in tumor cells’ survival and therefore represents a therapeutic target. Here a leukemia drug, 6-thioguanine, was found to be a potent inhibitor of USP2. Enzyme-kinetic and X-ray crystallographic data suggest that 6-thioguanine displays a noncompetitive and slow-binding inhibitory mechanism against USP2. Our study provides a clear rationale for the clinical evaluation of 6-thioguanine for USP2-upregulated cancers.
Introduction
Ubiquitination, a kind of post-translational modification, as well as deubiquitination, which together comprise the ubiquitination system, can regulate not only the turnover of cellular proteins but also, through various modes of mono- or polyubiquitination, receptor trafficking, DNA repair, cell-cycle progression, immune response, autophagy and apoptosis1. This versatility has raised interest in pinpointing components of the ubiquitin system as prime candidates for drug targets2. Among them, the pursuit of deubiquitinases (DUBs) as pharmaceutical targets is still in its infancy in that few inhibitors have entered clinical trials, albeit some broad-spectrum inhibitors have been patented and highly selective inhibitors have been developed3–6. The human genome encodes about 80–90 DUBs, of which the largest of the five DUB families is the ubiquitin-specific proteases (USPs)7. One of these, USP2, is able to remove ubiquitin from a range of protein substrates such as fatty acid synthase8, MDM29, cyclin D110 and Aurora-A11. Fatty acid synthase has been reported to be highly expressed in a variety of cancers, including cancers of the prostate, ovary, lung, colon, and breast12, while MDM2, cyclin D1 and Aurora-A play critical roles in p53 regulation, cell cycle progression and mitotic events, respectively9,10,13. All of these studies suggest that the inhibition of USP2 may be a good therapeutic approach against cancers. In 2016, Davis et al.6 reported that a small molecule, ML364, can induce cell cycle arrest and inhibit the growth of colorectal and mantle cell lymphoma cell lines via USP2 inhibition, albeit it is still not a clinical drug. In the present study, 6-thioguanine (6TG), an old drug still clinically used to treat leukemia, rheumatoid arthritis and bowel disease14, was identified as a potent USP2 inhibitor. Further studies using enzyme kinetics and X-ray crystallography to delineate its direct binding to USP2 were performed. Our findings support earlier results suggesting that 6TG has a broad spectrum of activity15–17, and have implications with respect to the need for direct clinical evaluation of its use against USP2-upregulated cancers.
Results and Discussion
Identification of inhibitors of human USP2
According to previous studies involving molecular docking experiments, the anti-leukemia and immunosuppressant drugs 6MP and 6TG, which act as inhibitors of SARS and MERS coronavirus papain-like proteases (PLpros), may also be able to bind to the active site of human USP2 as both kinds of enzymes share the palm-thumb-fingers structural scaffold and conserved catalytic triad Cys-His-Asp/Asn17–20. To validate the simulation results, the deubiquitination activity of the recombinant human USP2 catalytic domain (residues 258–605) was measured in the presence of various concentrations of 6MP or 6TG (Fig. 1A,B and Table 1). Interestingly, only 6TG but not 6MP shows a significant inhibitory effect with an IC50 of 40 ± 2.0 μM (Fig. 1A,B), whereas both compounds can inhibit coronaviral PLpro with an IC50 of 25 μM17,18. It is the first demonstration that besides serving as an anti-leukemia drug, 6TG might also be an effective chemotherapeutic agent against cancers dependent on USP2 upregulation8,21,22. Similar concept has been addressed by Davis et al.6. Using the strategies of high-throughput screening and chemical medicinal optimization, they identified a sulfamidobenzamide derivative, ML364, having an IC50 of 1.1 μM against USP2 (Table 1) and resulting in the growth arrest of several kinds of USP2-upregulated cancer cells. However, it is not a clinical medicine yet and the detailed inhibition mechanism is still unclear.
Figure 1.
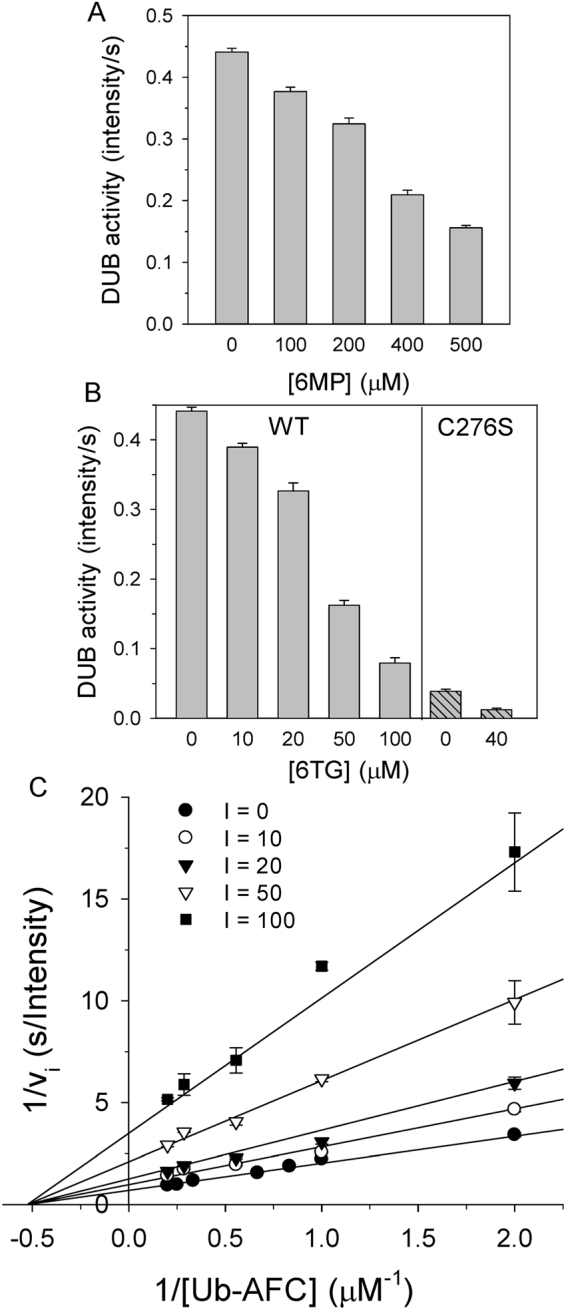
Inhibition of human USP2 deubiquitinating activity by 6MP or 6TG. (A) The activity of USP2 in the presence of various concentrations of 6MP was measured. (B) The activity of USP2 (left panel) or its C276S mutant (right panel) in the presence of various concentrations of 6TG was measured. In panel A and B, 1 μM Ub-AFC and 0.2 μM USP2 was used as the substrate and the enzyme, respectively. (C) Noncompetitive inhibition of USP2 by 6TG. Enzyme activity was measured under various concentrations of Ub-AFC (0.5–5 μM) and 6TG (0–100 μM), while protein concentration was held at 0.2 μM. The assays were performed in 20 mM phosphate (pH 7.6) at 30 °C and repeated to ensure reproducibility. The symbols and bars represent the mean and standard error, respectively. The solid lines represent the best global fit to the noncompetitive inhibition equation. Rsqr is 0.981 and the kinetic parameters from the best fit are shown in Table 1.
Table 1.
Kinetic parameters of 6MP and 6TG inhibition of human USP2 via deubiquitination assay.
| USP2 in | Structure | Kis (μM) | Kinact (μM)b |
|---|---|---|---|
| 6 MPa |
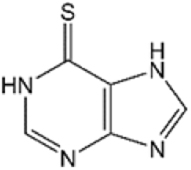
|
349.0 ± 23.7 | — |
| 6TGa |
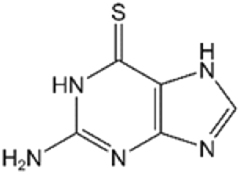
|
24.6 ± 1.8 | 15.6 ± 3.2 |
| ML364c |
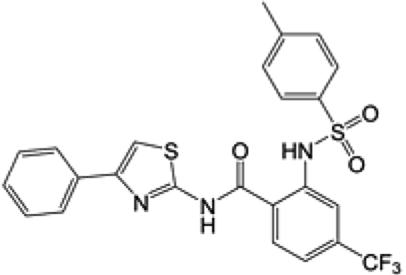
|
IC50: 1.1 μM |
aIn the presence of 6MP or 6TG, the apparent kinetic parameters were determined from the best fit of the data to a noncompetitive inhibition model (Eq. 1). The Rsqr were 0.980 and 0.981 for that of 6MP and 6TG, respectively.
bThe Kinact was determined from the best fit of the data to the saturation equation.
cML364 is a USP2 inhibitor and the IC50 value was from Davis et al.6.
Inhibition mechanism of 6TG
To understand the detailed kinetic mechanisms involved in the interaction of 6TG with USP2, the deubiquitination activity of the enzyme was measured under various combinations of inhibitor concentrations and Ub-AFC substrate concentrations. The inhibition data were globally fitted to all possible kinetic models including competitive, noncompetitive, uncompetitive and mixed inhibition models. Interestingly, the results suggest that a noncompetitive inhibition pattern can best describe the inhibition by 6TG with the double reciprocal plot showing all lines intercepting the x-axis and a Kis of 24.6 μM (Fig. 1C and Table 1). On the other hand, although 6MP also showed a noncompetitive inhibition pattern, the Kis of 6MP inhibition is 14-fold higher than that of 6TG (Table 1). The Kis for 6TG inhibition was 11-fold higher than the KM (2.3 ± 0.2 μM) of the deubiquitination reaction, whereas that for 6MP inhibition was 152-fold higher than the KM of the deubiquitination reaction. The latter Kis value indicates the ineffectiveness of 6MP for USP2 inhibition. In contrast, both 6TG and 6MP show a competitive inhibition pattern against coronaviral PLpro17,18.
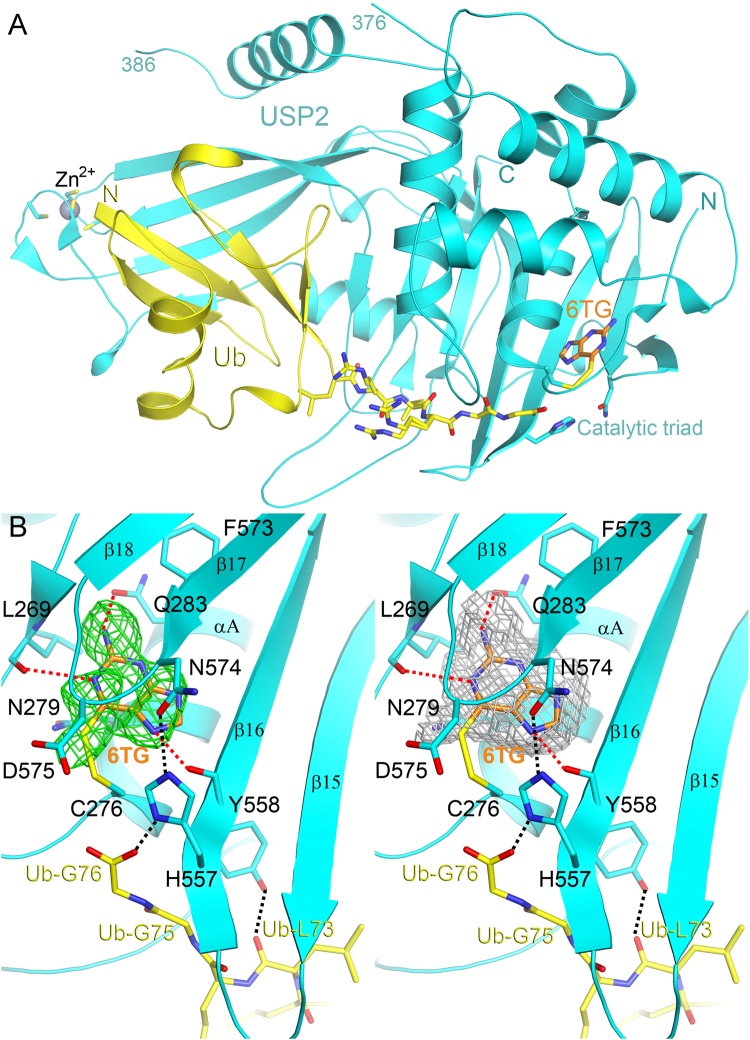
X-ray structure determination of human USP2 catalytic domain in complex with Ub and 6TG
Up to now, although USP2 inhibitors such as ML364 and chalcone-based small molecules5,6 have been discovered, no USP2-inhibitor complex structure has yet been solved. Moreover, as a noncompetitive inhibitor may show benefit in combination or adjuvant treatment with a competitive inhibitor, we are very interested in validating the binding mode of 6TG to USP2. Therefore, we tried to determine the structure of USP2 in complex with 6TG by X-ray crystallography. Although repeated attempts at crystallization of USP2 in complex with 6TG failed, we finally succeeded in the crystallization of the USP2 in complex with Ub and then soaked the crystals in the presence of 6TG. The crystal structure of the USP2-Ub-6TG complex was determined at 1.8 Å resolution (Table 2 and Fig. 2A). Fortunately, some residual electron density like 6TG molecule lied on one side of the catalytic triad of USP2, while Ub is located on the opposite side (Fig. 2B and extended data Fig. 1). There is no residual electron density like 6TG in the same place while refined using the diffraction data without 6TG soaking. However, after refinement, the temperature factor of 6TG is 65.2 Å2, higher than 38.9 Å2 of the protein. Decrease the occupancy to 50% can lower the temperature factor of the molecule to 40.9 Å2. It indicates that 6TG may not exist as a full but half occupancy in an asymmetric unit. We have tried to increase the soaking concentration or soaking time to enhance the occupancy or electron density but failed. The co-crystallization of the protein with 6TG also failed. Nevertheless, the structure indicates that 6TG has polar interactions with residues Leu269, Gln283 and Tyr558 and van der Waals contacts with nearby residues Asn279 and Phe573, while may also show a possible covalent bonding interaction between the sulfur atom of 6TG and the thiol group of residue Cys276 (Fig. 2B). Unlike 6TG, 6MP lacks an amide group (Table 1). This may explain the ineffectiveness of 6MP against USP2, as there is a hydrogen-bonding interaction between the amide of 6TG and the side-chain oxygen atom of the residue Gln283. Overall, the ternary complex structure of USP2-Ub-6TG suggests the existence of an enzyme-substrate-inhibitor (ESI) complex and confirms the noncompetitive mechanism of USP2 inhibition by 6TG.
Table 2.
X-ray diffraction data collection and refinement statistics.
| Human USP2-Ub-6TG complex | Human USP2 C276S mutant-Ub complex | |
|---|---|---|
| Data Collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 103.0, 54.7, 72.7 | 102.5, 54.0, 74.8 |
| β (°) | 107.5 | 107.8 |
| Resolutiona (Å) | 30–1.80 (1.86–1.80) | 30–1.24 (1.28–1.24) |
| Rmergeb (%) | 5.1 (46.6) | 3.4 (24.1) |
| I/σI | 19.9 (2.4) | 33.0 (5.7) |
| Completeness (%) | 97.9 (80.1) | 98.5 (96.9) |
| Redundancy | 3.7 (3.5) | 3.6 (3.4) |
| Refinement | ||
| Number of reflections | 33,397 (4,726) | 102,800 (14,652) |
| R factorc (%) | 17.9 (26.7) | 14.8 (15.9) |
| Free R factord (%) | 23.8 (31.0) | 16.4 (18.2) |
| Number of atoms | 3,568 | 3,761 |
| Protein | 3,285 | 3,404 |
| Ligand/ion | 11/11 | 0/1 |
| Water | 261 | 356 |
| B-factors (Å2) | ||
| Protein | 38.9 | 14.7 |
| Ligand/ion | 65.2 (40.7)e/57.9 | −/13.7 |
| Water | 50.9 | 23.9 |
| rmsd | ||
| Bond length (Å) | 0.012 | 0.007 |
| Bond angles (°) | 1.5 | 1.2 |
| Ramachandran analysis (%) | ||
| Favored | 91.6 | 92.6 |
| Allowed | 8.4 | 7.5 |
aThe numbers in parentheses are for the highest-resolution shell.
b, where Ihi is the integrated intensity of a given reflection and is the mean intensity of multiple corresponding symmetry-related reflections.
c, where and are the observed and calculated structure factors, respectively.
dFree R is R calculated using a random 5% of data excluded from the refinement.
eThe value in parentheses is calculated at 50% occupancy.
Figure 2.
Structure of human USP2 in complex with Ub and 6TG. (A) The overall structure of the USP2 258–605 fragment (cyan) in complex with ubiquitin (yellow) and 6TG (orange). In the structure, the residues from 377 to 385 (dashed line) are missing. The zinc ion, coordinated by four cysteine residues, is shown as a grey sphere while the catalytic triad (Cys276-His557-Asn574) and the final six C-terminal residues of Ub are shown as sticks. (B) Stereo view of the active site of USP2. The omit Fo–Fc electron-density map of 6TG at 1.8 Å, contoured at 2.5σ, is shown as green mesh. To improve the OMIT map quality, the polder map of 6TG (grey mesh) excluding bulk solvent contoured at 1.2σ was generated using PHENIX31,32. The dashed lines indicate hydrophilic interactions between USP2 and 6TG or Ub. One of the catalytic triad residues, Cys276, has a disulfide bonding interaction with 6TG. All structure figures in this paper were produced using PyMol (http://www.pymol.org/).
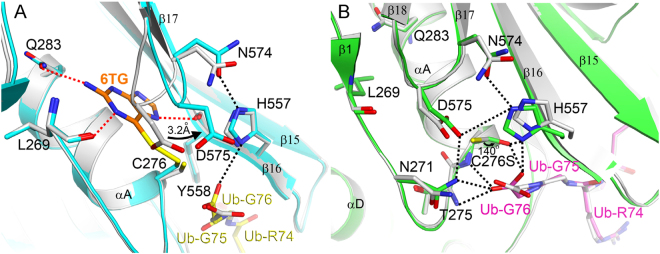
Structural comparison of human USP2-Ub in complex with or without 6TG
Because of the partial occupancy of 6TG in the complex structure, conformational change due to ligand binding can be used to further evaluate the binding effect. Here we compare the active site of USP2 in the presence or absence of 6TG (Fig. 3A). Interestingly, binding of 6TG leads to movement of the β17–β18 loop (residues 574–577), resulting in a 3.2 Å shift of the residue Asp575 toward His557. Previous studies suggested that the absolutely conserved residue Asp575 in the USPs plays a dual role in oxyanion hole formation and maintaining the correct alignment and protonation of His557 for catalytic competency, although there is no direct interaction between them in the crystal structure of USP2-Ub complex23,24. Accordingly, the movement of residue Asp575 following 6TG binding also results in the inhibition of USP2. Besides, in the present structure, the carboxyl group of Ub-Gly76 has polar interaction with residue His557 (Fig. 3A). This difference may be due to the change of the free thiol group of Cys276 to disulfide, which is less negative and then has less electrostatic repulsion with the carboxyl group of Ub-Gly76.
Figure 3.
Comparison with other structures of human USP2. Overlay of the active site of human USP2-Ub complex (grey; PDB code: 2hd5) with that of USP2-Ub-6TG complex (USP2: cyan; Ub: yellow; 6TG: orange) (A) or that of USP2 C276S mutant (green) in complex with Ub (magenta) (B). The dashed lines show hydrophilic interactions. The arrow in panel (A) indicates the movement of residue Asp575, while that in panel (B) shows the side-chain movement of residue 276, which has been mutated from cysteine to serine.
Structural comparison of human USP2-Ub and USP2 C276S-Ub
In an earlier study, the structure of wild-type USP2 in complex with Ub showed a misorientation of the thiol group of Cys276 due to electrostatic repulsion between the residue and the carboxyl group of Ub-Gly7623. To resolve this, in the present study, the crystal structure of the USP2 C276S mutant in complex with Ub was determined at 1.24 Å resolution (Table 2). Indeed, the hydroxyl group of Ser276 shows a 140° orientation change compared with the thiol group of Cys276 (Fig. 3B). This change results in the rescue of the hydrogen-bonding interaction between Ser276 and His557 and the reconnection of the catalytic triad even in the presence of Ub. Interestingly, but not surprisingly, the USP2 C276S mutant still maintains 10% deubiquitinating activity (Fig. 1B). This indicates that the C276S mutant is less active but not fully inactive. Furthermore, in the presence of 40 μM 6TG, the DUB activity of the USP2 C276S mutant decreases to 30%, suggesting that 6TG can still inhibit the enzyme without the disulfide-bonding interaction. Compared with those of wild-type USP2, the residues interacting with 6TG, such as Leu269, Gln283 and Tyr558, have no conformational change in C276S mutant, indicating the existence of 6TG binding pocket (Fig. 3B). The movement of residue Asp575 following 6TG binding may also result in the inhibition of C276S mutant. Moreover, in the present structure, one oxygen atom of the carboxyl group of Ub-Gly76 is located in the oxyanion hole consisting of the three amides from Asn271, Thr275 and Ser276 (Fig. 3B). Previous studies indicated that mutation of residue Asn271 (N271A) has no influence on kinetic parameters such as kcat and KM24. This indicates that the two main-chain amides from residues 275 and 276 are enough to stabilize oxyanion formation during catalysis. On the other hand, different to the USP2-Ub complex, USP2 C276S-Ub complex shows electrostatic interaction between residues Asp575 and His557 (Fig. 3B). It suggests that Asp575 may directly influence the protonation of His557, not via the interaction between Asn271 and Asp57524. Overall, the present structure provides a foundation for more understanding the molecular basis of USP2 catalysis.
Slow-binding inhibition of USP2 by 6TG
According to the ternary structure of USP2-Ub-6TG, the enzyme may have a covalent-bonding interaction with 6TG, albeit 6TG showed partial occupancy (Fig. 2B). Although rare, the disulfide-bonding formation of 6TG has been confirmed by van der Vlies et al. in 201225. In the present study, to validate the reversibility of the enzyme inactivation, USP2 was incubated with 1 mM 6TG for 2 h followed by removal of the small molecules using a Sephadex G-25 column. This treatment led to a 95% loss of activity, suggesting irreversible inhibition of the USP2 by 6TG. The irreversible inhibition of enzyme activity indicates that 6TG may be covalently associating with the enzyme26. Next, the 6TG-labelled USP2 was incubated with 2 or 20 mM βME for 10 min and then measured the activity to see if any re-activation. Although not very high, there is 14% and 20% of the enzyme activity restored after treated by βME, respectively. The rescuing effect of reductant suggests that the modification may be due to the disulfide bonding interaction between the enzyme and 6TG. Nevertheless, the above observations suggest that 6TG may be a “slow-binding” inhibitor26. Here we measured the time-resolved emission for 200 s under various concentrations of 6TG and found that the emission curve was curved downward, indicating a slow-binding mechanism (Fig. 4A). Different kinact at various concentrations of 6TG were determined by fitting the data to Eq. 2 and then plotted versus those various concentrations of 6TG (Fig. 4B). The saturation pattern and irreversible inhibition of enzyme activity suggests that slow binding may be due to affinity labeling, in accordance with Copland’s models26. Following this, fitting the data to the saturation equation, we obtained a Kinact of 15.6 μM (Fig. 4B and Table 2). This value is even lower than Kis, indicating that 6TG may undergo a covalent-bonding interaction with USP2 very soon after binding.
Figure 4.
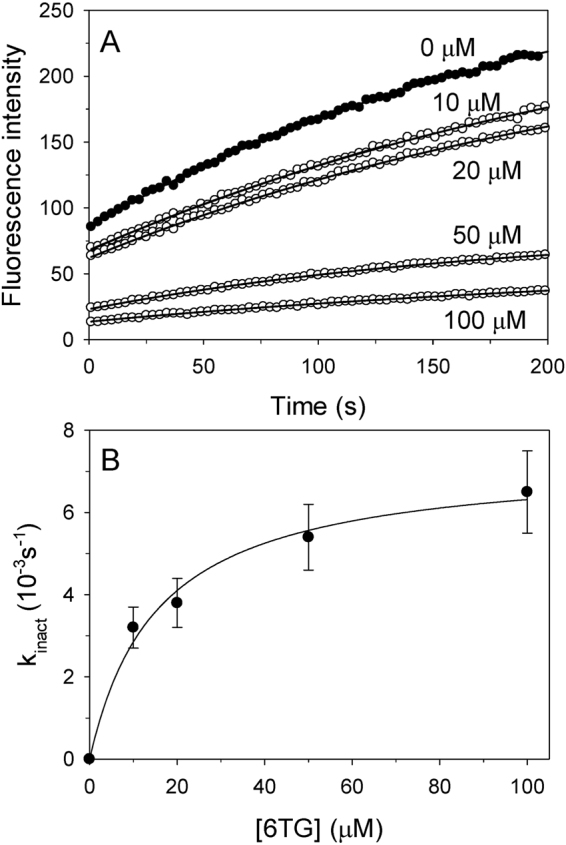
Time-dependent inactivation of USP2 by 6TG. (A) Different concentrations of 6TG (0 μM, closed circles; 10–100 μM, open circles) were incubated with USP2 and enzyme activity was measured for 200 s. Across all trials, Ub-AFC concentration was held at 0.5 μM and USP2 concentration was held at 0.2 μM. The solid lines show the best fit results when the data was fitted to the slow-binding equation. (B) The observed inactivation rate constants (kinact) from panel A were replotted against 6TG concentrations. The solid line represents the best fit of the data to the saturation equation. The apparent Kinact value is shown in Table 1.
Conclusion
Here we found that 6TG can noncompetitively inhibit human USP2. It also shows a slow-binding inhibitory effect against the same enzyme. The kinetic and catalytic mechanism was further confirmed by X-ray crystallography. The ternary complex structure of USP2-Ub-6TG can be interpreted as an enzyme-substrate-inhibitor complex, suggesting allosteric binding of inhibitor and substrate on the same enzyme. The binding of 6TG also leads to the movement of Asp575, an absolutely conserved residue playing an essential role in catalysis. This further explains why the binding of 6TG can inhibit USP2. Finally, the irreversible inhibition of the enzyme by the inhibitor and the slow-binding inhibition via affinity labeling suggested a possible covalent bonding interaction between 6TG and the residue Cys276 of USP2. Our findings once again revealed the broad spectrum of activity of 6TG and provides biochemical and structural evidence for evaluating the possible application of 6TG on the clinical or combinational treatment against those USP2-upregulated cancers.
Materials and Methods
Plasmid construction
The cDNA of human USP2 (Gene ID: 9099) catalytic domain (residues 258 to 605) was amplified by polymerase chain reaction with the primers 5′-GGCGCTAGCATGAATTCTAAGAGTGCCCAG-3′ and 5′-GGCCTCGAGCATTCGGGAGGGCGGGC-3′, using pOTB7-USP2 (MGC premier cDNA clone ID: BC002854) as a template. The PCR product was digested by NheI-XhoI and then inserted into the vector pET-24d with a 6x-His tag at the C-terminus (Novagen). The primer sequences for site-directed mutagenesis of the USP2 C276S mutant were 5′-CGAAACCTTGGGAACACGAGCTTCATGAACTCAATTCTG-3′ and 5′-CAGAATTGAGTTCATGAAGCTCGTGTTCCCAAGGTTTCG-3′. The reading frames of the above plasmids were verified by DNA sequencing.
Expression and purification of human USP2
In the present study, we used the catalytic domain (residues 258–605) since the full-length USP2 has not been obtained despite our effort to express the full-length protein by sumo-fusion strategy. The cloned plasmid was transformed into Escherichia coli Rosetta (DE3) cells (Novagen). Cultures of 0.8 l LB medium with 25 μg/ml kanamycin and 17 μg/ml chloramphenicol were grown at 37 °C for 4 h and then incubated at 20 °C for 20 h after adding 0.4 mM isopropyl-β-D-thiogalactopyranoside. After centrifugation, the cell pellets were resuspended in lysis buffer (20 mM Tris, pH 8.5, 250 mM NaCl, 5% glycerol, 0.2% Triton X-100 and 2 mM β-mercaptoethanol (βME)), sonicated and then centrifuged at 12,000 g at 4 °C for 20 min to remove the insoluble lysate. The supernatant was mixed with 1 ml Ni-NTA agarose (Qiagen) at 4 °C for 1 h, washed with buffer containing 20 mM Tris, pH 8.5, 250 mM NaCl, 8 mM imidazole and 2 mM βME and then eluted with buffer containing 20 mM Tris, pH 8.5, 30 mM NaCl, 150 mM imidazole and 2 mM βME. Next, the protein was loaded onto an S-100 gel-filtration column (GE Healthcare) using running buffer (15 mM sodium phosphate, pH 6.0, 100 mM NaCl and 2 mM dithiothreitol). The purity of the collected fraction was checked by SDS-PAGE and the protein was concentrated using an Amicon Ultra-4 30 kDa filter (Millipore). The final purified protein at 15 mg/ml with 5% glycerol was flash-frozen with liquid nitrogen and stored at −80 °C. The typical yield of protein was 15 mg for wild-type USP2 and 30 mg for C276S mutant per liter of cell culture.
Deubiquitination assays and steady-state kinetic analyses
Deubiquitination assays were performed in 20 mM phosphate buffer (pH 7.6) at 30 °C. The concentration of the fluorogenic substrate ubiquitin-7-amino-4-trifluoromethylcoumarin (Ub-AFC; Boston Biochem) was varied while the concentration of USP2 catalytic domain or its C276S mutant was held at 0.2 μM. Enzyme activity was measured by continuously monitoring the fluorescence emission increase at 485 nm upon substrate cleavage at excitation wavelength of 350 nm in a PerkinElmer LS 50B luminescence spectrometer. The increase in fluorescence was linear for the first 25 s. Kinetic parameters were obtained by fitting the initial velocities (vi) to the Michaelis-Menten equation. The program Sigmaplot 12 (Systat Software) was used for data analysis.
For the inhibition assays, concentrations of the substrate Ub-AFC and an inhibitor, 6-mercaptopurine (6MP) or 6TG, were varied while the concentration of the USP2 catalytic domain was held at 0.2 μM. The inhibition data for the inhibitors was found to best fit a noncompetitive inhibition model according to Eq. (1):
| 1 |
in which Vmax is the maximal initial velocity of the enzyme while [S] and [I] denote the concentrations of the substrate and inhibitor, respectively. KM is the Michaelis-Menten constant for the enzyme-substrate complex and Kis is the inhibition constant for the enzyme-inhibitor complex.
Slow-binding inhibition mechanism
To study slow-binding inhibition by 6TG, the USP2 catalytic domain at a concentration of 0.2 μM was incubated with different concentrations of 6TG in the presence of 0.5 μM Ub-AFC substrate. Enzyme activity was traced for 200 s and all of the progress curves showed exponential decay. The experimental data was analyzed according to the integrated rate equation (Eq. 2)18,26:
| 2 |
in which vs is the steady-state velocity, kinact is the apparent first-order rate constant for the interconversion between vi and vs and t is time.
Protein crystallization
Crystals of the human USP2 catalytic domain or C276S mutant in complex with Ub (Sigma-Aldrich) were obtained by the sitting-drop vapor-diffusion method. After incubating overnight, the USP2-Ub mixture (6.6:3.3 mg/ml; molar ratio 1:2) was crystallized in a reservoir solution consisting of 24.5% PEG 3350, 0.2 M MgCl2 and 0.1 M Bis-Tris (pH 7.0), while the USP2 C276S-Ub mixture was crystallized in a solution of 21% PEG 3350 and 8% Tacsimate (pH 8.0). Single diamond-shaped crystals 0.2–0.3 mm in size grew at 22 °C after a week. The USP2-Ub crystals were soaked in reservoir solution supplemented with 1 mM 6TG for 4 h and then flash-cooled in liquid nitrogen.
X-ray data collection, structure determination and refinement
X-ray diffraction data were collected at 100 K on beamlines BL13C1 and BL15A1 at the National Synchrotron Radiation Research Center, Taiwan, ROC, using an ADSC Q315r or Rayonix MX300HE CCD detector (X-ray wavelength 0.976 Å). The image data was processed and scaled with the HKL-2000 package27. Both kinds of crystals belong to space group C2; the cell dimensions of the USP2-Ub-6TG crystal were a = 103.0, b = 54.7, c = 72.7 Å, α = γ = 90° and β = 107.5°, while those of USP2 C276S-Ub were a = 102.5, b = 54.0, c = 74.8 Å, α = γ = 90° and β = 107.5°. The crystallographic asymmetric unit contained one USP2 catalytic domain and one Ub. The structures were solved by the molecular replacement method with Phaser28 using the structure of USP2-Ub (PDB code: 2hd523) as the model. Structural refinement was carried out using REFMAC29 and the atomic model was built using Coot30.
Accession codes
The final structure information has been deposited in the Protein Data Bank (PDB code: 5xu8 and 5xve for USP2-Ub-6TG and USP2 C276S-Ub, respectively).
Electronic supplementary material
Acknowledgements
This research was supported by grants from the Ministry of Science and Technology, Taiwan (104–2320-B-010–034, 105–2320-B-010–012 and 106–2320-B-010–013) to CYC and CGMH-NYMU joint research grant (CMRPG2F0431) to CYS and CYC. We are grateful for the experimental facilities and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility, which is supported by the National Core Facility Program for Biotechnology, Ministry of Science and Technology, Taiwan, ROC, and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, ROC.
Author Contributions
S.J.C. carried out most of the experiments and analyzed the kinetic data. S.C.C. carried out the construction of the expression vector as well as protein expression and purification. H.C.T. crystallized the USP2 C276S mutant in complex with Ub, collected the X-ray diffraction data and solved the structure. C.Y.S. participated in experimental design and scientific suggestions. C.Y.C. conceived the entire study, participated in experimental design and wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21476-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Guedat P, Colland F. Patented small molecule inhibitors in the ubiquitin proteasome system. BMC Biochem. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2091-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issaenko OA, Amerik AY. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle. 2012;11:1804–1817. doi: 10.4161/cc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MI, et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J Biol Chem. 2016;291:24628–24640. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 8.Graner E, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/S1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 9.Stevenson LF, et al. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36:469–476. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, et al. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J Biol Chem. 2011;286:38960–38968. doi: 10.1074/jbc.M111.231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/S0899-9007(99)00266-X. [DOI] [PubMed] [Google Scholar]
- 13.Andrews PD. Aurora kinases: shining lights on the therapeutic horizon? Oncogene. 2005;24:5005–5015. doi: 10.1038/sj.onc.1208752. [DOI] [PubMed] [Google Scholar]
- 14.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 15.Chan JF, et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munshi PN, Lubin M, Bertino JR. 6-thioguanine: a drug with unrealized potential for cancer therapy. Oncologist. 2014;19:760–765. doi: 10.1634/theoncologist.2014-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng KW, et al. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral research. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou CY, et al. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Chou CY, Chang GG. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antiviral chemistry & chemotherapy. 2009;19:151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 20.Chou CY, et al. Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease. Acta crystallographica. Section D, Biological crystallography. 2014;70:572–581. doi: 10.1107/S1399004713031040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gansler, T. S., Hardman, W. 3rd., Hunt, D. A., Schaffel, S. & Hennigar, R. A. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum Pathol28, 686–692 (1997). [DOI] [PubMed]
- 22.Hunt DA, Lane HM, Zygmont ME, Dervan PA, Hennigar RA. MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines. Anticancer Res. 2007;27:27–34. [PubMed] [Google Scholar]
- 23.Renatus M, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, et al. Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases. Biochemistry. 2011;50:4775–4785. doi: 10.1021/bi101958h. [DOI] [PubMed] [Google Scholar]
- 25.van der Vlies AJ, Hasegawa U, Hubbell JA. Reduction-sensitive tioguanine prodrug micelles. Mol Pharm. 2012;9:2812–2818. doi: 10.1021/mp3001183. [DOI] [PubMed] [Google Scholar]
- 26.Copeland, R. Enzymes: a practical introduction to structure, mechanism, and data analysis. 2nd edn, (Wiley-VCH Inc., 2000).
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebschner D, et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.