Summary
The GT‐1 cis‐element widely exists in many plant gene promoters. However, the molecular mechanism that underlies the response of the GT‐1 cis‐element to abiotic and biotic stresses remains elusive in rice. We previously isolated a rice short‐chain peptide‐encoding gene, Os2H16, and demonstrated that it plays important roles in both disease resistance and drought tolerance. Here, we conducted a promoter assay of Os2H16 and identified GT‐1 as an important cis‐element that mediates Os2H16 expression in response to pathogen attack and osmotic stress. Using the repeated GT‐1 as bait, we characterized an abscisic acid, stress and ripening 2 (ASR2) protein from yeast‐one hybridization screening. Sequence alignments showed that the carboxy‐terminal domain of OsASR2 containing residues 80–138 was the DNA‐binding domain. Furthermore, we identified that OsASR2 was specifically bound to GT‐1 and activated the expression of the target gene Os2H16, as well as GFP driven by the chimeric promoter of 2 × GT‐1‐35S mini construct. Additionally, the expression of OsASR2 was elevated by pathogens and osmotic stress challenges. Overexpression of OsASR2 enhanced the resistance against Xanthomonas oryzae pv. oryzae and Rhizoctonia solani, and tolerance to drought in rice. These results suggest that the interaction between OsASR2 and GT‐1 plays an important role in the crosstalk of the response of rice to biotic and abiotic stresses.
Keywords: bacterial blight, cis‐element, drought, defence response, rice, sheath blight disease
Introduction
Plants have developed complex mechanisms to defend against pathogen invasion. Upon pathogen recognition, various biochemical events occur in plant cells, such as reactive oxygen species burst, hypersensitive response, antimicrobial peptides and phytoalexins accumulation and callose deposition (Hammond‐Kosack and Jones, 1996; Muthamilarasan and Prasad, 2013). The transcriptional activation of numerous genes upon pathogen infection has been demonstrated. The signal transduction pathways in these activations are of particular interest. It is supposed that these activations are caused by both transcription factors (TFs) and corresponding cis‐elements in specific promoter regions (Buscaill and Rivas, 2014; Huang et al., 2012). Therefore, an important step of understanding gene activation mechanism is to identify the specific cis‐elements that are responsive to external signals.
Currently, studies have shown that many cis‐elements are required for the transcriptional regulation of defence genes under biotic stress. The GCC‐box (Muthamilarasan et al., 2015; Van der Does et al., 2013; Wang et al., 2013a) and the W boxes (Gao et al., 2016; Liu et al., 2016) are two well‐studied pathogen‐inducible cis‐elements. The GCC‐box (AGCCGCC) usually locates in the promoters of defence genes (Brown et al., 2003; Chakravarthy et al., 2003; Zarei et al., 2011). Two GCC‐like elements, such as JERE (AGACCGCC) and box S (AGCCACC), have been reported to regulate jasmonate‐ and elicitor‐responsive expression (Kirsch et al., 2001; Memelink et al., 2001). The W boxes [(T)TGAC(C/T)] are a major class of cis‐elements responsible for the induction of many plant genes by pathogen infection (Gao et al., 2016; Laloi et al., 2004; Lippok et al., 2007; Mohr et al., 2010; Yamamoto et al., 2004). Studies of the Arabidopsis transcriptome have also illustrated the importance of the W boxes during systemic acquired resistance (Maleck et al., 2000; Petersen et al., 2000). In addition to the above two cis‐elements, other pathogen‐inducible cis‐elements have been found in the defence gene promoters, including PRE2 and PRE4 (Cai et al., 2008), G‐box (Alves et al., 2013), E‐box (Miyamoto et al., 2012), GT‐1 (Park et al., 2004) and MYB recognition elements (MREs, Tao et al., 2015). However, the regulation mechanisms of these cis‐elements in response to pathogen attack remain elusive.
Bacterial blight disease, which is caused by biotrophic pathogen Xanthomonas oryzae pv. oryzae (Xoo), and sheath blight disease, which is caused by the necrotrophic fungus Rhizoctonia solani, are the two most common and economically important diseases of rice. To date, over 41 major resistance genes have been identified to against various strains of Xoo and some have been characterized (Cheema et al., 2008; Guo et al., 2010; Miao et al., 2010; Verdier et al., 2012; Wang et al., 2009, 2014; Zhang et al., 2014). Moreover, altering the expression of certain transcription factors, such as OsC3H12 (Deng et al., 2012), OsWRKY45 (Shimono et al., 2012) and TaCPK2‐A (Geng et al., 2013), significantly influenced rice resistance to Xoo. Compared with well‐documented studies on resistance against Xoo, limited progress has demonstrated that sheath blight resistance was only controlled by minor effect QTLs (Wang et al., 2012; Zuo et al., 2013). Only a few genes, such as OsACS2 (Helliwell et al., 2013), Os2H16 (Li et al., 2013a), OsJERF1 (Pan et al., 2013) and OsWRKY4 (Wang et al., 2015), were identified as resistant to R. solani. However, except an identified R. solani‐inducible cis‐element (Li et al., 2017a), knowledge regarding the regulation mechanisms of R. solani‐inducible genes is still very limited.
Previously, we identified a rice defence‐related gene, Os2H16, which encoded a short‐chain peptide of unknown function. Xoo, Xanthomonas oryzae pv. oryzicola (Xoc) and R. solani, as well as abiotic salt or drought stress rapidly induced the expression of this gene. Overexpressing Os2H16 significantly enhanced rice resistance to bacterial blight disease, sheath blight disease and drought, suggesting that the Os2H16 promoter is a multipathogen‐inducible and drought‐inducible promoter that contains one or more pathogen‐inducible and drought‐inducible cis‐elements (Li et al., 2013a). Here, we report that the GT‐1 cis‐element (GAAAAA) plays an important role in the Os2H16 promoter response to pathogen infection and osmotic stress. This finding suggested a novel function for GT‐1 in addition to its salt‐ and pathogen‐inducible activities (Park et al., 2004). Furthermore, we demonstrated that the GT‐1 cis‐element could be bound to a novel transcription factor, OsASR2. Finally, we showed that altering OsASR2 expression influences rice resistance to Xoo, R. solani and drought, which suggests that OsASR2 could modulate the response of rice to pathogen and drought by targeting the GT‐1 cis‐element.
Results
Putative cis‐elements prediction in the Os2H16 promoter
To characterize the regulatory mechanisms of the Os2H16 gene, we cloned its promoter region (−2197 to +60). According to the PLACE database (Higo et al., 1999), some putative cis‐elements are predicted in the Os2H16 promoter (Figure S1). The TATA‐box (5ˊ‐TATAA‐3ˊ) starts 89‐bp upstream of the ATG and 30‐bp upstream of the transcription start site (TSS). In addition, there was no obvious CAAT box next to the TATA‐box on either strand. The Os2H16 promoter also contains two GT‐1 elements, GAAAAA (Park et al., 2004) on the negative strand, three auxin‐responsive elements, CTTTA and GTCTC (Mironova et al., 2014), two GA responsive elements, CCTTTT (Woodger et al., 2003) and four ABA responsive elements, ACACG, ACCCG and ACGTG (Li et al., 2013b). On the opposite strand of the Os2H16 promoter, we found one element that is complementary to the TGACG element in association with the as‐1 element (Bacha et al., 2015; Krawczyk et al., 2002) and six CANNTG elements that are recognized by the helix‐loop‐helix (bHLH) TF superfamily (Wang et al., 2013c).
Tissue‐specific and pathogen‐inducible expression patterns of the Os2H16 promoter
First, we investigated the tissue expression patterns of the Os2H16 promoter in transgenic rice. As shown in Figure S2, the Os2H16 promoter was expressed primarily in young root, anther and endosperm. The GFP fluorescence pattern in transgenic rice plants was similar to the previously reported expression pattern of the Os2H16 gene (Li et al., 2013b).
We then examined the effect of Xoo, Xoc and R. solani on GFP reporter gene expression in transgenic rice leaves. Treatment with Xoo strain PXO99 activated GFP fluorescence starting at 4 h postinoculation (h p.i.), and this fluorescence was maximized at 12 h p.i. compared with the control. Treatment with Xoc strain RS105 resulted in similar GFP fluorescence patterns. R. solani strain YWK196 induced GFP expression at 12 h p.i., and this expression continued to increase until 24 h p.i. (Figure S3). These results validated that the Os2H16 promoter could respond to multiple phytopathogens.
Deletion analysis of the Os2H16 promoter
To determine the pathogen‐inducible regions in the Os2H16 promoter, a series of 5ˊ deletions were made in the Os2H16 promoter (Figure 1a). Each construct was introduced into rice plants by Agrobacterium tumefaciens‐mediated transformation, and GFP fluorescence was quantified at 24 h p.i. with Xoo strain PXO99 or R. solani strain YWK196; no infection was used as a control. The deletion constructs containing up to −1742 (pCXGFP D1), −1259 (pCXGFP D2) and −720 (pCXGFP D3) showed GFP induction almost equal to that of the pC1381 D0 construct. In contrast, GFP inducible activity was nearly lost in the pCXGFP D4 construct containing a deletion up to −309 (Figure 1b,c). These results indicated that the deleted region −720 to −310 contained candidate elements that are essential for the pathogen‐responsive expression of the Os2H16 promoter.
Figure 1.
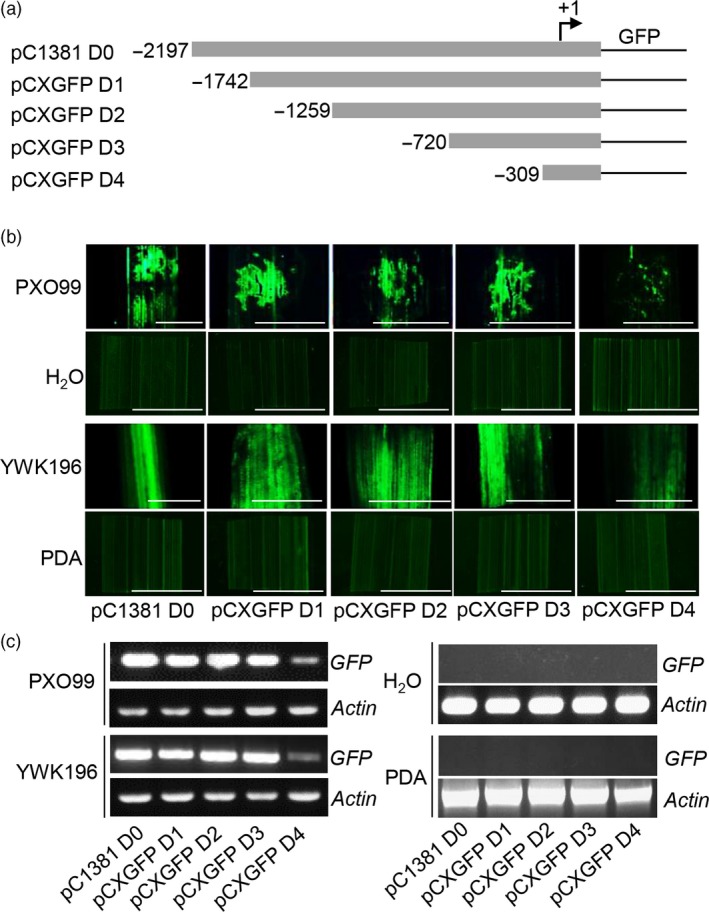
Fluorometric assays for GFP driven by various Os2H16 promoter deletion constructs. (a) Diagram of various deletion derivatives of the Os2H16 promoter. Deletion end points are indicated in bp from the transcription start site. All promoter derivatives were fused to a GFP reporter vector, pCXGFP‐P. (b) GFP fluorescence of the DNA constructs prepared in (a) in the transgenic rice plants. The rice leaves were inoculated with Xoo strain PXO99 and R. solani strain YWK196 for 24 h, and no infection was used as control. Bars = 5 mm. (c) GFP expression of the DNA constructs prepared in (a) in the transgenic rice plants by RT‐PCR.
To mine the cis‐elements within the −720 to −310 region that are responsible for pathogen induction, a series of 5ˊ deletions were generated in this region (Figure 2a). Then, each of construct was tested in transient expression assays in tobacco leaves at 24 h p.i. with YWK196, and no infection was used as a control. The deletion constructs containing up to −614 and −513 (pCXGFP D5 and pCXGFP D6) showed GFP induction almost equal to that of the pCXGFP D3 construct. The construct containing the −411 to +60 region (pCXGFP D7) showed an approximately one‐half reduction in GFP induction compared with the pCXGFP D3, pCXGFP D5 and pCXGFP D6 constructs. However, the deletion of the sequence between −411 and −309 resulted in a significant loss of GFP accumulation (Figure 2b,c). These results demonstrated that two regions, −513 to −412 and −411 to −309, play important roles in the pathogen‐inducible expression of the Os2H16 promoter, and the −411 to −309 region contained elements that exhibited a more significant effect.
Figure 2.
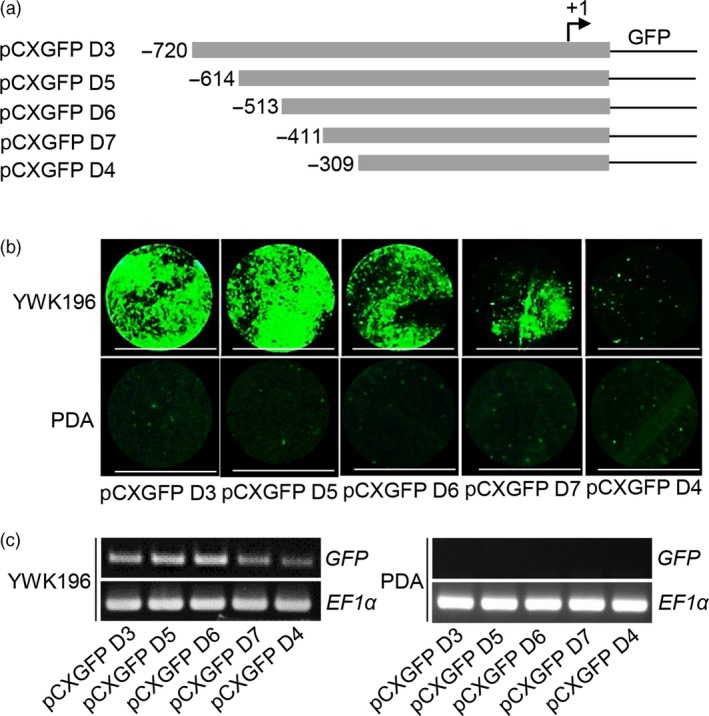
Fluorometric assays for GFP driven by deletion constructs in the −720 to −309 region of the Os2H16 promoter. (a) Diagram of deletion constructs in the −720 to −309 region of the Os2H16 promoter. All promoter derivatives were fused to a GFP reporter vector, pCXGFP‐P. (b) GFP fluorescence of the DNA constructs prepared in (a) in a tobacco transient expression system. The tobacco leaves were inoculated with R. solani strain YWK196 for 24 h, and no infection was used as control. Bars = 5 mm. (c) GFP expression of the DNA constructs prepared in (a) in a tobacco transient expression system by RT‐PCR.
The GT‐1 cis‐element functions importantly in the Os2H16 promoter response to Xoo and R. solani
In vivo GFP assays of the Os2H16 promoter in rice and N. benthamiana leaves identified two regions, −513 to −412 and −411 to −309, that primarily mediate the pathogen response; the −411 to −309 region more strongly affected the pathogen response than the −513 to −412 region. The −411 to −309 region contained a pathogen‐inducible cis‐element, GT‐1 (GAAAAA), which was reported to be responsive to Pseudomonas syringae in Arabidopsis (Park et al., 2004). To test whether GT‐1 plays an important role in the response of the Os2H16 promoter to Xoo or R. solani, we made the GT‐1 deletion construct (pCXGFP‐ΔGT‐1) and transformed it into the rice cultivar Zhonghua 11. Transgenic rice plants were then inoculated with PXO99 and YWK196 for 24 h, and no infection was used as control. The GFP gene was strongly induced by PXO99 or YWK196 in pC1381 D0 construct, but this induction was significantly reduced in the pCXGFP‐ΔGT‐1 construct (Figure 3a). Therefore, the GT‐1 cis‐element plays an important role in the response of the Os2H16 promoter to Xoo and R. solani.
Figure 3.
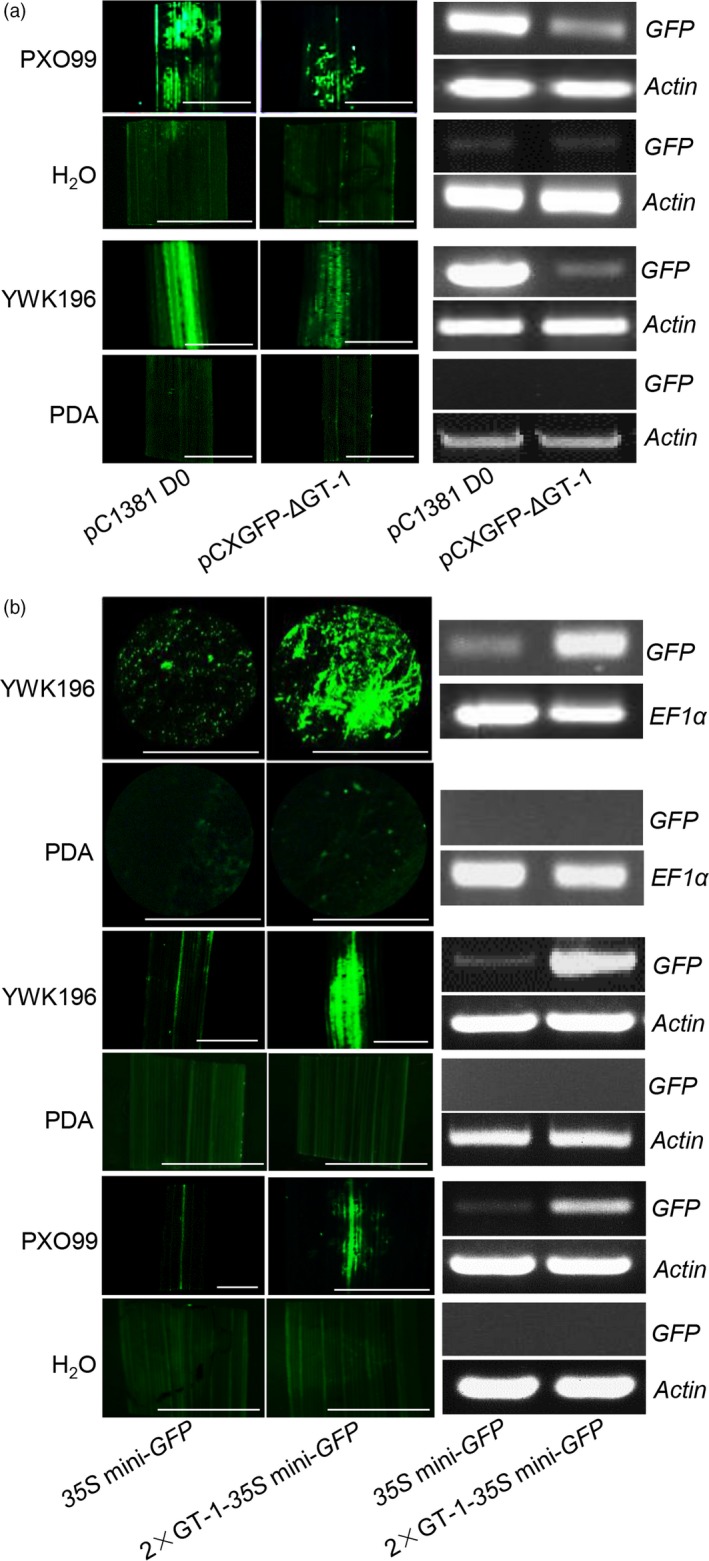
Identification of the GT‐1 cis‐element involved in response to pathogens. (a) The effect of deletion of the GT‐1 cis‐element within the Os2H16 promoter on GFP expression in the transgenic rice plants. The rice leaves were inoculated with Xoo strain PXO99 and R. solani strain YWK196 for 24 h and subjected to fluorescence examination and RNA extraction. Bars = 5 mm. (b) The effect of the individual GT‐1 cis‐element on GFP expression in a tobacco transient expression system and the transgenic rice plants. The tobacco and rice leaves were inoculated with Xoo strain PXO99 and R. solani strain YWK196 for 24 h and subjected to fluorescence examination and RNA extraction. Bars = 5 mm.
To further determine whether the GT‐1 cis‐element could respond to Xoo and R. solani independently, we produced a construct in which two tandem repeats of the GT‐1 cis‐element were fused with the 35S minimum promoter‐GFP (2 × GT‐1‐35S mini‐GFP). The empty vector (35S mini‐GFP) was used as a control. First, by transient assay in N. benthamiana leaves, a large amount of GFP accumulation was observed in leaves expressing the 2 × GT‐1‐35S mini‐GFP construct, while weak GFP fluorescence was observed in leaves expressing the empty vector (Figure 3b). Furthermore, transgenic rice plants carrying the 2 × GT‐1‐35S mini‐GFP or 35S mini‐GFP constructs were also generated. These plants were inoculated with PXO99 and YWK196 for 24 h. Compared with the 35S mini‐GFP construct, the GFP expression was strongly induced in plants carrying the 2 × GT‐1‐35S mini‐GFP construct (Figure 3b). These results demonstrated that the GT‐1 cis‐element could independently respond to Xoo and R. solani in rice.
The GT‐1 cis‐element confers drought response
Previous study showed that polyethylene glycol (PEG) could induce the expression of the Os2H16 gene (Li et al., 2013a). To test whether the GT‐1 cis‐element could respond to osmotic stress, rice plants transformed with the pCXGFP‐ΔGT‐1 and 2 × GT‐1‐35S mini‐GFP constructs were subjected to PEG8000 treatment, and no treatment was used as control. As shown in Figure 4a, strong activation of the GFP gene was detected in the PEG8000 (15% w/v)‐treated leaves of Os2H16 promoter transgenic rice, while approximately three quarters of reduction in GFP induction was detected in the pCXGFP‐ΔGT‐1 transgenic rice. This result indicated that the GT‐1 cis‐element is partly responsible for the Os2H16 promoter response to osmotic stress. Moreover, relatively high (threefold) induction of 2 × GT‐1 suggested that the individual GT‐1 cis‐element could also respond to osmotic stress (Figure 4b).
Figure 4.
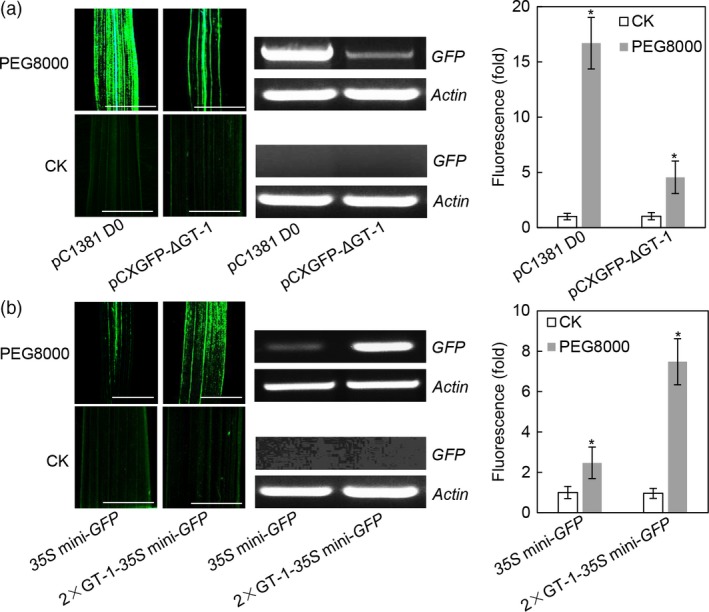
Identification of the GT‐1 cis‐element involved in response to drought. (a) The effect of deletion of the GT‐1 cis‐element within Os2H16 promoter on GFP expression in the transgenic rice plants. The rice leaves were treated with PEG8000 (15% w/v) for 24 h and subjected to fluorescence examination and quantification. GFP quantification was calculated relative to CK of the pC1381 D0 construct. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis. Bars = 5 mm. (b) The effect of the individual GT‐1 cis‐element on GFP expression in the transgenic rice plants. The rice leaves were treated with PEG‐8000 (15% w/v) for 24 h and subjected to fluorescence examination and quantification. GFP quantification was calculated relative to CK of the 35S mini‐GFP construct. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis. Bars = 5 mm.
Characterization of a GT‐1 interaction protein
A trihelix DNA‐binding factor, AtGT‐3b, has been reported to bind to the GT‐1 cis‐element and regulate gene expression in response to pathogen and salt treatment in Arabidopsis (Park et al., 2004). To characterize the binding protein of the GT‐1 cis‐element in rice, we performed a yeast one‐hybrid (Y1H) analysis to screen the rice cDNA library with GT‐1 as bait. Interestingly, no trihelix DNA‐binding factors were identified from the library, while an abscisic acid, stress and ripening 2 (ASR2) protein was identified and temporarily named OsASR2. OsASR2 contains 138 amino acid residues with a calculated molecular mass of 15.5 kD. Structural analysis showed that it contains an abscisic acid/water deficit stress (ABA/WDS) domain from residues 78 to 128 (Figure S4a). A phylogenetic tree of OsASR2 and other ASR proteins from different plant species showed that it was a highly conserved protein (Figure S4b).
This interaction was firstly verified with a yeast selection system. The yeast cells were transformed with OsASR2 cDNA fused to the GAL4 activation domain and the GT‐1 upstream of the iso‐1‐cytochrome C minimal promoter. The OsASR2 protein and the GT‐1 cis‐element conferred Ura− and Leu− selection in the presence of 150 ng/mL aureobasidin A (AbA). In contrast, yeast cells carrying empty vector did not grow on medium lacking Ura and Leu in the presence of 150 ng/mL AbA (Figure 5a).
Figure 5.
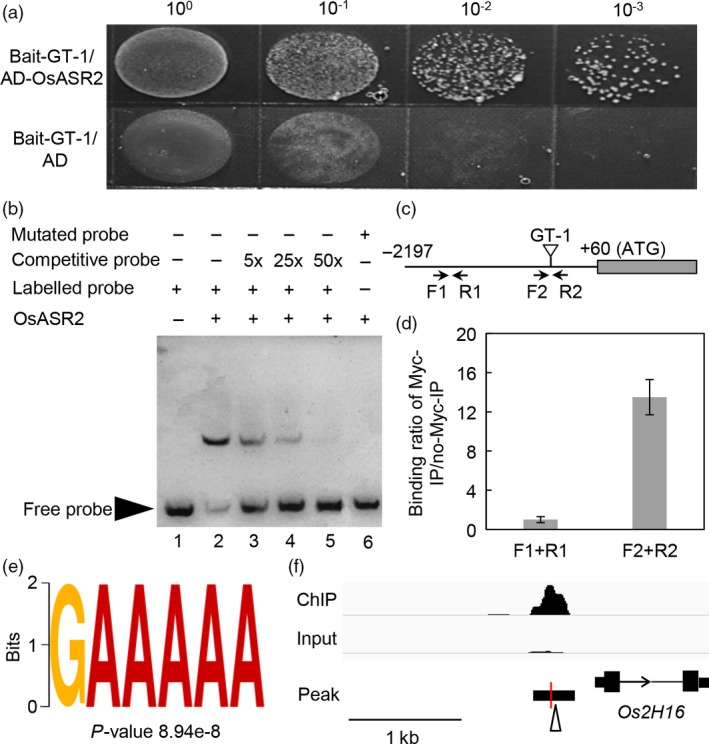
Interaction of OsASR2 with the GT‐1 cis‐element. (a) Interaction of OsASR2 with the GT‐1 cis‐element by the yeast selection system. Y1HGold yeast strains carrying empty vector pGADT7 or pGADT7‐OsASR2 were grown overnight at 30 °C. Cultures were diluted to OD 600 = 1.0, and then, serial 10‐fold dilutions were spotted onto medium lacking Ura and Leu in the presence of 150 ng/mL AbA. (b) EMSA using the recombinant OsASR2 protein and the GT‐1 cis‐element. Lane 1 contained only the free probe. Lane 2 contained the purified OsASR2 protein and the probe. For the competitive EMSA, the purified OsASR2 protein was pre‐incubated with 5 (lane 3)‐, 25 (lane 4)‐ or 50 (lane 5)‐fold molar excess of unlabelled GT‐1 cis‐element before the addition of the biotin‐labelled GT‐1 cis‐element. For the mutant EMSA, the purified OsASR2 protein was incubated with the labelled mutant GT‐1 cis‐element. (c) The positions of primers used in the ChIP‐qPCR experiment. (d) Binding of OsASR2 to the GT‐1 cis‐element in a ChIP‐qPCR assay. (e) The GT‐1 binding motif identified in OsASR2 binding peaks in ChIP‐seq assay. (f) OsASR2 biding profile in the promoter of Os2H16. The open arrowhead refers to the GT‐1 around the peak summit. The red vertical line denotes the peak summit.
Then, this interaction was further analysed in vitro. We expressed Myc‐tagged OsASR2 protein in Escherichia coli and purified the recombinant protein. The ability of the recombinant OsASR2‐Myc fusion protein binding to the GT‐1 cis‐element (5′‐GATTAAGATTTTTCCTACCCTA‐3′) was validated by EMSA. As shown in Figure 5b, the protein in lane 2 bound to the oligo DNA molecule with the GT‐1 cis‐element. Lanes 3, 4 and 5, which contained fivefold to 50‐fold molar excesses of the unlabelled GT‐1 cis‐element, showed that the intensities of the combined bands weakened. To demonstrate whether the GT‐1 cis‐element was essential for the sequence‐specific binding activity, we tested a labelled mutant probe in which the TTTTTC sequence was changed to CCCCCA. No binding band was detected (Figure 5b, lane 6).
To confirm the in vivo binding of OsASR2 to the GT‐1 cis‐element, the recombinant OsASR2‐Myc fusion protein and the pC1381 D0 construct were co‐transformed into N. benthamiana leaves. After 5 days, chromatin was harvested and immunoprecipitations were performed using the antibody specific to Myc. In addition, normal mouse IgG was used as nonspecific control. The primers were designed to amplify the sequence of the Os2H16 promoter with the GT‐1 cis‐element. As shown in Figure S5, OsASR2 bound to the promoter region containing the GT‐1 cis‐element. Importantly, the immunoprecipitation was specific to OsASR2, because the promoter region was not detected when a nonspecific antibody was used for immunoprecipitation. The above results provide strong evidence for an interaction between OsASR2 and the GT‐1 cis‐element.
OsASR2 is a functional transcription factor
Transcription factors bind to promoter sequences to regulate the expression of downstream genes, and most of these transcription factors are located in the nucleus. To identify the localization of OsASR2, we constructed an OsASR2‐GFP fusion protein and demonstrated that it exclusively localized to the nuclei of N. benthamiana epidermal cells (Figure 6a).
Figure 6.
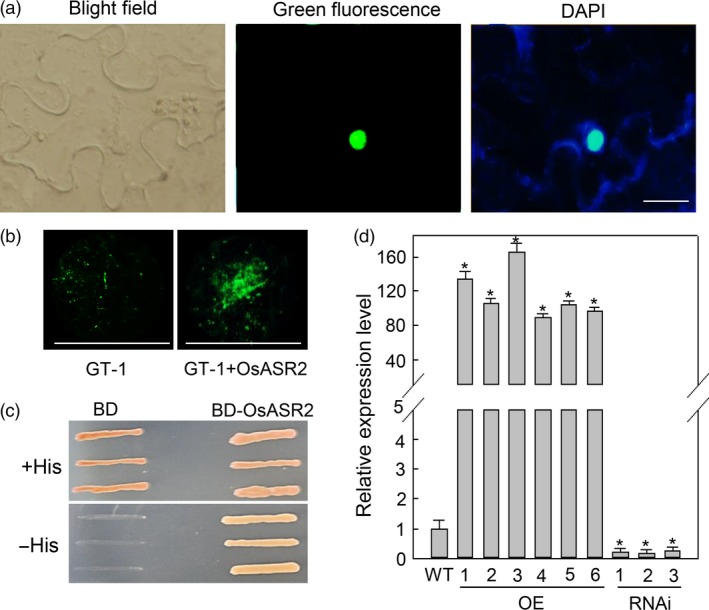
OsASR2 is a transcription factor. (a) Subcellular localization of OsASR2. OsASR2 was fused to GFP to yield OsASR2‐GFP recombinant protein. The construct was transiently expressed in tobacco leaves, and subcellular localization was determined by GFP assay and DAPI staining. Bars = 10 μm. (b) Activation of the GT‐1 cis‐element by OsASR2 in vivo. GT‐1 alone or GT‐1 and OsASR2 were transformed into tobacco leaves, and GFP was examined by fluorimetric assays. Bars = 5 mm. (c) Transcriptional activation activity of OsASR2. Yeast cultures were diluted and then streaked onto nonselective medium (+His) and selective medium (−His) plates and incubated at 30 °C for 3 days before examination. (d) Expression levels of Os2H16 in OsASR2 OE and RNAi lines. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis.
The above results showed that OsASR2 could interact with the GT‐1 cis‐element, but the ability of OsASR2 to activate the GT‐1 cis‐element remains unknown. To test the specific activation activity of OsASR2, the 2 × GT‐1‐35S mini‐GFP construct and the pCXUN‐OsASR2‐Myc construct were co‐transformed into N. benthamiana leaves. A large amount of GFP accumulation was detected in leaves expressing both the GT‐1 cis‐element and OsASR2 compared with the leaves only expressing the GT‐1 cis‐element at 5 dpi (Figure 6b). Therefore, OsASR2 could induce the expression of GFP derived under the 2 × GT‐1‐35S mini promoter.
To test the transcriptional activation activity of OsASR2, we performed a transcriptional activity assay using a Y1H system. pGBKT7 carrying the completed OsASR2 cDNA and empty pGBKT7 serving as a negative control were transformed into the yeast strain AH109. The activity of the HIS gene was tested on a medium lacking histidine. In the absence of histidine, the yeast transformed with the construct containing OsASR2 survived, while yeast containing the empty vector did not (Figure 6c). These results suggested that OsASR2 has transcriptional activation activity.
We constructed six overexpression (OE) and three RNA interference (RNAi) T0 lines of OsASR2 to explore whether OsASR2 could regulate the expression of Os2H16, the putative target gene carrying the GT‐1 cis‐element in rice. The basal levels of OsASR2 expression were analysed in 5‐week‐old T0 plants using quantitative real‐time PCR (qRT‐PCR) and Western blotting (Figure S6). As shown in Figure 6d, Os2H16 expression was significantly induced in OE lines and suppressed in RNAi lines compared with WT plants. Therefore, OsASR2 could regulate the expression of Os2H16 by interacting with the GT‐1 cis‐element.
Additionally, the interaction between OsASR2 and the GT‐1 cis‐element was validated in transgenic rice through ChIP‐qPCR. Chromatin was harvested, and immunoprecipitations were performed using c‐Myc Tag antibody. In addition, no‐Myc immunoprecipitation was used as a negative control. The primers were designed to amplify the promoter sequences with or without the GT‐1 cis‐element (Figure 5c). As shown in Figure 5d, the GT‐1 region was enriched 13.5‐fold compared with the region without the GT‐1 cis‐element. To further investigate the OsASR2 binding motifs, the ChIP‐seq assay was performed using the transgenic lines (Figure S7, Accession No. SRP112505). Nine motifs including the GT‐1 cis‐element were identified (Figure 5e, Figure S8), and the peak summit for OsASR2 binding site was located 369‐bp upstream of the Os2H16 TSS (Figure 5f). These results further indicated that OsASR2 could bind to the GT‐1 cis‐element.
Analysis of the OsASR2 Expression in Rice
To analyse the involvement of OsASR2 in rice basal defence, we examined its expression in response to PXO99 and YWK196. OsASR2 expression was significantly induced in leaves from 4 h p.i. and peaked at 12 h p.i. but declined to normal levels at 24 h p.i. for PXO99 inoculation. After YWK196 infection, OsASR2 expression was markedly induced at 12 h p.i. and then declined to a low level but remained higher than that of the control (Figure S9a). Also, treatment with PEG‐8000 (15% v/v) induced OsASR2 expression at 3 h, which peaked at 6 h and then slightly decreased at 12 h (Figure S9b). This expression pattern indicated that OsASR2 is involved in the pathogen response and osmotic stress.
Modulating OsASR2 expression influences rice responses to Xoo and Rhizoctonia solani
To assess the function of OsASR2 in rice disease resistance, we first examined the responses of WT and OsASR2 transgenic plants to Xoo in the T0 generation. After 21 dpi with PXO99, six OE lines were enhanced resistance to PXO99, with lesion lengths ranging from 2.2 cm to 4.2 cm, compared with 10.5 cm for WT plants (Figure 7a,b). Three RNAi lines were more susceptible to PXO99, with lesion length ranging from 15.4 cm to 18.2 cm, compared with 9.5 cm for WT plants (Figure 7c,d). The lesion length significantly correlated with the OsASR2 expression level in OE and RNAi lines, with correlation coefficients of −0.968 (P = 0.006) and −0.942 (P = 0.009), respectively.
Figure 7.
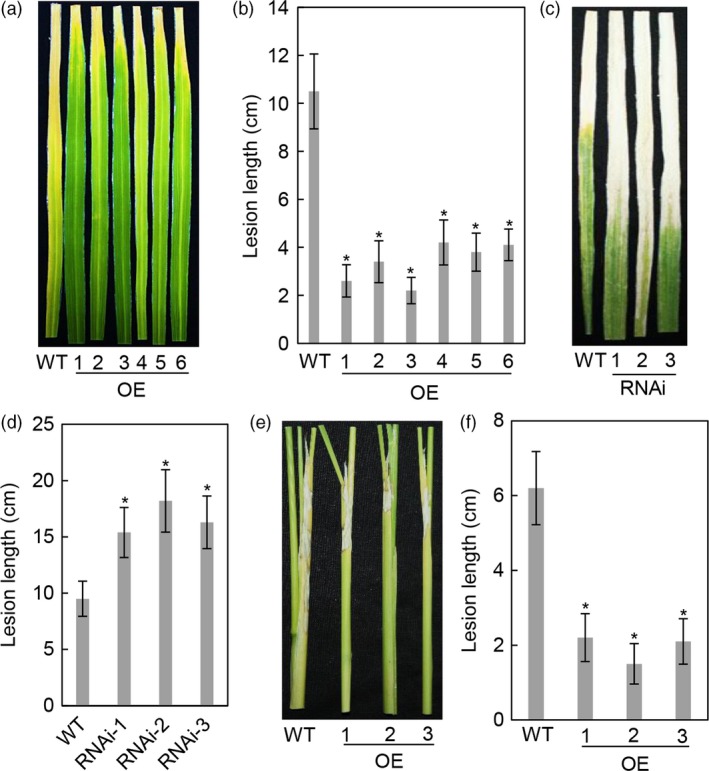
Responses of OsASR2 transgenic lines to Xoo strain PXO99 and R. solani strain YWK196. (a) Rice bacterial blight symptoms on WT and T0 OsASR2 OE lines after 21 days of inoculation with Xoo strain PXO99. (b) Lesion length after 21 days of inoculation with Xoo strain PXO99. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis. (c) Rice bacterial blight symptoms on WT and T0 OsASR2 RNAi lines after 21 days of inoculation with Xoo strain PXO99. (d) Lesion length after 21 days of inoculation with Xoo strain PXO99. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis. (e) Rice sheath blight symptoms on WT and T1 OsASR2 OE lines after 7 days of inoculation with R. solani strain YWK196. (f) Lesion length after 7 days of inoculation with R. solani strain YWK196. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis.
Because R. solani could induce OsASR2 expression, we investigated the role of OsASR2 in the resistance to R. solani. Three T1 OE lines were inoculated with YWK196 and investigated further. After 7 days of inoculation, the OE lines showed reductions in lesion length of 4.0–4.7 cm, compared with 6.2 cm for WT plants (Figure 7e,f). These results demonstrated that OsASR2 confers resistance to both leaf blight bacterium and sheath blight fungus.
OsASR2 positively regulates rice tolerance to drought stress
Due to the induction of OsASR2 by osmotic stress, we wondered whether OsASR2 function in drought tolerance. Above three OE lines and WT plants were assayed. When water was cut off, the WT plants exhibited delayed growth compared with the OE lines, which continued growing normally. After drought treatment for 21 days, almost all of the WT plants wilted, while the OE lines exhibited less wilting than WT plants. Upon re‐watering for 7 days, all OE lines recovered but the WT plants did not (Figure 8a). Additionally, the OE lines exhibited a 30%–36% reduction in water loss compared with 51% for WT plants (Figure 8b).
Figure 8.
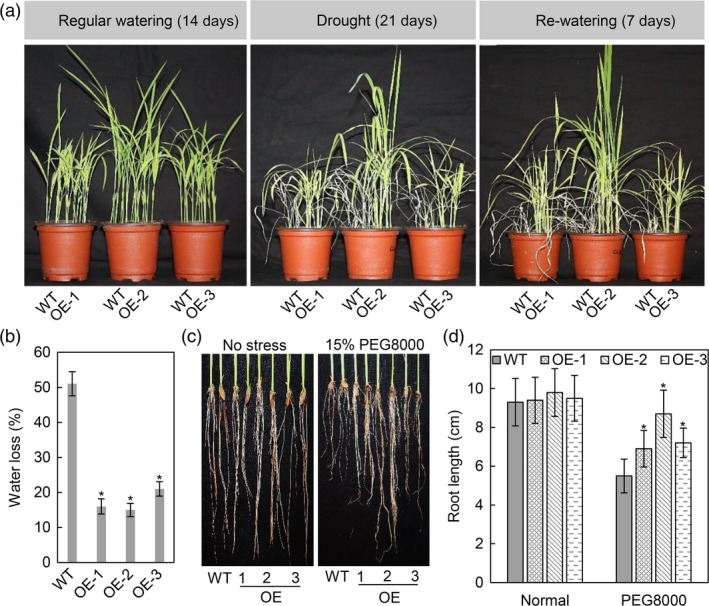
Drought stress tolerance assessment in OsASR2 transgenic rice plants. (a) Phenotypes of WT and OE lines under regular watering, drought stress and re‐watering conditions. (b) Water loss of three T1 OE lines. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis. (c) Phenotypes of roots from WT and OE lines under normal and osmotic stress. (d) Root length in response to the osmotic stress. Error bars indicate the SD (n = 3). Asterisks indicate P < 0.05 (*) in Student's t‐test analysis.
It has been reported that osmotic stress could inhibit root growth (Zhu, 2002). Here, we assessed the root elongation of WT and transgenic seedlings in high osmotic conditions. All OE lines showed little difference in root length compared with WT plants under normal conditions. However, the OE lines displayed longer root lengths than WT plants after treatment with PEG8000 (15% w/v) for 7 days (Figure 8c,d). These results demonstrated that OsASR2 plays a positive role in regulating rice response to drought stress.
Discussion
Small peptides are the smallest biological molecules mainly containing 5–75 amino acids. They play diverse roles in plant growth, development, reproduction, symbiotic interactions and stress responses (Albert, 2013; Czyzewicz et al., 2013; Matsubayashi, 2014). Previously, we identified a small peptide gene Os2H16, which was involved in resistance to two important diseases and tolerance to drought (Li et al., 2013a). However, the regulatory mechanism of Os2H16 in the crosstalk of disease and drought resistance remains unclear. In this study, using a promoter deletion analysis, two promoter regions, −513 to −412 and −411 to −309, were proven to be involved in the response of Os2H16 to pathogen infection (Figure 2). A database search did not identify any known pathogen‐inducible cis‐elements in the −513 to −412 region, while a GT‐1 cis‐element (GAAAAA) was located in the −411 to −309 region. Moreover, deletion of the GT‐1 cis‐element in −411 to −309 region significantly affected the induction levels of the Os2H16 promoter by pathogens (Figure 3a), suggesting that the GT‐1 cis‐element plays a major role in the response of the Os2H16 promoter to pathogens. Interestingly, deleting the −2197 to −1742 region, which contained the other GT‐1 cis‐element (−2137 to −2132), did not affect the pathogen‐inducible activity of the Os2H16 promoter (Figure 1), implying that the pathogen‐inducible activity of the GT‐1 cis‐element might be related to the distance from the TSS.
The cis‐element of GT‐1 was first found in the promoter of the ribulose 1, 5‐bisphosphate carboxylase/oxygenase (Rubisco) small subunit gene and identified as the box II elemenτ (Green et al., 1987). The GT‐1 cis‐elements positively or negatively affect transcription depending on the promoter structure. One common feature of the GT‐1 cis‐elements is a core sequence defined as 5ˊ‐G‐Pu‐(T/A)‐A‐A‐(T/A) (Zhou, 1999). The high degeneracy of the GT‐1 cis‐element is thought to partly explain its diverse functions except for its light‐specific regulatory function. For example, the GT‐1 cis‐element (GGTTAA) is involved in cell‐type‐specific transcriptional regulation (Villain et al., 1996), while the GT‐1 cis‐element (GAAAAA) responds to P. syringae treatment (Park et al., 2004). Our results showed that except for P. syringae, the GT‐1 cis‐element could also be activated by Xoo and R. solani (Figure 3b), suggesting that it has a broad spectrum of pathogen responses and might be involved in plant basal defence. Additionally, the GT‐1 cis‐element reportedly responds to salt stress (Park et al., 2004). We found that drought stress could also activate the GT‐1 cis‐element (Figure 4b), suggesting a new function for the GT‐1 cis‐element in plant response to abiotic stress. Taken together, multiple roles of the GT‐1 cis‐element imply its complex mechanisms in plants responding to biotic and abiotic stresses.
In general, binding sites within the promoter and their regulatory proteins control gene expression. The currently identified GT‐1 binding proteins are collectively called GT‐factors, which contain one to three trihelix DNA‐binding motifs (Du et al., 2016; Fang et al., 2010; Osorio et al., 2012). Here, we identified a novel GT‐1 binding protein OsASR2 by yeast one‐hybrid screening, and the interaction between OsASR2 and the GT‐1 cis‐element was determined both in vitro and in vivo (Figure 5, Figure S5). ASR proteins widely exist in plant kingdom, ranging from ancient gymnosperms to monocots and dicots. These proteins are usually nucleic localization and function as transcription factors (Frankel et al., 2006; González and Iusem, 2014). Moreover, the DNA‐binding domains of the ASR proteins locate at the carboxy‐terminal end (González and Iusem, 2014) and it has been reported that the amino residues 61–115 of the tomato ASR1 have a DNA‐binding activity (Rom et al., 2006). In this study, OsASR2 could activate the expression of the GT‐1 cis‐element and the Os2H16 gene in N. benthamiana and rice, respectively, and it was localized in the nucleus and exhibited transcriptional activation activity (Figure 6). Sequence alignments showed that the C terminal (residues 87–138) domain of OsASR2 displayed 65% identities to the DNA‐binding domain of the tomato ASR1 (Figure S10), implying that the amino residues 87–138 might be the DNA‐binding domain of OsASR2. These results strongly suggested that OsASR2 is a transcription factor to regulate the Os2H16 gene expression by targeting the GT‐1 cis‐element.
To date, most studies have focused on the involvement of ASR proteins in sugar transport and ABA response (Dominguez et al., 2013; Joo et al., 2013; Saumonneau et al., 2012), and the roles in improving drought tolerance have also been well characterized (Dai et al., 2011; Hu et al., 2013, 2014; Li et al., 2017b; Liu et al., 2010; Philippe et al., 2010). However, the involvement of ASR proteins in pathogen response is poorly understood, and the related reports are very rare (Liu et al., 2010; Zhu et al., 2012). Here, we showed that OsASR2 could be induced by Xoo, R. solani and PEG8000 treatments (Figure S9), and overexpressing OsASR2 significantly enhanced rice resistance to Xoo, R. solani and drought (Figures 7, 8). Additionally, it has been reported that ASR proteins are crucial during the pollen‐drying stage (González and Iusem, 2014; Wang et al., 2013b), and we found that suppressing OsASR2 expression by RNAi could result in rice abortion in the T0 generation. Therefore, we could not test R. solani resistance and drought tolerance using RNAi lines in the T1 generation. Moreover, a total of 224 genes containing the GT‐1 cis‐elements in their promoters were identified and GO analysis showed 83 genes could respond to biotic and abiotic stimulus (Table S2). These results suggest that OsASR2 might positively regulate rice disease resistance and drought tolerance by activating genes containing the GT‐1 cis‐element in their promoters.
In summary, we determined the new functions of the GT‐1 cis‐element in the response to biotic and abiotic stresses and identified a novel GT‐1 binding protein, OsASR2. Furthermore, we proved that OsASR2 could also respond to pathogens and drought stress and mediate rice resistance to pathogens and drought. The data obtained from this study lead to a model to elucidate the roles played by DNA‐protein interactions in the regulation of pathogen and drought‐responsive genes with GT‐1 cis‐element in their promoters, including Os2H16, during rice defence responses (Figure 9). First, the OsASR2 transcription factor perceived both signals of phytopathogens and drought; then, it activated the expression of the GT‐1 containing genes, such as Os2H16, and finally resulted in disease resistance and drought tolerance.
Figure 9.
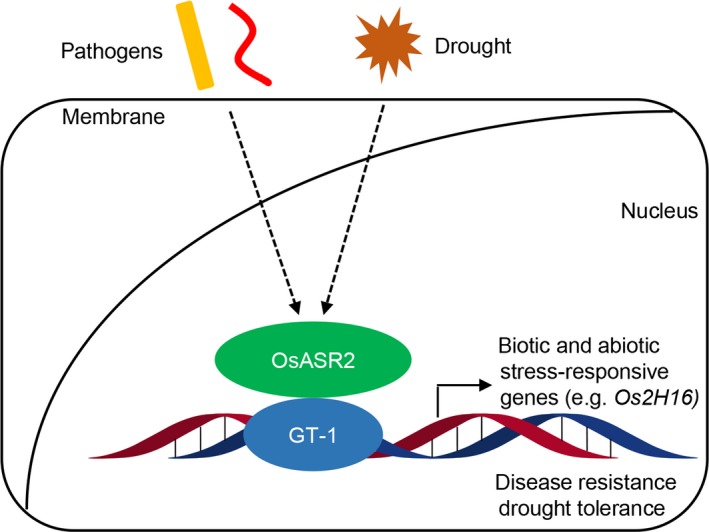
Model for the roles played by DNA–protein interactions in the Os2H16 gene expression during rice defence responses. First, the OsASR2 transcription factor perceived the both signals of phytopathogens and drought. Then, it activated the expression of the GT‐1 containing genes, such as the Os2H16, and finally resulted in disease resistance and drought tolerance.
Experimental procedures
Plant materials and treatments
Rice plants (IRBB13 and Zhonghua 11) and N. benthamiana were grown at 28 °C and 25 °C with a 16/8‐h light/dark cycle, respectively. For tissue‐specific analysis of the Os2H16 promoter, tissues from different growth periods of transgenic rice plants were harvested for GFP assay and total RNA extraction. For pathogen‐inducible activity analysis of the Os2H16 promoter, Xanthomonas oryzae strains PXO99 and RS105 were cultured on polypeptone‐sucrose‐agar medium at 28 °C for 2 days and then suspended by sterile water to OD600 = 0.5. Plants were inoculated with PXO99 and RS105 suspension by syringe infiltration at seedling stage. YWK196 was cultured on potato‐dextrose‐broth medium at 25 °C for 3 days. Leaves of rice were inoculated with YWK196 and harvested at 4, 8, 12 and 24 h p.i. for GFP assay and RNA extraction (Li et al., 2017a). For drought‐induced OsASR2 expression analysis, roots of 7‐day‐old rice plants were treated with PEG‐8000 (15% w/v). Roots were harvested at 3, 6, 12 and 24 h after treatment for total RNA isolation.
RT‐PCR and qRT‐PCR analysis
Total RNA was extracted using TRI reagent (Sigma‐Aldrich St. Louis, Missouri, USA) according to the manufacturer's instructions. First‐strand cDNA synthesis was conducted with SuperQuickRT MasterMix Kit (CWBIO, Beijing, China). Quantitative real‐time PCR was performed in an UltraSYBR Mixture (CWBIO) with an ABI QuantStudio™ 6 Flex real‐time PCR detection system. OsActin were used to normalize the expression of each gene. Expression changes were calculated using the ΔΔ Ct method. The primers used are listed in Table S1.
Disease assays
As previously reported (Li et al., 2013b), after 21 dpi with Xoo, lesion length was measured on T0 transgenic lines and WT plants. Lesion length was measured on infected sheaths from transgenic and WT plants at 7 dpi with R. solani YWK196. Three independent T1 transgenic lines and over ten plants for each line were assessed.
Drought assays
WT plants and transgenic lines were normally watered for 14 days. Then, water was cut off for 21 days and re‐watering for 7 days (Li et al., 2013a). The weights of the OE and WT plants were measured before and after the drought treatment. The water loss was quantified using the following formula, water loss (%) = (fresh weight–dry weight)/fresh weight × 100 (Chen et al., 2014). For the root elongation test, the WT and OE seedlings were cultured normally for 14 days. Seedlings were then treated with or without PEG8000 (15% w/v), and the roots length were measured 7 days later.
Yeast one‐hybrid cDNA library assay, electrophoretic mobility shift assay and chromatin immunoprecipitation sequencing
Yeast one‐hybrid assay was performed with Matchmaker Gold Yeast One‐Hybrid (Y1H) Library Screening System (Clontech, Mountain View, CA) according to the manufacturer's instructions. The probe containing GT‐1 element was synthesized artificially and labelled with dUTP using terminal deoxynucleotidyl transferase (Thermo, Rockford Illinois, USA) and incubated with the purified OsASR2 protein (1 μg per reaction). Subsequently, EMSA was conducted using a LightShift Chemiluminescent EMSA kit according to the manufacturer's instructions (Thermo, Rockford). Chromatin immunoprecipitation (ChIP) was performed with anti‐myc tag antibody‐ChIP grade (Abcam, Cambridge, MA). Library construction and sequencing was performed by Wuhan IGENEBOOK Biotechnology Co., Ltd (http://www.igenebook.com/index.asp). Illumina sequencing libraries were constructed with the OsASR2‐Myc DNA samples according to the manufacturer's instructions. The end of DNA fragments were repaired and ligated to an adaptor. Then, DNA fragment of ~200 bp was selected using SPRIselect beads and amplified by PCR for 18–20 cycles. The amplified DNA products were collected and sequenced with Hiseq2000.
Statistical analysis
All data analyses were repeated three times with three replicate experiments independently. Standard deviations were indicated by error bars, and the statistical significances were determined by one‐way variance analysis. The mean differences were compared using Student's t‐test, and P values <0.05 were considered significant.
Other methods
Details of the methods for plasmid construction and plant transformation, subcellular localization, yeast one‐hybrid, prokaryotic expression, ChIP and transactivation assays are available in supplementary methods (Appendix S1) at PBJ online.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Schematic map of the Os2H16 promoter with putative promoter elements.
Figure S2 Tissue expression patterns of the Os2H16 promoter.
Figure S3 Pathogen‐inducible expression patterns of the Os2H16 promoter.
Figure S4 Analysis of amino acid sequence of OsASR2.
Figure S5 Binding of OsASR2 to the GT‐1 cis‐element in a ChIP assay.
Figure S6 Verification of OsASR2 transgenic lines.
Figure S7 Overview of ChIP‐seq data.
Figure S8 Binding motifs identified in OsASR2 binding peaks in the ChIP‐seq assay.
Figure S9 Time causes of OsASR2 expression in response to pathogen and drought treatments.
Figure S10 Sequence alignments of OsASR2 with the tomato ASR1.
Table S1 PCR primers used.
Table S2 GO analysis of genes containing the GT‐1 cis‐element in the ChIP‐seq data.
Appendix S1 Supplementary methods.
ACKNOWLEDGEMENTS
This work was supported by the National Key Research and Development Program of China (2016YFD0100903), the National Natural Science Foundation of China (31601279), the Shandong Modern Agricultural Technology & Industry system (SDAIT‐17‐06), Promotive Research Fund for Excellent Young and Middle‐Aged Scientists of Shandong Province (BS2014BSB01127) and Funds of Shandong ‘Double Tops’ Program (2017).
Accession numbers of sequence data: the Os2H16 promoter (KC710214), Os2H16 (KC710213), OsASR2 (EF576160), OsActin (GQ339773), NbEF1α (AF120093), OsASR3 (XM_015761796), ZmASR (Y12330), SiASR1 (XM_004977238), BdASR3 (XM_003577763), PeASR3 (XM_011045937), CsASR (KF880380), MaASR (GU134777), NnASR2 (XM_010262679) and AtGT‐3b (NM_129382).
Contributor Information
Xinhua Ding, Email: xhding@sdau.edu.cn.
Zhaohui Chu, Email: zchu@sdau.edu.cn.
References
- Albert, M. (2013) Peptides as triggers of plant defence. J. Exp. Bot. 64, 5269–5279. [DOI] [PubMed] [Google Scholar]
- Alves, M.S. , Dadalto, S.P. , Gonçalves, A.B. , De Souza, G.B. , Barros, V.A. and Fietto, L.G. (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacha, S. , Khatoon, A. , Asif, M. , Yuan, J. and Bashir, A. (2015) Identification and analysis of an efficient dicot constitutive promoter from tomato. Pak. J. Bot. 47, 1115–1120. [Google Scholar]
- Brown, R.L. , Kazan, K. , McGrath, K.C. , Maclean, D.J. and Manners, J.M. (2003) A role for the GCC‐box in jasmonate‐mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill, P. and Rivas, S. (2014) Transcriptional control of plant defense responses. Curr. Opin. Plant Biol. 20, 35–46. [DOI] [PubMed] [Google Scholar]
- Cai, M. , Qiu, D. , Yuan, T. , Ding, X. , Li, H. , Duan, L. , Xu, C. et al (2008) Identification of novel pathogen‐responsive cis‐elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant, Cell Environ. 31, 86–96. [DOI] [PubMed] [Google Scholar]
- Chakravarthy, S. , Tuori, R.P. , D'Ascenzo, M.D. , Fobert, P.R. , Després, C. and Martin, G.B. (2003) The tomato transcription factor Pti4 regulates defense‐related gene expression via GCC box and non‐GCC box cis elements. Plant Cell, 15, 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema, K. , Grewal, N. , Vikal, Y. , Sharma, R. , Lore, J.S. , Das, A. , Bhatia, D. et al (2008) A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. Camb. 90, 397–407. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Wang, Y. , Lv, B. , Li, J. , Luo, L. , Lu, S. , Zhang, X. et al (2014) The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. [DOI] [PubMed] [Google Scholar]
- Czyzewicz, N. , Yue, K. , Beeckman, T. and De Smet, I. (2013) Message in bottle: small signaling peptide outputs during growth and development. J. Exp. Bot. 64, 5281–5296. [DOI] [PubMed] [Google Scholar]
- Dai, J. , Liu, B. , Feng, D. , Liu, H. , He, Y. , Qi, K. , Wang, H. et al (2011) MpAsr encodes an intrinsically unstructured protein and enhances osmotic tolerance in transgenic Arabidopsis . Plant Cell Rep. 30, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Deng, H. , Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012) A CCCH‐type zinc finger nucleic acid‐binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 158, 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, P.G. , Frankel, N. , Mazuch, J. , Balbo, I. , Iusem, N. , Fernie, A.R. and Carrari, F. (2013) ASR1 mediates glucose‐hormone cross talk by affecting sugar trafficking in tobacco plants. Plant Physiol. 161, 1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Huang, M. and Liu, L. (2016) The genome wide analysis of GT transcription factors that respond to drought and waterlogging stresses in maize. Euphytica, 208, 113–122. [Google Scholar]
- Fang, Y. , Xie, K. , Hou, X. , Hu, H. and Xiong, L. (2010) Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genomics, 283, 157–169. [DOI] [PubMed] [Google Scholar]
- Frankel, N. , Carrari, F. , Hasson, E. and Iusem, N. (2006) Evolutionary history of the Asr gene family. Gene, 378, 74–83. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Jia, S. , Wang, C. , Wang, F. , Wang, F. and Zhao, K. (2016) BjMYB1, a transcription factor implicated in plant defence through activating BjCHI1 chitinase expression by binding to a W‐box‐like element. J. Exp. Bot. 67, 4647–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, S. , Li, A. , Tang, L. , Yin, L. , Wu, L. , Lei, C. , Guo, X. et al (2013) TaCPK2‐A, a calcium‐dependent protein kinase gene that is required for wheat powdery mildew resistance enhances bacterial blight resistance in transgenic rice. J. Exp. Bot. 64, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, R.M. and Iusem, N.D. (2014) Twenty years of research on Asr (ABA‐stress‐ripening) genes and proteins. Planta, 239, 941–949. [DOI] [PubMed] [Google Scholar]
- Green, P.J. , Kay, S.A. and Chua, N.H. (1987) Sequence‐specific interactions of a pea nuclear factor with light‐responsive elements upstream of the rbcS‐3A gene. EMBO J. 6, 2543–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. , Zhang, D. and Lin, X. (2010) Identification and mapping of a novel bacterial blight resistance gene Xa35(t) originated from Oryza minuta . Sci. Agric. Sin. 43, 2611–2618. [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J.D.G. (1996) Resistance gene‐dependent plant defense responses. Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, E.E. , Wang, Q. and Yang, Y. (2013) Transgenic rice with inducible ethylene production exhibits broad‐spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani . Plant Biotechnol. J. 11, 33–42. [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W. , Huang, C. , Deng, X. , Zhou, S. , Chen, L. , Li, Y. , Wang, C. et al (2013) TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant, Cell Environ. 36, 1449–1464. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Yang, X. , Li, X. , Yu, X. and Li, Q. (2014) The SlASR gene cloned from the extreme halophyte Suaeda liaotungensis K. enhances abiotic stress tolerance in transgenic Arabidopsis thaliana . Gene, 549, 243–251. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Ma, S. , Bai, L. , Zhang, L. , Ma, H. , Jia, P. , Liu, J. et al (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. [DOI] [PubMed] [Google Scholar]
- Joo, J. , Lee, Y.H. , Kim, Y.K. , Nahm, B.H. and Song, S.L. (2013) Abiotic stress responsive rice ASR1 and ASR3 exhibit different tissue‐dependent sugar and hormone‐sensitivities. Mol. Cell, 35, 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, C. , Logemann, E. , Lippok, B. , Schmelzer, E. and Hahlbrock, K. (2001) A highly specific pathogen‐responsive promoter element from the immediate‐early activated CMPG1 gene in Petroselinum crispum . Plant J. 26, 217–227. [DOI] [PubMed] [Google Scholar]
- Krawczyk, S. , Thurow, C. , Niggeweg, R. and Gatz, C. (2002) Analysis of the spacing between the two palindromes of activation sequence‐1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 30, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi, C. , Mestres‐Ortega, D. , Marco, Y. , Meyer, Y. and Reichheld, J.P. (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W‐box‐mediated response to pathogen elicitor. Plant Physiol. 134, 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Kong, L. , Zhou, W. , Zhang, X. , Wei, S. , Ding, X. and Chu, Z. (2013a) Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell, Tissue Organ Cult. 115, 429–441. [Google Scholar]
- Li, J. , Sima, W. , Ouyang, B. , Luo, Z. , Yang, C. , Ye, Z. and Li, H. (2013b) Identification and expression pattern of a ZPR1 gene in wild tomato (Solanum Pennellii). Plant Mol. Biol. Rep. 31, 409–417. [Google Scholar]
- Li, N. , Chen, J. , Yang, F. , Wei, S. , Kong, L. , Ding, X. and Chu, Z. (2017a) Identification of two novel Rhizoctonia solani‐inducible cis‐acting elements in the promoter of the maize gene, GRMZM2G315431 . Sci. Rep. 7, 42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Li, Y. , Yin, Z. , Jiang, J. , Zhang, M. , Guo, X. , Ye, Z. et al (2017b) OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signaling in rice. Plant Biotechnol. J. 15, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok, B. , Birkenbihl, R.P. , Rivory, G. , Brümmer, J. , Schmelzer, E. , Logemann, E. and Somssich, I.E. (2007) Expression of AtWRKY33 encoding a pathogen‐or PAMP‐responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20, 420–429. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Dai, J. , Feng, D. , Liu, B. , Wang, H. and Wang, J. (2010) Characterization of a novel plantain Asr gene, MpAsr, that is regulated in response to infection of Fusarium oxysporum f. sp. cubense and abiotic stresses. J. Integr. Plant Biol. 52, 315–323. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Xu, W. , Hu, X. , Liu, H. and Lin, Y. (2016) W‐box and G‐box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 6, 20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K. , Levine, A. , Eulgem, T. , Morgen, A. , Schmid, J. , Lawton, K.A. , Dangl, J.L. et al (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. (2014) Posttranslationally modified small‐peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Memelink, J. , Verpoorte, R. and Kijne, J.W. (2001) Organization of jasmonate‐responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6, 212–219. [DOI] [PubMed] [Google Scholar]
- Miao, L. , Wang, C. , Zheng, C. , Che, J. , Gao, Y. , Wen, Y. , Li, G. et al (2010) Molecular mapping of a new gene for resistance to rice bacterial blight. Sci. Agric. Sin. 43, 3051–3058. [Google Scholar]
- Mironova, V.V. , Omelyanchuk, N.A. , Wiebe, D.S. and Levitsky, V.G. (2014) Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genom. 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, K. , Shimizu, T. , Lin, F. , Sainsbury, F. , Thuenemann, E. , Lomonossoff, G. , Nojiri, H. et al (2012) Identification of an E‐box motif responsible for the expression of jasmonic acid‐induced chitinase gene OsChia4a in rice. J. Plant Physiol. 169, 621–627. [DOI] [PubMed] [Google Scholar]
- Mohr, T.J. , Mammarella, N.D. , Hoff, T. , Woffenden, B.J. , Jelesko, J.G. and McDowell, J.M. (2010) The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant Microbe Interact. 23, 1303–1315. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan, M. and Prasad, M. (2013) Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 38, 433–449. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan, M. , Zeng, X. , Iyer, N.J. , Klein, P. and Mahalingam, R. (2015) A GCC‐box motif in the promoter of nudixhydrolase 7 (AtNUDT7) gene plays a role in ozone response of Arabidopsis ecotypes. Genomics, 105, 31–38. [DOI] [PubMed] [Google Scholar]
- Osorio, M.B. , Bücker‐Neto, L. , Castilhos, G. , Turchetto‐Zolet, A.C. , Wiebke‐Storhm, B. , Bodanese‐Zanettini, M.H. and Margis‐Pinheiro, M. (2012) Identification and in silico characterization of soybean trihelix‐GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 35, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Li, Y. , Zhang, H. , Huang, R. , Liu, W. , Ming, J. , Liu, S. et al (2013) Expression of signalling and defence‐related genes mediated by over‐expression of JERF1, and increased resistance to sheath blight in rice. Plant. Pathol. 63, 109–116. [Google Scholar]
- Park, H.C. , Kim, M.L. , Kang, Y.H. , Jeon, J.M. , Yoo, J.H. , Kim, M.C. , Park, C.Y. et al (2004) Pathogen‐ and NaCl‐induced expression of the SCaM‐4 promoter is mediated in part by a GT‐1 box that interacts with a GT‐1‐like transcription factor. Plant Physiol. 135, 2150–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M. , Brodersen, P. , Naested, H. , Andreasson, E. , Lindhart, U. , Johansen, B. , Nielsen, H.B. et al (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell, 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Philippe, R. , Courtois, B. , McNally, K.L. , Mournet, P. , El‐Malki, R. , Le Paslier, M.C. , Fabre, D. et al (2010) Structure, allelic diversity and selection of Asr genes, candidate for drought tolerance, in Oryza sativa L. and relatives. Theor. Appl. Genet. 121, 769–787. [DOI] [PubMed] [Google Scholar]
- Rom, S. , Gilad, A. , Kalifa, Y. , Konrad, Z. , Karpasas, M.M. , Goldgur, Y. and Bar‐Zvi, D. (2006) Mapping the DNA‐ and zinc‐binding domains of ASR1 (abscisic acid stress ripening), an abiotic‐stress regulated plant specific protein. Biochimie, 88, 621–628. [DOI] [PubMed] [Google Scholar]
- Saumonneau, A. , Laloi, M. , Lallemand, M. , Rabot, A. and Atanassova, R. (2012) Dissection of the transcriptional regulation of grape ASR and response to glucose and abscisic acid. J. Exp. Bot. 63, 1495–1510. [DOI] [PubMed] [Google Scholar]
- Shimono, M. , Koga, H. , Akagi, A. , Hayashi, N. , Goto, S. , Sawada, M. , Kurihara, T. et al (2012) Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y. , Wang, F. , Jia, D. , Li, J. , Zhang, Y. , Jia, C. and Wang, D. (2015) Cloning and functional analysis of the promoter of a stress‐inducible gene (ZmRXO1) in maize. Plant Mol. Biol. Rep. 33, 200–208. [Google Scholar]
- Van der Does, D. , Leon‐Reyes, A. , Koornneef, A. , Van Verk, M.C. , Rodenburg, N. , Pauwels, L. , Goossens, A. et al (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell, 25, 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier, V. , Vera Cruz, C. and Leach, J.E. (2012) Controlling rice bacterial blight in Africa: needs and prospects. J. Biotechnol. 159, 320–328. [DOI] [PubMed] [Google Scholar]
- Villain, P. , Mache, R. and Zhou, D. (1996) The mechanism of GT element‐mediated cell type‐specific transcriptional control. J. Biol. Chem. 271, 32593–32598. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Wen, G. , Lin, X. , Liu, X. and Zhang, D. (2009) Identification and fine mapping of the new bacterial blight resistance gene, Xa31(t), in rice. Eur. J. Plant Pathol. 123, 235–240. [Google Scholar]
- Wang, Y. , Pinson, S.R.M. , Fjellstrom, R.G. and Tabien, R.E. (2012) Phenotypic gain from introgression of two QTL, qSB9‐2 and qSB12‐1, for rice sheath blight resistance. Mol. Breed. 30, 293–303. [Google Scholar]
- Wang, P. , Du, Y. , Zhao, X. , Miao, Y. and Song, C. (2013a) The MPK6‐ERF6‐ROS‐responsive cis‐acting element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 161, 1392–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.S. , Hsu, S.W. and Hsu, Y.F. (2013b) New insights into desiccation‐associated gene regulation by Lilium longiflorum ASR during pollen maturation and in transgenic Arabidopsis. Int. Rev. Cell Mol. Biol. 301, 37–94. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Hu, J. , Qian, Q. and Xue, H. (2013c) LC2 and OsVIL2 promote rice flowering by photoperiod‐induced epigenetic silencing of OsLF . Mol. Plant, 6, 514–527. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Fan, Y. , Zheng, C. , Qin, T. , Zhang, X. and Zhao, K. (2014) High‐resolution genetic mapping of rice bacterial blight resistance gene Xa23 . Mol. Genet. Genomics, 289, 745–753. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Meng, J. , Peng, X. , Tang, X. , Zhou, P. , Xiang, J. and Deng, X. (2015) Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. [DOI] [PubMed] [Google Scholar]
- Woodger, F.J. , Millar, A. , Murray, F. , Jacobsen, J.V. and Gubler, F. (2003) The role of GAMYB transcription factors in GA‐regulated gene expression. J. Plant Growth Regul. 22, 176–184. [Google Scholar]
- Yamamoto, S. , Nakano, T. , Suzuki, K. and Shinshi, H. (2004) Elicitor‐induced activation of transcription via W box‐related cis‐acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochim. Biophys. Acta, 1679, 279–287. [DOI] [PubMed] [Google Scholar]
- Zarei, A. , Körbes, A.P. , Younessi, P. , Montiel, G. , Champion, A. and Memelink, J. (2011) Two GCC boxes and AP2/ERF‐domain transcription factor ORA59 in jasmonate/ethylene‐mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Zhuo, D. , Zhang, F. , Huang, L. , Wang, W. , Xu, J. , Vera Cruz, C. et al (2014) Xa39, a novel dominant gene conferring broad‐spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant. Pathol. 64, 568–575. [Google Scholar]
- Zhou, D. (1999) Regulatory mechanism of plant gene transcription by GT‐elements and GT‐factors. Trends Plant Sci. 4, 210–214. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Huang, X. , Liu, T. , Gao, S. and Chen, J. (2012) Cloning and function analysis of a drought‐inducible gene associated with resistance to Curvularia leaf spot in maize. Mol. Biol. Rep. 39, 7919–7926. [DOI] [PubMed] [Google Scholar]
- Zuo, S. , Yin, Y. , Pan, C. , Chen, Z. , Zhang, Y. , Gu, S. , Zhu, L. et al (2013) Fine mapping of qSB‐11 LE, the QTL that confers partial resistance to rice sheath blight. Theor. Appl. Genet. 126, 1257–1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Schematic map of the Os2H16 promoter with putative promoter elements.
Figure S2 Tissue expression patterns of the Os2H16 promoter.
Figure S3 Pathogen‐inducible expression patterns of the Os2H16 promoter.
Figure S4 Analysis of amino acid sequence of OsASR2.
Figure S5 Binding of OsASR2 to the GT‐1 cis‐element in a ChIP assay.
Figure S6 Verification of OsASR2 transgenic lines.
Figure S7 Overview of ChIP‐seq data.
Figure S8 Binding motifs identified in OsASR2 binding peaks in the ChIP‐seq assay.
Figure S9 Time causes of OsASR2 expression in response to pathogen and drought treatments.
Figure S10 Sequence alignments of OsASR2 with the tomato ASR1.
Table S1 PCR primers used.
Table S2 GO analysis of genes containing the GT‐1 cis‐element in the ChIP‐seq data.
Appendix S1 Supplementary methods.


