Summary
Wood production is dependent on the activity of the vascular cambium, which develops from the fascicular and interfascicular cambia. However, little is known about the mechanisms controlling how the vascular cambium is developed in woody species. Here, we show that PtrHB4, belonging to the Populus HD‐ZIP III family, plays a critical role in the process of vascular cambium development. PtrHB4 was specifically expressed in shoot tip and stem vascular tissue at an early developmental stage. Repression of PtrHB4 caused defects in the development of the secondary vascular system due to failures in interfascicular cambium formation. By contrast, overexpression of PtrHB4 induced cambium activity and xylem differentiation during secondary vascular development. Transcriptional analysis of PtrHB4 repressed plants indicated that auxin response and cell proliferation were affected in the formation of the interfascicular cambium. Taken together, these results suggest that PtrHB4 is required for interfascicular cambium formation to develop the vascular cambium in woody species.
Keywords: cambium activity, HD‐ZIP III, interfascicular cambium, secondary growth, Populus
Introduction
The plant vascular system, which enabled green plants to successfully colonize terrestrial land, is not only a pipeline for the transport of water, nutrients, signalling molecules and other materials over long distances, but also a skeleton to provide mechanical support for vertical growth. The primary vascular system comprises of a group of discrete vascular bundles containing fascicular cambium, primary phloem and primary xylem. The primary vascular bundles are originated from procambium cells at the peripheral region of the rib zone of the shoot apical meristem (SAM). In perennial woody plants, fascicular cambium located at the centre of primary vascular bundles undergoes extension into the interfascicular region and generates interfascicular cambium tangentially to form a ring of vascular cambium. Then, the meristematic activity of the vascular cambium gives rise to the continuous production of cylindrical secondary vascular tissue (wood), which is a large source of sustainable energy and a sink for atmospheric carbon dioxide.
To date, molecular understanding regarding how the ring of vascular cambium is developed in woody species is limited due to challenges faced in performing forward genetic analysis in trees. The herbaceous species Arabidopsis has been used in several studies as a research model to screen for mutants or to induce secondary growth with hormone treatment (Chaffey et al., 2002; Davin et al., 2016; Ko et al., 2004; Zhang et al., 2011). A number of genes has been identified for their role in regulating Arabidopsis cambium activity (Agusti et al., 2011; Parker et al., 2003; Pineau et al., 2005; Suer et al., 2011). WOX4, a WUSCHEL‐related HOMEOBOX gene, regulated by CLE41/44 (CLAVATA3/ESR‐related 41/44)/TDIF (tracheary element differentiation inhibitory factor) peptide and its receptor PXY (PHLOEM INTERCALATED WITH XYLEM)/TDR (TDIF receptor), is required to promote cambial cells division (Baurle and Laux, 2005; Etchells and Turner, 2010; Hirakawa et al., 2008, 2010; Suer et al., 2011). HCA2 (high cambial activity), a nuclear‐localized DNA binding with one finger (Dof) transcription factor Dof5.6 promotes interfascicular cambium formation without alternating the organization of the vascular bundles in Arabidopsis inflorescence stems (Guo et al., 2009). MOL1 (MORE LATERAL GROWTH1) negatively regulates cambium activity by acting antagonistically to the CLE41/PXY/WOX4 cascade (Agusti et al., 2011; Gursanscky et al., 2016). Other receptor‐like kinases such as REDUCED IN LATERAL GROWTH1 (RUL1) act as an opposing regulator of cambium activity to MOL1 (Agusti et al., 2011). However, there are some developmental characteristics of secondary growth in woody species that may not be characterized using the Arabidopsis system. Secondary growth in Arabidopsis usually occurs at the basal part of inflorescence stems. The wall‐thickened interfascicular fibre cells, which are differentiated from interfascicular parenchyma cells, contribute to most of the basal secondary growth tissue. In contrast, in woody species, secondary growth originates from the meristematic activity of the vascular cambium, which forms vertically below the SAM via connecting the discrete fascicular/interfascicular cambia together (Larson, 1994; Nieminen et al., 2015; Philipson et al., 1971; Romberger et al., 1993; Sehr et al., 2010). Vascular cambium in woody plants produces secondary vascular tissue, which is precisely organized with vessel elements, fibre cells and ray parenchyma cells (Little et al., 2002; Mazur et al., 2014; Parker et al., 2003; Pineau et al., 2005). Thus, woody species are believed to have evolved specific molecular mechanisms to regulate the development of secondary growth such as controlling interfascicular cambium formation that have yet to be elucidated.
HD‐ZIP III gene family has been shown to act in both distinctive and redundant manners to regulate meristem function, organ polarity and vascular development (Emery et al., 2003; Izhaki and Bowman, 2007; McConnell et al., 2001; Prigge et al., 2005). HD‐ZIP III genes along with auxin, auxin polar transporter PINs and auxin response factor MP/ARF5, form an integrated feedback loop that is essential for the formation of the procambium during the development of the Arabidopsis embryo, leaf and root (Donner et al., 2009; Jouannet et al., 2015; Muller et al., 2016). During secondary growth in trees, auxin peaks are present in the cambium zone and developing xylem in a lateral gradient manner (Tuominen et al., 1997). Meanwhile, Populus PIN genes show the same expression pattern in the cambial zone (Schrader et al., 2003). PoptrMP1, a homologue of the MP/ARF5 gene in Populus, is expressed specially in developing secondary xylem and its overexpression increases the expression of HD‐ZIP III genes (Johnson and Douglas, 2007). These data suggest that a conserved HD‐ZIP III‐auxin‐PIN‐MP/ARF5 signalling pathway may be shared between procambium formation and vascular cambium establishment.
In this study, a HD‐ZIP III gene PtrHB4 (Potri.001G372300), which was observed to be highly expressed in vascular tissues in Populus (Zhu et al., 2013), was investigated for its role in the development of secondary growth in Populus. The results suggest PtrHB4 is required for interfascicular cambium formation, likely via a mechanism which influences the process of auxin response during vascular cambium development in woody species.
Results
PtrHB4 expression is correlated with the process of vascular cambium formation
PtrHB4 expression was examined from the top SAM tissue successively down to internodes (IN) undergoing secondary growth in the Populus stem. RT‐qPCR analysis indicated that expression of PtrHB4 was prominent in shoot tip and young stem (IN2, IN4), but dramatically decreased in regions of the stem undergoing secondary growth (IN8, IN10 and IN12) (Figure 1a). A similar expression pattern was observed in PtrHB4 promoter (PtrHB4pro:GUS) transgenic plants, in which strong GUS stain appeared in shoot tip as well as at the early development stage of vascular tissues (Figure 1b), while the GUS stain was barely detected in secondary vascular tissues (Figure S1a). To further analyse the location of PtrHB4 expression, PtrHB4‐specific antibodies were generated (Figure S1b‐e) and immunolocalization analyses were performed within the early development stage of vascular tissues. PtrHB4 was detected at the centre of tip including SAM with procambium cells and its surrounded region (Figure 1c and Figure S1f and g). As the vascular tissue expanded longitudinally, PtrHB4 protein was gradually restricted to primary vascular bundles in IN1 (Figure 1d–f, Figure S1f and g and Figure S2a and b). In IN2, PtrHB4 was localized in primary xylem, phloem and interfascicular parenchyma cells which can be induced to initiate cell division (Figure 1g–i and Figure S2e and f). Consistent with RT‐qPCR and the GUS assay, expression of the PtrHB4 protein was not found in the vascular tissues of IN12 undergoing secondary growth (Figure 1j). The expression pattern of PtrHB4 suggests it may play a role associated with the process of vascular cambium development.
Figure 1.
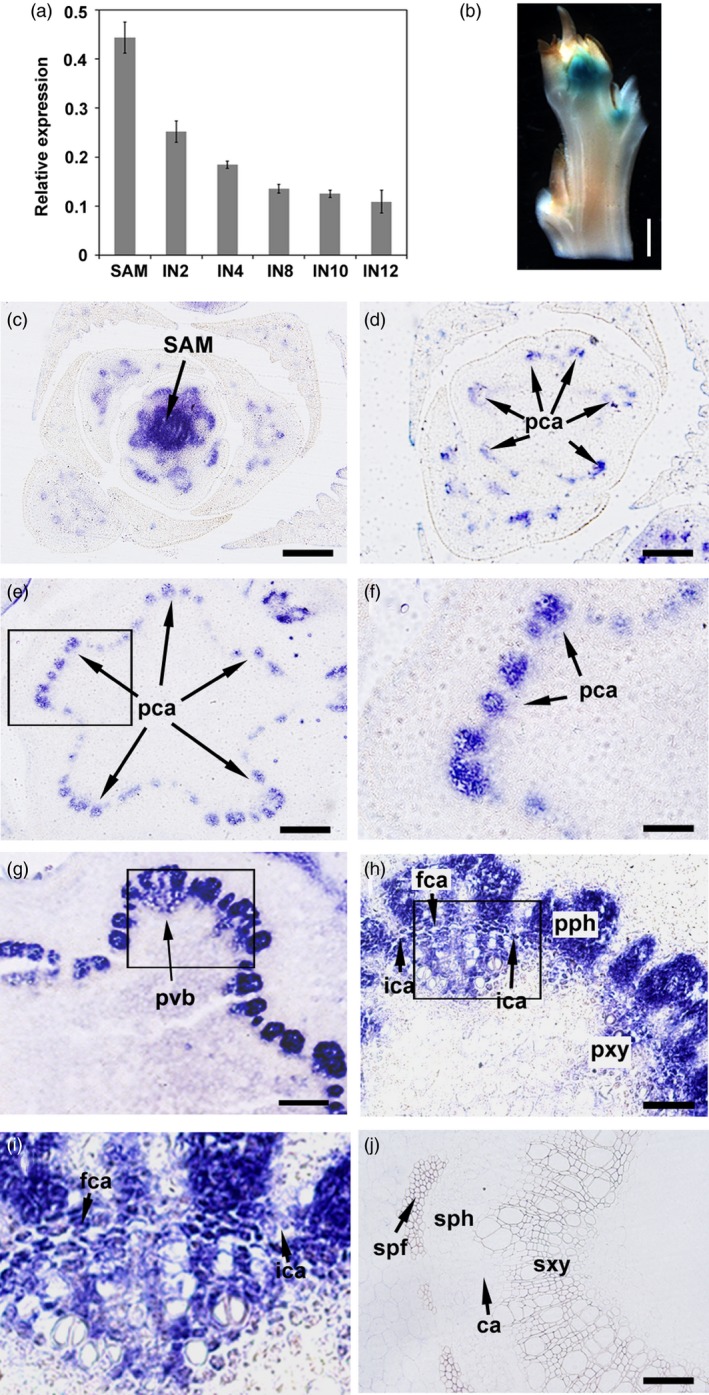
Expression pattern of PtrHB4 during vascular cambium formation. (a) Expression of PtrHB4 in Populus stem analysed by RT‐qPCR. PtrActin1 was used as a reference gene. Bars are means ± SD of n = 3 biological replicates. (b) Histochemical analysis of GUS activity in shoot tip of PtrHB4pro:GUS transgenic plants. (c‐j) Immunolocalization analysis of PtrHB4 in shoot apex (c), in IN1 (d and e, indicating continual sections from top to bottom), (f) magnification of the framed section in (e). In IN2 (g), (h) magnification of the framed section in (g), (i) magnification of the framed section in (h) and in IN12 (j). IN, internode; SAM, shoot apical meristem; pca, procambium; ca, cambium; fca, fascicular cambium; ica, interfascicular cambium; ipc, interfascicular parenchyma cells; pvb, primary vascular bundle; pph, primary phloem; pxy, primary xylem; sph, secondary phloem; sxy, secondary xylem; spf, secondary phloem fibre. Bars: 1 mm in (b), 200 μm in (c), (d), (e) and (g), 100 μm in (f), (h) and (j), 20 μm in (i).
Repression of PtrHB4 resulted in changes to secondary vascular tissue formation due to defects in interfascicular cambium development
To investigate the function of PtrHB4 in Populus, a PrtHB4 repressor was generated (PtrHB4SRDX) based on the chimeric repressor silencing technology (CRES‐T) (Figure S3) for circumventing the effects of HD‐ZIP III redundant genes (Hiratsu et al., 2003; Zhong et al., 2011). A total of thirty independent transgenic lines were generated and eighteen lines with increased PtrHB4 expression (Figure 2c) showed similar phenotypes which were different from the wild type (WT) plant. PtrHB4SRDX plants displayed large, downward curling leaves (Figure 2a), although the number of leaves per PtrHB4SRDX plant was the same as WT plants in the same growth period (Figure 2a). PtrHB4SRDX plants grew shorter internodes and thicker stems compared to WT plants (Figure 2d and e). The stems of PtrHB4SRDX plants were polygonal prism shaped vs. WT plants which had cylindrical stems (Figure 2b). Cross sections of IN12 of PtrHB4SRDX plants showed that the vascular bundle‐like tissues were formed in an isolated manner, while WT plants had developed a cylindrical secondary vascular tissue (Figure 2f and g). Phloroglucinol‐HCl staining further indicated that each isolated vascular bundle exhibited closed xylem tissue while phloem tissue was formed surrounding the bundle structure (Figure 2h and i).
Figure 2.
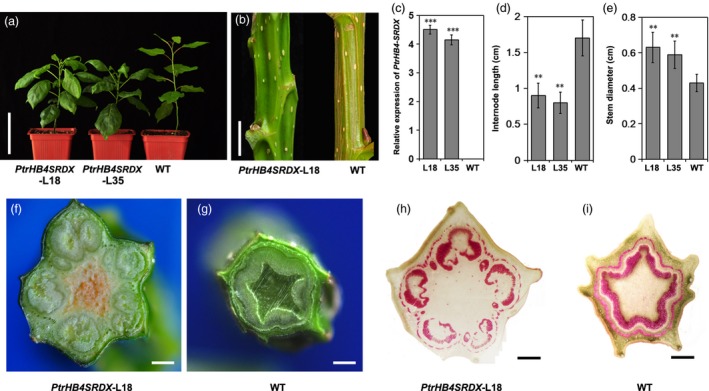
Repression of PtrHB4 disrupted secondary vascular tissue development. (a) Morphological phenotype of PtrHB4SRDX (Line 18 and Line 35) and WT plants. (b) Stems of PtrHB4SRDX and WT plants. (c) Expression levels of PtrHB4 in transgenic and WT plants. (d) Internode length and (e) stem diameter. Bars in (c), (d) and (e) are means ± SD of n = 3 biological replicates. Significance testing was conducted using the two‐sample t‐test (*P < 0.05, **P < 0.01 and ***P < 0.001). (f) and (g) Secondary vascular pattern in IN12 of PtrHB4SRDX and WT plants. (h) and (i) Cross sections of IN12 in PtrHB4SRDX and WT plants stained by phloroglucinol‐HCl. Bars: 10 cm in (a), 2 cm in (b), 5 mm in (f) and (g), 500 μm in (h) and (i).
PtrHB4 was expressed as early as cambium initiation, and developmental defects were observed in the process of vascular cambium formation in PtrHB4 repressed plants. Primary vascular bundles of both PtrHB4SRDX and WT plants were observed in IN2, but the distance between adjacent vascular bundles was larger in PtrHB4SRDX (Figure 3a and b) than in WT plants (Figure 3c and d). As early as IN3, the individual vascular bundles in WT plants had started to join each other through interfascicular cambium formation (Figure 3g and h). However, far to IN6 in PtrHB4SRDX plants, vascular bundles still failed to link together (Figure 3e and f), while at this stage, WT plants had started the formation of secondary vascular tissue (Figure 3k and l). Upon closer look, fascicular cambium cells appeared normal (Figure 3j), but the direction of cell division of parenchyma cells at the edge of fascicular cambium was changed in PtrHB4SRDX plants (Figure 3i). Consequently, the isolated vascular bundles developed into closed bundle tissues within IN16 of PtrHB4SRDX plants (Figure S4a, b, c and e). In addition, secondary xylem fibre cells were defective in terms of secondary cell wall formation and vessel elements showed smaller sizes with normal secondary cell wall deposition (Figure S4d and f). These results suggest that repression of PtrHB4 affected cell division and led to failure in interfascicular cambium development.
Figure 3.
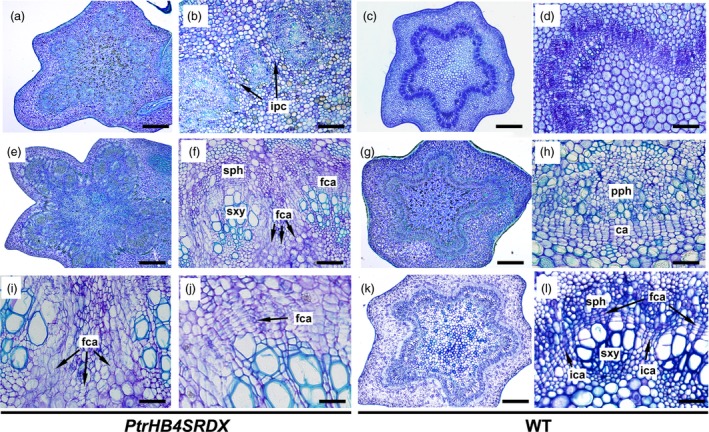
Repression of PtrHB4 affected interfascicular cambium formation. (a) and (c) Cross section of the IN2 in PtrHB4SRDX and WT. (b) and (d) Magnification of (a) and (c). (e) and (k) Cross section of IN6 of PtrHB4SRDX and WT plants. (f) and (l) Magnification of (e) and (k). (g) Cross section of the IN3 in WT. (h) Magnification of (g). (i) Abnormal parenchyma cell division at the edge of fascicular cambium in PtrHB4SRDX. (j) Normal dividing activity of fascicular cambium in PtrHB4SRDX plants. ca, Cambium; fca, fascicular cambium; ipc, interfascicular parenchyma cells; sph, secondary phloem; sxy, secondary xylem. Bars: 500 μm in (a), (c), (e), (g) and (k), 100 μm in (b), (d), (f), 20 μm in (h), (i), (j) and (l).
PtrHB4 induced cambium activity and xylem cell differentiation
To obtain further evidence to aid in understanding of PtrHB4 function, PtrHB4 was mutated (PtrHB4mt) by replacing four nucleotides within the miR165/166 target sequence to avoid miRNA regulation. Then, the PtrHB4mt gene was transformed into Populus under the control of the CaMV35S promoter (Figure S5) (Mallory et al., 2004). Twenty‐six independent transgenic lines were identified with high expression of PtrHB4mt and similar phenotypic changes (Figure 4a and c). Mature leaves of PtrHB4mt plants were up‐curled and smaller than WT plants, while the number of leaves in each PtrHB4mt plant was similar with WT plants in the same growth period (Figure 4a and b). PtrHB4mt plants had thinner stems with twisted and shortened internodes (Figure 4a, d and e). Anatomical analysis showed that parenchyma cells, within the cortex and pith of IN2 in PtrHB4mt plants, displayed dividing activity, indicated by induction of cell division (Figure 5a, b and c). Within IN12, the vascular tissues appeared normal in PtrHB4mt plants (Figure S6a and b). Several layers of ectopic cambium cells were formed from parenchyma cells in cortex (Figure 5d and e). Further, the newly produced ectopic cambium cells were differentiated into xylem‐like cells (Figure 5d–f), which are morphologically similar to fibre cells and vessel elements as indicated by secondary cell wall staining (Figure 5g–i). Here, the results indicate that overexpression of PtrHB4mt led to induction of ectopic cambium and secondary xylem‐like cells in cortex parenchyma cells.
Figure 4.

Morphological changes in overexpression of PtrHB4mt Populus. (a) Morphological phenotype of PtrHB4mt and WT plants. (b) Mature leaves of PtrHB4mt and WT plants. (c) Expression levels of PtrHB4mt in transgenic plants and WT plants. (d) Internode length and (e) stem diameter of PtrHB4mt and WT plants. Bars in (c), (d) and (e) are means ± SD of n = 3 biological replicates. Significance testing was conducted using the two‐sample t‐test (*P < 0.05, **P < 0.01 and ***P < 0.001). Bars: 10 cm in (a), 5 cm in (b).
Figure 5.
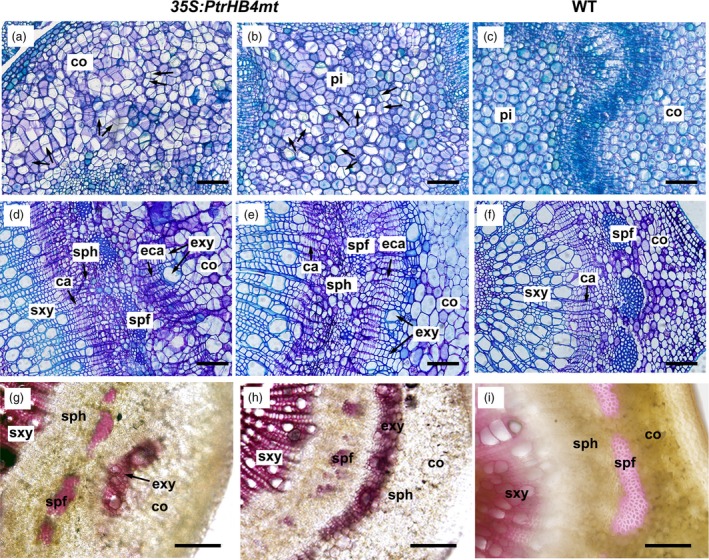
Overexpression of PtrHB4mt induced ectopic vascular cambium and xylem cells. Cross section of the IN2 in PtrHB4mt (a, cortex and b, pith) and WT plants (c). Cross section of the IN12 in PtrHB4mt (d and e, indicating different areas of cortex) and WT plants (f). Secondary cell wall staining by phloroglucinol‐HCl in the cortex of PtrHB4mt (g and h, indicating different areas of cortex) and WT plants (i). All sections were sampled from three‐month‐old plants. ca, Cambium; eca, ectopic cambium; sxy, secondary xylem; exy, ectopic xylem; sph, secondary phloem; spf, secondary phloem fibre; co, cortex. Bars: 20 μm in (a), (b) and (c), 100 μm in (d), (e), (f), (g), (h) and (i).
PtrHB4 may associate with the auxin signalling pathway during cambium formation
To investigate how PtrHB4 affects molecular processes during vascular cambium formation (Figure 1b), RNA sequencing of the shoot tip and IN1‐IN4 tissues from PtrHB4SRDX and WT plants was performed. Sequencing analysis showed that a total of 994 genes were differentially expressed (DE) due to PtrHB4 repression (PtrHB4SRDX versus WT), among which 498 genes were down‐regulated and 496 genes were up‐regulated (5 ≤ log2 ratio (RPKM of PtrHB4SRDX/RPKM of WT) ≤−5 and P value ≤0.05) (Table S1 and S2). Gene ontology (GO) enrichment analysis indicated that the DE genes were associated with meristem development, postembryonic development and anatomical structure development. The GO categories for cell differentiation, protein localization and regulation of transcription were overrepresented among down‐regulated genes, while GO categories for cell division and regulation of signal transduction were enriched among up‐regulated genes (Figure 6a).
Figure 6.
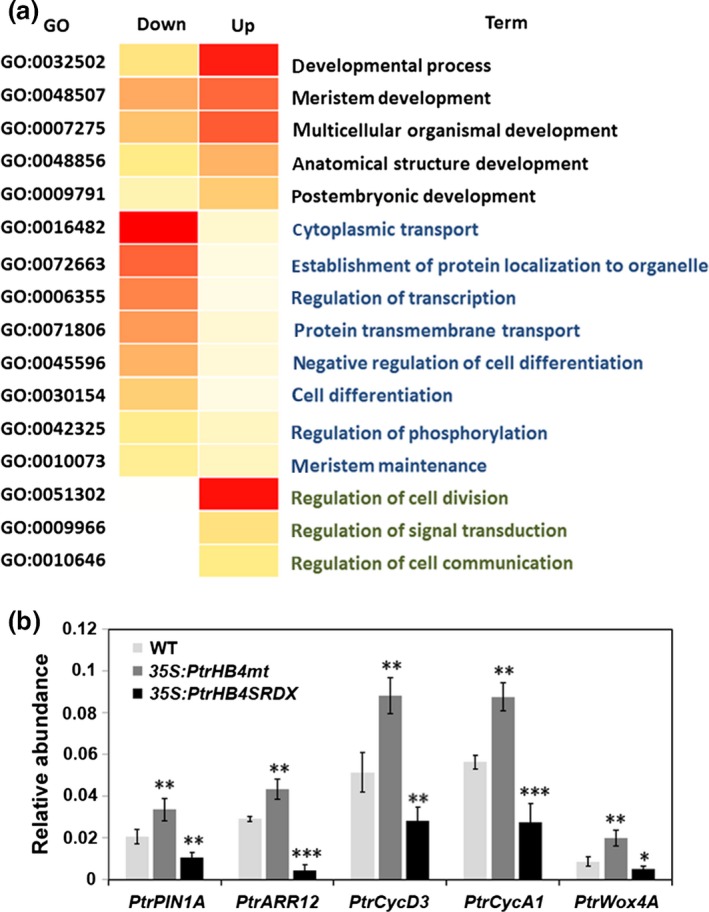
GO enrichment of differential expression genes and expression levels of genes related to PtrHB4 signalling pathway. (a) Significantly overrepresented GO biological process terms for DE genes. (b) Expression analysis of PtrPIN1A, PtrARR12, PtrCycD3, PtrCycA1, PtrWox4A in PtrHB4SRDX, PtrHB4mt and WT plants. PtrActin1 was used as a reference gene. Bars are means ± SD of n = 3 biological replicates of two independent lines of PtrHB4SRDX and PtrHB4mt. Significance testing was conducted using the two‐sample t‐test (*P ≦ 0.05, **P < 0.01 and ***P < 0.001).
Genes related to auxin response were down‐regulated in PtrHB4SRDX plants, including TIR1 (Transport Inhibitor Response1, Potri.004G033900), AUX1 (Auxin Resistant1, Potri.016G113600) and PIN1 (PINFORMED1, Potri.015G038700). Moreover, cyclin genes, cyclin‐dependent kinase genes and ARR12 (Potri.018G021300) were down‐regulated in PtrHB4SRDX plants. In addition, the CLV/WUS signalling pathway‐related genes were down‐regulated in PtrHB4SRDX plants, including one ortholog of CLV1 (Potri.005G241500), one ortholog of WOX4 (Potri.014G025300), POL (Poltergeist, Potri.002G185000), PLL4 (Poltergeist like4, Potri.001G239300) and BRAD1 (Breast Cancer Associated Ring1, Potri.002G259000). An orthologue of KANADI2 (Potri.003G096300) was up‐regulated in PtrHB4SRDX plants. Expression alternations of genes related to both auxin and cytokinin signalling pathways were further verified in both PtrHB4SRDX and PtrHB4mt plants by RT‐qPCR analysis (Figure 6b). Together, transcriptional analysis suggests that PtrHB4 may associate with auxin and cytokinin signalling pathways, which in turn may be involved in PtrHB4‐mediated interfascicular formation processes during vascular cambium development.
Discussion
Secondary growth in woody species is dependent on the meristematic activity of the vascular cambium. During the formation of the cylindrical vascular cambium, discrete primary vascular bundles need to connect to each other through interfascicular cambium development. Generally, interfascicular cambium is initiated by interfascicular parenchyma cells undergoing periclinal cell division adjacent to the fascicular cambium and then merged with the fascicular cambium to form a ring of vascular cambium (Eames and MacDaniels, 1947; Little et al., 2002; Mazur et al., 2014). However, little is known regarding how vascular cambium development is molecularly regulated. In this study, PtrHB4, a HD‐ZIP III gene in Populus, was showed to regulate interfascicular cambium formation in a developmental‐specific manner.
To develop vascular cambium, the procambium in the rib zone right beneath SAM undergoes vertical division contributing to the initiation of the fascicular cambium in primary vascular bundles (Medford, 1992). As illustrated in Figure 7, interfascicular cambium is formed through periclinal division of parenchyma cells at the edge of fascicular cambium, which makes individual vascular bundles linking together to form a ring of vascular cambium system. Then, vascular cambium undergoes anticlinal division and differentiates secondary xylem and phloem (Larson, 1994; Little et al., 2002; Mazur et al., 2014; Philipson et al., 1971; Romberger et al., 1993). Repression of PtrHB4 led to defective establishment of a ring of vascular cambium from fascicular and interfascicular cambia, likely due to the failure in initiation of interfascicular cambium, suggesting that PtrHB4 was required for vascular cambium development. On the other hand, by overexpression of PtrHB4 in Populus, normal ring of vascular cambium was retained, but ectopic cambium and xylem‐like cells were initiated from parenchyma cells in cortex, suggesting a PtrHB4 role in induction of cambium activity. The results indicate that interfascicular cambium formation may be due to the capability of fascicular cambium activity regulated by PtrHB4.
Figure 7.
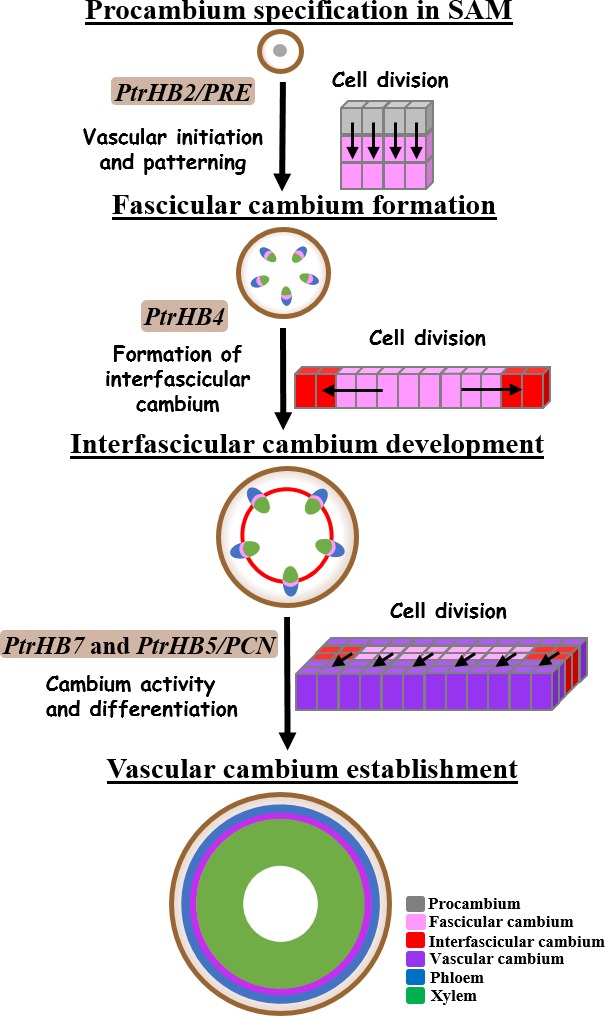
Functions of HD‐ZIP III genes during vascular development in Populus. A schematic view of distinctive roles of HD‐ZIP III genes in regulating vascular development process in the stem of Populus. Arrows represented the areas with cells undergoing vertical, periclinal or anticlinal division, characterized for following stages of cambium development.
HD‐ZIPIII gene family has indicated its ancestral role in vascular development and organ initiation (Prigge and Clark, 2006). It appears more specific function that HD‐ZIP III genes have displayed in the process of secondary growth in woody plants (Robischon et al., 2011; Zhu et al., 2013). Eight HD‐ZIP III genes (PtrHB1 and PtrHB2, PtrHB3 and PtrHB4, PtrHB5 and PtrHB6 and PtrHB7 and PtrHB8) in Populus are divided into four clades and their distinctive expression patterns in Populus imply their function diversity in association with vascular development (Du et al., 2011; Zhu et al., 2013). As indicated in Figure 7, PtrHB2/PRE plays a major role in regulating initiation of cambium and patterning of vascular tissue (Medford, 1992; Robischon et al., 2011). PtrHB7 promotes cambium activity and xylem differentiation (Zhu et al., 2013). PtrHB5/PCN inhibits cambium activity and xylem differentiation during secondary growth (Du et al., 2011; Zhu et al., 2013). Our evidence supports that PtrHB4 plays a role in regulating the process of interfascicular cambium formation. On the other hand, overexpression of PtrHB2/PRE, PtrHB4 and PtrHB7 all induced ectopic cambium activity in cortex cells (Robischon et al., 2011), indicating that PtrHB2/PRE, PtrHB4 and PtrHB7 may have overlapping functions in regulating vascular development.
The maximum auxin content has been detected in the cambium zone and developing xylem of Populus (Schrader et al., 2003; Tuominen et al., 1997). Meanwhile, the auxin response factor MP/ARF5 has been shown to activate expression of HD‐ZIP III genes in vascular tissue of Populus (Donner et al., 2009; Johnson and Douglas, 2007). Auxin diffusing laterally from the fascicular cambium, which is mediated by polar auxin transport protein PINs, has been suggested to be a key process stimulating formation of the interfascicular cambium (Little et al., 2002). HD‐ZIP III genes have been shown to affect the expression pattern of PINs during vasculature initiation in leaf, root and lateral growth (Benjamins and Scheres, 2008; Vernoux et al., 2010). Expression of PtrPIN1 and PtrWOX4, which was suggested for stimulating cambium activity in an auxin‐dependent manner (Suer et al., 2011), was down‐regulated by repression of PtrHB4 and up‐regulated by overexpression of PtrHB4. These results suggest that PtrHB4 regulation of the interfascicular cambium formation may go through auxin signalling process. However, more detailed characterization is required to elucidate the exact mechanism that PtrHB4 conducts in interfascicular cambium initiation.
Development of vascular cambium is essential for plant secondary growth which is a critical biological process for sustaining woody plants to grow a long‐life span. Through secondary growth, a large number of photosynthetic products are accumulated in secondary xylem tissue to provide wood, fibre and chemical materials for meeting demands of human society. Finding of the PtrHB4 role in interfascicular cambium initiation would improve our understanding of the molecular mechanisms underlying vascular cambium formation as well as help develop new strategies to engineer the secondary growth for ideal wood and fibre production.
Experimental procedures
Cloning and plant transformation
The full coding sequence of PtrHB4 and the 2.5 kb promoter of PtrHB4 of Populus trichocarpa were cloned as previous study using the primers in (Table S3) (Zhu et al., 2013). PtrHB4pro:GUS constructs were generated by replacing the cauliflower mosaic virus 35S promoter of pBI121 binary vector to the PtrHB4 promoter. The stop codon of PtrHB4 was replaced by nucleotide sequence of the SRDX domain using PCR amplification with the reverse primers (Table S3), and then the amplicon of PtrHB4SRDX was cloned into pBI121 vector to generate 35S:PtrHB4SRDX construct. The overexpression construct 35S:PtrHB4mt was generated as previous study using one pair of overlapping primers PtrHB4mt‐F and PtrHB4mt‐R (Table S3). All constructs were transformed into Populus×euramericana cv. ‘Nanlin895’ by Agrobacterium‐mediated transformation according to the protocol adopted in our laboratory (Li et al., 2003). For each transformation, at least 25 independent lines of transgenic plants were generated. After transgenic plants were identified and verified by examining transgene expression, the transgenic plants were multiplied through micro‐cutting propagation and used as biological repeats. Among the independent lines which showed morphological phenotype, 2–3 lines with the highest expression of PtrHB4SRDX or PtrHB4mt were selected for detail characterization (Figures 2c and 5c). All plants in the first 2 months were grown in a phytotron with a light and dark cycle of 16 h and 8 h at 22 °C under a light density of 150 μE/m2/s and then moved into a greenhouse with the same light and dark cycle and supplementary light of 200 μE/m2/s. Measurement of stem diameter and internode length was taken from the 30th internode from 3‐month‐old plants. Two‐sample t‐test was used to determine statistical significance between wild type and individual transgenic lines.
Gene expression analysis
Total RNA was isolated from tissues of SAM, shoot tip, different stem internodes, and leaves for examining the tissue specificity of PtrHB4 expression using modified CTAB method (Richards et al., 2001). After treatment with DNase I, total RNA was used for first‐strand cDNA synthesis using Hifair™ II 1st Strand cDNA Synthesis SuperMix (Yeasen, http://www.yeasen.com) and followed by RT‐qPCR analysis using gene‐specific primers showing in (Table S3). RT‐qPCR was performed using UNICON™ SYBR Green® Real‐Time PCR Master Mix (Yeasen, http://www.yeasen.com) and an iQ5™ Real‐Time PCR Detection System (Bio‐Rad, http://www.bio-rad.com/) according to the manufacturer's instructions. All reactions were performed with at least three biological repeats and three technical repeats. Statistical analyses (two‐sample t‐test) were preformed to evaluated statistical significance between wild type and individual transgenic lines.
Histochemical and histological analyses
Shoot tips and series of internodes of stem from PtrHB4pro: GUS transgenic plants were hand‐sectioned and then incubated in 50% acetone (v/v) for 10 min on ice, and incubated in GUS stain solution (100 mm sodium phosphate (pH7.0), 10 mm EDTA, 0.5 mm ferricyanide, 0.5 mm ferrocyanide, 0.1% Triton X‐100, 20% methanol and 2 mm X‐Gluc) at 37 °C for 2 h. Following staining, sections were cleared by 75% ethanol and photographed. Shoot tips and stem segments with 3 mm length at different internodes from transgenic and wild type plants were fixed in formaldehyde‐acetic acid solution (formaldehyde:glacial acetic acid:ethanol [1 : 1 : 18]) for 24 h, dehydrated in graded ethanol series, and embedded into paraplast. The samples were sectioned to 10 μm thick using rotary microtome of Leica RM2235 (Leica, http://www.leica-microsystems.com/products). The sections were stained with toluidine blue and observed under a light microscope of OLYMPUS BX51 (OLYMPUS). For lignin staining, sections were immersed in 1% phloroglucinol (w/v) in 12% HCl for 5 min and immediately observed with a light microscope.
Immunolocalization
An N‐terminal‐specific peptide of PtrHB4 (SKDKHMDSSKYVRY) was synthesized and injected into rabbits to raise antibodies (Abmart, Shanghai, China) (Figure S1b). Rabbit serum was collected and purified using protein‐A/G Sepharose. The purified antibodies were diluted into a concentration of 1 μg/μL for later use. The N‐terminal 150 amino acids length of PtrHB4 protein was cloned into pET28 vector and then expressed in E. coli. (BL21). Total proteins of cell lysate after IPTG induction were separated by SDS‐PAGE to examination expression of protein (Figure S1c). Western blot was performed using total proteins of cell lysate and from shoot tip of wild type plants against PtrHB4 antibody (diluted at 1 : 1000) to analysis antibody specificity according to the previous protocol (Figure S1d and e) (Song et al., 2010). The secondary antibodies (linked with alkaline phosphatase, Santa Cruz, CA) were diluted in 1 : 5000. The shoot tip and internodes of wild plants were embedded and sliced into 10‐μm‐thick sections for immunolocalization according to the previous protocol (Song et al., 2010). The first antibodies were diluted in 1 : 200. The secondary antibodies were diluted in 1 : 1000. After colour development, the sections were gradually dehydrated with alcohols, cleared with xylene and observed under an OLYMPUS BX51 light microscope (Olympus, NY).
RNA sequencing and analysis
Total RNAs were isolated from the shoot tips using modified CTAB method, which were collected from three independent lines of the transgenic and WT plants. The quality of the total RNAs was determined by OD260/OD280 ratios and agarose gel electrophoresis. The concentration of the total RNAs was >400 ng/μL and the total quantity was >20 μg per samples. The RNA sequencing libraries were constructed after combination three biological repeats of transgenic or WT plants as our previous protocol (Zhu et al., 2013). The library products were sequenced via Illumina HiSeq™ 2000 with the paired‐end‐100 bp reads. The RNA‐Seq data have been submitted to NCBI Sequence Read Archive (accession number SUB2483520). The raw sequence data were analysed using Illumina HiSeq™ 2000 software. The raw reads were filtered to generate clean reads and then mapped to the Populus trichocarpa genome using SOAPalibner/soap2 (Li et al., 2009). The reads with no more than two bases of mismatch were used for alignment. Gene annotation was on the basis of the Arabidopsis genome database (TAIR10). The transcript level was calculated using the RPKM (Reads Per kb per Million reads) method (Mortazavi et al., 2008). The expression difference between PtrHB4SRDX and WT was examined with the threshold of P‐value in multiple tests (Audic and Claverie, 1997; Benjamini et al., 2001). A total of 45 786 334 and 47 587 472 cDNA reads were identified from samples of PtrHB4SRDX and WT plants. 81.27% and 81.42% of these reads were mapped to the Populus trichocarpa genome and 47 445 and 47 703 genes were detected in the two samples, respectively. GO enrichment analysis was performed as described in (Xue et al., 2013).
Supporting information
Figure S1 Expression of PtrHB4 during vascular cambium development.
Figure S2 Interfascicular cambium development in stems of Populus.
Figure S3 Construct of 35S:PtrHB4SRDX.
Figure S4 Repression of PtrHB4 affected xylem development.
Figure S5 Mutations in the miRNA166 target sites.
Figure S6 Overexpression of PtrHB4mt transformed vascular bundles pattern to amphivasal in Arabidopsis.
Table S1List of the down‐regulated genes in PtrHB4SRDX plants.
Table S2 List of the up‐regulated genes in PtrHB4SRDX plants.
Table S3 List of primers used in this study.
Acknowledgements
We thank Liangjiao Xue (University of Geogia) for GO enrichment analysis. This work was supported by National Nature Science Foundation of China (31630014 and 31300500), the Chinese Academy of Sciences (XDPB0402) and Ministry of Science and Technology of China (2016YFD0600104). The authors declare no conflict of interest.
References
- Agusti, J. , Lichtenberger, R. , Schwarz, M. , Nehlin, L. and Greb, T. (2011) Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet. 7, e1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic, S. and Claverie, J.M. (1997) The significance of digital gene expression profiles. Genome Res. 7, 986–995. [DOI] [PubMed] [Google Scholar]
- Baurle, I. and Laux, T. (2005) Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell, 17, 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , Drai, D. , Elmer, G. , Kafkafi, N. and Golani, I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284. [DOI] [PubMed] [Google Scholar]
- Benjamins, R. and Scheres, B. (2008) Auxin: The Looping Star in Plant Development. Annu. Rev. Plant Biol. 59, 443–465. [DOI] [PubMed] [Google Scholar]
- Chaffey, N. , Cholewa, E. , Regan, S. and Sundberg, B. (2002) Secondary xylem development in Arabidopsis: a model for wood formation. Physiol. Plant. 114, 594–600. [DOI] [PubMed] [Google Scholar]
- Davin, N. , Edger, P.P. , Hefer, C.A. , Mizrachi, E. , Schuetz, M. , Smets, E. , Myburg, A.A. et al (2016) Functional network analysis of genes differentially expressed during xylogenesis in soc1ful woody Arabidopsis plants. Plant J. 86, 376–390. [DOI] [PubMed] [Google Scholar]
- Donner, T.J. , Sherr, I. and Scarpella, E. (2009) Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development, 136, 3235–3246. [DOI] [PubMed] [Google Scholar]
- Du, J. , Miura, E. , Robischon, M. , Martinez, C. and Groover, A. (2011) The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE, 6, e17458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames, A.J. and MacDaniels, L.H. (1947) An Introduction to Plant Anatomy. New York: McGraw‐Hill. [Google Scholar]
- Emery, J.F. , Floyd, S.K. , Alvarez, J. , Eshed, Y. , Hawker, N.P. , Izhaki, A. , Baum, S.F. et al (2003) Radial patterning of Arabidopsis shoots by class III HD‐ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Etchells, J.P. and Turner, S.R. (2010) The PXY‐CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development, 137, 767–774. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Qin, G. , Gu, H. and Qu, L.J. (2009) Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell, 21, 3518–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursanscky, N.R. , Jouannet, V. , Grünwald, K. , Sanchez, P. , Laaber‐Schwarz, M. and Greb, T. (2016) MOL1 is required for cambium homeostasis in Arabidopsis. Plant J. 86, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, Y. , Shinohara, H. , Kondo, Y. , Inoue, A. , Nakanomyo, I. , Ogawa, M. , Sawa, S. et al (2008) Non‐cell‐autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA, 105, 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, Y. , Kondo, Y. and Fukuda, H. (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell, 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K. , Matsui, K. , Koyama, T. and Ohme‐Takagi, M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Izhaki, A. and Bowman, J.L. (2007) KANADI and class III HD‐Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell, 19, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L.A. and Douglas, C.J. (2007) Populus trichocarpa MONOPTEROS/AUXIN RESPONSE FACTOR5 (ARF5) genes: comparative structure, sub‐functionalization, and Populus –Arabidopsis microsynteny This article is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Can. J. Bot., 85, 1058–1070. [Google Scholar]
- Jouannet, V. , Brackmann, K. and Greb, T. (2015) (Pro)cambium formation and proliferation: two sides of the same coin? Curr. Opin. Plant Biol. 23, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.H. , Han, K.H. , Park, S. and Yang, J. (2004) Plant body weight‐induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole‐transcriptome profiling. Plant Physiol. 135, 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, P.R. (1994) The Vascular Cambium: Development and Structure. Berlin, Germany: Springer‐Verlag. [Google Scholar]
- Li, L. , Zhou, Y. , Cheng, X. , Sun, J. , Marita, J.M. , Ralph, J. and Chiang, V.L. (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA, 100, 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Yu, C. , Li, Y. , Lam, T.W. , Yiu, S.M. , Kristiansen, K. and Wang, J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics, 25, 1966–1967. [DOI] [PubMed] [Google Scholar]
- Little, C.A. , MacDonald, J.E. and Olsson, O. (2002) Involvement of Indole‐3‐acetic acid in fascicular and interfascicular cambial growth and interfascicular extraxylary fiber differentiation in arabidopsis thaliana inflorescence stems. Int. J. Plant Sci. 163, 519–529. [Google Scholar]
- Mallory, A.C. , Reinhart, B.J. , Jones‐Rhoades, M.W. , Tang, G. , Zamore, P.D. , Barton, M.K. and Bartel, D.P. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur, E. , Kurczynska, E.U. and Friml, J. (2014) Cellular events during interfascicular cambium ontogenesis in inflorescence stems of Arabidopsis. Protoplasma, 251, 1125–1139. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R. , Emery, J. , Eshed, Y. , Bao, N. , Bowman, J. and Barton, M.K. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature, 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Medford, J.I. (1992) Vegetative apical meristems. Plant Cell, 4, 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Muller, C.J. , Valdes, A.E. , Wang, G. , Ramachandran, P. , Beste, L. , Uddenberg, D. and Carlsbecker, A. (2016) PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in arabidopsis. Plant Physiol. 170, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen, K. , Blomster, T. , Helariutta, Y. and Mahonen, A.P. (2015) Vascular cambium development. Arabidopsis Book, 13, e0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, G. , Schofield, R. , Sundberg, B. and Turner, S. (2003) Isolation of COV1, a gene involved in the regulation of vascular patterning in the stem of Arabidopsis. Development, 130, 2139–2148. [DOI] [PubMed] [Google Scholar]
- Philipson, W.R. , Ward, J.M. and Butterfield, B.G. (1971) The Vascular Cambium: Its Development and Activity. London: Chapman and Hall. [Google Scholar]
- Pineau, C. , Freydier, A. , Ranocha, P. , Jauneau, A. , Turner, S. , Lemonnier, G. , Renou, J.P. et al (2005) hca: an Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant J. 44, 271–289. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J. and Clark, S.E. (2006) Evolution of the class III HD‐Zip gene family in land plants. Evol. Dev. 8, 350–361. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J. , Otsuga, D. , Alonso, J.M. , Ecker, J.R. , Drews, G.N. and Clark, S.E. (2005) Class III homeodomain‐leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell, 17, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E. , Reichardt, M. and Rogers, S. (2001) Preparation of genomic DNA from plant tissue. Curr. Protoc. Mol. Biol. 27:I:2.3:2.3.1‐2.3.7 [DOI] [PubMed] [Google Scholar]
- Robischon, M. , Du, J. , Miura, E. and Groover, A. (2011) The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 155, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberger, J.A. , Hejnowicz, Z. and Hill, J.F. (1993) Plant Structure: Function and Development: A Treatise on Anatomy and Vegetative Development, with Special Reference to Woody Plants. Berlin; New York: Springer‐Verlag. [Google Scholar]
- Schrader, J. , Baba, K. , May, S.T. , Palme, K. , Bennett, M. , Bhalerao, R.P. and Sandberg, G. (2003) Polar auxin transport in the wood‐forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc. Natl Acad. Sci. 100, 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehr, E.M. , Agusti, J. , Lehner, R. , Farmer, E.E. , Schwarz, M. and Greb, T. (2010) Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, D. , Shen, J. and Li, L. (2010) Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol. 187, 777–790. [DOI] [PubMed] [Google Scholar]
- Suer, S. , Agusti, J. , Sanchez, P. , Schwarz, M. and Greb, T. (2011) WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell, 23, 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen, H. , Puech, L. , Fink, S. and Sundberg, B. (1997) A radial concentration gradient of indole‐3‐acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 115, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux, T. , Besnard, F. and Traas, J. (2010) Auxin at the shoot apical meristem. Cold Spring Harb. Perspect. Biol. 2, a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L.‐J. , Guo, W. , Yuan, Y. , Anino, E.O. , Nyamdari, B. , Wilson, M.C. , Frost, C.J. et al (2013) Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state but not growth in transgenic Populus . Plant Cell, 25, 2714–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Elo, A. and Helariutta, Y. (2011) Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22, 293–299. [DOI] [PubMed] [Google Scholar]
- Zhong, R. , McCarthy, R.L. , Lee, C. and Ye, Z.H. (2011) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Song, D. , Sun, J. , Wang, X. and Li, L. (2013) PtrHB7, a class III HD‐Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus . Mol Plant, 6, 1331–1343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression of PtrHB4 during vascular cambium development.
Figure S2 Interfascicular cambium development in stems of Populus.
Figure S3 Construct of 35S:PtrHB4SRDX.
Figure S4 Repression of PtrHB4 affected xylem development.
Figure S5 Mutations in the miRNA166 target sites.
Figure S6 Overexpression of PtrHB4mt transformed vascular bundles pattern to amphivasal in Arabidopsis.
Table S1List of the down‐regulated genes in PtrHB4SRDX plants.
Table S2 List of the up‐regulated genes in PtrHB4SRDX plants.
Table S3 List of primers used in this study.


