Abstract
Mechanistic target of rapamycin (mTOR) resides as two complexes within skeletal muscle. mTOR complex 1 [mTORC1–regulatory associated protein of mTOR (Raptor) positive] regulates skeletal muscle growth, whereas mTORC2 [rapamycin-insensitive companion of mTOR (Rictor) positive] regulates insulin sensitivity. To examine the regulation of these complexes in human skeletal muscle, we utilized immunohistochemical analysis to study the localization of mTOR complexes before and following protein-carbohydrate feeding (FED) and resistance exercise plus protein-carbohydrate feeding (EXFED) in a unilateral exercise model. In basal samples, mTOR and the lysosomal marker lysosomal associated membrane protein 2 (LAMP2) were highly colocalized and remained so throughout. In the FED and EXFED states, mTOR/LAMP2 complexes were redistributed to the cell periphery [wheat germ agglutinin (WGA)-positive staining] (time effect; P = 0.025), with 39% (FED) and 26% (EXFED) increases in mTOR/WGA association observed 1 h post-feeding/exercise. mTOR/WGA colocalization continued to increase in EXFED at 3 h (48% above baseline) whereas colocalization decreased in FED (21% above baseline). A significant effect of condition (P = 0.05) was noted suggesting mTOR/WGA colocalization was greater during EXFED. This pattern was replicated in Raptor/WGA association, where a significant difference between EXFED and FED was noted at 3 h post-exercise/feeding (P = 0.014). Rictor/WGA colocalization remained unaltered throughout the trial. Alterations in mTORC1 cellular location coincided with elevated S6K1 kinase activity, which rose to a greater extent in EXFED compared with FED at 1 h post-exercise/feeding (P < 0.001), and only remained elevated in EXFED at the 3 h time point (P = 0.037). Collectively these data suggest that mTORC1 redistribution within the cell is a fundamental response to resistance exercise and feeding, whereas mTORC2 is predominantly situated at the sarcolemma and does not alter localization.
Keywords: mTORC1, mTORC2, Raptor, Rictor, lysosome
resistance exercise and protein ingestion are potent anabolic stimuli, elevating muscle protein synthesis (MPS) (5, 20) resulting in a positive net protein balance (NPB) (5). Such elevations in MPS are underpinned by the activation of the conserved serine/threonine kinase, mechanistic target of rapamycin (mTOR). This kinase can both augment MPS (17) and offset muscle protein breakdown (MPB)(19). In skeletal muscle, mTOR resides in two distinct complexes distinguishable by the composition of proteins within each. For example, complex 1 (mTORC1) contains mTOR, regulatory associated protein of mTOR (Raptor), G protein β-subunit-like (GβL), proline-rich Akt1 substrate 1 of 40 kDa (PRAS40), and DEP domain-containing mTOR-interacting protein (DEPTOR) (4), and is believed to activate protein synthetic machinery (1), whereas complex 2 (mTORC2) is composed of mTOR, rapamycin-insensitive companion of mTOR (Rictor), DEPTOR, GβL, stress-activated protein kinase interacting protein 1 (Sin1), and protein observed with Rictor (Protor) and is implicated in insulin sensitivity and actin cytoskeleton dynamics (4). Because of the critical role mTORC1 plays in regulating protein synthesis, this complex has received the most detailed examination in relation to resistance exercise and protein feeding. Acute resistance exercise, protein ingestion, or combinations of such stimuli are consistently reported to elevate mTORC1 activity (5, 20), with effects maintained for up to 24 h (6). Furthermore, the acute inhibition of mTORC1 with rapamycin administration ablates any effect of anabolic stimuli on MPS (8, 9).
As mTORC1 activity seems to be directly implicated in the stimulation of MPS, research has focused on understanding the mechanism by which mTORC1 is activated. Sancak et al. (21), identified the interaction of mTORC1 with the lysosome to be of particular importance to the activation of the kinase complex in vitro. A similar mechanism has also been reported in rodent skeletal muscle, where eccentric contractions of the tibialis anterior muscle induce mTOR-lysosome colocalization (13) in parallel to increases in mTORC1 activity [inferred by the phosphorylation of ribosomal protein S6 kinase 1 threonine 389 (S6K1Thr389)]. Together these data infer an importance of mTOR-lysosome colocalization in the activation of molecular pathways implicated in protein synthesis. Recently, however, Korolchuk et al. (15) reported that the cellular localization of these mTOR/lysosomal complexes plays a pivotal role in mTOR activation. In support of this hypothesis, we recently reported that a single bout of resistance exercise initiated mTOR/lysosome translocation to the cell periphery, and occurred in parallel to an increase in mTOR activity and interaction between mTOR and proteins involved in translation initiation (23).
While the use of immunofluorescence approaches allowed us to study the cellular localization of mTOR, this approach did not enable us to distinguish between mTOR complexes. Consequently, we were unable to conclude whether the movement of mTOR following anabolic stimuli was mTORC1 or mTORC2 specific. Further, given the parallel group design we employed (23), we were unable to assess whether mTOR translocation was amplified by feeding. Therefore, the aim of the current study was to evaluate whether mTOR translocation following resistance exercise and/or protein-carbohydrate feeding is specific to mTORC1. In addition, we utilized a within-subject design to evaluate whether a synergistic effect of exercise and feeding exists. We hypothesized that exercise plus protein-carbohydrate feeding would elicit a greater mTOR/LAMP2 translocation to the cell periphery compared with feeding alone. Further, we hypothesized this translocation would be specific to mTORC1.
METHODS
Subjects.
Eight young, healthy, recreationally active males (age = 22.5 ± 3.1 yr, BMI = 24.6 ± 2.2 kg/m2, body fat = 17.6 ± 4.8%, mean ± SD) volunteered to partake in the study. Potential participants were informed about all experimental procedures to be undertaken and any risks involved before written informed consent was obtained. The study was approved by the Hamilton Integrated Research Ethics Board (REB 14-736) and adhered to the ethical standards outlined by the Canadian tri-council policy statement regarding the use of human participants in research as well as the principles according to the Declaration of Helsinki as revised in 2008.
Experimental design.
Following initial assessment for one-repetition maximum (1RM) on leg extension 7 days previously, participants reported to the laboratory at ~7.00 AM after a 10-h overnight fast. Participants then rested in a semisupine position on a bed, and an initial skeletal muscle biopsy was taken from the vastus lateralis using a modified bergstrom needle. Following this biopsy, participants performed four sets of unilateral leg extension (Atlantis, Laval, QC, Canada) at 70% 1RM until volitional failure interspersed by 2 min recovery. Immediately following the cessation of the final set of leg extension all participants consumed a commercially available beverage (Gatorade Recover, Chicago, IL) that provided 20, 44, and 1 g of protein, carbohydrate, and fat, respectively. Subsequent bilateral skeletal muscle biopsies were obtained from the vastus lateralis at 1 h and 3 h after beverage ingestion to examine mTORC1-related signaling and associated localization.
Skeletal muscle immunohistochemistry.
Skeletal muscle immunohistochemical preparation and staining were conducted as described previously (23). All samples from each subject were sectioned onto the same slide, in duplicate, to ensure accurate comparisons between time points could be made.
Antibodies.
The mouse monoclonal anti-mTOR (no. 05-1592) antibody was purchased from Merck Chemical (Nottingham, UK). The corresponding conjugated secondary antibody to this was goat anti-mouse IgGγ1 Alexa 594 (no. R37121, ThermoFisher). Antibodies targeting LAMP2 (no. AP1824d, Abgent), Rictor (CST no. 53A2, Cell Signaling Technology) and Raptor (no. ab40768, Abcam, Cambridge, UK) were visualized using goat anti-rabbit IgG(H+L) Alexa 488 secondary antibodies (no. A11008, ThermoFisher). Finally, wheat germ agglutinin (WGA-350, no. 11263, ThermoFisher) was used to identify the sarcolemmal membrane of muscle fibers.
Antibody validation.
The specificity of Rictor (CST no. 53A2) and Raptor (Abcam no. ab40768) primary antibodies were tested utilizing skeletal muscle samples from the gastrocnemius of muscle-specific knockout (mKO) mice for each protein respectively (3). Wild-type (WT), littermate muscle samples for each mouse model were used as controls. Primary antibodies were also omitted from a subset of samples on slides to examine any background staining from the secondary antibody utilized. The fluorescence intensity of each image was then calculated using ImageJ software (version 1.51 for Windows; National Institutes of Health, Bethesda, MD).
Image capture.
Prepared slides were imaged as described previously (23). DAPI UV (340–380 nm) filter was used to view WGA-350 (blue) signals, and mTOR proteins tagged with Alexa Fluor 594 fluorophores (red) were visualized under the Texas red (540–580 nm) excitation filter. The FITC (465–495 nm) excitation filter was used to capture signals of mTOR-complex proteins and LAMP2, which were conjugated with Alexa Fluor 488 fluorophores. On average, 8 images were captured per section, and each image contained ~8 muscle fibers such that around 120 fibers per time point (per subject) were used for analysis. Image processing and analysis was undertaken on ImagePro Plus 5.1 (Media Cybernetics), and all factors, i.e., exposure time and despeckling, were kept constant between all images on each individual slide. Image signals generated by WGA were used to estimate cell membrane borders, which were merged with the corresponding target protein images to identify the association between the protein of interest and the plasma membrane. Pearson’s correlation coefficient (Image-Pro software) was used to quantify colocalization with the plasma membrane and mTOR-associated proteins. This process was also completed to quantify the localization of mTOR with complex-associated proteins (Raptor and Rictor) and a marker of the lysosomal membrane (LAMP2).
Akt and S6K1 kinase activity assays.
At each time point during the experimental trial, a separate piece of muscle tissue was blotted and freed from any visible adipose or connective tissue. The tissue was then frozen in liquid nitrogen and stored at −80°C. The kinase activity of Akt and S6K1 was determined via [-γ-32P] ATP kinase assays following immunoprecipitation of the target protein, as previously described (18).
Statistical analysis.
All statistical analysis was conducted on SPSS (version 22 for Windows; SPSS, Chicago, IL). Differences in staining intensity between mKO, WT, and primary omitted (CON) muscle sections were analyzed using a one-way analysis of variance (ANOVA). Differences in kinase activity, fluorescence intensity, and staining colocalization were analyzed using a two-factor mixed-model ANOVA with two within subject factors (time; three levels: PRE vs. 1 h vs. 3 h and condition; two levels: FED vs. EXFED), with Bonferroni correction for multiple comparisons. Pairwise comparisons were conducted when a significant main/interaction effect was found. Significance for all variables analyzed was set at P ≤ 0.05. Data are presented as means ± SE unless otherwise stated.
RESULTS
Rictor and Raptor antibodies are specific to their target proteins.
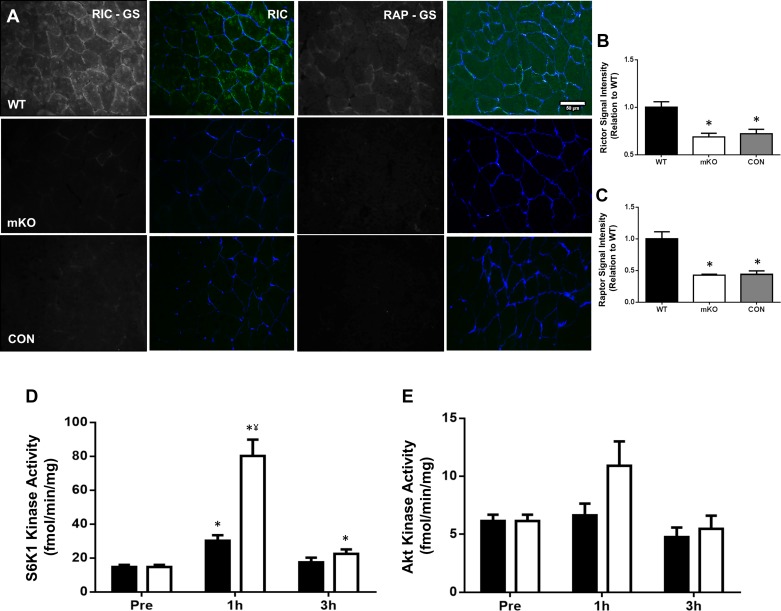
Rictor protein staining intensity in Rictor mKO tissue was significantly lower than that in littermate WT controls (P < 0.001, Fig. 1B). Furthermore, the staining intensity in this tissue was comparable to when the primary antibody was omitted in both mKO and WT tissue (P > 0.999, Fig. 1B). Raptor protein staining intensity in Raptor mKO tissue was also significantly lower than that noted in littermate WT controls (P < 0.001, Fig. 1C), with this staining intensity again similar to when the primary antibody was omitted in either tissue (P > 0.999, Fig. 1C). Therefore, we take this as evidence that the Rictor (CST no. 53A2) and Raptor (Abcam no. ab40768) antibodies are specific to their target protein.
Fig. 1.
Rictor and Raptor antibody validation and ribosomal protein S6 kinase 1 (S6K1) and Akt kinase activity. A–C: immunofluorescent staining of each protein was performed in muscle-specific knockout (mKO) and littermate wild-type (WT) samples, in addition to staining of each sample with primary antibodies omitted (CON). Rictor/Raptor is displayed in green and wheat germ agglutinin (WGA; cell membrane) is stained in blue. Representative images of staining in each condition are displayed (A) alongside the corresponding quantification for Rictor (B) and Raptor (C). Scale bars, 50 µm. Data presented as means ± SE. *Significantly different WT (P < 0.001). D and E: S6K1 (D) and Akt (E) kinase activity following unilateral resistance exercise and/or protein-carbohydrate feeding (FED). Black bars denote FED condition and open bars denote resistance exercise plus protein-carbohydrate feeding (EXFED) condition. Data presented as means ± SE. *Significantly different from baseline (P < 0.05), ¥significant difference between conditions at this time point (P < 0.001).
S6K1 and Akt kinase activity.
A significant condition by time effect was observed for S6K1 activity (P < 0.001). S6K1 activity rose above baseline in both conditions at 1 h post-exercise/feeding (FED-P = 0.015, EXFED-P < 0.001), and kinase activity at this time point was 165% greater in the EXFED condition (P < 0.001, Fig. 1D). At 3 h post-exercise/feeding, kinase activity only remained above baseline values in the EXFED condition (52.8% greater than baseline, P = 0.037, Fig. 1D). A significant main effect for time was noted for Akt kinase activity (P = 0.023, Fig. 1E). Pairwise comparisons displayed a trend toward an increase in Akt kinase activity 1 h post-intervention, when conditions were combined, compared with 3 h post-intervention (P = 0.073).
Lysosomal content and colocalization with mTOR.
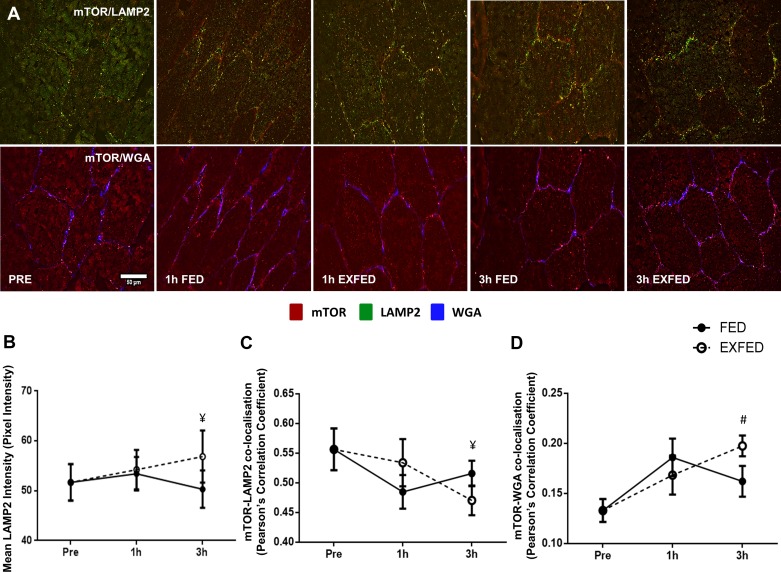
LAMP2 fluorescence intensity was unchanged from baseline in either condition; however, a significantly greater intensity was noted in the EXFED condition, compared with FED, at 3 h post-exercise/feeding (P = 0.41, Fig. 2B). A significant condition × time effect was observed for mTOR-LAMP2 colocalization (P = 0.004). Consistent with our previous work (23), mTOR and LAMP2 were highly localized in basal skeletal muscle (Fig. 2C). The colocalization of these two proteins did not change from baseline in either condition over the 3 h post-exercise/feeding period. However, at the 3 h time point, the colocalization of the proteins was greater in the FED condition compared with the EXFED condition [0.51(FED) vs. 0.47 (EXFED), P = 0.011, Fig. 2C].
Fig. 2.
The effect of resistance exercise and/or protein carbohydrate feeding on mechanistic target of rapamycin (mTOR)-lysosomal associated membrane protein 2 (LAMP2) and mTOR-WGA colocalization. A: representative images of mTOR-LAMP2 and mTOR-WGA colocalization at rest, and following resistance exercise and/or protein-carbohydrate feeding. Orange/yellow regions denote areas of mTOR localization with the marker of the lysosome in images on the top row. mTOR-positive staining is shown in red, LAMP2-positive in green, and WGA-positive in blue. B–D: quantification of LAMP2 fluorescence intensity (B), mTOR-LAMP2 colocalization (C), and mTOR-WGA colocalization (D) at each time point. Scale bars, 50 µm. Data presented as means ± SE. ¥Significant difference between conditions at this time point (P < 0.05), #significantly different compared with baseline when conditions combined (P = 0.008).
mTOR/lysosome translocation to the cell membrane.
Significant main effects of condition (P = 0.05) and time (P = 0.025) were observed for mTOR colocalization with the cell membrane (WGA-positive staining). The significant main effect of condition suggests that, when all time points are combined, mTOR-WGA was greater in the EXFED condition compared with the FED condition. Subsequent pairwise comparisons also display that when both conditions were combined, mTOR colocalization with the cell membrane was greater at 3 h post-exercise/feeding compared with baseline values (P = 0.008, Fig. 2C). Further comparisons also displayed a trend toward a difference between mTOR-WGA colocalization between conditions at the 3 h time point [0.16(FED) vs. 0.19(EXFED), P = 0.085]. This pattern of colocalization was mirrored when analyzing LAMP2-WGA colocalization (main effect of time, P = 0.031, data not shown.), reiterating the constant colocalization of mTOR and the lysosome.
Rictor colocalization with mTOR and WGA.
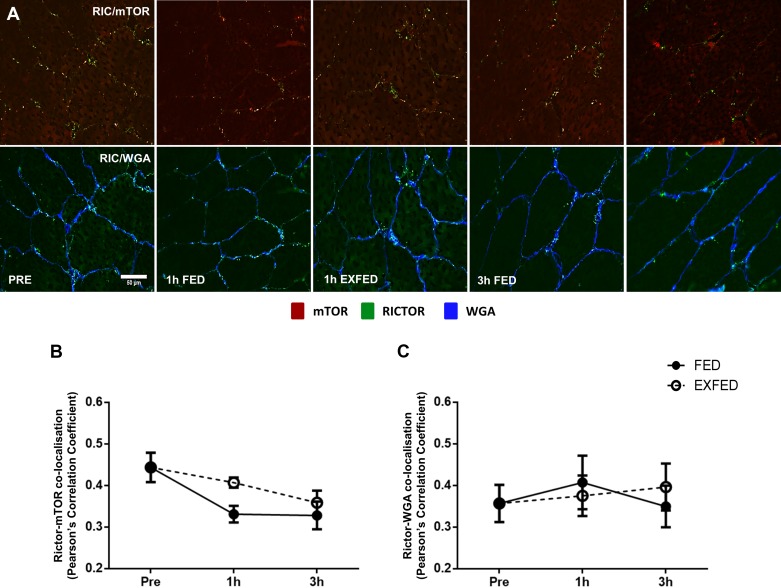
Significant main effects of group (P = 0.046) and time (P = 0.035) were noted for Rictor colocalization with mTOR proteins (Fig. 3B). Overall, there was a greater colocalization of these two proteins in the EXFED condition compared with the FED condition. Following pairwise comparisons, there was no difference in the colocalization between Rictor and mTOR between any time points (P > 0.05, Fig. 3B). Furthermore, Rictor colocalization with WGA did not change from baseline at any time point in either condition (Fig. 3C), suggesting that post-exercise translocation is specific to mTORC1.
Fig. 3.
The effect of resistance exercise and/or protein carbohydrate feeding on Rictor-mTOR and Rictor-WGA colocalization. A: representative images of Rictor-mTOR and Rictor-WGA colocalization at rest, and following resistance exercise and/or protein-carbohydrate feeding. Orange/yellow regions denote areas of Rictor localization with mTOR on top row. mTOR-positive staining is shown in red, Rictor-positive in green, and WGA-positive in blue. B and C: quantification of Rictor-mTOR (B) and Rictor-WGA (C) colocalization at each time point. Scale bars, 50 µm. Data presented as means ± SE.
Raptor colocalization with mTOR and WGA.
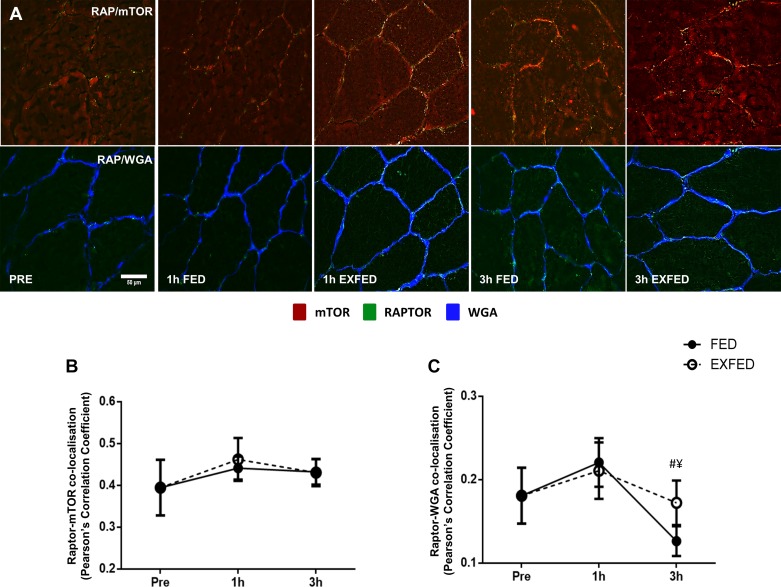
The colocalization of Raptor and mTOR proteins did not change in either group, at any time point, suggesting that any alterations in subcellular location of either protein occurred concurrently (Fig. 4B). A significant condition × time effect was observed for Raptor colocalization with WGA (P = 0.029). Here, Raptor colocalization with WGA rose to a similar extent to the previously reported increase in mTOR-WGA colocalization at 1 h post-exercise/feeding in both conditions. At the 3 h time point, Raptor-WGA colocalization in the FED group dropped below baseline and 1 h post-exercise/feeding levels (P = 0.007, Fig. 4C), and colocalization at this time point was greater in the EXFED condition [0.12(FED) vs. 0.17(EXFED), P = 0.014, Fig. 4C].
Fig. 4.
The effect of resistance exercise and/or protein carbohydrate feeding on Raptor-mTOR and Raptor-WGA colocalization. A: representative images of Raptor-mTOR and Raptor-WGA colocalization at rest, and following resistance exercise and/or protein-carbohydrate feeding. Orange/yellow regions denote areas of Raptor localization with mTOR. mTOR-positive staining is shown in red, Raptor-positive in green, and WGA-positive in blue B and C: quantification of Raptor-mTOR (B) and Raptor-WGA (C) colocalization at each time point. Scale bar, 50 µm. Data presented as means ± SE. ¥Significant difference between conditions at this time point (P = 014), #significantly different compared with baseline when conditions combined (P = 0.007).
DISCUSSION
Utilizing a within-subject design, we report that a combination of unilateral resistance exercise and protein-carbohydrate feeding elicits a greater mTOR translocation toward the cell membrane than feeding alone. This observation is consistent with previous findings from our laboratory in which we reported that mTOR associates with the lysosome in basal skeletal muscle, with mTOR/lysosomal complexes translocating to the cell periphery following mTOR activation (23). Utilizing immunofluorescent approaches to distinguish between mTORC1 and mTORC2, the present study extends this observation, suggesting that mTORC1 seems to be the predominant mTOR complex translocating in human skeletal muscle following anabolic stimuli, with mTORC2 in constant association with the cell membrane.
In addition to mTORC1 translocation to the cell periphery, we report a greater colocalization of mTOR and LAMP2 in the FED condition, compared with the EXFED condition, at the 3 h time point. This finding was unexpected and contrasted our previous research using a parallel group design (23). The greater association of mTORC1 with lysosomes in the FED condition would infer greater mTORC1 activity in this leg (13, 21); however, this was not apparent in our S6K1 kinase activity data. A possible explanation for this difference is the increased lysosomal content (LAMP2 fluorescence intensity) noted in the EXFED condition at this time point (Fig. 2B). It is possible that the acute resistance exercise bout may have elicited an increase in chaperone assisted selective autophagy as a stress response to the strenuous exercise, as previously reported (24). This may have increased the free-lysosomal pool (24) and altered the ratio of mTOR-LAMP2 association. As this is only a proxy measure of lysosomal content, further research directed towards lysosomal biogenesis in response to physiological stimuli would be needed to address this mechanism.
Previous research from our laboratory has shown an elevation in mTOR association with the cell membrane in response to resistance exercise, in both the fed and fasted state (23). This association coincided with an increase in S6K1 kinase activity, suggesting that mTOR trafficking is associated with an increase in intrinsic mTOR activity. Consistent with this hypothesis, here we report that mTORC1-cell membrane association increased 1 h post-intervention, in both FED and EXFED conditions, and the increment was similar to that noted in our previous work (23). However, in contrast to our previous results, mTOR-WGA colocalization in the FED condition returned close to baseline values at 3 h and colocalization in the EXFED condition displayed a continued elevation. In addition to the main effects of time (P = 0.025) and condition (P = 0.05) apparent here, a trend toward greater colocalization in the EXFED condition (P = 0.085) was noted at the 3 h time point. This greater colocalization is suggestive of retention of mTOR at the cell periphery when resistance exercise is followed with protein/carbohydrate ingestion, inferring a synergistic effect of resistance exercise and protein-carbohydrate feeding, an observation previously reported for MPS (5).
The mTORC1 and mTORC2 protein complexes are involved in varying metabolic signaling processes in skeletal muscle, and as such are suggested to reside in distinct cellular locations (4). As mTOR-lysosome translocation has been previously associated with mTORC1 activation in response to amino acids in vitro (15), we sought to determine whether mTORC1 is the principal mTOR complex translocating in human skeletal muscle as we have previously reported (23). The colocalization of Raptor with WGA increased at 1 h post-intervention in both conditions, and to a similar extent to that noted in mTOR-WGA colocalization, suggesting that mTORC1 is a spatially regulated mTOR complex in human skeletal muscle. Further to this notion, a disparity between conditions became apparent at the 3 h time point, with Raptor-WGA colocalization enhanced in the EXFED condition (P = 0.014). This is in agreement with the data regarding mTOR-WGA colocalization where a trend toward EXFED eliciting greater membrane colocalization compared with FED is reported. Raptor colocalization with mTOR itself was not altered at any time point, or between conditions; however, we did observe a reduction in raptor association with WGA at 3 h in both FED and EXFED. We are currently unable to explain this result; however, it could be due to an increase in free Raptor content (14) or increased Raptor degradation (12), both potential mechanisms proposed to regulate mTORC1 activity. In contrast, costaining of Rictor with WGA suggested that mTORC2 localizes with the cell membrane in basal tissue, with this colocalization unaffected by resistance exercise or protein/carbohydrate ingestion. This finding has also been replicated using in vitro models, where a large proportion of mTORC2 activity was noted at the plasma membrane of HEK293 cells (10).
Our data are congruent with both in vitro (15) and in vivo (23) studies suggesting that mTORC1 cellular colocalization is linked to mTORC1 activity. While this observation is in contrast to previous in vitro studies (21, 25), where mTOR translocation to the lysosome is deemed essential, we believe the increase in autophagy/MPB in post-absorptive skeletal muscle prevents the disassociation of mTOR and the lysosome noted in previous in vitro studies, where a complete amino acid withdrawal protocol is utilized. Further, many physiological mechanisms occur at the cell periphery suggesting that the redistribution of mTORC1/lysosomal complexes to the cell periphery is physiologically relevant. mTORC1 is known to stimulate MPS, which, through the use of the SUnSET technique (22) and immunohistochemical staining methods, is purported to occur primarily in peripheral regions of muscle fibers (11). Consistent with this, we previously identified mTOR to interact with Rheb, eukaryotic translation initiation factor 3 subunit F (eIF3F), and the microvasculature at the cell periphery following resistance exercise in the fed state (23). Collectively, these data therefore suggest that both upstream regulators and downstream substrates of mTORC1 are membrane-associated in skeletal muscle (11). Further, given we observed that mTORC1 association with the cell periphery was prolonged with feeding (a scenario of heightened MPS), we propose that maintaining mTORC1 at the cell periphery may provide a mechanistic explanation as to why exercise in the fed state results in prolonged increases in MPS in human skeletal muscle compared with exercise or feeding in isolation (6).
In summary, our data show that mTOR-lysosome translocation in response to resistance exercise and feeding is driven primarily by mTORC1, and occurs in parallel to increases in S6K1 activity. Further, we report that resistance exercise combined with protein-carbohydrate feeding sustains this response, compared with feeding alone, suggesting a synergistic effect of these two stimuli. Collectively, these data add further support to the importance of spatial regulation of mTORC1 in response to anabolic stimulation. Further research should now examine the relevance of mTORC1 colocalization in clinical scenarios, i.e., aging (7) or obesity (2). Finally, the tools described herein to study mTORC2 localization could be used to examine the regulation of skeletal muscle glucose uptake and insulin sensitivity, factors thought to be under the direct control of mTORC2 (16).
GRANTS
This study was funded in part by Biotechnology and Biological Sciences Research Council (BBSRC) New Investigator Award BB/L023547/1 to A. Philp and a University of Birmingham “Exercise as Medicine” doctoral training studentship to N. Hodson. Work in the laboratory of M. A. Rüegg was supported by the Swiss National Science Foundation and the University of Basel. Work in the laboratory of D. L. Hamilton was funded by a Society for Endocrinology Early Career Grant. Work in the laboratory of S. M. Phillips was funded by the National Science and Engineering Research Council of Canada and the Canada Research Chairs Program.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
N.H., C.M., S.Y.O., S.J., Z.S., M.A.R., and D.L.H. performed experiments; N.H., S.J., Z.S., and D.L.H. analyzed data; N.H. and A.P. interpreted results of experiments; N.H. prepared figures; N.H. and A.P. drafted manuscript; N.H., C.M., S.Y.O., S.J., M.A.R., D.L.H., S.M.P., and A.P. edited and revised manuscript; N.H., C.M., S.Y.O., S.J., Z.S., M.A.R., D.L.H., S.M.P., and A.P. approved final version of manuscript; C.M., S.M.P., and A.P. conceived and designed research.
REFERENCES
- 1.Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol Cell Physiol 276: C120–C127, 1999. [DOI] [PubMed] [Google Scholar]
- 2.Beals JW, Sukiennik RA, Nallabelli J, Emmons RS, van Vliet S, Young JR, Ulanov AV, Li Z, Paluska SA, De Lisio M, Burd NA. Anabolic sensitivity of postprandial muscle protein synthesis to the ingestion of a protein-dense food is reduced in overweight and obese young adults. Am J Clin Nutr 104: 1014–1022, 2016. doi: 10.3945/ajcn.116.130385. [DOI] [PubMed] [Google Scholar]
- 3.Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Rüegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab 8: 411–424, 2008. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol 203: 563–574, 2013. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biolo G, Tipton KD, Klein S, Wolfe RR. An abundant supply of amino acids enhances the metabolic effect of exercise on muscle protein. Am J Physiol Endocrinol Metab 273: E122–E129, 1997. [DOI] [PubMed] [Google Scholar]
- 6.Burd NA, West DW, Moore DR, Atherton PJ, Staples AW, Prior T, Tang JE, Rennie MJ, Baker SK, Phillips SM. Enhanced amino acid sensitivity of myofibrillar protein synthesis persists for up to 24 h after resistance exercise in young men. J Nutr 141: 568–573, 2011. doi: 10.3945/jn.110.135038. [DOI] [PubMed] [Google Scholar]
- 7.Cuthbertson D, Smith K, Babraj J, Leese G, Waddell T, Atherton P, Wackerhage H, Taylor PM, Rennie MJ. Anabolic signaling deficits underlie amino acid resistance of wasting, aging muscle. FASEB J 19: 422–424, 2005. doi: 10.1096/fj.04-2640fje. [DOI] [PubMed] [Google Scholar]
- 8.Dickinson JM, Fry CS, Drummond MJ, Gundermann DM, Walker DK, Glynn EL, Timmerman KL, Dhanani S, Volpi E, Rasmussen BB. Mammalian target of rapamycin complex 1 activation is required for the stimulation of human skeletal muscle protein synthesis by essential amino acids. J Nutr 141: 856–862, 2011. doi: 10.3945/jn.111.139485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol 587: 1535–1546, 2009. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebner M, Sinkovics B, Szczygieł M, Ribeiro DW, Yudushkin I. Localization of mTORC2 activity inside cells. J Cell Biol 216: 343–353, 2017. doi: 10.1083/jcb.201610060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25: 1028–1039, 2011. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han K, Xu X, Xu Z, Chen G, Zeng Y, Zhang Z, Cao B, Kong Y, Tang X, Mao X. SC06, a novel small molecule compound, displays preclinical activity against multiple myeloma by disrupting the mTOR signaling pathway. Sci Rep 5: 12809, 2015. doi: 10.1038/srep12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs BL, You JS, Frey JW, Goodman CA, Gundermann DM, Hornberger TA. Eccentric contractions increase the phosphorylation of tuberous sclerosis complex-2 (TSC2) and alter the targeting of TSC2 and the mechanistic target of rapamycin to the lysosome. J Physiol 591: 4611–4620, 2013. doi: 10.1113/jphysiol.2013.256339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Pajvani UB. “Free” Raptor – a novel regulator of metabolism. Cell Cycle 15: 1174–1175, 2016. doi: 10.1080/15384101.2016.1159835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O’Kane CJ, Deretic V, Rubinsztein DC. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13: 453–460, 2011. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, Guertin DA, Sabatini DM, Baur JA. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335: 1638–1643, 2012. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcotte GR, West DW, Baar K. The molecular basis for load-induced skeletal muscle hypertrophy. Calcif Tissue Int 96: 196–210, 2015. doi: 10.1007/s00223-014-9925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGlory C, White A, Treins C, Drust B, Close GL, Maclaren DP, Campbell IT, Philp A, Schenk S, Morton JP, Hamilton DL. Application of the [γ-32P] ATP kinase assay to study anabolic signaling in human skeletal muscle. J Appl Physiol (1985) 116: 504–513, 2014. doi: 10.1152/japplphysiol.01072.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer AJ, Lorin S, Blommaart EF, Codogno P. Regulation of autophagy by amino acids and MTOR-dependent signal transduction. Amino Acids 47: 2037–2063, 2015. doi: 10.1007/s00726-014-1765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273: E99–E107, 1997. [DOI] [PubMed] [Google Scholar]
- 21.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303, 2010. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277, 2009. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 23.Song Z, Moore DR, Hodson N, Ward C, Dent JR, O’Leary MF, Shaw AM, Hamilton DL, Sarkar S, Gangloff Y-G, Hornberger TA, Spriet LL, Heigenhauser GJ, Philp A. Resistance exercise initiates mechanistic target of rapamycin (mTOR) translocation and protein complex co-localisation in human skeletal muscle. Sci Rep 7: 5028, 2017. doi: 10.1038/s41598-017-05483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Höhfeld J. Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy 11: 538–546, 2015. doi: 10.1080/15548627.2015.1017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334: 678–683, 2011. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]