Abstract
DNAzymes have been shown as a promising platform for metal ions detection and a few DNAzyme-based sensors have been reported to detect metal ions inside cells. However, these methods required an influx of metal ions to increase their concentrations for detection. To address this major issue, we herein report the design of a catalytic hairpin assembly (CHA) reaction to amplify the signal from photocaged Na+-specific DNAzyme to detect endogenous Na+ inside cells. Upon light activation and in the presence of Na+, NaA43 DNAzymes cleave the substrate strands and release initiator DNA that trigger the followed CHA amplification reaction. This strategy has allowed detection of endogenous Na+ inside cells, which has been demonstrated by both fluorescent imaging of individual cells and flow cytometry of the whole cell population. This method can be generally applied to detect other endogenous metal ions and thus contribute to deeper understanding of the role of metal ions in biological systems.
Keywords: biosensors, DNAzymes, photo-activation, nucleic acid amplification
Graphical Abstract
DNAzyme-Catalytic Hairpin Assembly (DzCHA) Probe: we designed a catalytic hairpin assembly (CHA) reaction to amplify the signal from photocaged Na+-specific DNAzyme cleavage to detect endogenous Na+ inside cells. Upon light activation and in the presence of Na+, NaA43 DNAzymes cleave the substrate strands and release initiator DNA that can trigger the CHA signal amplification reaction.
Metal ions play critical roles in numerous biological processes.[1] Metal ions both in excess and deficient can be detrimental to their normal functions. To gain a better understanding of the functions of these metal ions in biology, it is important to detect and monitor their concentrations inside cells. Towards this goal, a number of fluorescent probes, such as small molecule-based probes[2] and protein-based probes,[3] have been reported to detect metal ions inside cells. Despite the progress made, most current sensors can detect only a limited number of metal ions, such as Ca2+, Mg2+, and Zn2+. And methods that can be readily applied to detect a wide number of other metal ions remain a challenge.
In order to meet this challenge, we and others have taken advantages of DNAzymes, which are DNA molecules that can catalyze chemical reactions in the presence of metal ions.[4] DNAzymes with high selectivity for almost any metal ion can be obtained from a large DNA library containing up to 1015 different sequences.[4–5] In the past decades, a number of DNAzymes have been selected and converted into sensors for metal ions, including Zn2+,[5,6] Pb2+,[7] UO2+,[8] Cu2+,[9] Hg2+,[10] and other metal ions.[11] Recently, a few DNAzyme-based sensors have been applied to image metal ions in living cells.[12] However, a significant issue is that these reported methods need an additional influx of metal ions to increase their intracellular concentration to detect a response. To our knowledge, no current DNAzyme-based sensing method is capable of detecting endogenous intracellular metal ions, which is necessary to understand the roles of metal ions in biological systems.
To address the above issue, we herein report a novel method of amplifying the signal from DNAzyme-catalyzed cleavage using catalytic hairpin assembly (called DzCHA hereafter) to detect the low level of endogenous metal ions inside cells. CHA which was first reported by Pierce et al in 2008,[13] is an enzyme-free signal amplification method. Unlike other signal amplification methods using enzymes, such as rolling circle amplification (RCA),[14] and strand displacement amplification (SDA),[15] enzyme-free amplification methods achieve the amplification simply through DNA hybridization and strand displacement, making them ideally suitable for nucleic acids amplification in living cells.[16] To demonstrate the use of this DzCHA probe for detection of endogenous metal ions inside cells, we chose the Na+-specific DNAzyme (NaA43) as a proof of concept. Sodium ions, one of the most abundant metal ions inside cells, are involved in many biological functions, including heart contraction, and signal transduction pathways.[17] Though the concentration of free Na+ in cells is range from 9~19 mM,[17a] the current NaA43 DNAzyme sensor cannot detect the endogenous Na+ inside cells.[12c] Therefore, it is necessary to develop a method which can realize the detection of endogenous Na+ in living cells.
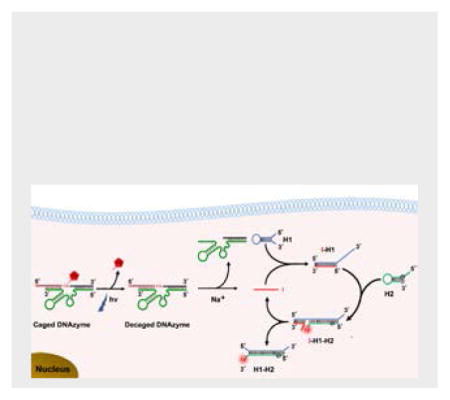
As shown in Scheme 1A, the DzCHA probe consists of a Na+-specific NaA43 DNAzyme and CHA amplification triggered by the cleaved product of the DNAzyme. In the presence of Na+, the DNAzyme cleaves the substrate strand at the rA position, and releases the cleaved fragment (in red) from the substrate strand. The red fragment becomes the initiator DNA (I) for the CHA amplification using two hairpin DNA molecules (H1 and H2). In the absence of the free initiator DNA, the H1 and H2 remain intact because the cross-reactivity is blocked by their intramolecular hybridization to form loops. Once the initiator DNA (I) is released, it will bind to H1 through toehold-assisted hybridization to open H1. The generated single stranded portion on H1 then serves as a second toehold to initiate strand displacement reaction to open H2, generating the fluorescence signal turn-on. As a result of forming a stronger duplex between H1 and H2, I fragment is then released from H1 and catalyze the next round of CHA reaction, thus one I fragment is able to generate multiple signal outputs, realizing the goal of signal amplification for endogenous Na+ detection.
Scheme 1.
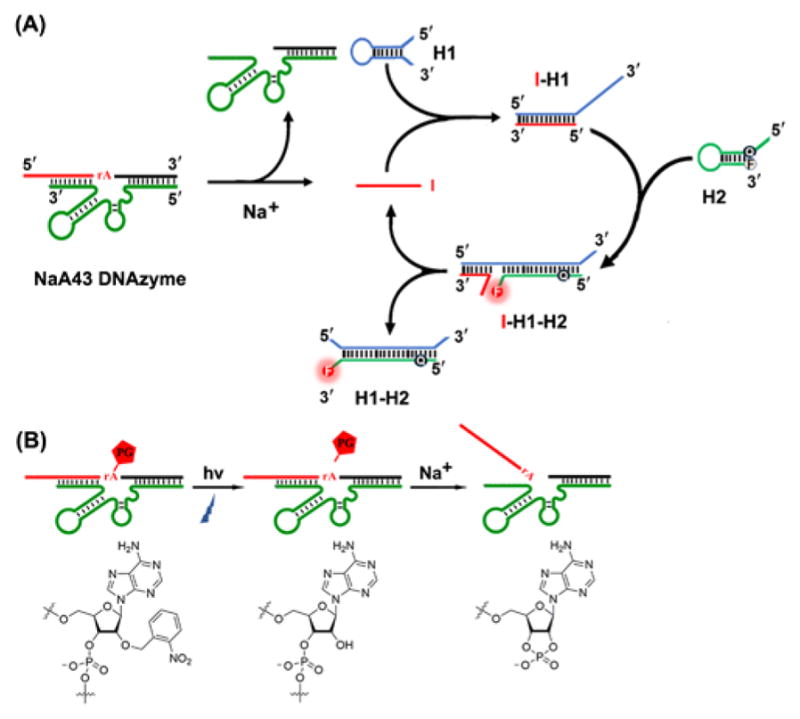
Design of a DNAzyme-Catalytic Hairpin Assembly (DzCHA) Probe for Endogenous Sodium Ions Detection.
The feasibility of DzCHA probe was first verified in buffer. As shown in Figure 1A, in the absence of Na+, the DzCHA probe displayed low background fluorescence signal (pink line). In the presence of 100 mM Na+, the fluorescence intensity increased by ~4-fold (green line), indicating the Na+-specific cleavage and followed CHA reaction. Probes with a non-cleavable NaA43 DNAzyme (blue line) or control H1 (black line), used as negative controls, showed low signals. A study of this DzCHA probe using 12% native polyacrylamide gel electrophoresis (PAGE) further confirmed the above results. (Figure S1).
Figure 1.
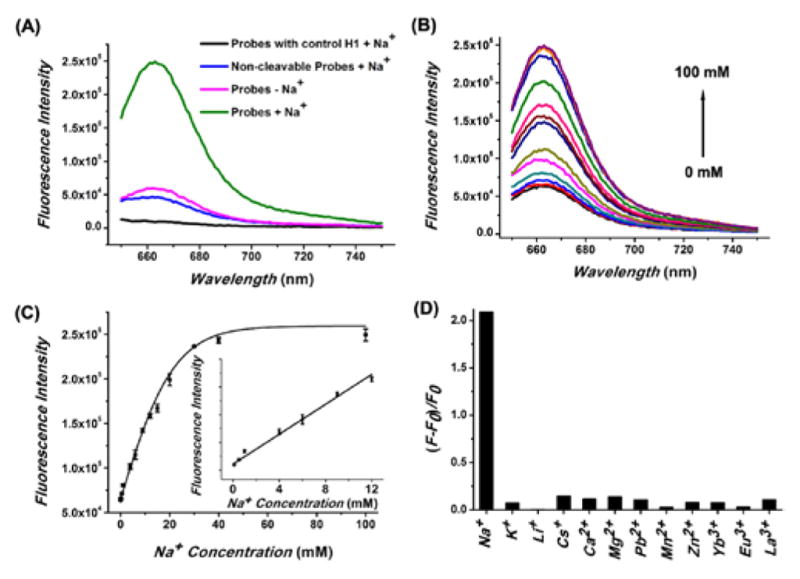
Feasibility, sensitivity and selectivity of the NaA43 DzCHA probe. (A) Fluorescence spectra of the DzCHA probe with control H1 (black), non-cleavable NaA43 DzCHA probe (blue), DzCHA probe in buffer without Na+ (pink), and DzCHA probe in buffer with 100 mM Na+ (green). (B) Fluorescence signal increase of the sensor over different concentration of Na+, ranging from 0, 0.1, 0.5, 1, 4, 6, 9, 12, 15, 20, 30, 40, 100 mM. (C) Plot of fluorescence intensity versus Na+ concentration. The inset shows a magnification of the linear region. Error bars represent the SD from three independent experiments. (D) Response of the probe to different competing metal ions. The mono-, di-, and trivalent metal ion is 30 mM, 1 mM, and 0.1 mM, respectively.
To determine if the DzCHA probe could detect Na+ in buffer quantitatively, we measured its fluorescence spectra in different concentrations of Na+. As shown in Figure 1B, the fluorescent signals increased with increasing Na+ from 0.1 mM to 100 mM. The fluorescent signal intensity at 661 nm increased linearly with the target Na+ concentration ranging from 0.1 mM to 12 mM (Figure 1C). From the data, a limit of detection (LOD) of 14 μM was obtained (3σ/slope rule). This high sensitivity suggested that this probe has the potential to detect endogenous Na+ inside cells. The selectivity of this DzCHA was next investigated. As shown in Figure 1D, the fluorescence signal in presence of Na+ was at least ~15 times higher than those in the presence of other metal ions, demonstrating excellent selectivity for Na+ against other metal ions. Furthermore, the DzCHA probe showed an excellent specificity to the released initiator DNA strand with a capability of differentiating single nucleotide polymorphisms (Figure S2). A further investigation of real-time fluorescence responses revealed that initial rates of fluorescence increase were also dynamically correlated to Na+ concentrations (Figure S3). Because the initial rate of fluorescence increase could be obtained within 5 min, these rates might also be used as a measure for quantifying Na+ concentrations or following the concentration changes.
In order to prevent cleavage of the NaA43 DNAzyme during the cellular delivery process and achieve controlled activation of the DzCHA probe, the substrate strand of the NaA43 DNAzyme was photocaged,[12b,c] in which the adenosine ribonucleotide (rA) at the scissile site was replaced by a photocaged group (PG), 2′-O-nitrobenzyl adenosine (Figure S4–S5). Upon light irradiation at 365 nm, the PG was removed from the substrate strand, restoring the 2′ hydroxide of the ribonucleotide and thus allowing the substrate to be cleaved by the DNAzyme (Scheme 1B). The effectiveness of this photo-caging strategy was demonstrated by fluorescence spectra (Figure S6).
To test the performance of this probe in a complex matrix, the probe was incubated with cell lysate at 37 °C for 3 h. As shown in Figure S7, the decaged NaA43 DzCHA probe showed a ~3-fold fluorescence signal increase over the caged NaA43 DzCHA. In contrast, both the non-cleavable and inactive NaA43 DzCHA showed minimal fluorescence signal increase, suggesting not only high selectivity of the DzCHA probe, but also good stability for applications inside the cells.
Having demonstrated the ability of the DzCHA probe in buffer, we next investigated its application to detect endogenous Na+ ions in living cells. A cationic polypeptide named G8 that belongs to the α-helical cationic polypeptide family,[12c,18] with an ability to escape from endosomes or lysosomes, was chosen as the transfection agent for the DNA delivery in this work. To confirm localization of the probe inside cells, only Cy5 labeled probe was used and delivered using the G8 polypeptides. As shown in Figure 2, the fluorescence of Cy5 highly overlapped with ER Tracker and did not co-localized well with Lyso Tracker Red, suggesting that the DzCHA probe was mainly located in the cytosol. A Z-stack imaging analysis was further performed to confirm that the DzCHA probe is located inside the cells (Figure S8).
Figure 2.
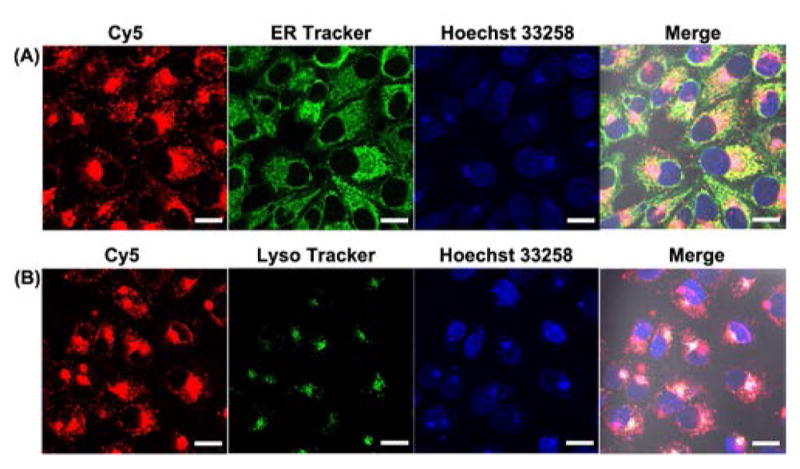
Intracellular co-localization of DzCHA probe with ER Tracker (A) and Lyso Tracker (B). Scale Bar: 20μmm.
Given the fact that G8 polypeptides serve as an efficient DNA transfection agent, we used it to transfect the caged NaA43 DzCHA probe into HeLa cells for endogenous Na+ detection. After decaging, the fluorescence insides cells increased after a 3 h reaction (Figure 3A). Under the same condition and during the same period, the non-cleavable DzCHA probe showed no obvious fluorescence increase inside the cells (Figure 3B). To ensure that the fluorescence increase was solely due to the Na+ specific response, an inactive caged NaA43 DzCHA probe was used as a negative control. As shown in Figure S9, after 3 h reaction, no obvious fluorescence signal increase was observed, confirming that the fluorescence signal increase observed from the caged active DzCHA probe was due to the Na+ specific DNAzyme cleavage and followed by CHA amplification reaction.
Figure 3.
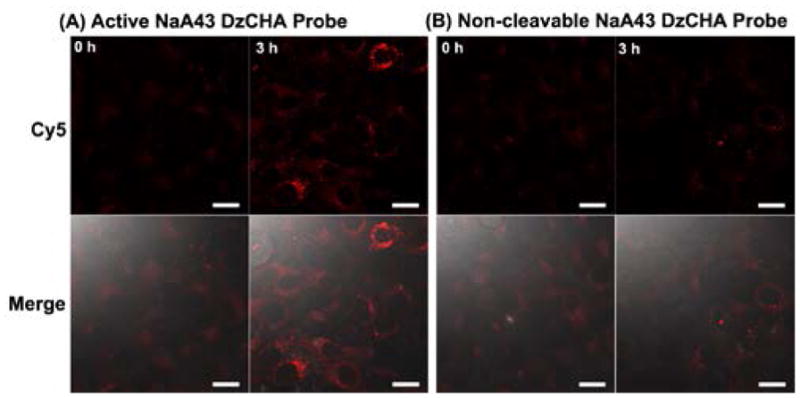
Intracellular imaging of HeLa cells transfected with active caged DzCHA probe (A) and non-cleavable DzCHA probe (B). Scale Bar: 20μmm.
To ensure the results from confocal microscopy imaging apply to the entire cell population, we examined the fluorescence of the DzCHA probe in a large cell population using analytical flow cytometry. As shown in Figure S10, the active caged NaA43 DzCHA probe exhibited a fluorescence signal peak shift, while the inactive caged NaA43 DzCHA or non-cleavable NaA43 DzCHA probes showed no obvious peak shift. We also calculated the mean fluorescence intensities of these three DzCHA probes. The caged active NaA43 DzCHA probe displayed a fluorescence signal increase over the caged inactive DzCHA (3.5-fold) and the non-cleavable DzCHA probe (6.3-fold). Therefore, the flow cytometry results were in strong agreement with the confocal imaging results, suggesting the successful detection of endogenous Na+ by the NaA43 DzCHA probe.
To further verify that this probe could be used to monitor Na+ concentration change inside cells, we used gramicidin D, monensin, and ouabain to increase the sodium concentration inside cells. The use of these molecules in combination is known to equilibrate the intracellular and extracellular Na+ concentration in a short time.[19] Compared to HeLa cells without influx of Na+, cells with influx of Na+ displayed an obvious increase of fluorescence intensity with increasing Na+ concentrations (Figure 4A–B and Figure S11C). Cells with influx of Na+ showed ~1.9-fold average fluorescence intensity increase (Figure 4C). The results were further confirmed using flow cytometry assay on a large population of cells (Figure 4D and Figure S11A), and the peak intensities of the flow cytometry profiles were correlated to the concentrations of Na+ inside a series of cell standards (Figure S11B). Together, these results demonstrated that this DzCHA probe can be used as an effective strategy for not only quantifying endogenous Na+ concentrations, but also monitoring Na+ concentration changes in living cells.
Figure 4.
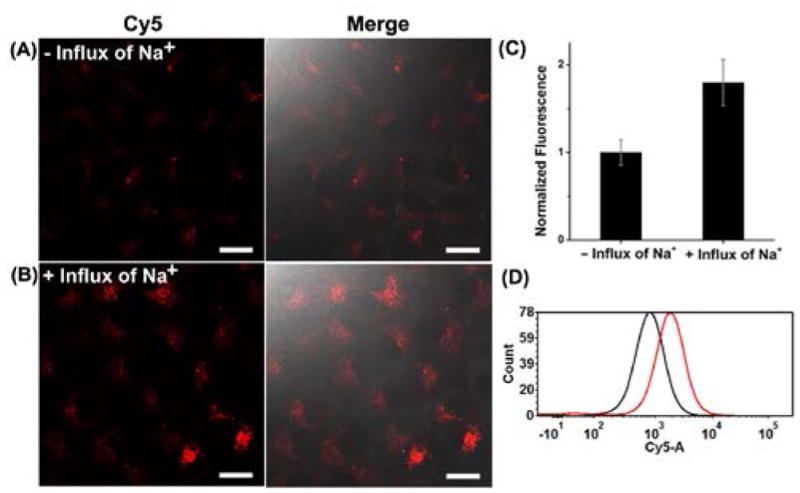
Confocal images of HeLa cells transfected with active caged NaA43 DzCHA probe without (A) or with (B) influx of 50 mM Na+. (C) The mean fluorescence intensity was measured using 10 confocal images for each condition. (D) Flow cytometry assay of the cells without or with influx of 50 mM Na+. 10,000 cells were counted for each sample. Black line represents the sample without influx of Na+, red line represents the sample with influx of Na+.
In conclusion, we have developed and demonstrated a DzCHA probe to detect endogenous metal ions in living cells by combining the Na+-specific DNAzyme with CHA reaction. To our knowledge, this work is the first design of a signal-amplified DNAzyme sensor for living cell imaging and the first application of DNAzyme sensor for imaging endogenous intracellular metal ions. Since DNAzymes selective for other metal ions have been reported and these DNAzymes often share a similar secondary structure and reaction mechanism,[4,11] the method demonstrated here potentially can be applied as a general strategy to improve the sensitivity of any DNAzyme-based sensor for detection of metal ions at low concentrations. As a result, the application of DNAzymes can be expanded for imaging many metal ions in biology, which will provide deeper understanding of the role of metal ions in biological systems.
Supplementary Material
Acknowledgments
This material is based on work supported by the US National Institutes of Health (NIH) under Award R21MH110975 (to Y.L.) and NSFC (Grants 21527810, 21521063 to J-H. Jiang). Z. Wu acknowledges support from China Scholarship Council. The authors thank Dr. Jingjing Zhang and Claire E. McGhee for experiment discussion. The authors thank Dr. Jianjun Cheng for kindly providing the G8 polypeptides.
Footnotes
Conflict of interest
The authors declare no conflict of interests.
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Zhenkun Wu, State Key Laboratory of Chemeo/Bio-Sensing and Chemometrics, Institute of Chemical Biology and Nanomedicine and College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
Huanhuan Fan, State Key Laboratory of Chemeo/Bio-Sensing and Chemometrics, Institute of Chemical Biology and Nanomedicine and College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
Nitya Sai Reddy Satyavolu, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
WenJing Wang, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States, State Key Laboratory of Analytical Chemistry for Life Science and School of Chemistry & Chemical Engineering, Nanjing University, Nanjing, 210093, P. R. China.
Ryan Lake, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
Prof. Jian-Hui Jiang, State Key Laboratory of Chemeo/Bio-Sensing and Chemometrics, Institute of Chemical Biology and Nanomedicine and College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China
Prof. Yi Lu, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States
References
- 1.a) Holm RH, Kennepohl P, Solomon EI. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]; b) Outten CE, O’Halloran TV. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]; c) Guengerich FP. J Biol Chem. 2009;284:709. doi: 10.1074/jbc.R800060200. [DOI] [PubMed] [Google Scholar]
- 2.a) Williams DA, Fogarty KE, Tsien RY, Fay FS. Nature. 1985;318:558–561. doi: 10.1038/318558a0. [DOI] [PubMed] [Google Scholar]; b) Komatsu H, Iwasawa N, Citterio D, Suzuki Y, Kubota T, Tokuno K, Kitamura Y, Oka K, Suzuki K. J Am Chem Soc. 2004;126:16353–16360. doi: 10.1021/ja049624l. [DOI] [PubMed] [Google Scholar]; c) Taki M, Wolford JL, O’Halloran TV. J Am Chem Soc. 2004;126:712–713. doi: 10.1021/ja039073j. [DOI] [PubMed] [Google Scholar]; d) Domaille DW, Que EL, Chang CJ. Nat Chem Biol. 2008;4:168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]; e) Qian F, Zhang C, Zhang Y, He W, Gao X, Hu P, Guo Z. J Am Chem Soc. 2009;131:1460–1468. doi: 10.1021/ja806489y. [DOI] [PubMed] [Google Scholar]; f) Marbella L, Serli-Mitasev B, Basu P. Angew Chem Int Ed. 2009;48:3996–3998. doi: 10.1002/anie.200806297. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2009;121:4056–4058. [Google Scholar]; g) Bagchi P, Morgan MT, Bacsa J, Fahrni CJ. J Am Chem Soc. 2013;135:18549–18559. doi: 10.1021/ja408827d. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Radford RJ, Lippard SJ. Curr Op Chem Biol. 2013;17:129–136. doi: 10.1016/j.cbpa.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikurak M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]; b) Park JG, Qin Y, Galati DF, Palmer AE. ACS Chem Biol. 2012;7:1636–1640. doi: 10.1021/cb300171p. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen Z, Ai H-w. Anal Chem. 2016;88:9029–9036. doi: 10.1021/acs.analchem.6b01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Breaker RR, Joyce GF. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]; b) Li Y, Sen D. Biochemistry. 1997;36:5589–5599. doi: 10.1021/bi962694n. [DOI] [PubMed] [Google Scholar]; c) Li Y, Breaker RR. Curr Opin Struct Biol. 1999;9:315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]; d) Lu Y. Chem Eur J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]; e) Joyce GF. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 5.a) Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schlosser K, Li Y. Biochemistry. 2004;43:9695–9707. doi: 10.1021/bi049757j. [DOI] [PubMed] [Google Scholar]
- 6.a) Kim HK, Rasnik I, Liu J, Ha T, Lu Y. Nat Chem Biol. 2007;3:763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Endo M, Takeuchi Y, Suzuki Y, Emura T, Hidaka K, Wang F, Willner I, Sugiyama H. Angew Chem Int Ed. 2015;54:10550–10554. doi: 10.1002/anie.201504656. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2015;127:10696–10700. [Google Scholar]
- 7.a) Li J, Lu Y. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]; b) Xiao Y, Rowe AA, Plaxco KW. J Am Chem Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]; c) Lan T, Furuya K, Lu Y. Chem Comm. 2010;46:3896–3898. doi: 10.1039/b926910j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc Natl Acad Sci USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee JH, Wang Z, Liu J, Lu Y. J Am Chem Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Liu J, Lu Y. J Am Chem Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]; b) Wang S, Liu C, Li G, Sheng Y, Sun Y, Rui H, Zhang J, Xu J, Jiang D. ACS Sensors. 2017;2:364–370. doi: 10.1021/acssensors.6b00667. [DOI] [PubMed] [Google Scholar]
- 10.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. Angew Chem Int Ed. 2008;47:4346–4350;. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2008;120:4418–4422. [Google Scholar]
- 11.a) Liu J, Cao Z, Lu Y. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xiang Y, Lu Y. Nat Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cha TG, Pan J, Chen H, Salgado J, Li X, Mao C, Choi JH. Nat Nanotechnol. 2014;9:39–43. doi: 10.1038/nnano.2013.257. [DOI] [PubMed] [Google Scholar]; d) Zhang J, Cheng F, Li J, Zhu JJ, Lu Y. Nano Today. 2016;11:309–329. doi: 10.1016/j.nantod.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Huang PJ, MV, Liu J. Biochemistry. 2016;55:2518–2525. doi: 10.1021/acs.biochem.6b00132. [DOI] [PubMed] [Google Scholar]; f) Hwang K, Hosseinzadeh P, Lu Y. Inorg Chim Acta. 2016;452:12–24. doi: 10.1016/j.ica.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Wu P, Hwang K, Lan T, Lu Y. J Am Chem Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y, Lu Y. Angew Chem Int Ed. 2014;53:13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2014;126:14018–14022. [Google Scholar]; c) Torabi S-F, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y. Proc Natl Acad Sci USA. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhou W, Liang W, Li D, Yuan R, Xiang Y. Biosens Bioelectron. 2016;85:573–579. doi: 10.1016/j.bios.2016.05.058. [DOI] [PubMed] [Google Scholar]; e) Wang W, Satyavolu NSR, Wu Z, Zhang J-R, Zhu J-J, Lu Y. Angew Chem Int Ed. doi: 10.1002/anie.201701325. in press. [DOI] [Google Scholar]
- 13.Yin P, Choi HM, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 14.a) Ali MM, Li Y. Angew Chem Int Ed. 2009;48:3512–3515. doi: 10.1002/anie.200805966. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2009;121:3564–3567. [Google Scholar]; b) Yan J, Song S, Li B, Huang Q, Zhang Q, Zhang H, Fan C. Small. 2010;6:2520–2525. doi: 10.1002/smll.201001220. [DOI] [PubMed] [Google Scholar]; c) Liu M, Zhang Q, Chang D, Gu J, Brennan JD, Li Y. Angew Chem Int Ed. 2017;56:1–6. doi: 10.1002/anie.201700054. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2017;129:1–6. [Google Scholar]
- 15.a) Zhang H, Li F, Dever B, Li XF, Le XC. Chem Rev. 2013;113:2812–2841. doi: 10.1021/cr300340p. [DOI] [PubMed] [Google Scholar]; b) Tian L, Cronin TM, Weizmann Y. Chem Sci. 2014;5:4153–4162. [Google Scholar]
- 16.a) Cheglakov Z, Cronin TM, He C, Weizmann Y. J Am Chem Soc. 2015;137:6116–6119. doi: 10.1021/jacs.5b01451. [DOI] [PubMed] [Google Scholar]; b) Wu C, Cansiz S, Zhang L, Teng IT, Qiu L, Li J, Liu Y, Zhou C, Hu R, Zhang T, Cui C, Cui L, Tan W. J Am Chem Soc. 2015;137:4900–4903. doi: 10.1021/jacs.5b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wu Z, Liu GQ, Yang XL, Jiang JH. J Am Chem Soc. 2015;137:6829–6836. doi: 10.1021/jacs.5b01778. [DOI] [PubMed] [Google Scholar]; d) Li L, Feng J, Liu H, Li Q, Tong L, Tang B. Chem Sci. 2016;7:1940–1945. doi: 10.1039/c5sc03909f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Yan L, Hui J, Liu Y, Guo Y, Liu L, Ding L, Ju H. Biosens Bioelectron. 2016;86:1017–1023. doi: 10.1016/j.bios.2016.07.102. [DOI] [PubMed] [Google Scholar]
- 17.a) Efendiev R, Budu CE, Cinelli AR, Bertorello AM, Pedemonte CH. J Biol Chem. 2003;278:28719–28726. doi: 10.1074/jbc.M303741200. [DOI] [PubMed] [Google Scholar]; b) Baartscheer A, Schumacher CA, van Borren MM, Belterman CN, Coronel R, Opthof T, Fiolet JW. Cardiovasc Res. 2005;65:83–92. doi: 10.1016/j.cardiores.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Zheng N, Song Z, Yin L, Cheng J. Biomaterials. 2014;35:3443–3454. doi: 10.1016/j.biomaterials.2013.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MK, Lim CS, Hong JT, Han JH, Jang HY, Kim HM, Cho BR. Angew Chem Int Ed. 2010;49:364–367. doi: 10.1002/anie.200904835. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2010;122:374–377. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


