Abstract
Current methods to quantify in vivo RNA dynamics are limited. Here, we developed a novel stable isotope (D2O) methodology to quantify RNA synthesis (i.e., ribosomal biogenesis) in cells, animal models, and humans. First, proliferating C2C12 cells were incubated in D2O-enriched media and myotubes ±50 ng/ml IGF-I. Second, rat quadriceps (untrained, n = 9; 7-wk interval-“like” training, n = 13) were collected after ~3-wk D2O (70 atom %) administration, with body-water enrichment monitored via blood sampling. Finally, 10 (23 ± 1 yr) men consumed 150-ml D2O followed by 50 ml/wk and undertook 6-wk resistance exercise (6 × 8 repetitions, 75% 1-repetition maximum 3/wk) with body-water enrichment monitored by saliva sampling and muscle biopsies (for determination of RNA synthesis) at 0, 3, and 6 wk. Ribose mole percent excess (r-MPE) from purine nucleotides was analyzed via GC-MS/MS. Proliferating C2C12 cell r-MPE exhibited a rise to plateau, whereas IGF-I increased myotube RNA from 76 ± 3 to 123 ± 3 ng/μl and r-MPE by 0.39 ± 0.1% (both P < 0.01). After 3 wk, rat quadriceps r-MPE had increased to 0.25 ± 0.01% (P < 0.01) and was greater with running exercise (0.36 ± 0.02%; P < 0.01). Human muscle r-MPE increased to 0.06 ± 0.01 and 0.13 ± 0.02% at 3/6 wk, respectively, equating to synthesis rates of ~0.8%/day, increasing with resistance exercise to 1.7 ± 0.3%/day (P < 0.01) and 1.2 ± 0.1%/day (P < 0.05) at 3/6 wk, respectively. Therefore, we have developed and physiologically validated a novel technique to explore ribosomal biogenesis in a multimodal fashion.
Keywords: ribosomal biogenesis, D2O, RNA synthesis, muscle
cellular protein content is under constant renewal to maintain cellular homeostasis. Typically, the balance between protein synthesis and breakdown remains relatively stable, yet under conditions of growth, atrophy, or cellular proliferation, rapid and significant changes in protein content and cell size occur (5). With protein synthesis rates determined by the number (translational capacity) and activity (translational efficiency) of ribosomes (27, 39), the ribosome provides a primary point of control in cellular homeostasis; yet our understanding of dynamic ribosome metabolism is poorly understood. Ribosomal biogenesis is the product of the coordinated synthesis of multiple ribosomal RNAs (rRNA) and proteins. In being an energy-consuming process, ribosomal biogenesis is tightly regulated by multiple signaling pathways responsive to nutrition, hormones, and mechanical activity (22). However, basal rates of ribosomal biogenesis and ribosome half-life across tissues are for the most part largely undescribed. With the demands to modulate and maintain protein content varying considerably across cell types, such as rapidly dividing single cells, to the coordinated maintenance repair of multicellular organs, regulation of ribosome pools is likely to be tightly linked with protein metabolism (10). Furthermore, when the coordinated control of ribosome biogenesis becomes unregulated, it can be the source of many conditions such as cancer (30).
Skeletal muscle is one of the most plastic tissues of the body, undergoing substantial and rapid hypertrophy or atrophy under conditions of functional overload, disuse, or malnutrition (7a, 9, 17). Understanding these processes is of great importance as preservation of muscle mass and function throughout life is crucial in preventing disability and maintaining quality of life, particularly in advanced aging (16, 31). In being a postmitotic tissue, muscle mass is controlled by the balance between muscle protein synthesis (MPS) and muscle protein breakdown. Many acute changes in MPS (<5 h) are accompanied by the activation or suppression of proteins in the mammalian target of rapamycin complex 1 pathway (1), modulating translational efficiency rather than translational capacity (6). However, prolonged exposure to muscle loading modifies RNA content, increasing with hypertrophy (3, 12, 34) and decreasing with atrophy (15). As such, ribosomal biogenesis is thought to be central to muscle-mass regulation.
With rRNA comprising 80% of total RNA, changes in RNA concentration are thought to be indicative of changes in the balance of ribosome synthesis and breakdown. However, in addition to relying on long-term interventions, efficient extraction, and normalization to muscle weight all introducing variability, measures of concentration do not inform on dynamic RNA metabolism, and importantly changes in RNA synthesis will naturally precede those of content. Past measures of RNA synthesis have typically relied on the incorporation of modified nucleotides such as [3H]uridine or 5-bromouridine. However, the use of these techniques is limited and generally cannot be used in whole animals due to their mutagenic nature. Alternatively, stable isotope tracers offer a safe method for use in humans, and measures of RNA synthesis using these have been made (8, 14). However, their applicability in human research has been limited due to numerous caveats, including variable and complex salvage pathways, infusions, and time limits (<24 h), resulting in a lack of methods to determine RNA synthesis rates, particularly in tissues with slow renewal rates (like skeletal muscle). Heavy water (D2O) provides alternate routes in the measurement of substrate metabolism and can overcome some limitations associated with other stable isotope methods. In being easily administered and with the precursor pool being maintained over weeks to months, we (40) and others (32) have made long-term cumulative measures of muscle protein synthesis with many other substrates measured in a range of different tissues including DNA (23, 29). Deuterium is incorporated via nucleotide de novo synthesis, overcoming previous limitations of nucleotide analogs, and thus similarly creates a viable route in the measurement of RNA synthesis. Here, we developed sensitive GC-MS/MS universally applicable methods for the measurement of RNA synthesis including in slower turning over tissues requiring only minimal D2O consumption; we validate these methods in cell cultures, preclinical models, and humans and in a cell type of contemporary interest, i.e., skeletal muscle.
MATERIALS AND METHODS
Cell culture.
Murine C2C12 myoblasts [passages 5–7; European Collection of Authenticated Cell Cultures (ECACC), Salisbury, United Kingdom] were seeded and maintained in Dulbecco’s modified Eagle’s medium as previously described (7) containing 10% fetal bovine serum, amphotericin B (1%), penicillin-streptomycin (1%), and 4 mM l-glutamine (Sigma-Aldrich, Poole, United Kingdom). Sterile 70% deuterium and [U-13C]glucose were added to DMEM at required enrichments and distributed among wells for labeling consistency. In proliferating cells, media were changed every 48 h, and cells were scraped at required time points. At 90% confluency, cells were differentiated by reducing serum concentrations to 2% with RNA synthesis stimulated 6 days after differentiation with 50 ng/ml IGF-I.
Animals.
Mixed population of female and male (n = 22) high responders to training rats for aerobic training were used for the study. Rats originated from the generations 17 and 18 of selection for their training response and were 9.2 ± 3.0 mo old at the start of the experiment (21). All experimental procedures described in this study protocol were approved by the Animal Care and Use Committee of Southern Finland, license number ESAVI-2010-07989/Ym-23, STH 534A (21.9.2010) and complements ESAVI/1968/04.10.03/2011, PH308A (30.3.2011) and ESAVI/722/04.10.07/2013, PH275A (1.3.2013). All experiments were conducted in accordance with the Guidelines of the European Community Council Directive 86/609/EEC. Rats were kept in air-conditioned rooms single-housed at an ambient temperature of 21 ± 2°C and relative humidity at 50 ± 10%. Artificial lighting provided light cycles of 12:12-h light-total darkness. Commercially available pelleted rodent diet (R36; Labfor; Lantmännen, Malmö, Sweden) and tap water (from the municipal water system of Jyväskylä, Finland) was available ad libitum for rats throughout the study. The energy content of the feed was 1,260 kJ/100 g (300.93 kcal/100 g). The feed contained 18.5% raw protein, 4.0% raw fat, 55.7% nitrogen-free extracts, 3.5% fiber, 6.3% ash, and <12% water. Rats received a gavage of 7.2 ml/kg 70% D2O for the remaining 3 wk of the 7-wk training period, with drinking water enriched to 2% to maintain body-water enrichment. Body-water enrichment was determined from blood samples collected at necropsy and used to represent the average enrichment throughout; although variability may occur over time, enriched drinking water minimizes these effects. Interval training consisted of warm-up for 5 min at 50–60% of maximum speed (individual speed for each rat) and running for 15 min, 3 min at 85–90% and 2-min pauses at 50%, repeated three times, inclination 15° uphill. Training was done three times per week with 1-day rest between (if possible). Forty-eight hours after the last training bout, animals were anesthetized with carbon dioxide and killed by cardiac puncture and thereafter immediately necropsied. Left quadriceps were rapidly exposed, removed, and immediately frozen by complete immersion in liquid nitrogen.
Subject characteristics and ethics.
Ten healthy younger (23 ± 1 yr, body mass index: 24 ± 1) men were recruited as previously described (3). All subjects provided their written, informed consent to participate after all procedures and risks (in relation to muscle biopsies, blood sampling, et cetera) were explained. Following inclusion in the study, subjects were studied over a 6-wk period. After baseline bilateral biopsies, subjects provided a saliva and blood sample and then consumed 150 ml of D2O (70 atom %; Sigma-Aldrich) to label the body-water pool to ~0.2% atom percent excess (APE), which was maintained with weekly top-up boluses (50 ml/wk). Thereafter, subjects performed progressive unilateral resistance exercise training (RET) 3/wk at 75% one-repetition maximum with additional bilateral biopsies taken at 3 and 6 wk to monitor RNA incorporation. Blood was collected at 0, 3, and 6 wk to follow deuterium incorporation into peripheral blood mononuclear cells (PBMCs), isolated using Histopaque (Sigma-Aldrich). For the temporal monitoring of body-water enrichment, each participant provided a saliva sample on RET visits >30 min after their last meal or drink, with further samples taken ~3 h after weekly 50-ml boluses to ensure that body-water enrichment was accurately represented. Samples were collected in sterile plastic tubes and immediately cold-centrifuged at 16,000 g to remove any debris that might be present; they were then aliquoted into 2-ml glass vials and frozen at −20°C until analysis. This study was approved by the University of Nottingham Research Ethics Committee, complied with studies conducted in accordance with the Declaration of Helsinki, and registered at https://clinicaltrials.gov/ (NCT02152839).
Media and body-water enrichment.
The deuterium enrichment was measured in media collected from cell culture plates and plasma from rats by incubating 100 µl of each sample with 2 µl of 10 M NaOH and 1 µl of acetone for 24 h at room temperature. Following incubation, the acetone was extracted into 200 µl of n-heptane, and 0.5 µl of the heptane phase was injected into the GC-MS/MS for analysis. A standard curve of known D2O enrichment was run alongside the samples for calculation of enrichment. Human body-water enrichment was extracted by heating 100-µl saliva in an inverted 2-ml autosampler vial for 4 h at 100°C. Vials were then placed upright on ice to condense extracted body water and transferred to a clean autosampler vial ready for injection. A total of 0.1-µl body water was injected into a high-temperature conversion elemental analyzer (Thermo Finnigan, Thermo Scientific, Hemel Hempstead, United Kingdom) connected to a Delta V Advantage Isotope Ratio Mass Spectrometer (Thermo Finnigan, Thermo Scientific).
Protein-bound alanine muscle fraction enrichment and calculation of fractional synthesis rate.
Myofibrillar protein was isolated from human vastus lateralis (VL) muscle biopsies and rat quadriceps by homogenizing 30–50 mg of muscle in ice-cold homogenization buffer and rotated for 10 min, and the supernatant was collected after centrifugation at 13,000 g for 5 min at 4°C. The myofibrillar pellet was solubilized in 0.3 M NaOH and separated from the insoluble collagen by centrifugation, and the myofibrillar protein was precipitated using 1 M perchloric acid. Protein-bound amino acids were released using acid hydrolysis by incubating in 0.1 M HCl in Dowex H+ resin slurry overnight before being eluted from the resin with 2 M NH4OH and evaporated to dryness; amino acids were then derivatized as their N-methoxycarbonyl methyl esters. Dried samples were suspended in 60 µl of distilled water and 32 µl of methanol, and following vortex, 10 µl of pyridine and 8 µl of methyl chloroformate were added. Samples were vortexed for 30 s and left to react at room temperature for 5 min. The newly formed N-methoxycarbonyl methyl ester amino acids were then extracted into 100 µl of chloroform. A molecular sieve was added to each sample for ~20 s before being transferred to a clean glass gas chromatography insert, removing any remaining water by size exclusion absorption. Human protein-bound alanine enrichment was determined by gas chromatography-pyrolysis-isotope ratio mass spectrometry (Delta V Advantage; Thermo Scientific) with rat protein-bound alanine enrichment determined by gas chromatography-tandem mass spectrometry (TSQ 8000; Thermo Scientific) alongside a standard curve of known dl-alanine-2,3,3,3-d4 enrichment to validate measurement accuracy of the machine. Myofibrillar MPS was calculated from the incorporation of deuterium-labeled alanine into protein using the enrichment of body water [corrected for the mean number of deuterium moieties incorporated per alanine (3.7) and the dilution from the total number of hydrogens in the derivative (i.e., 11)] as the surrogate precursor labeling between subsequent biopsies. The equation used was:
where APEala equals deuterium enrichment of protein-bound alanine, APEp indicates mean precursor enrichment over the time period, and t is the time between biopsies.
RNA extraction, digestion, and derivatization.
To extract RNA, approximately 20–30 mg of muscle was homogenized in extraction buffer (5 µl/mg) containing 0.1 M Tris·HCl, pH 8, 0.01 M EDTA, pH 8, and 1 M NaCl. Proteinase K was added to a final concentration of 50 µg/µl and placed at 55°C for ~2 h with occasional mixing until complete digestion had occurred. For cell culture, each well was scraped in 200 µl of extraction buffer, and PBMCs were homogenized in 200 µl of extraction buffer. To the extractions, an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added and inverted several times to mix, and the upper aqueous layer was removed to a clean Eppendorf after centrifugation at 13,000 rpm for 10 min. To remove additional protein, an equal volume of chloroform-isoamyl alcohol (24:1) was added to the aqueous layer and repeated as above. To precipitate RNA, an equal volume of isopropanol was added to the aqueous layer, inverted several times, and centrifuged at 13,000 rpm for 20 min. The pellet was washed three times in 70% ethanol, air-dried, resuspended in 22 µl of molecular biology, digested with 5 µl of 375 mM sodium acetate (pH 4.8) and 750 µM ZnSO4 containing 0.5 units of nuclease S1 and 0.25 units of potato acid phosphatase, and placed at 37°C overnight. Hydrolysates were then reacted with 10 µl of O-benzylhydroxylamine (2% wt/vol) and 7.5 µl of acetic acid at 100°C for 30 min. Samples were allowed to cool at room temperature before the addition of 10 µl of 1-methylimidazole and 100 µl of acetic anhydride. The reactions were transferred to a boiling tube and quenched by the addition of 2 ml of double-distilled water. The newly formed derivatives were extracted by the addition of 750 µl of dichloromethane (DCM) vortex mixed, and phases were allowed to separate. By prewetting the tip with DCM, the lower layer was removed to a clean boiling tube, and the procedure was repeated. DCM extracts were then dried and resuspended in 40 µl of ethyl acetate for GC-MS/MS analysis.
GC-MS/MS instrument conditions and fractional synthesis rate calculation.
To measure RNA enrichment, 2 µl of sample was injected into a TRACE 1310 Gas Chromatograph connected to TSQ 8000 triple quadrupole GC-MS/MS (Thermo Scientific). Samples were injected on splitless mode with inlet temperature at 280°C. GC ramp conditions were 120°C for 1 min, ramp to 280°C at 10°C/min, and hold for 3 min. Selected reaction monitoring (SRM) was performed for the mass-to-charge ratios of 273.1:111.1 and 274.1:112.1 representing the M and M+1 ions with a collision-induced dissociation energy of 6. Enrichment was calculated as M+1/(M + M+1) with the mole percent excess (MPE) expressed as difference from unlabeled D2O free samples. Fractional synthesis rates (FSR) were calculated as FSR (%/day) = (r-MPE)/[(p-MPE) × t] × 100, where r-MPE is the excess enrichment of bound ribose, p-MPE is the mean precursor enrichment over the time period, and t is the time between samples. In cell culture and rat studies, p-MPE was calculated as the water enrichment multiplied by the amplification factor of 2.098 determined in cells. In human studies, p-MPE was taken as the ribose PBMC enrichment measured over the labeling period. Samples were run in triplicate alongside standard curves of known ribose standards, and the average of both peaks was used in the results. Additionally, unlabeled samples were injected in different quantities to determine abundance effects.
Statistical analysis.
Descriptive statistics were produced for all data sets to check for normal distribution (accepted if P > 0.05) using a Kolmogorov-Smirnov test. All data are presented as means ± SE. Differences between the effects of interval training and control on RNA synthesis in rates were analyzed by t-test. All other data sets were analyzed by repeated-measures one-way or two-way ANOVA with a Bonferroni correction using GraphPad Prism 5 software (La Jolla, CA). Correlations were assessed using Pearson product moment correlation coefficient. The α-level of significance was set at P < 0.05.
RESULTS
GC-MS/MS chromatography and SRM transitions.
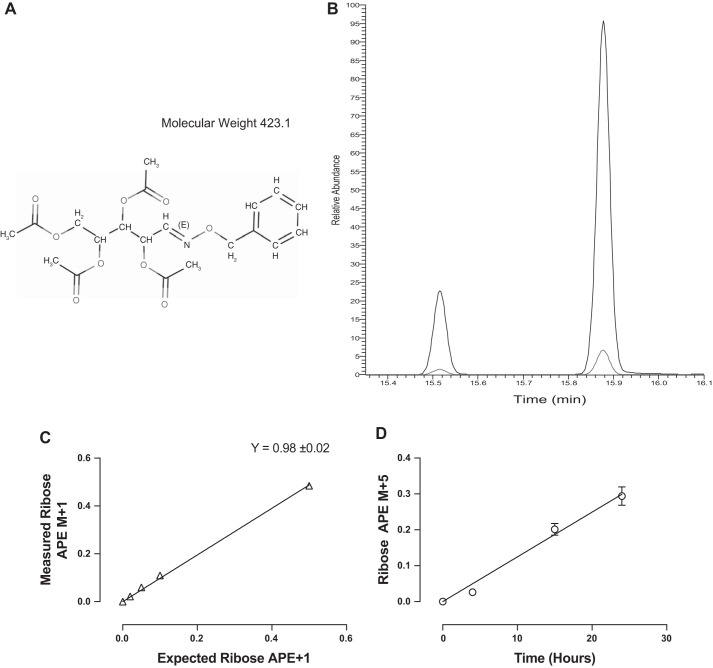
Addition of O-benzylhydroxylamine and four acetyl groups to ribose produces a derivative with a molecular weight of 423.1 (Fig. 1A). On gas chromatography, the derivative produces two peaks representing the cis-trans isomers formed due to the anomeric carbon of ribose (Fig. 1B). Full-scan MS analysis of the derivative produces a most abundant fragment with best chromatography of 273.1, with second fragmentation producing a most abundant fragment of 111.1. Analysis of this transition is highly selective and produces GC-MS/MS spectra with very low background, detecting standard enrichments as little as 0.02 APE (Fig. 1C). Furthermore, this SRM encompasses all backbone carbons of ribose confirmed by +5 enrichment from [U-13C]glucose incorporation (Fig. 1D).
Fig. 1.
A: structure and mass of the ribose benzylhydroxylamine tetra-acetate derivative. B: typical GC-MS/MS chromatogram of the ribose derivative on a DB-17 column for the SRM transitions of 273.1–111.1. Higher line represents the M, and lower line the M+1. C: standard curve of 0.02, 0.05, 0.1, and 0.5 [1-13C]ribose. D: measurement of the +5 isotopomer of RNA bound ribose from C2C12 myotubes incubated in [U-13C]glucose enriched media.
Deuterium incorporation into RNA bound ribose.
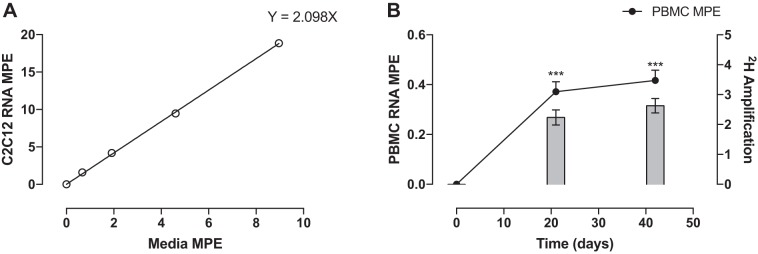
The MPE of ribose (Fig. 2A) extracted from purine nucleotides of RNA from C2C12 cells increased linearly with increasing concentrations of media D2O enrichment being 0, 1.6 ± 0.08, 4.1 ± 0.1, 9.5 ± 0.15, and 18.8 ± 0.18% at media concentrations of 0, 0.67, 1.9, 4.6, and 8.9%, respectively. Linear regression revealed that on average 2.1 2H are incorporated into purine ribose during synthesis of new RNA. PBMCs from human subjects showed an increase in MPE to 0.37 ± 0.04 and 0.42 ± 0.04% (both P < 0.01), revealing the average accessible hydrogens to be 2.6 ± 0.2 (Fig. 2B).
Fig. 2.
A: MPE of RNA from maximally labeled C2C12s vs. D2O media MPE. B: the amplification of deuterium into PBMCs from human subjects. ***Significantly different from baseline, P < 0.001.
Validation of deuterium incorporation in RNA in vitro.
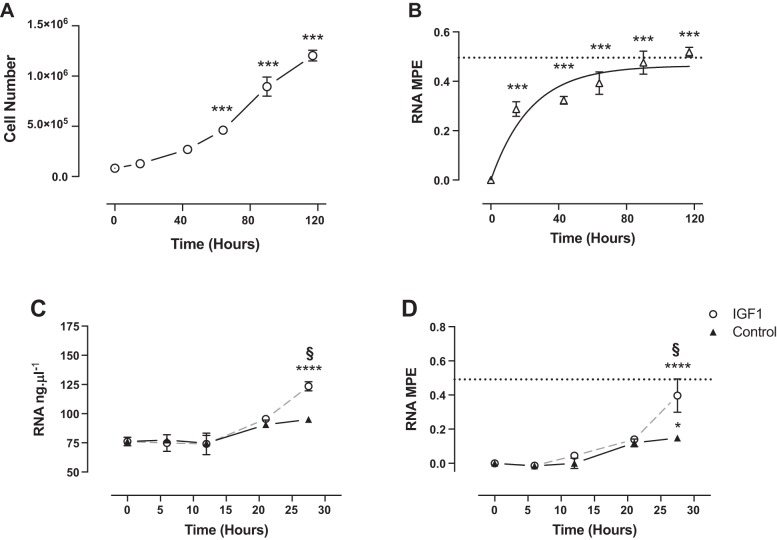
C2C12 cell number increased from 0.083 ± 0.001 million to 1.2 ± 0.03 million per well after 117 h of proliferation (Fig. 3A), whereas RNA MPE followed a rise to plateau relationship from 0.28 ± 0.03% after 15 h to 0.52 ± 0.02% by 117 h (Fig. 3B). In response to IGF-I treatment, RNA content increased by 27.5 h from 76.1 ± 3 to 123.4 ± 3 ng/μl (Fig. 3C). Similarly, RNA MPE significantly increased in control to 0.15 ± 0.01% at 27.5 h, with the increase in IGF-I treatment being significantly greater (Fig. 3D).
Fig. 3.
Time course of cell number (A) and MPE of RNA (B) in proliferating C2C12s. Time courses in the concentration of RNA (C) and MPE of RNA (D) in nontreated condition (Control) and in responses to IGF-I are shown. Dotted line represents plateau enrichment. Significantly different from baseline, *P < 0.05, ***P < 0.001, ****P < 0.0001. §Significantly different from control at that time point, P < 0.01.
RNA synthesis in rat muscle in vivo.
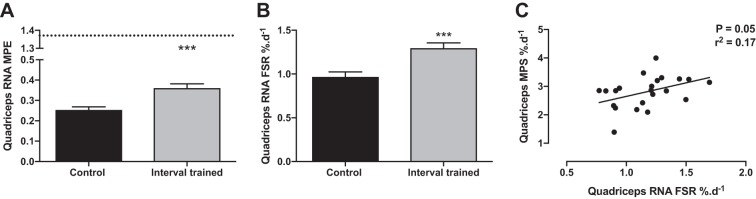
After 3 wk of continuous D2O administration, RNA MPE significantly increased to 0.25 ± 0.01% in control and was significantly greater with interval training to 0.36 ± 0.01% (P < 0.001; Fig. 4A). The calculated RNA FSR was 0.97 ± 0.05%/day with a significant increase in response to interval training to 1.3 ± 0.05%/day (P < 0.001; Fig. 4B). Rat quadriceps MPS showed a correlation with quadriceps RNA synthesis percentage of r2 = 0.17 and P = 0.05 (Fig. 4C).
Fig. 4.
A: MPE of RNA bound ribose from control and exercised rat quadriceps. B: FSR of RNA from control and exercised rat quadriceps. C: correlation between quadriceps in MPS%/day and RNA in FSR%/day. Dotted line represents plateau enrichment. ***Significantly different from control, P < 0.001. d, Day.
RNA synthesis in human muscle.
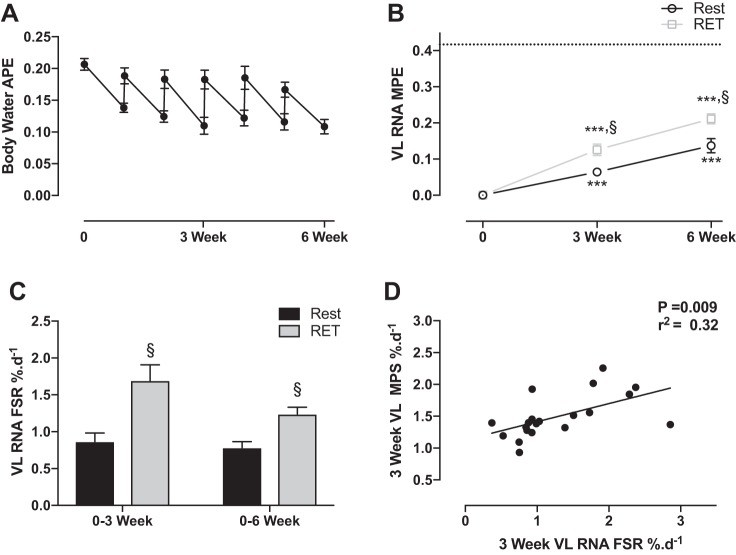
The body-water enrichment over the 6 wk is shown in Fig. 5A. The MPE from RNA bound ribose increased in rest legs to 0.064 ± 0.01 and 0.137 ± 0.02% at 3 and 6 wk, respectively (P < 0.001). In RET legs, the MPE increased to 0.125 ± 0.02 and 0.211 ± 0.01% at 3 and 6 wk, respectively (P < 0.001), being greater than rest at both time points (P < 0.005; Fig. 5B). Corrected for PBMC RNA enrichment, FSR of 0.86 ± 0.1 and 0.78 ± 0.1%/day were determined in rest legs at 3 and 6 wk, respectively (Fig. 5C). In RET legs, the RNA FSR was significantly greater than rest, being 1.69 ± 0.2%/day (P < 0.01) and 1.24 ± 0.1%/day (P < 0.05) at 3 and 6 wk, respectively. Human VL MPS was highly correlated with VL RNA synthesis P = 0.009 and an r2 = 0.32 (Fig. 5D).
Fig. 5.
A: time course of body-water enrichment measured through saliva samples over the 6 wk of labeling. B: MPE of RNA bound ribose from human VL in rest and RET legs. Dotted line represents the average plateau PBMC enrichment. C: FSR of RNA bound ribose from human VL in rest and RET legs. D: correlation between VL MPS%/day and RNA FSR %/day. ***Significantly different from baseline, P < 0.001. §Significantly different from control at that time point, P < 0.05.
DISCUSSION
We have developed and validated D2O-based methods for the measurement of in vitro and in vivo RNA synthesis that can be used safely and effectively in humans and with the potential for application to any tissue. This is a step forward from current practices in providing methods for long-term measures of RNA synthesis in humans, particularly those of slow turnover pools such as skeletal muscle. RNA content is closely linked to cellular metabolism, with ribosomal biogenesis being required for cellular proliferation (10) and growth (34). Currently, changes in RNA content are primarily determined by crude measures of RNA concentrations, with limited methods in place to determine dynamic rates of RNA synthesis in vivo. Our approach will provide insight into the workings of dynamic ribosomal biogenesis in vitro, in animal models, and in humans, across cell types.
Method development and validation of in vitro RNA synthesis in skeletal muscle cells.
Nucleotide synthesis involves complex precursor pools with variable nucleotide salvage (23), making the incorporation of stable isotope-labeled compounds difficult to interpret (13). The advantage of using D2O is that deuterium becomes incorporated into the backbone hydrogen of ribose during nucleotide synthesis, with deoxyribose from purine deoxyribonucleosides primarily synthesized via de novo synthesis, as such, providing a reliable input of isotope (23, 29). As deoxyribonucleotides are reduced from ribonucleotides, this further creates a viable method for measures of RNA synthesis, and for the first time we have shown a constant incorporation of deuterium across a range of media concentrations into purine ribose.
Total RNA encompasses rRNA, tRNA, and mRNA that will have variable turnover rates (33). The quickest of these will be tRNA and mRNA that will contribute to early increases in detectable enrichment. However, in making up <20% of total cellular RNA, these pools become quickly saturated, and deuterium incorporation follows a rise to plateau in proliferating cells reflecting the required expansion of rRNA for cell division (Fig. 3B; Refs. 8, 10). Furthermore, as initial validation using established stimulators of in vitro myotube hypertrophy and ribosomal biogenesis (i.e., IGF-I; Ref. 7), we were able to detect simultaneous increases in both RNA content and deuterium incorporation into RNA. Therefore, deuterium incorporation into RNA was reflective of newly synthesized RNA.
To use precursor product calculations, a measure of the precursor, or a proxy thereof, is required (38). Alternatively, when using D2O, an amplification factor can be used to represent the amount of accessible hydrogen in the precursor that can incorporate deuterium and be multiplied by the body-water enrichment (4). Nucleotide precursor pools are difficult to measure, with continuous input of unlabeled substrates such that the maximal theoretical plateau is never achieved (24). To investigate the number of accessible hydrogen in ribose in vitro, murine C2C12 skeletal muscle cells were repeatedly passaged in a range of D2O media enrichments, revealing a constant incorporation of ~2.1 deuteriums out of a total 6. Previously, values of ~3.1 have been reported for deoxyribose out of a total 7 (26, 29), expectedly higher due to the additional hydrogen that exchanges with ribonucleotide reductase.
Validation of in vivo RNA synthesis in an animal model.
Compared with many tissues, skeletal muscle has a relatively slow habitual protein renewal rate, with little to no active DNA synthesis (11); in contrast, actual RNA synthesis rates are practically unknown and will vary considerably across tissues. As D2O can be simply administered by oral consumption and easily maintained, D2O can be used to capture a vast range of synthesis rates. Recently, D2O has been used to measure ribosome renewal in mouse liver, although in using GC-MS this requires high levels of enrichment (5% APE) and fast rates of turnover (~10%) that can be burdensome and limit applications (25). Applying our validated in vitro methods to rodents, to our knowledge, we made the first long-term measures of RNA synthesis in skeletal muscle. In doing so, we demonstrated there is active renewal of RNA pools of ~1%/day. Furthermore, using an exercise stimulus to activate ribosomal biogenesis (39), we validated the existence of a significant increase in deuterium incorporation into RNA, demonstrating increased RNA synthesis. Intriguingly, RNA synthesis rates were correlated with MPS, which we speculate is due to a coordinate regulation in response to exercise.
RNA synthesis in human muscle and the effect of resistance exercise.
Presumably, most tissues will have a constant level of rRNA synthesis to maintain functional ribosomes for cellular protein synthesis. That said, ribosome biogenesis consumes considerable energy and will therefore likely be maintained at minimal requirements. Using the methods described here, to our knowledge, we report the first measures of RNA synthesis in human muscle, showing a constant synthesis rate of ~0.8%/day during “habitual activity.” Furthermore, to assess precursor enrichment, we measured the plateau enrichment of a population of cells 100% replenished (PBMCs) over the labeling period (29). This accounts for individual variability in the number of accessible hydrogens, and further we showed on average 2.6 deuteriums were incorporated, similar to our in vitro measures.
Skeletal muscle RNA content is highly responsive to functional overload (28, 37), and here we have shown that in response to RET, RNA synthesis was significantly increased after 3 and 6 wk of exercise training in humans. Once again, RNA synthesis was correlated with MPS, which further validates that in muscle, ribosome and protein metabolism are likely to be inextricably related [likely via mammalian target of rapamycin complex 1 (18)]. Similarly, although there are little other data for us to compare our results with, whole body rRNA turnover determined by breakdown products in urine have been estimated ~2.5% (36). Furthermore, this showed a strong relationship between whole body protein degradation rates of r2 = 0.7, supporting that these are closely linked processes in muscle homeostasis.
Further application of methods for RNA synthesis.
Previously, measures of RNA synthesis have been made in humans using [6,6-2H2]glucose, however, this requires large amounts of tracer to be consumed (1 g/kg) and is generally limited to fast turnover cells (8). Furthermore, achieving high levels of enrichment to perform GC-MS analysis in humans is costly, requiring high levels of D2O consumption that are burdensome on the individual and may potentially cause adverse effects such as nausea and vertigo (19). Additionally, rates of RNA synthesis will vary considerably across tissues, making the detection of slow turnover pools such as muscle using GC-MS techniques difficult. Here, by combining sensitive GC-MS/MS techniques (detection limits of ≥0.02% MPE) and the ability to administer D2O from days to weeks, this method creates opportunities to measure RNA synthesis over a range of rates and tissues. Such measures can be employed through simple D2O administration and access to tissue samples or blood, with some prior expectation of synthesis helpful. For instance, human body-water enrichment can be simply maintained approximately 0.15–0.2% APE using an initial bolus of 150 ml of D2O, followed by weekly doses of 50 ml (3). In this situation, sampling from a tissue after 5 days with an RNA turnover rate of ~10%/day would result in an easily detectable product enrichment using GC-MS/MS of approximately 0.075–0.1%, whereas an RNA turnover rate of ~1%/day would result in an undetectable enrichment of approximately 0.0075–0.01%. This is not to say these measures cannot be made by other means. Raising body-water enrichment will increase end-point enrichment, and body-water enrichments as high as 2% would make GC-MS techniques an option. However, D2O consumption of such high levels is costly and burdensome on subjects. As such, the methods used here can be readily applied to many situations.
Conclusion.
In summary, we have developed and validated the use of D2O in measurement of RNA synthesis both in vitro and in vivo. With many RNA synthesis rates unknown, these new methods will have a significant impact in being able to measure a wide range of RNA turnover rates in varied tissues. Furthermore, ribosomal biogenesis has been the interest of recent publications in muscle-adaptive mechanisms (12, 20, 34, 35) and will likely play a significant role elucidating muscle metabolism at rest and in response to hypertrophic/atrophic conditions.
GRANTS
This work was supported by Medical Research Council (MRC) Grant MR/K00414X/1 and Arthritis Research UK (ARUK) Grant 19891 as part of the MRC-ARUK Centre for Musculoskeletal Ageing Research; the Physiological Society (awarded to P. J. Atherton and K. Smith); Dunhill Medical Trust Grant R264/1112 (to K. Smith, P. J. Atherton, and D. J. Wilkinson); and a MRC Confidence in Concept Award (CIC12019; to P. J. Atherton, P. L. Greenhaff, N. J. Szewczyk, and K. Smith). The founding low- and high-exercise response (LRT/HRT) rat model system was funded by the Office of Research Infrastructure Programs Grant P40-OD-021331 (to L. G. Koch and S. L. Britton) from the National Institutes of Health. Rat models for LRT and HRT are maintained as an international resource with support from the Department of Anesthesiology at the University of Michigan, Ann Arbor, Michigan (see http://koch-britton.med.umich.edu/ for information). Rat tissues used in this development work were derived as part of ongoing (i.e., not involving new studies) METAPREDICT studies, a European Union Seventh Framework Program (HEALTH-F2-2012-277936 to H. Kainulainen).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.S.B., D.J.W., N.J.S., H.K., S.L., L.G.K., S.L.B., P.L.G., K.S., and P.J.A. conceived and designed research; M.S.B., D.J.W., W.K.M., B.E.P., H.K., and S.L. performed experiments; M.S.B., D.J.W., B.E.P., K.S., and P.J.A. analyzed data; M.S.B., D.J.W., K.S., and P.J.A. interpreted results of experiments; M.S.B., D.J.W., and P.J.A. prepared figures; M.S.B., D.J.W., W.K.M., J.L.L., B.E.P., N.J.S., H.K., S.L., L.G.K., S.L.B., P.L.G., K.S., and P.J.A. drafted manuscript; M.S.B., D.J.W., W.K.M., J.L.L., B.E.P., N.J.S., H.K., S.L., L.G.K., S.L.B., P.L.G., K.S., and P.J.A. edited and revised manuscript; M.S.B., D.J.W., W.K.M., J.L.L., B.E.P., N.J.S., H.K., S.L., L.G.K., S.L.B., P.L.G., K.S., and P.J.A. approved final version of manuscript.
ACKNOWLEDGMENTS
We are grateful for the clinical, technical, and administrative support of Margaret Baker, Amanda Gates, and Tanya Fletcher.
REFERENCES
- 1.Atherton PJ, Etheridge T, Watt PW, Wilkinson D, Selby A, Rankin D, Smith K, Rennie MJ. Muscle full effect after oral protein: time-dependent concordance and discordance between human muscle protein synthesis and mTORC1 signaling. Am J Clin Nutr 92: 1080–1088, 2010. doi: 10.3945/ajcn.2010.29819. [DOI] [PubMed] [Google Scholar]
- 3.Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, Greenhaff PL, Smith K, Atherton PJ. Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide-derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J 29: 4485–4496, 2015. doi: 10.1096/fj.15-273755. [DOI] [PubMed] [Google Scholar]
- 4.Busch R, Kim YK, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta 1760: 730–744, 2006. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 5.Cheek DB. Human Growth. Body Composition, Cell Growth, Energy, and Intelligence. Philadelphia, PA: Lea & Febiger, 1968. [Google Scholar]
- 6.Chesley A, MacDougall JD, Tarnopolsky MA, Atkinson SA, Smith K. Changes in human muscle protein synthesis after resistance exercise. J Appl Physiol (1985) 73: 1383–1388, 1992. [DOI] [PubMed] [Google Scholar]
- 7.Crossland H, Kazi AA, Lang CH, Timmons JA, Pierre P, Wilkinson DJ, Smith K, Szewczyk NJ, Atherton PJ. Focal adhesion kinase is required for IGF-I-mediated growth of skeletal muscle cells via a TSC2/mTOR/S6K1-associated pathway. Am J Physiol Endocrinol Metab 305: E183–E193, 2013. doi: 10.1152/ajpendo.00541.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.De Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, Maffulli N, Movin T, Narici MV, Rennie MJ. The temporal responses of protein synthesis, gene expression and cell signalling in human quadriceps muscle and patellar tendon to disuse. J Physiol 585: 241–251, 2007. doi: 10.1113/jphysiol.2007.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defoiche J, Zhang Y, Lagneaux L, Pettengell R, Hegedus A, Willems L, Macallan DC. Measurement of ribosomal RNA turnover in vivo by use of deuterium-labeled glucose. Clin Chem 55: 1824–1833, 2009. doi: 10.1373/clinchem.2008.119446. [DOI] [PubMed] [Google Scholar]
- 9.DeFreitas JM, Beck TW, Stock MS, Dillon MA, Kasishke PR 2nd. An examination of the time course of training-induced skeletal muscle hypertrophy. Eur J Appl Physiol 111: 2785–2790, 2011. doi: 10.1007/s00421-011-1905-4. [DOI] [PubMed] [Google Scholar]
- 10.Derenzini M, Montanaro L, Chillà A, Tosti E, Vici M, Barbieri S, Govoni M, Mazzini G, Treré D. Key role of the achievement of an appropriate ribosomal RNA complement for G1-S phase transition in H4-II-E-C3 rat hepatoma cells. J Cell Physiol 202: 483–491, 2005. doi: 10.1002/jcp.20144. [DOI] [PubMed] [Google Scholar]
- 11.Drake JC, Bruns DR, Peelor FF 3rd, Biela LM, Miller RA, Miller BF, Hamilton KL. Long-lived Snell dwarf mice display increased proteostatic mechanisms that are not dependent on decreased mTORC1 activity. Aging Cell 14: 474–482, 2015. doi: 10.1111/acel.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ. Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. Am J Physiol Endocrinol Metab 309: E72–E83, 2015. doi: 10.1152/ajpendo.00050.2015. [DOI] [PubMed] [Google Scholar]
- 13.Grimble GK, Malik SB, Boza JJ. Methods for measuring tissue RNA turnover. Curr Opin Clin Nutr Metab Care 3: 399–408, 2000. doi: 10.1097/00075197-200009000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Grimble GK, Millward DJ. The measurement of ribosomal ribonucleic acid synthesis in rat liver and skeletal muscle in vivo. Biochem Soc Trans 5: 913–916, 1977. doi: 10.1042/bst0050913. [DOI] [PubMed] [Google Scholar]
- 15.Haddad F, Baldwin KM, Tesch PA. Pretranslational markers of contractile protein expression in human skeletal muscle: effect of limb unloading plus resistance exercise. J Appl Physiol (1985) 98: 46–52, 2005. doi: 10.1152/japplphysiol.00553.2004. [DOI] [PubMed] [Google Scholar]
- 16.Hirani V, Blyth F, Naganathan V, Le Couteur DG, Seibel MJ, Waite LM, Handelsman DJ, Cumming RG. Sarcopenia is associated with incident disability, institutionalization, and mortality in community-dwelling older men: the concord health and ageing in men project. J Am Med Dir Assoc 16: 607–613, 2015. doi: 10.1016/j.jamda.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Houston DK, Nicklas BJ, Ding J, Harris TB, Tylavsky FA, Newman AB, Lee JS, Sahyoun NR, Visser M, Kritchevsky SB; Health ABC Study . Dietary protein intake is associated with lean mass change in older, community-dwelling adults: the Health, Aging, and Body Composition (Health ABC) Study. Am J Clin Nutr 87: 150–155, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Iadevaia V, Liu R, Proud CG. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol 36: 113–120, 2014. doi: 10.1016/j.semcdb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Jones PJ, Leatherdale ST. Stable isotopes in clinical research: safety reaffirmed. Clin Sci (Lond) 80: 277–280, 1991. doi: 10.1042/cs0800277. [DOI] [PubMed] [Google Scholar]
- 20.Kirby TJ, Lee JD, England JH, Chaillou T, Esser KA, McCarthy JJ. Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J Appl Physiol (1985) 119: 321–327, 2015. doi: 10.1152/japplphysiol.00296.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch LG, Pollott GE, Britton SL. Selectively bred rat model system for low and high response to exercise training. Physiol Genomics 45: 606–614, 2013. doi: 10.1152/physiolgenomics.00021.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 556: 27–34, 2015. doi: 10.1016/j.gene.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Macallan DC, Fullerton CA, Neese RA, Haddock K, Park SS, Hellerstein MK. Measurement of cell proliferation by labeling of DNA with stable isotope-labeled glucose: studies in vitro, in animals, and in humans. Proc Natl Acad Sci USA 95: 708–713, 1998. doi: 10.1073/pnas.95.2.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini WZ, Chinkes DL, Wolfe RR. Quantification of DNA synthesis from different pathways in cultured human fibroblasts and myocytes. Metabolism 53: 128–133, 2004. doi: 10.1016/j.metabol.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Mathis AD, Naylor BC, Carson RH, Evans E, Harwell J, Knecht J, Hexem E, Peelor FF 3rd, Miller BF, Hamilton KL, Transtrum MK, Bikman BT, Price JC. Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol Cell Proteomics 16: 243–254, 2017. doi: 10.1074/mcp.M116.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, Murphy EJ, Koduru P, Ferrarini M, Zupo S, Cutrona G, Damle RN, Wasil T, Rai KR, Hellerstein MK, Chiorazzi N. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest 115: 755–764, 2005. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millward DJ, Garlick PJ, James WP, Nnanyelugo DO, Ryatt JS. Relationship between protein synthesis and RNA content in skeletal muscle. Nature 241: 204–205, 1973. doi: 10.1038/241204a0. [DOI] [PubMed] [Google Scholar]
- 28.Nakada S, Ogasawara R, Kawada S, Maekawa T, Ishii N. Correlation between ribosome biogenesis and the magnitude of hypertrophy in overloaded skeletal muscle. PLoS One 11: e0147284, 2016. doi: 10.1371/journal.pone.0147284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, Hoh R, Antelo F, Strawford A, McCune JM, Christiansen M, Hellerstein MK. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci USA 99: 15345–15350, 2002. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: current evidence and emerging paradigms. Semin Cancer Biol 37–38: 36–50, 2016. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Rizzoli R, Reginster JY, Arnal JF, Bautmans I, Beaudart C, Bischoff-Ferrari H, Biver E, Boonen S, Brandi ML, Chines A, Cooper C, Epstein S, Fielding RA, Goodpaster B, Kanis JA, Kaufman JM, Laslop A, Malafarina V, Mañas LR, Mitlak BH, Oreffo RO, Petermans J, Reid K, Rolland Y, Sayer AA, Tsouderos Y, Visser M, Bruyère O. Quality of life in sarcopenia and frailty. Calcif Tissue Int 93: 101–120, 2013. doi: 10.1007/s00223-013-9758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MM, Turner SM, Hellerstein MK, Hamilton KL, Miller BF. Long-term synthesis rates of skeletal muscle DNA and protein are higher during aerobic training in older humans than in sedentary young subjects but are not altered by protein supplementation. FASEB J 25: 3240–3249, 2011. doi: 10.1096/fj.11-186437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross J. mRNA stability in mammalian cells. Microbiol Rev 59: 423–450, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. Am J Physiol Endocrinol Metab 310: E652–E661, 2016. doi: 10.1152/ajpendo.00486.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stec MJ, Mayhew DL, Bamman MM. The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J Appl Physiol (1985) 119: 851–857, 2015. doi: 10.1152/japplphysiol.00489.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topp H, Fusch G, Schöch G, Fusch C. Noninvasive markers of oxidative DNA stress, RNA degradation and protein degradation are differentially correlated with resting metabolic rate and energy intake in children and adolescents. Pediatr Res 64: 246–250, 2008. doi: 10.1203/PDR.0b013e31817cfca6. [DOI] [PubMed] [Google Scholar]
- 37.von Walden F, Casagrande V, Östlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol 302: C1523–C1530, 2012. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- 38.Watt PW, Lindsay Y, Scrimgeour CM, Chien PA, Gibson JN, Taylor DJ, Rennie MJ. Isolation of aminoacyl-tRNA and its labeling with stable-isotope tracers: use in studies of human tissue protein synthesis. Proc Natl Acad Sci USA 88: 5892–5896, 1991. doi: 10.1073/pnas.88.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West DW, Baehr LM, Marcotte GR, Chason CM, Tolento L, Gomes AV, Bodine SC, Baar K. Acute resistance exercise activates rapamycin-sensitive and -insensitive mechanisms that control translational activity and capacity in skeletal muscle. J Physiol 594: 453–468, 2016. doi: 10.1113/JP271365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson DJ, Franchi MV, Brook MS, Narici MV, Williams JP, Mitchell WK, Szewczyk NJ, Greenhaff PL, Atherton PJ, Smith K. A validation of the application of D2O stable isotope tracer techniques for monitoring day-to-day changes in muscle protein subfraction synthesis in humans. Am J Physiol Endocrinol Metab 306: E571–E579, 2014. doi: 10.1152/ajpendo.00650.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]