Impaired growth hormone-mediated signaling is observed in ethanol-exposed hepatocytes and is explained by differential effects of alcohol dehydrogenase (ADH)- and cytochrome P450 2E1 (CYP2E1)-mediated ethanol metabolism on the Jak2/STAT5B pathway.
Keywords: WIF-B cells; ethanol, hepatotoxicity; CYP2E1
Abstract
The liver metabolizes alcohol using alcohol dehydrogenase (ADH) and cytochrome P450 2E1 (CYP2E1). Both enzymes metabolize ethanol into acetaldehyde, but CYP2E1 activity also results in the production of reactive oxygen species (ROS) that promote oxidative stress. We have previously shown that microtubules are hyperacetylated in ethanol-treated polarized, hepatic WIF-B cells and livers from ethanol-fed rats. We have also shown that enhanced protein acetylation correlates with impaired clathrin-mediated endocytosis, constitutive secretion, and nuclear translocation and that the defects are likely mediated by acetaldehyde. However, the roles of CYP2E1-generated metabolites and ROS in microtubule acetylation and these alcohol-induced impairments have not been examined. To determine if CYP2E1-mediated alcohol metabolism is required for enhanced acetylation and the trafficking defects, we coincubated cells with ethanol and diallyl sulfide (DAS; a CYP2E1 inhibitor) or N-acetyl cysteine (NAC; an antioxidant). Both agents failed to prevent microtubule hyperacetylation in ethanol-treated cells and also failed to prevent impaired secretion or clathrin-mediated endocytosis. Somewhat surprisingly, both DAS and NAC prevented impaired STAT5B nuclear translocation. Further examination of microtubule-independent steps of the pathway revealed that Jak2/STAT5B activation by growth hormone was prevented by DAS and NAC. These results were confirmed in ethanol-exposed HepG2 cells expressing only ADH or CYP2E1. Using quantitative RT-PCR, we further determined that ethanol exposure led to blunted growth hormone-mediated gene expression. In conclusion, we determined that alcohol-induced microtubule acetylation and associated defects in microtubule-dependent trafficking are mediated by ADH metabolism whereas impaired microtubule-independent Jak2/STAT5B activation is mediated by CYP2E1 activity.
NEW & NOTEWORTHY Impaired growth hormone-mediated signaling is observed in ethanol-exposed hepatocytes and is explained by differential effects of alcohol dehydrogenase (ADH)- and cytochrome P450 2E1 (CYP2E1)-mediated ethanol metabolism on the Jak2/STAT5B pathway.
INTRODUCTION
The liver is the major site of ethanol metabolism and, accordingly, is most susceptible to injury from chronic alcohol consumption. Although the progression of alcoholic liver disease is well described clinically, little is known about the molecular mechanisms that promote alcohol-induced hepatotoxicity. However, alcohol metabolites and enhanced oxidative stress have been linked to promoting liver injury. Alcohol is metabolized by two enzymes, alcohol dehydrogenase (ADH) and cytochrome P450 2E1 (CYP2E1). ADH-mediated metabolism results in the production of acetaldehyde, a highly reactive intermediate that can form stable, covalent modifications on other macromolecules (mainly proteins) (reviewed in Ref. 39). CYP2E1-mediated metabolism not only produces acetaldehyde but also highly reactive oxygen and hydroxyethyl radicals (39). These oxygen radicals contribute to enhanced oxidative stress and can covalently modify lipids (i.e., lipid peroxidation), which in turn promotes their degradation and the production of two other highly reactive intermediates, malondialdehyde and 4-hydroxy-2-nonenal. Like acetaldehyde, all of these CYP2E1-generated metabolites can form stable, covalent modifications on proteins, lipids, DNA, and other macromolecules (5, 10, 19, 20, 29, 39, 41). Thus one hypothesis for alcohol-induced hepatotoxicity is that the accumulated covalent modifications during chronic alcohol consumption disrupt the normal function of hepatic proteins, lipids and DNA leading to hepatic cell dysfunction and injury. However, the targets for modification and how the modifications alter proper cell function are not well understood.
Our ultimate goal is to identify the molecular basis of alcoholic liver injury by identifying and defining the mechanisms that lead to alcohol-induced defects in hepatic protein trafficking described decades ago in livers from ethanol-fed rats or mice and inferred in patients with alcoholic liver disease (reviewed in Ref. 35). Over a dozen years ago, we discovered that microtubules are more highly acetylated and more stable in ethanol-treated polarized hepatic WIF-B cells and liver slices and in livers from ethanol-fed rats (18). In subsequent years, we have shown that the defects in microtubule-dependent protein trafficking, including post-Golgi delivery of secretory cargo to the cell surface (16), basolateral-to-apical transcytosis (11), and the nuclear translocation of a subset of transcription factors (9) can be explained by increased microtubule acetylation. We have further shown that increased microtubule acetylation may likely lead to impaired vesicle motility by impeding microtubule-based motor translocation and processivity along the filamentous tracks (11). We have also correlated alcohol-induced global protein acetylation with specific impairment of clathrin-mediated internalization (37, 38). In this case, enhanced protein acetylation leads to impaired dynamin recruitment to the necks of invaginated-coated pits resulting in decreased vesicle fission and impaired endocytosis (38).
Our early characterization of microtubule modifications in ethanol-treated cells revealed that hyperacetylation displayed saturable ethanol dose-dependent and time-dependent kinetics (18), implying that this posttranslational modification was ultimately dependent on ethanol metabolism. More direct evidence for the metabolism dependence of microtubule acetylation and increased stability came from experiments using 4-methyl pyrazole (4-MP; an ADH inhibitor that leads to decreased acetaldehyde levels; Refs. 30, 31) and cyanamide (an aldehyde dehydrogenase inhibitor that leads to increased acetaldehyde levels; Ref. 23). 4-MP coincubation prevented tubulin acetylation whereas cyanamide potentiated the effect indicating that ADH-mediated ethanol metabolism is indeed required for increased microtubule acetylation and stability and that these effects are likely mediated by acetaldehyde (18). Since then, similar studies with 4-MP have also shown that impaired secretion, nuclear translocation, and clathrin-mediated internalization are also dependent on ADH-mediated ethanol metabolism (9, 16, 38). To date, the role of CYP2E1-mediated ethanol metabolites or reactive oxygen species (ROS) on microtubule acetylation or on these alcohol-induced protein trafficking defects has not been examined.
The purpose of these studies was to determine if CYP2E1-mediated alcohol metabolism is required for alcohol-induced microtubule acetylation and associated defects in microtubule-dependent protein trafficking in fully differentiated, polarized, hepatic WIF-B cells (34). Importantly, we determined that the WIF-B cells express endogenous ADH and CYP2E1 and exhibit ADH, ALDH, and CYP2E1 activities comparable with intact hepatocytes (24, 33). They produce a time- and dose-dependent increase in acetaldehyde, ROS, and oxidized proteins upon ethanol exposure, also comparable with that observed in intact hepatocytes (24, 33). The WIF-B cells display other aspects of alcohol-induced liver injury including increased triglyceride accumulation (e.g., fatty liver) and an increased lactate/pyruvate redox ratio (24, 33) and exhibit alcohol-induced impairments in protein trafficking as described in situ or in isolated hepatocytes (8, 16, 18, 38) such that they serve as an excellent model system for the studies described here. The approach was straightforward; we coincubated cells with ethanol and with two well-described and long-used pharmacological agents: diallyl sulfide (DAS; a CYP2E1 inhibitor in use for almost 30 yr; Refs. 3, 4) or N-acetyl cysteine (NAC; a glutathione precursor used as an antioxidant first described in 1960; Refs. 28, 40, 45). For these studies, we used the same experimental design we first described and characterized in 2009 (24) and assayed for selected protein-trafficking steps. Confirmatory experiments were performed in ethanol-exposed HepG2 cells expressing only ADH or only CYP2E1. Finally, quantitative RT-PCR was used to determine if impaired STAT5B activation and nuclear translocation blunted growth hormone (GH)-mediated gene expression.
MATERIALS AND METHODS
Reagents and antibodies.
DAS, NAC, and monoclonal antibodies against α-tubulin or acetylated α-tubulin were purchased from Sigma-Aldrich (St. Louis, MO). HEPES was purchased from HyClone (Logan, UT). Alexa-conjugated secondary antibodies were purchased from Life Technologies (Carlsbad, CA). Antibodies against STAT5B (C-17) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-pSTAT5B (C7E5), Jak2 (D2e12), and pJak2 (C80) antibodies were purchased from Cell Signaling Technologies (Beverly, MA). Recombinant GH was purchased from Shenandoah Biotechnology (Warwick, PA). Polyclonal antibodies against asialoglycoprotein receptor (ASGP-R) and rat serum albumin were generously provided by Dr. A. Hubbard (Johns Hopkins University School of Medicine, Baltimore, MD).
Cell culture.
WIF-B cells were grown in a humidified 7% CO2 incubator at 37°C as described previously (34). Briefly, cells were grown in F12 medium (Sigma-Aldrich), pH 7.0, supplemented with 5% fetal bovine serum (Gemini, Woodland, CA), 10 μM hypoxanthine, 40 nM aminopterin, and 1.6 μM thymidine. Cells were seeded onto glass coverslips at 1.3 × 104 cells/cm2 and grown for 8–12 days until they reached maximum polarity. Cells were treated on day 7 with 50 mM ethanol in the absence or presence of 100 μM DAS or 5 mM NAC in medium buffered with 10 mM HEPES, pH 7.0 for 72 h as described previously (32). On day 9, cells were reincubated in fresh buffered medium in the continued absence or presence of alcohol and/or the pharmacological agents. VA-13 and E47 cells (provided by Dr. D. Clemens, University of Nebraska Medical Center, Omaha, NE) were grown in a 5% CO2 incubator at 37°C in DMEM containing 10% FBS and 400 μg/ml of zeocin or G418 as described previously (7). In general, cells were treated on day 2 with 50 mM ethanol for 24 h in medium buffered with 10 mM HEPES, pH 7.0. For some experiments, cells were additionally treated with 50 nM (1.11 ng/ml) GH for up to 4 h at 37°C as indicated (9).
Immunofluorescence microscopy.
In general, WIF-B, E47 or VA-13 cells were fixed on ice with PBS containing 4% paraformaldehyde for 1 min and permeabilized with methanol for 10 min as described previously (14). To detect acetylated tubulin, cells were fixed and permeabilized with methanol at −20°C for 5 min. Cells were processed for indirect immunofluorescence using anti-α-tubulin (1:400), anti-acetylated-α-tubulin (1:250), anti-STAT5B (1:300), anti-pSTAT5B (1:100) monoclonal antibodies (Sigma-Aldrich), or anti-ASGP-R (1:1000) polyclonal antibodies. Alexa-488 or 568-conjugated secondary antibodies (Molecular Probes, Eugene, OR) were used at 5 μg/ml. Labeled cells were visualized at room temperature by epifluorescence with an Olympus BX60 Fluorescence Microscope (OPELCO, Dulles, VA) using an UPlanFl ×60/NA 1.3, phase 3, oil immersion objective. Images were taken with an HQ2 CoolSnap digital camera (Roper Scientific) and Metamorph Imaging software (Molecular Devices, Sunny Vale, CA). Adobe Photoshop (Adobe Systems, Mountain View, CA) was used to process images and to compile figures. To quantitate the relative distributions of STAT5B or phospho-STATB, random fields were visualized by epifluorescence and digitized. From micrographs, the average pixel intensity of selected regions of interest (ROI) placed in the nucleus or cytoplasm of the same cell was measured using the Measure ROI tool of the ImageJ software (National Institutes of Health) as described previously (25, 27). The ratio of nuclear vs. cytoplasmic fluorescence intensities was determined. Typically, values were determined from at least 3 independent experiments where 5–10 random fields were acquired for each condition that contained 15–30 polarized cells each. To monitor oxidative stress, WIF-B, E47, or VA-13 cells were incubated with CellROX Green Reagent (Invitrogen) according to the manufacturer’s instructions and fixed with 3.7% paraformaldehyde at room temperature. Labeled cells were visualized as described above.
Immunoblotting.
Proteins were separated using SDS-PAGE, transferred to nitrocellulose and immunoblotted with antibodies against total STAT5B (1:5,000–10,000) or Jak2 (1:1,000) diluted in PBS containing 5% (wt/vol) milk and 0.1% (vol/vol) Tween-20. Antibodies specific to phospho-STAT5B or phospho-Jak2 were diluted in TBS containing 1% (wt/vol) BSA and 0.1% (vol/vol) Tween-20 (both 1:1000). Incubations were performed overnight at 4°C. Immunoreactivity was detected using enhanced chemiluminescence (PerkinElmer, Crofton, MD). Relative expression levels were determined by densitometric analysis of immunoreactive bands.
Secretion assays.
Control or treated cells were rinsed five times with prewarmed serum-free medium and then reincubated in serum-free medium. At 0, 5, 15, or 30 min after reincubation, aliquots of media were collected and analyzed for albumin secretion by immunoblotting as described previously (16). The cell lysates were collected by solubilization directly into SDS-PAGE sample buffer. Samples were processed for Western blotting and densitometric analysis of immunoreactive bands. The albumin levels secreted for control or DAS- or NAC-treated cells at 15 min were set to 100% to which the other corresponding values were normalized.
Quantitative RT-Jak/STAT signaling pathway PCR arrays.
The quantitative RT-PCR analysis was performed by SABiosciences (Frederick, MD). Briefly, mature RNA was extracted from control or ethanol-treated WIF-B cells additionally treated with GH for 0, 30, or 240 min. RNA quality was spectrophotometrically determined. Only RNA that met with the appropriate quality standards was further reverse transcribed. The resultant cDNA was used to probe RT2 Profiler Rat Jak/STAT signaling PCR Arrays (96-well format) purchased from Qiagen (Ventura, CA) along with RT2 Sybr Green qPCR Mastermix. Fold changes in gene expression were calculated using the ΔΔ-cycle threshold (ΔΔCT) method. The ΔCT was calculated between the gene of interest and an average of reference house-keeping genes. The ΔΔCT was calculated by taking the difference of the ΔCT for the test group and the ΔCT for the control group. Fold changes were calculated using the 2ΔCT formula. The CT cut-off was set to 35. Three independent experiments were performed for each comparison. Values were considered only if similarities in fold change were observed in at least two out of the three experiments.
RESULTS
Ethanol-induced protein acetylation and corresponding defects in protein trafficking are not mediated by ROS or CYP2E1 ethanol metabolites.
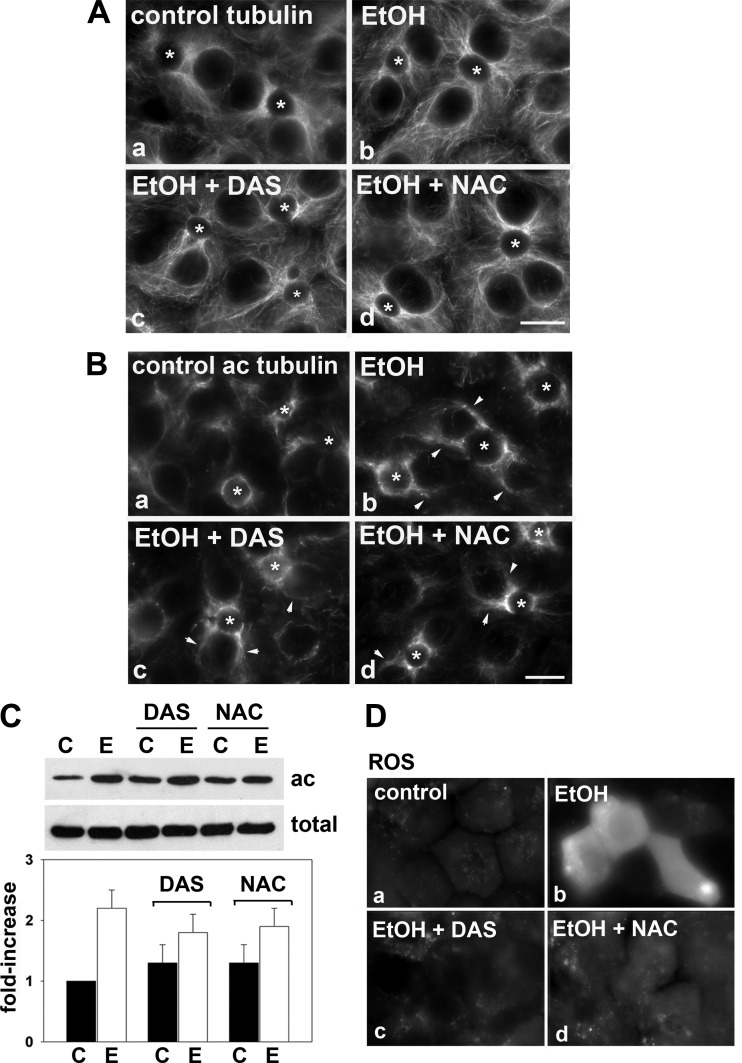
Microtubules emanate from centrosomal structures located near the canalicular surfaces in WIF-B cells as they do in hepatocytes in situ (Fig. 1Aa). Although this orientation is maintained in ethanol-treated cells, the microtubules undergo a large morphological change and become much shorter, more discrete, and more gnarled (Fig. 1Ab). Because this gnarled phenotype is a hallmark of stable microtubules (22, 26, 42), we stained ethanol-treated cells for acetylated α-tubulin, a posttranslational modification present on stable microtubules. As shown in Fig. 1B and as we have shown previously (18), the specific anti-acetylated-α-tubulin antibodies recognized microtubules at or near the bile canaliculi in both control and ethanol-treated cells. However, the staining was more intense in ethanol-treated cells and was observed on microtubules more distal from the bile canaliculi (Fig. 1Bb, marked with arrows).
Fig. 1.
Addition of diallyl sulfide (DAS) or N-acetyl cysteine (NAC) does not prevent ethanol-induced microtubule acetylation. A and B: WIF-B cells were treated in the absence (a) or presence of 50 mM ethanol (EtOH) for 72 h (b–d) in the additional presence of 100 μM DAS (c) or 5 mM NAC (d) as indicated. In A, cells were labeled for total α-tubulin and in B, for acetylated α-tubulin. *Selected bile canaliculi. Arrows in Bb–Bd indicate the enhanced acetylated microtubule labeling present in long filaments emanating from the bile canaliculi. Bar = 10 μm. C: WIF-B cells were treated in the absence (C) or presence of 50 mM ethanol (E) in the additional presence of 100 μM DAS or 5 mM NAC as indicated. Cells were immunoblotted for acetylated (ac) or total α-tubulin as indicated. The fold increase in tubulin acetylation was determined by densitometric analysis of immunoreactive bands normalized to total tubulin levels. The untreated control values were set to 1.0. Values represent the means ± SE from at least 3 independent experiments. D: WIF-B cells were treated in the absence (a) or presence of 50 mM ethanol (b) for 72 h. Ethanol-treated cells were coincubated with 100 μM DAS (c) or 5 mM NAC (d) and labeled with CellROX Green reagent to monitor oxidative stress by epifluorescence. Only ethanol-exposed cells (b) exhibited reactive oxygen species (ROS).
These morphological changes persisted in ethanol-treated cells that were coincubated with DAS or NAC. Similarly shorter, more discrete, and gnarled microtubules were observed as in cells treated with ethanol alone (Fig. 1, Ac and Ad). Also as in ethanol-treated cells, increased levels of acetylated tubulin staining were observed in cells treated with either DAS or NAC that were more distal from the canalicular surface (Fig. 1, Bc and Bd, marked with arrows). These results were confirmed biochemically by immunoblotting cell lysates with specific antibodies to detect the total α-tubulin pool or the acetylated population (Fig. 1C). Importantly, no change in the total amount of α-tubulin was detected in any of the treated samples. Also importantly, DAS or NAC treatment alone had no significant effect on acetylated tubulin levels. Consistent with the morphological analysis and our previous results (18), ethanol treatment induced α-tubulin acetylation greater than twofold over control levels (Fig. 1C). Also consistent with the morphological analysis, additional treatment with DAS or NAC did not prevent the ethanol-induced tubulin acetylation. In both cases, approximately twofold greater acetylation levels were observed. Together, these results indicate that ethanol-induced microtubule acetylation is not mediated by ROS or CYP2E1-mediated metabolites and further implicates the reactive ethanol metabolite acetaldehyde as the important mediator of enhanced acetylation (see discussion).
To confirm the effects of DAS and NAC on ethanol-treated WIF-B cells as we have previously described (24), we monitored oxidative stress by labeling cells with CellROX Green reagent. As expected, addition of ethanol led to a significant increase in labeling indicative of enhanced oxidative stress (Fig. 1Db). Also as expected, virtually no oxidative stress was observed in cells additionally treated with DAS (Fig. 1Dc) or NAC (Fig. 1Dd), indicating they effectively prevented ROS production thereby confirming our system.
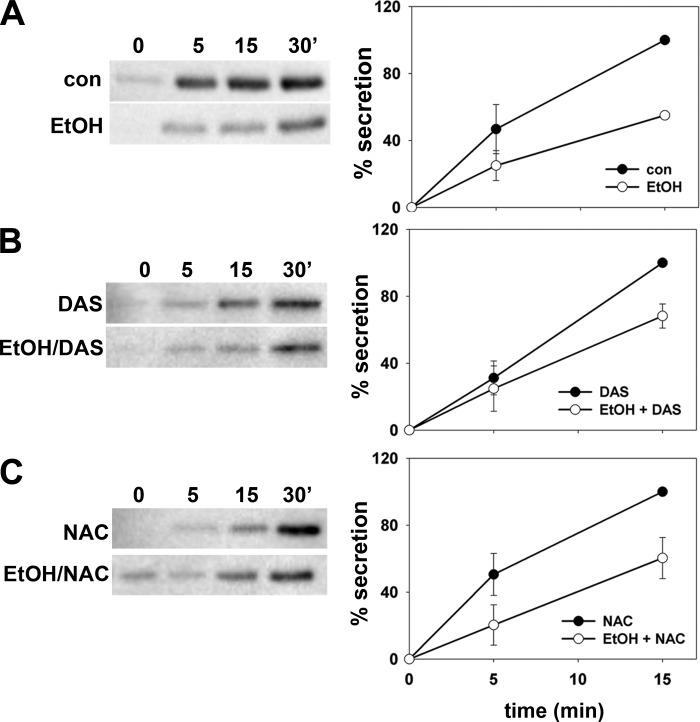
An ethanol-induced defect in hepatic secretion has been well established in animal models and inferred in patients with alcoholic liver disease (reviewed in Ref. 35). Our more recent studies have provided strong evidence that ethanol-induced microtubule acetylation is at the heart of the impairment likely by interfering with vesicle-mediated surface delivery along microtubule tracks (11, 16). If enhanced microtubule acetylation can indeed explain impaired secretion, the prediction is that DAS or NAC treatment should not prevent the ethanol-induced defect. To measure secretion, we monitored albumin release into the medium by immunoblotting with an assay we developed (16). As shown in Fig. 2A and as we have previously described, decreased basolateral albumin release was observed in ethanol-treated cells (Fig. 2A, left). When quantitated, we found that secretion was decreased to only 60% of control levels (62.9 ± 11.7%) in ethanol-treated cells consistent with our earlier findings (16). In ethanol-treated cells coincubated with DAS (Fig. 2B) or NAC (Fig. 2C), similar levels of impaired albumin secretion were observed (68.2 ± 7.2 and 60.4 ± 12.3 of control, respectively). These results indicate that impaired secretion is not mediated by ROS or CYP2E1-mediated metabolites and further confirm that enhanced microtubule acetylation can explain the defect.
Fig. 2.
Addition of DAS or NAC does not prevent the ethanol-induced impairment of secretion. A: WIF-B cells were treated in the absence or presence of 50 mM ethanol (EtOH) for 72 h as indicated. B: cells were treated with 100 μM DAS in the absence or presence of ethanol. C: cells were treated with 5 mM NAC were incubated in the absence or presence of ethanol. Cells were rinsed 5 times with prewarmed serum-free medium and then reincubated in serum-free medium. At 0, 5, 15, and 30 min after reincubation, aliquots of media were collected and analyzed for albumin secretion by immunoblotting (left). The percent albumin secreted was calculated from densitometric analysis of immunoreactive bands and plotted in the graphs shown on the right. Values are expressed as the means ± SE from 3 independent experiments.
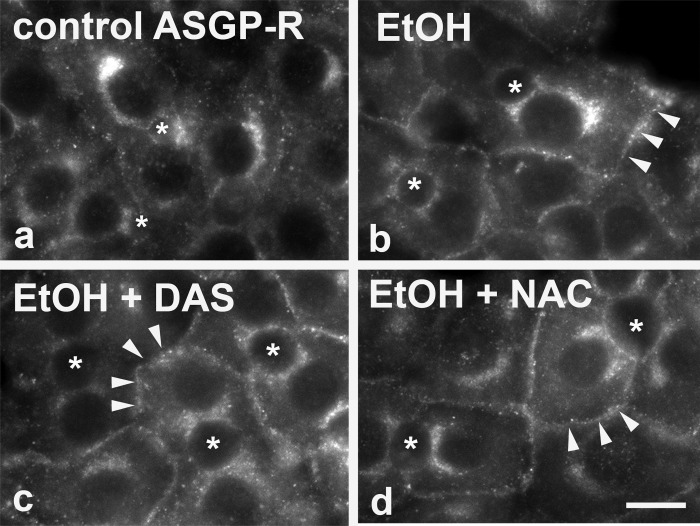
From a proteomics screen performed in livers from ethanol-fed rats, we determined that ethanol led to global increases in protein acetylation (including cortactin, actin, and tubulin) (37) and that this enhanced acetylation strongly correlated with impaired clathrin-mediated internalization (38). If enhanced protein acetylation can explain impaired internalization, we further predicted that DAS and NAC should not prevent the ethanol-induced defect in endocytosis. To monitor changes in internalization, we immunolabeled control and treated cells for asialoglycoprotein receptor (ASGP-R), a receptor known to be internalized by clathrin-mediated endocytosis. Consistent with our earlier reports (38), ethanol treatment led to the redistribution of ASGP-R from intracellular early endosomes to the cell surface into discrete puncta that we have identified as invaginated-coated pits (38) (Fig. 3b). Similar redistributions were observed in cells coincubated with DAS (Fig. 3c) or NAC (Fig. 3d) supporting our hypothesis thereby further indicating that impaired clathrin-mediated internalization is also not mediated by ROS or CYP2E1-mediated metabolites.
Fig. 3.
Addition of DAS or NAC does not prevent the ethanol-induced impairment of clathrin-mediated endocytosis. WIF-B cells were treated in the absence (a) or presence of 50 mM ethanol (EtOH) for 72 h (b–d) in the additional presence of 100 μM DAS (c) or 5 mM NAC (d) as indicated. Cells were labeled for asialoglycoprotein receptor (ASGP-R). *Selected bile canaliculi. Arrows in b–d indicate the enhanced ASGP-R basolateral staining observed in treated cells. Bar = 10 μm.
Decreased Jak2/STAT5B activation requires CYP2E1-mediated ethanol metabolism.
We have also strongly correlated ethanol-induced microtubule acetylation with impaired nuclear translocation of a subset of transcription factors (9), so the next prediction was that DAS and NAC addition should also not prevent this defect. As we have shown previously, ethanol treatment led to the cytosolic redistribution of STAT5B at steady state (Fig. 4Ac) implying impaired nuclear translocation. Semiquantitation of STAT5B nuclear vs. cytoplasmic fluorescence intensities confirmed these observations. At steady state (0 min), the ratio of nuclear to cytoplasmic STAT5B staining was significantly decreased in ethanol-treated cells (Fig. 4B, left). Upon addition of GH for only 5 min, STAT5B nearly completely redistributed to the nucleus in control cells, but this translocation was blunted in ethanol-treated cells (Fig. 4A, compare Ab and Ad). Although less pronounced than at steady state, the ratio of nuclear to cytoplasmic fluorescence intensity was also decreased in ethanol-treated cells after GH addition (Fig. 4B, right).
Fig. 4.
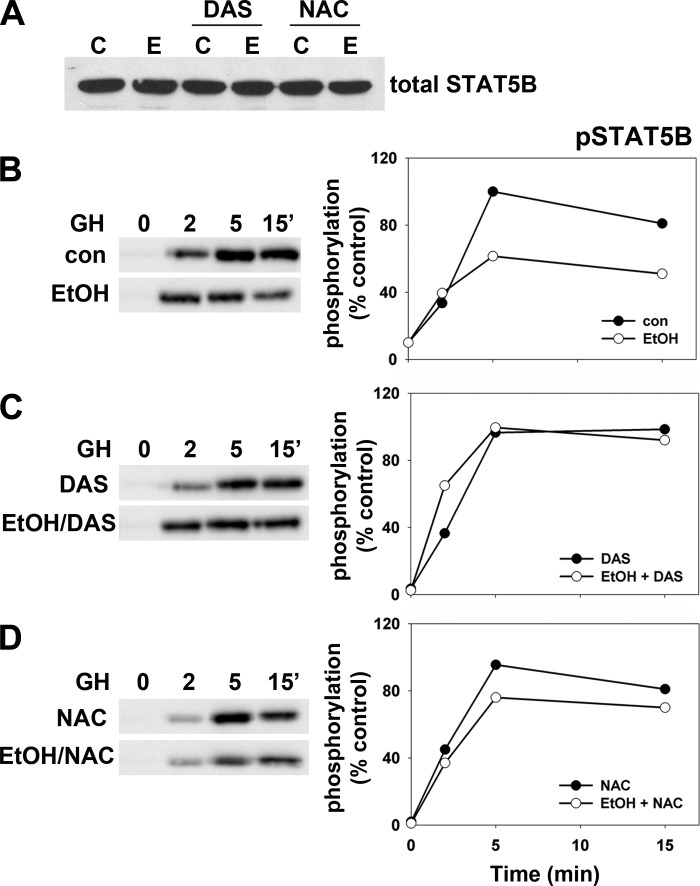
Addition of DAS or NAC prevents the ethanol-induced impairment of STAT5B nuclear translocation. A: WIF-B cells were treated in the absence (a and b) or presence of 50 mM ethanol (EtOH) for 72 h (c–h) in the additional presence of 100 mM DAS (e and f) or 5 mM NAC (g and h) as indicated. Cells were incubated for 0 or 5 min with growth hormone (GH) and labeled for total STAT5B. Note the enhanced cytosolic staining in cells treated only with ethanol (c and d). Bar = 10 μm. B: WIF-B cells were treated in the absence (C) or presence of 50 mM ethanol (E) in the additional presence of 100 μM DAS or 5 mM NAC as indicated. The ratio of nuclear-to-cytoplasmic fluorescence intensities was calculated for STAT5B at steady state (0 min; left) or after 5 min of GH addition (right). Values represent the means ± SE from at least 3 independent experiments.
Somewhat surprisingly, in ethanol-treated cells coincubated with DAS or NAC, the distributions of STAT5B were indistinguishable from control cells with little cytosolic labeling observed at steady state (Fig. 4, Ae and Ag) and with nearly identical ratios of nuclear to cytoplasmic labeling to that of control (Fig. 4B, left). Similar results were observed for STAT5B distributions after GH addition. Within 5 min, near complete nuclear translocation was observed in DAS- and NAC-treated cells (Fig. 4, Af and Ah) and ratios were restored to control levels (Fig. 4B, right). Similar results were obtained when labeling for activated and phosphorylated STAT5B (pSTAT5B) distributions after GH addition. When quantitated, all ratios of nuclear to cytosolic staining were similar to control levels except for cells treated with ethanol only, which were decreased to 75% of control (Table 1).
Table 1.
Addition of DAS or NAC prevents the ethanol-induced impairment of pSTAT5B nuclear translocation
| Treatment | Nuclear/Cytoplasmic Fluorescence Intensity, %Control |
|---|---|
| Control | 100.0 ± 0.0 |
| DAS | 96.0 ± 3.6 |
| NAC | 103.7 ± 8.2 |
| EtOH | 76.3 ± 1.3 |
| EtOH + DAS | 111.0 ± 16.5 |
| EtOH + NAC | 98.0 ± 6.1 |
Values represent the means ± SE from at least 3 independent experiments. WIF-B cells were treated in the absence or presence of 50 mM ethanol (EtOH) for 72 h in the additional presence of 100 µM diallyl sulfide (DAS) or 5 mM N-acetyl cysteine (NAC) as indicated. Cells were incubated for 5 min with growth hormone (GH) and labeled for phospho-STAT5B (pSTAT5B). The ratio of nuclear-to-cytoplasmic fluorescence intensities was calculated for pSTAT5B and the untreated control value set to 100%.
From our previous studies using pharmacological agents that promote global protein acetylation (trichostatin A) or tubulin-specific acetylation (taxol at low concentrations) in the absence of ethanol, we determined that microtubule acetylation impairs the microtubule-dependent translocation of STAT5B to the nucleus after its activation by Jak2 (9). However, these agents did not impair Jak2 or STAT5B activation/phosphorylation after GH addition as was observed in ethanol-treated cells (9). Thus, to reconcile the disparate results described above, we examined the events in STAT5B signaling that are independent of microtubules yet impaired by alcohol administration.
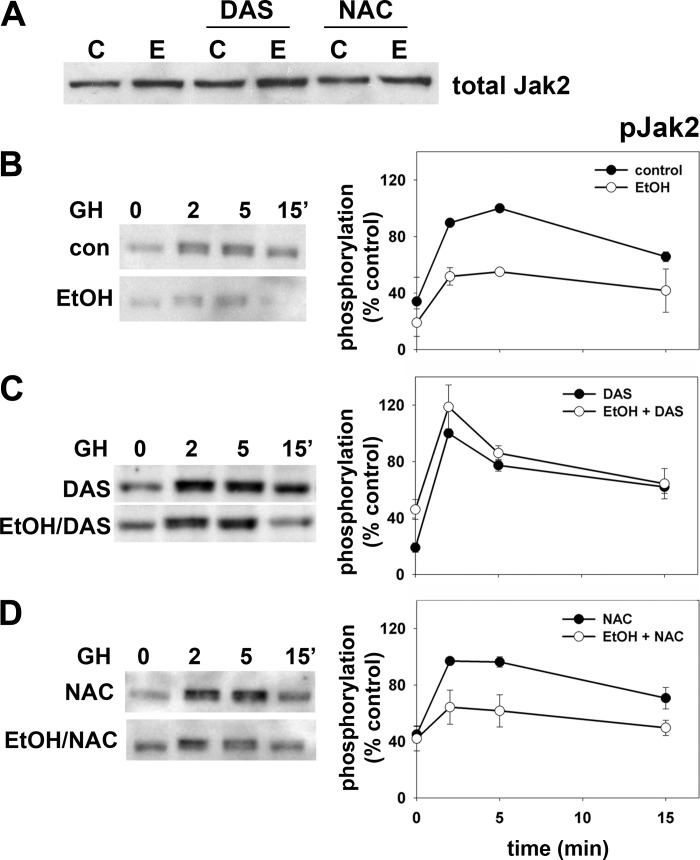
Upon GH addition, the receptor-associated tyrosine kinase Jak2 is rapidly activated by cross phosphorylation and in turn phosphorylates the GH receptor (GH-R). The receptor/Jak2 complex then recruits and phosphorylates STAT5B thereby activating it allowing for subsequent microtubule-mediated nuclear translocation. To first examine Jak2 activation, we immunoblotted cell lysates with antibodies specific for total or phospho-Jak2 (pJak2) in control or treated cells. Importantly, addition of DAS or NAC either alone or in combination with ethanol did not change total levels of Jak2 expression (Fig. 5A). In control cells, Jak2 was rapidly phosphorylated reaching peak activation within 2–5 min after GH addition returning to basal levels by ~15 min (Fig. 5B, top left). As we have reported earlier (9), ethanol treatment significantly blunted peak Jak2 activation at 2 min (Fig. 5B, bottom left) to half that seen in control cells (Fig. 5B, right). In ethanol-treated cells with added DAS, the impairment is nearly completely prevented (Fig. 5C, left) with activation kinetics mirroring those observed in control cells (Fig. 5C, right) whereas NAC addition partially prevented the Jak2 phosphorylation defect (Fig. 5D).
Fig. 5.
Addition of DAS or NAC prevents the ethanol-induced impairment of Jak2 phosphorylation. A: WIF-B cells were treated in the absence or presence of 50 mM ethanol in the additional presence of 100 μM DAS or 5 mM NAC as indicated. Samples were immunoblotted for total Jak2 expression. B: WIF-B cells were treated in the absence or presence of 50 mM ethanol (EtOH) for 72 h as indicated. C: cells were treated with 100 μM DAS were treated in the absence or presence of ethanol. D: cells were treated with 5 mM NAC were incubated in the absence or presence of ethanol. Cells were incubated for up to 15 min with GH and lysates immunoblotted for phospho-Jak2 (pJak2) (left). Immunoreactivity was measured by densitometry, normalized to total Jak2 levels and plotted as the percentage of maximal phosphorylation (right). Values represent the means ± SE from 3 independent experiments.
To examine subsequent STAT5B activation by Jak2, we immunoblotted control and treated lysates with antibodies specific for total or pSTAT5B. Importantly, addition of DAS or NAC alone or in combination with ethanol did not change total levels of STAT5B expression (Fig. 6A). In control cells and as expected, STAT5B activation lags behind that of Jak2 with peak phosphorylation observed between 5 and 15 min (Fig. 6B, top left). As we have previously shown (9), ethanol treatment significantly blunts peak STAT5B activation at 5 min (Fig. 6B, bottom left) to ~60% of that seen in control cells (Fig. 6B, right). In contrast, in ethanol-treated cells with added DAS or NAC, the impairment was nearly completely prevented (Fig. 6, C and D, left) with activation kinetics mirroring those observed in control cells (Fig. 6, C and D, right). Together these results indicate that the microtubule-independent activation of Jak2 and STAT5B is ultimately impaired by a CYP2E1-mediated mechanism.
Fig. 6.
Addition of DAS or NAC prevents the ethanol-induced impairment of STAT5B phosphorylation. A: WIF-B cells were treated in the absence or presence of 50 mM ethanol in the additional presence of 100 μM DAS or 5 mM NAC as indicated. Samples were immunoblotted for total STAT5B expression. B: WIF-B cells were treated in the absence or presence of 50 mM ethanol (EtOH) for 72 h as indicated. C: cells were treated with 100 μM DAS were treated in the absence or presence of ethanol. D: cells were treated with 5 mM NAC were incubated in the absence or presence of ethanol. Cells were incubated for up to 15 min with GH and lysates immunoblotted for phospho-STAT5B (pSTAT5B) (left). Immunoreactivity was measured by densitometry, normalized to total STAT5B levels and plotted as the percentage of maximal phosphorylation (right). Results from a representative experiment from at least 3 independent experiments are shown for each.
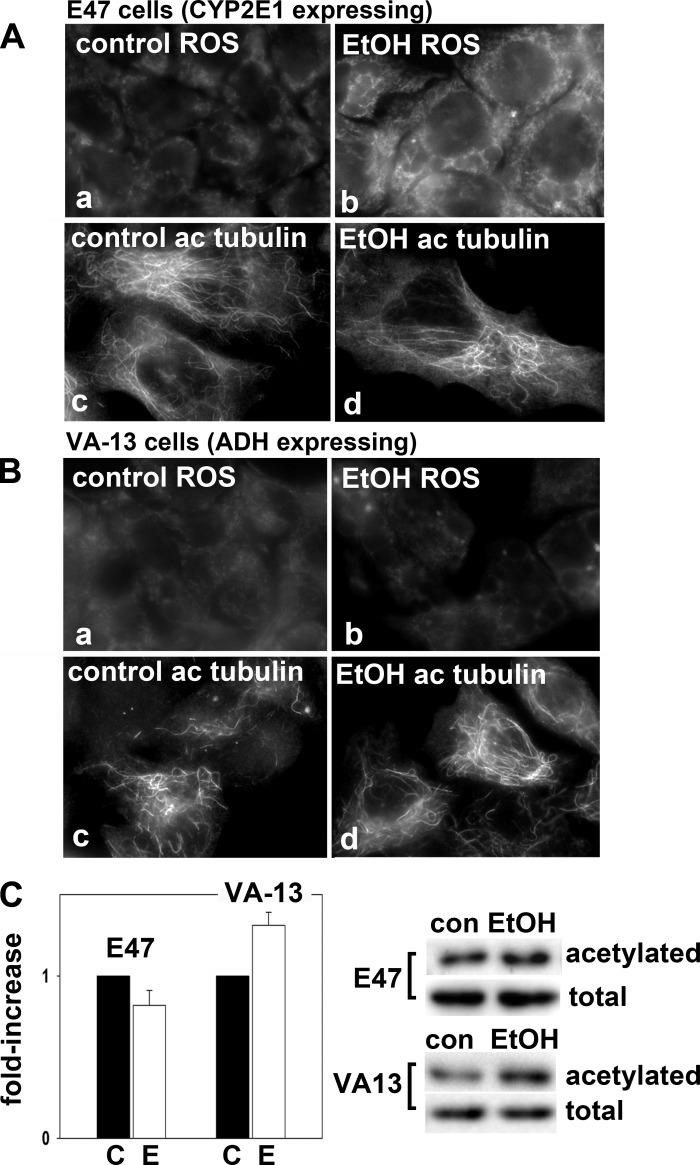
To confirm the metabolite-specific responses to alcohol exposure, we continued our studies in the well-characterized HepG2-derived E47 and VA-13 cell lines that stably express either CYP2E1 or ADH, respectively (reviewed in Ref. 6). As predicted and as previously shown, oxidative stress was observed in ethanol-treated E47 cells (Fig. 7Ab) confirming CYP2E1-mediated ethanol metabolism was occurring. In contrast, but as also predicted, oxidative stress was not observed in VA-13 cells in the absence (Fig. 7Ba) or presence of ethanol (Fig. 7Bb) consistent with only ADH expression in these cells. To further confirm our inhibitor studies, we also labeled cells for acetylated tubulin. In these cell lines, microtubules display the characteristic arrangement found in nonpolarized cells emanating from a juxta-nuclear microtubule organizer center. As predicted, only the VA-13 cells displayed enhanced microtubule acetylation morphologically (Fig. 7Bd) and biochemically (Fig. 7C). Although the morphological changes were not as dramatic as observed for WIF-B cells, when examined closely, enhanced labeling is observed for each cell both with respect to overall intensity and to numbers of labeled filaments (compare Fig. 7, Bc and Bd). When quantitated, tubulin acetylation was enhanced ~1.5-fold in ethanol-treated VA-13 cells. Although more modest than in WIF-B cells, the increase was reproducible and expected as these cells are less differentiated than the fully polarized WIF-B cells. In contrast, but also as predicted, no increased tubulin acetylation was observed in E47 cells morphologically (compare Fig. 7, Ac and Ad) or biochemically (Fig. 7C).
Fig. 7.
Only CYP2E1-expressing cells display ethanol-induced ROS while only alcohol dehydrogenase (ADH)-expressing cells display alcohol-induced microtubule acetylation. E47 (A) or VA-13 (B) cells were treated in the absence (a and c) or presence of 50 mM ethanol (b and d) for 24 h. Cells were labeled for CellROX Green to monitor oxidative stress (a and b) or immunolabeled for acetylated tubulin (c and d). C: cells were immunoblotted for acetylated or total α-tubulin as indicated. The fold increase in tubulin acetylation was determined by densitometric analysis of immunoreactive bands normalized to total tubulin levels. The untreated control values were set to 1.0. Values represent the means ± SE from at least 3 independent experiments.
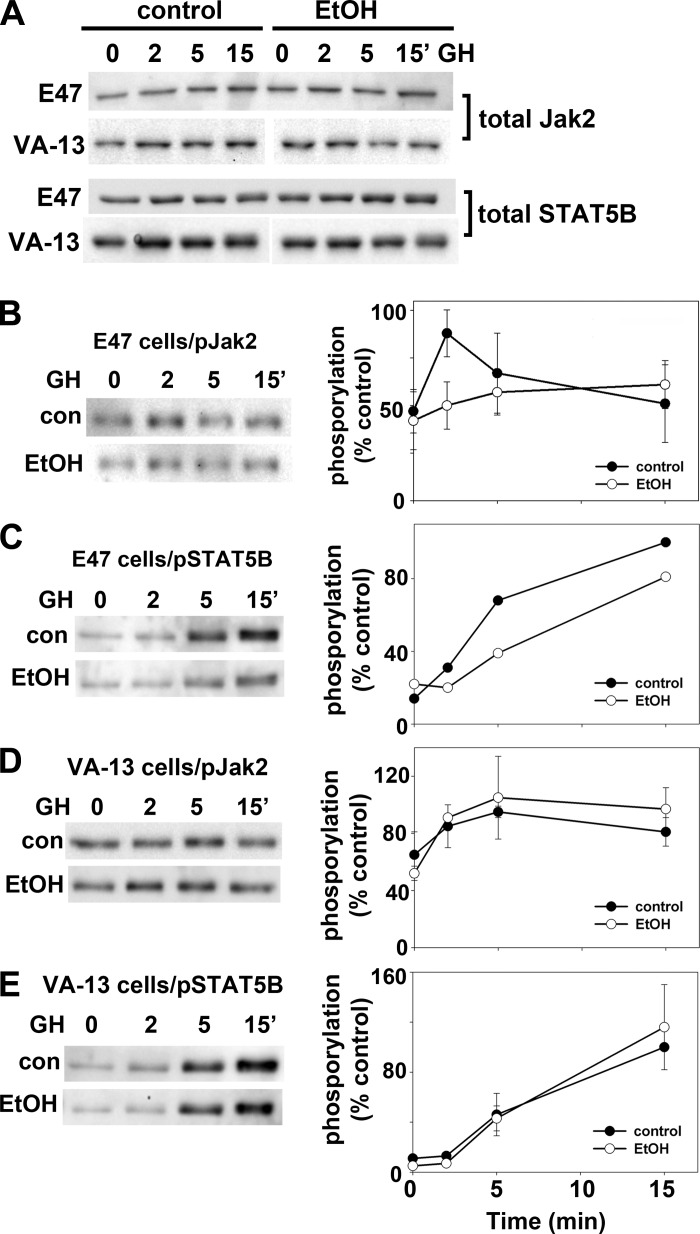
To confirm that CYP2E1 ethanol metabolism specifically impairs Jak2 and STAT5B activation, we immunoblotted lysates from E47 or VA-13 cells treated with GH for total and phospho-Jak2 or STAT5B. Importantly, addition of ethanol did not change total levels of Jak2 or STAT5B expression in either cell type (Fig. 8A). In control E47 cells, Jak2 was rapidly phosphorylated reaching peak activation within 2 min after GH addition returning to basal levels by ~15 min (Fig. 8B). Ethanol treatment significantly blunted peak Jak2 activation at 2 min to nearly half that seen in control cells (Fig. 8B). In control E47 cells and as expected, STAT5B activation lagged behind that of Jak2 with peak phosphorylation varying between 5 or 15 min that was blunted in ethanol-treated cells (Fig. 8C). Although a more modest impairment was observed in E47 cells than in WIF-B cells, when quantitated, Jak2 phosphorylation was reproducibly decreased to ~80% of control (82.0 ± 4.0%, P ≤ 0.004) after 15 min of GH exposure. In control VA-13 cells, Jak2 activation was also observed after GH addition but was somewhat delayed peaking at 5 min (Fig. 8D). No change in Jak2 phosphorylation was observed in ethanol-treated VA-13 cells, and the activation kinetics mirrored those observed in control cells (Fig. 8D). Similarly, STAT5B phosphorylation displayed delayed activation in both control and ethanol-treated VA-13 cells, but no apparent difference in activation kinetics was observed (Fig. 8E). Together these results confirm our findings that that the microtubule-independent activation of Jak2 and STAT5B is ultimately impaired by a CYP2E1-mediated mechanism.
Fig. 8.
Only CYP2E1-expressing cells display ethanol-induced impairments in Jak2 and STAT5B activation. A: E47 or VA-13 cells were treated in the absence or presence of 50 mM ethanol for 24 h. Cells were incubated for up to 15 min with GH and lysates immunoblotted for total Jak2 and STAT5B as indicated. A representative immunoblot from at least 3 independent experiments is shown for each. E47 (B and C) and VA-13 (D and E) cells were treated in the absence or presence of 50 mM ethanol (EtOH) for 24 h as indicated. Cells were incubated for up to 15 min with GH and lysates immunoblotted for phospho-Jak2 (B and D) or phospho-STAT5B as indicated (C and E). Immunoreactivity was measured by densitometry, values were normalized to total Jak2 or STAT5B levels, and maximum values were set to 100%. Values represent the means ± SE from at least 3 independent experiments in B–E. Due to variability in peak activation time, one representative immunoblot and graph out of 4 independent experiments are shown in C.
Jak/STAT signaling is blunted and not sustained in ethanol-treated hepatic cells.
These results indicate that ethanol metabolism leads to impaired Jak2/STAT5B signaling at two steps: Jak2/STAT5B activation upon GH binding to its receptor and subsequent microtubule-dependent STAT5B nuclear translocation. To determine if these impairments impact hepatocyte-specific GH-mediated signaling, we performed quantitative RT-PCR on 96-well arrays of 84 known Jak/STAT signaling pathway genes and 4 house-keeping genes (for normalization) using cDNA prepared from control or ethanol-treated WIF-B cells. We first assayed for changes in steady state gene expression in ethanol-treated cells relative to control. As shown in Table 2, 29 genes showed reproducibly altered expression in ethanol-treated cells with over 70% (21 of 29) of these genes showing decreased expression suggesting GH/Jak2/STAT5-mediated signaling is blunted in ethanol-treated cells. Of these downregulated genes, over 70% (15 of 21) are known STAT5 target genes or code for proteins known to interact with/activate GH-R, Jak2, or STAT5 (2, 12, 13, 17), confirming the validity of our screen. Interestingly, the changes in gene expression associated with the cell cycle are all consistent with decreased cell division. Also, the identification of the many cytokines, cytokine-receptors, and other immune response modulators is consistent with known alterations in inflammation associated with alcoholic liver disease.
Table 2.
Steady-state Jak/STAT signaling is blunted in ethanol-treated cells
| Gene | n | Fold Change | Comments |
|---|---|---|---|
| Jak kinases | |||
| Jak1 | 3 | −1.50 ± 0.09† | |
| Jak2 | 3 | −1.57 ± 0.19† | Activated by growth hormone |
| Jak3 | 3 | −1.40 ± 0.18† | |
| Tyk2 | 2 | −1.57† | |
| STATs | |||
| Stat3 | 3 | −1.42 ± 0.25† | |
| Stat5 | 3 | −1.72 ± 0.35† | Activated by growth hormone |
| Jak/STAT receptors | |||
| Csfr1 | 3 | 1.80 ± 0.53 | Colony-stimulating factor receptor; ligand binding activates Jak2/STAT5 |
| Epor | 3 | 5.44 ± 2.92 | Erythropoietin-receptor; hepatoprotective; ligand binding activates Jak2/STAT5 |
| Prlr | 3 | −2.93 ± 0.91† | Prolactin-receptor; ligand binding activates Jak2/STAT5; can be activated by GH; known STAT5 target gene |
| Nr3c1 | 2 | −1.60† | Glucocorticoid-receptor; coactivator of GH-mediated responses via STAT5 binding |
| Jak/STAT modulators | |||
| Ptpn1 | 2 | 1.49 | Protein tyrosine phosphatase; can interact with GH recetpro, STAT5, and Jak2 |
| Socs2 | 2 | −1.47† | STAT signaling inhibitor molecule; known STAT5 target gene |
| Cytokines, cytokine receptors, and other secreted proteins | |||
| Crp | 2 | 2.39 | C-reactive protein; made in liver is response to inflammation; acute phase response to IL-6 |
| Cxcl9 | 3 | 2.22 ± 0.64 | Chemokine (CXC motif) ligand 9; T cell attractant induced by IFNγ |
| A2M | 3 | −2.58 ± 0.67† | α2-Macroglobulin; inactivates circulating proteases |
| Il20 | 3 | −2.58 ± 0.69† | Interleukin-20 |
| Osm | 2 | −1.89† | Oncostatin M; inhibits proliferation and production of other cytokines; known STAT5 target |
| Ifnar | 2 | −1.50† | Interferon-α receptor; known pan-STAT target |
| Il10ra | 3 | −4.74 ± 2.88† | Interleukin-10 receptor A |
| Other immune response modulators | |||
| Isg15 | 2 | 1.84 | Interferon-induced 15-kDa Protein; Ub-like protein that attaches to lysine |
| B2M | 2 | 1.52 | β2-Microglobulin; major histocompatibility complex class 1 component |
| Cebpb | 3 | −1.53 ± 0.25† | CCAAT-binding protein-β; transcription factor; promotes cytokine expression; can interact with STAT5 in liver |
| Cebpd | 2 | −1.79† | CCAAT-binding protein-δ; transcription factor; promotes cytokine expression; can interact with STAT5 in liver |
| Irf1 | 3 | −1.48 ± 0.17† | Inteferon regulatory factor 1; activates interferon transcription; known STAT5 target gene |
| Irf9 | 3 | −1.69 ± 0.21† | Interferon regulatory factor 9; activates interferon transcription; known pan-STAT target |
| Nfkb | 3 | −1.62 ± 0.23† | NF-κB; proinflammatory transcription factor |
| Cell cycle regulators | |||
| Cdkn1 | 2 | 1.71 | Cyclin-dependent kinase inhibitor 1A/p21; blocks cell cycle progression; known STAT5 target |
| Akt | 2 | −1.70† | Akt/protein kinase B; Ser/Thr kinase; stimulates proliferation |
| Myc | 3 | −1.45 ± 0.22† | Transcription factor that promotes cell cycle entry |
Values represent the means ± SE from 3 independent experiments (n = 3) or the average from 2 (n = 2) as indicated. Brief descriptions of gene functions are listed. Known associations between the gene and GH/STAT5-mediated signaling are also provided. Jak/STAT signaling pathway PCR arrays were probed with cDNAs prepared from WIF-B cells treated in the presence or absence of ethanol (see materials and methods). The fold change in gene expression for the 84 genes on the array in ethanol-treated cells relative to control was determined. Three independent experiments were performed.
Values that indicate those genes with decreased expression.
We next identified the genes that are differentially expressed in control or ethanol-treated cells after acute (30 min) or prolonged (4 h) GH addition. We first identified those genes with differential expression at both time points relative to 0 min. As shown in Table 3, addition of GH for 30 min led to similar changes in Jak/STAT-mediated gene expression in ethanol and control cells (marked with asterisks). Of the 11 genes identified with altered expression in ethanol-treated cells, almost half shared similar changes as those observed for control cells. However, the GH-mediated response in ethanol-treated cells was not sustained. After 4 h, only two genes showed sustained altered expression (Egfr and Ptpn1) in ethanol-treated cells with only one value shared with control cells (Epor). This trend is also apparent when changes in gene expression in ethanol-treated cells are compared with those in control for each time point. As shown in Table 4, GH addition for 30 min transiently restored expression to control levels in the majority of genes (15 of 21) with impaired expression in ethanol-treated cells. However, after 4 h, expression levels returned to steady-state levels (compare values for 0 min and 4 h). Interestingly, four genes showed altered expression levels independent of GH addition in ethanol-treated cells (Prlr, Cebpb, Cebpd, and Irf9), which may reflect other STAT-specific signaling defects. Nonetheless, these results indicate that overall GH/Jak2/STAT5/-mediated signaling is blunted and not sustained in ethanol-treated cells consistent with the defects in activation and translocation we observed.
Table 3.
Long-term GH-induced Jak/STAT signaling is attenuated in ethanol-treated cells
| Gene | n | Fold Change | n | Fold Change |
|---|---|---|---|---|
| 30-min Control | 30-min Ethanol | |||
| A2M | — | 3 | 1.49 ± 0.30 | |
| Cdkn1* | 2 | 1.72 | 2 | 1.74 |
| Csfr1* | 3 | 2.82 ± 0.61 | 3 | 1.93 ± 0.47 |
| Fas | 2 | 2.75 | — | |
| Il2ra | — | 2 | 2.17 | |
| Mcl1 | 2 | 1.85 | — | |
| Socs3* | 2 | 2.06 | 2 | 1.97 |
| Socs5 | — | 2 | 2.19 | |
| Egfr | — | 3 | −1.48 ± 0.41† | |
| Gbp1* | 3 | −2.76 ± 1.04† | 3 | 1.74 ± 0.36 |
| Il10ra | 2 | −3.20† | — | |
| Il20 | 2 | −5.5† | — | |
| Il2rg* | 2 | −3.41† | 2 | −3.72† |
| Pias1 | — | 3 | −1.57 ± 0.29† | |
| Ptpn1 | — | 3 | −1.48 ± 0.39† | |
| 4-h Control | 4-h Ethanol | |||
| Cdkn1 | 3 | 2.24 ± 0.14 | NSC | |
| Csfr1 | 3 | 3.41 ± 1.06 | NSC | |
| Irf1 | 2 | 1.7 | NSC | |
| Junb | 2 | 2.12 | NSC | |
| Egfr | — | 2 | −2.06† | |
| Epor* | 2 | −3.10† | 3 | −2.82 ± 0.46† |
| Il10ra | 2 | −4.33† | — | |
| Myc | 2 | −1.98† | — | |
| Ptpn1 | — | 2 | −2.13† | |
| Spi1 | — | 2 | −1.73† |
Values represent the means ± SE from 3 independent experiments (n = 3) or the average from 2 (n = 2) as indicated. Jak/STAT signaling pathway PCR arrays were probed with cDNAs prepared from control or ethanol-treated WIF-B cells after 0, 30, or 240 min of additional treatment with GH (see materials and methods). The fold change in gene expression for the 84 genes on the array in cells treated for 0 min relative to those treated for 30 min (top) or 4 h (bottom table) was determined for both control and ethanol-treated cells. Three independent experiments were performed. Mcl1, myeloid cell leukemia 1 (a BH-3 family member antiapoptotic protein); Egfr, epidermal growth factor receptor (a known STAT5 target); Gbp1, interferon-induced guanylate binding protein; Pias1, protein inhibitor of activated STAT; Spi1, Spi-1 proto-oncogene. NSC, no sustained changes in upregulated genes.
These genes had gene expression changes shared in both control and ethanol-treated cells.
These values represent the fold change in genes with decreased expression.
Table 4.
GH transiently restores gene expression in ethanol-treated cells
| 0 min |
30 min |
240 min |
||||
|---|---|---|---|---|---|---|
| Gene | n | Fold change | n | Fold change | n | Fold change |
| Jak kinases | ||||||
| Jak1 | 3 | −1.50 ± 0.09 | ||||
| Jak2 | 3 | −1.57 ± 0.19 | 3 | −1.82 ± 0.45 | ||
| Jak3 | 3 | −1.40 ± 0.18 | 3 | −1.48 ± 0.29 | ||
| Tyk2 | 2 | −1.57 | 2 | −2.00 | ||
| Usf1 | 3 | −1.84 ± 0.26 | ||||
| STATs | ||||||
| Stat3 | 3 | −1.42 ± 0.25 | ||||
| Stat5a | 3 | −1.72 ± 0.35 | ||||
| Jak/STAT receptors | ||||||
| Egfr | 3 | −1.79 ± 0.06 | 3 | −1.90 ± 0.43 | ||
| Ghr | 3 | −1.82 ± 0.28 | ||||
| Prlr* | 3 | −2.93 ± 0.91 | 3 | −1.59 ± 0.29 | 3 | −2.98 ± 0.58 |
| Nr3c1 | 2 | −1.60 | ||||
| Jak/STAT modulators | ||||||
| Socs2 | 2 | −1.47 | ||||
| Socs3 | 3 | −1.83 ± 0.22 | ||||
| Signaling adaptor proteins | ||||||
| Grb2 | 3 | −1.65 ± 0.36 | ||||
| Sh2b1 | 2 | −1.95 | ||||
| Crk | 3 | −1.67 ± 0.43 | ||||
| Cytokines, cytokine receptors, and other secreted proteins | ||||||
| A2M | 3 | −2.58 ± 0.67 | 3 | −2.21 ± 0.31 | ||
| Ifng | 3 | −1.47 ± 0.17 | ||||
| Il20 | 3 | −2.58 ± 0.69 | 3 | −3.21 ± 0.50 | ||
| Osm | 2 | −1.89 | ||||
| Ifnar | 2 | −1.50 | ||||
| Il2rg | 2 | −4.45 | 2 | −3.02 | ||
| Il10ra | 3 | −4.74 ± 2.88 | ||||
| Other immune modulators | ||||||
| Cebpb* | 3 | −1.53 ± 0.25 | 2 | −2.18 | 2 | −1.80 |
| Cebpd* | 2 | −1.79 | 3 | −2.11 ± 0.27 | 2 | −3.72 |
| Gbp1 | 3 | −2.40 ± 0.78 | ||||
| Irf1 | 3 | −1.48 ± 0.17 | 2 | −1.77 | ||
| Irf9* | 3 | −1.69 ± 0.21 | 3 | −1.54 ± 0.24 | 3 | −1.56 ± 0.25 |
| Nfkb | 3 | −1.62 ± 0.23 | ||||
| Cell cycle regulators | ||||||
| Akt | 2 | −1.70 | ||||
| Junb | 2 | −2.85 | ||||
| Myc | 3 | −1.45 ± 0.22 | ||||
| Src | 3 | −1.91 ± 0.43 | ||||
Values represent the means ± SE from 3 independent experiments (n = 3) or the average from two (n = 2) as indicated. Jak/STAT signaling pathway PCR arrays were probed with cDNAs prepared from control or ethanol-treated WIF-B cells after 0, 30, or 240 min of additional treatment with GH (see materials and methods). The fold change in gene expression in ethanol-treated cells relative to control for each time point was determined. Only genes showing decreased expression levels are shown. Three independent experiments were performed. Ghr, growth hormone receptor (known Jak2/STAT5 receptor); Grb2, growth factor receptor-bound protein 2 (a Tyr kinase signaling adaptor protein); SH2B, SH2 domain-containing protein 2B (a known Jak2/GH-receptor adaptor protein); CRK, CRK proto-oncogene (a tyr kinase signaling adaptor protein).
These genes had changes in gene expression that are independent of GH addition.
DISCUSSION
To date, the role of CYP2E1-mediated ethanol metabolites or ROS on microtubule acetylation or on alcohol-induced protein trafficking defects has not yet been examined. To determine if CYP2E1-mediated alcohol metabolism is required for the observed impairments, we took a straightforward approach. To inhibit CYP2E1-mediated ethanol metabolism, we coincubated cells with ethanol and DAS, and to decrease ROS production, we coincubated cells with NAC. Addition of either agent failed to prevent microtubule hyperacetylation or prevent acetylation-dependent defects in protein secretion or clathrin-mediated internalization. However, somewhat surprisingly, both agents prevented the alcohol-induced defect in Jak2/STAT5B activation before STAT5B microtubule-dependent translocation. These results were confirmed in HepG2 cells expressing only ADH or only CYP2E1. Importantly, quantitative RT-PCR using Jak/STAT signaling arrays revealed that GH-mediated hepatic signaling was blunted and attenuated in ethanol-treated cells such that the hepato-protective effects of GH are decreased thereby contributing to hepatic injury.
Alcohol-induced microtubule acetylation and associated defects in microtubule-dependent protein trafficking are mediated by acetaldehyde.
Our earlier studies determined that increased microtubule acetylation and stability displayed both ethanol time and dose dependence (18). Furthermore, microtubule hyperacetylation and stability were prevented by 4-MP and potentiated by cyanamide, indicating that ADH-mediated ethanol metabolism to acetaldehyde was required (18). Since then, similar studies with 4-MP have also shown that impaired secretion, nuclear translocation, and clathrin-mediated internalization are also dependent on ADH-mediated ethanol metabolism (9, 16, 38). The studies reported here confirm those conclusions and further establish that ethanol metabolism by ADH to acetaldehyde only (not via other CYP2E1-generated metabolites) is responsible for enhanced microtubule acetylation and associated defects in protein trafficking.
How acetaldehyde promotes microtubule acetylation is not completely understood, but a possible answer might come from our studies on the distributions, levels, and biochemical properties of histone deacetylase 6 (HDAC6), the major tubulin deacetylase. Although ethanol treatment did not alter HDAC6 subcellular distributions in polarized WIF-B cells, it led to a 25% decrease in its protein levels (36). We also determined that HDAC6 binding to endogenous microtubules was impaired in ethanol-treated cells while its ability to bind or deacetylate exogenous tubulin did not change, suggesting that tubulin from ethanol-treated cells was modified thereby preventing HDAC6 binding (36). Because both impaired HDAC6 microtubule binding and tubulin hyperacetylation require ethanol metabolism by ADH (but not CYP2E1) (18, 36), and because tubulin can be acetaldehyde-adducted (15, 44), we propose that tubulin-acetaldehyde adducts impede HDAC6-tubulin binding thereby preventing deacetylation.
Decreased Jak2/STAT5B activation requires CYP2E1-mediated ethanol metabolism ROS.
Unlike for STAT5B translocation, ethanol metabolism by CYP2E1 is responsible for impaired Jak2/STAT5B activation. The mechanism whereby the CYP2E1-generated metabolites impair signaling is not yet known, but clues may come from earlier studies on GH-mediated signaling in liver slices from animals after acute or chronic ethanol administration (1, 21, 43). Despite decreased levels of GH target hepatic gene expression in slices from treated animals, no changes in GH-R numbers, receptor binding, or receptor affinity for GH were observed (1, 21, 43). Our studies have further shown that total STAT5B or Jak2 protein levels are not changed by ethanol administration in WIF-B, VL-17A, VA-13, or E47 cells or in livers from ethanol-fed rats (9). Thus blunted gene expression results from impairments in the signal transduction pathway itself, which is fully consistent with our findings that decreased Jak2/STAT5B activation and STAT5B nuclear translocation are observed in ethanol-treated cells. These results further suggest that accumulated CYP2E1-generated reactive ethanol metabolites form detrimental adducts on critical residues that alter Jak2 tyrosine kinase activity directly or disrupt Jak2/STAT5B recruitment to GH-R upon ligand binding. Future studies are needed to sort out these and other details.
Hepatocyte-specific responses to GH-mediated signaling are blunted and attenuated.
The primary target organ for GH-mediated signaling is the liver. In general, GH binds its cognate receptor, GH-R, and signaling is mediated by sequential phosphorylation/activation of the tyrosine kinase Jak2 and then the transcription factor STAT5A/B. Phosphorylated STAT5A/B translocates to the nucleus and binds γ-interferon-activation sites (GAS) sites thereby promoting expression of specific targets. Because STAT5B protein is present at levels 20-fold greater than STAT5A in liver, the majority of the hepatocyte target genes are mediated by STAT5B signaling (13). The STAT5 target genes regulate a host of hepatic responses including cell growth, cell cycle progression, and the metabolism of lipids, steroids, bile acids, and drugs (2). Thus it comes as no surprise that defects in STAT5B signaling have been associated with a number of liver diseases including hepatocellular carcinoma, steatosis and fibrosis (2).
As expected, many of the genes with altered expression in ethanol treated cells at steady state are known STAT5B targets (Prlr, Socs2, Osm, Ifnar, Irf1, Irf9, and Cdkn1) (2, 12, 13, 17). We also noticed that a number of the downregulated genes encode proteins that interact with the GH-R/Jak2/STAT5 signaling machinery (Csfr, Epor, and Egfr), are known tyrosine-binding adaptor proteins (Grb2, CRK, and SH2B), or coordinate with STAT5B to regulate gene expression (Nr3c1, Cebpb, and Cebpd) (2, 12, 13, 17). Whether these represent direct STAT5B target genes is not yet known, but their decreased expression is consistent with the observed decrease in GH-mediated signaling. Interestingly, one of the few upregulated genes identified at steady state is Ptpn1, a known tyrosine phosphatase. This finding is also consistent with decreased Jak2/STAT5 phosphorylation in ethanol-treated cells. Although we have previously confirmed decreased EGF-R protein levels in livers from ethanol-fed rats (9), our efforts are now aimed at confirming other targets at the protein level and examine their possible roles in impaired GH-mediated signaling.
GRANTS
This work was supported by the National Institute of Alcohol Abuse and Alcoholism Grant R01-AA-17626 (to P. L. Tuma).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.J.T. and P.L.T. conceived and designed research; J.L.G. and P.L.T. prepared figures; E.E.D., J.L.G., J.R.W., B.M.F., and D.J.F. performed experiments; E.E.D., J.L.G., J.R.W., B.M.F., D.J.T., D.J.F., and P.L.T. interpreted results of experiments; E.E.D., J.L.G., J.R.W., B.M.F., D.J.T., D.J.F., and P.L.T. edited and revised manuscript; J.L.G., D.J.T., D.J.F., and P.L.T. analyzed data; P.L.T. drafted manuscript; P.L.T. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Ann Hubbard (Johns Hopkins University School of Medicine) for providing for the anti-albumin and anti-ASGP-R antibodies used in these studies. We also thank Dr. Dahn Clemens (University of Nebraska Medical School) for providing the E47 and VA-13 cells.
REFERENCES
- 1.Badger TM, Ronis MJ, Frank SJ, Chen Y, He L. Effects of chronic ethanol on hepatic and renal CYP2C11 in the male rat: interactions with the Janus-kinase 2-signal transducer and activators of transcription proteins 5b pathway. Endocrinology 144: 3969–3976, 2003. doi: 10.1210/en.2002-0163. [DOI] [PubMed] [Google Scholar]
- 2.Baik M, Yu JH, Hennighausen L. Growth hormone-STAT5 regulation of growth, hepatocellular carcinoma, and liver metabolism. Ann N Y Acad Sci 1229: 29–37, 2011. doi: 10.1111/j.1749-6632.2011.06100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady JF, Ishizaki H, Fukuto JM, Lin MC, Fadel A, Gapac JM, Yang CS. Inhibition of cytochrome P-450 2E1 by diallyl sulfide and its metabolites. Chem Res Toxicol 4: 642–647, 1991. doi: 10.1021/tx00024a008. [DOI] [PubMed] [Google Scholar]
- 4.Brady JF, Wang MH, Hong JY, Xiao F, Li Y, Yoo JS, Ning SM, Lee MJ, Fukuto JM, Gapac JM, Yang CS. Modulation of rat hepatic microsomal monooxygenase enzymes and cytotoxicity by diallyl sulfide. Toxicol Appl Pharmacol 108: 342–354, 1991. doi: 10.1016/0041-008X(91)90123-V. [DOI] [PubMed] [Google Scholar]
- 5.Brooks PJ. DNA damage, DNA repair, and alcohol toxicity–a review. Alcohol Clin Exp Res 21: 1073–1082, 1997. [PubMed] [Google Scholar]
- 6.Clemens DL. Use of cultured cells to study alcohol metabolism. Alcohol Res Health 29: 291–295, 2006. [PMC free article] [PubMed] [Google Scholar]
- 7.Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol 38: 92–101, 2006. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez DJ, McVicker BL, Tuma DJ, Tuma PL. Ethanol selectively impairs clathrin-mediated internalization in polarized hepatic cells. Biochem Pharmacol 78: 648–655, 2009. doi: 10.1016/j.bcp.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez DJ, Tuma DJ, Tuma PL. Hepatic microtubule acetylation and stability induced by chronic alcohol exposure impair nuclear translocation of STAT3 and STAT5B, but not Smad2/3. Am J Physiol Gastrointest Liver Physiol 303: G1402–G1415, 2012. doi: 10.1152/ajpgi.00071.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraenkel-Conrat H, Singer B. Nucleoside adducts are formed by cooperative reaction of acetaldehyde and alcohols: possible mechanism for the role of ethanol in carcinogenesis. Proc Natl Acad Sci USA 85: 3758–3761, 1988. doi: 10.1073/pnas.85.11.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groebner JL, Fernandez DJ, Tuma DJ, Tuma PL. Alcohol-induced defects in hepatic transcytosis may be explained by impaired dynein function. Mol Cell Biochem 397: 223–233, 2014. doi: 10.1007/s11010-014-2190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrington J, Smit LS, Schwartz J, Carter-Su C. The role of STAT proteins in growth hormone signaling. Oncogene 19: 2585–2597, 2000. doi: 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- 13.Hosui A, Hennighausen L. Genomic dissection of the cytokine-controlled STAT5 signaling network in liver. Physiol Genomics 34: 135–143, 2008. doi: 10.1152/physiolgenomics.00048.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol 123: 1761–1775, 1993. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennett RB, Saffari-Fard A, Sorrell MF, Smith SL, Tuma DJ. Increased covalent binding of acetaldehyde to calmodulin in the presence of calcium. Life Sci 45: 1461–1466, 1989. doi: 10.1016/0024-3205(89)90036-2. [DOI] [PubMed] [Google Scholar]
- 16.Joseph RA, Shepard BD, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Microtubule acetylation and stability may explain alcohol-induced alterations in hepatic protein trafficking. Hepatology 47: 1745–1753, 2008. doi: 10.1002/hep.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of signal transducers and activators of transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics 14: 4, 2013. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannarkat GT, Tuma DJ, Tuma PL. Microtubules are more stable and more highly acetylated in ethanol-treated hepatic cells. J Hepatol 44: 963–970, 2006. doi: 10.1016/j.jhep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Kenney WC. Acetaldehyde adducts of phospholipids. Alcohol Clin Exp Res 6: 412–416, 1982. doi: 10.1111/j.1530-0277.1982.tb05000.x. [DOI] [PubMed] [Google Scholar]
- 20.Kenney WC. Formation of Schiff base adduct between acetaldehyde and rat liver microsomal phosphatidylethanolamine. Alcohol Clin Exp Res 8: 551–555, 1984. doi: 10.1111/j.1530-0277.1984.tb05728.x. [DOI] [PubMed] [Google Scholar]
- 21.Lang CH, Liu X, Nystrom G, Wu D, Cooney RN, Frost RA. Acute effects of growth hormone in alcohol-fed rats. Alcohol 35: 148–158, 2000. doi: 10.1093/alcalc/35.2.148. [DOI] [PubMed] [Google Scholar]
- 22.LeDizet M, Piperno G. Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J Cell Biol 103: 13–22, 1986. doi: 10.1083/jcb.103.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchner H, Tottmar O. A comparative study on the effects of disulfiram, cyanamide and 1-aminocyclopropanol on the acetaldehyde metabolism in rats. Acta Pharmacol Toxicol (Copenh) 43: 219–232, 1978. doi: 10.1111/j.1600-0773.1978.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 24.McVicker BL, Tuma PL, Kharbanda KK, Lee SM, Tuma DJ. Relationship between oxidative stress and hepatic glutathione levels in ethanol-mediated apoptosis of polarized hepatic cells. World J Gastroenterol 15: 2609–2616, 2009. doi: 10.3748/wjg.15.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyasae LK, Hubbard AL, Tuma PL. Transcytotic efflux from early endosomes is dependent on cholesterol and glycosphingolipids in polarized hepatic cells. Mol Biol Cell 14: 2689–2705, 2003. doi: 10.1091/mbc.E02-12-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol 104: 289–302, 1987. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramnarayanan SP, Cheng CA, Bastaki M, Tuma PL. Exogenous MAL reroutes selected hepatic apical proteins into the direct pathway in WIF-B cells. Mol Biol Cell 18: 2707–2715, 2007. doi: 10.1091/mbc.E07-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reas HW. The use of N-acetylcysteine in the treatment of cystic fibrosis. South Med J 56: 1271–1278, 1963. doi: 10.1097/00007611-196311000-00016. [DOI] [PubMed] [Google Scholar]
- 29.Ristow H, Obe G. Acetaldehyde induces cross-links in DNA and causes sister-chromatid exchanges in human cells. Mutat Res 58: 115–119, 1978. doi: 10.1016/0165-1218(78)90103-9. [DOI] [PubMed] [Google Scholar]
- 30.Salaspuro MP, Lindros KO, Pikkarainen P. Ethanol and galactose metabolism as influenced by 4-methylpyrazole in alcoholics with and without nutritional deficiencies. Preliminary report of a new approach to pathogenesis and treatment in alcoholic liver disease. Ann Clin Res 7: 269–272, 1975. [PubMed] [Google Scholar]
- 31.Salaspuro MP, Pikkarainen P, Lindros K. Ethanol-induced hypoglycaemia in man: its suppression by the alcohol dehydrogenase inhibitor 4-methylpyrazole. Eur J Clin Invest 7: 487–490, 1977. doi: 10.1111/j.1365-2362.1977.tb01640.x. [DOI] [PubMed] [Google Scholar]
- 32.Schaffert CS, Todero SL, Casey CA, Thiele GM, Sorrell MF, Tuma DJ. Chronic ethanol treatment impairs Rac and Cdc42 activation in rat hepatocytes. Alcohol Clin Exp Res 30: 1208–1213, 2006. doi: 10.1111/j.1530-0277.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 33.Schaffert CS, Todero SL, McVicker BL, Tuma PL, Sorrell MF, Tuma DJ. WIF-B cells as a model for alcohol-induced hepatocyte injury. Biochem Pharmacol 67: 2167–2174, 2004. doi: 10.1016/j.bcp.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Shanks MR, Cassio D, Lecoq O, Hubbard AL. An improved polarized rat hepatoma hybrid cell line. Generation and comparison with its hepatoma relatives and hepatocytes in vivo. J Cell Sci 107: 813–825, 1994. [DOI] [PubMed] [Google Scholar]
- 35.Shepard BD, Fernandez DJ, Tuma PL. Alcohol consumption impairs hepatic protein trafficking: mechanisms and consequences. Genes Nutr 5: 129–140, 2010. doi: 10.1007/s12263-009-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepard BD, Joseph RA, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Alcohol-induced alterations in hepatic microtubule dynamics can be explained by impaired histone deacetylase 6 function. Hepatology 48: 1671–1679, 2008. doi: 10.1002/hep.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Exp Res 34: 280–291, 2010. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepard BD, Tuma DJ, Tuma PL. Lysine acetylation induced by chronic ethanol consumption impairs dynamin-mediated clathrin-coated vesicle release. Hepatology 55: 1260–1270, 2012. doi: 10.1002/hep.24785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuma DJ, Casey CA. Dangerous byproducts of alcohol breakdown–focus on adducts. Alcohol Res Health 27: 285–290, 2003. [PMC free article] [PubMed] [Google Scholar]
- 40.Webb WR. Clinical evaluaton of a new mucolytic agent, acetyl-cysteine. J Thorac Cardiovasc Surg 44: 330–343, 1962. [PubMed] [Google Scholar]
- 41.Wehr H, Rodo M, Lieber CS, Baraona E. Acetaldehyde adducts and autoantibodies against VLDL and LDL in alcoholics. J Lipid Res 34: 1237–1244, 1993. [PubMed] [Google Scholar]
- 42.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol 4: 938–947, 2003. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 43.Xu X, Ingram RL, Sonntag WE. Ethanol suppresses growth hormone-mediated cellular responses in liver slices. Alcohol Clin Exp Res 19: 1246–1251, 1995. doi: 10.1111/j.1530-0277.1995.tb01607.x. [DOI] [PubMed] [Google Scholar]
- 44.Yoon Y, Török N, Krueger E, Oswald B, McNiven MA. Ethanol-induced alterations of the microtubule cytoskeleton in hepatocytes. Am J Physiol Gastrointest Physiol 274: G757–G766, 1998. [DOI] [PubMed] [Google Scholar]
- 45.Ziment I. Acetylcysteine: a drug that is much more than a mucokinetic. Biomed Pharmacother 42: 513–519, 1988. [PubMed] [Google Scholar]