Abstract
It is well established that transforming growth factor-β (TGFβ) switches its function from being a tumor suppressor to a tumor promoter during the course of tumorigenesis, which involves both cell-intrinsic and environment-mediated mechanisms. We are interested in breast cancer cells, in which SMAD mutations are rare and interactions between SMAD and other transcription factors define pro-oncogenic events. Here, we have performed chromatin immunoprecipitation (ChIP)-sequencing analyses which indicate that the genome-wide landscape of SMAD2/3 binding is altered after prolonged TGFβ stimulation. De novo motif analyses of the SMAD2/3 binding regions predict enrichment of binding motifs for activator protein (AP)1 in addition to SMAD motifs. TGFβ-induced expression of the AP1 component JUNB was required for expression of many late invasion-mediating genes, creating a feed-forward regulatory network. Moreover, we found that several components in the WNT pathway were enriched among the late TGFβ-target genes, including the invasion-inducing WNT7 proteins. Consistently, overexpression of WNT7A or WNT7B enhanced and potentiated TGFβ-induced breast cancer cell invasion, while inhibition of the WNT pathway reduced this process. Our study thereby helps to explain how accumulation of pro-oncogenic stimuli switches and stabilizes TGFβ-induced cellular phenotypes of epithelial cells.
INTRODUCTION
The signaling pathways triggered by the transforming growth factor β (TGFβ) family members control a wide range of cellular processes. TGFβ signals via heterotetrameric complexes of type I and type II serine/threonine kinase receptors. The activated receptor complex initiates intracellular signaling by phosphorylating receptor-regulated (R-) SMAD proteins (SMAD2 and SMAD3). The activated R-SMADs form heteromeric complexes with SMAD4, which accumulate in the nucleus and control expression of target genes (1–3). However, SMADs have relatively weak affinity for DNA and in many cases interact with so called master transcription factors to achieve high affinity and target-gene specificity (4,5). These interactions alter the intensity, duration and specificity of the TGFβ-signaling response, in a context- and cell-type-specific manner (6–8).
TGFβ plays a dual role in tumor progression. In normal or premalignant cells TGFβ functions as a tumor suppressor by inhibiting cell proliferation and inducing apoptosis. However, in late stages of tumor development, TGFβ instead acts as a tumor promoter by stimulating cell motility, invasion, metastasis and tumor stem cell maintenance. This is reflected by the observation that specific types of cancers are insensitive to the cytostatic effect of TGFβ due to inactivation of core components in the TGFβ pathway (9,10). On the other hand, in breast cancer and certain other cancers, defects in the TGFβ/SMAD signaling itself are relatively uncommon; instead tumor promoting effects of TGFβ/SMAD signaling dominates (reviewed in (11,12)). In line with this, TGFβ is frequently overexpressed in breast cancer and its expression correlates with poor prognosis and metastasis (13). The influence of TGFβ on tumor growth is also affected by crosstalk between the TGFβ signaling pathway and a wide variety of signal transduction pathways. For example, the Ras-MAP-kinase (MAPK) pathway (14) regulates cell migration and invasion synergistically with TGFβ (8,11,15,16). Interestingly, transcriptome-wide analysis of mouse primary hepatocytes treated with TGFβ revealed that the early TGFβ response was characterized by expression of genes involved in cell cycle arrest and apoptosis, while the late gene signature was associated with an aggressive and invasive tumor phenotype that effectively identified clinical relevant subgroups of hepatocellular carcinoma (17).
We previously reported that prolonged stimulation with TGFβ induces mesenchymal and invasion-associated genes through interaction between SMAD and activator protein (AP)1 components, in particular JUNB (16). AP1 transcription factors are targeted by many signal transduction pathways and regulate a magnitude of cellular processes, including cell proliferation, survival, differentiation, invasion and carcinogenesis, depending on their dimer composition (18–20). SMAD and AP1 members interact at different levels. For example, TGFβ induces the expression of specific AP1 components and reporter assays suggested that the AP1 components JUN and JUNB cooperate with SMAD2/3 to activate TGFβ-induced promoters regulated by AP1 binding sites (21,22), while antagonizing DNA binding of the same SMADs on promoters controlled by SMAD binding sites (23). However, little is known about the SMADs and AP1 crosstalk at the genome-wide level.
Identification and characterization of signaling molecules that switch TGFβ/SMAD signaling from tumor suppression to tumor promotion is critical for the development of therapies targeting the TGFβ pathway (24). To identify SMAD complexes and target genes involved in tumor progression on a genome-wide scale, we performed SMAD2/3 chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) and RNA sequencing analyses, both early and late after TGFβ stimulation. Our results indicate that most of SMAD2/3 is redirected to different sites on the genome after prolonged TGFβ treatment. De novo motif analyses predicted enrichment of binding motifs for AP1 and SMAD, or the SMAD Binding Element (SBE) consensus sequence CAGA, in SMAD2/3 binding regions. Moreover, our results suggest that TGFβ-induced expression of JUNB via a positive feed-forward mechanism enables a switch of the early TGFβ transcriptional program to a late, invasion-mediating program. Furthermore, we found that genes related to WNT signaling pathways are enriched among the late TGFβ-target genes. Consistently, modulation of the WNT signaling pathway aggravated TGFβ-induced breast cancer cell invasion and metastasis. Our study thereby helps to explain how accumulation of oncogenic stimuli switches TGFβ responsiveness in epithelial cells.
MATERIALS AND METHODS
Cell culture
Human breast epithelial MCF10A MII cells were obtained from Dr Fred Miller (Barbara Ann Karmanos Cancer Institute, Detroit, USA) and maintained at 37°C and 5% CO2 in DMEM/F12 (Gibco), supplemented with 5% fetal bovine serum (FBS) (HyClone), 20 ng/ml epidermal growth factor (EGF) (PeproTech), 100 ng/ml cholera toxin (Sigma-Aldrich), 0.5 μg/ml hydrocortisone (Sigma-Aldrich), 10 μg/ml insulin (Sigma-Aldrich). MCF10A MII cells are derived from MCF10A cells by transformation with Ha-Ras. Human breast cancer MDA-MB-231 cells and human lung cancer A549 cells were obtained from ATCC and maintained at 37°C and 5% CO2 in DMEM (Sigma-Aldrich), supplemented with 10% FBS (Bio West). Breast cancer Hs578T and BT-549 cells were obtained from ATCC, and maintained as recommended. Briefly, Hs578T cells were cultured at 37°C and 5% CO2 in DMEM (Gibco) supplemented with 10% FBS (HyClone), and 10 μg/ml insulin (Gibco), and BT-549 cells were maintained in RPMI-1640 (Gibco), supplemented with 10% FBS (HyClone), and 0.023 IU/ml insulin (Gibco).
Lentiviral transduction
MCF10A MII cells were infected with lentivirus encoding an shRNA sequence against human JUNB (TRCN0000014943, TRCN0000014946, TRNC0000014947) selected from the MISSION shRNA library (Sigma-Aldrich). As a control an empty pLKO vector was used. Virus transduction was performed overnight and the infected cells were selected using culture medium containing Puromycin.
Reagents and antibodies
Recombinant human TGFβ3 (a generous gift of Dr K. Iwata, OSI Pharmaceuticals, Inc, New York, USA, or purchased from R&D Systems) was used for stimulation of cells. Epithelial cells that express betaglycan respond similarly to the three TGFβ isoforms. Recombinant human WNT7A was from PeproTech. The TGFβ type I kinase receptor (TGFβRI) inhibitor SB505124 (ALK5i) and IWP-2 (WNTi), which is an inhibitor of WNT processing and secretion, were purchased from Sigma-Aldrich and Merck Millipore, respectively. Puromycin was purchased from Invivogen and used at a concentration of 0.5 μg/ml. For siRNA-mediated knockdown, Dharmacon On Target Plus pools of four oligonucleotides (GE Healthcare Life Sciences) was transfected using siLentFect (Bio-Rad) transfection reagent according to manufacturer's instructions at 25 nM final concentration.
Antibodies against the following proteins were used: ERK1/2 (#4695, Cell Signaling Technology), phospho-Thr202/Tyr204-ERK1/2 (#4370, Cell Signaling Technology), FN1 (F3648, Sigma-Aldrich), JUN (#9164, Cell Signaling Technology), JUNB (sc-8051, Santa Cruz), FOS (sc-52, Santa Cruz), FOSB (#2251, Cell Signaling Technology), FOSL1 (sc-22794, Santa Cruz), FOSL2 (sc-604, Santa Cruz), MYC (sc-40, Santa Cruz), SMAD2/3 (#610843, BD Transduction Laboratories), phospho-Ser465/467-SMAD2 (#3108, Cell Signaling Technology), phospho-Ser423/425-SMAD3 (#9520, Cell Signaling Technology), SMAD4 (sc-7966, Santa Cruz), α-TUBULIN (sc-8035, Santa Cruz) and WNT7B (AF3460, R&D Systems). A custom-made JUND antibody was raised in chicken against a synthetic polypeptide CQLLPQHQVPAY, corresponding to the unique C-terminal part of JUND (Immune Systems).
Plasmid construction
WNT7A and WNT7B cDNAs were kindly provided by Dr Brad St. Croix. For stable cell line establishment, cDNAs were cloned into an episomal expression vector pPyCAG-IRES-Puro, which contains polyoma Ori and can be propagated episomally in cells (25).
Western blot analysis
MCF10A MII cells were seeded in 6-well-plates (2.5 × 105 cells/well), and starved the following day for 16 h in 0.2% FBS, and cells were then stimulated with 5 ng/ml of TGFβ3 for indicated time-periods. Cells were lysed in 2× SDS Laemmli sampler buffer (5% SDS, 25% glycerol, 150 mM Tris–HCl pH 6.8, 0.01% bromophenol blue, 100 mM dithiothreitol (DTT)). Samples were separated by SDS-PAGE, blotted onto nitrocellulose membrane (Amersham Protran, GE Healthcare Life Science), and the chemiluminescent signal was detected using the Immobilon Western kit (Merck Millipore).
3D spheroid collagen invasion assay
One thousand cells, of the indicated cell line, were trypsinized, re-suspended in medium containing 2.4 mg/ml methylcellulose (Sigma-Aldrich) and added into each well of a U-bottom 96-well-plate (Greiner Bio One) allowing the formation of one spheroid per well. Two days after plating, a U-bottom 96-well-plate was coated with neutralized bovine collagen-I (PureCol, Advanced BioMatrix) according to manufacturer's protocol. Spheroids were harvested and embedded in a 1:1 mix of neutralized collagen and medium supplemented with 12 mg/ml of methylcellulose and allowed to polymerize on the top of the neutralized collagen. TGFβ3 and/or recombinant WNT7A were directly added to the embedding solution. After polymerization, medium supplemented with 1.6% FBS was added to the top of the collagen. SB505124 and IWP-2 were added in the medium. Pictures were taken at day 0 and day 2 after embedding and quantified by measuring the area occupied by cells using Adobe Photoshop CS3 software.
Zebrafish maintenance
This study was approved by The Institutional Committee for Animal Welfare of the Leiden University Medical Center (LUMC). Zebrafish and embryos were maintained according to standard procedures. The transgenic fish line Tg(fli1:GFP) was used in this study as described before (26,27). All experiments were performed in accordance with approved guidelines and regulations.
Embryo preparation and tumor cell implantation
Tg(fli1:GFP) zebrafish embryos were dechorionated at 2 days post fertilization (dpf). Single cell suspensions of mCherry labelled MCF10A MII, MDA-MB-231 or A549 cells were re-suspended in PBS and kept at 4°C before injection. Cell suspensions were loaded into borosilicate glass capillary needles (1 mm O.D. × 0.78 mm I.D.; Harvard Apparatus). Injections were performed with a Pneumatic Picopump and a manipulator (WPI). Dechorionated embryos were anaesthetized with 0.003% tricaine (Sigma) and mounted on 10-cm Petri dishes coated with 1% agarose. Approximately 400 cells were injected at the duct of Cuvier (DOC). Injected zebrafish embryos were maintained at 34°C. All the experiments were repeated at least two times and at least 30 embryos were analyzed per group.
Microscopy and analysis
Six days post infection (dpi) embryos were fixed with 4% paraformaldehyde at 4°C overnight. Fixed embryos were analyzed and imaged in PBS with a Leica SP5 STED confocal microscope (Leica). The numbers of clusters formed in caudal hematopoietic tissue (CHT) of each embryo were counted. Confocal stacks were processed for maximum intensity projections with matched software LAS AF Lite. Brightness and contrast of images were adjusted as well.
RNA isolation, cDNA synthesis and quantitative real time-PCR (qRT-PCR)
Total RNA was isolated by RNeasy Kit (Qiagen). cDNA was prepared by using iScript kit (Bio-Rad) using 0.5 μg of total RNA, according the manufacturer's instructions. The cDNA samples were diluted 10 times in water. qRT-PCR was performed using KAPA SYBR FAST qPCR kit Master Mix (KAPA Biosystems) and BioRad CFX96 real-time PCR detection system according the manufacturer's instructions. qRT-PCR reactions were performed as follow: one cycle of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 30 s, followed by one cycle of 95°C for 15 s and 65°C for 5 s. Relative gene expression was determined using the ΔΔCt method. The expression was normalized to the GAPDH gene and quantified relative to the control condition. The complete primers list can be found in Supplementary Table S1 in the Supplementary Data.
Chromatin immunoprecipitation (ChIP)
Cells were cultured in 10-cm plates to ∼80–90% confluence, and one plate was used per immunoprecipitation. Cells were fixed in 1% formaldehyde for 10 min at room temperature with swirling. Glycine was added to a final concentration of 0.125 M, and the incubation was continued for an additional 5 min. Cells were washed twice with ice-cold phosphate-buffered saline, harvested by scraping, pelleted, and resuspended in 1 ml of SDS lysis buffer (50 mM Tris–HCl, pH 8.0, 1% SDS, 10 mM EDTA, protease inhibitors (Complete EDTA-free protease inhibitors; Roche Diagnostics)). Samples were sonicated three times for 30 s each time (output H) at intervals of 30 s with a Diagenode Bioruptor sonicator. Samples were centrifuged at 14 000 rpm at 4°C for 10 min. After removal of a control aliquot (whole-cell extract), supernatants were diluted 10-fold in ChIP dilution buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100). Samples were incubated at 4°C overnight in 2-methacryloyloxyethyl phosphorylcholine polymer-treated 15-ml polypropylene tubes (Assist, Japan) with anti-mouse IgG-Dynabeads that had been preincubated with 5 μg of anti-SMAD2/3 antibody in phosphate buffered saline, 0.5% bovine serum albumin. The beads were then moved to 1.7-ml siliconized tubes (#3207; Corning) and washed five times with ChIP wash buffer (50 mM HEPES–KOH, pH 7.0, 0.5 M LiCl, 1 mM EDTA, 0.7% deoxycholate, 1% Igepal CA630) and once with TE buffer, pH 8.0. Immunoprecipitated samples were eluted and reverse cross-linked by incubation overnight at 65°C in elution buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS). Genomic DNA was then extracted with a PCR purification kit (Qiagen). The immunoprecipitated DNA was analyzed by qRT-PCR using locus specific primers (the complete primers list can be found in Supplementary Table S2 in the Supplementary Data) and normalized to input DNA. Relative fold enrichment corresponded to the SMAD2/3 enrichment in each locus divided by the enrichment in the negative control regions (hemoglobin β (HBB) promoter and HPRT1 first intron) and quantified relative to the control- or the siNTC-condition as indicated.
ChIP-sequencing (ChIP-seq) and data analysis
Chromatin isolation, sonication and immunoprecipitation using anti-SMAD2/3 antibody were performed essentially as described (28,29). The library was prepared using NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (New England Biolabs), KAPA DNA Library Preparation Kits for Illumina (KAPA Biosystems), or IonXpress Plus Fragment Library Kit (Thermo Fisher Scientific). High-throughput sequencing of the ChIP fragments was performed using Genome Analyzer IIx or HiSeq 2000 (Illumina) or Ion Proton sequencer (Thermo Fisher Scientific) following the manufacturer's protocols. Reference files of the human reference sequence assembly (NCBI Build 37/hg19, February 2009) and GTF annotation file were obtained from iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.html). All ChIP-seq data sets were aligned using Bowtie (version 1.1.0) (30) with the command ‘-S -a –best –strata -v 1 -m 1’. SMAD2/3 binding regions were identified using MACS software (Model based analysis of ChIP-seq) (version 1.4.2) (31) with a P-value threshold of 1e-5. Assigning a binding site to the nearest gene within 100 kb from a peak was performed using CisGenome ver2 (32). De novo motif prediction was performed by MEME-ChIP with a slight modification of the default settings (maximum width: 10) (MEME-ChIP version 4.10; http://meme.nbcr.net/meme/cgi-bin/meme-chip.cgi) (33). The logo plots were generated using the R package seqLogo. Mapping of TFBSs to the specific genomic regions were calculated by the CisGenome. Gene Ontology (GO) enrichment analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.7; http://david.abcc.ncifcrf.gov) (34). Biological functions associated with the SMAD2/3 binding sites were predicted using GREAT (Genomic Regions Enrichment of Annotations Tool) (35). The ChIP-Seq data of H3K4me1, H3K4me3 and corresponding control input DNA of MCF10A cells (SRA045635) (36) were obtained from the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra). The ChIP-Seq data of H3K4me1 and H3K4me3 of HMEC were generated and available from ENCODE consortium (37).
RNA-sequencing (RNA-seq) and data analysis
RNA-seq libraries were prepared essentially as described (38). In short, mRNA was isolated from 1 μg total RNA using Dynabeads Oligo(dT)25 (Life Technologies) and fragmented to 150–200 nt in first strand buffer for 3 min at 94°C. Random hexamer primed first strand was generated in presence of dATP, dGTP, dCTP and dTTP. Second strand was generated using dUTP instead of dTTP to tag the second strand. Subsequent steps to generate the sequencing libraries were performed with the NEBNext kit for Illumina sequencing (New England Biolabs) with minor modifications; after indexed adapter ligation to the dsDNA fragments, the library was treated with USER (Uracil-Specific Excision Reagent) Enzyme (New England Biolabs) in order to digest the second strand derived fragments. After amplification of the libraries, samples with unique sample indexes were pooled and sequenced using HiSeq 2000 with TruSeq SBS Kit v3 reagent or HiSeq 2500 with TruSeq SBS Kit v4 reagent (Illumina) following the manufacturer's protocols.
Gene expression levels in fragments per kilobase of exon per million fragments mapped (FPKM) were estimated using Tophat/Cufflinks (version 2.0.13 and 2.2.1, respectively) with the default parameter settings (39). For the analysis and visualization of the data generated by Cufflinks, we used the R package cummeRbund.
Analysis of Breast Cancer clinical datasets
For the analysis of patient datasets from Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (40), all statistical tests were performed using R software (version 3.2.5, https://www.r-project.org/) as described previously (41). Z-scored expression values of mRNA were obtained from cBioPortal (42,43) in September 2017. Patients were divided into low and high expressers using the median values of mRNA expression. The overall survival was estimated with the Kaplan-Meier method and differences between groups were evaluated by the log-rank test, using the R package cmprsk. P-values were calculated using Welch's t-test, or unequal variance t-test (*P < 0.05, **P < 0.01, ***P < 0.001).
Meta-analysis of Breast Cancer datasets were performed using KM plotter (http://kmplot.com) (44) with default settings; all subtypes, n = 3557; ER+ subjects, n = 2036; ER- subjects, n = 807; luminal A subtype, n = 2069; luminal B subtype, n = 1166; HER2-subtype, n = 239; basal-like subtype, n = 668), and the data sets includes E-MTAB-365, GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE16446, GSE17705, GSE17907, GSE19615, GSE20194, GSE20271, GSE2034, GSE20685, GSE20711, GSE21653, GSE2603, GSE26971, GSE2990, GSE31448, GSE31519, GSE3494, GSE5327, GSE6532, GSE7390 and GSE9195.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) analyses were performed using the tool available at http://www.broadinstitute.org/gsea/index.jsp (45). In brief, fold change (log2) in gene expression from two experimental conditions were calculated and the list was then used as a ranked list in the Pre-Ranked function of the GSEA software.
Statistical analysis
For ChIP-qPCR and qRT-PCR at least three independent experiments were performed and results are shown by dot plot chart. The differences between experimental groups were analyzed using Welch's t-test, with *P < 0.05, **P < 0.01 and ***P < 0.001 being considered significant. Collagen invasion assays contained n ≥ 6 spheroids for each condition, and was repeated at least twice with similar results. Data are presented as means ± SD. The differences between experimental groups were analyzed using Welch's t-test, with *P < 0.05, **P < 0.01 and ***P < 0.001 being considered significant. For the zebrafish experiments statistical analysis was performed using Prism 4 software (GraphPad La Jolla, USA). Results are expressed as the mean ± SEM. Student's t-test or one-way analysis of variance (ANOVA) were performed followed by the Tukey's method for multiple comparison. P < 0.05 was considered to be statistically significant (*0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001).
RESULTS
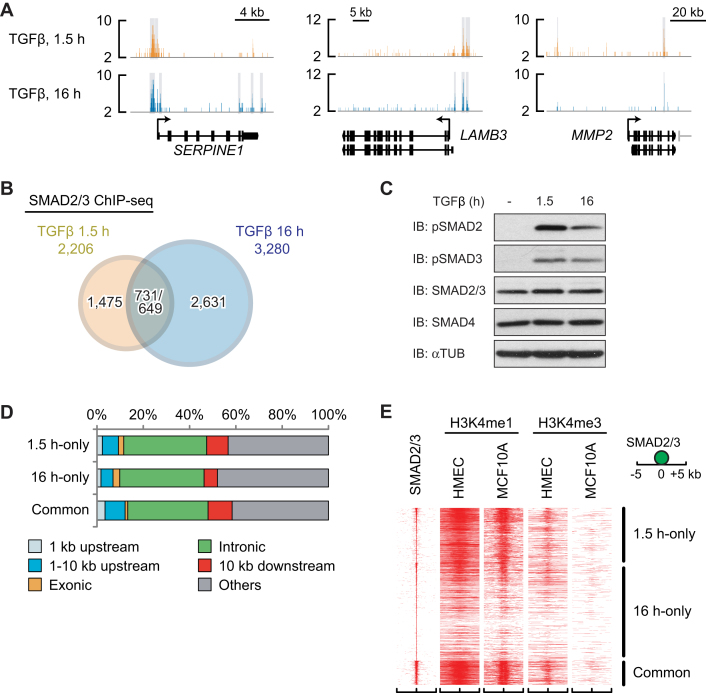
SMAD2/3 are redirected to different sites after prolonged TGFβ treatment
To identify both early and late SMAD-containing complexes and target genes involved in tumor progression, we first conducted SMAD2/3 ChIP-seq in MCF10A MII breast cancer cells after 1.5 and 16 h of TGFβ treatment. Analysis of three well known TGFβ/SMAD target genes, SERPINE1, laminin β (LAMB)3 and matrix metalloprotease (MMP)2, as expected, showed enriched SMAD2/3 binding in specific regions of the gene loci, including the SMAD2/3 binding site that was previously identified in the SERPINE1 promoter in HaCaT keratinocytes (46) (Figure 1A). TGFβ-dependent SMAD2/3 binding to these three genes was also detected by ChIP-qPCR analysis (Supplementary Figure S1A). Interestingly, at the late time point SMAD2/3 was found to bind to different regions of the SERPINE1 and LAMB3 loci, whereas in the MMP2 gene locus SMAD2/3 binding to the binding site located 40 kb upstream of the transcription start site (TSS) was lost (Figure 1A). Moreover, overall SMAD2/3 recognized more target sites after 16 h of TGFβ stimulation (3280 sites) compared to 1.5 h stimulation (2206 sites), and only ∼700 SMAD2/3 binding sites overlapped between the two time points (Figure 1B), suggesting that the activated SMAD2/3 proteins (Figure 1C) were redirected to different binding sites over the genome at the late time point. Furthermore, there were no differences in preferences of SMAD2/3 binding sites on the genome between the two conditions; ∼35% of the SMAD2/3 binding sites were located in the introns of known genes and ∼10% in the promoter regions within 10 kb upstream of known TSSs (Figure 1D).
Figure 1.
SMAD2/3 are redirected to different sites in MCF10A MII after prolonged TGFβ treatment. (A) Genomic loci of SERPINE1 (plasminogen activator inhibitor 1, or PAI-1), MMP2 and LAMB3 genes are shown together with the results of SMAD2/3 ChIP-seq data. The direction of transcription is shown by the arrow beginning at the transcription start site (TSS). Statistically significant regions are marked by a gray-colored box. (B) A Venn diagram indicating overlap of SMAD2/3 binding sites of MCF10A MII cells after 1.5 and 16 h TGFβ (5 ng/ml) treatment. The numbers of overlapped regions are not identical, since some of the peaks are not on a one-by-one correspondence. (C) Western blots for phospho-SMAD2/3 in MCF10A MII cells after 0, 1.5 and 16 h TGFβ (5 ng/ml) treatment. (D) Distribution of SMAD2/3 binding sites in MCF10A MII cells relative to known genes in the human genome (hg19). (E) Heat map representation of the location of the indicated histone marks in breast HMEC and MCF10A epithelial cells within the 10-kb region surrounding the center of the SMAD2/3 peaks. SMAD2/3 binding sites were ordered based on the strength of binding (y axis). The presence of epigenetic marker (36,37) is displayed.
We next compared our SMAD2/3 binding data with previously reported enhancer data in non-stimulated normal human mammary epithelial cells (HMEC) and parental MCF10A cells (36,37). The SMAD2/3 binding sites shared between cells stimulated 1.5 and 16 h overlapped well with the previously identified enhancer regions characterized by H3K4me1 (Figure 1E and Supplementary Figure S1B). The 1.5 h-only sites also overlapped with these H3K4me1 marks, but the 16 h-only sites did not (Figure 1E and Supplementary Figure S1B). In contrast, fewer SMAD2/3 peaks overlapped with the previously reported promoter regions characterized by H3K4me3. This could mean that after 1.5 h TGFβ stimulation, SMAD2/3 preferentially binds to enhancer regions already accessible in non-stimulated normal mammary epithelial cells, but after 16 h prefers different regions. In fact, distinct gene ontologies (GOs) were enriched in the genes associated with 16 h-only sites compared with those of 1.5 h-only sites (Supplementary Figure S1C).
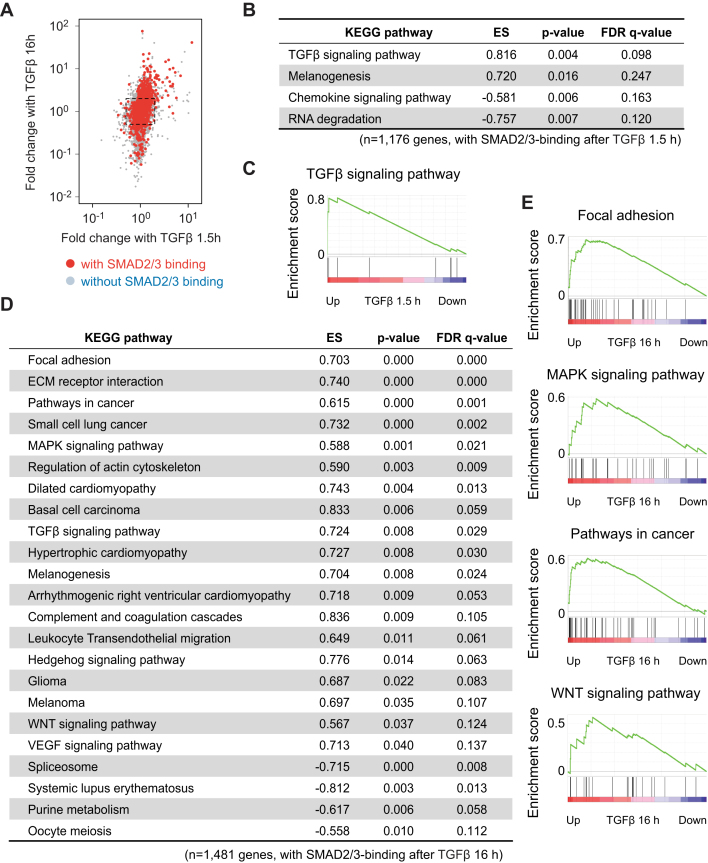
To validate whether the changes in SMAD2/3 binding indeed result in changes in target gene programs, we performed RNA-seq transcriptome analysis after short (1.5 h) and long (16 h) periods of TGFβ stimulation of MCF10A MII cells and compared with unstimulated cells. Consistent with the SMAD2/3 binding profiles, RNA-seq data revealed that more genes were strongly induced at the late time point compared to the early time point (Figure 2A). Gene set enrichment analysis (GSEA) based on Kyoto encyclopedia genes and genomes (KEGG)-defined pathways confirmed that genes associated with GOs like the TGFβ signaling pathway were enriched among the early TGFβ target genes with SMAD2/3 binding sites, whereas genes within Focal adhesion and MAPK signaling pathways were enriched among the late TGFβ target genes (Figure 2B–E).
Figure 2.
Identification of a late TGFβ target gene signature. (A) Scatter plot representing fold change after TGFβ (5 ng/ml) treatment. Each point represents values of a gene. Genes with a SMAD2/3 binding within 50 kb from gene bodies after 16 h TGFβ treatment are colored red. A dot square represents 2-fold change of gene expression. (B–E) Gene set enrichment analysis (GSEA) of expression changes of SMAD2/3 target genes after 1.5 h (B and C) and 16 h (D and E) of TGFβ (5 ng/ml) treatment. The SMAD2/3 target genes were pre-rank-ordered according to their fold change (log2) after TGFβ treatment for the indicated time periods, and analyzed based on KEGG signaling pathway enrichment. Gene sets with a P-value < 5% and an FDR q-value < 25% were considered significant. (C and E) Enrichment score (ES) is plotted on the y axis.
JUNB is a critical AP1 component for SMAD2/3 binding after TGFβ stimulation
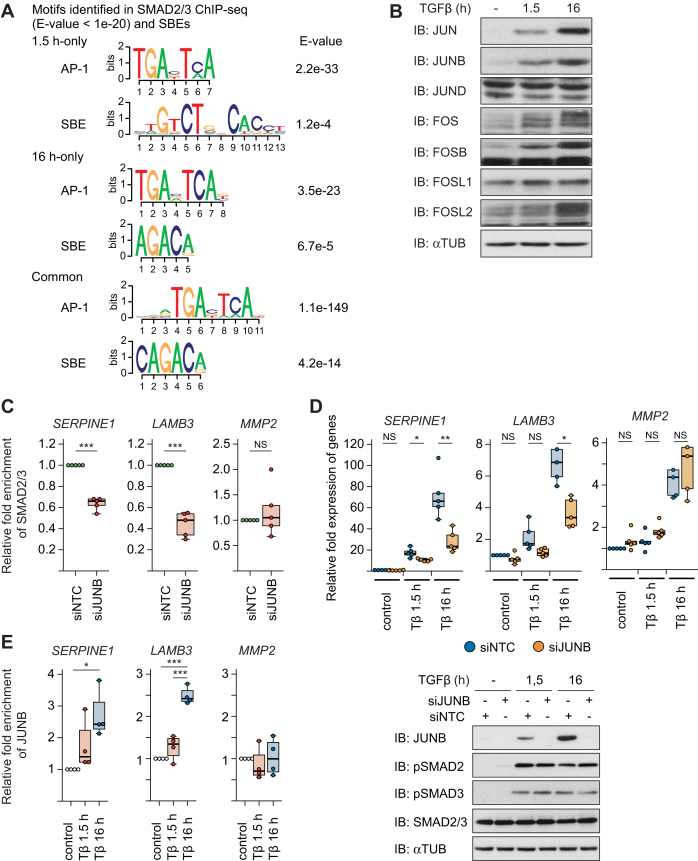
An explanation for the changes in SMAD2/3 binding at 16 h might be that DNA binding factors that are modulated by TGFβ-SMAD signaling at early time points subsequently redirect SMAD2/3 to different binding sites on the genome as a part of a feed-forward loop, e.g. by interacting with SMAD2/3 and/or affecting its chromatin accessibility. To obtain more clues on this, we performed de novo motif prediction analysis. Interestingly, AP1 binding motifs were identified as the major recognition elements among both the early and late sites, with higher significance than SBEs (Figure 3A).
Figure 3.
JUNB is a critical AP1 component for SMAD2/3 binding after TGFβ stimulation. (A) Motifs enriched in the SMAD2/3 binding sites. Motifs which resemble the motif of AP1 were identified as well as SBE. (B) Western blots of various AP1 components in MCF10A MII cells after no TGFβ treatment (–), or TGFβ (5 ng/ml) treatment for 1.5 or 16 h. (C) ChIP-qPCR showing SMAD2/3 binding to the indicated gene loci in MCF10A MII cells transfected with non-targeting control (siNTC) or specific JUNB siRNA and stimulated for 16 h with TGFβ (5 ng/ml). Results of five independent experiments are shown by dot plot chart; ***P < 0.001 versus siNTC. (D) qRT-PCR analysis (top) and Western blot control (bottom) to investigate the role of JUNB in TGFβ-induced gene expression. MCF10A MII cells were transfected with non-targeting control (siNTC) or specific JUNB siRNA and stimulated for 1.5 or 16 h with TGFβ (5 ng/ml). Results of five independent experiments are shown by dot plot chart; *P < 0.05, **P < 0.01. (E) ChIP-qPCR showing time-dependent recruitment of JUNB to the indicated gene loci in MCF10A MII cells before (–) or after TGFβ treatment (1.5 or 16 h). Results of three independent experiments are shown by dot plot chart; *P < 0.05, ***P < 0.001.
We next analyzed the expression profiles of AP1 at protein and mRNA levels (Figures 3B and Supplementary Figure S2A). Both JUN, JUNB, FOS, FOSB and FOSL2 were strongly induced after TGFβ treatment, while FOSL1 was suppressed at the mRNA level but unaffected at the protein level, in line with our previous findings (16). Moreover, in these cells JUNB was most critical for TGFβ-induced invasion as well as induction of some invasion-associated genes (16). It is also of note that JUNB gene amplification occurred in 1–14% of breast cancer patients (Supplementary Figure S2B) (40,42,43). In addition, patients with JUNB amplification had a trend of poorer prognosis (Supplementary Figure S2C), although this was not statistically significant because of the small number of cases. We therefore decided to functionally assess the role of JUNB in the recruitment of SMAD2/3 to the late TGFβ-induced gene program.
We first analyzed again the three well known TGFβ/SMAD target genes, SERPINE1, LAMB3, and MMP2. Knock-down of JUNB strongly inhibited the recruitment of SMAD2/3 to the SERPINE1 and LAMB3 gene loci after 16 h of TGFβ stimulation (Figure 3C and Supplementary Figure S2D), while SMAD2/3 recruitment to the MMP2 gene locus was not affected. Moreover, knock-down of JUNB inhibited TGFβ-induced mRNA expression of SERPINE1 and LAMB after prolonged TGFβ stimulation, but not of MMP2, and phosphorylation of SMAD 2 and 3 was hardly influenced (Figure 3D). The late JUNB-dependent binding of SMAD2/3 to the SERPINE1 and LAMB3 gene loci (Figure 3C and Supplementary Figure S1A), correlated with enhanced binding of JUNB to the same gene loci (Figure 3E). Based on these results, we hypothesized that JUNB may determine the target- and time-specificity of SMAD complexes as a co-binding factor for a specific subset of invasion genes.
A JUNB-mediated feed-forward mechanism regulates genes associated with cell adhesion and invasion, and controls invasion in zebrafish xenograft models
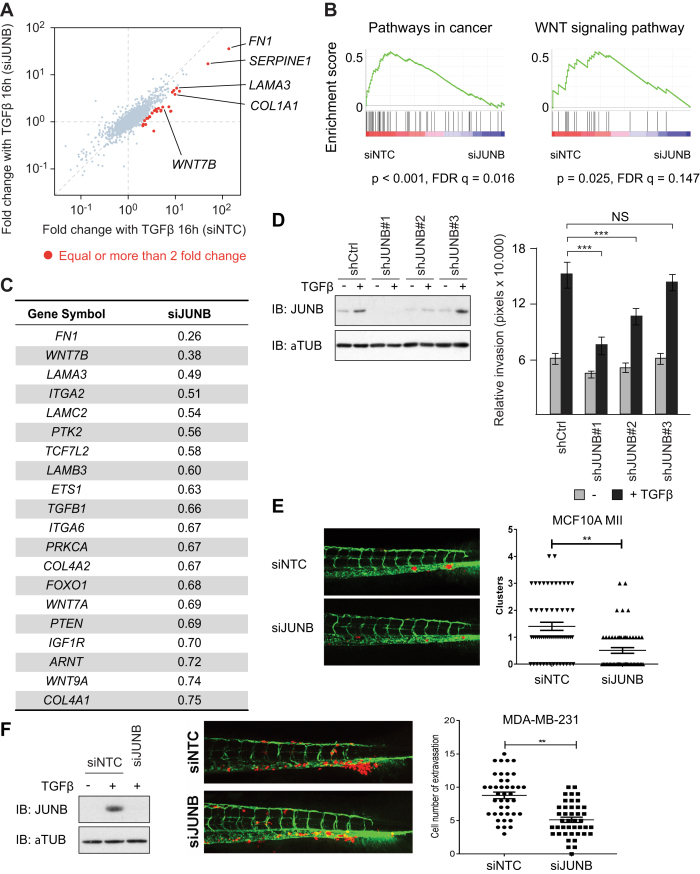
To characterize the significance of JUNB for TGFβ-SMAD-target genes on a genome-wide scale, we performed RNA-seq transcriptome analysis in JUNB-knock-down MCF10A MII cells (Figure 4A and Supplementary Figure S3A). We found that several well-characterized TGFβ-SMAD-target genes associated with cell adhesion, invasion and mesenchymal phenotype, e.g.fibronectin (FN)1 and integrin α (ITGA)2, were dependent on JUNB-induction (Supplementary Figure S3B), which was also confirmed by GO analysis (Supplementary Figure S3C). Interestingly, 20 genes appeared in the core-enriched genes of the pathway ‘Pathways in cancer’ in GSEA analysis (Figure 4B and C), at least 8 of which, FN1, ITGA2, ITGA6, LAMA3, LAMB3, LAMC2, collagen (COL)4A1, and COL4A2, are known target genes of TGFβ (8,47–49). In addition, genes in the WNT signaling pathway were enriched, which is discussed.
Figure 4.
A JUNB-mediated feed-forward mechanism regulates genes associated with cell adhesion, invasion and controls invasion in a zebrafish model. (A) Scatter plot representing fold change after TGFβ (5 ng/ml) treatment. Each point represents values of a gene. Genes whose induction after 16 h TGFβ (5 ng/ml) treatment was attenuated more than 50% with siJUNB treatment are colored red. (B) GSEA of expression changes of SMAD2/3 target genes after manipulation of JUNB expression. The SMAD2/3 target genes were pre-rank-ordered according to their fold change (log2) between siNTC and siJUNB, and analyzed based on KEGG signaling pathway enrichment. Gene sets with P-value < 5% and FDR q-value < 25% were considered significant. Enrichment score (ES) is plotted on the y axis. (C) A list of core-enriched genes of the pathway ‘Pathways in cancer’, which contribute most to the enrichment score of the pathway. (D) Stable knock-down of JUNB in MCF10A MII cells with three distinct shJUNB expressing lentiviral vectors. Whereas #1 is efficient, #3 does not inhibit JUNB expression. Left: Western blot analysis. Right: collagen invasion of MCF10A MII spheroids stably expressing the sh control (Ctrl) or three distinct shJUNB lentiviral constructs. Spheroids were embedded in collagen in the absence or presence of TGFβ (5 ng/ml) as indicated. Relative invasion was quantified as the mean area that the spheroids occupied 36 h after being embedded in collagen. Data represent means ± SD (n ≥ 6 spheroids per condition) and are representative of three independent experiments; ***P < 0.001. (E and F) MCF10A MII (E) or MDA-MB-231 (F) mCherry cells transfected with non-targeting control (siNTC) or specific JUNB siRNA (siJUNB) were injected into the ducts of Cuvier (DoC) of 48 h post-fertilization (hpf) zebrafish embryos. Left: representative images of zebrafish at 6 days post-injection (dpi). Right: quantification of invasive cell cluster numbers in non-targeting and JUNB knock-down cells injected zebrafish larvae. (F) Most left, western blot control of knock-down efficiency.
Taken together, the gene set analysis presented above, and the observation that JUNB is required for efficient expression of selected TGFβ-SMAD-target genes associated with cell invasion and mesenchymal phenotype ((16), Figures 3D and 4C), suggest that a late SMAD/JUNB-induced gene program is critical for TGFβ-induced invasion and cancer progression. In line with this hypothesis, we previously found transient siRNA-mediated knock-down of JUNB to result in strongly reduced TGFβ-induced invasion of MCF10A MII spheroids in collagen (16). To further validate these data, we stably knocked down JUNB with lentiviral vectors, which showed that decreased levels of JUNB correlate with decreased collagen invasion (Figure 4D). To examine the importance of JUNB in breast cancer cell invasion in vivo, we used an embryonic zebrafish xenograft invasion model (27). We have previously demonstrated that TGFβ signaling is critical for MCF10A MII invasion in this model (50). Importantly, knock-down of JUNB with siRNA resulted in reduced invasion compared to non-targeting siRNA control groups (Figure 4E). Moreover, knock-down of JUNB also resulted in reduced zebrafish invasion of the TGFβ-dependent metastatic human breast cancer cell line MDA-MB-231 (51,52) (Figure 4F). These results confirm that JUNB is important for breast cancer invasion.
Since tumorigenesis is a long-term event, we next verified whether a more extended TGFβ exposure, up to 72 h, results in a similar ‘late-stage’ TGFβ-induced gene expression program as 16 h treatment. As exemplified in Supplementary Figure S4A, the data obtained for these later time points were consistent with the data obtained at 16 h. In addition, since we identified the mesenchymal marker fibronectin as one of the main JUNB-dependent genes (Figure 4A, C and Supplementary Figure S3B), we examined the effect of JUNB depletion in the human pulmonary adenocarcinoma cancer cell line A549, which undergoes epithelial-mesenchymal transition (EMT) in response to prolonged TGFβ stimulation. The expression of various TGFβ-induced mesenchymal and/or EMT controlling genes was severely reduced by JUNB knock-down in these pulmonary adenocarcinoma cells (Supplementary Figure S4B), and JUNB was also found to be critical for invasion of A549 cells in the zebrafish xenograft model (Supplementary Figure S4C), This further confirms the pro-oncogenic protential of JUNB in TGFβ induced invasion.
Activation of the WNT signaling pathway strengthens the TGFβ-induced migratory phenotype
Interestingly, we also found that genes related to the WNT signaling pathway were enriched among the late TGFβ target genes, in addition to the genes associated with adhesion and invasion (Figures 2E and 4B). We therefore focused on the most prominent JUNB-dependent WNT pathway and breast cancer associated gene in the list, WNT7B, and examined its importance in TGFβ-induced cell migration and invasion. Our SMAD2/3 ChIP-seq and -qPCR analysis showed enhanced TGFβ-induced binding of SMAD2/3 to the WNT7B locus in a time-dependent manner (Figure 5A and Supplementary Figure S5A). In line with this, WNT7B expression was preferably induced after prolonged TGFβ-treatment (Figure 5B). Moreover, WNT7B was induced after prolonged TGFβ stimulation in a SMAD4- and JUNB-dependent manner (Figure 5C). The late JUNB-dependent expression of WNT7B and the time-dependent recruitment of SMAD2/3 to the WNT7B locus (Figure 5A), correlated with enhanced binding of JUNB to the same gene locus after 16 h of TGFβ stimulation (Figure 5D). Together, these results identify WNT7B as a JUNB-mediated late TGFβ-SMAD-target gene.
Figure 5.
Activation of the WNT signaling pathway strengthens the TGFβ-induced migratory phenotype. (A) ChIP-qPCR showing time-dependent recruitment of SMAD2/3 binding to the WNT7B gene locus in MCF10A MII before (–) or after TGFβ (5 ng/ml) treatment (1.5 and 16 h). Results of four independent experiments are shown by dot plot chart; *P < 0.05. (B) qRT-PCR analysis showing time-dependent WNT7B mRNA expression in MCF10A MII before (–) or after TGFβ (5 ng/ml) treatment (1.5 or 16 h). Results of six independent experiments are shown by dot plot chart; **P < 0.01. (C) Left: qRT-PCR analysis of WNT7B mRNA expression in MCF10A MII cells transfected with the indicated control (siNTC) or JUNB and SMAD4 specific siRNAs, and stimulated for 16 h with TGFβ (5 ng/ml). Results of four independent experiments are shown by dot plot chart; **P < 0.01 versus siNTC TGFβ 16 h. Right: Western blot control of knock-down efficiency. (D) ChIP-qPCR showing time-dependent recruitment of JUNB to the WNT7B gene locus in MCF10A MII before (–) or after TGFβ (5 ng/ml) treatment (1.5 and 16 h). (E) Collagen invasion assay of MCF10A MII spheroids stably expressing control GFP or ectopic WNT7B-MYC. Spheroids were embedded in collagen in the absence or presence of TGFβ, the TGFβRI inhibitor (ALK5i) SB505124 (2.5 μM) or the WNT inhibitor (WNTi) IWP-2 (5 μM), as indicated. Left: representative pictures of spheroids taken 36 h after being embedded in collagen. Right: relative invasion was quantified as the mean area that the spheroids occupied 36 h after being embedded in collagen. Data represent means ± SD (n ≥ 6 spheroids per condition) and are representative of three independent experiments; ***P < 0.001. (F) Western blot analysis of the MCF10A MII cells stably expressing control GFP or ectopic WNT7B-MYC. Cells were treated for 12 h with TGFβ (5 ng/ml) in the absence or presence of DMSO control, the TGFβRI inhibitor (ALK5i) SB505124 (2.5 μM) or the WNT inhibitor (WNTi) IWP-2 (5 μM), as indicated. (G) qRT-PCR target gene analysis of the cells shown in E and F, treated for 16 h with TGFβ (5 ng/ml) as indicated. A representative results of three independent experiments is shown.
To directly test if WNT7B is important for TGFβ-induced invasion, we performed collagen invasion assays. Addition of the TGFβ type I kinase receptor (TGFβRI) inhibitor SB505124 almost completely blocked TGFβ-induced collagen invasion of MCF10A MII spheroids, as expected (Figure 5E). Addition of the general WNT-inhibitor IWP-2 (53) also significantly inhibited TGFβ-induced invasion. To directly evaluate the role of WNT7B, we generated MCF10A MII cells stably expressing WNT7B (Supplementary Figure S5B). Exogenous expression of WNT7B enhanced both basal and TGFβ-induced invasion (Figure 5E). Consistent with this finding, addition of recombinant WNT7A, which was also one of the late TGFβ target genes (Figure 4C) and shares 82% amino acid identity with WNT7B, or expression of WNT7A, enhanced both basal and TGFβ-induced invasion (Supplementary Figure S5C and S5D). Interestingly, addition of the TGFβRI inhibitor SB505124 strongly inhibited TGFβ-induced invasion also in WNT7B expressing cells (Figure 5E), suggesting that WNT7B stabilizes the TGFβ-induced migratory phenotype of epithelial cells, rather than merely functioning as a downstream mediator of TGFβ signaling. In line with this notion, we found enhanced levels of TGFβ-induced phospho-SMAD2 and 3 in WNT7B overexpressing cells, whereas the general WNT-inhibitor IWP-2 reduced this phosphorylation, and also in the parental cells (Figure 5F). In addition, the WNT7B overexpressing cells contained increased levels of activated phosphorylated ERK1/2 and the expression of various TGFβ/SMAD-induced invasion genes was enhanced (Figure 5G). This indicates that WNT7B increases invasion/migration to a large extent by enhancing TGFβ type I receptor mediated signaling.
WNT7B promotes breast cancer cell invasion
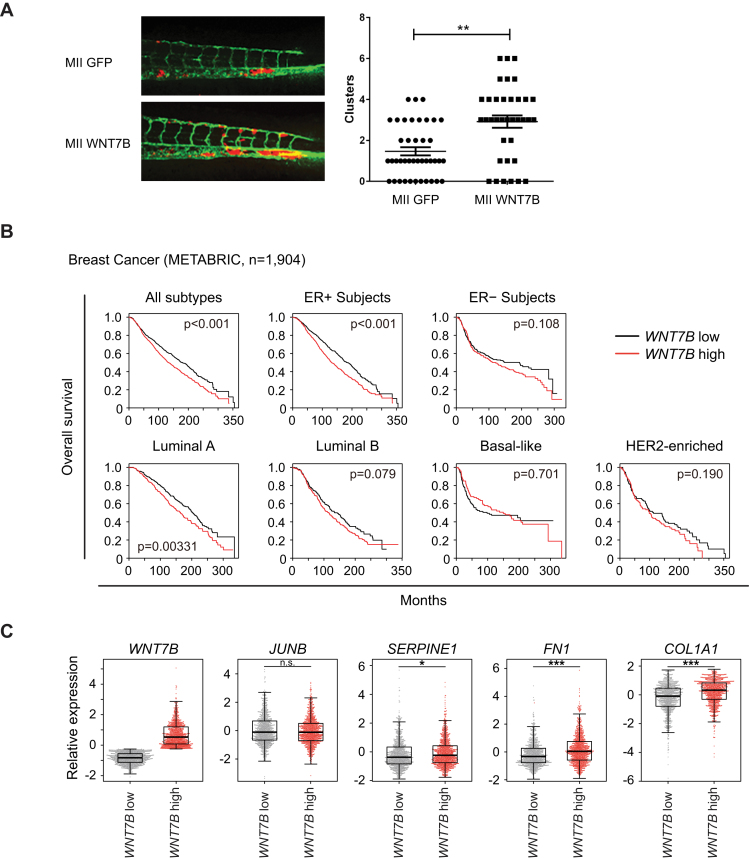
To investigate the role of WNT7B in invasion and metastasis in vivo, we again used the zebrafish embryo xenograft model. Embryos injected with MCF10A MII cells stably expressing WNT7B showed a significant increase in invasive cell numbers compared to control cells (Figure 6A). This result demonstrates that WNT7B expression stimulates MCF10A MII invasion in zebrafish.
Figure 6.
WNT7B promotes breast cancer cell invasion. (A) MCF10A MII mCherry stably expressing control GFP (MII GFP) or ectopic WNT7B-MYC (MII WNT7B) were injected into the DoC of 48-hpf zebrafish embryos. Left: representative images of zebrafish at 6 days post-injection (dpi). Right: quantification of invasive cell cluster numbers in GFP or WNT7B-MYC expressing MCF10A MII cells injected zebrafish larvae. (B) Kaplan-Meyer analysis of overall survival of breast cancer datasets from Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (40); all subtypes, n = 1904; ER+ subjects, n = 1445; ER− subjects, n = 429; luminal A subtype, n = 679; luminal B subtype, n = 461; HER2- subtype, n = 220; basal-like subtype, n = 199). Survival analysis was performed using a log-rank test. (C) Z-scored expression values of mRNA were obtained with cBioPortal (42,43). (***P < 0.001; n.s. not significant, Welch's t-test).
To further address the clinical significance of WNT7B expression in breast cancers, we analyzed patient datasets from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (40). We found that higher expression of the WNT7B gene was linked with shorter overall survival (Figure 6B). Moreover, high expression of WNT7B correlated with poorer prognosis in a cohort of ER+ tumors, especially in those of luminal type, but not of basal-like or triple negative breast cancers (TNBC). The WNT7B-high subgroup had higher mRNA expression of FN1 and COL1A1, well-established markers for the mesenchymal phenotype or tumor invasiveness (Figures 4A and 6C). In addition, we performed in silico meta-analysis of published microarray datasets using the Kaplan-Meier plots website (44), which also indicated that mRNA expression of WNT7B predicted poorer outcome especially in ER+ patients (Supplementary Figure S6A).
To verify whether ER-negative tumor cells have a similar genome-wide SMAD2/3 binding landscape as ER+ cells, we performed SMAD2/3 ChIP-seq analysis in the TNBC lines Hs-578-T and BT-549 (Supplementery Figure S6B). In Hs-578-T cells SMAD2/3 did not bind the WNT7B locus (Supplementery Figure S6B), while SMAD2/3 binding was observed in the WNT7B locus of BT-549 cells. However, in contrast to MCF10A MII cells, the number of SMAD2/3 binding sites was higher at 1.5 h than at 16 h with about 50% overlap (Supplementary Figure S6C). Moreover, although the AP1 motif was enriched in the SMAD2/3 binding sites in BT-549 (Supplementary Figure S6D), the data suggests that there is no JUNB-mediated redirection of SMAD2/3 in BT-549. Thus, our data showed heterogeneity among the TNBC cell lines.
The selective association in the ER+ group may be explained by the finding that TGFβ mainly functions as a tumor suppressor in the ER+ group, but as a tumor promoter in the ER- group of the breast cancer patients (13). Our data thus suggest that inhibition of the JUNB-mediated feed-forward loop may restore the tumor suppressive roles of TGFβ. It also implies that the feed-forward loop and/or activation of WNT7B signaling pathway may be a biomarker for the use of TGFβ inhibitors for tumor treatment.
DISCUSSION
It is well established that during the later stages of tumorigenesis TGFβ promotes tumor progression by enhancing migration, invasion and survival of tumor cells, by stimulating extracellular matrix deposition and tissue fibrosis, perturbing immune surveillance, stimulating angiogenesis and promoting EMT (8,11,15). One of the contributing factors is the effect of TGFβ on the tumor microenvironment, which in turn affects the tumor cells. In addition, sequential acquisition of genomic mutations changes the TGFβ responsiveness of cancer cells in a cell-intrinsic manner (54). For instance, in pancreatic cancer where SMAD4 mutations are common, loss of SMAD4 enables escape from cytostatic TGFβ effects or lethal effects associated with TGFβ-induced-EMT (55). In breast cancer cells, however, SMAD mutations are rare (56,57). This suggests that DNA-binding co-factors for SMADs, including JUNB, cause quantitative and/or qualitative changes in SMAD signaling and thereby play essential roles in the switch of the cancer-associated functions of TGFβ, from cytostasis/apoptosis to tumor-promotion.
We have previously demonstrated that SMAD3, SMAD4 and the AP1 components JUN, JUNB, FOS and FOSL1 cooperatively regulate several established TGFβ-target genes with a known function in EMT and invasion, including MMP1, MMP9, SNAI1 and SERPINE1, and enhance TGFβ-induced collagen invasion of MCF10A MII spheroids (16). The ChIP-seq and RNA-seq analyses in the current study show that the strong and prolonged induction of JUNB by TGFβ redirects SMAD2/3 to different target sites and thereby plays a major role in the activation of late TGFβ target genes as critical component of a feed-forward regulatory network. Interestingly, AP1 has previously been reported to potentiate chromatin accessibility of the glucocorticoid receptor (GR) in a murine mammary epithelial cells (58), and in human breast cancer cells to colocalize on the genome with YAP/TAZ/TEAD, Hippo pathway transducers and transcription factors (59). Since critical roles of AP1 components in breast cancer have been well documented, especially in the aggressive clinical subtype TNBC (60), induction of AP1 by TGFβ may potentiate aggressive phenotypes of breast cancer cells through other signaling pathways in vivo, in addition to the feed-forward network of TGFβ.
Interestingly, our list of late TGFβ target genes was enriched with signaling components of the WNT pathway (Figures 2E and 4B). It has been reported that a small portion of breast cancers (∼10%) express 30-fold higher levels of WNT7B compared with normal or benign breast tissues (61). In addition, recent data suggest that WNT7B is associated with anchorage-independent growth of breast cancer cells (62). The importance of crosstalk between TGFβ and WNT signaling pathways has been established (63,64). For acquisition of mesenchymal phenotypes in the breast TGFβ and WNT signaling pathways (both canonical and non-canonical) collaborate to activate mesenchymal genes and function in an autocrine fashion (65). Similarly, activation of canonical WNT signaling is required for TGFβ-mediated fibrosis (66). Furthermore, it was recently shown that WNT7A is secreted by breast tumor cells that promote fibroblast recruitment and conversion to a cancer-associated fibroblast (CAF) phenotype, which promotes metastasis (67). WNT7A-mediated CAF activation was mediated via enhanced TGFβ receptor signaling and not via classical WNT receptor signaling. This suggests that the JUNB-mediated feed-forward network of TGFβ is further stabilized by WNT ligands, resulting in more migratory and mesenchymal cell phenotypes. In line with this, we found enhanced ERK1/2 and SMAD2/3 phosphorylation, and enhanced TGFβ target gene expression in cells stably expressing WNT7B (Figure 5F and G), indicating that WNT7B increases invasion/migration in part by enhancing TGFβ type I receptor mediated signaling.
It should be noted that when we examined the role of canonical WNT signaling, as measured by TCF/LEF-dependent transcriptional reporter activity, we only found less than a two-fold increase by WNT7B (Supplementary Figure S5E). However, MII cells show autocrine TGFβ (-related) signaling (16,68) and our RNA sequencing analysis showed that both WNT7A, WNT7B and WNT9A besides being induced by TGFβ (Figure 4C) already show relatively high basal expression.
In accordance with our analysis, high expression of WNT7B mRNA was associated with poorer outcomes of ER+ breast cancer patients in a recent large-scale clinical study and meta-analysis (Figure 6B and C, and Supplementary Figure S6A). In line with this, SMAD2/3 ChIP-seq analysis in the TNBC lines Hs-578-T and BT-549 (Supplementary Figure S6B and C) showed that the binding patterns of SMAD2/3 in these TNBC cell lines are different from MII cells and, in addition, heterogeneity among the TNBC cell lines.
In summary, our study presents a model how JUNB mediates a TGFβ signaling feed-forward network in which WNT7B plays an effector role in specific breast cancer subtypes to promote breast cancer invasion (Supplementary Figure S7).
AVAILABLITY
ChIP- and RNA-seq raw data are available in Gene Expression Omnibus under accession number GSE83788.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues, in particular Aristidis Moustakas and Oleksander Voytyuk, for their valuable discussion, Martijn Rabelink, Sijia Liu, Kaori Shiina and Hiroko Meguro for technical assistance, the ENCODE Consortium for data use, and Leiden Genome Technology Center in Leiden University Medical Center for sequencing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Cancer Foundation [09 0773, 10 0452, 2016/445]; Swedish Research Council [2015-02757]; Kanae Foundation for Research Abroad; ITO Genboku and SAGARA Chian Memorial Scholarship (to M.M.); The Japan Society for the Promotion of Science (JSPS) (to N.K.); KAKENHI Grants-in-Aid for Scientific Research (S) [15H05774] (to K.M.). Funding for open access charge: Swedish Research Council [2015-02757].
Conflict of interest statement. None declared.
REFERENCES
- 1. Heldin C.H., Miyazono K., ten Dijke P.. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997; 390:465–471. [DOI] [PubMed] [Google Scholar]
- 2. Kang J.S., Liu C., Derynck R.. New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 2009; 19:385–394. [DOI] [PubMed] [Google Scholar]
- 3. Shi Y., Massagué J.. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003; 113:685–700. [DOI] [PubMed] [Google Scholar]
- 4. Massagué J., Seoane J., Wotton D.. Smad transcription factors. Genes Dev. 2005; 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- 5. Ross S., Hill C.S.. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 2008; 40:383–408. [DOI] [PubMed] [Google Scholar]
- 6. Akhurst R.J., Padgett R.W.. Matters of context guide future research in TGFβ superfamily signaling. Sci. Signal. 2015; 8:re10. [DOI] [PubMed] [Google Scholar]
- 7. Ikushima H., Miyazono K.. TGF-β signal transduction spreading to a wider field: a broad variety of mechanisms for context-dependent effects of TGF-β. Cell Tissue Res. 2012; 347:37–49. [DOI] [PubMed] [Google Scholar]
- 8. Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012; 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roberts A.B., Wakefield L.M.. The two faces of transforming growth factor β in carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:8621–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akhurst R.J., Derynck R.. TGF-β signaling in cancer–a double-edged sword. Trends Cell Biol. 2001; 11:S44–S51. [DOI] [PubMed] [Google Scholar]
- 11. Ikushima H., Miyazono K.. TGFβ signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010; 10:415–424. [DOI] [PubMed] [Google Scholar]
- 12. Padua D., Massagué J.. Roles of TGFβ in metastasis. Cell Res. 2009; 19:89–102. [DOI] [PubMed] [Google Scholar]
- 13. Moses H., Barcellos-Hoff M.H.. TGF-β biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 2011; 3:a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garraway L.A., Lander E.S.. Lessons from the cancer genome. Cell. 2013; 153:17–37. [DOI] [PubMed] [Google Scholar]
- 15. Sundqvist A., en Dijke P., van Dam H.. Key signaling nodes in mammary gland development and cancer: Smad signal integration in epithelial cell plasticity. Breast Cancer Res. 2012; 14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sundqvist A., Zieba A., Vasilaki E., Herrera Hidalgo C., Soderberg O., Koinuma D., Miyazono K., Heldin C.H., Landegren U., en Dijke P. et al. Specific interactions between Smad proteins and AP-1 components determine TGFβ-induced breast cancer cell invasion. Oncogene. 2013; 32:3606–3615. [DOI] [PubMed] [Google Scholar]
- 17. Coulouarn C., Factor V.M., Thorgeirsson S.S.. Transforming growth factor-β gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008; 47:2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Dam H., Castellazzi M.. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001; 20:2453–2464. [DOI] [PubMed] [Google Scholar]
- 19. Ozanne B.W., Spence H.J., McGarry L.C., Hennigan R.F.. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007; 26:1–10. [DOI] [PubMed] [Google Scholar]
- 20. Shaulian E., Karin M.. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002; 4:E131–E136. [DOI] [PubMed] [Google Scholar]
- 21. Qing J., Zhang Y., Derynck R.. Structural and functional characterization of the transforming growth factor-β-induced Smad3/c-Jun transcriptional cooperativity. J. Biol. Chem. 2000; 275:38802–38812. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., Feng X.H., Derynck R.. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998; 394:909–913. [DOI] [PubMed] [Google Scholar]
- 23. Javelaud D., Mauviel A.. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis. Oncogene. 2005; 24:5742–5750. [DOI] [PubMed] [Google Scholar]
- 24. Akhurst R.J., Hata A.. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012; 11:790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niwa H., Masui S., Chambers I., Smith A.G., Miyazaki J.. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol. 2002; 22:1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lawson N.D., Weinstein B.M.. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002; 248:307–318. [DOI] [PubMed] [Google Scholar]
- 27. He S., Lamers G.E., Beenakker J.W., Cui C., Ghotra V.P., Danen E.H., Meijer A.H., Spaink H.P., Snaar-Jagalska B.E.. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol. 2012; 227:431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Isogaya K., Koinuma D., Tsutsumi S., Saito R.A., Miyazawa K., Aburatani H., Miyazono K.. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res. 2014; 24:994–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morikawa M., Koinuma D., Mizutani A., Kawasaki N., Holmborn K., Sundqvist A., Tsutsumi S., Watabe T., Aburatani H., Heldin C.H. et al. BMP sustains embryonic stem cell self-renewal through distinct functions of different Krüppel-like factors. Stem Cell Rep. 2016; 6:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H.. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008; 26:1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 35. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hong C.P., Choe M.K., Roh T.Y.. Characterization of chromatin structure-associated histone modifications in breast cancer cells. Genomics Inform. 2012; 10:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A.. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009; 37:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016; 7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vasilaki E., Morikawa M., Koinuma D., Mizutani A., Hirano Y., Ehata S., Sundqvist A., Kawasaki N., Cedervall J., Olsson A.K. et al. Ras and TGF-β signaling enhance cancer progression by promoting the ΔNp63 transcriptional program. Sci. Signal. 2016; 9:ra84. [DOI] [PubMed] [Google Scholar]
- 42. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z.. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010; 123:725–731. [DOI] [PubMed] [Google Scholar]
- 45. Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 46. Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., Aburatani H.. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol. Cell. Biol. 2009; 29:172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roberts A.B., Sporn M.B., Assoian R.K., Smith J.M., Roche N.S., Wakefield L.M., Heine U.I., Liotta L.A., Falanga V., Kehrl J.H. et al. Transforming growth factor type β: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:4167–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ignotz R.A., Massagué J.. Transforming growth factor-β stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 1986; 261:4337–4345. [PubMed] [Google Scholar]
- 49. Pickup M., Novitskiy S., Moses H.L.. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer. 2013; 13:788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drabsch Y., He S., Zhang L., Snaar-Jagalska B.E., ten Dijke P.. Transforming growth factor-β signalling controls human breast cancer metastasis in a zebrafish xenograft model. Breast Cancer Res. 2013; 15:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Petersen M., Pardali E., van der Horst G., Cheung H., van den Hoogen C., van der Pluijm G., en Dijke P.. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 2010; 29:1351–1361. [DOI] [PubMed] [Google Scholar]
- 52. Deckers M., van Dinther M., Buijs J., Que I., Löwik C., van der Pluijm G., ten Dijke P.. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006; 66:2202–2209. [DOI] [PubMed] [Google Scholar]
- 53. Chen B., Dodge M.E., Tang W., Lu J., Ma Z., Fan C.W., Wei S., Hao W., Kilgore J., Williams N.S. et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009; 5:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A. Jr, Kinzler K.W.. Cancer genome landscapes. Science. 2013; 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. David C.J., Huang Y.H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., Massagué J.. TGF-β tumor suppression through a lethal EMT. Cell. 2016; 164:1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C., Nik-Zainal S., Martin S., Varela I., Bignell G.R. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012; 486:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.H., Trump S., Lightman S.L. et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011; 43:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., Piccolo S.. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015; 17:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao C., Qiao Y., Jonsson P., Wang J., Xu L., Rouhi P., Sinha I., Cao Y., Williams C., Dahlman-Wright K.. Genome-wide profiling of AP-1-regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res. 2014; 74:3983–3994. [DOI] [PubMed] [Google Scholar]
- 61. Huguet E.L., McMahon J.A., McMahon A.P., Bicknell R., Harris A.L.. Differential expression of human Wnt genes 2, 3, 4, and 7B in human breast cell lines and normal and disease states of human breast tissue. Cancer Res. 1994; 54:2615–2621. [PubMed] [Google Scholar]
- 62. Ni M., Chen Y., Lim E., Wimberly H., Bailey S.T., Imai Y., Rimm D.L., Liu X.S., Brown M.. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011; 20:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labbé E., Letamendia A., Attisano L.. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and Wnt pathways. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:8358–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nishita M., Hashimoto M.K., Ogata S., Laurent M.N., Ueno N., Shibuya H., Cho K.W.. Interaction between Wnt and TGF-β signalling pathways during formation of Spemann's organizer. Nature. 2000; 403:781–785. [DOI] [PubMed] [Google Scholar]
- 65. Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L. et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011; 145:926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Akhmetshina A., Palumbo K., Dees C., Bergmann C., Venalis P., Zerr P., Horn A., Kireva T., Beyer C., Zwerina J. et al. Activation of canonical Wnt signalling is required for TGF-β-mediated fibrosis. Nat. Commun. 2012; 3:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Avgustinova A., Iravani M., Robertson D., Fearns A., Gao Q., Klingbeil P., Hanby A.M., Speirs V., Sahai E., Calvo F. et al. Tumour cell-derived Wnt7a recruits and activates fibroblasts to promote tumour aggressiveness. Nat. Commun. 2016; 7:10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wiercinska E., Naber H.P., Pardali E., van der Pluijm G., van Dam H., ten Dijke P.. The TGF-β/Smad pathway induces breast cancer cell invasion through the up-regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Res. Treat. 2011; 128:657–666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.