Abstract
DNA recombination is required for effective segregation and diversification of genomes and for the successful completion of meiosis. Recent studies in various species hybrids have demonstrated a genetic link between DNA recombination and speciation. Consistent with this, we observed a striking suppression of recombination in the hybrids between two nematodes, the hermaphroditic Caenorhabditis briggsae and the gonochoristic C. nigoni. To unravel the molecular basis underlying the recombination suppression in their hybrids, we generated a C. nigoni genome with chromosome-level contiguity and produced an improved C. briggsae genome with resolved gaps up to 2.8 Mb. The genome alignment reveals not only high sequence divergences but also pervasive intra- and inter-chromosomal sequence re-arrangements between the two species, which are plausible culprits for the observed suppression. Comparison of recombination boundary sequences suggests that recombination in the hybrid requires extensive sequence homology, which is rarely seen between the two genomes. The new genomes and genomic libraries form invaluable resources for studying genome evolution, hybrid incompatibilities and sex evolution for this pair of model species.
INTRODUCTION
Genome stability is essential for maintaining species identity. Different populations of the same species may possess a certain level of sequence divergence, but in most cases they can still effectively exchange their genetic information through crossing and DNA recombination (1). However, this exchanges of genetic information between populations may be severely compromised or lost between species, a phenomenon called reproductive isolation, which ultimately leads to speciation. Reproductive isolation can take place before or after mating, which are referred to as pre- and post-zygotic hybrid incompatibility, respectively. The prevailing explanation for the post-zygotic hybrid incompatibility is that the differential divergence of genes between separate populations leads to disrupted interactions, resulting in hybrid sterility or death (2). A handful of such gene pairs have been identified whose interactions cause hybrid sterility or lethality (3,4).
Recent studies in hybrid mice support a genetic link between DNA recombination and speciation (5). For example, the hybrid sterility locus on chromosome X was found to regulate the meiotic recombination rate in mice. Recombination is required for effective segregation and diversification of the genomes during meiosis (6). Compromised recombination severely affects fertility. It is well established that chromosome rearrangements such as inversion or translocation inhibit DNA recombination (7,8). It is also possible that chromosome sequences demand substantial homology to allow chromosome pairing or crossover during meiosis. During chromosome pairing, the homologous copies of each chromosome find each other through an active search process that enables chromosomes to distinguish ‘self versus non-self’ and assume a side-by-side alignment (6). This pairing is essential for crossover formation and thus the successful completion of meiosis. However, the precise requirements for effective recombination remain elusive, especially in the case of hybrids.
We have recently characterized the post-zygotic hybrid incompatibilities by genome-wide introgression between two closely related nematode species, C. briggsae and C. nigoni (9,10), whose hybrids are partially viable (11). Backcrossing the C. briggsae X chromosome into an otherwise C. nigoni background frequently leads to hybrid male sterility. The gene cbr-him-8 was identified as a possible recessive maternal-effect suppressor of F1 hybrid male-specific lethality in crosses with C. briggsae as the mother (12), although these results remain controversial (13). To develop a molecular understanding of this male sterility, we previously generated a C. nigoni draft genome using Illumina synthetic long reads (SLRs) with a contig N50 size of 57 kb (14). We demonstrated that spermatogenesis genes were significantly downregulated in the sterile males compared to the wild type. Intriguingly, the down-regulated genes were targeted by a subset of 22G RNAs that were significantly upregulated in the sterile males compared with the wild-type males (15). To facilitate genome wide comparisons, we generated so called C. nigoni pseudo chromosomes by chaining the contigs together using the homology to C. briggsae genome. However, this approach could not identify genome rearrangements between the two species because this step forces the C. nigoni to have a similar syntenic block to that of C. briggsae, but does not improve the actual size of the contigs themselves. Therefore, C. nigoni genome is rather fragmented given its small size of contig N50.
Although the recombination frequency shows variations along chromosome arms and between autosome and sex chromosome in C. briggsae (16), the overall frequency does not exhibit a significant drop compared with other phylogenetic groups. We referred to recombination frequency drop as large chromosomal regions being refractory to recombination rather than the reduction of recombination frequency to less than one recombination event per meiosis. Surprisingly, the recombination frequency is severely compromised in the hybrids between the two species, compared with the intraspecific crosses in C. briggsae. In addition, the two species use different mating systems, with C. briggsae being androdioecious (coexistence of males and hermaphrodites) and C. nigoni being gonochoristic (11). The mechanism through which recombination becomes suppressed in the hybrids remains largely unknown. However, the widespread recombination suppression in these hybrids between C. briggsae and C. nigoni provides an excellent opportunity to unravel the sequence requirements for effective recombination. The key to addressing these questions relies on the existence of high quality of genome sequences for both parental species. Here we first produced an improved C. briggsae genome using SLRs and high-throughput chromosome conformation capture (Hi-C) data. We next generated a C. nigoni genome assembly with chromosome-level contiguity using a combination of SLRs, Hi-C, Nanopore sequencing and mate-pair sequencing of the ends from both fosmid and BAC genomic libraries. Alignment between these two high-quality genomes allowed us to pinpoint the molecular bases of recombination suppression in the hybrids between C. briggsae and C. nigoni.
MATERIALS AND METHODS
Strains, maintenance and backcrossing
All strains were maintained at 25°C on NGM plates seeded with OP50 as food source. Strain AF16 and JU1421 were used as a wild isolate for C. briggsae and C. nigoni, respectively, for genomic DNA preparation. The strain JU1421 was subject to inbreeding for at least 70 generations by crossing a single young adult male with a single L4 female in three replicates before extraction of genomic DNA for preparation of sequencing or genomic libraries. Backcrossing was performed as described previously (9).
Generation of SLRs for C. briggsae genome
Genomic DNA was extracted from mixed-staged C. briggsae animals using PureLink® Genomic DNA Mini Kit (Invitrogen). The sequencing libraries were prepared according to Illumina's protocol as described previously (14). 500 ng high quality genomic DNA was sheared in g-Tube by Covaris sonicator and went through end repair, dA-tailing and adaptor ligation. Products after ligation were size-selected for 8–10 kb by agarose gel followed by qPCR quantification. In the first stage, the sequencing and PCR errors of the reads were corrected and read depths were normalized across fragments. In the second stage, the String Graph Assembler (SGA) was used to construct a string graph based on the pre-processed reads, which was used to produce an initial set of contigs with the paired-end information from the short reads. In the third stage, the contigs were further scaffolded to resolve repeats and fill gaps. After examination of possible sequence errors and mis-assemblies, the assembled long-reads were generated as FASTQ files and used in the subsequent analysis.
Construction of C. nigoni fosmid and BAC library
For construction of fosmid genomic library, DNA extraction was performed by using PureLink® Genomic DNA Mini Kit (Invitrogen). Fosmid library was prepared according to the protocol of NxSeq® 40 kb Mate Pair Cloning Kit (Lucigen) with modifications. The genomic DNAs were end-repaired and run at 0.7% agarose gel at 40 V overnight. The end-repaired genomic DNAs were size-selected for fragments pf around 40 kb. Size-selected genomic DNAs were recovered by using GELase Enzyme and GELase 50X Buffer provided in CopyControl™ Fosmid Library Production Kit with pCC1FOS™ Vector (Epicentre). The recovered genomic DNAs were ligated into pNGS FOS Cloning Vector provided in NxSeq® 40 kb Mate Pair Cloning Kit (Lucigen), which contains NGS primer binding sites. The recombinant pNGS FOS fosmid clones were packaged into MaxPlax Lambda Packing Extract provided in CopyControl™ Fosmid Library Production Kit with pCC1FOS™ Vector (Epicentre). The packed fosmid clones were transfected into Replicator FOS strain provided in NxSeq® 40 kb Mate Pair Cloning Kit (Lucigen) and spread on LB agar plates with 12.5 μg/ml chloramphenicol. The clones were inoculated into 96-well plates with LB broth and 12.5 μg/ml chloramphenicol, cultured and then stored in –80°C. Construction of BAC genomic library was outsourced to Genome Resource Laboratory of Huazhong Agricultural University using the methods described previously (17).
Paired-end sequencing of C. nigoni fosmid or BAC library by NGS or nanopore sequencing
The fosmid clones were inoculated into 96-well plates with LB broth and 12.5 μg/ml chloramphenicol. They were cultured in 37°C for 16 h and pooled with a four-dimension strategy by columns, rows, plates with the same last digit and plates with the same second last digit. The fosmids were extracted from those pools and subject to CviQI enzyme digestion. The digested DNAs were run at 0.7% agarose gel at 85 V for 50 min and were size-selected for 8–9 kb. The recovered fosmid backbones carrying two ends of genomic DNA insertions were self-circularized and subject to PCR amplification with barcoded Illumina compatible primers (Supplementary Table S1). The PCR products were sequenced on MiSeq at 2 × 250 bp mode or Nanopore sequencing and the end sequences generated were used in the subsequent analysis. Mate-pair sequencing of BAC ends and its data analysis were performed as described previously (17).
C. briggsae genome repair and de novo genome assembly
15× SLRs were aligned against C. briggsae genome (‘cb4’) using BWA-MEM (18). If at least four SLR showed 100% sequence identity and spanned both ends of a gap with any number of ‘N’, the ‘Ns’ were replaced and the missing sequences within the gap were added with the sequences within the SLR. If the end of a gap could be aligned to two separate SLRs, the gap was filled as described previously (14). The process was reiterated for 12 times till no more ‘Ns’ could be replaced by any SLRs, which generated an assembly called ‘cb4_improve’. De novo genome assembly using SLRs was also performed basically as described (14). Scaffolding of the SLR-derived contigs using Hi-C data was described below to generate an assembly called ‘cb_SLR_Hi-C’.
Hi-C sequencing and data analysis
Hi-C sequencing was performed as described previously (19) with modifications. Briefly, approximately 10 000 synchronized L1 animals were fixed with 2% formaldehyde, snap frozen in liquid nitrogen for one hour, then quenched by glycine at room temperature followed by brief centrifugation. The remaining DNA ligation and Illumina sequencing library preparation steps were performed basically according to those described earlier (19), without size selection.
For Hi-C data analysis, a duplicate-free list of paired alignments of Hi-C reads against a given contig assembled with SLRs was generated using Juicer pipeline (20). The paired alignments were used as input for 3D de novo assembly (3D-DNA) pipeline (20) with the following parameters: haploid model, three rounds of mis-join correction and six expected chromosomes. To validate the genome assembly using the Hi-C reads, a de novo assembled C. briggsae genome was generated using ∼100× Hi-C data and 15× contigs generated using SLRs with an N50 of 61 kb. Comparison of chromosomal collinearity between the de novo generated C. briggsae assembly (cb_SLR_Hi-C) and the existing C. briggsae genome assembly ‘cb4’ was performed to provide confidence level of the newly generated genome with Hi-C data (Figure 2D). Approximately 100X C. nigoni Hi-C data were used in the similar way as that of C. briggsae using the contigs we produced earlier using Illumina SLRs with an N50 size of 57 kb (14). Further scaffolding process was performed for C. nigoni genome using mate-pair sequencing data of the ends of fosmid (∼3×) and BAC (about 6X), with an average insertion size of 37 and 140 kb, respectively, using SSpace-3.0 (21) with default parameters.
Figure 2.
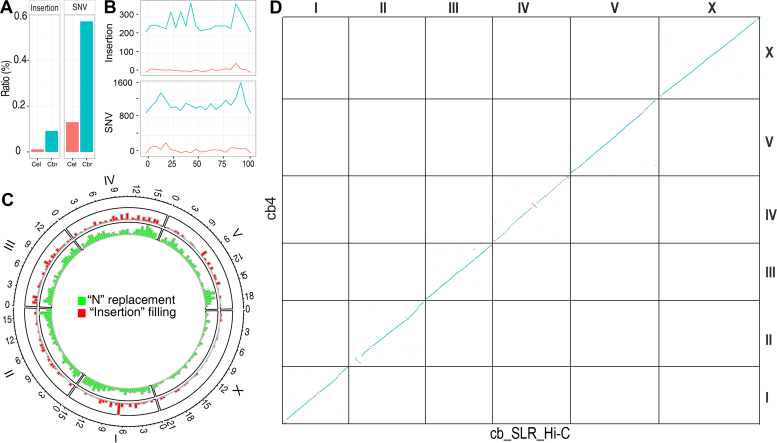
C. briggsae genome repairing and de novo assembly using SLRs and Hi-C sequencing. (A and B) Comparison of sequence discrepancies between the SLRs and C. elegans (brown) or C. briggsae (cyan) genome. (A) Ratios of insertion or SNV (single nucleotide variation) identified in the SLRs when compared with the two genomes. (B) Distribution of insertions (top) or SNVs (bottom) across the relative length (percentile) of chromosome III with a bin size of 100 kb. (C) Distribution of ‘repaired’ sequences across C. briggsae genome. Recovered missing sequences in the current assembly (‘cb4’) is shown in red as ‘insertion filling’ and replacement of gap sequences (i.e. those present as ‘N’ in ‘cb4’) with actual base pairs is shown in green as ‘N’ replacement for each chromosome. (D) Dotplot of DNA sequences between ‘cb4’ and our C. briggsae de novo genome assembly using SLRs and Hi-C data (see Materials and Methods).
Nanopore sequencing and data analysis
Genomic DNAs were prepared from mix-staged and starved C. nigoni animals using Qiagen DNeasy Blood & Tissue Kit followed by shearing to 8–10 kb using Covaris G-TUBE. Nanopore sequencing was performed with MinION using Ligation Sequencing Kit 1D from Oxford Nanopore Technologies according to the manufacturer's description. Around 30× reads (assuming C. nigoni genome size of ∼130 Mb) were generated and were subject to self-correction using Canu assembler (22). The corrected reads were used for de novo genome assembly using Canu or scaffolding using SSpace-3.0 (21).
Analysis of synteny between C. briggsae and C. nigoni
One-to-one orthologous gene pairs were defined by mutual best BLASTP hits. The orthologous pairs were used as an input to MCScan package (23), to scan for syntenic block, which demanded at least ten contiguous orthologous genes as an seed. At most five gene rearrangements between the two seeds were allowed for extending the syntenic block.
C. nigoni gene prediction
The gene prediction were carried out using Marker 3.0 with default EVidenceModeler (EVM) parameters (24). Three ab initio gene prediction tools, the Augustus (25), SNAP (26) and GeneMark (27) were incorporated into the Maker pipeline. De novo mRNA assembly with Trinity and the pre-trained Augustus parameters were inherited from our previous study. The smallest size of a single exonic gene was set as ten amino acids for SNAP. Protein and CDS sequences from C. briggsae, C. elegans, C. sinica and C. remanei were used for sequence alignment using BLASTX or BLASTN, which were input for Maker pipeline.
Identification of orthologs and phylogenetic analysis
Protein coding genes among 10 Caenorhabditis species from Wormbase (WS252) was used to identify ortholog groups using OrthoMCL (v.2.0.9) (28) based on all-vs-all BLASTP with an E-value cutoff of 1 × e–5. The aligned protein sequences of a total of 248 single-copy orthologs among the 10 species were used for phylogenetic analysis. The maximum-likelihood (ML) analysis was performed by using the bootstrap RAxML pipeline (29) implemented in ETE3 package (30). The divergent time estimation was calculated using RelTime method (31), with calibration by including two established pair-wise divergence time between species (32).
Gene family (Pfam domain) analysis
All protein coding genes from all 14 species were scanned against Pfam A domains using ‘hmmscan’ (hmmer.org). A domain with multiple copies in a protein was counted only once. The Pfam domain counts in C. nigoni were compared against C. briggsae. Significance of the difference between the two species were compared with Fisher's exact test in R (V3.1.1). Only gene families with a P value <0.05 were considered as significantly expanded/contracted. The gain or loss of a domains in the evolution history of all the 10 species were calculated using Count (33). The numbers of gene gain or loss on each branches of the phylogenetic tree were calculated using Wagner parsimony (34).
Analysis of conservation of the sequences flanking recombination sites
A total of 19 introgression boundaries were mapped with NGS previously (9). 12 out of the 19 boundaries were able to be mapped with up to 250-bp resolution (approximately the read size of MiSeq sequencer), which were used in this analysis. Mapping resolution for the remaining boundaries was too poor to be used for the same analysis. The poor resolution was mainly due to the low sequencing coverage (<3×) in the relevant regions.
The 6-kb flanking genomic regions centering on the 12 mapped boundaries were retrieved from C. briggsae genome. Corresponding 6-kb syntenic regions from C. nigoni were retrieved using the C. briggsae 6-kb sequences as a query. Pairwise Smith-Waterman (SW) alignment was implemented using SIMD Smith–Waterman Library (35) with default parameters. SW alignment scores were calculated using the following parameters, i.e. match = 2, mismatch = –1, gap open = –3, gap extend = –1.
For permutation analysis, one thousand 6-kb genomic regions without any gap were randomly sampled from C. briggsae chromosomes, which were used to retrieve their syntenic regions in C. nigoni genome. Pairwise alignment and its score calculation were performed in the similar way as that for the boundary sequences. Statistical difference in alignment scores between the boundary and permutated sequences was inferred with Wilcoxon test implemented in R package.
RESULTS
Recombination suppression in the hybrids between C. briggsae and C. nigoni
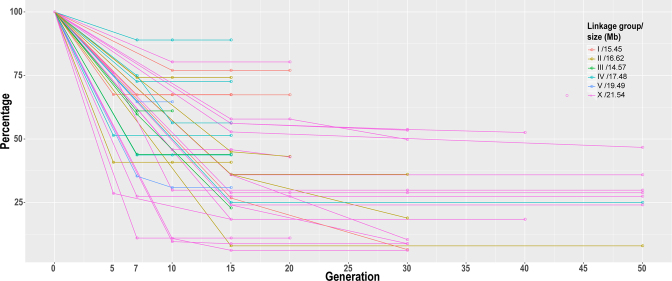
We previously performed backcrossing of 96 chromosomally integrated GFP markers in the C. briggsae genome into C. nigoni using the GFP as a selection marker (9). We observed an unusually large introgression size after 15 generations of backcrossing compared with that expected from intraspecific crossing in C. briggsae (16). The number of backcrossing generations was arbitrarily chosen based on Drosophila backcrossing data (36). To examine whether more backcrossing steps could reduce the introgression size, we chose a subset of these strains carrying a GFP-labeled introgression fragment for further backcrossing followed by genotyping with single-worm PCR. Unfortunately, most of these strains demonstrated little or no reduction in introgression size even after 50 generations of backcrossing (Figure 1). Many of the introgressions retained over one-fourth of the original chromosome, and approximately half of them contain nearly half of the entire tagged chromosome after 15 or more generations of backcrossing, indicating that recombination became more difficult for the marked DNA fragment in the hybrids than in its parental species, and the size of the introgression fragment could not be reduced simply by more backcrossing steps. However, the mechanism behind the suppressed recombination was not clear.
Figure 1.
Change of C. briggsae introgression size in percentage in C. nigoni background over backcrossing generations. Shown are 40 independent introgression lines, each carrying a GFP insertion on individual C. briggsae chromosomes that are differentially color-coded. Y axis denotes the percentage of remaining C. briggsae chromosome after backcrossing with C. nigoni and X axis the crossing generation. C. briggsae chromosome sizes in Mb are indicated on top right. Each dot indicates timing for genotyping with single-worm PCR or NGS.
Highly accurate SLRs allow gap filling in the existing C. briggsae genome
To unravel the molecular basis of the recombination suppression in the hybrids, we set out to improve the existing C. briggsae genome (‘cb4’), which contains 5997 gaps represented by an arbitrary number of ‘Ns’ on six chromosomes and 361 ‘unassigned contigs’ that were not anchored to any chromosome (16). We previously used the SLRs to generate a de novo assembly of the C. elegans genome (14). The SLRs were generated from next-generation sequencing (NGS) reads but with a much higher read length, i.e. ∼9.2 kb. We demonstrated that the reads were highly accurate and were able to recover most types of repetitive sequences except tandem repeats. Here, we generated approximately 15X C. briggsae (AF16) SLRs with an N50 size of ∼8.8 kb by a similar method to that used for C. elegans (14). As expected, the overall alignment of the C. elegans-derived SLRs (14) against the C. elegans genome (37) showed a very low ratio of discrepancies, whereas a similar alignment of the C. briggsae-derived SLRs against the C. briggsae genome (‘cb4’) revealed a much higher level of discrepancy (Figure 2A and B), indicating the presence of many more errors in the ‘cb4’ genome assembly than in the C. elegans genome. The N50 size of the contigs assembled with the SLRs was expected to be significantly improved compared with those obtained by de novo genome assembly using NGS reads only (38,39).
We previously demonstrated that the SLRs were not reliable in detecting extra inserted sequences that were incorrectly included in the C. elegans reference genome, but they were highly accurate at the nucleotide level and were fairly reliable in identifying missing sequences in this genome (14). We therefore decided to repair the C. briggsae genome in a similar manner to fill the gaps (i.e., those represented by ‘Ns’ in the ‘cb4’ assembly) or recover missing sequences using the SLRs. In total, we closed a total of 2304 out of the 5997 gaps by replacing a total of roughly 240 kbs on six chromosomes (Supplementary Table S2). We also partially closed gaps in ‘unassigned contigs’ by identifying and filling ‘self-mapped’ or ‘cross-mapped’ gaps as we did previously (14), which allowed extension of roughly 800-kb sequences. In total, we filled gaps by replacing or adding a total of 1040 kb of sequences across the ‘cb4’ genome using the SLRs (Figure 2C). We validated a subset of five cases in which the missing sequences were more 100 bps in length by PCR as we did previously (14). All the missing sequences were found to be real as judged by the expected sizes of the PCR products (data not shown). We next repaired the C. briggsae genome by anchoring a total of 124 out of 361 ‘unassigned’ contigs listed in the ‘cb4’ assembly with a cumulative size of ∼1.8 Mb back to individual chromosomes. This was achieved by alignment between ‘cb4’ assembly and the de novo assembly produced by a combination of SLRs and Hi-C data as described below. Seven out of the 124 were merged with our updated genome with DDtools (https://jgi.doe.gov/data-and-tools/bbtools/) using the cutoff of 98% sequence identity (Supplementary Table S3). In summary, we were able to update a total of 2.8 Mb of the C. briggsae genome, including the replaced ‘Ns’ or the recovered missing sequences and the ‘unassigned’ contigs that were anchored back to chromosome (Table 1).
Table 1. Statistics of newly assembled C. nigoni genome (‘cn2’) and improved C. briggsae genome ‘cb4_improve’.
| C. nigoni (‘cn2’) | C. briggsae (‘cb4_improve’) | C. briggsae (‘cb4’) | ||||||
|---|---|---|---|---|---|---|---|---|
| LG | Size (bp)* | Gene count | % repeat | Size (bp) | Updated sequence (bp)# | Size (bp) | Gene count | % repeat |
| I | 14 110 143 | 2454 | 30.5 | 15 452 308 | 661 459 | 15 455 979 | 3076 | 26.1 |
| II | 20 338 624 | 3520 | 31.2 | 16 619 993 | 307 295 | 16 627 154 | 3324 | 27.0 |
| III | 15 784 304 | 2554 | 35.8 | 14 574 343 | 355 551 | 14 578 851 | 2884 | 26.2 |
| IV | 22 101 115 | 2584 | 33.2 | 17 474 118 | 476 300 | 17 485 439 | 3271 | 23.8 |
| V | 24 289 761 | 4470 | 33.3 | 19 485 256 | 799 850 | 19 495 157 | 4799 | 22.5 |
| X | 27 349 973 | 4053 | 26.8 | 21 532 320 | 224 578 | 21 540 570 | 3744 | 17.6 |
| Un | 11 770 734 | 2827 | 27.8 | 1 378 321 | NA | 3 201 015 | 765 | 30.4 |
| Total | 135 744 654 | 22 462 | 31.1 | 107 902 949 | 2 825 033 | 108 384 165 | 21 863 | 23.7 |
*including ‘Ns’ estimated with BAC or fosmid interval. # including replaced ‘Ns’, newly added sequences and ‘unassigned’ contigs anchored back to chromosome. Number of the ‘Ns’ in some gaps seems overestimated as judged by the sequences present in the SLRs covering these gaps. LG, linkage group. NA, not applicable.
To independently validate the repaired sequences by SLRs, we examined two cases that were located within an existing gene model and caused gene model change after sequence repairing (Supplementary Figure S1). To verify the corrected gene models, we mapped the existing RNA-seq data (15,40,41) against the revised sequence. We observed perfect alignments between some RNA-seq reads and the newly added sequences, indicating a missing exon and its flanking sequences in the existing gene model.
Evaluation of Hi-C data for contig scaffolding
We previously produced a C. nigoni draft genome using approximately 22X SLRs (15). As the SLRs could not further extend a contig when they encounter tandem repeats, the overall contig size of the assembly was relatively small with an N50 size of 57 kb. To facilitate the molecular characterization of the recombination suppression in the hybrids and empower the species pair as a model for speciation genetics and sex evolution, we decided to generate a C. nigoni genome assembly with chromosome-level contiguity using a combination of data consisting of the previous SLRs, Hi-C, Nanopore sequencing reads and the mate-pair end-sequences of both fosmid and BAC genomic libraries.
To gain confidence of the Hi-C data for de novo genome assembly, we first produced C. briggsae Hi-C data with around 100X coverage. We then used these data to perform de novo C. briggsae genome assembly by using the SLRs. The resulting assembly was 108 Mb in size (Table 1), which was comparable with the size of ‘cb4’. We called this assembly ‘cb_SLR_Hi-C’ (Figure 2C). The total size of contigs that could be assembled into chromosome was only 95 Mb, in contrast to the 105 Mb of sequences assigned to chromosomes in the ‘cb4’ assembly, leading to 13 Mb of contigs that could not be assigned onto any chromosome (referred to as ‘unassigned’) (data not shown). This relatively large portion of ‘unassigned contigs’ was likely due to the tandem repeats present at their ends.
The alignment of the newly generated C. briggsae assembly against ‘cb4’ revealed an excellent collinearity, albeit with some obvious inversions on the chromosomes II, IV and X (Figure 2D), suggesting potential errors in the ‘cb4’ assembly. Importantly, the N50 size of the scaffolded contigs increased from 57 kb to over 10 Mb after incorporation of the Hi-C data (Figure 3B). These results indicate that Hi-C data are highly reliable for scaffolding of SLR-derived contigs. The comparison of ‘cb_SLR_Hi-C’ with ‘cb4’ allowed a total of 124 ‘unassigned’ contigs with a combined size of 1.8 Mb present in the ‘cb4’ assembly to be anchored back to individual chromosomes (Supplementary Table S3). As a result, we generated an improved C. briggsae genome assembly called ‘cb4_improve’ after the incorporation of both SLRs and the Hi-C data. It is worthy of note that the ‘cb4_improve’ retains the ‘cb4’ scaffolds, but with updated sequences by SLRs and a subset of ‘unassigned’ contigs anchored back to individual chromosomes (see Materials and Methods).
Figure 3.
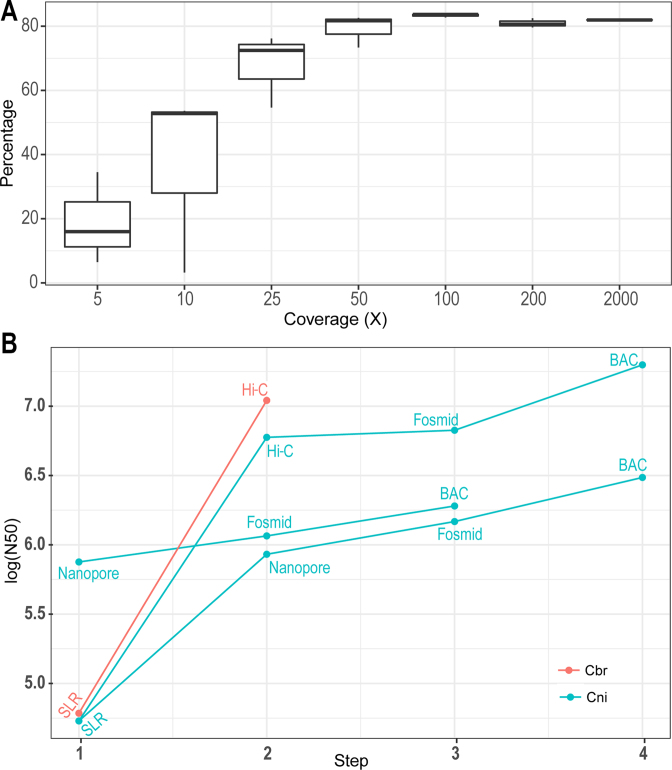
C. nigoni genome assembly strategies. (A) Evaluation of the effect of Hi-C sequencing depth on scaffolding of contigs assembled with SLRs. Shown is plotting of the contig percentage that can be incorporated into scaffold (Y axis) against genomic coverage of Hi-C data (X axis). (B) Comparison of scaffold N50 sizes with different assembly strategies. Shown are plottings of log10 scale of N50 size (bp) achieved with various assembly method. SLR: contigs assembled with Illumina Synthetic long reads only. cbr, C. briggsae; cni, C. nigoni.
A high-quality of C. nigoni genome with chromosome-level of contiguity
We next produced Hi-C data using C. nigoni synchronized L1 animals (see Materials and Methods) with a genome coverage of up to 2000× of its genome. Modeling analysis showed that the contribution of the Hi-C data to the scaffolding of the SLR-derived contigs reached a plateau with approximately 50X Hi-C reads (Figure 3A), demonstrating that an 100X coverage of Hi-C data is sufficient for scaffolding purposes when used with SLR-derived contigs. Given the relatively small proportion of the contigs that could be assigned into individual chromosomes compared with that using genetic mapping data in C. briggsae, i.e. 95 versus 105 Mb (16), to further improve scaffolding results, we produced roughly 15× fosmid and 10× BAC (Bacterial Artificial Chromosome) genomic libraries using C. nigoni DNAs, respectively. We were able to obtain unambiguous mate-pair end sequences for about 3× fosmid (∼6133 clones) (Supplementary Table S4) and 8× BAC (6442 clones) libraries (Supplementary Table S5). In addition, we generated around 30× Nanopore sequencing reads with MinION to further facilitate scaffolding (see Materials and Methods).
Starting from the SLR derived contigs we produced earlier (15), we first performed scaffolding using the 100× Hi-C data, resulting in a sharp increase in the scaffold N50 size from 57 kb to ∼7 Mb, which is approaching a chromosome-level of contiguity (Figure 3B). As a result, we were able to obtain six large contigs, each representing an individual chromosome. Despite the modest contribution of the fosmid end-sequences to the contig N50 size, the incorporation of the BAC end-sequences more than doubled the N50 size, from 7 to 17 Mb, which is essentially the chromosome size of the ‘cb4’ assembly (16). As an alternative genome assembly strategy, we performed scaffolding of the SLR-derived contigs first using the Nanopore sequencing data. This resulted in an increase in the N50 size from 57 kb to approximately 1 Mb. Further scaffolding with the fosmid and BAC end-sequences led to a linear increase in the N50 size, to approximately 1.5 and 3.2 Mb, respectively (Figure 3B). We also explored another strategy by performing de novo genome assembly first with the Nanopore sequencing reads followed by scaffolding using the fosmid and BAC end sequences. Using this method, we were able to achieve an N50 size of approximately 1.8 Mb. Given the different scaffold sizes achieved with the various assembly strategies, we decided to use the C. nigoni genome assembly with the first strategy with a genome size of approximately 135 Mb (Table 1) for the subsequent analysis, which was termed as ‘cn2’. The assembly consisted of 124.0 Mb of chromosomal contigs and 11.8 Mb of small contigs that could not be assigned to any chromosome (Table 1). The total size was ∼27 Mb bigger than that of the C. briggsae genome (‘cb4’) (Table 1). To examine whether the size difference is contributed by repetitive sequence, we defined repetitive sequences in both genomes using RepeatMasker (repeatmasker.org). We found extra repetitive sequences in C. nigoni genome accounted for roughly 17 out of the 27 Mb genomic sequences, while the remaining 10 Mb genome sequences could not be explained by repetitive sequences. However, it is plausible that these regions may encode quite a few genes, but they are missed in our gene list due to under-prediction as described below.
A total of 22 462 protein-coding genes were predicted in the newly assembled genome (Table 1), compared with the 21 863 C. briggsae genes annotated in Wormbase (WS252) (42). It should be noted that the genes were likely be under-predicted in C. nigoni. This was because the EVM parameters we used to finalize the gene model demanded a satisfaction score from all inputs, including ab initio gene prediction results from all independent gene-prediction algorithms, and the results from de novo assembled mRNAs as well as gene-model conservation across species (see Materials and Methods). Therefore, the predicted genes may only represent the highly reliable ones. However, there might be a substantial number of genes that were missed out in our prediction, for example, those highly divergent ones or those with simple gene structure. Comparison of one-to-one orthologs (mutual best hits) allowed us to infer a phylogenetic distance of between Caenorhabditis. Using an estimated divergence time of ∼30 million years between C. briggsae and C. elegans (32) as a reference, we estimated the divergence time between C. briggsae and C. nigoni is ∼3.7 million years (Supplementary Figure S2). This is roughly comparable with what was estimated previously between the two species using synonymous site divergence (43).
Chromosomes of C. briggsae and C. nigoni show overall collinearity but demonstrate substantial sequence divergences and pervasive rearrangements
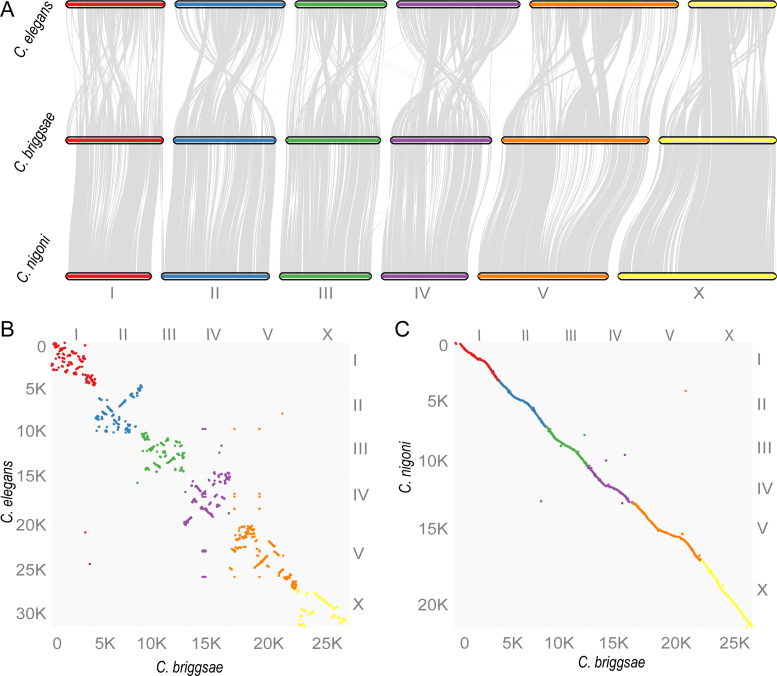
To pinpoint the molecular basis of the recombination suppression, we first examined the overall DNA sequence conservation between the two species. We then evaluated the synteny between the two genomes using one-to-one orthologous genes. Specifically, we performed genome-wide sequence alignment between the two genomes (see Materials and Methods) and extracted the best hits as their corresponding genomic regions for visualization of genomic collinearity and arrangement. In principle, most of the repetitive sequences should be excluded from the orthologous regions because few of them could find a single best hit. The results show that overall genome sequences were alignable and are located within the corresponding chromosomal parts (Figure 4A and C). However, considering that their female hybrid progeny were fertile, the level of sequence divergence between the two genomes was surprisingly high. For example, the coding sequences of their one-to-one orthologs exhibited only around a 90% identity, and most of their introns and intergenic regions were not alignable (Figure 4A and data not shown).
Figure 4.
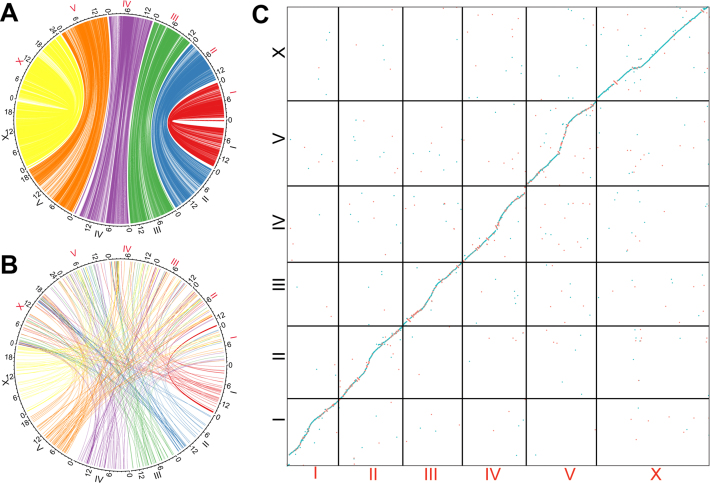
Genomic collinearity and rearrangement between C. briggsae and C. nigoni. (A) Diagram showing corresponding sequences defined by mutual best hits using LASTAL with a size cutoff of one Kb. Note the white regions are not alignable. Names of C. briggsae and C. nigoni chromosomes are labeled in black and red, respectively. (B) Diagram showing inter-chromosomal translocations that contain sequence over 1 kb in size between the two species. Chromosome names are similarly labelled as in (A). (C) Dotplot of corresponding chromosomes between C. briggsae (‘cb4_improve’, black) and C. nigoni (‘cn2’).
To examine the genomic collinearity and rearrangement, we extracted all of the best hits between the two genomes that were >1 kb in size. We observed over 400 inter-chromosomal translocations across the genomes (Figure 4B). Many more translocations were found within a single chromosome than between chromosomes (data not shown). We postulated that the presence of these pervasive genomic rearrangements and widespread unalignable sequences prevents effective chromosome crossover, thus inhibiting recombination in the hybrids. Given that hybrid females between C. briggsae and C. nigoni are fertile, it is unexpected that the two genomes have undergone such a high level of sequence divergence and rearrangement.
Despite the apparent recombination suppression in the hybrids between C. briggsae and C. nigoni, we were able to produce homozygous viable introgressions representing ∼28% of the C. briggsae genome (9), indicating that over one quarter of the genome was interchangeable between the two species. To examine whether the inviability of the remaining C. briggsae genome in the C. nigoni background as homozygote was caused by gene rearrangements leading to abnormal gene loss or gain, we produced one-to-one orthologous gene pairs between C. briggsae and C. nigoni, as defined by the mutual best hits using BLASTP. This resulted in a total of 15,157 orthologous gene pairs (Supplementary Table S6). To contrast the genic synteny between C. briggsae and C. nigoni and that between C. briggsae and C. elegans, we retrieved C. briggsae’s one-to-one orthologous gene pairs with C. elegans from WormBase (WS252). The pairwise comparisons between C. briggsae and C. nigoni revealed an excellent genic synteny between the two species. In contrast, frequent intrachromosomal rearrangements were observed between C. briggsae and C. elegans (Figure 5). These results demonstrated that the large C. briggsae chromosome fragments that were not viable in the C. nigoni background were unlikely to be caused by major inversions or translocations that prevent chromosome alignment, but possibly by local rearrangements or sequence divergences that compromise the inheritance of orthologous gene pairs.
Figure 5.
Comparison of synteny consisting of gene blocks (see Materials and Methods) between C. briggsae and C. nigoni or C. elegans. A total of 15 157 (Supplementary Table S6) and 7679 (WS252) orthologous gene pairs were used for comparison between C. briggsae and C. nigoni or C. elegans, respectively, with a bin size of 30 genes. (A) Syntenic view. (B and C) Dotplot view. Gene count is indicated on each axis along with its associated chromosome (differentially color coded).
In addition to the one-to-one orthologs, many genes were found to have undergone substantial species-specific amplification or loss (Supplementary Table S7). Therefore, these could not be unambiguously assigned as orthologous pairs. Many of the C. nigoni-specific genes seemed to originate from transposable elements (Supplementary Figures S3A and S4). Others encoded putative proteins but with unknown functions. In contrast, many of the C. briggsae-specific genes encoded transcription factors or proteins containing domains of unknown function (Figures S3B and S4). It is not clear why these transcription factors underwent specific amplification in C. briggsae. Despite the species-specific amplification or loss of genes, the overall number of protein domains gained or lost was roughly comparable for the two species (Supplementary Figure S5). The C. nigoni genome contained a substantially higher proportion of repeats than the C. briggsae genome (Table 1). The difference in repeat content accounted for 57% of the differences in genome size difference between the two species.
Recombination in hybrids appears to demand an extended region of high sequence identity
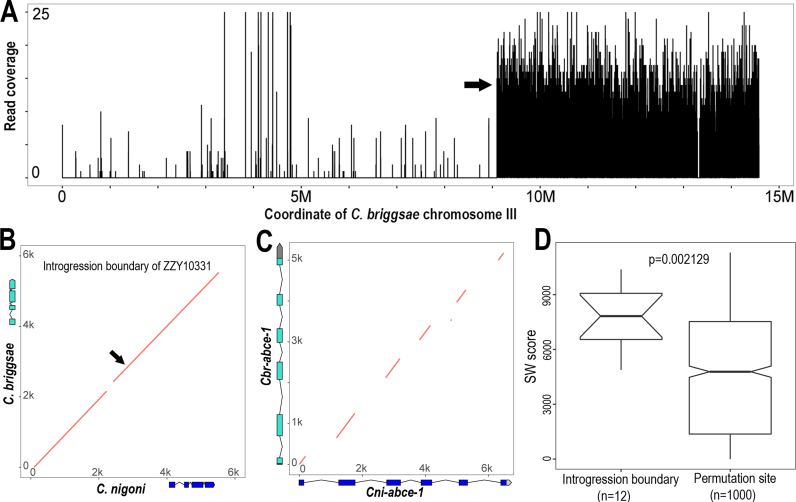
Availability of precise collinearity of the genomic sequences between C. briggsae and C. nigoni permitted pinpointing of the detailed sequence requirements for recombination in the hybrids. We previously mapped the recombination boundaries for all of the homozygous introgressions using NGS with up to 100-bp resolution (9). We first focused on one particular introgression, ZZY10331, for which the NGS data had the highest coverage (∼13×) (Figure 6A) to characterize the boundary sequences of recombination site. This introgression was mapped onto the right arm of C. briggsae chromosome III, with its left and right boundaries extending to the middle and the very end of the chromosome, respectively. We retrieved the sequence of a 6-kb C. briggsae genomic fragment centered on the mapped recombination site of the left boundary. All of the sequence and syntenic information between C. briggsae and C. nigoni can be found at http://158.182.16.70:8080/. Most parts of the sequences within the region were alignable against their C. nigoni syntenic regions, albeit with a 100-bp gap (Figure 6B). Surprisingly, in addition to the coding sequences of the gene present within the region, the intergenic sequences and introns were also alignable, which seems contradicting our prediction that most of these regions are not alignable. We next asked whether the intronic sequences were conserved between orthologous pair. To this end, we performed alignment of syntenic 6-kb regions between C. briggsae and C. nigoni that each contained an orthologue of abce-1, an extremely conserved and essential gene in C. elegans. This gene was found to be essential in protein translation and its depletion produced 100% embryonic lethality. Importantly, it was present as a single copy across metazoan genomes (10,44). As expected from our genome-wide sequence alignment, we observed excellent alignment only between exonic regions, but not between intronic regions (Figure 6C), demonstrating that most of the intronic regions were barely alignable. We finally assess whether the recombination boundary sequences are more likely to be conserved than genomic background systematically. To this end, we performed pairwise alignments of the 6-kb syntenic regions centering on the 12 recombination sites which were mapped to resolution up to 100 bp. We calculated their alignment scores using SIMD Smith–Waterman Library (35). To calculate alignment score for background sequences, we performed permutation analysis by randomly choosing one thousand 6-kb syntenic regions between the two species followed by alignment score calculation. We observed a significantly higher conservation level in recombination sites than in genomic background (P = 0.002129, Wilcoxon test) (Figure 6D). The results suggested that the conservation of sequences extending up to 6 kb in length in the recombination site was likely to be an exception in the non-coding region, and that such a conserved block of multiple kbs in length was possibly the minimal requirement for recombination to take place in a hybrid.
Figure 6.
Conservation of sequences flanking recombination site between C. briggsae and C. nigoni. (A) The left and right boundaries of an introgression (ZZY10331) were mapped onto the right arm of C. briggsae chromosome III by NGS. Shown is the coverage of sequencing reads (Y axis) derived from hybrid mapped against C. briggsae chromosome III (X axis). The left boundary is indicated with an arrow and the right boundary is mapped to the very end of the chromosome. (B) Dotplot of the 6-kb syntenic sequences flanking the recombination site (indicated with arrow) between C. briggsae and C. nigoni. Gene models within the region are shown in scale. (C) Dotplot of the 6-kb homologous sequences spanning the genomic regions of a highly conserved gene, abce-1 between C. briggsae and C. nigoni. abce-1 gene models are shown in scale for both species. Note that only exons are conserved. (D) Boxplot of alignment scores for the 6-kb syntenic genomic DNAs of recombination regions (left) or randomly sampled genomic regions (right). SW score, Smith–Waterman alignment score.
DISCUSSION
DNA recombination is required for the correct segregation of genetic material and successful completion of meiosis. It is regulated by both cis-acting elements and trans-acting factors, which rapidly evolve across species (45). The highly divergent cis elements and trans factors presumably serve to reduce the recombination compatibility between species. Indeed, recent studies in mice and other species indicate a genetic link between recombination and speciation (46). Consistent with this, we found DNA recombination was severely suppressed in the hybrids between C. briggsae and C. nigoni compared with that in the parental species (16). The availability of high-quality genomes for both species allowed us to pinpoint the molecular basis behind the suppressed recombination in the hybrids.
First, the cis elements and trans factors might be too divergent to be compatible with each other for recombination to take place in the hybrids between C. briggsae and C. nigoni. Consistent with this, there are only 15 157 one-to-one orthologs between the two, although a total of 21 863 (WS252) and 22 462 genes (Table 1, this study) were predicted in the genomes of C. briggsae (47) and C. nigoni, respectively. Second, most parts of the two genomes might be too divergent to allow the pairing or crossover of homologous chromosomes that is critical for recombination to takes place (48). For example, the overall sequence identity of the coding sequences from the one-to-one orthologous genes was <90%, and the vast majority of intronic and intergenic regions were not alignable (Figures 4A and 6C). Third, despite the overall synteny of orthologous protein-coding genes between the two species, there are pervasive intra- and inter-chromosomal sequence rearrangements between the two genomes, which are expected to impair recombination. Fourth, the size of C. nigoni genome was substantially larger than that of C. briggsae, i.e. 136 versus 108 Mb, although some residual heterozygosity could have contributed to overestimation of the C. nigoni genome size. Some of the divergent alleles could be erroneously treated as paralogs, especially for those sequences present as ‘unassigned’ contigs, which are highly enriched for repetitive fragments. Genome shrinkage in a selfing species (e.g. C. briggsae) versus its outcrossing relative, for example, C. nigoni, seems not uncommon (49). Nearly half of the reduced genome in C. briggsae could be explained by its lost repeats relative to the C. nigoni genome. These differences in genome size and repeat content are expected to create gaps in the sequence alignment or make the syntenic sequences not alignable, thus decreasing recombination efficiency. Nevertheless, it remains possible that bombardment may have introduced some balanced chromosomal rearrangements into our GFP lines that are expected to prevent effective recombination (7,8) but are not detected by our genotyping steps. Alternatively, the presence of the GFP transgene may disrupt synteny completely in an unnatural way, which could exert some sort of ‘shadow’ of recombination suppression around itself that would not be observed in unmarked DNA.
Genome instability was proposed to be intimately linked to speciation and evolution (50). The newly generated C. nigoni genome and the improved C. briggsae genome will facilitate the studies of genome evolution and speciation genetics. They should also facilitate the studies of sex evolution given the distinct reproductive modes of the two species. Our end-sequenced fosmid and BAC libraries provide invaluable resources for the functional characterization of hybrid incompatible loci and other DNA elements. The availability of boundary-resolved introgressions in the context of syntenic chromosomal regions should form a foundation for further studying the molecular and genetic control of recombination frequency.
Nanopore sequencing is expected to perform better in de novo genome assembly than it did in our study. One potential issue is that the read coverage was not deep enough. We produced only 30× reads of the C. nigoni genome. Given the high error-rate of its read sequence, some remaining errors in its self-corrected reads may prevent it from effective scaffolding and contig assembly. In addition, we performed size selection for the DNA library, which was optimized to recover genomic fragments of ∼ 8–10 kb in length. This would constrain the power of their reads for scaffolding or de novo sequencing. The generation of a higher coverage of reads and size selection of fragments with greater size may boost their performance in de novo genome assembly.
We did not expect that Hi-C data to be more valuable than Nanopore reads for scaffolding the SLR-derived contigs (Figures 2D and 3). The superior performance relative to the Nanopore reads was likely the result of the higher coverage (∼100×) and the highly accurate read sequences generated by NGS. In summary, the improved C. briggsae genome and the new C. nigoni genome provide insights into the unusual low rate of recombination for the large C. briggsae DNA fragments in C. nigoni background. The availability of both genomes forms invaluable resources for elucidation of the molecular mechanisms underlying the hybrid incompatibilities between the two species.
AVAILABILITY
C. briggsae SLRs and Hi-C data and C. briggsae genome assembles were deposited into SRA database under BioProject number PRJNA396670. C. nigoni Hi-C, Nanopore sequencing data, BAC and fosmid end-sequencing data as well as C. nigoni genome assembly ‘cn2’ and its annotation files were deposited into SRA database under BioProject number PRJNA306403. Synteny and genomic positions of BAC and fosmid clones can be accessed under designated website, http://158.182.16.70:8080/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr. Chung Wai Shing for the logistic support and the members of Zhao's lab for helpful discussion and comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
General Research Funds [HKBU12103314, HKBU12123716] from Hong Kong Research Grant Council; HKBU Strategic Development Fund (SDF 15-1012-P04) to Z.Z.; National Natural Science Foundation of China [91231109] To X.L.; National Natural Science Foundation of China [61472205, 81630103], China’s Youth 1000-Talent Program and Beijing Advanced Innovation Center for Structural Biology to J.Y. Funding for open access charge: HKBU12123716.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dey A., Chan C.K.W., Thomas C.G., Cutter A.D.. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11056–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coyne J.A., Orr H.A.. Speciation. 2004; Sunderland, MA: Sinauer Associates; 545. [Google Scholar]
- 3. Presgraves D.C. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 2010; 11:175–180. [DOI] [PubMed] [Google Scholar]
- 4. Maheshwari S., Barbash D.A.. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 2011; 45:331–355. [DOI] [PubMed] [Google Scholar]
- 5. Balcova M., Faltusova B., Gergelits V., Bhattacharyya T., Mihola O., Trachtulec Z., Knopf C., Fotopulosova V., Chvatalova I., Gregorova S. et al. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLOS Genet. 2016; 12:e1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hillers K.J., Jantsch V., Martinez-Perez E., Yanowitz J.L.. Villeneuve A, Greenstein D. Meiosis. WormBook. The C. elegans Research Community. 2017; WormBook; doi:10.1895/wormbook.1.178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKim K.S., Howell A.M., Rose A.M.. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988; 120:987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zetka M.C., Rose A.M.. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics. 1992; 131:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bi Y., Ren X., Yan C., Shao J., Xie D., Zhao Z.. A Genome-wide hybrid incompatibility landscape between Caenorhabditis briggsae and C. nigoni. PLoS Genet. 2015; 11:e1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan C., Bi Y., Yin D., Zhao Z.. A method for rapid and simultaneous mapping of genetic loci and introgression sizes in nematode species. PLoS One. 2012; 7:e43770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woodruff G.C., Eke O., Baird S.E., Felix M.A., Haag E.S.. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics. 2010; 186:997–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ragavapuram V., Hill E.E., Baird S.E.. Suppression of F1 male-specific lethality in Caenorhabditis hybrids by cbr-him-8. G3 (Bethesda). 2016; 6:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ryan L.E., Haag E.S.. Revisiting Suppression of Interspecies Hybrid Male Lethality in Caenorhabditis Nematodes. G3; Genes|Genomes|Genetics. 2017; 7:1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li R., Hsieh C.-L., Young A., Zhang Z., Ren X., Zhao Z.. Illumina Synthetic Long Read Sequencing Allows Recovery of Missing Sequences even in the ‘Finished’ C. elegans Genome. Sci. Rep. 2015; 5:10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li R., Ren X., Bi Y., Ho V.W.S., Hsieh C., Young A., Zhang Z., Lin T., Zhao Y., Miao L. et al. Specific downregulation of spermatogenesis genes targeted by 22G RNAs in hybrid sterile males associated with an X-Chromosome introgression. Genome Res. 2016; 26:1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ross J.A., Koboldt D.C., Staisch J.E., Chamberlin H.M., Gupta B.P., Miller R.D., Baird S.E., Haag E.S.. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011; 7:e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei X., Xu Z., Wang G., Hou J., Ma X., Liu H., Liu J., Chen B., Luo M., Xie B. et al. pBACode: a random-barcode-based high-throughput approach for BAC paired-end sequencing and physical clone mapping. Nucleic Acids Res. 2016; 45:gkw1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013; arXiv:1303.3997v1. [Google Scholar]
- 19. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., Shamim M.S., Machol I., Lander E.S., Aiden A.P. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017; 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W.. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011; 27:578–579. [DOI] [PubMed] [Google Scholar]
- 22. Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017; 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T., Jin H., Marler B., Guo H. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Campbell M.S., Law M., Holt C., Stein J.C., Moghe G.D., Hufnagel D.E., Lei J., Achawanantakun R., Jiao D., Lawrence C.J. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014; 164:513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stanke M., Morgenstern B.. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005; 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004; 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Besemer J., Borodovsky M.. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005; 33:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Stoeckert C.J., Roos D.S.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huerta-Cepas J., Serra F., Bork P.. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016; 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura K., Battistuzzi F.U., Billing-Ross P., Murillo O., Filipski A., Kumar S.. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cutter A.D. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008; 25:778–786. [DOI] [PubMed] [Google Scholar]
- 33. Csurös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010; 26:1910–1912. [DOI] [PubMed] [Google Scholar]
- 34. Farris J.S. Methods for computing Wagner trees. Syst. Biol. 1970; 19:83–92. [Google Scholar]
- 35. Zhao M., Lee W.-P., Garrison E.P., Marth G.T.. SSW library: an SIMD Smith-Waterman C/C++ library for use in genomic applications. PLoS One. 2013; 8:e82138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masly J.P., Presgraves D.C.. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 2007; 5:e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998; 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- 38. Mortazavi A., Schwarz E.M., Williams B., Schaeffer L., Antoshechkin I., Wold B.J., Sternberg P.W.. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res. 2010; 20:1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar S., Koutsovoulos G., Kaur G., Blaxter M.. Toward 959 nematode genomes. Worm. 2012; 1:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uyar B., Chu J.S., Vergara I.A., Chua S.Y., Jones M.R., Wong T., Baillie D.L., Chen N.. RNA-seq analysis of the C. briggsae transcriptome. Genome Res. 2012; 22:1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas C.G., Li R., Smith H.E., Woodruff G.C., Oliver B., Haag E.S.. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 2012; 22:2167–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C. et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016; 44:D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thomas C.G., Wang W., Jovelin R., Ghosh R., Lomasko T., Trinh Q., Kruglyak L., Stein L.D., Cutter A.D.. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 2015; 25:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Z., Fang L.L., Johnsen R., Baillie D.L.. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2004; 323:104–111. [DOI] [PubMed] [Google Scholar]
- 45. Wagner C.R., Kuervers L., Baillie D.L., Yanowitz J.L.. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature. 2010; 467:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Payseur B.A. Genetic links between recombination and speciation. PLoS Genet. 2016; 12:e1006066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A. et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003; 1:E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gong W.J., McKim K.S., Hawley R.S.. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 2005; 1:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fierst J.L., Willis J.H., Thomas C.G., Wang W., Reynolds R.M., Ahearne T.E., Cutter A.D., Phillips P.C.. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLOS Genet. 2015; 11:e1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dion-Côté A.-M., Barbash D.A.. Beyond speciation genes: an overview of genome stability in evolution and speciation. Curr. Opin. Genet. Dev. 2017; 47:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.