Abstract
Graded levels of molecular oxygen (O2) exist within developing mammalian embryos and can differentially regulate cellular specification pathways. During differentiation, cells acquire distinct epigenetic landscapes, which determine their function, however the mechanisms which regulate this are poorly understood. The demethylation of 5-methylcytosine (5mC) is achieved via successive oxidation reactions catalysed by the Ten-Eleven-Translocation (Tet) enzymes, yielding the 5-hydroxymethylcytosine (5hmC) intermediate. These require O2 as a co-factor, and hence may link epigenetic processes directly to O2 gradients during development. We demonstrate that the activities of Tet enzymes display distinct patterns of [O2]-dependency, and that Tet1 activity, specifically, is subject to differential regulation within a range of O2 which is physiologically relevant in embryogenesis. Further, differentiating embryonic stem cells displayed a transient burst of 5hmC, which was both dependent upon Tet1 and inhibited by low (1%) [O2]. A GC-rich promoter region within the Tet3 locus was identified as a significant target of this 5mC-hydroxylation. Further, this region was shown to associate with Tet1, and display the histone epigenetic marks, H3K4me3 and H3K27me3, which are characteristic of a bivalent, developmentally ‘poised’ promoter. We conclude that Tet1 activity, determined by [O2] may play a critical role in regulating cellular differentiation and fate in embryogenesis.
INTRODUCTION
Early embryonic development requires the establishment of complex and diverse cellular and tissue systems, characterized by tightly-controlled, spacio-temporal patterns of gene expression. However, the precise molecular mechanisms which direct this asymmetry within the early embryo remain poorly understood. The importance of gradients of ‘morphogens’ in this process has long been suggested (1). These are biological substances that diffuse between cells, and act to generate specific responses, dependent upon their concentrations (reviewed in (2)). Before the establishment of the circulatory system, mammalian development occurs under relatively low levels of O2 (estimated to be between 2% and 8%; reviewed in (3,4)), and the availability of O2 is determined by its diffusion. This will give rise to micro-gradients of O2 and more hypoxic niches within the early embryo (5,6). The functional significance of the levels of O2 in the regulation of cellular differentiation has been demonstrated and found to be both concentration- and context-dependent. Thus, several studies have demonstrated that the levels of available O2 can act to promote the differentiation of certain types of stem or progenitor cells and yet inhibit the differentiation of others (reviewed in (4)). Therefore O2 can be considered to be a developmental morphogen that can influence cell fate in a manner akin to gradients of secreted growth factors such as members of the Transforming Growth Factor-β superfamily (7).
The term ‘epigenetics’ has come to be defined as changes in gene expression, and hence cell function, that are not determined by DNA sequence (reviewed in (8)), and to a large extent this involves the reorganisation of chromatin structure to mediate the accessibility of specific gene loci to their cognate transcription factors. During development, changes to the chromosomal architecture act to determine gene expression patterns and ultimately determine cell fate (8). These chromosomal changes are primarily facilitated by two dynamically regulated processes; the methylation/demethylation of cytosine, primarily at CpG dinucleotides (9) and the post-translational modification of histone tails (10). The establishment of cell specificity and fate (and therefore the initiation of epigenetic changes) originates from the first stages of embryogenesis. Intriguingly, epigenetic modifying enzymes which act to demethylate both histones and 5-methylcytosine (5mC) are 2-oxoglutarate-(2-OG)-dependent, Fe2+-dependent dioxygenases, which display an absolute requirement for molecular oxygen (11). In the case of DNA demethylation, this is achieved by the successive oxidation of 5mC, by the Ten-Eleven-Translocation family of dioxygenases (Tets) resulting in the formation of 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (12–14). DNA repair mechanisms can subsequently excise 5fC and 5caC which then become replaced by unmethylated cytosine, in an ‘active’ demethylation process. Alternatively, the oxidised methylcytosines can be lost in a replication-dependent, ‘passive’ mechanism (15). However, 5hmC is not committed to spontaneous subsequent oxidation and demethylation, and hence also represents a stable epigenetic modification with specific regulatory functions which are increasingly being elucidated (16,17).
There are three Tet enzymes (13), which display tissue-specific and developmental-stage specific patterns of expression in mammalian cells (18–20). Consistently, significant expression of Tet1 and Tet2 has been reported in undifferentiated embryonic stem cells (ESCs) (13,21–24), while Tet3 expression is low in undifferentiated ESCs, becomes induced during ESC differentiation (18,21,25), and is known to be enriched in both developing and mature neuronal cell lineages (20,25,26). Loss-of-function studies both in vitro and in vivo have demonstrated these enzymes to serve both overlapping and distinct roles in the maintenance of ESC pluripotency (13,27–30) and also in cellular specification and transcriptional fidelity during development (13,21,25,31–36). However, the possibility that O2 gradients may mediate distinct cellular differentiation pathways, regulated in part via the differential regulation of these epigenetic modifiers, has not been investigated.
We here demonstrate that during the earliest stages (3 days) of mouse ESC (mESC) differentiation in vitro, a transient burst of (global) 5hmC, mediated specifically by Tet1, is apparent under atmospheric O2 levels, but is inhibited in O2-poor (1%) culture conditions, which are physiologically relevant within the early developing embryo. Unexpectedly, the most significant target of the [O2]-dependent 5mC- hydroxylation during these early stages of ESC differentiation was identified as a promoter region of Tet3. Culture of differentiating ESCs under 1% O2 acted to prevent 5mC-hydroxylation at this Tet3 promoter, and inhibited significantly the induction of Tet3 mRNA transcription later during ESC differentiation, concomitant with a loss in some molecular markers of neural lineages. These data therefore demonstrate the potential role of O2 gradients within the early embryo to regulate epigenetic changes and cellular differentiation via the differential modulation of the activity of Tet1.
MATERIALS AND METHODS
Cell culture
Human Embryonic kidney cells (HEK-293T) were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Sigma) and 2 mM l-glutamine, 100 U/ml penicillin, streptomycin 100 μg/ml solution (Sigma). Cells were transfected with Tet-overexpression plasmids were gifts from Anjana Rao, obtained from Addgene (Tet1:49792, Tet2:41710, Tet3:49446) and pcDNA™ 3.1 (Invitrogen) using Lipofectamine™-2000 (Invitrogen). All plasmids encode full-length, unmutated human (Tet1 and Tet3) or mouse (Tet2) proteins which have been fully sequenced (22,37,38). R1 mouse embryonic stem cells (mESCs), (kindly provided by Dr. Shukry Habib), were maintained undifferentiated on 0.1% gelatin coated flasks in DMEM/F12 (Invitrogen) containing 8 μM 2-mercaptomethanol, 2 mM GlutaMAX-I™ (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen), supplemented with 10% EmbryoMax FBS (Millipore), 20 ng/ml leukemia inhibitory factor (LIF; Millipore), 3 μM CHIR-99021 (Sigma), 1 μM PD0325901 (Sigma) and 5μg/ml plasmocin prophylactic (InvivoGen). Stable knockdown mESCs were generated from MISSION® shRNA lentiviral transduction particles (Sigma): Tet1 (TRCN0000341849), Tet2 (TRCN0000192770), Tet3 (TRCN0000376843) and MISSION® pLKO.1-puro non-targeted shRNA control (SHC016V-1). Cells were infected in the presence of 10 μg/ml polybrene (Sigma) and selected and maintained with 1 μg/ml puromycin (Invitrogen), 72-h post-infection. Unbiased mESC-induction to embryoid bodies was performed by culture in non-adherent petri dishes in KnockOut DMEM (Invitrogen) containing 15% KnockOut serum replacement (Invitrogen), 0.1 mM MEM amino acid solution (Invitrogen) and 2 mM GlutaMax-I (ThermoFisher). Cells were exposed to low [O2] in a ProOx C21 regulated C-chamber (BioSpherix) positioned inside a 37°C humidified incubator. Desired [O2] was achieved by N2 balance, in 5% [CO2]. 1 mM Dimethyloxaloylglycine (DMOG) (Sigma) was used as hypoxia-mimetic.
RNA purification and qPCR analysis
Total RNA was extracted using the ReliaPrep™ RNA Tissue miniprep system (Promega). For the mouse cell lineage identification, RT2 profiler PCR array (Qiagen), triplicate samples were pooled and 400 ng RNA reverse transcribed with M-MLV RT (Promega). Relative gene expression using SYBR green (PCR Biosystems) was performed on a ViiA™ 7 system (Applied Biosystems) and quantified using the comparative Ct method. β-Actin was selected for normalization. For other analyses, 1 μg RNA was reverse transcribed and quantified on a StepOnePlus™ system (Applied Biosystems). Canx was used for normalisation, selected using the geNorm™ reference selection kit (Primerdesign). qPCR primer sequences are shown in Table 1.
Table 1. Primer list.
| qPCR | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| Tet1 | GAGCCTGTTCCTCGATGTGG | CAAACCCACCTGAGGCTGTT |
| Tet2 | TGTTGTTGTCAGGGTGAGAATC | TCTTGCTTCTGGCAAACTTACA |
| Tet3 | CCGGATTGAGAAGGTCATCTAC | AAGATAACAATCACGGCGTTCT |
| Oct4 | GTTGGAGAAGGTGGAACCAA | CTCCTTCTGCAGGGCTTTC |
| Nanog | AAGGATGAAGTGCAAGCGGT | GGTGCTGAGCCCTTCTGAAT |
| Gdf3 | CTTCTCCCAGACCAGGGTTTT | TCTAGAGTCAGCTGGGCCAT |
| Lefty1 | CAGCTCGATCAACCGCCAGT | GGCTGGCATGGCTGTGTT |
| Dnmt3b | CCCTCCCCCATCCATAGT | TCTGCTGTCTCCCTTCATTGT |
| Sox7 | AGATGCTGGGAAAGTCATGG | AGAGGGAGCTGAGGAGGAAG |
| Hnf4a | ACACCACCCTGGAGTTTGAA | GCCCAGGCTGTTGGATGAAT |
| Gata2 | CTCCAGCTTCACCCCTAAGC | ACCACAGTTGACACACTCCC |
| Hand1 | CGGAAAAGGGAGTTGCCTCA | GGTGCGCCCTTTAATCCTCT |
| Nppa | CAACACAGATCTGATGGATTTCA | CCTCATCTTCTACCGGCATC |
| Myh7 | AGCAGCAGTTGGATGAGCGACT | CCAGCTCCTCGATGCGTGCC |
| Hes5 | CCCAAGGAGAAAAACCGACT | TGCTCTATGCTGCTGTTGATG |
| Sox2 | GCACATGAACGGCTGGAGCAACG | TGCTGCGAGTAGGACATGCTGTAGG |
| Dcx | ACGACCAAGACGCAAATGGA | CTTGTGCTTCCGCAGACTTC |
| Fabp7 | AACCAGCATAGATGACAGAAACTG | ACTTCTGCACATGAATGAGCTT |
| Canx | TTCCAGACCCTGATGCAGA | TCCCATTCTCCGTCCATATC |
| hMeDIP | ||
| Tet3 | GAGAGGGCATAGCGGACTTG | GCAGACTGCAGATGAGTGGA |
| TrueMethyl | ||
| Tet3 | GATTTTTTTTAGAAGAGAAATTTGTTTAAG | CAAACCAAATCAATCCTCCCTA |
| ChiP | ||
| Tet3 | GGGTCATCTGGTGGATCTTC | GACACCGCTAGAACACAGCA |
Antibodies
Antibodies used: 5hmC (Active Motif, 39770), 5mC (Active Motif, 39649), H3K27me3 (Active Motif, 39155), and H3K4me3 (Cell Signalling, 9751), Tet1 (Millipore, 09-872), FLAG (Sigma, F3165), α-tubulin (Sigma, T5168).
DNA extraction and immuno-dot-blot analyses
Genomic DNA was prepared by 350 μg/ml proteinase K (Sigma) digestion in 15 mM NaCl, 1%SDS, 100 mM EDTA, 50 mM Tris–HCl pH 8 at 55°C for 4 h. Samples were sonicated (Branson 150 sonfier) to obtain 200–1000 bp fragments and RNaseA (140 μg/ml; Qiagen) digested at 55°C for 30 min. DNA was phenol/chloroform extracted and ethanol precipitated. 2 μg of DNA was applied to Hybond™-N (GE Healthcare) using a dot-blot hybridisation manifold (Cleaver Scientific) as described in (39). Known DNA standards (Active Motif) were included as a measure of antibody specificity. Membranes were blocked in 10% milk PBS/T or Odyssey Blocking Buffer (PBS) (Li-Cor) and probed with 5hmC or 5mC antibodies respectively overnight. Blots were developed and quantified using an Odyssey® CLx imaging system (Li-Cor).
Mass spectrometry
One microgram of genomic DNA was digested with DNA degradase plus (Zymo Research) as described previously (40) Samples were four times diluted and injected into Agilent 1100 LC system interfaced directly to Waters Quattro LC triple quadrupole mass spectrometer for C, 5mC and 5hmC detection.
1H NMR assessment of metabolites
HEK-293Ts were washed in ice-cold PBS and scraped into metabolite extraction buffer (methanol:choloroform:H2O; 1:1:1). The methanol/H2O phase was evaporated under a SpeedVac™ concentrator (Thermo Scientific) at 30°C. Dried extracts were reconstituted in 100 mM sodium monophosphate buffer (pH 7.0) containing 500 μM TMSP (sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4), 1.5 mM NaN3, 0.5 mM EDTA and 100% D2O. A 700 MHz Bruker spectrometer equipped with cryogenic probe was used for 1D 1H NMR data acquisition with suppressed water resonance. 1H NMR spectra were acquired using 9.3 kHz spectral width and 32 K data points with acquisition time of 1.67 s, relaxation delay of 5 s and 128 scans. Resulting spectra were processed to 65 536 data point and corrected for phasing and zero referencing using NMRLab (41). Resonance assignments and quantification were made with reference to Chenomx NMR Suite 7.1 (Chenomx). Samples were normalised to total protein, determined by a Bradford assay (Thermo Scientific).
5hmC DNA-Immunoprecipitation (hMeDIP)-Seq/PCR
hMeDIP (Active Motif) was performed on 1 μg of fragmented genomic DNA, pooled from triplicate samples, according to manufacturer's instructions. Libraries were prepared from the test sample (day 3-differentiated mESCs grown under atmospheric O2; 2.5 ng), and equivalent amount of control, total genomic DNA using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs; E7645), following the manufacturer's protocol. DNA fragments were end-repaired and ligated to adaptors. Fragments with 200 bp inserts were size selected using Agencourt Ampure XP Beads (Beckman Coulter), eluted in 10 mM Tris, and indexed and amplified by PCR for 12 cycles. The libraries were quantified by qPCR using NEBNext Library Quant Kit for Illumina (New England Biolabs; E7630), and pooled at 4 nM concentration. The library pool was sequenced on the MiSeq single read for 50 cycles. Data was analysed using BaseSpace (Illumina) and Galaxy (https://usegalaxy.org/).
PCR amplification of the Tet3 promoter region (primer sequences shown in Table 1) from day 3-differentiated mESCs exposed to atmospheric and 1% O2 was performed in a 50 μl reaction volume using Q5® high fidelity DNA polymerase (New England Biolabs), as per the manufacturer's protocol, with the addition of Enhancer buffer. Samples were amplified for 30 cycles using a 64°C annealing temperature. PCR products were visualized by agarose gel-electrophoresis.
TrueMethyl genome analysis (bisulfite and oxidative-bisulfite sequencing)
Four hundred nanogram genomic DNA from undifferentiated and day 3-differentiated mESCs were processed through the TrueMethyl® Whole Genome kit (CEGX) protocol. Samples were split evenly to enable parallel quantification of 5mC and 5hmC using bisulfite and oxidative-bisulfite chemistry respectively. The Tet3 promoter fragment (primer sequences shown in Table 1, designed using epidesigner.com) was amplified using KAPA HiFi HS Uracil PCR ready mix (Kapa Biosystems) under manufacture's conditions. Samples were amplified for 35 cycles using a 60°C annealing temperature. PCR products were analysed by agarose gel-electrophoresis and extracted for Sanger Sequencing (Source Bioscience).
Chromatin-immunoprecipitation (ChIP)
Undifferentiated and day 3-differentiated mESCs were fixed in 1% formaldehyde (Sigma), harvested, lysed and sonicated as described for immuno-dot-blot analyses. ChIP assays were performed using the EZ-Magna ChIP™ (Millipore) kit according to manufacturer's instructions, using negative control IgG and antibodies to H3K27me3, H3K4me3 or Tet1. PCR amplification of the Tet3 promoter region (primer sequences shown in Table 1) was performed in 25 μl reaction volumes using Q5® high fidelity DNA polymerase, according to the manufacturer's instructions, with added Enhancer buffer. H3K27me3 and H3K4me3 were amplified for 28 cycles and Tet1 for 30 cycles, at a 66°C annealing temperature and 30 s extension time.
Western blotting
Protein samples were prepared as described previously (42). 30 μg were loaded onto 7.5% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (GE Healthcare). Blots were blocked in 10% milk TBS/T and probed overnight with a FLAG antibody at the supplier's recommended conditions. Blots were visualised and quantified on an Odyssey® CLx imaging system (Li-Cor). Blots were probed with α-tubulin (1/10 000 dilution) to evidence equal loading (Supplementary Figure S1).
RESULTS
Lowered [O2] changes the pattern of mESC differentiation
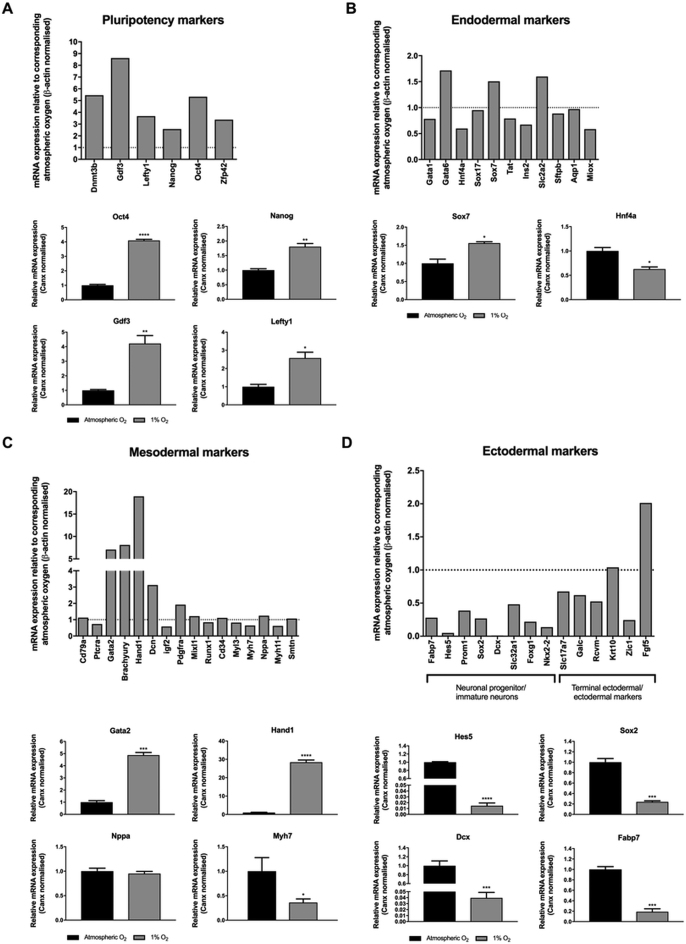
Although precise levels of O2 are challenging to measure directly in vivo, most estimates suggest the mammalian uterine environment under which the embryo develops to be in the range of 2–8% (reviewed in (3)). Further, before the development of the circulatory system, cells within some embryonic compartments likely experience [O2] significantly below this, and, indeed, [O2] within some stem cell niches have been reported to be as low as 1% (6). We sought to determine the effect of O2-poor culture conditions (1% O2), upon the determination of cell-fate of a population of mESCs, allowed to differentiate into all three germ layers without bias. mESCs were grown for 7 days under atmospheric (∼21%) or 1% O2 and the mRNA expression profiles of a panel of genes, representative of pluripotency and all three germ layers was assessed using a cell-lineage qPCR array (Figure 1). The mRNA levels of a representative selection of these markers were further validated by independent qPCR. Overall, the expression patterns of all endodermal markers tested displayed little sensitivity to altered O2 levels (less than 2-fold change in any case; Figure 1B), while some early mesodermal markers, such as Hand1 (43) and Gata2 (44) showed a marked increase (>5-fold; Figure 1C) in 1% O2. The expression of later mesodermal markers, including the cardiac markers Myh7 and Nppa (45), were not increased and remained low at this early differentiation time-point. By contrast, many early ectodermal markers, and in particular markers of neuronal progenitor cells, such as Dcx (46), displayed a marked decrease in their expression, in response to lowered O2 levels (Figure 1D). Finally, the expression levels of all the genes tested which are characteristic of pluripotent ESCs, including Oct4 and Nanog (47), were significantly increased in response to lowered [O2] (Figure 1A). These data suggest that low [O2] acts both to maintain the undifferentiated state, and additionally skews the differentiation of a population of mESCs away from more ectodermal (and specifically neuronal) lineages, towards a more mesodermal fate.
Figure 1.
1% O2 changes the pattern of mESC differentiation. mESCs were differentiated for 7 days in atmospheric and 1% O2 and a cell lineage qPCR profiler array was performed upon pooled triplicate mRNA samples to give relative quantification of expression of (A) pluripotency genes, (B) endodermal-expressed genes, (C) mesodermal-expressed genes and (D) ectodermal-expressed genes (including neuronal progenitor markers). Data represent mRNA levels from 1% O2-cultured mESCs, as a fold-change relative to corresponding expression in atmospheric O2, depicted by the dotted line (set at 1 in each case). Independent qPCR-validation of a representative selection of genes is displayed as the mean ± SEM, analysed by an unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Tet enzymes are potential oxygen sensors
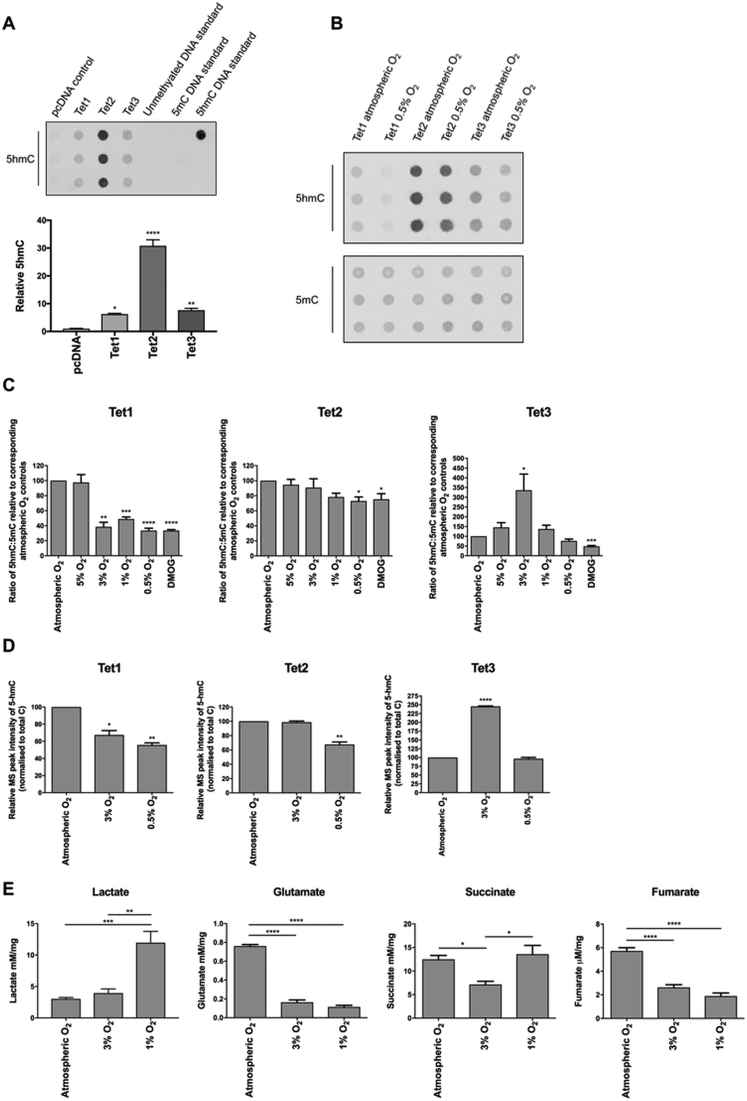
We sought to determine whether the changes in mRNA expression, induced by low (1%) [O2] in the differentiating mESCs might in part be mediated via Tet(s). It was thus necessary to establish whether the activities of the Tet enzyme(s) might be influenced by changes in [O2] over a range considered physiological in differentiating ESCs. Plasmid constructs expressing Tet1, Tet2 or Tet3 were transiently transfected into human embryonic kidney cells (HEKs), and subsequently cultured under atmospheric conditions, together with a graded set of different O2 levels, ranging from 0.5% to 5%. In addition, as a control, cells (at atmospheric [O2]) were incubated with 1 mM dimethyloxalylglycine (DMOG), a competitive inhibitor of 2-OG-dependent dioxygenases. The relative global levels of 5hmC and 5mC were measured by immuno-dot-blot to give a measure of Tet activity in each case (Figure 2B and C). In addition, specific samples were additionally assessed for absolute levels of 5hmC, 5mC and C by mass spectrometry (Figure 2D). As shown in Figure 2A, the levels of 5hmC in HEKs (which were barely detectable) were shown to increase significantly upon ectopic expression of each Tet (under atmospheric conditions), although the activity resulting from Tet2 overexpression appeared significantly higher than that resulting from Tet1 or Tet3. Successful transfection and expression of each Tet enzyme was additionally demonstrated by western-blot analyses, which also indicated that the expression of Tet2, relative to Tet1 or Tet3 was not increased in these experiments, further suggesting that the catalytic activity of Tet2 was significantly higher, or more promiscuous than Tet1 or Tet3 (Supplementary Figure S1).
Figure 2.
Tet1/2/3 are differentially regulated by [O2]. (A) Immuno-dot-blot and quantification of levels of 5hmC resulting from HEKs transfected with pcDNA control, or Tet1/2/3-containing expression plasmids and cultured under atmospheric O2 for 48 h. Known DNA standards were included to validate antibody specificity. Histograms depict levels of 5hmC, relative to control levels (arising from pcDNA-transfected cells). (B) Representative immuno-dot-blot depicting 5hmC and 5mC levels in HEKs after Tet1/2/3-overexpression (as indicated) and culture under atmospheric or 0.5% O2 for 48 h. C: Quantification of immuno-dot-blot analyses of HEK cells after Tet1/2/3-overexpression and culture for 48 h under atmospheric O2, and under O2 concentrations ranging from 0.5% to 5% as indicated. Cells grown under atmospheric O2 in 1 mM DMOG served as a positive control. Histograms depict the ratio of 5hmC:5mC, in each case shown relative to the level observed after culture under atmospheric O2 (100%). (D) Mass-spectrometry analyses of 5hmC levels in HEK cells cultured under atmospheric, 3% and 0.5% O2 for 48 h. Histograms depict MS peak intensity of 5hmC, normalized to total C, shown in each case relative to the level observed after culture under atmospheric O2 (100%). (E) 1H NMR analyses of levels of metabolites (as indicated) in HEKs after culture under atmospheric, 3% and 1% O2 for 48 h. Data are expressed as the mean ± SEM, and either analysed by a t-test or one-way ANOVA with Tukey's post-hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The data in Figure 2C and D clearly demonstrate that the activity of Tet1 was not altered by culture in 5%, compared to atmospheric, O2, but was inhibited significantly upon culture at 3%, and lower O2 levels. Contrasting with this, the activity of Tet2 displayed no inhibition of activity at either 5% or 3% O2, and in fact was only marginally affected during culture under O2 levels as low as 0.5%, or by DMOG-inhibition. Surprisingly, although not significantly changed at 5% O2 (compared to atmospheric conditions), the activity of Tet3 was found to be enhanced at 3% O2, but then decreased during culture under 1% and 0.5% O2. It is, however, well established that the activities of the Tet enzymes have a requirement for 2-OG and can also be antagonised by high levels of sterically-similar TCA metabolites; succinate and fumarate (48). Culturing cells under different levels of O2 will affect their metabolic fluxes, and consequently the intracellular levels of all these TCA metabolites. To further investigate a potential cause of the unexpected increase in Tet3 activity under more hypoxic conditions, a metabolic profile of HEKs was determined by 1H NMR after culture in 3% and 1% O2, and compared to that seen under atmospheric conditions (Figure 2E). Lactate levels increased under lowered O2 conditions, consistent with the activation of glycolysis, and decreased oxidative phosphorylation and TCA cycle activity. In addition, levels of glutamate (from which 2-OG can be derived, to have an anaplerotic effect on the TCA cycle) were reduced in both 3% and 1% O2. Together, these findings suggest a decrease in levels of 2-OG, which might be expected to inhibit further the activities of the Tets under enhanced hypoxia. However, both succinate and fumarate levels were significantly decreased in 3%, compared to atmospheric, O2, which could in part account for increased Tet3 activity at this point. However, when cultured at 1% O2, succinate levels in HEKs had increased back to atmospheric O2 levels (although fumarate levels remained low) and this may have acted in part to inhibit further the activity of Tet3 at 1% O2, compared to 3% O2, as we observed.
Irrespective of the precise mechanism underlying the biphasic response of Tet3 activity to increasing levels of hypoxia, we conclude that the activities of the Tet enzymes are differentially regulated by O2 levels. We also conclude that Tet1 activity, specifically, is potentially differentially inhibited by low, graded levels of O2 which are physiologically relevant within the developing mammalian embryo.
Global levels of 5hmC are dynamically regulated during early mESC differentiation and are inhibited by low [O2]
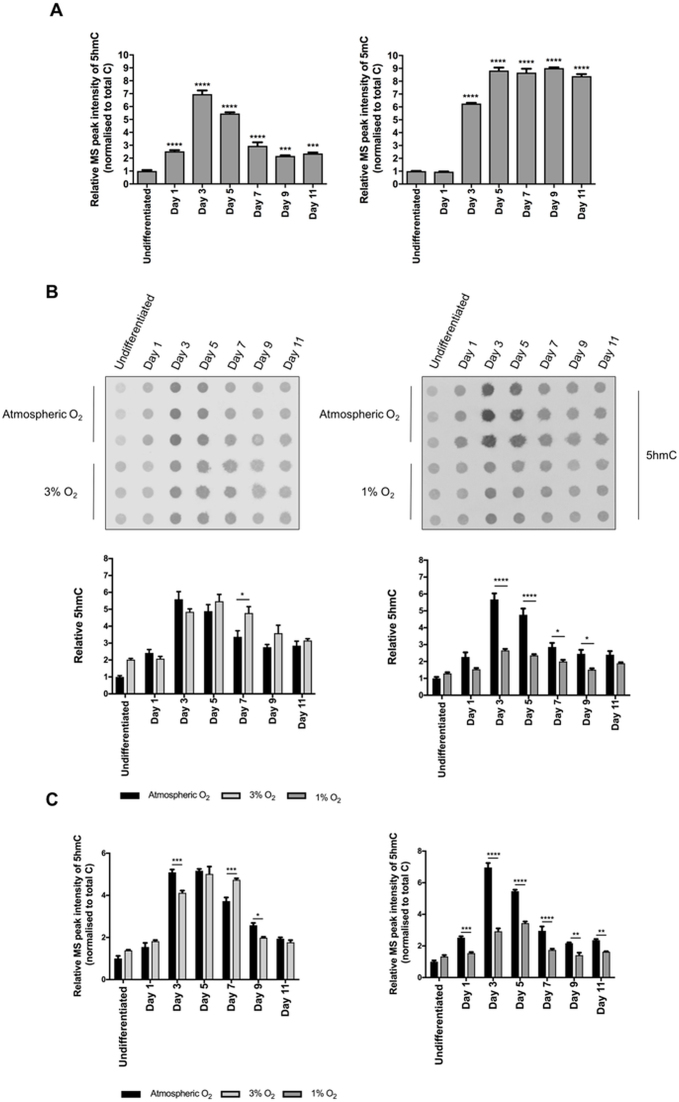
The levels of 5mC and 5hmC within undifferentiated ESCs are known to be highly dependent upon the conditions under which they are cultured, and consequently there are differing reports of these levels within the literature (21,22,49–51). In particular, inhibition of MEK and Gsk3β by two small molecule inhibitors (‘2i’ conditions, as employed in these studies), results in a hypomethylated state, with consequent low levels of 5hmC. It has been shown that this better represents the epigenetic state of inner cell mass cells of the very early (preimplantation) blastocyst, compared to ESCs classically-grown without 2i (49–51). We determined the genome-wide levels of both 5mC and 5hmC in undifferentiated mESCs grown under 2i conditions, and subsequently in these mESCs induced to differentiate under atmospheric conditions over a time-course of 11 days. We demonstrated that 5hmC levels were low in these undifferentiated mESCs, but upon differentiation they exhibited a ‘burst’ of 5hmC, which was apparent by day 1 and which peaked at day 3 before subsequently subsiding to day 9 (Figure 3A). By contrast, the 5mC levels increased sharply at day 3 and subsequently reached a plateau by day 5. Thus, perhaps surprisingly, the increase in the stable genome-wide levels of 5mC did not appear to precede the burst of 5hmC. This might suggest that this burst of hydroxymethylation occurs at already existing 5mC residues, or that the methylation and hydroxymethylation reactions are tightly coupled at this specific stage of mESC differentiation. To determine the effect of [O2] upon this burst of 5hmC generation, mESCs were induced to differentiate over the same time scale (0–11 days) in 3% and 1% O2. Intriguingly, this transient burst of genome-wide 5hmC was not affected at 3% O2, but was significantly blunted by culture under 1% O2 (Figure 3B and C).
Figure 3.
Levels of 5-hmC are dynamically regulated during mESC differentiation. (A) 5hmC and 5mC levels detected by mass-spectrometry in mESCs differentiated over 11 days, cultured under atmospheric O2 at time points as indicated. Histograms depict MS peak intensities of 5hmC or 5mC, normalized to total C, shown in each case relative to the level observed in undifferentiated cells. Data expressed as the mean ± SEM and analysed by a one-way ANOVA with Dunnett's post-hoc test to levels in undifferentiated mESCs. (B) Immuno-dot-blot analyses of 5hmC levels in mESCs differentiated over 11 days at time points as indicated, cultured under atmospheric, 3% or 1% O2. Histograms depict levels of 5hmC, in each case shown relative to the level observed in undifferentiated mESCs in atmospheric O2. (C) Mass spectrometry analyses of samples as in (B). Histograms depict MS peak intensity of 5hmC, normalized to total C, shown in each case relative to the level observed in undifferentiated cells in atmospheric O2. Data expressed as the mean ± SEM and analysed by a two-way ANOVA with Bonferroni's post-hoc test.*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The burst of 5hmC in differentiating ESCs is mediated by Tet1
We assessed the mRNA levels of each Tet enzyme over this time-course of early mESC differentiation, in order to determine which Tet isoform may be catalysing the observed burst of ([O2]-dependent) 5hmC at day3. However, consistent with other reports (21,22), none of the expression profiles of the Tets appeared to mirror the dynamic changes in global 5hmC levels observed. Indeed, mRNA expression levels of both Tet1 and Tet2 were observed to decline significantly, concomitant with the increase in 5hmC levels, while the expression of Tet3 remained low until after the burst in 5hmC had subsided (Figure 4A). However, at day 3 (when 5hmC levels were highest) Tet1 was significantly (approximately 40-fold) more highly expressed than Tet2, which was itself ∼5-fold more highly expressed than Tet3 (Figure 4B).
Figure 4.
Tet1 mediates the burst of 5hmC in differentiating mESCs at day 3. (A) qPCR analyses of Tet1, Tet2 and Tet3 mRNA expression in mESCs over a time course of differentiation in atmospheric O2 as indicated. Levels are normalised to Canx, and shown relative to the levels observed in undifferentiated mESCs in each case. (B) Relative levels of Tet 1–3 mRNA expression, normalised to Canx, in mESCs at day 3 of differentiation. Levels are shown relative to Tet1 mRNA expression. (C) Immuno-dot-blot analyses of 5hmC in undifferentiated or day 3-differentiated mESCs cultured under atmospheric O2, stably-infected with control, or shRNAs directed to Tet 1–3, to generate Tet1–3 knockdown (KD) mESCs. Histograms depict levels of 5hmC, in each case shown relative to the level observed in undifferentiated, control mESCs. (D) Mass-spectrometry analyses of samples as in (C). Histograms depict MS peak intensity of 5hmC, normalised to total C, shown in each case relative to the level observed in undifferentiated, control mESCs. E: Immuno-dot-blot analyses of 5hmC levels in undifferentiated or day 3-differentiated, control or Tet1 KD mESCs, and in control mESCs, differentiated for 3 days under 1% O2. Histograms depict levels of 5hmC, in each case shown relative to that observed in undifferentiated, control mESCs under atmospheric O2. Data expressed as the mean ± SEM and analysed by either a one-way ANOVA with Dunnett's or Tukey post-hoc test or two-way ANOVA with Dunnett's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine which Tet enzyme was functional in mediating this transient increase in 5hmC, stably transformed mESCs were generated in which the expression of Tet1, Tet2 or Tet3 was down-regulated by shRNA-expressing lentiviral infection. In each case successful gene silencing was demonstrated by qPCR (Supplementary Figure S2). Downregulation of Tet1, specifically, acted to decrease significantly the levels of 5hmC, both in undifferentiated mESCs and at day 3 of mESC differentiation to the levels seen in 1% O2-cultured cells (Figure 4C–E). We thus conclude that the burst of genome-wide 5hmC, observed upon the induction of mESCs to differentiate, is mediated via Tet1-dependent activity, in an [O2]-dependent and time-dependent manner.
Downregulation of Tet1 acts to inhibit mESC differentiation
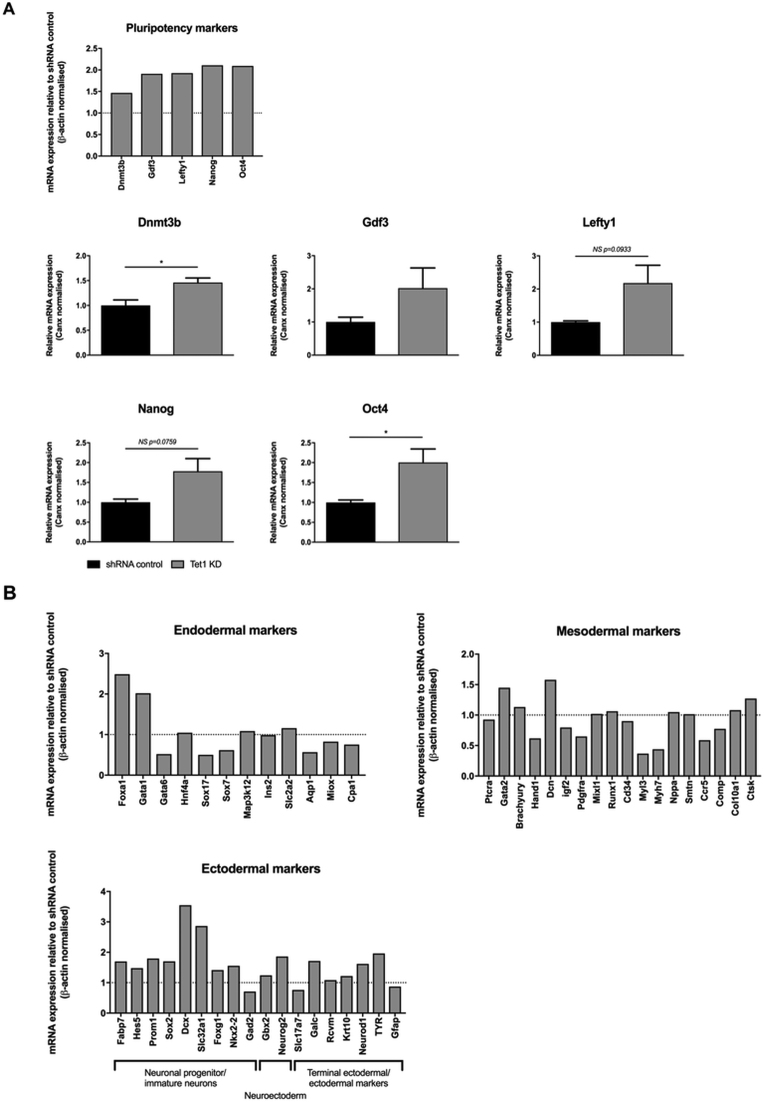
The effect of Tet1-downregulation upon the mRNA expression of pluripotent and lineage-specific genes was investigated. Tet1-depleted mESCs were induced to differentiate for 7 days, and their profiles of expression were compared to control shRNA-infected cells. As shown in Figure 5A, silencing of Tet1 acted to increase the expression of all the pluripotency genes analysed, as was observed when mESCs were cultured in 1% O2, although the relative changes in gene expression were less marked, and did not always reach significance. Tet1-depletion also acted to change the relative expression levels of many early differentiation markers, but it did not skew differentiation away from the ectodermal/neuronal lineages and towards mesodermal lineages, (as was evident in mESCs grown under 1% O2). Rather, the changes in gene expression of lineage-specific genes were highly variable, and did not demonstrate a consistent trend towards (or away from) a profile characteristic of any specific germ layer or cell lineage (Figure 5B).
Figure 5.
Tet1 ablation increases pluripotency marker expression. Control shRNA and Tet1 KD mESCs were differentiated for 7 days under atmospheric O2 and a cell lineage qPCR profiler array was performed upon pooled triplicate mRNA samples to give relative quantification of expression of (A) Pluripotency genes, (B) Endodermal-expressed genes, Mesodermal-expressed genes, and Ectodermal-expressed genes, (including neuronal progenitor markers). Data represent mRNA levels from Tet1 KD mESCs, expressed as a fold-change relative to corresponding expression in control mESCs, depicted by the dotted line (set at 1 in each case). Independent qPCR-validation of the mRNA levels of all pluripotency genes assessed genes are displayed as the mean ± SEM, analysed by an unpaired t-test. *P < 0.05.
Tet3 is a genomic target of [O2]-regulated 5mC-hydroxylation during early mESC differentiation
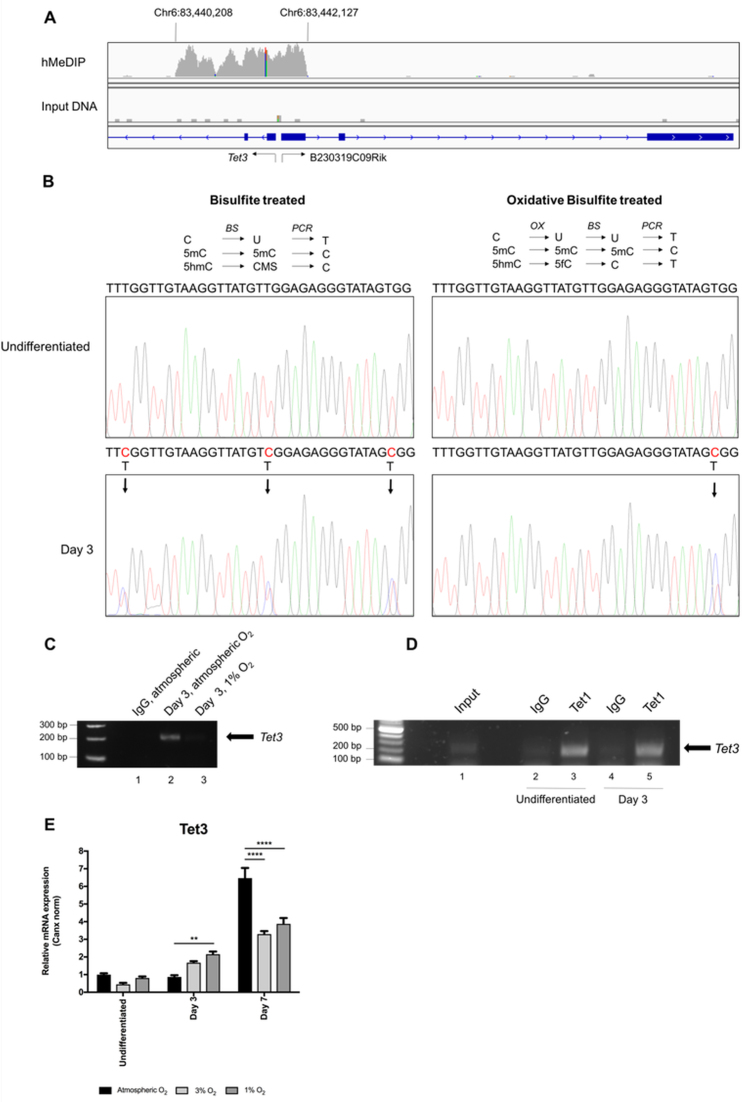
We sought to determine the genomic targets of the burst of 5hmC in differentiating mESCs at day 3 (cultured under atmospheric O2) by performing 5hmC DNA-immunoprecipitation (hMeDIP) followed by high throughput sequencing (hMeDIP-seq). We identified 610 gene loci which were >2-fold enriched for 5hmC, compared to total genomic input (control) DNA (Supplementary Table S1). Functional annotation clustering of the enriched gene list, using The DAVID Gene Functional Annotation Tool (http://david.ncifcrf.gov), revealed the most enriched cluster, containing 55 genes, was associated with developmental and cell differentiation pathways. These sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE107204. Unexpectedly, the most highly enriched sequences identified in these analyses mapped to a GC-rich promoter region of the Tet3 gene locus which also comprises the expressed sequence tag, B230319C09Rik (Figure 6A). Strikingly, the number of sequencing reads at this region was greater than 7-fold higher than that at any other identified genomic locus (Supplementary Table S1). The mouse Tet3 gene contains (at least) three distinct promoter regions (52), and the enriched sequences identified in this screen correspond to the most downstream of these three. To confirm this finding and to determine whether the levels of 5hmC at this locus were a result of de novo methylation and/or hydroxymethylation, during the mESC differentiation process, we performed bisulfite and oxidative-bisulfite sequencing on genomic DNA from both undifferentiated mESCs and day3-differentiated mESCs. We investigated the 5mC and 5hmC status of three specific CpG dinucleotides, identified within a 200 bp GC-rich genomic sequence of this Tet3 promoter sequence. As shown in Figure 6B, in undifferentiated ESCs, all 3 CpG dinucleotides within this genomic region were (perhaps surprisingly) devoid of either 5mC or 5hmC epigenetic marks. However, at day 3 of mESC differentiation, all 3C residues demonstrated significant conversion to 5mC/5hmC, as determined by bisulfite sequencing (in each case, ∼60% conversion within the PCR amplicon, generated from the whole mESC population; arrowed in Figure 6B). Further, two of these CpG dinucleotides appeared hydroxymethylated, whereas the remaining CpG di-nucleotide appeared predominately methylated (arrowed in Figure 6B). The appearance of 5hmC at the Tet3 promoter locus at day 3 of mESC differentiation was also confirmed by hMeDIP-PCR (Figure 6C) and was shown to be lost by culturing mESCs in 1% O2. We further assessed the occupancy of Tet1 at this locus by ChIP, and (by contrast to the appearance of the 5hmC mark), we found Tet1 to be bound to the Tet3 promoter region both in undifferentiated and in day 3-differentiated mESCs (Figure 6D).
Figure 6.
Tet3 is the predominant target of the 5hmC burst in early-differentiating mESCs. (A) hMeDIP-seq was performed on day 3 mESC samples. A Tet3 promoter region, co-incident with the expressed sequence tag, B230319C09Rik, was found to have the largest 5hmC enrichment, visualised using the Integrative Genome Viewer (IGV). (B) Bisulfite and oxidative-bisulfite chemistries confirmed the gain of 5hmC from undifferentiated to day 3 mESCs within the Tet3 promoter region. Arrows after bisulfite treatment show the gain of methylation/hydroxymethylation on CpGs (5mC and 5hmC) at day 3. After oxidative-bisulfite treatment, the failure to convert cytosine to thymine is indicative of 5hmC at these CpGs sites. The remaining arrow demonstrates that 5mC is present on that CpG. (C) hMeDIP-PCR analyses of 5hmC enrichment on the Tet3 promoter in mESC after 3 days of differentiation in atmospheric or 1% O2. (D) ChIP-PCR analyses of Tet1 occupancy at the Tet3 promoter region in undifferentiated and day 3-differentiated mESCs, cultured under atmospheric O2, shown in each case relative to the respective IgG negative control. (E) qPCR analyses of Tet3 expression, normalised to that of Canx, in mRNA isolated from mESCs cultured under 1%, 3% and atmospheric O2, for times as indicated. Data expressed are expressed as the mean ± SEM and analysed by a two-way ANOVA with Tukey post-hoc test. **P < 0.01, ****P < 0.0001.
We next determined the effect of [O2] upon Tet3 mRNA expression in differentiating mESCs. Many studies have suggested that the 5hmC epigenetic mark (which is reduced at the Tet3 promoter region as a result of culture under low [O2]) to be involved in gene transcriptional activation (17,53). mESCs induced to differentiate over a time course of 7 days were cultured in atmospheric, 3% or 1% O2 and Tet3 mRNA levels were compared at specific time points. As shown in Figure 6E, the levels of Tet3 at day 3 of mESC differentiation were, perhaps surprisingly, not reduced as a result of the more hypoxic culturing conditions (and in fact were increased at this point). However, by day 7, Tet3 mRNA levels were significantly decreased in cells grown in both 1% and 3% O2. Taken together, these data suggest that the [O2]-dependent 5mC-hydroxymethylation of the Tet3 promoter region may act to ‘mark’ it for increased expression later during cellular differentiation and development.
Tet1 occupancy associates with both activating and repressive histone epigenetic marks at the Tet3 promoter in ESCs
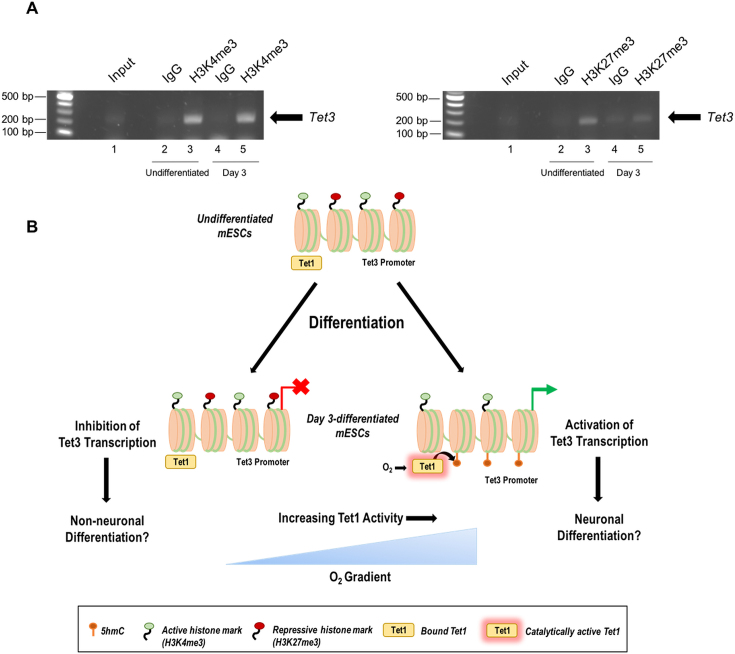
The Tet3 promoter region, identified here as a target of Tet1-mediated hydroxymethylation, comprises a highly GC-rich, CpG island (reviewed in (54). Virtually, all such promoters have been shown in ESCs to be marked by the activating, histone epigenetic mark; H3K4me4 (55). A subset of these gene promoters, termed ‘bivalent’ gene promoters, have additionally been shown to exhibit the repressive, H3K27me3 mark (55). Genes associated with such bivalent promoter regions typically include developmentally regulated genes, which are silenced in undifferentiated ESCs, but are poised to become activated as required at the appropriate stage of (cell type-specific) differentiation (56). Tet1 occupancy at regulatory CpG islands has been implicated in the generation of both the H3K4me3 and H3K27me3 histone marks, via the recruitment of the associated lysine methyltransferases; SET1A/B or mixed lineage leukemia (MLL), and Polycomb repressive complex 2 (PRC2), respectively (30,57–59). We therefore investigated the occurrence of these histone modifications at the identified Tet3 promoter. As shown in Figure 7A, both histone marks were clearly apparent at this locus in undifferentiated mESCs. At day 3 of differentiation, the H3K4me3 mark persisted (and the corresponding band representing this remained equally intense), whereas the H3K27me3 mark, although still present, appeared significantly decreased. These data therefore identify this Tet3 gene locus as a Tet1-associated, bivalent promoter, and further suggest that the (partial) loss of the H3K27me3 mark at this locus is coincident with [O2]-dependent, Tet1-mediated 5mC-hydroxylation in differentiating mESCs (Figure 7B).
Figure 7.
The Tet3 promoter region is associated with activating and repressive histone marks. (A) ChIP-PCR analyses of the occurrence of H3K4me3 and H3K27me3 at the Tet3 promoter region in undifferentiated and day 3-differentiated mESCs, cultured under atmospheric O2, shown in each case relative to the respective IgG negative control. (B) Schematic representation of proposed mechanism of [O2]-dependent regulation of Tet3 and cellular differentiation. In undifferentiated mESCs, a GC-rich, Tet3 promoter region is associated with Tet1. It is marked as a bivalent promoter by both active (H3K4me3) and repressive (H3K27me3) histone modifications and CpG dinucleotides are not methylated or 5mC-hydroxylated. As mESCs are induced to differentiate, de novo methylation of CpG dinucleotides provides the substrate for the catalytic activity of Tet1. Tet1 activity is induced in a graded manner, dependent upon the available [O2], and acts to generate 5hmC at CpG dinucleotides within the Tet3 promoter. Concomitant with this the repressive H3K27me3 histone mark becomes lost, while the activating, H3K4me3 mark remains. Tet3 is thus ‘marked’ to become expressed specifically in more O2-rich microenvironments (promoting neural differentiation).
DISCUSSION
The focus of this study was to investigate the potential role of Tets in mediating [O2]-dependent effects upon the differentiating mESC phenotype. Consistent with other studies in which low-O2 conditions were shown to skew cellular differentiation (reviewed in (4,6,60)) we show here that culturing ESCs in 1% O2, compared to atmospheric (approximately 21%) O2, for 7 days resulted in the promotion of cells acquiring a mesodermal cell-lineage fate, and a concomitant inhibition of cells acquiring an ectodermal (and specifically neuronal) fate. The more hypoxic conditions also acted to promote the expression of pluripotency markers in the mESCs.
It is evident that epigenetic changes in chromatin architecture and DNA methylation underlie the determination of cell fate during cellular differentiation (reviewed in (61)). Further, it is well documented that hypoxia can be sufficient to induce such epigenetic changes, notably as has been demonstrated in the progression of many cancers (reviewed in (62,63)). The absolute requirement of Tet enzymatic activity for O2 led us to determine whether Tets might, in part, mediate the O2-dependent regulation of mESC function and differentiation, via modulation of DNA methylation.
Tet1 is a potential O2-sensor in developing mammalian embryos
We demonstrated that in HEK cells, Tet1 activity is differentially regulated at the low levels of O2 which are considered physiologically relevant in mammalian embryogenesis (0.5–5%; (4,6)). Consistent with our findings, the Km value of (recombinant, purified) Tet1 for O2 was determined in vitro to be 30 μM, indicating that significant catalytic activity would be maintained at the relatively hypoxic environment of 3% O2, (48), while a separate in vivo study has demonstrated inhibition of Tet activity at 0.5% O2 (64). Intriguingly, we also demonstrated that the activities of the three individual Tet isoforms display distinct patterns of sensitivity to levels of both O2 and the TCA metabolites, fumarate and succinate, whose levels themselves may be dependent upon O2 availability. This is a potentially significant finding, which may in part underlie both the regulation of Tet-specific functions in other systems, and the cell-type-specific sensitivity to O2.
We analysed changes in global 5hmC (and 5mC) during the earliest stages of mESC differentiation, both by immuno-dot-blot analyses and by mass spectrometry. The results obtained from these separate analyses correlated extremely well, supporting the validity of these data. 5hmC levels within the mESCs were found here to be present at about 12% of the level of 5mC, and approximately 0.08% of that of total C (data not shown), in broad agreement with a previous study (22). We observed a novel, transient burst of 5hmC which peaked at day3 and was inhibited significantly by culturing the differentiating mESCs in 1% O2. Tet1 was identified as the enzyme which primarily mediated this burst of 5mC-hydroxylation. We therefore propose that the activity of Tet1 within the early-developing embryo may be graded, dependent upon the cellular availability of O2, and thus propose Tet1 to act as an O2 sensor (65).
Temporal regulation of Tet activity during mESC differentiation
A clear anomaly in our data is the observation that Tet1 (and Tet2) expression levels are decreasing at a time when the catalytic capacity is seen to increase (at day 3 of differentiation). Several studies have suggested Tet1 to serve non-catalytic functions, involved in the recruitment and maintenance of other chromatin modelling enzymes to specific regulatory DNA regions (30,59). Perhaps consistent with this, Tet1 was found here to be bound to the Tet3 promoter region in undifferentiated mESCs, in the absence of 5hmC at this locus (Figure 6D). It is also likely that the lack of 5hmC at the Tet3 locus is a reflection of the low global levels of the Tet substrate; 5mC, in the mESCs, due to their culture under 2i conditions (49–51). As the cells are induced to differentiate in the absence of 2i, 5mC levels then rise, providing the substrate for Tet activity. However, it should be noted that another (CpG-poor) gene loci; Orm1, which has previously been reported to be constitutively methylated in mESCs (66), was also found to be methylated (but not hydroxymethylated) in our undifferentiated mESCs (data not shown), consistent with methylation remaining in some loci within ESCs cultured under 2i-conditions (67).
In addition, the activities of the Tets have been shown to be upregulated significantly by ascorbic acid (vitamin C; AA), by virtue of their requirement for Fe2+ (68,69). AA acts to recycle (oxidised) Fe3+ to the reduced Fe2+ form, and we note that the addition of AA to the undifferentiated mESCs was sufficient to induce a small yet significant increase in 5hmC levels in these cells, again suggesting the occurrence of detectable levels of existing 5mC (Supplementary Figure S3).
We also investigated whether changes in cellular AA levels (potentially resulting from the KnockOut Serum Replacement (68), in which the mESCs were induced to differentiate) might also be responsible for the increased Tet activity seen at day 3 of differentiation. However, the addition of the ascorbate transporter inhibitor, phloretin (70) did not decrease the levels of 5hmC in the differentiating mESCs (Supplementary Figure S3). Further, when the undifferentiated cells were cultured in KnockOut serum (rather than EmbryoMax), the levels of 5hMC were still seen to significantly increase upon differentiation at day 3 (in KnockOut serum; Supplementary Figure S4).
The precise molecular mechanisms which underlie the increased Tet activity, as Tet expression levels fall, therefore remain unclear. To our knowledge, this transient increase in 5hmC levels has not previously been reported in early differentiating mESCs in vitro. However, recently a Tet1-dependent transient burst of 5mC-hydroxylation was observed in vivo, in the mouse E6.5 epiblast (33). Further, in this study, Tet1 was demonstrated to serve a non-redundant role in the early embryo, as genetic ablation of Tet1 resulted in embryonic defects at gastrulation. The possibility that this function of Tet1 may in part be [O2]-dependent at this early stage in the embryo is clearly intriguing and will be the focus of future studies.
Tet3 is a target of [O2]- dependent 5hmC-hydroxylation
A particularly interesting (and unexpected) finding of this study was that the most significant target of 5mC-hydroxylation in the early-differentiating mESCs was a promoter region within the Tet3 gene locus. We demonstrated that this hydroxymethylation occurred de novo, after the cells were induced to differentiate, consistent with its occurrence as a part of the ‘burst’ of 5mC-hydroxylation apparent at the day 3 time point. Perhaps surprisingly, we found no evidence of methylation at the CpG dinucleotides investigated, in undifferentiated mESCs. It is therefore possible that the methylation and hydroxymethylation reactions are closely coupled in these cells, as suggested above.
Levels of 5hmC at the Tet3 promoter locus were lost when the mESCs were differentiated under more hypoxic culture conditions (1% O2). Further, we demonstrated by ChIP that this region is associated with Tet1, and that this association was also apparent in undifferentiated mESCs. However, it should be noted that we have been unable to demonstrate a loss of 5hmC at this site in the Tet1-ablated mESCs, after 3 days of differentiation (at atmospheric O2). We suggest that this may result from functional compensation by Tet2 (which is also abundantly expressed in the mESCs) and/or functional redundancy between Tet1 and Tet2, as has been reported previously in mESCs (71,72). None-the-less, together, our data strongly support that in wild-type (wt) mESCs, the 5mC-hydroxylation of the Tet3 promoter is predominantly mediated by Tet1, and is differentially regulated within a gradient of [O2].
The Tet3 promoter region, targeted for 5mC-hydroxylation, displays the characteristics of a GC-rich, bivalent promoter, being associated with both activating (H3K4me27) and repressive (H3K27me3) histone epigenetic marks. Tet1 occupancy and activity has been demonstrated previously at such promoters (59,73,74), which are suggested to maintain developmental stage- and tissue-specific genes in a silent state in ESCs, yet poised for subsequent activation upon differentiation (reviewed in (58). Consistent with this, our data suggest that Tet3 mRNA expression is low in undifferentiated, wt mESCs and is strongly induced between day 3 and day 7 of differentiation (in culture under atmospheric O2). Inhibition of 5mC-hydoxylation at the Tet3 promoter region, by culture in low (1% or 3%) O2, resulted in a significant blunting of this transcriptional activation. Several studies have demonstrated the critical role of Tet3 in neural development (25,75), and it is therefore potentially significant that the culture of differentiating mESCs in 1% O2 was also shown here to inhibit the expression of early neuronal markers.
Roles of Tet1 in neural development and function have also been demonstrated (25,35,36), and data presented here suggest a functional interdependence between two Tet enzymes, in which the activity of Tet1 potentiates the transcriptional activation of Tet3. The specific region within Tet3, targeted for 5hmC-hydroxylation, corresponds to the promoter region spanning the transcriptional start site(s) for an isoform of Tet3 which lacks the N-terminal, CXXC, DNA-binding domain (52). Several studies have demonstrated the different functional requirements for this truncated protein, compared the longer isoform, in particular in neuronal specification and phenotype (52,76). It is thus possible that the Tet1-mediated hydroxymethylation described here may play a role in the relative transcription patterns of the different Tet3 isoforms, in differentiating mESCs.
The downregulation of Tet1, however was not sufficient to completely mirror the effects of low [O2] upon neuronal expression during mESC differentiation (Figure 5B), Again, functional redundancy between Tet1 and Tet2 may in part explain this (71,72), and in addition other O2-dependent processes are clearly likely also to function in the regulation of cellular differentiation, such as histone demethylation by members of the Jumonji protein family (77). Never-the-less, both low O2 (1%) and Tet1-downregulation resulted in some increased expression of all pluripotency markers (which, in the cases of Dnmt3b and Oct4, reached significance) in differentiating mESCs, suggesting a non-redundant (potentially [O2]-dependent) role for Tet1 in mESC self-renewal.
To conclude, we demonstrate here the potential for the catalytic function of Tet1 to be differentially activated in response to a gradient of O2, at (low) levels which are physiologically relevant to the early developing embryo (0.5–5%). We demonstrate that differentiating mESCs exhibit an early burst of [O2]-regulated 5mC-hydroxylation that is predominantly mediated by Tet1. In addition, we identify a GC rich, bivalent Tet3 promoter region as a target of this [O2]-regulated 5mC-hydroxylation. Based on these data we propose that gradients of available O2 result in the non-uniform catalytic activity of Tet1 within the early embryo. As a consequence, this Tet3 promoter region becomes 5mC-hydroxylated, and ‘marked’ for transcriptional activation (later in differentiation) only in specific regions of the embryo with adequate available O2. This [O2]-dependent transcriptional regulation of Tet3 may be an important mediator of cellular fate, and in particular neuronal differentiation, during the earliest stages of embryonic development (Figure 7B).
ACCESSION NUMBERS
Gene Expression Omnibus (GEO) database under accession number GSE107204.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank John Pizzey for critical reading of the manuscript, Daniel Martin for technical assistance and discussion, and Han Lu for assistance in manuscript preparation. All authors read and approved the article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
British Heart Foundation programme [RG/13/11/30384, PG/15/119/31970, PG/15/27/31374]; British Heart Foundation studentship [FS/13/55/30643]. Funding for open access charge: British Heart Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Turing A.M. The chemical basis of morphogenesis. 1953. Bull. Math. Biol. 1990; 52:153–197. [DOI] [PubMed] [Google Scholar]
- 2. Briscoe J., Small S.. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015; 142:3996–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takahashi M. Oxidative stress and redox regulation on in vitro development of mammalian embryos. J. Reprod. Dev. 2012; 58:1–9. [DOI] [PubMed] [Google Scholar]
- 4. Simon M.C., Keith B.. The role of oxygen availability in embryonic development and stem cell function. Nat. Rev. Mol. Cell Biol. 2008; 9:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maltepe E., Simon M.C.. Oxygen, genes, and development: an analysis of the role of hypoxic gene regulation during murine vascular development. J. Mol. Med. (Berl.). 1998; 76:391–401. [DOI] [PubMed] [Google Scholar]
- 6. Mohyeldin A., Garzon-Muvdi T., Quinones-Hinojosa A.. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010; 7:150–161. [DOI] [PubMed] [Google Scholar]
- 7. Beyer T.A., Narimatsu M., Weiss A., David L., Wrana J.L.. The TGFbeta superfamily in stem cell biology and early mammalian embryonic development. Biochim. Biophys. Acta. 2013; 1830:2268–2279. [DOI] [PubMed] [Google Scholar]
- 8. Hitchler M.J., Domann F.E.. An epigenetic perspective on the free radical theory of development. Free Radic. Biol. Med. 2007; 43:1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 10. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salminen A., Kauppinen A., Kaarniranta K.. 2-Oxoglutarate-dependent dioxygenases are sensors of energy metabolism, oxygen availability, and iron homeostasis: potential role in the regulation of aging process. Cell. Mol. Life Sci. 2015; 72:3897–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y.. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010; 466:1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. An J., Rao A., Ko M.. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 2017; 49:e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franchini D.M., Schmitz K.M., Petersen-Mahrt S.K.. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu. Rev. Genet. 2012; 46:419–441. [DOI] [PubMed] [Google Scholar]
- 17. Vasanthakumar A., Godley L.A.. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 2015; 208:167–177. [DOI] [PubMed] [Google Scholar]
- 18. Szwagierczak A., Bultmann S., Schmidt C.S., Spada F., Leonhardt H.. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010; 38:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams K., Christensen J., Helin K.. DNA methylation: TET proteins-guardians of CpG islands. EMBO Rep. 2011; 13:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wen L., Tang F.. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014; 104:341–346. [DOI] [PubMed] [Google Scholar]
- 21. Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G. et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011; 8:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K.. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011; 473:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S. et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011; 42:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Yue X., Pastor W.A., Lin L., Georges R., Chavez L., Evans S.M., Rao A.. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E8267–E8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011; 14:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dawlaty M.M., Ganz K., Powell B.E., Hu Y.C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.W., Page D.C. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011; 9:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Freudenberg J.M., Ghosh S., Lackford B.L., Yellaboina S., Zheng X., Li R., Cuddapah S., Wade P.A., Hu G., Jothi R.. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012; 40:3364–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Y., Chavez L., Chang X., Wang X., Pastor W.A., Kang J., Zepeda-Martinez J.A., Pape U.J., Jacobsen S.E., Peters B. et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H., D’Alessio A.C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y.. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011; 473:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dawlaty M.M., Breiling A., Le T., Raddatz G., Barrasa M.I., Cheng A.W., Gao Q., Powell B.E., Li Z., Xu M. et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013; 24:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kang J., Lienhard M., Pastor W.A., Chawla A., Novotny M., Tsagaratou A., Lasken R.S., Thompson E.C., Surani M.A., Koralov S.B. et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4236–E4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khoueiry R., Sohni A., Thienpont B., Luo X., Velde J.V., Bartoccetti M., Boeckx B., Zwijsen A., Rao A., Lambrechts D. et al. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet. 2017; 49:1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li C., Lan Y., Schwartz-Orbach L., Korol E., Tahiliani M., Evans T., Goll M.G.. Overlapping requirements for Tet2 and Tet3 in normal development and hematopoietic stem cell emergence. Cell Rep. 2015; 12:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rudenko A., Dawlaty M.M., Seo J., Cheng A.W., Meng J., Le T., Faull K.F., Jaenisch R., Tsai L.H.. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013; 79:1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang R.R., Cui Q.Y., Murai K., Lim Y.C., Smith Z.D., Jin S., Ye P., Rosa L., Lee Y.K., Wu H.P. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013; 13:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ko M., An J., Bandukwala H.S., Chavez L., Aijo T., Pastor W.A., Segal M.F., Li H., Koh K.P., Lahdesmaki H. et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013; 497:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010; 468:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown T. Dot and slot blotting of DNA. Curr. Protoc. Mol. Biol. 2001; doi:10.1002/0471142727.mb0209bs21. [DOI] [PubMed] [Google Scholar]
- 40. Le T., Kim K.P., Fan G., Faull K.F.. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Anal. Biochem. 2011; 412:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ludwig C., Gunther U.L.. MetaboLab–advanced NMR data processing and analysis for metabolomics. BMC Bioinformatics. 2011; 12:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murray T.V., Smyrnias I., Shah A.M., Brewer A.C.. NADPH oxidase 4 regulates cardiomyocyte differentiation via redox activation of c-Jun protein and the cis-regulation of GATA-4 gene transcription. J. Biol. Chem. 2013; 288:15745–15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2003; 258:1–19. [DOI] [PubMed] [Google Scholar]
- 44. Minegishi N., Suzuki N., Yokomizo T., Pan X., Fujimoto T., Takahashi S., Hara T., Miyajima A., Nishikawa S., Yamamoto M.. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood. 2003; 102:896–905. [DOI] [PubMed] [Google Scholar]
- 45. Christoffels V.M., Habets P.E., Franco D., Campione M., de Jong F., Lamers W.H., Bao Z.Z., Palmer S., Biben C., Harvey R.P. et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000; 223:266–278. [DOI] [PubMed] [Google Scholar]
- 46. Couillard-Despres S., Winner B., Schaubeck S., Aigner R., Vroemen M., Weidner N., Bogdahn U., Winkler J., Kuhn H.G., Aigner L.. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005; 21:1–14. [DOI] [PubMed] [Google Scholar]
- 47. Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006; 38:431–440. [DOI] [PubMed] [Google Scholar]
- 48. Laukka T., Mariani C.J., Ihantola T., Cao J.Z., Hokkanen J., Kaelin W.G. Jr, Godley L.A., Koivunen P.. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 2016; 291:4256–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013; 13:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013; 13:360–369. [DOI] [PubMed] [Google Scholar]
- 51. Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013; 20:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jin S.G., Zhang Z.M., Dunwell T.L., Harter M.R., Wu X., Johnson J., Li Z., Liu J., Szabo P.E., Lu Q. et al. Tet3 reads 5-carboxylcytosine through Its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016; 14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pastor W.A., Aravind L., Rao A.. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013; 14:341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deaton A.M., Bird A.. CpG islands and the regulation of transcription. Genes Dev. 2011; 25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 57. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K. et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013; 49:645–656. [DOI] [PubMed] [Google Scholar]
- 58. Voigt P., Tee W.W., Reinberg D.. A double take on bivalent promoters. Genes Dev. 2013; 27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Neri F., Incarnato D., Krepelova A., Rapelli S., Pagnani A., Zecchina R., Parlato C., Oliviero S.. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013; 14:R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Keith B., Simon M.C.. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007; 129:465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen T., Dent S.Y.. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsai Y.P., Wu K.J.. Epigenetic regulation of hypoxia-responsive gene expression: focusing on chromatin and DNA modifications. Int. J. Cancer. 2014; 134:249–256. [DOI] [PubMed] [Google Scholar]
- 63. Melvin A., Rocha S.. Chromatin as an oxygen sensor and active player in the hypoxia response. Cell Signal. 2012; 24:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thienpont B., Steinbacher J., Zhao H., D’Anna F., Kuchnio A., Ploumakis A., Ghesquiere B., Van Dyck L., Boeckx B., Schoonjans L. et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016; 537:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaelin W.G. Jr, Ratcliffe P.J.. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008; 30:393–402. [DOI] [PubMed] [Google Scholar]
- 66. Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B., Bibel M., Schubeler D.. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008; 30:755–766. [DOI] [PubMed] [Google Scholar]
- 67. von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. et al. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Mol Cell. 2016; 62:848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013; 500:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z. et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013; 45:1504–1509. [DOI] [PubMed] [Google Scholar]
- 70. Minor E.A., Court B.L., Young J.I., Wang G.. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013; 288:13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013; 495:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu H., Zhang Y.. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011; 10:2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mantsoki A., Devailly G., Joshi A.. CpG island erosion, polycomb occupancy and sequence motif enrichment at bivalent promoters in mammalian embryonic stem cells. Sci. Rep. 2015; 5:16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Park J.L., Kim H.J., Seo E.H., Kwon O.H., Lim B., Kim M., Kim S.Y., Song K.S., Kang G.H., Kim H.J. et al. Decrease of 5hmC in gastric cancers is associated with TET1 silencing due to with DNA methylation and bivalent histone marks at TET1 CpG island 3′-shore. Oncotarget. 2015; 6:37647–37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li T., Yang D., Li J., Tang Y., Yang J., Le W.. Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Mol. Neurobiol. 2015; 51:142–154. [DOI] [PubMed] [Google Scholar]
- 76. Perera A., Eisen D., Wagner M., Laube S.K., Kunzel A.F., Koch S., Steinbacher J., Schulze E., Splith V., Mittermeier N. et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015; 11:283–294. [DOI] [PubMed] [Google Scholar]
- 77. Hancock R.L., Dunne K., Walport L.J., Flashman E., Kawamura A.. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015; 7:791–811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.