Abstract
Unfolded protein response (UPR) is triggered by the accumulation of unfolded proteins in the endoplasmic reticulum (ER), which is accomplished by a dramatic induction of genes encoding ER chaperones. Activation of these genes involves their rapid transcription by Hac1p, encoded by the HAC1 precursor transcript harboring an intron and a bipartite element (3′-BE) in the 3′-UTR. ER stress facilitates intracellular targeting and recruitment of HAC1 pre-mRNA to Ire1p foci (requiring 3′-BE), leading to its non-spliceosomal splicing mediated by Ire1p/Rlg1p. A critical concentration of the pre-HAC1 harboring a functional 3′-BE element is governed by its 3′→5′ decay by the nuclear exosome/DRN. In the absence of stress, pre-HAC1 mRNA undergoes a rapid and kinetic 3′→5′ decay leading to a precursor pool, the majority of which lack the BE element. Stress, in contrast, causes a diminished decay, thus resulting in the production of a population with an increased abundance of pre-HAC1 mRNA carrying an intact BE, which facilitates its more efficient recruitment to Ire1p foci. This mechanism plays a crucial role in the timely activation of UPR and its prompt attenuation following the accomplishment of homeostasis. Thus, a kinetic mRNA decay provides a novel paradigm for mRNA targeting and regulation of gene expression.
INTRODUCTION
The nuclear exosome selectively degrades a diverse spectrum of aberrant messages in the nucleus of Saccharomyces cerevisiae and thereby safeguards the cells from their detrimental effects (1–6). These aberrant transcripts include transcription elongation assembly-defective transcripts (7–12), intron-containing splice-defective messages (13–17), transcription termination-defective 3′-extended read-through transcripts (18,19), and export-defective global and individual mRNAs (20,21). The exosome is a large multi-protein complex consisting of eleven 3′→5′ exoribonucleases and a few associated nuclear and cytoplasmic cofactors (4,5,22–24). Of these eleven subunits, nine core subunits are arranged in a barrel (known as EXO9), which consists of two stacked-ring structures—a ‘trimeric’ cap (composed of Rrp4p, Rrp40p and Csl4p) placed on the top of a ‘hexameric’ ring (consisting of Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p and Mtr3p) with a central channel. Accordingly, EXO9 contacts two catalytically active subunits, a processive tenth subunit (Dis3p/Rrp44p, possessing both endo- and 3′→5′ exoribonuclease activity) and a distributive eleventh subunit (Rrp6p, 3′→5′ exoribonuclease) from opposite sides to form EXO11Dis3p+Rrp6p (24–26). Notably, EXO11Dis3p+Rrp6p requires ancillary cofactors to target and degrade a specific class of substrate RNA since it lacks the catalytic specificity. Consequently, a set of ancillary cofactors, referred to as the exosome-cofactors, collectively modulates and enhances exosomal function to target the appropriate substrate RNA class among the plethora of RNA substrates in a highly selective manner (27). The TRAMP complex in S. cerevisiae provides the best-characterized example of such exosome-cofactor, which consists of a noncanonical poly(A) polymerase, Trf4p (Pap2p)/Trf5p, a DExH box RNA helicase, Mtr4p (6,28,29); and the Zn-knuckle RNA binding proteins, Air1p/2p (6,29). Moreover, DRN (Decay of RNA in the nucleus) defines another nuclear mRNA decay apparatus, which consists of the nuclear mRNA cap-binding protein Cbc1p/2p (18,30), Rrp6p (20) and two nucleocytoplasmic shuttling proteins, Upf3p and Tif4631p (30). Previous genetic data and a body of recent findings indicate that DRN also acts as an additional exosomal cofactor by modulating the nuclear exosome function and by assisting the exosome to selectively target and degrade a few distinct classes of aberrant nuclear mRNA substrates (31).
A global transcriptomic analysis revealed an enhancement in the stability and the steady-state levels of nearly two hundred normal messages including HAC1 mRNA in both cbc1-Δ and rrp6-Δ yeast strains, both of which are deficient in the nuclear exosome/DRN function (32). Notably, HAC1 encodes the key transcription factor (Hac1p) implicated in the activation of the unfolded protein response (UPR) pathway in Saccharomyces cerevisiae, which is triggered in response to endoplasmic reticulum (ER) stress (33–35). Thus, the enhancement of HAC1 mRNA in cbc1-Δ and rrp6-Δ strains suggests that the nuclear exosome in association with DRN targets this transcript, thus leading to a provocative proposal that the nuclear exosome machinery may control the activity of the UPR via the regulation of the normal/physiological ‘repertoire’ of HAC1 mRNA. Notably, the induction of the UPR in Saccharomyces is principally accomplished by non-spliceosomal splicing of a HAC1 precursor transcript that contains an intron. In the absence of stress, the precursor HAC1 mRNA harboring the intron remains translationally inactive via the formation of a secondary structural bridge formed between the intron and 5′-UTR of the transcript body (36). ER stress eliminates the intron via a non-spliceosomal splicing reaction mediated by the ER-resident kinase-endoribonuclease Ire1p and the tRNA ligase Rlg1p, thereby reinstating the translation of mature HAC1 mRNA to produce an enormous amount of Hac1p (36–39). A crucial cis-acting bipartite element (3′-BE) in the 3′-UTR of HAC1 pre-mRNA/mRNA plays an instrumental role in recruiting and targeting the precursor HAC1 mRNA to the specific intracellular foci consisting of active Ire1p, which rapidly cluster in response to stress (40). Furthermore, activation of the UPR is also accompanied by (i) a massive reduction of global protein synthesis, (ii) augmentation of ER protein-folding capacity, and (iii) stimulation of the degradation of misfolded proteins (33)—all of which eventually restore ER homeostasis. Strikingly, the accomplishment of homeostasis in a timely fashion is extremely crucial, since failure to achieve it leads to the triggering of the programmed cell death (33,41,42). It is therefore vital to avert an inappropriate activation of the UPR following a basal-level stimulus. Moreover, attenuation of the UPR after the activation of ER homeostasis is equally crucial to avoid eliciting programmed cell death. Findings from recent research implicate two important mechanisms that efficiently disable the UPR once protein folding homeostasis is restored in the ER: (i) the binding of the ER chaperone BiP to the ER luminal domain of Ire1p following favorable folding conditions, which promotes its desensitization and subsequent deactivation of the UPR (43), and (ii) the Ire1-catalyzed phosphoryl-transfer reaction, which promptly disassembles Ire1 signaling complexes (44) and attenuates its RNase activity (45).
Activation of the UPR in Saccharomyces cerevisiae in the laboratory is typically mimicked by the addition of several drugs and reducing agents, of which tunicamycin is the most specific and potent (46). Notably, this drug inhibits the N-linked glycosylation of the nascent polypeptide chains (47), thereby stimulating protein unfolding within the ER lumen. In a narrow window of drug concentration (1 μg/ml - 2.5 μg/ml), the wild-type cells having functional UPR machinery grow normally, while the strains with compromised UPR activity (such as the hac1-Δ and ire1-Δ strains) are unable to grow (48). Consequently, the growth ability of a given yeast strain in the presence of tunicamycin is a precise indicator of the level of UPR function. Reduced growth of a strain in the presence of the drug reflects a compromised UPR, whereas robust growth in such medium indicates an enhancement in the signaling process.
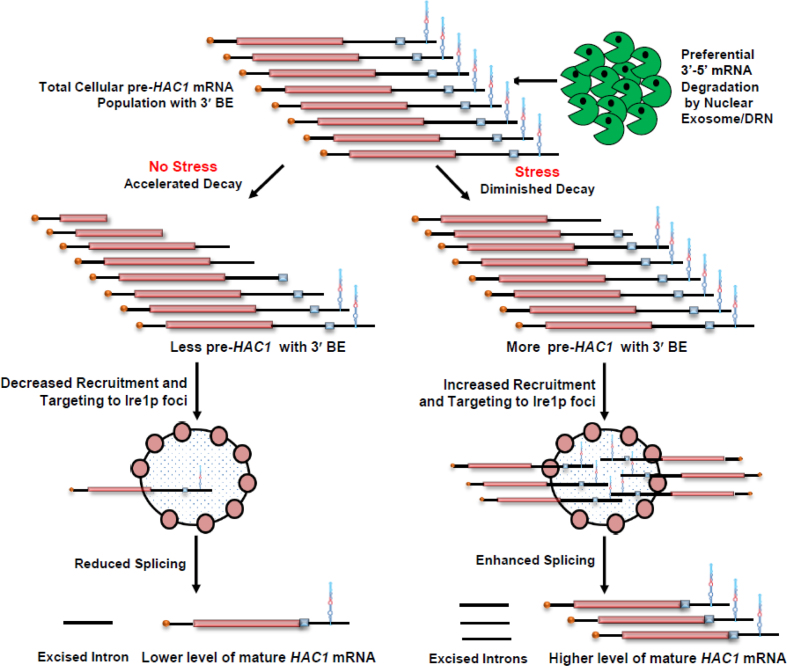
Enhanced abundance and half-life of HAC1 mRNA in the exosome/DRN-deficient yeast cells, as observed in the previous microarray data, prompted us to address whether (i) this message undergoes a selective nuclear decay by the exosome/DRN and (ii) the decay mechanism plays any regulatory role in the function of the UPR pathway. In this report, we establish that the exosome and DRN regulate the function of the UPR in S. cerevisiae, which is accomplished by a preferential and kinetic 3′→5′ decay of HAC1 pre-mRNA. Our findings imply that decay of the HAC1 pre-mRNA by the nuclear exosome/DRN is subject to tight regulation under different circumstances. In the absence of stress, this tight control leads to an accelerated and rapid decay of the pre-mRNA in a 3′→5′ direction, thus resulting in the production of a population of precursor message with a heterogeneous 3′-termini, most of which lack a functional 3′-BE. Consequently, this pool of pre-HAC1 transcript fails to be targeted and recruited efficiently to the Ire1p foci in the absence of ER stress. Under ER stress, however, an optimum repertoire of precursor HAC1 message is produced, which still carries a spectrum of heterogeneous 3′-termini, but the most of this pool harbors a functional and intact BE due to a diminished 3′→5′ decay of the transcript body. Subsequently, this critical concentration of pre-HAC1 mRNA with an intact and functional BE promotes a more efficient targeting and recruitment of this precursor message to the Ire1p clusters. The nuclear exosome/DRN thus exerts a fine control on the functioning of UPR elements, which is vital for its timely activation in the presence of an appropriate stress signal and termination upon removal of the ER stress.
MATERIALS AND METHODS
Yeast strains, plasmids and oligonucleotides
All strains and plasmids and oligonucleotides are listed in Supplementary Tables S1, S2 and S3, respectively.
Cell viability assay
Cell Viability Assay was performed according to the protocol of Back et al. (48). Briefly, each strain was grown overnight, and the culture was then subsequently diluted to 1 × 107 cells/ml. This stock was further diluted 10 fold serially to achieve 1 × 104, 1 × 103 and 1 × 102 cells/ml respectively. Three microliters of each stock was then spotted onto the surface of YPD plate (Yeast Extract 1%, Peptone 2%, Dextrose 2%, Agar 2%) without and with 1 μg/ml tunicamycin. Plates were then incubated for 48–72 h at 30°C. For liquid growth assay, specific strains of S. cerevisiae were grown in triplicate until the O.D.600 reached 0.6 when either DMSO (mock treated, –Tm/–DTT) or 1 μg/ml tunicamycin (+Tm) or 5 mM DTT (+DTT) was added to one of them. Aliquots of each culture were collected at different times (unless otherwise mentioned) after addition of DMSO/tunicamycin/DTT, appropriately diluted and subsequently spread onto YPD plates. Plates were then incubated for 48–72 h at 30°C.
RNA analyses and determination of steady-state and decay rate of mRNAs
Total RNA was isolated as described earlier (30) by harvesting appropriate yeast strains followed by extracting the cell suspension in the presence of phenol–chloroform–IAA (25:24:1) and glass bead. Following the extraction, the RNA was recovered by precipitation with RNAase-free ethanol. Northern blot analyses of the total RNA was done as described previously using 10–20 μg of total RNA (30). The RNA samples following electrophoresis were transferred onto a positively charged nylon membrane (Brightstar-Plus, Ambion Inc.). The blot was subsequently hybridized with either HAC1 or SCR1 probe, which was prepared by psoralen-biotin labeling method using ULTRAhyb® (Ambion Inc.) at 42°C overnight as recommended by the manufacturer. The hybridized blot was developed using the Brightstar®Biodetect™ Kit (Ambion Inc.).
For the preparation of cDNA, total RNA samples were first treated with 1 μg RNAase free DNase I (Fermentas Inc.) at 37°C for 30 min followed by first strand cDNA synthesis using Superscript Reverse Transcriptase (Invitrogen) using Random Primer (Bioline Inc.) by incubating the reaction mixture at 50°C for 30 min. Real Time qPCR analyses with 2–3 ng of cDNA samples were used to determine the steady-state levels of HAC1, RLG1, IRE1 and SCR1, etc. were performed as described previously (30).
Stabilities of a specific mRNA was determined by the inhibition of global transcription with transcription inhibitor 1,10-phenanthroline (Sigma-Aldrich) at 30°C, as described previously (30,49). Briefly, the specific strain was grown at 30°C till mid-logarithmic phase, when either an appropriate amount of DMSO (unstressed) or 1 mg/ml tunicamcycin (ER Stress) was added to the culture and continued to grow for another 3 h. 1,10-Phenanthroline was then added to the growing culture at 20 μg/ml final concentration followed by withdrawal of a 25 ml of aliquots of culture at various times after transcription shut off. Messenger RNA levels were quantified from cDNA by real-time PCR analysis and the signals associated with the specific messages were normalized against SCR1 signals. The decay rates and half-lives of specific mRNAs were estimated with the regression analysis program (Origin 8) using a single exponential decay formula (assuming mRNA decay follows a first order kinetics), y = 100e−bx was used.
Protein analyses by western blot
Total protein was isolated from specific yeast strains grown overnight at 30°C in YPD broth. Following centrifugation at 5000 rpm for 7 min the cell pellets were quickly frozen in liquid nitrogen and stored at –70°C. Frozen pellets were thawed on ice and resuspended in 1 ml of Buffer A (50 mM Tris–Cl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, 1 mM PMSF) supplemented with protease inhibitor (Invitrogen Inc. Carlsbad, CA, USA) and the cells were lysed by vortexing 10–15 times with glass beads followed by clarification of the particulate fraction by centrifugation. Supernatants were collected by centrifugation at 10 000 rpm for 20 min and saved as the total soluble protein fraction for further analysis. Protein concentration was determined by Bradford reagent assay kit (Bio-Rad Inc., Valencia, CA, USA). For Western Analysis, 30–50 μg of total protein was used, which was resolved either in 8% or 10% SDS-polyacrylamide gel. The separated proteins were transferred to PVDF membrane at 100 mA for 16 h for immunobloting. Blots were blocked with 5% milk in Tris-buffered saline (10 mM Tris, 150 mM NaCl, 0.1% Tween 20) and incubated with primary antibodies for specific proteins for 1 h at room temperature in following dilutions: rabbit polyclonal anti-Hac1 (1:2000), rabbit polyclonal anti-Kar2 (1:2000), rabbit polyclonal anti-Cbc1 (1:1000), and mouse monoclonal anti-Tubulin (1:3000). Blots were further washed in TBS with 0.1% Tween 20 and incubated in HRP-conjugated secondary anti-rabbit or anti-mouse antibody diluted at 1:5000 in wash buffer for 1 h at room temperature. Immunoreactive bands were developed and detected by chemiluminescence (ECL imager kit, Abcam) and the images were captured either by X-ray film or myECL Chemidoc Imager (Thermo Scientific, USA).
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation assay was performed using ChIP assay kit (Upstate Biotechnology, Lake Placid, NY, USA) according to the recommendation of the manufacturer. Appropriate strains were grown in YPD followed by treatment with 1 μg/ml tunicamycin for 2 h at 30°C. Crosslinking, sonication and antibody incubations were done according to manufacturer's protocol. Anti-RNA Polymerase II antibody was included in the kit, and Anti-Histone3 antibody was purchased from Abcam® (USA).
Galactose induction of CBC1 gene
The native promoter of CBC1 gene in wild type strain is replaced by pGAL1/GAL10 promoter by homologous recombination and confirmed using genomic PCR. This strain, harboring pGAL1/GAL10::CBC1 was grown in non-inducible non-repressible 2% raffinose sucrose medium till 0.6 O.D. Galactose was then added to the culture at 2% final concentration followed by harvesting of cells at different times post-induction. Total RNA or protein samples were isolated from cells harvested at different time points as described above for further downstream analysis.
Confocal microscopy and image processing
The co-localization imaging of HAC1 mRNA with Ire1 proteins in the live cell was done by using three plasmids (50). The reporter plasmid carries an engineered version of HAC1 mRNA in which a 22-nucleotide RNA module, identical to the nucleolin recognition element (NRE) was inserted at the 3′-UTR (HAC1NRE). The second plasmid expressed a chimeric protein consisting of an NRE-binding nucleolin domain (ND) which is fused to the monomeric form of the green fluorescent protein 2 (GFP2). In the third plasmid, Ire1p is conjugated with red fluorescent protein (RFP) (51). All these plasmids were co-transformed into appropriate yeast strain and the transformed cells were grown till 0.6 O.D. 1 μg/ml of tunicamycin was then added to the cultures, which were then incubated for another 1 hour at 30°C. An aliquot of the cell was used for in vivo imaging studies. Images were captured using OLYMPUS FV10-ASW confocal microscope equipped with HCX PL APO 63×/1.2 NA water immersion objective lens. The GFP and YFP were excited at 458 and 514 nm respectively, and emissions were recorded at 466–526 nm and 530–650 nm respectively. Image processing was done using OLYMPUS FLUOVIEW Ver.2.0b Viewer software. Co-localization Index is measured by ImageJ software (NIH, USA). The intensities of GFP and RFP signals were determined independently using the software and ratio of GFP to RFP were estimated in various strains. Pearson's correlation coefficient (52) (PCC) was determined from the relationship given below for a typical image consisting of red and green channels.
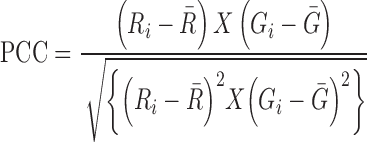 |
where Ri and Gi refer to the fluorescence pixel intensity of the red and green channels of sample ‘i’, 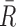 &
& 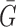 mean fluorescence intensities of the red and green channels, respectively. PCC values range from +1 to –1 for two images. +1 value indicates fluorescence intensities are linearly related to each other and perfect image registration. Whereas –1 indicates total lack of overlap between pixels from the images and fluorescence intensities are perfectly, but inversely, related to one another.
mean fluorescence intensities of the red and green channels, respectively. PCC values range from +1 to –1 for two images. +1 value indicates fluorescence intensities are linearly related to each other and perfect image registration. Whereas –1 indicates total lack of overlap between pixels from the images and fluorescence intensities are perfectly, but inversely, related to one another.
Flow cytometry-based FRET
Flow Cytometry-based Fluorescence Resonance Energy Transfer (FRET) measurement was done by using the protocol as described earlier (53). The amount of signal quenched in the presence of acceptor from a donor is considered as the indicator of quantitative FRET and donor-acceptor pair should have significant overlap of excitation spectra. In our case, GFP2 as part of the bound HAC1NRE-GFP-nucleolin module and RFP as part of the Ire1-RFP fluorophores served as the donor and acceptor respectively. The appropriate strain harboring either all three plasmids i.e. HAC1NRE, GFP2-Nucleolin, and Ire1-RFP or only GFP2-Nucleolin were grown till 0.6 OD in the selective medium along with unlabelled cell. 1 μg/ml of Tunicamycin was added to each culture and aliquots of cells were harvested at indicated times. Fluorescence intensities of each sample were determined by single laser GFP filter (FL1) of BD Accuri™ C6 Flow Cytometer. Mean fluorescence intensity was calculated by BD Accuri™ C6 software by taking 50,000 cells/sample. Average FRET efficiency (E) of the measured population according to the following equation:
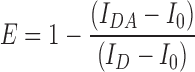 |
IDA is fluorescence intensity of donor-acceptor sample, ID is fluorescence intensity of donor only sample and I0 is fluorescence intensity of unlabeled sample.
Statistical analyses
All the quantitative experiments reported in this paper (mRNA steady-state levels, decay rates, chromatin immunoprecipitation experiments, quantitative growth experiments in liquid media) were done from at least three independent sample size (biological replicates) (n = 3). However, in some instances, the sample sizes are four or five. For each biological replicate, a given yeast strain was grown and treated under the same conditions independently before a given experiment was conducted. Technical replicate, in contrast, involved repetition/analyses of the same biological replicate sample for many times. A technical replicate was used to establish the variability (experimental error) involved the analysis, thus allowing one to set the confidence limits for what is significant data. The values, which varied too much from the rest of the values in the particular group were excluded keeping in mind that the variation could be the result of handling and instrumental errors. All the statistical parameters such as mean, standard deviations, P-values, standard error of the mean were estimated using OriginPro 8 SRO, version 8.0724 (OriginLab Corporation, Northampton, MA, USA). P-values were determined using Student's two-tailed t-test (unpaired).
RESULTS
The nuclear exosome and DRN influence the UPR pathway in Saccharomyces cerevisiae
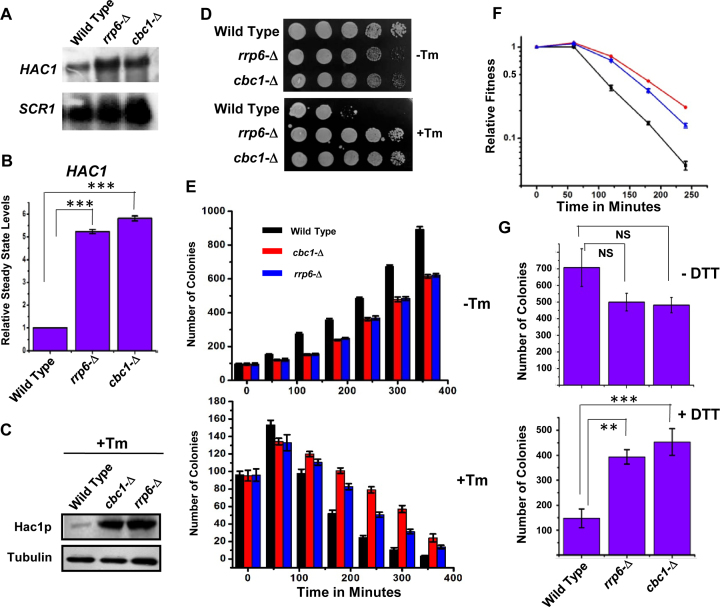
To validate the previous transcriptomic data, we first determined whether the steady-state level of HAC1 mRNA is altered by any of the mutations in the exosome and DRN components. The abundance of this mRNA in wild-type and exosome- and DRN-defective rrp6-Δ and cbc1-Δ strains was consequently determined in the absence of tunicamycin by northern blot and qRT-PCR analyses. The steady-state level of the HAC1 precursor message was enhanced significantly in both rrp6-Δ and cbc1-Δ yeast strains as shown in Figure 1A (northern blot) and B (qRT-PCR). This observation preliminarily validates the microarray data and indicates that perhaps the precursor-HAC1 message undergoes an enhancement in these exosome/DRN-defective yeast strains (since the experiment was conducted in absence of ER stress). We attributed the augmentation in the level of HAC1 pre-mRNA to the diminished Rrp6p/Cbc1p-dependent mRNA degradation since none of the rrp6-Δ and cbc1-Δ alleles have any conceivable influence in the transcriptional induction (18). To further test whether the Hac1p protein level is affected by any of these alleles, we determined the level of Hac1p in rrp6-Δ and cbc1-Δ yeast strains by western blot analysis using an anti-Hac1p antibody (a gift from Prof. P. Walter, the University of California at San Francisco, USA). The Hac1p protein was barely detectable in the wild-type strain, as well as in the strains depleted of Cbc1p and Rrp6p proteins (data not shown) in the unstressed condition. However, the level of Hac1p was found to be enhanced in the wild-type strain in the presence of tunicamycin compared to the unstressed condition (data not shown). Interestingly, both strains depleted of Rrp6p and Cbc1p showed a striking increase in the abundance of Hac1p protein (Figure 1C). This finding, together with that from the previous experiment, predicts that the exosome/DRN-deficient strains accumulate higher levels of HAC1 pre-mRNA and Hac1p protein when ER stress is imposed. Observations from these experiments therefore prompted us to ask whether the yeast strains depleted of the nuclear exosome/DRN function (rrp6-Δ and cbc1-Δ strains) would sustain ER stress more efficiently. We answered the above question by evaluating the growth of isogenic wild-type, rrp6-Δ and cbc1-Δ yeast strains in the presence and absence of two different kinds of ER stress inducers—tunicamycin in both solid and liquid medium and DTT in liquid medium. As shown in Figure 1D and Supplementary Figure S1A, both rrp6-Δ and cbc1-Δ yeast strains grew much better compared to the wild-type strain in the solid YPD medium containing various concentrations of tunicamycin. The results of the growth experiment carried out in liquid medium in the absence and presence of tunicamycin (Figure 1E) revealed that the viability of all three strains decreased in the presence of tunicamycin. Following the induction of ER stress, the mortality rate of the wild-type strain was found to be much higher compared to those of the rrp6-Δ and cbc1-Δ strains (Figure 1E, bottom histogram). A similar estimate of their relative fitness revealed that DRN-deficient rrp6-Δ and cbc1-Δ strains possess a better fitness in the presence of strong ER stress as depicted in Figure 1F. The T50 values (time required to reach 50% of initial cell population) for the cbc1-Δ and rrp6-Δ strains were found to be nearly twice as much as that of the wild-type strain in the presence of 1 μg/ml tunicamycin. Data from the growth experiment in the presence and absence of DTT revealed that the DRN-deficient yeast strains sustained much better growth in the presence of 5 mM DTT, thus revealing that exosome/DRN-defective yeast strains are probably more resistant to ER stress (Figure 1G). We then addressed whether the enhanced growth ability of the exosome/DRN-deficient yeast strains in tunicamycin is associated with the diminution of HAC1 pre-mRNA decay. To evaluate this issue, we tested the growth pattern of an rrp6-Δ strain transformed with an empty plasmid and plasmids carrying either a wild-type RRP6 or rrp6-3 allele (a gift from Prof. J. Scott Butler, the University of Rochester, USA). The rrp6-3 allele harbors a catalytically inactive point mutation in the exonuclease I domain (D238A) of the Rrp6p protein (54). As shown in Supplementary Figure S1B, the rrp6-3 was unable to complement the growth defect phenotype of the rrp6-Δ strain, and the strain grew like an rrp6-Δ strain in the presence of ER stress-inducer tunicamycin. This result strongly suggests that the enhanced growth of the yeast strains in the presence of tunicamycin in strains deficient in functional nuclear exosome is attributable to the exonucleolytic activity of the nuclear exosome component, Rrp6. Thus, the findings from our preliminary experiment strongly argue in favor of the view that the nuclear exosome/DRN may potentially influence the activity of the UPR in budding yeast.
Figure 1.
Inactivation of DRN causes alteration of the UPR activity in a reciprocal manner. Northern blot (A) and histogram obtained from the qPCR analysis (B) revealing the levels of HAC1 pre-mRNA in a wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in the absence of ER stress. The northern blot was hybridized with a DNA fragment encompassing both the intronic and exonic sequences of the HAC1 gene. For qRT-PCR analysis, the amplicon used to quantify the HAC1 species encompasses the intronic sequence of HAC1. SCR1 RNA in these strains was used as internal loading control. Three independent cDNA preparations (biological replicates, n = 3) were used to determine the levels of HAC1 mRNA. Normalized value of HAC1 mRNA from normal samples was set to 1. (C) Relative steady state level of Hac1p protein in wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in presence of 1 μg/ml tunicamycin. α-tubulin (lower panel) was used as the loading control. Cells of indicated strains were grown in absence and in the presence of tunicamycin, harvested and total protein extract were prepared as described in Materials and Methods. 50 μg of total cellular protein was loaded in each lane, resolved in 12% SDS-PAGE and probed with anti-Hac1p and anti-tubulin antibodies to detect these proteins as described in Materials and Methods. (D) Relative growth of wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in YPD solid growth media in the absence (–Tm) and presence of 1 μg/ml tunicamycin (+Tm). (E) Relative growth of Wild type (yBD-117) (Black Bar), cbc1-Δ (yBD-131) (Red Bar), rrp6-Δ (yBD-129) (Blue Bar) strains in YPD liquid growth media in the absence (–Tm) and the presence of 1 μg/ml tunicamycin (+Tm). The histogram represents the mean colony numbers obtained from three independent experiments (n = 3) (biological replicates), and the standard error of means was plotted as error bars. (F) Relative fitness of sustenance of Wild type (yBD-117) (Black), cbc1-Δ (yBD-131) (red), rrp6-Δ (yBD-129) (blue) strains in presence tunicamycin. Relative index values were estimated from the experiment described in panel E, which is defined as the ratio of no. colonies of a given strain in the presence of tunicamycin to the number of colonies in the absence of tunicamycin. (G) Histogram showing the relative growth efficiency of wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in liquid YPD medium in absence and presence of 5 mM DTT. Cells of indicated strains were grown in YPD liquid medium till the A600 of the cultures reach 0.6 as mentioned in the Materials and Methods. Either water (–DTT) or 5 mM DTT (+DTT) was then added to them, and they were continued to grow for another 240 min. The relative viability count was determined as described in the Materials and Method section.
To bolster this notion, we then addressed whether the alteration of the nuclear exosome and DRN functions leads to an oscillation in the output of the UPR pathway. We tested this possibility by measuring the messenger RNA and protein levels of ER stress markers in rrp6-Δ and cbc1-Δ yeast strains under ER stress. Examination of the steady-state levels of two ER chaperone messages, BiP/KAR2 and PDI, revealed a consistent 2- to 2.5-fold upregulation in the presence of tunicamycin in the exosome/DRN-deficient rrp6-Δ and cbc1-Δ strains (Figure 2A). Furthermore, the level of Kar2p protein displayed a robust enhancement in the rrp6-Δ and cbc1-Δ yeast strains compared to the wild-type strain grown in the presence of tunicamycin (Figure 2B). These collective findings led us to conclude that the nuclear exosome in association with DRN may exert a functional control on the activity of UPR in Saccharomyces cerevisiae.
Figure 2.
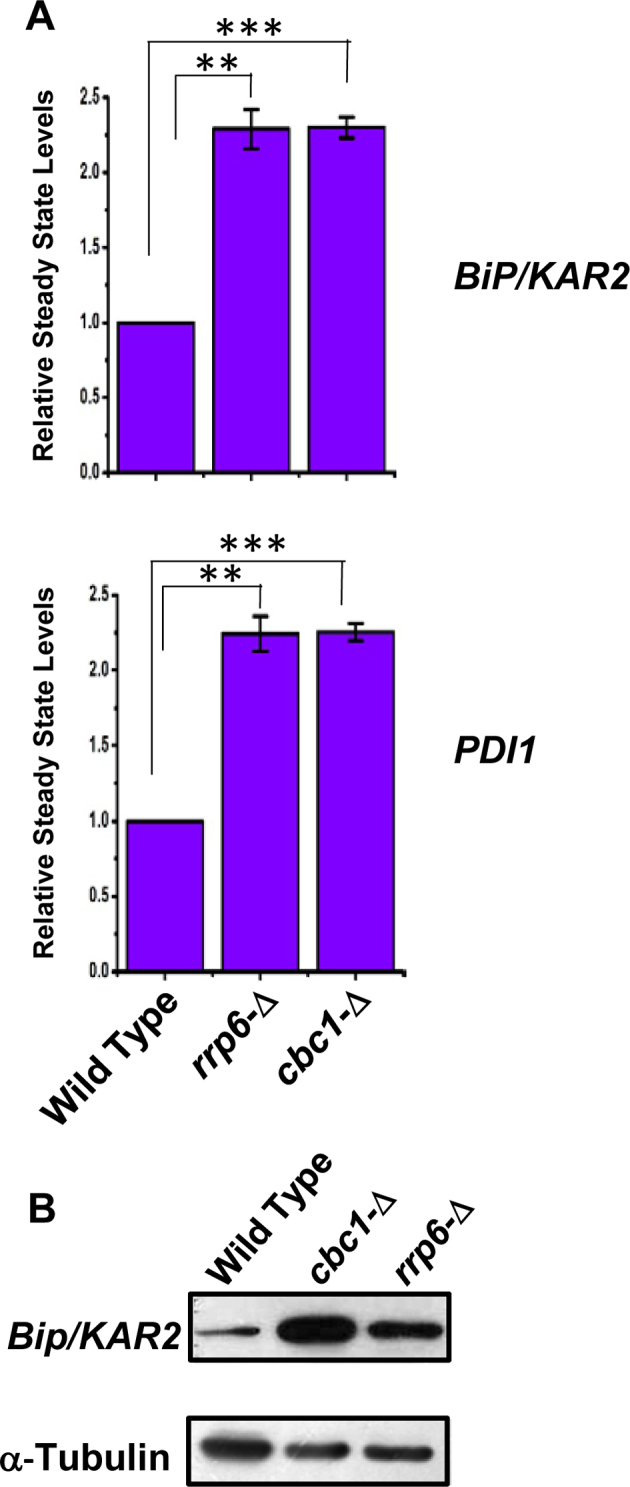
DRN deficient strains display elevated Levels of UPR marker transcripts and protein: (A) Histogram showing the abundance of transcripts encoding ER chaperones BiP and PDI in a Wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in presence tunicamycin at 30°C. Three independent cDNA preparations (biological replicates, n = 3) were used to determine the levels of these mRNAs. Normalized value of individual mRNA from normal samples was set to 1. The statistical significance of difference as reflected in the ranges of P-values estimated from Student's two-tailed t-tests for a given pair of test strains for each message are presented with following symbols, ∗P < 0.05, ∗∗P < 0.005 and ∗∗∗P < 0.001, NS, not significant. (B) Relative steady state level of Bip/KAR2 protein in wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in the presence of 1 μg/ml tunicamycin (+Tm) at 30°C. α-Tubulin (lower panel) was used as the loading control. Cells of indicated strains were grown in the absence and in the presence of tunicamycin, harvested and total protein extract were prepared as described in Materials and Methods. 50 μg of total cellular protein was loaded in each lane, resolved in 12% SDS-PAGE and probed with anti-Bip and anti-tubulin antibodies to detect these proteins as described in Materials and Methods.
The nuclear exosome and DRN target precursor HAC1 mRNA
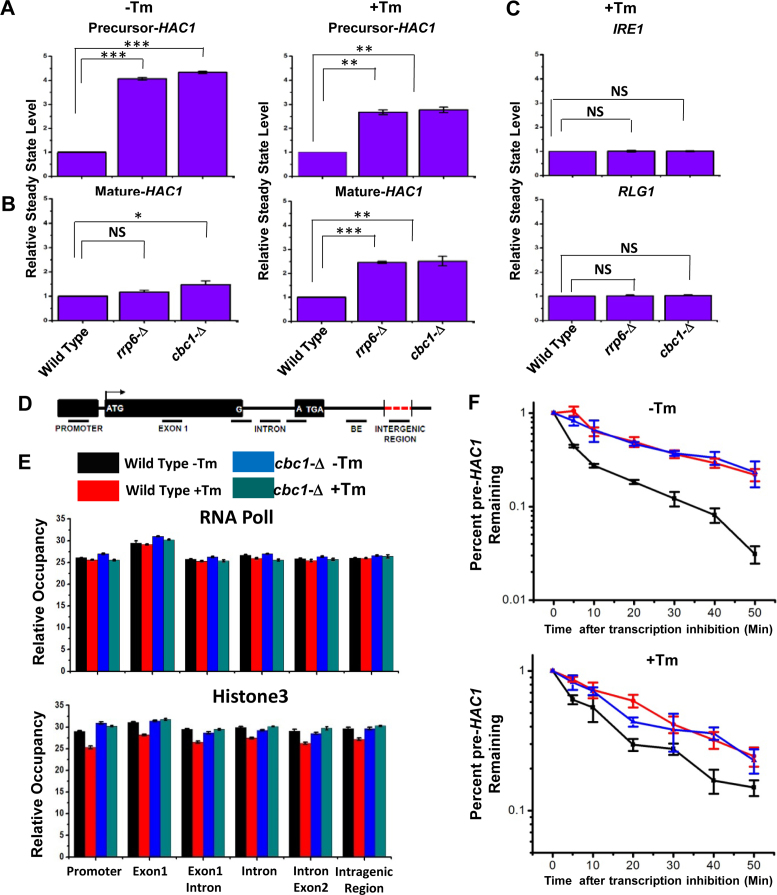
UPR activation is accomplished via the regulated expression of the HAC1 gene and the concerted actions of Ire1p and Rlg1p in a sequential manner, which culminate in a massive production of Hac1p and ER chaperones. Thus, it is conceivable that the enhancement in the level of HAC1 mRNA/pre-mRNA during ER stress is an indirect consequence, which might have resulted from the degradation of mRNA(s) encoding any other component(s) of the UPR. Therefore, we address which particular pre-mRNA(s)/ mRNA(s) encoding UPR machinery is/are targeted and degraded by the nuclear exosome/DRN. Consequently, the steady-state levels of each of the precursor and mature HAC1, IRE1, and RLG1 messages were evaluated in wild-type, rrp6-Δ and cbc1-Δ strains in the absence and presence of ER stress. As shown in Figure 3A, the abundance of the precursor HAC1 mRNA displayed a steady-state increase of approximately ≅4- and ≅2.5-fold in the rrp6-Δ and cbc1-Δ strains in the absence and presence of ER stress, respectively. The levels of the HAC1 mature message, whose copy number is extremely low in the absence of stress, exhibited only a modest increase of 1.5-fold (Figure 3B). In the presence of stress, however, the mature HAC1 mRNA showed a 2.5-fold enhancement in both the rrp6-Δ and cbc1-Δ strains (Figure 3B). On the other hand, the levels of IRE1 and RLG1 mRNAs did not alter appreciably in either of these strains (Figure 3C). Importantly, the extent of enhancement of both precursor and mature HAC1 mRNAs is the same in the presence of ER stress (approximately 2.5-fold). These results support the argument that the actual target of the nuclear exosome/DRN is the precursor HAC1 mRNA. This finding was further corroborated by the observation that in an ire1-Δ strain background (splicing of pre-HAC1 is abolished in this strain with the concomitant accumulation of pre-HAC1), the level of pre-HAC1 mRNA displayed a more substantial increase (by ∼6- to 7-fold) upon inactivation of exosome/DRN function (Supplementary Figure S1C). Thus, these observations unequivocally identified the precursor HAC1 mRNA as the principal target of the nuclear exosome and DRN. Interestingly, the extent of enhancement of the precursor HAC1 message upon inactivation of the exosome and DRN displayed a differential effect in the presence and absence of ER stress. In the absence of the stress, the extent of enhancement of the above message in rrp6-Δ and cbc1-Δ strains was almost twice as much as that obtained in the presence of stress (Figure 3A&B). A relatively lower enhancement of this transcript in the presence of ER stress possibly implicates a diminution of decay activity by the nuclear exosome/DRN. We favor a view that this diminished decay of pre-HAC1 mRNA may reflect a regulation of exosome/DRN-dependent decay during stress to maintain a better supply line of this precursor message downstream to the Ire1p/Rlg1p-mediated splicing apparatus (see next section and Discussion).
Figure 3.
Elevated levels of UPR activity in rrp6-Δ and cbc1-Δ strains is accomplished by the selective and targeted decay of pre-HAC1 mRNA by the nuclear exosome and DRN. Histogram showing the steady state levels of pre-HAC1 (A) and mature HAC1 (B) transcripts in the absence (–Tm) and the presence of 1 μg/ml tunicamycin (+Tm) in a normal, cbc1-Δ, and rrp6-Δ strains (n = 3). Three independent cDNA preparations (biological replicates, n = 3) were used to determine the levels of these mRNAs. Normalized value of individual mRNA from normal samples was set to 1. (C) Histogram depicting the abundance of IRE1 and RLG1 mRNAs in the presence of 1 μg/ml tunicamycin (+Tm) in indicated strains as determined from three independent experiments (n = 3). Three independent cDNA preparations (biological replicates, n = 3) were used to determine the levels of these mRNAs. Normalized value of individual mRNA from normal samples was set to 1. (D) Schematic presentation of HAC1 genomic locus (not up to the scale) showing the location of different primer pairs used to determine the RNA PolII and Histone H3 occupancy as well as the exon 1 and BE specific amplicons for experiment described in Figure 4A and B. (E) Histogram depicting the relative occupancy of RNA Pol II and Histone H3 at different regions along HAC1 genomic locus in a normal (–Tm, black bar and +Tm, red bar) and cbc1-Δ strains (–Tm, blue bar and +Tm, green bar) using chromatin immunoprecipitation (ChIP) technique using specific antibodies against RNA Pol II and Histone H3. Mean ChIP signal obtained from three independent experiments (biological replicates) is presented as means ± SE (n = 3 for each strain). (F) The decay rate of pre-HAC1 mRNA in a Wild type (black line), rrp6-Δ (red line) and cbc1-Δ strains (blue line) in absence and presence of 1 μg/ml tunicamycin at 30°C. Decay rates were determined from three independent experiments (biological replicates) by qRT-PCR analysis (using primer sets of HAC1 intronic sequences) and the intronic signals were normalized to SCR1 RNA and normalized signals (mean values ± SD) were presented as the fraction of remaining RNA (with respect to normalized signals at 0 min) versus time of incubation in the presence of 1,10-phenanthroline. The statistical significance of difference as reflected in the ranges of p-values estimated from Student's two-tailed t-tests for a given pair of test strains for each message are presented with following symbols, ∗P < 0.05, ∗∗P < 0.005 and ∗∗∗P < 0.001, NS, not significant.
Next, we addressed whether the observed increase in the level of HAC1 pre-mRNA in rrp6-Δ and cbc1-Δ strains is due to diminution of its degradation or to stimulation of its transcription or to both. Consequently, we evaluated the transcriptional activity around the HAC1 locus in wild-type, rrp6-Δ and cbc1-Δ strains in the presence and absence of tunicamycin by measuring the genomic occupancies of RNA polymerase II (RNAPII) and histone H3 by chromatin-immunoprecipitation (ChIP) technique (55). Accordingly, their occupancies were determined in various regions of the HAC1 locus that included the promoter, exon 1, exon1-intron junction, intron, intron-exon 2 junction and the intergenic region between HAC1 and the AGX1 gene (located next to HAC1 on chromosome VI) (Figure 3D). As shown in Figure 3E, none of the RNAPII or H3 occupancies displayed any dramatic alterations in the wild-type or cbc1-Δ strains, either in the absence or the presence of ER stress. This result thus indicated that the alteration of the transcription rate is not associated with the observed increase in the level of HAC1 pre-mRNA in the exosome/DRN-deficient strains. Notably, our observation contradicts earlier findings, where the induction of the HAC1 message was shown to involve transcriptional stimulation (56,57). We believe that this inconsistency is associated with the difference in the methods used to determine transcriptional activity in two experimental setups. While the previous studies utilized an indirect method to determine HAC1 promoter activity using a reporter gene (55), we employed a much more direct and precise technique to estimate it. To gain further support in favor of our view, we determined the decay rates of precursor HAC1 mRNA in wild-type, cbc1-Δ, and rrp6-Δ strains in the presence and absence of stress after shutting off their transcription with 1,10-phenanthroline (49). As shown in Figure 3F, while the decay rate of HAC1 pre-mRNA is very rapid in the absence of ER stress (the half-life of HAC1 pre-mRNA was 4.8 min, upper graph, black line, Table 1), it is substantially slower in the presence of ER stress (half-life of HAC1 pre-mRNA was 9.8 min, lower graph, black line, Table 1). Most significantly, the decay rate and stability of this message were dramatically enhanced in both the cbc1-Δ (blue line) and rrp6-Δ (red line) strains in the absence as well as in the presence of tunicamycin. The half-life values were found to be 25.25 and 20.05 min in cbc1-Δ and rrp6-Δ strains, respectively, in the absence of tunicamycin and 28.63 and 22.75 min in cbc1-Δ and rrp6-Δ strains, respectively, in the presence of the drug (Table 1). Interestingly, the half-life values of this precursor RNA were found to be twice as much as that obtained in the presence of ER stress (half-life values 4.8 and 9.8 min in the absence and presence of stress, respectively) (see Figure 3F, Table 1). These values are in good agreement with the decreased stabilization of this precursor transcript in exosome/DRN deficient strains in the presence of stress (because a smaller fraction of the precursor message undergoes decay) (see Figure 3A). Therefore, the extent of its steady-state enhancement could be directly correlated to the increase in its half-life under the same conditions, thus sustaining the view that the steady-state alteration of this transcript in rrp6-Δ and cbc1-Δ strains involve a change in the nuclear exosome and DRN-dependent mRNA decay rate. Thus, the evidence described in this section strongly implies that the enhancement of HAC1 pre-mRNA in DRN-deficient rrp6-Δ and cbc1-Δ strains is due to the rapid and selective decay of the precursor-HAC1 transcript by the nuclear exosome and DRN. This decay probably becomes diminished in response to stress to accomplish an increased rate of supply of this precursor (see next section for additional experimental support).
Table 1. Half Life values of HAC1 pre-mRNA in different strains in presence and absence of ER stress.
| Pertinent genotype | Half-lives (in minutes) | |
|---|---|---|
| Without ER stress | With ER stress | |
| WT | 4.8 | 9.8 |
| cbc1-Δ | 25.25 | 28.63 |
| rrp6-Δ | 20.05 | 22.75 |
The half-lives were determined from three independent mRNA Decay Experiment (biological replicates) in which relevant yeast strains were grown till mid-log phase followed by the induction of ER Stress, shutting off the transcription with 1,10-phenanthroline, measuring the specific mRNA levels by Real-time quantitative PCR analyses and plotting the data as percent mRNA remaining as a function of time as described in Materials and Methods.
The nuclear exosome and DRN exert a negative regulatory influence on the UPR pathway
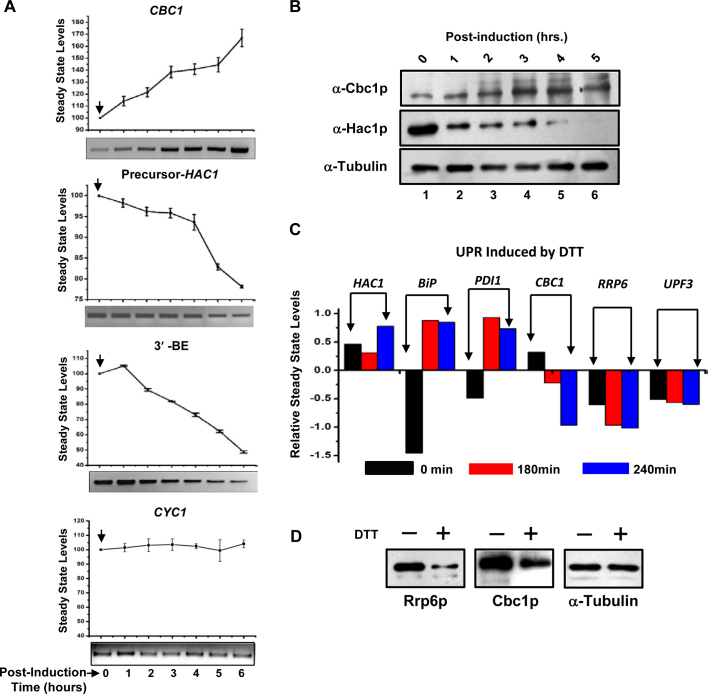
Involvement of the nuclear exosome and DRN in the preferential degradation of HAC1 precursor mRNA prompted us to ask whether this degradation employs a regulatory impact on the UPR output. Consequently, we reasoned that a regulatory influence of the exosome/DRN should lead to a reciprocal relationship between steady-state levels of precursor-HAC1 and CBC1 mRNAs. Accordingly, we verified this prediction by determining the steady-state levels of precursor-HAC1 (induced by tunicamycin) and CBC1 messages (differentially expressed from a pGAL1-GAL10 promoter following induction with 2% galactose) in a wild-type strain. In support of our prediction, the precursor-HAC1 mRNA level was found to decline sharply during post-induction growth (Figure 4A, second graph from the top) when the steady-state level of CBC1 mRNA increased following the galactose induction (Figure 4A, top graph). As a control, we also determined the abundance of CYC1 mRNA under similar conditions, which remained unaltered at different times post-induction (Figure 4A, bottom-most graph). To further support our view, we determined the levels of Hac1p and Cbc1p proteins by immunoblotting using anti-Hac1p and anti-Cbc1p antibodies, respectively, under identical conditions of induction by tunicamycin and 2% galactose (as mentioned above). Most significantly, the relative profile of the steady-state levels of these proteins (Figure 4B) indeed reflected an identical reciprocal pattern as revealed by the levels of these mRNAs under the same conditions (Figure 4A). Thus, an inverse relationship between the exosome/DRN and UPR component is indeed sustained in a wild-type yeast cell at the RNA and protein levels (Figure 4A and B). Collective findings thus substantiate the view that the nuclear exosome and DRN inversely regulate the function of the UPR in baker's yeast by maintaining an optimum supply line of precursor HAC1 mRNA under both the unstressed and ER-stressed conditions, which is mediated by a selective decay of pre-HAC1 mRNA.
Figure 4.
Cellular levels of UPR activity and CBC1 mRNA/protein levels bear a reciprocal relationship. (A) Relative Levels of CBC1 mRNA and precursor HAC1 mRNA/HAC1-3′-BE bears a reciprocal relationship. Levels of CBC1 mRNA expressed under the control of a pGAL1-GAL10 promoter, and precursor-HAC1 mRNA were determined by qPCR using RNA harvested from the wild-type yeast strain harboring the engineered pGAL1-GAL10::CBC1 allele at different times after induction with 2% galactose and are presented as means ± SE (n = 3, biological replicates). ER stress was previously imposed with 1 μg/ml tunicamycin, which was added to the culture 3 h before the addition of 2% galactose. The bottom panel of each graph shows the representative gels revealing the relative levels of respective transcripts at different times post-induction as determined by semi-quantitative PCR analysis using cDNA samples prepared from these cells. The arrow denotes the point of induction with 2% galactose. (B) Relative Levels of Cbc1p protein and Hac1p protein bears an inverse relationship. Levels of Cbc1p and Hac1p proteins expressed in wild-type yeast cells expressing a CBC1 allele under the control of pGAL1-GAL10 promoter as determined by western blot analyses using total cellular proteins harvested from a normal strain harboring the engineered CBC1 allele at different times after the induction with 2% galactose. Cells were harvested at different times post-induction as indicated above the panel and total cellular protein extracts were prepared as described and resolved at 12% SDS-PAGE followed by the immunodetection using anti-Cbc1p and anti-Hac1p antibodies. As before, the α-tubulin served as the loading control. (C) Relative levels of transcripts encoding UPR pathway and nuclear exosome/DRN machinery in response to stress induced by DTT (58). The data was retrieved from whole genome expression profile database (www.yeastgenome.org). (D) Relative steady state levels of Cbc1p and Rrp6p proteins in wild type (yBD-117) strains in the absence (–) and in the presence of 5mM DTT (+). α-Tubulin was used as the loading control. Cells were grown in the absence and in the presence of DTT, harvested and total protein extract were prepared as described in Materials and Methods. 50 μg of total cellular protein was loaded in each lane, resolved in 12% SDS-PAGE and probed with anti-Cbc1p, anti-Rrp6p, and anti-tubulin antibodies as described in Materials and Methods.
Although we demonstrated a reciprocal relationship between the cellular levels of HAC1 pre-mRNA and Hac1p protein and those of the CBC1 mRNA and Cbc1p protein, the physiological relevance of this putative control remains an open question. To resolve this issue, we further investigated whether the inverse relationship between the exosome/DRN and UPR components is also maintained under normal physiological conditions. Interestingly, the genome-wide expression profile database (www.yeastgenome.org) revealed that the abundance of the transcript levels of the UPR components, HAC1, BiP/KAR2 and PDI, increased dramatically following the activation of the UPR by DTT (58) (Figure 4C). In contrast, transcript abundance of exosome/DRN components, RRP6, CBC1, UPF3, displayed a gradual decline under identical conditions (Figure 4C). Moreover, levels of the mRNAs encoding the exosome/DRN components also appear to maintain a reciprocal relationship with HAC1 pre-mRNA/mRNA in both the wild-type and the ire-Δ yeast strains during ER stress induced by either DTT or tunicamycin (Supplementary Figure S2A) (58). This reciprocal relationship between the levels of messages encoding the exosome/DRN and the UPR components, however, is not exhibited during other kinds of stress, such as those induced by H2O2 or nitrogen depletion (Supplementary Figure S2B) (59). Thus, the transcriptomic data imparts a physiological relevance to our findings and supports the idea that the nuclear exosome/DRN exerts a regulatory influence on the function of the UPR. To further strengthen our model, we tested whether the steady-state levels of Cbc1p and Rrp6p proteins alter in response to ER stress created by 5 mM DTT. As shown in Figure 4D, both the nuclear exosome/DRN players displayed a significant reduction in their levels when ER stress was imposed with 5 mM DTT. Together, these findings are in good agreement with the view that the nuclear exosome and DRN impose a negative regulatory influence on the activity of the UPR in S. cerevisiae, which operates at the post-transcriptional level via preferential mRNA degradation.
The nuclear exosome and DRN generate an optimum pool of precursor-HAC1 transcripts harboring an intact and functional 3′-BE and thereby govern its recruitment to functional Ire1p foci
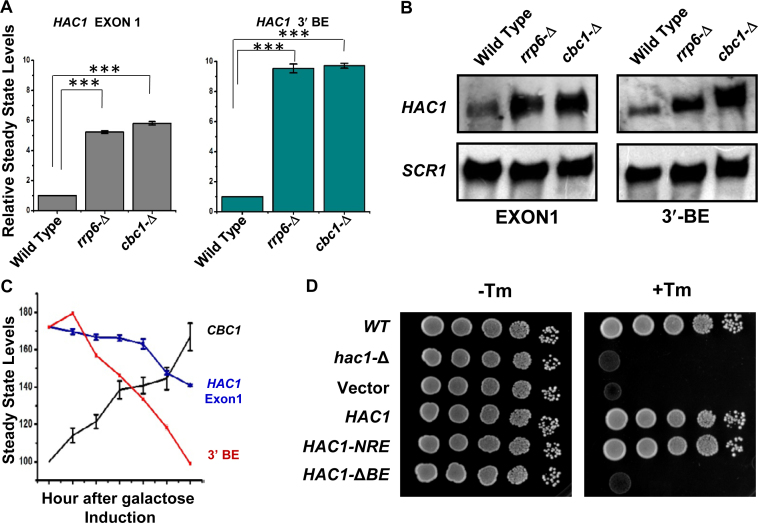
The nuclear exosome and DRN degrade the transcript body from the 3′ to 5′ direction, which appears to be the major mechanism of mRNA decay in the nucleus (1–3,5,6,20), and accordingly, the precursor HAC1 mRNA is expected to undergo a decay from the 3′→5′ direction. Consequently, a transcript segment located closer to the 3′-terminus of the message is expected to display stronger stabilization in the nuclear exosome/DRN-deficient strain than the one located more towards the 5′-end. To verify this prediction, we evaluated the relative abundance of two different segments of the pre-HAC1 transcript body in wild-type, rrp6-Δ and cbc1-Δ strains in the absence of stress. The first segment corresponds to the exon 1 sequence (determined using Primer set EXON1, location shown in Figure 3D), and the second one encompasses a specific cis-acting element in the 3′-UTR of the message, termed 3′-BE (determined using Primer set BE, location shown in Figure 3D) (40). As predicted, these two fragments displayed different extents of stabilization in strains deficient in the nuclear exosome and DRN. The RNA segment corresponding to exon 1 (5′-segment) showed a 4- to 5-fold increase in its steady-state level in both rrp6-Δ and cbc1-Δ strains (Figure 5A, gray histogram). The 3′-end segment encompassing the BE element, in contrast, exhibited a much higher enhancement in its steady-state level (∼9- to 10-fold) in these strains (Figure 5A, green histogram). The relative steady-state levels of these transcript segments as determined by northern blot analysis in the wild type, rrp6-Δ and cbc1-Δ strains, also revealed a very similar profile—the relative level of enhancement of the 3′-BE RNA segment was more dramatic compared to that of the Exon 1 segment (Figure 5B). These findings imply that the 3′-terminus of the HAC1 precursor transcript undergoes more accelerated and active degradation by the nuclear exosome and the DRN. Furthermore, the decline of the signal of a transcript segment corresponding to the 3′-BE is much sharper compared to that encompassing the HAC1 Exon 1 segment following the induction of a pGAL1-GAL10::CBC1 allele by tunicamycin and 2% galactose (Figure 5C). Together, these findings are consistent with the idea that the nuclear exosome and DRN-dependent degradation of the precursor HAC1 message initiates from the 3′-terminus and proceeds towards the 5′-end. Consequently, we hypothesized that this preferential 3′ to 5′ decay generates a mixed pool of precursor HAC1 messages with heterogeneous 3′-termini at any given time, some of which would harbor the 3′-BE, while others would lack it. Thus, it is conceivable that an accelerated decay can result in the formation of a precursor HAC1 mRNA pool, most of which lack the BE in unstressed cells, whereas a diminished decay of this transcript during ER stress can lead to the generation of another pool, the majority of which carry the BE element. Notably, the 3′-BE is implicated in the targeting and recruitment of precursor HAC1 message to Ire1p foci and its subsequent splicing in response to ER stress (40). Hence, this mechanism, in principle, may result in the formation of a decisive concentration of the precursor HAC1 transcript population with or without an intact 3′-BE element, which in turn governs its intracellular targeting to Ire1p foci (see Figure 7). We addressed this issue and evaluated this possibility by determining the relative intracellular distribution of HAC1 messages and Ire1p proteins in the absence and in presence of ER stress using confocal imaging techniques (see below).
Figure 5.
Relative levels of transcript segments corresponding to HAC1 Exon 1 and 3′-BE in absence of ER-stress. (A) Histogram showing the relative abundance of transcript segments corresponding to HAC1 exon 1 and HAC1 3′-BE element in normal, cbc1-Δ, and rrp6-Δ strains in the absence of tunicamycin as determined from three independent experiments (biological replicates) and are presented as means ± SE (n = 3 for each strain). Normalized value of individual amplicon from normal strain sample was set to 1. (B) Northern blot revealing the levels of transcript segments corresponding to HAC1 exon 1 and HAC1 3′-BE level in a wild type (yBD-117), cbc1-Δ (yBD-131), rrp6-Δ (yBD-129) strains in the absence of ER stress. The northern blot was hybridized with a DNA oligonucleotide encompassing either an HAC1-Exon1 sequence or HAC1-3′-BE element sequence respectively. (C) Composite graph revealing the relative abundance of the CBC1 message, HAC1 Exon 1 and 3′-BE element in a wild type yeast strain in the absence of ER-stress harboring an engineered allele of CBC1 gene expressed under pGAL1-GAL10 promoter at different times post-induction with 2% galactose. (D) Relative growth of wild type (yBD-117), hac1-Δ (yBD-118), hac1-Δ pRS316 (yBD-404) hac1-Δ pHAC1 (yBD-405), hac1-Δ pHAC1-NRE (yBD-406) and hac1-Δ pHAC1-BE (yBD-407) strains in YPD solid growth media in absence (–Tm) and in presence of 1 μg/ml tunicamycin (+Tm).
Figure 7.
Model of DRN mediated regulation of UPR activity in Saccharomyces cerevisiae. Precursor HAC1 mRNA with 3′-BE element is subjected to kinetic and preferential decay activity by DRN from 3′ to 5′ direction. In the absence of ER stress the precursor HAC1 mRNA undergoes an accelerated decay from 3′ to 5′ direction in a kinetic manner thus producing a higher population of precursor transcripts lacking the 3′-BE element resulting in a less efficient recruitment to Ire1p foci. Note that the degradation machinery does not stop after degrading the 3′-BE element, but continues to degrade the transcript body progressively in the 3′-5′ direction. This progressive degradation produces a heterogeneous population of pre-HAC1 transcripts having a heterogeneity at their 3′-termini most of which lack the 3′-BE. The absence of 3′-BE in these pre-HAC1 population would lead to reduced splicing by Ire1p/Rlg1p thereby resulting in the accumulation of precursor HAC1 transcript in the absence of ER stress. When ER stress is induced, degradation activity of DRN is diminished, which consequently generates a much higher population of precursor HAC1 mRNA population, most of which carry the functional BE element and is thereby targeted and recruited more efficiently to Ire1p foci. Increased recruitment of HAC1 precursor to Ire1p cluster results in increased splicing by Ire1p/Rlg1p thereby producing mature HAC1 mRNA and Hac1p for successful and efficient induction of UPR activity.
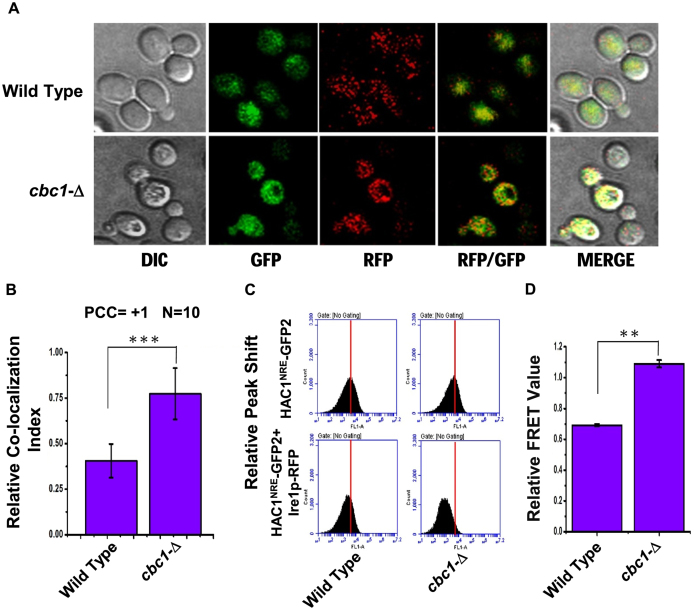
First, we tested whether the presence of the 3′-BE is crucial for the observed 3′→5′ decay of HAC1 pre-mRNA by nuclear exosome/DRN. To test this hypothesis, we evaluated the steady-state level of the HAC1 allele that lacks the 3′-BE element (HAC1-ΔBE) in wild-type, cbc1-Δ, and rrp6-Δ strains in the absence of ER stress. As shown in Supplementary Figure S3A, the stability profiles of native HAC1 harboring an intact 3′-BE element and one lacking it (HAC1-ΔBE) were found to be very similar and comparable. The steady-state levels of both of these transcripts were found to be enhanced similarly in the DRN-deficient cbc1-Δ yeast strains, thus indicating that the nuclear exosome/DRN does not specifically target the BE element present in the HAC1 transcript. Moreover, this HAC1-ΔBE allele fails to complement the hac1-Δ defect (Figure 5D), thus verifying the functional authenticity of the construct used in the stability experiment described above. These collective findings thus completely ruled out the possibility that the 3′-BE element plays any role in directing the decay of the HAC1 precursor RNA dependent on the exosome/DRN. Next, we asked whether the nuclear exosome/DRN contributes to a regulatory decay mechanism that may dictate the efficiency of targeting and recruitment of precursor HAC1 mRNA to the Ire1p sites in response to appropriate cues. Accordingly, our model predicts more efficient recruitment of pre-HAC1 to cellular Ire1p sites in a cbc1-Δ strain owing to a higher abundance of precursor message carrying an intact and functional 3′-BE. We verified this speculation by determining the intracellular distribution of HAC1 mRNA and Ire1p protein sites in wild-type and cbc1-Δ strains, both of which co-express HAC1NRE chimeric mRNA, a Nucleolin-GFP fusion protein (50) and Ire1p-RFP (Ire1p protein tagged with RFP) (51) using confocal imaging technique (Figure 6A). The in situ distribution of HAC1NRERNA and Ire1p-RFP in wild-type cells (Figure 6A, top panel) shows that GFP (representing the HAC1 message) and RFP (representing Ire1p) signals appear to be considerably dispersed. Although an overlap between these signals was detected, the extent of their overlap/colocalization (represented by the yellow spots in the merged panel) was small (CI = 0.43±08) (Figure 6A&B, co-localization data provided in Supplementary Table S4). A careful examination of their distribution in the cbc1-Δ strain (Figure 6A, bottom panel) revealed a more intense, frequent colocalization of the GFP signal with the RFP clusters (CI = 0.85 ± 0.15) (Figure 6A&B, colocalization data provided in Supplementary Table S4). This observation indicates a higher and more efficient targeting and recruitment of the precursor-HAC1 mRNA to the intracellular Ire1p sites as predicted by our hypothesis. To reinforce this observation, we verified the functionality of the HAC1NRE and Ire1p-RFP constructs. In the first experiment, we tested the ability of the HAC1NRE construct to complement the hac1-Δ defect. As shown in Figure 5D, this construct could complement the hac1-Δ defect as assessed by the ability of the hac1-Δ strain transformed with HAC1NRE constructs to grow in the presence of tunicamycin. Furthermore, we measured the stability of the HAC1NRE mRNA produced from the construct in wild-type and cbc1-Δ strains in the absence of ER stress, which revealed that the HAC1NRE pre-mRNA displayed a similar enhancement in a cbc1-Δ strain as shown by native HAC1 pre-mRNA (Supplementary Figure S3A). Moreover, we also tested the specificity of the HAC1NRE construct by carrying out a control experiment in which the relative distribution of a version of HAC1 mRNA lacking the NRE module was tested with that of the Ire1p-RFP. As shown in Supplementary Figure S3B, the data revealed that there is hardly any colocalization between the GFP and RFP signals, indicating that the GFP signal, which is not tethered to the HAC1 message (owing to the absence of the NRE module), does not co-localize with RFP. The authenticity of the Ire1p-RFP construct was also evaluated in multiple ways. First, the ability of the Ire1p-RFP construct to complement the ire1-Δ defect was verified by the ability of the ire1-Δ strain transformed with an Ire1p-RFP construct to grow in the presence of tunicamycin (Supplementary Figure S4A). In addition, multiple Ire1p foci could be detected in both wild-type (Supplementary Figure S4B) and cbc1-Δ (data not shown) strains transformed with the Ire1-RFP when strong ER stress was imposed with 5 mM DTT for 30 min (Supplementary Figure S4B), which was not detectable in these strains in the unstressed condition.
Figure 6.
Precursor HAC1 mRNA is recruited more efficiently to Ire1p foci in DRN deficient cbc1-Δ strains due to the diminished decay of its 3′-BE. (A) Localization of Ire1p-RFP and HAC1NRE mRNA decorated with NRE-GFP following the induction of ER stress by 1 μg/ml tunicamycin in a wild type and cbc1-Δ strain. The images were captured and processed as described in Materials and methods. (B) Histogram depicting the co-localization of HAC1NRE-GFP-nucleolin mRNA into Ire1p-RFP foci at 10 minutes after ER stress by 1 μg/ml tunicamycin in a wild type and cbc1-Δ strain. Co-localization index for HAC1NRE-GFP-nucleolin recruitment into Ire1 foci was expressed in arbitrary units. Means and standard error of the mean were determined from n = 17. Co-localization index (Pearson correlation coefficient, PCC) were determined as described in the method section. (C) Relative fluorescence resonance energy transfer (FRET) between HAC1-GFP-nucleolin and Ire1p-RFP in a wild type and cbc1-Δ strain. The bottom panel shows the quenching of donor HAC1- GFP-nucleolin mRNA in the presence of Ire1p-RFP (acceptor) in normal and cbc1-Δ strains. The top panel is the negative control of this experiment showing no donor (HAC1-GFP-nucleolin) quenching in the absence of Ire1p-RFP in these strains. (D) Histogram depicting the Relative FRET value (in arbitrary units) from donor HAC1NRE-GFP-nucleolin mRNA into Ire1p-RFP foci at 10 minutes after ER stress by 1 μg/ml tunicamycin in a normal and cbc1-Δ strain. Donor (HAC1NRE-GFP-nucleolin mRNA) quenching as an indicator of relative FRET value was measured for HAC1NRE-GFP-nucleolin recruitment into Ire1p foci. Means and standard error of the mean were determined from n = 5 (biological replicates). Donor quenching was determined by Flow Cytometry from 50 000 cells of each strain were determined as described in the method section using BD Accuri™ C6 software. The statistical significance of difference as reflected in the ranges of P-values estimated from Student's two-tailed t-tests for a given pair of test strains for each message are presented with following symbols, ∗P < 0.05, ∗∗P < 0.005 and ∗∗∗P < 0.001, NS, not significant.
To further corroborate this finding and bolster our model, we evaluated the relative proximity of the HAC1NRE-Nucleolin-GFP module and Ire1p-RFP moiety by determining the fluorescence resonance energy transfer (FRET) from HAC1NRE-Nucleolin-GFP (donor) to Ire1p-RFP fluorophore (acceptor). Fluorescence quenching of the donor HAC1NRE-Nucleolin-GFP was determined as an indicator of FRET in wild-type and cbc1-Δ strains at different times after addition of tunicamycin (Supplementary Figure S5 and Supplementary Table S5). As shown in Figure 6C and D, a 2-fold higher donor HAC1NRE-Nucleolin-GFP quenching due to a higher FRET from HAC1NRE-Nucleolin-GFP to Ire1p-RFP was detected in the cbc1-Δ strain compared to the wild-type strain at 10 min after ER stress was imposed (Figure 6D). This finding strongly suggests a closer physical proximity between the HAC1NRE-Nucleolin-GFP and Ire1p-RFP in an exosome/DRN deficient cbc1-Δ strain, which in turn implies that HAC1 precursor mRNA is more efficiently recruited onto the active Ire1p clusters in a cbc1-Δ strain. Together, these data support the conclusion that the abundance of the precursor-HAC1 mRNA harboring an intact 3′-BE element is dictated by a preferential and regulatory decay mechanism by the nuclear exosome/DRN. This selective decay is crucial for its proper translocation, recruitment to Ire1p foci and its subsequent splicing. Exosome/DRN thus regulates the UPR by imparting a preferential and kinetic decay of precursor HAC1-mRNA in a 3′→5′ direction, which controls its recruitment to Ire1p foci and thereby governs the rate of supply of HAC1 precursor mRNA to Ire1p/Rlg1p mediated splicing (See discussion below and Figure 7).
DISCUSSION
In this study, we present evidence of the existence of an additional layer of regulation in the function of the UPR pathway in Saccharomyces cerevisiae. This regulation is accomplished by a preferential 3′→5′ decay of precursor HAC1 mRNA, which, in turn, generates an ‘optimum’ physiological ‘repertoire’ of the pre-HAC1 message carrying heterogeneous 3′-termini. At any given instance, a fraction of this repertoire harbors an intact BE at the 3′-end of the message, whereas the other fraction lacks it. The status of ER stress dictates the relative abundance of each of these pools by controlling the rate of pre-HAC1 mRNA decay. A higher fraction of the pre-HAC1 message lacking an intact BE is predominantly present in the unstressed cell, which results from an accelerated decay by nuclear exosome/DRN. ER stress, in contrast, prompts a diminution of the decay activity, thus leading to the production of a higher fraction of the pre-HAC1 message carrying an intact BE (Figure 7). This regulatory decay mechanism of precursor HAC1 mRNA thus controls its targeting/recruitment to the active Ire1p foci at an optimum level and thereby governs the subsequent rate of its downstream splicing reaction. The nuclear exosome/DRN therefore finely tunes the output of the UPR in S. cerevisiae.
We began our investigation with the validation of our previous microarray data to establish that both the precursor HAC1 mRNA and Hac1p protein levels increased dramatically in the yeast strains defective in the components of the nuclear exosome and DRN (Figure 1A–C, Figure 3A and B). Consistent with this finding, these strains sustained ER stress more efficiently when challenged either with 1 μg/ml tunicamycin or 5 mM DTT (Figure 1D–G, Supplementary Figure S1A and B). Moreover, ER chaperone markers were also enhanced dramatically in the presence of ER stress in exosome/DRN-deficient strains, thus further supporting our model (Figure 2). We then demonstrated that augmentation of HAC1 mRNA in the rrp6-Δ and cbc1-Δ strains was directly correlated to the stabilization of HAC1 precursor mRNA (Figure 3F, Table 1) and was not associated with a transcriptional induction of the HAC1 promoter (Figure 3E). Notably, the stability of the HAC1 precursor message in both the rrp6-Δ and cbc1-Δ strains did not change in the presence or absence of ER stress, because the exosome/DRN decay apparatus is impaired in such strains. An altered stability of precursor-HAC1 mRNA in the wild-type strain in the absence and presence of ER stress (Table 1) strongly implied a regulation in the HAC1 pre-mRNA decay rate during ER stress. Consequently, we corroborated the regulation in the decay rate by correlating the diminution of decay with the reduced levels of the DRN components, Cbc1p, and Rrp6p, during ER stress (Figure 4D). Notably, two previous studies reported apparently contradictory findings concerning this issue. One study revealed no difference in the stability of the HAC1 mRNA decay rate in the presence of ER stress induced by DTT (57), which contrasts with our data. We argue that this discrepancy reflects the difference in the mode of determination of half-life values in two different experimental systems. While these researchers determined the HAC1 signal using an exon 1-specific probe, we used an intron-specific probe to determine the decay/abundance of precursor-HAC1 mRNA more precisely during half-life measurement in our study. Interestingly, a more recent study reported that the stability of HAC1 pre-mRNA is altered in the presence of ER stress (60). While the decay rate of HAC1 pre-mRNA is faster in the absence of ER stress, it displayed a diminished decay when ER stress was imposed. These researchers showed the alteration of stability of the HAC1 precursor message is probably caused by a physical association of the RNA with the Rab-GTPase Ytp1p. However, the mechanistic insight of the alteration of the stability of the HAC1 message during ER stress was unclear (60). This finding thus poses an interesting question about the connection between the association of the pre-HAC1 message with Ytp1p (and a few other proteins) and its regulated degradation by the nuclear exosome/DRN, which will be the focus our future work. Consistent with the idea of a dynamic regulation of mRNA decay during ER stress, the cellular levels of CBC1 mRNA and Cbc1p protein level were found to bear a reciprocal relationship with the levels of HAC1 pre-mRNA and Hac1p protein (Figure 4A and B). Our data mining analyses of whole genome expression profiles also revealed an inverse relationship between the cellular levels of mRNAs encoding the components of the nuclear exosome/DRN and the UPR during ER stress (Figure 4C and Supplementary Figure S2A). Collectively, these findings strongly argue that the nuclear exosome/DRN exert a fine control on the activity of the UPR.
Finally, we presented data that are consistent with the view that the nuclear exosome/DRN-dependent degradation of HAC1 precursor mRNA is possibly exerted in the 3′→5′ direction. This preferential decay plays an instrumental role in the generation of two kinds of pre-HAC1 mRNA populations—one that possesses the 3′-BE and the other that lacks it (Figure 5A–C). Each of these two populations themselves is heterogeneous at their 3′-termini. Presumably, the population that lacks the BE element (predominant species in the absence of ER stress) is targeted to Ire1p foci less efficiently than the one that harbors the BE (predominant during ER stress). This conclusion received further support from the observation that HAC1 precursor mRNA is indeed targeted and recruited to Ire1p foci more efficiently in a strain lacking a functional DRN (Figure 6). Thus, our model is consistent with the view that the precursor HAC1 mRNA undergoes an accelerated degradation in the absence of stress leading to an increased elimination of 3′-BE from the HAC1 pre-mRNA population. Loss of the BE element, in turn, results in the reduced targeting and recruitment of the resulting precursor message to Ire1p foci (supported by the data presented in Figure 3A, B and F and Figure 4A–D), thus generating a lower supply of HAC1 precursor RNA for downstream splicing reactions. ER stress, in contrast, prompts a diminution in the exosome-dependent decay, which permits an increased retention of the 3′-BE element in the resulting pre-HAC1 transcript population (supported by the data presented in Figure 3A, B and F and Figure 4A–D). Consequently, an increased abundance of precursor HAC1 mRNA population harboring an intact BE led to improved targeting to the Ire1p foci under ER stress, thus causing its enhanced splicing (supported by the data presented in Figure 6A–D for cbc1-Δ strain). We thus favor a view that the nuclear exosome and DRN-dependent decay can be altered in a context-dependent manner to generate an optimum delivery of HAC1 precursor mRNA to the splicing pipeline. To our understanding, exosome/DRN-dependent circuitry certainly qualifies as a suitable candidate for a rate-limiting regulatory step to optimally control the output of the UPR in S. cerevisiae.
In this context, we would like to note that the nuclear exosome/DRN is highly processive and does not selectively clip off the 3′-Bipartite Elements present in the HAC1 pre-mRNA. In contrast, our model predicts that the exosome/DRN does not halt after clipping off the transcript segment encompassing the BE element. Instead, the decay mechanism will continue to degrade the rest of the transcript body from the 3′-termini in a progressive manner. Consistent with this view, the stability of the native HAC1 pre-mRNA (carries the BE) and the one that lacks it (HAC1-ΔBE) were found to be very similar in wild-type, cbc1-Δ/rrp6-Δ yeast strains (Figure 5D and Supplementary Figure S3A). Notably, the unwinding of the strong stem-loop structure of BE must precede the degradation of the rest of the transcript body of HAC1 pre-mRNA. Typically, RNA secondary structures consisting of stems and loops (such as a polyG18 track) confer a partial block to both 5′→3′ and 3′→5′ exoribonucleases (61–64) and thus pose a challenge to the cellular decay machinery. However, both the cytoplasmic and nuclear exosome is equipped with powerful ATP-dependent RNA helicases such as Ski2p and Mtr4p (components of cytoplasmic and nuclear exosome, respectively) (65,66). In addition, the nuclear exosome-associated trimeric NNS complex (Nrd1p/Nab3p/Sen1p) was found to participate in the decay of the nuclear mRNA surveillance by the nuclear exosome/DRN and displays an epistatic relationship with Rrp6p and Cbc1p (P. Singh and B. Das, unpublished data). The Sen1p component encodes an ATP-dependent RNA/DNA helicase that participates in various events during transcription termination and RNA processing (67,68). Ski2p/Mtr4p are essential for the degradation of various RNA substrates and intermediates of preribosomal RNA-processing events containing potentially strong secondary structures with multiple stems and loops (such as ITS2) (69,70). Although we did not specifically address this issue here, it would not be unreasonable to assume that either of the nuclear Dob1p/Mtr4 or Sen1p in principle may play a crucial role in unwinding the secondary structure of 3′-BE and subsequent recruitment of the RNA transcript to the nuclear exosome/DRN machinery.
We did not specifically address the feature(s)/determinant(s) present in the HAC1 pre-mRNA, which qualify it as a privileged substrate for the nuclear exosome/DRN. Interestingly, however, our previous work suggests that the HAC1 belongs to a group of cellular messages referred to as the ‘special’ mRNAs, all of which display a high susceptibility to the nuclear exosome/DRN (See Introduction) (32). Some of these messages were shown to have an intrinsically slow nuclear export and a long intranuclear dwelling time, unlike the bulk-normal typical messages (32). It was proposed that the selectivity and susceptibility of the ‘special’ messages (including HAC1 pre-mRNA) to the nuclear exosome/DRN principally depend on their prolonged nuclear retention—a distinctive property of the ‘special’ mRNAs (32). Thus, it is reasonable to assume that the selectivity and susceptibility of HAC1 pre-mRNA/mRNA may result from its relatively longer nuclear dwelling time. However, further research is required to establish whether HAC1 pre-mRNA poses an intrinsically slow export and longer nuclear dwelling time. Interestingly, employing a post-transcriptional mechanism such as mRNA decay to regulate a stress response pathway such as UPR is undoubtedly more beneficial than using a transcriptional mode. A sharp and stepwise output can be generated more efficiently from the regulatory circuit that is controlled post-transcriptionally by the nuclear exosome/DRN using a decay mechanism, and this stepwise output enables the cells to respond more promptly to ER stress. Moreover, this means of control is crucial to avert an undesired stimulation of UPR in response to innocuous stimuli as well as to attenuate it once ER homeostasis is accomplished. Although the exact mechanism of the alteration of the decay rate of pre-HAC1 is not known at the molecular level, data from genome-wide expression profile analysis (58) (Figure 4C) and western blot analysis (Figure 4D) strongly suggest a change in the level of Cbc1p and Rrp6p as the underlying principle. Our data support a view that a possible mechanism of the attenuation of the decay rate of HAC1 pre-mRNA during ER stress may involve a decline in the levels of the proteins encoding exosome and DRN components. In this context, we would like to mention that insight into the association of HAC1 pre-mRNA with the Rab-GTPase Ytp1p should be explored to reveal further novel mechanistic insight into this regulatory decay mechanism.
Notably, cellular Hac1p levels are also regulated at translation-initiation and via protein degradation in addition to the Ire1p/Rlg1p-dependent splicing pathway, both of which are dependent on the HAC1 intron (71), thereby implying the significance of the multifaceted regulatory circuitry in defining the final output of a crucial cellular effector. In the light of this central paradigm, our findings added another regulatory layer upstream of the Ire1p-dependent splicing mechanism. We favor the view that the preferential decay of HAC1 pre-mRNA exerts a fine tuning of the UPR gain in Saccharomyces cerevisiae. It did not escape our notice that the exosome/DRN may also potentially degrade other mRNA targets encoding export, translation, and protein degradation factors (such as a component which preferentially degrades Hac1p protein). This large-scale degradation may exert indirect means of regulation of the HAC1 mRNA/Hac1p protein levels via additional mechanisms. Consistent with this view, the decay of Ire1α mRNA was recently shown to shape the UPR in mammalian cells, thus implicating a relevance and significance of post-transcriptional control in the regulation of the stress response (72). Furthermore, the nuclear exosome has been demonstrated to control a number of cellular responses and processes. These include the cellular iron response and sensitivity to oxidative stress (73), silencing of heterochromatic (74) and meiotic (75) genes, as well as regulation of cellular levels of mRNAs carrying decay-promoting introns (76). To our knowledge, the evidence presented here possibly provides the first example of selective and kinetic mRNA degradation as a central paradigm, which controls intracellular mRNA targeting and thereby regulates the activity of a signaling pathway. Future research will focus on (i) the delineation of the molecular determinant(s) present in the HAC1 transcript, which permits its recognition as a privileged DRN substrate, and (ii) the mechanistic aspect of the fine-tuning of the degradative activity, which kinetically speeds up or slows down the selective decay of this precursor mRNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Profs. Kazutoshi Mori, (Kyoto University, Japan), Peter Walter (University of California San Francisco, USA), Martin Schroeder (University of Durham, UK), Drs Madhusudan Dey and Anish Anshu (University of Wisconsin, Milwaukee, USA), Prof. J. Scott Butler (University of Rochester, Rochester, USA) for various yeast strains and plasmids. Kind assistance to use the Confocal Microscope facility of the University of Calcutta from Drs Arindam Bhattacharyya, Subir Biswas, Sougata Roy Chowdhury (University of Calcutta, India) and Subhadeep Das (Das Laboratory, Jadavpur University) is gratefully acknowledged. We also thank Ritabrata Ghosh (IISER-Kolkata) for his help in capturing confocal imaging and processing of the images. We are grateful to all the reviewers for their critical comments and constructive criticisms. We acknowledge Prof. Nitai C. Mandal (Former Professor, Bose Institute, Kolkata, India), Dr Satarupa Das (Jadavpur University, India), Prof. J. Scott Butler (University of Rochester, Rochester, USA), Prof. Randall H. Morse (Wadsworth Center, Albany, USA) and members of Das laboratory for critically reading the manuscript, and for useful comments and suggestions.
Authors contributions: D.S. and S.P. conceived and conducted the experiments and wrote the manuscript. B.D. conceived and designed all the experiments, directed the research program and organized and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology, Govt. of India [BT/PR6078/BRB/10/1114/2012 to BD]; Innovative Research Program, Jadavpur University (to B.D.); Council of Scientific and Industrial Research [09/096(0660)/2010-EMR-I to D.S.]; DST-PURSE Program, Jadavpur University (to S.P.). Funding for open access charge: Institutional Funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Houseley J., LaCava J., Tollervey D.. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006; 7:529–539. [DOI] [PubMed] [Google Scholar]
- 2. Doma M.K., Parker R.. RNA quality control in eukaryotes. Cell. 2007; 131:660–668. [DOI] [PubMed] [Google Scholar]
- 3. Fasken M.B., Corbett A.H.. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009; 6:237–241. [DOI] [PubMed] [Google Scholar]
- 4. Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012; 191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das S., Das B.. mRNA quality control pathways in Saccharomyces cerevisiae. J. Biosci. 2013; 38:615–640. [DOI] [PubMed] [Google Scholar]
- 6. Kilchert C., Wittmann S., Vasiljeva L.. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016; 17:227–239. [DOI] [PubMed] [Google Scholar]
- 7. Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H.. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell Biol. 2002; 22:8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R. et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002; 417:304–308. [DOI] [PubMed] [Google Scholar]
- 9. Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F.. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 2002; 22:8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen T.H., Dower K., Libri D., Rosbash M.. Early formation of mRNP: license for export or quality control. Mol. Cell. 2003; 11:1129–1138. [DOI] [PubMed] [Google Scholar]
- 11. Thomsen R., Libri D., Boulay J., Rosbash M., Jensen T.H.. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA. 2003; 9:1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vinciguerra P., Stutz F.. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004; 16:285–292. [DOI] [PubMed] [Google Scholar]
- 13. Moore M.J., Schwartzfarb E.M., Silver P.A., Yu M.C.. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol. Cell. 2006; 24:903–915. [DOI] [PubMed] [Google Scholar]
- 14. Kong K.Y., Tang H.M., Pan K., Huang Z., Lee T.H., Hinnebusch A.G., Jin D.Y., Wong C.M.. Cotranscriptional recruitment of yeast TRAMP complex to intronic sequences promotes optimal pre-mRNA splicing. Nucleic Acids Res. 2014; 42:643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bousquet-Antonelli C., Presutti C., Tollervey D.. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000; 102:765–775. [DOI] [PubMed] [Google Scholar]
- 16. Gudipati R.K., Xu Z., Lebreton A., Seraphin B., Steinmetz L.M., Jacquier A., Libri D.. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell. 2012; 48:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneider C., Kudla G., Wlotzka W., Tuck A., Tollervey D.. Transcriptome-wide analysis of exosome targets. Mol. Cell. 2012; 48:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Das B., Guo Z., Russo P., Chartrand P., Sherman F.. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 2000; 20:2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torchet C., Bousquet-Antonelli C., Milligan L., Thompson E., Kufel J., Tollervey D.. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002; 9:1285–1296. [DOI] [PubMed] [Google Scholar]
- 20. Das B., Butler J.S., Sherman F.. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003; 23:5502–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Das B., Das S., Sherman F.. Mutant LYS2 mRNAs retained and degraded in the nucleus of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:10871–10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butler J.S. The yin and yang of the exosome. Trends Cell Biol. 2002; 12:90–96. [DOI] [PubMed] [Google Scholar]
- 23. Liu Q., Greimann J.C., Lima C.D.. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006; 127:1223–1237. [DOI] [PubMed] [Google Scholar]
- 24. Wasmuth E.V., Januszyk K., Lima C.D.. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014; 511:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dziembowski A., Lorentzen E., Conti E., Seraphin B.. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007; 14:15–22. [DOI] [PubMed] [Google Scholar]
- 26. Makino D.L., Schuch B., Stegmann E., Baumgartner M., Basquin C., Conti E.. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015; 524:54–58. [DOI] [PubMed] [Google Scholar]
- 27. Schmidt K., Butler J.S.. Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip. Rev. RNA. 2013; 4:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halbach F., Reichelt P., Rode M., Conti E.. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013; 154:814–826. [DOI] [PubMed] [Google Scholar]
- 29. Hardwick S.W., Luisi B.F.. Rarely at rest: RNA helicases and their busy contributions to RNA degradation, regulation and quality control. RNA Biol. 2013; 10:56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das S., Saha U., Das B.. Cbc2p, Upf3p and eIF4G are components of the DRN (Degradation of mRNA in the Nucleus) in Saccharomyces cerevisiae. FEMS Yeast Res. 2014; 14:922–932. [DOI] [PubMed] [Google Scholar]
- 31. Maity A., Chaudhuri A., Das B.. DRN and TRAMP degrade specific and overlapping aberrant mRNAs formed at various stages of mRNP biogenesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2016; 16:fow088. [DOI] [PubMed] [Google Scholar]
- 32. Kuai L., Das B., Sherman F.. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13962–13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walter P., Ron D.. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011; 334:1081–1086. [DOI] [PubMed] [Google Scholar]
- 34. Bernales S., Papa F.R., Walter P.. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006; 22:487–508. [DOI] [PubMed] [Google Scholar]
- 35. Kawahara T., Yanagi H., Yura T., Mori K.. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell. 1997; 8:1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruegsegger U., Leber J.H., Walter P.. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001; 107:103–114. [DOI] [PubMed] [Google Scholar]
- 37. Lee K.P., Dey M., Neculai D., Cao C., Dever T.E., Sicheri F.. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008; 132:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sidrauski C., Walter P.. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997; 90:1031–1039. [DOI] [PubMed] [Google Scholar]
- 39. Kohno K., Normington K., Sambrook J., Gething M.J., Mori K.. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993; 13:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aragon T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A., Walter P.. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009; 457:736–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J.H., Walter P. et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell. 2009; 33:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P.. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007; 318:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pincus D., Chevalier M.W., Aragon T., van Anken E., Vidal S.E., El-Samad H., Walter P.. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010; 8:e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubio C., Pincus D., Korennykh A., Schuck S., El-Samad H., Walter P.. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 2011; 193:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chawla A., Chakrabarti S., Ghosh G., Niwa M.. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J. Cell Biol. 2011; 193:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heifetz A., Keenan R.W., Elbein A.D.. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979; 18:2186–2192. [DOI] [PubMed] [Google Scholar]
- 47. Barnes G., Hansen W.J., Holcomb C.L., Rine J.. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol. Cell. Biol. 1984; 4:2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Back S.H., Schroder M., Lee K., Zhang K., Kaufman R.J.. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005; 35:395–416. [DOI] [PubMed] [Google Scholar]
- 49. Parker R., Herrick D., Peltz S.W., Jacobson A.. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991; 194:415–423. [DOI] [PubMed] [Google Scholar]
- 50. Anshu A., Mannan M.A., Chakraborty A., Chakrabarti S., Dey M.. A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing, and translation. Mol. Cell. Biol. 2015; 35:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mannan M.A., Shadrick W.R., Biener G., Shin B.S., Anshu A., Raicu V., Frick D.N., Dey M.. An ire1-phk1 chimera reveals a dispensable role of autokinase activity in endoplasmic reticulum stress response. J. Mol. Biol. 2013; 425:2083–2099. [DOI] [PubMed] [Google Scholar]
- 52. Dunn K.W., Kamocka M.M., McDonald J.H.. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011; 300:C723–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagy P., Vereb G., Damjanovich S., Matyus L., Szollosi J.. Measuring FRET in flow cytometry and microscopy. Curr. Protoc. Cytom. 2006; doi:10.1002/0471142956.cy1208s38. [DOI] [PubMed] [Google Scholar]
- 54. Phillips S., Butler J.S.. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003; 9:1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sandoval J., Rodriguez J.L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., Lopez-Rodas G.. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004; 32:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ogawa N., Mori K.. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells. 2004; 9:95–104. [DOI] [PubMed] [Google Scholar]
- 57. Leber J.H., Bernales S., Walter P.. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2004; 2:E235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O.. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000; 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P.. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000; 101:249–258. [DOI] [PubMed] [Google Scholar]
- 60. Tsvetanova N.G., Riordan D.P., Brown P.O.. The yeast Rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 RNA. PLoS Genet. 2012; 8:e1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Beelman C.A., Parker R.. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994; 269:9687–9692. [PubMed] [Google Scholar]
- 62. Decker C.J., Parker R.. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993; 7:1632–1643. [DOI] [PubMed] [Google Scholar]
- 63. Muhlrad D., Decker C.J., Parker R.. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994; 8:855–866. [DOI] [PubMed] [Google Scholar]
- 64. Muhlrad D., Decker C.J., Parker R.. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995; 15:2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Anderson J.S., Parker R.P.. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998; 17:1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de la Cruz J., Kressler D., Tollervey D., Linder P.. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998; 17:1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim H.D., Choe J., Seo Y.S.. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999; 38:14697–14710. [DOI] [PubMed] [Google Scholar]
- 68. Ursic D., Himmel K.L., Gurley K.A., Webb F., Culbertson M.R.. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997; 25:4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Joseph N., Krauskopf E., Vera M.I., Michot B.. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999; 27:4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Allmang C., Mitchell P., Petfalski E., Tollervey D.. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000; 28:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Di Santo R., Aboulhouda S., Weinberg D.E.. The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. Elife. 2016; 5:e20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Karam R., Lou C.H., Kroeger H., Huang L., Lin J.H., Wilkinson M.F.. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015; 16:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsanova B., Spatrick P., Jacobson A., van Hoof A.. The RNA exosome affects iron response and sensitivity to oxidative stress. RNA. 2014; 20:1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tucker J.F., Ohle C., Schermann G., Bendrin K., Zhang W., Fischer T., Zhang K.. A novel epigenetic silencing pathway involving the highly conserved 5′-3′ exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet. 2016; 12:e1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Egan E.D., Braun C.R., Gygi S.P., Moazed D.. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA. 2014; 20:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kilchert C., Wittmann S., Passoni M., Shah S., Granneman S., Vasiljeva L.. Regulation of mRNA levels by decay-promoting introns that recruit the exosome specificity factor Mmi1. Cell Rep. 2015; 13:2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.