Summary
Peptidoglycan is the predominant stress‐bearing structure in the cell envelope of most bacteria, and also a potent stimulator of the eukaryotic immune system. Obligate intracellular bacteria replicate exclusively within the interior of living cells, an osmotically protected niche. Under these conditions peptidoglycan is not necessarily needed to maintain the integrity of the bacterial cell. Moreover, the presence of peptidoglycan puts bacteria at risk of detection and destruction by host peptidoglycan recognition factors and downstream effectors. This has resulted in a selective pressure and opportunity to reduce the levels of peptidoglycan. In this review we have analysed the occurrence of genes involved in peptidoglycan metabolism across the major obligate intracellular bacterial species. From this comparative analysis, we have identified a group of predicted ‘peptidoglycan‐intermediate’ organisms that includes the Chlamydiae, Orientia tsutsugamushi, Wolbachia and Anaplasma marginale. This grouping is likely to reflect biological differences in their infection cycle compared with peptidoglycan‐negative obligate intracellular bacteria such as Ehrlichia and Anaplasma phagocytophilum, as well as obligate intracellular bacteria with classical peptidoglycan such as Coxiella, Buchnera and members of the Rickettsia genus. The signature gene set of the peptidoglycan‐intermediate group reveals insights into minimal enzymatic requirements for building a peptidoglycan‐like sacculus and/or division septum.
Introduction
Peptidoglycan structure and function
Peptidoglycan (also called murein) is one of the largest macromolecules in a bacterial cell, typically forming a mesh‐like structure called the peptidoglycan sacculus that encases the cytoplasmic membrane (Vollmer et al., 2008a; Weidel and Pelzer, 1964). In Gram‐negative (or, more precisely, diderm) bacteria, this peptidoglycan sacculus resides in the periplasm between the cytoplasmic and outer membrane, whilst in Gram‐positive (monoderm) species the peptidoglycan layer is thicker and connected with other major cell wall polymers such as wall teichoic acid, capsular polysaccharide and the S‐layer (Weidenmaier and Peschel, 2008; Silhavy et al., 2010). Peptidoglycan is structurally distinct from cell wall components in archaea and single celled eukaryotes (with the exception of certain plant and algae chloroplasts), and has no homolog in multicellular eukaryotic organisms. Peptidoglycan has at least three major functions. First, it enables the bacterial cell to sustain the high turgor, which results from the difference between the high osmolarity of the bacterial cytoplasm and the comparatively low osmolarity of the external environment. Second, peptidoglycan maintains the shape of a bacterial cell. Third, it provides rigidity to envelope‐spanning surface structures such as flagella and retractile pili that exert force and require a solid support to push or pull against. The essentiality of peptidoglycan for survival of bacteria in a hypoosmolar environment along with its role in anchoring cell surface appendages that are often important virulence determinants makes it an attractive antibiotic target, and multiple classes of clinically successful antibiotics target various aspects of peptidoglycan synthesis, for example beta‐lactams and glycopeptides (Silver, 2013).
Peptidoglycan is composed of polysaccharide chains made up of alternating ß‐1,4‐linked N‐acetylglucosamine (GlcNAc) and N‐acetylmuramic acid (MurNAc) residues which are connected via short peptides (Fig. 1) (Weidel and Pelzer, 1964; Vollmer et al., 2008a). These peptides contain D‐amino acids such as D‐alanine or D‐glutamate, as well as unusual non‐proteinogenic amino acids, such as meso‐diaminopimelic acid (meso‐DAP). The length of individual glycan chains, the amino acid sequence of the peptides and the structure of cross‐links are variable between species and may differ at distinct stages of growth in one species (Vollmer and Höltje, 2004; Vollmer, 2008; Vollmer and Seligman, 2010). Chemical modifications are found in the glycan backbone or peptides, and these may have emerged in response to selective pressure on peptidoglycan from peptidoglycan‐targeting enzymes or antibiotics (Vollmer and Tomasz, 2002; Vollmer, 2008; Figueiredo et al., 2012). Given the importance of a structurally intact sacculus on bacterial cell integrity, the polymerisation and insertion of new peptidoglycan strands is a complex and robustly regulated process (Typas et al., 2012; Pazos et al., 2017) and this is particularly critical in the context of bacterial cell division (Egan and Vollmer, 2013).
Figure 1.
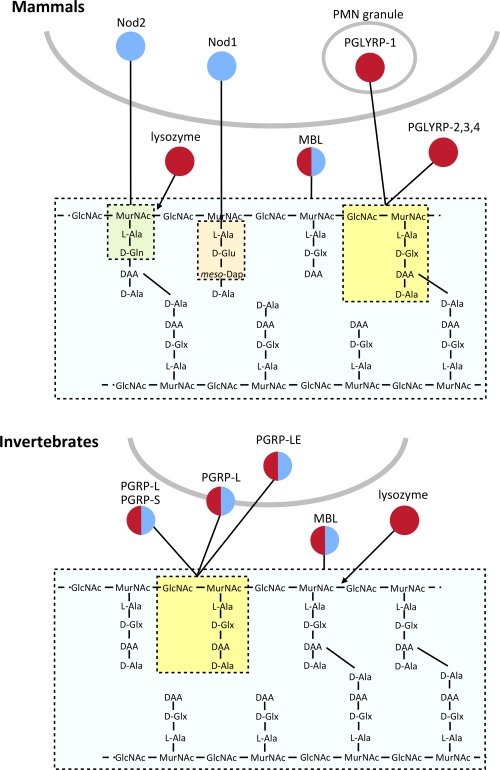
Summary of peptidoglycan recognition proteins in invertebrates and mammals. An overview of peptidoglycan recognition proteins in invertebrates and mammals. Some proteins degrade peptidoglycan (shown in red) whilst others induce downstream signalling pathways (shown in blue). Polymerised peptidoglycan is shown, with fragments recognised by different PGRPs indicated by dotted lines/boxes.
Immune responses to peptidoglycan
The innate immune response of vertebrates and invertebrates has evolved a repertoire of components to detect and destroy invading bacteria. This is achieved through recognition of characteristic non‐self structures, called pathogen associated molecular patterns (PAMPs). Peptidoglycan is an important PAMP, due to its presence in virtually all bacteria and its almost complete absence in higher eukaryotes. Specific fragments released from peptidoglycan are recognised by various peptidoglycan recognition proteins (Fig. 1 and Table 1) (Sukhithasri et al., 2013; Neyen and Lemaitre, 2016). The sacculus is hidden from the innate immune system by the presence of an outer membrane in intact diderm bacteria, but soluble peptidoglycan turnover products can be released from intact cells either directly into the surrounding milieu or within outer membrane vesicles. Furthermore, bacterial cells undergoing lysis release peptidoglycan fragments into the extracellular environment. (Goodell and Schwarz, 1985; Uehara and Park, 2008; Schwechheimer et al., 2013). Some peptidoglycan recognition proteins directly hydrolyse peptidoglycan through their amidase or muramidase activity, whilst others have no enzymatic activity but activate a downstream signaling pathway in response to binding peptidoglycan fragments which can then lead to immune cell maturation or the release of proinflammatory cytokines (Fig. 1) (Boneca, 2005; Chaput and Boneca, 2007). Whilst the aim of this immune response is to clear the bacterial infection, overstimulation of the inflammatory response can lead to fatal conditions such as septic shock (Calandra, 2001; Neyen and Lemaitre, 2016). Peptidoglycan recognition proteins are located in multiple organs and tissues throughout the body, and can be located extracellularly, intracellularly or attached to the surface of a host cell (Girardin and Philpott, 2004; Royet and Dziarski, 2007; Dziarski and Gupta, 2010). This ensures the detection of invading pathogens in almost every possible location (Fig. 1 and Table 1).
Table 1.
Overview of peptidoglycan recognition proteins in mammals and invertebrates
| Peptidoglycan recognition protein | Peptidoglycan fragment detected | Main effect | Main tissue distribution | Cellular localisation |
|---|---|---|---|---|
| MAMMALS (H. sapiens) | ||||
| PGLYRP 2 | GlcNAc‐MurNAc‐tetrapeptide | Amidase activity | Liver, skin, oral, intestinal | Soluble |
| PGLYRP 1,3,4 | GlcNAc‐MurNAc‐tetrapeptide | Bactericidal | PMN granules, skin, sweat glands, sebaceous glands, mouth, intestinal tract, eyes | Soluble (PGLYRP 3,4), PMN granules (PGLYRP 1) |
| Nod1 | Tripeptide: L‐Ala‐D‐Glu‐DAP | Activation of pro‐inflammatory pathway via NF‐κB signalling | Ubiquitous | Cytoplasm |
| Nod2 | GlcNAc‐MurNAc‐Ala‐Glu | Activation of pro‐inflammatory pathway via NF‐κB signalling | Monocytes | Cytoplasm |
| Lysozyme | MurNAc‐GlcNAc glycosidic bond | Muramidase activity; bactericidal effects | Phagocytic granules, serum, body secretions | Soluble |
| C‐type lectins (e.g. MBL, RegIII) | Glycan polymer | Complement activation; bactericidal effects | Serum | Soluble |
| INVERTEBRATES (D. melanogaster) | ||||
| PGRP‐L (multiple) | GlcNAc‐MurNAc‐tetrapeptide (Lys‐type) | Amidase activity; induction of antimicrobial peptides via activation of Imd pathway; phagocytosis | Haemocytes, fat body, gut, trachea, haemolymph | Cell surface, cytoplasm and soluble |
| PGRP‐S (multiple) | GlcNAc‐MurNAc‐tetrapeptide (DAP‐type) | Amidase activity; induction of antimicrobial peptides via activation of Toll pathway; phagocytosis; general bactericidal activity | Haemocytes, fat body, gut, trachea, epidermis, haemolymph | Soluble |
| Lysozyme | MurNAc‐GlcNAc glycosidic bond | Muramidase activity; general bactericidal activity | Gut, salivary glands | Soluble |
| C‐type lectins | Glycan polymer | Encapsulation; melanisation; bactericidal activity | Haemolymph, fat body | Cell surface and soluble |
Obligate intracellular bacteria
Bacterial species have evolved to exploit an enormous diversity of environmental niches for their growth and replication. One of the most specialised replicative niches is the interior of a living eukaryotic cell. Some bacterial pathogens adopt this niche at some point during their lifecycle (facultative intracellular bacteria) whilst others have become so adapted to this environment that they have lost the ability to replicate in the absence of their cellular host (obligate intracellular bacteria). These intracellular bacteria typically replicate either free in the eukaryotic cytoplasm, or within specialised vacuoles. It is hypothesised that an early mutualistic adaptation between bacteria and archaea led to the emergence of modern mitochondria and chloroplasts (Bonen et al., 1977; Kuntzel et al., 1981; Carvalho et al., 2015). The major groups of obligate intracellular bacteria are the phylogenetically distinct Chlamydiales and Rickettsiales orders, as well as Coxiella, Buchnera and Mycobacterium leprae (Fig. 2 and Table 2). Whilst most cause human or animal disease, a small number (including Wolbachia, Buchnera and Protochlamydia amoebophila) are not known to cause disease and have a mutualistic relationship with their eukaryotic hosts. A common feature of most diverse species of obligate intracellular bacteria is their significantly reduced genome (compared to most free‐living bacteria), which is a consequence of the adaptation to the stable, intracellular environment and of their parasitic lifestyle. An overview of representative obligate intracellular bacteria, their pathogenesis and their cellular lifestyles, is given in Table 2.
Figure 2.
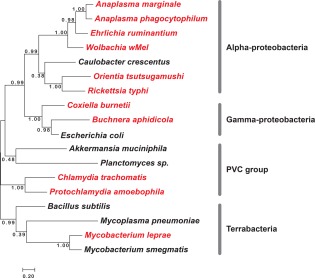
Phylogenetic tree showing relationship between selected obligate intracellular and free‐living bacteria discussed in this review. Obligate intracellular bacteria are shown in red, and free‐living bacteria are shown in black.
Table 2.
Lifestyle and pathogenesis of selected obligate intracellular bacteria
| LPS (LpxA) | Peptidoglycan (Class A PBPs) | Peptidoglycan (Class B PBPs) | Bacteria | Human disease | Animal reservoir(s) | Vector/ spread | Cellular niche | Major cellular tropism (in human infections) | Primary tissue tropism (in human infections) |
|---|---|---|---|---|---|---|---|---|---|
| CHLAMYDIALES | |||||||||
| + | – | + | Chlamydia pneumoniae | Pneumonia | – | Aerosol | Vacuole | Epithelial cells, endothelial cells, macrophages | Lungs, heart |
| + | – | + | Chlamydia trachomatis | Urethritis, pneumonia, trachoma | – | Direct contact | Vacuole | Epithelial cells, monocytes/ macrophages | Genitourinary tract, lungs, eyes |
| + | – | + | Waddlia chondrophila | Associated with miscarriage | – | Unknown | Vacuole | Placental cells | Placenta |
| + | – | + | Simkania negevensis | Pneumonia | – | Unknown | Vacuole | Epithelial cells | Respiratory tract |
| + | – | + | Protochlamydia amoebophila | – | – | Amoeba | Vacuole | – | – |
| RICKETTSIALES | |||||||||
| – | – | + | Orientia tsutsugamushi | Scrub typhus | Rodents | Mite | Cytoplasm | Endothelial cells, dendritic cells, monocytes/macrophages | Vascular endothelium |
| + | + | + | Rickettsia prowazekii | Epidemic typhus and Brill‐Zinsser disease | Flying squirrels | Louse | Cytoplasm | Endothelial cells | Vascular endothelium |
| + | + | + | Rickettsia typhi | Murine typhus | Rats | Flea | Cytoplasm | Endothelial cells | Vascular endothelium |
| + | + | + | Rickettsia rickettsii | Rocky mountain spotted fever | Dogs, rabbits, birds | Tick | Cytoplasm | Endothelial cells | Vascular endothelium |
| + | + | + | Rickettsia akari | Rickettsialpox | House mice, rats | Mite | Cytoplasm | Monocytes/macrophages | Blood |
| + | + | + | Rickettsia conorii | Mediterranean spotted fever | Rodents, dogs | Tick | Cytoplasm | Endothelial cells | Vascular endothelium |
| – | – | + | Anaplasma marginale | – | Cattle, wild ruminants | Tick | Vacuole | Erythrocytes | Blood |
| – | – | – | Anaplasma phagocytophilum | Human granulocytic anaplasmosis (HGE) | Deer, cats, dogs, ruminants, rodents | Tick | Vacuole | Neutrophils | Blood |
| – | – | – | Ehrlichia chaffeensis | Human monocytic ehrlichiosis (HME) | Dogs, deer | Tick | Vacuole | Monocytes/macrophages, endothelial cells | Blood, vascular endothelium |
| – | – | – | Ehrlichia ruminatum | – | Cattle, wild ruminants | Tick | Vacuole | Monocytes/macrophages, endothelial cells | Blood, vascular endothelium |
| – | – | – | Neorickettsia sennetsu | Sennetsu fever | Fish | Trematode | Vacuole | Monocytes/macrophages | Blood |
| – | – | + | Wolbachia WMel | – | – | Arthropod, insect, nematode | Vacuole | – | – |
| OTHER | |||||||||
| + | + | + | Coxiella burnettii | Q fever | Ruminants | Tick and aerosol | Vacuole | Monocytes/macrophages | Liver, lungs, heart |
| – | + | + | Buchnera aphidicola | – | – | Pea aphid | Vacuole | – | – |
| – | + | + | Mycobacterium leprae | Leprosy | Armadillos | Aerosol | Vacuole | Histiocytes, nerve cells, macrophages, epithelial cells | Skin |
The unique lifecycle of obligate intracellular bacteria, while shielding them from extracellular innate immune surveillance, has resulted in particular selective pressures on their peptidoglycan, especially with respect to peptidoglycan‐sampling immune surveillance mechanisms (e.g., NOD1/2) located in the cytoplasm of host cells (Chaput and Boneca, 2007). Their location within the isotonic eukaryotic cell confers osmotic protection, whilst their constant proximity to host cell immune receptors means that they are under pressure to reduce recognition of this key PAMP. In this review, we explore the ability of obligate intracellular bacteria to assemble the peptidoglycan transiently during the cell cycle, at reduced levels and/or to synthesize a chemically modified version of peptidoglycan.
The peptidoglycan of obligate intracellular bacteria
Chlamydiales
The Chlamydiales are a large and diverse order of bacteria that include both human and animal pathogens, as well as non‐pathogenic environmental species. The two major human pathogens are C. trachomatis and C. pneumoniae. However, some of the lesser‐known so‐called ‘environmental’ chlamydia (sometimes referred to as ‘chlamydia‐like’ bacteria), which live inside amoeba, have recently been described as putative human pathogens, including Waddlia chondrophila and possibly Simkania negevensis (Ammerdorffer et al., 2017; Vouga et al., 2017). Chlamydiae are tropic for endothelial, epithelial and monocyte/macrophage cells and even primitive macrophage‐like cells such as amoebae (Kebbi‐Beghdadi and Greub, 2014). They secrete effector proteins through a type 3 secretion system to trigger dramatic rearrangement of the host cytoskeleton resulting in engulfment of the bacterium (Nans et al., 2015). Once phagocytosed by the host cell, Chlamydiae remodel the phagocytic vacuole, resulting in a specialised membrane‐surrounded organelle, called inclusion, that provides a protected environment for replication (Nans et al., 2015). The chlamydial cell type that is engulfed is the dispersal form known as the elementary body, a non‐replicative and poorly metabolically active cell type. Upon uptake, the elementary body differentiates within the inclusion into the replicative reticulate body which is no longer infectious (Nans et al., 2015). Chlamydiales are amongst the few known bacteria that lack FtsZ, the bacterial tubulin homolog that organizes the divisome complex at midcell to direct septal peptidoglycan synthesis and facilitates cytokinesis (Busiek and Margolin, 2015). Evidence has been provided that the bacterial actin complex, composed of MreB actin and its regulator RodZ, partially substitute for the role of FtsZ in cytokinesis and spatial regulation of septal peptidoglycan synthesis (Jacquier et al., 2014; Kemege et al., 2015; Liechti et al., 2016). After a complete replication cycle re‐differentiation into elementary bodies takes place in response to unknown signals, and new elementary bodies are released from the cell by either lysis or extrusion.
For many years, the chlamydial anomaly described the paradox of a non‐detectable peptidoglycan, despite the susceptibility of these organisms towards β‐lactams, which target peptidoglycan synthesis (Ghuysen and Goffin, 1999). Notably, unlike for other bacteria, β‐lactams are not bactericidal for chlamydia, but lead to a persistent infection of polyploid aberrant bodies (Skilton et al., 2009). The bactericidal action of β‐lactams in most bacteria is due to a lethal uncoupling of peptidoglycan synthetic and remodelling activities during growth and cell division, followed by lysis (Tomasz and Waks, 1975; Kohlrausch and Höltje, 1991; Cho et al., 2014). Although chlamydia can survive in the osmoprotective, intracellular environment, they cannot multiply without peptidoglycan synthesis, suggesting that peptidoglycan is required for chlamydial division (Henrichfreise et al., 2009; Skilton et al., 2009 ; Jacquier et al., 2014; 2015). The chlamydial anomaly has recently been resolved through the use of highly sensitive mass spectrometry techniques and newly developed fluorescent probes based on the peptidoglycan‐specific D‐Ala‐D‐Ala dipeptide (Liechti et al., 2013; Pilhofer et al., 2013). It is now known that several Chlamydiae possess peptidoglycan‐like structures, although the composition and arrangement seems to vary throughout the order. Complete peptidoglycan sacculi have been isolated and observed by cryoelectron tomography in some (but not all) environmental isolates (Pilhofer et al., 2013). However, the human pathogen C. trachomatis has no peptidoglycan sacculus but a discrete and transient peptidoglycan ring structure, which constricts together with the septum of dividing cells (Liechti et al., 2016; Packiam et al., 2015). Hence, even in the absence of a need for osmoprotection C. trachomatis cells maintain a rudimentary and β‐lactam‐sensitive peptidoglycan structure for cytokinesis. The elementary bodies of this species appears to maintain cell envelope integrity by a network of outer membrane proteins cross‐linked via disulphide bonds (Hatch et al., 1986).
Chlamydia are a member of the PVC superphylum (Planctomycetes‐Verrucomicrobia‐Chlamydiae). Whilst many of these genera are not obligate intracellular bacteria they will be briefly discussed here because they are unusual in having multiple members that are free living bacteria predicted to lack a peptidoglycan cell wall. The Verrucomicrobia, including Akkermansia muciniphila, possess a classical cell wall and are described as being diderm (Gram‐negative) species. In contrast, the Planctomycetes were long described as universally lacking peptidoglycan and having a proteinaceous cell wall instead. Two recent reports, however, have demonstrated the detection of a peptidoglycan‐like substance in some members of the planctomycetes (Jeske et al., 2015; van Teeseling et al., 2015). These include Kuenenia stuttgartiensis Planctomyces limnophilus, Gemmata obscuriglobus and Rhodopirellula baltica. Similar to the Chlamydiae, the peptidoglycan in these organisms was difficult to detect and present at low abundance.
Rickettsiales
The order Rickettsiales contains two families: the Rickettsiaceae and the Anaplasmataceae (Eremeeva et al., 2005). Significant differences in their lifecycles and cell tropism may have resulted in distinct selective pressures on their peptidoglycan, and they will be discussed separately in this section. The order Rickettsiales is evolutionarily related to the predicted precursor of modern mitochondria (Emelyanov, 2001).
The Rickettsiaceae are a group of obligate intracellular, vector‐borne bacteria that cause a range of typhus‐like diseases in humans (Parola and Raoult, 2006; Walker, 2007). They primarily target the vascular endothelium, but R. akari is tropic to monocytes/macrophages (Radulovic et al., 2002) and Orientia tsutsugamushi is also found in monocytes/macrophages and dendritic cells (Moron et al., 2001; Paris et al., 2012). Unlike the Chlamydiales, the Rickettsiaceae cannot spread directly between infected individuals but are transferred via a mite, tick, louse or flea vector, likely due to a different tropism and/or poor survival outside cells compared to chlamydial elementary bodies. With the exception of R. prowazekii, which can spread directly between infected individuals via the body louse, most Rickettsiaceae are maintained through a range of animal reservoirs (Eremeeva and Dasch, 2015). Some Rickettsiaceae, such as O. tsutsugamushi, are also able to be transmitted transovarially to vector offspring, bypassing the absolute requirement for an intermediate animal reservoir (Shin et al., 2014; Takhampunya et al., 2016). Rickettsiaceae use a zipper‐like mechanism for uptake into the target cell (Ihn et al., 2000; Lee et al., 2008; Cho et al., 2010a). Once inside the cell, they escape from membrane‐enclosed vacuoles in the endolysosomal pathway and undergo growth and replication directly in the host cytosol (Chu et al., 2006). The bacteria are therefore directly exposed to autophagy machinery (Choi et al., 2013; Ko et al., 2013) and cytosolic immune receptors such as Nod1 and Nod2 (Cho et al., 2010b), and this has likely put selective pressure on these organisms to minimise receptor activation and downstream effectors.
With the exception of Orientia, which is a distinct genus within the Rickettsiaceae, the Rickettsiaceae are thought to possess a complete peptidoglycan structure and are sensitive to penicillin when grown in cultured cells (Silverman and Wisseman, 1978; Wisseman et al., 1982). Orientia is insensitive to ß‐lactams and was historically thought to lack any peptidoglycan structures (Amano et al., 1987), despite the presence of an apparently complete peptidoglycan biosynthetic pathway encoded in its genome (Cho et al., 2007; Min et al., 2008; Nakayama et al., 2008). Recent work has provided evidence that O. tsutsugamushi may possess a minimal peptidoglycan‐like structure and is sensitive to a range of non‐ß‐lactam peptidoglycan‐targeting antibiotics such as D‐cycloserine and phosphomycin, but remains insensitive to all ß‐lactams tested, which might be explained by an intrinsic insensitivity by the class B PBPs of this organism (Atwal et al., 2017) or an unknown beta‐lactamase. O. tsutsugamushi was also shown to possess a disulphide cross‐linked protein network on the outer membrane, analogous to the Chlamydiae (Atwal et al., 2017).
The Anaplasmataceae comprise a group of tick‐borne human and veterinary pathogens. This family also contains the nematode‐borne pathogen Neorickettsia sennetsu (Dittrich et al., 2015) and the prolific and promiscuous insect symbiont Wolbachia (Sicard et al., 2014). This family is unlike the Rickettsiaceae in primarily residing in erythrocytes, neutrophils and monocytes/macrophages and having a vacuolar cellular localisation (Carlyon and Fikrig, 2003; Rikihisa, 2003; Munderloh et al., 1999). Similar to the Chlamydiales, Anaplasmataceae exhibit a morphologically distinct biphasic life cycle, transitioning between non‐replicative and infectious dense‐core particles and replicating reticulate cells (Troese and Carlyon, 2009). Anaplasma phagocytophilum and Ehrlichia chaffeensis have been reported to lack both peptidoglycan and LPS, and this is supported by the absence of genes required for their synthesis (Lin and Rikihisa, 2003). Peptidoglycan has never been detected in Wolbachia, however, it has been shown that lipid II is essential for cell division (Vollmer et al., 2013) and Wolbachia contains a functional peptidoglycan amidase (Wilmes et al., 2017). There are no reports of the analysis or isolation of peptidoglycan in Anaplasma marginale. Anaplasmataceae have the ability to incorporate host‐derived lipids and sterols such as sphingolipids or cholesterol into their membranes, conferring some degree of structural rigidity in the absence of peptidoglycan (Lin and Rikihisa, 2003).
Coxiella burnettii
C. burnettii is an intracellular γ‐proteobacterium and the causative agent of the human disease Q fever (van Schaik et al., 2013). It is generally described as an obligate intracellular bacterium, but specific growth media has recently been developed that supports growth in the absence of living host cells (Omsland et al., 2009). Similar to the Anaplasmataceae and Chlamydiales, Coxiella occupies a replicative niche within a membrane‐enclosed vacuole of an infected cell (Kohler and Roy, 2015). The Coxiella‐containing vacuole, however, is acidified, and this organism has developed mechanisms to survive and proliferate under these conditions (van Schaik et al., 2013).
Coxiella differentiates between the replicative large cell variant (LCV) form, and the non‐replicative short cell variant (SCV). Whilst both are capable of infecting cultured cells, the extracellular SCV form is the predominate agent of transmission in the environment. The SCV form of Coxiella is incredibly stable and able to withstand harsh environmental conditions such as extended desiccation and heat. It is highly infectious, with only 10 particles sufficient to cause disease in humans, making Coxiella a potential bioterrorism threat (Azad, 2007; Oyston and Davies, 2011).
Structural rigidity in SCV‐form Coxiella is partially conferred by a thick peptidoglycan, and it has recently been shown that this is characterised by an abundance of LD‐transpeptidase‐mediated 3‐3 peptide cross‐links (Sandoz et al., 2016).
Mycobacterium leprae
M. leprae is a member of the Mycobacteriaceae family within the class Actinobacteria, and is the causative agent of the human disease leprosy (Rodrigues and Lockwood, 2011). Mycobacteria possess an almost impermeable, waxy cell surface with an unusual second membrane that is rich in mycolic acids and attached to a thick peptidoglycan layer via the polysaccharide arabinogalactan (Jankute et al., 2015; Nataraj et al., 2015). With 3.3 Mbp and 1,604 predicted proteins, M. leprae has a small genome compared with 4.4 Mbp and 3,924 predicted proteins in M. tuberculosis (Cole et al., 2001; Gutierrez et al., 2009), and extensive attempts to culture it in the laboratory have been unsuccessful (Lagier et al., 2015). It is therefore considered to be unique amongst mycobacteria in having an obligate intracellular lifecycle. M. leprae is predominantly localised in histiocytes and nerve cells, but is also found in macrophages and epithelial cells (Rodrigues and Lockwood, 2011). The peptidoglycan of M. leprae is comparable with that of other mycobacteria in that there is a high percentage of 3‐3‐cross‐links, but it lacks the N‐glycolation on muramic acid residues found in some mycobacteria, and it contains glycine in place of L‐alanines in a fraction of the peptides (Mahapatra et al., 2008).
Buchnera
Buchnera aphidicola is the primary endosymbiont of the pea aphid Acyrthosiphon pisum (Douglas et al., 2011). Unlike the other obligate intracellular bacteria discussed here this organism is an obligate endosymbiont, and the insect host cannot survive in its absence. Buchnera is found within specialised polyploidy cells in the aphid body cavity, called bacteriocytes, where bacteria live within membrane‐enclosed vacuoles called symbiosomes. Buchnera is a γ‐proteobacterium, but lacks genes required for synthesising LPS in its outer membrane. Bacteria are spherical or oval in shape, are sensitive to penicillin and have a meso‐DAP‐containing peptidoglycan sacculus (Griffiths and Beck, 1974; Houk et al., 1977). Genome evolution of B. aphidicola has been intensively studied and it has been proposed to have evolved from an E. coli‐like genome by means of gene removal (Silva et al., 2001).
Overview of this work
In the current study, we have sought to explore the relationship between the presence of enzymatic activities involved in peptidoglycan biosynthesis and turnover, and the presence of peptidoglycan in different obligate intracellular bacterial species. We have selected representatives of all major known groups of obligate intracellular bacteria, and have included a closely related free‐living organism as an outgroup for each species or group of species. This includes C. crescentus for the alpha‐proteobacteria (Rickettsiales); E. coli for the gamma‐proteobacteria (Coxiella and Buchnera); B. subtilis and M. smegmatis for the terrabacteria (Mycobacterium leprae) and Akkermansia muciniphila and Planctomyces limnophilus as planctomycetes and verrucomicrobia members of the PVC group respectively (Chlamydiae). These species were selected based on the availability of complete and annotated genomes and experimental evidence on the presence or absence of peptidoglycan (where available). We used a combination of KEGG database analysis and protein homology blast searches to identify proteins involved in different stages of peptidoglycan biosynthesis, and these are shown in Figs 4–7. Organisms are coloured according to their peptidoglycan status, with black indicating the complete absence of any peptidoglycan biosynthetic capacity (Mycoplasma pneumoniae, Anaplasma phagocytophilum, Ehrlichia ruminatum), red indicating a demonstrated low level or incomplete peptidoglycan sacculus (Protochlamydia amoebophila, Chlamydia trachomatis, Planctomyces limnophilus and Orientia tsutsugamushi) and green indicating the presence of a classical peptidoglycan sacculus (Mycobacterium leprae, Mycobacterium smegmatis, Buchnera aphidicola, Rickettsia typhi, Caulobacter crescentus, Coxiella burnettii, Bacillus subtilis and Escherichia coli). Where the peptidoglycan status is unknown, organisms are coloured in orange (Wolbachia strain wMel, Anaplasma marginale). In all dendrograms the organisms are grouped according to their similarity in protein profile across all genes considered in this study (Table 3 and full list shown in Supporting Information Table S1) and the proteins are grouped according to their similarity across all organisms considered in this work (Supporting Information Table S1).
Table 3.
Summary of all genes included in this study
| Gene name | KEGG number | Protein function |
|---|---|---|
| alr | K01775 | Alanine racemase |
| amiA,B,C | K01448 | N‐Acetylmuramoyl‐L‐alanine amidase |
| amiD | K11066 | N‐Acetylmuramoyl‐L‐alanine amidase |
| ampG | K08218 | MFS transporter, PAT family, beta‐lactamase induction signal transducer |
| ampH | K18988 | Serine‐type D‐Ala‐D‐Ala carboxypeptidase/endopeptidase |
| anmK | K09001 | Anhydro‐N‐acetylmuramic acid kinase |
| argD | K00821 | Acetylornithine/N‐succinyldiaminopimelate aminotransferase |
| asd | K00133 | Aspartate‐semialdehyde dehydrogenase |
| aspC | K10206 | Aspartate aminotransferase |
| bacA (uppP) | K06153 | Undecaprenyl‐diphosphatase |
| dacA | K01286 | DD‐Carboxypeptidase PBP5 |
| dacB | K07259 | DD‐Carboxy‐/endopeptidase PBP4 |
| dacC | K07258 | D‐Alanyl‐D‐alanine carboxypeptidase; penicillin‐binding protein 6a |
| dacD | K07258 | D‐Alanyl‐D‐alanine carboxypeptidase; penicillin‐binding protein 6b |
| dapA | K01714 | 4‐Hydroxy‐tetrahydrodipicolinate synthase |
| dapC (argD) | K14267 | N‐Succinyldiaminopimelate aminotransferase |
| dapD | K00674 | 2,3,4,5‐Tetrahydropyridine‐2‐carboxylate N‐succinyltransferase |
| dapE (msgB) | K01439 | Succinyl‐diaminopimelate desuccinylase |
| dapF | K01778 | Diaminopimelate epimerase |
| ddl | K01921 | D‐Alanine‐D‐alanine ligase |
| ftsA | K03590 | Cell division protein FtsA |
| ftsB | K05589 | Cell division protein FtsB |
| ftsE | K09812 | Cell division transport system ATP‐binding protein |
| ftsI (pbpB) | K03587 | Transpeptidase involved in septal peptidoglycan synthesis (DD‐transpeptidase) |
| ftsK (spoIIIE) | K03466 | DNA segregation ATPase |
| ftsL (divIC) | K03586 | Cell division protein FtsL |
| ftsN | K03591 | Cell division protein FtsN |
| ftsP (sufl) | K04753 | Suppressor of FtsI |
| ftsQ | K03589 | Cell division protein FtsQ |
| ftsW (rodA, spoVE) | K03588 | Cell division protein FtsW, SEDS protein |
| ftsX | K09811 | Cell division transport system permease protein |
| ftsZ | K03531 | Cell division protein FtsZ (tubulin homolog) |
| glmM | K03431 | Phosphoglucosamine mutase |
| glmS | K00820 | Glucosamine‐‐fructose‐6‐phosphate aminotransferase |
| glmU | K04042 | Bifunctional UDP‐N‐acetylglucosamine pyrophosphorylase / Glucosamine‐1‐phosphate N‐acetyltransferase |
| glyA | K00600 | Glycine hydroxymethyltransferase, SHMT |
| ldt (erfK, srfK) | K16291 | LD‐transpeptidase |
| lpxT (yeiU) | K19803 | Lipid A 1‐diphosphate synthase; undecaprenyl pyrophosphate:lipid A 1‐phosphate phosphotransferase |
| lysC (apk) | K00928 | Aspartate kinase |
| metC | K01760 | Cystathionine beta‐lyase |
| mepA | K07261 | Murein DD‐endopeptidase |
| mepH | K19303 | Murein DD‐endopeptidase |
| mepM | K19304 | Murein DD‐endopeptidase |
| mepS | K13694 | Murein DD‐endopeptidase |
| mltB | K08305 | Membrane‐bound lytic murein transglycosylase B |
| mltC | K08306 | Membrane‐bound lytic murein transglycosylase C |
| mltD | K08307 | Putative membrane‐bound lytic murein transglycosylase D |
| mltE | K08308 | Lytic murein endotransglycosylase E |
| mltF | K18691 | Membrane‐bound lytic transglycosylase F, murein hydrolase |
| mltG | K07082 | Endolytic murein transglycosylase, septation protein, ampicillin sensitivity |
| mraY | K01000 | Phospho‐N‐acetylmuramoyl‐pentapeptide‐transferase |
| mrcA (ponA) | K05366 | Penicillin‐binding protein 1A/PBP1A (glycosyltransferase/DD‐transpeptidase) |
| mrcB (ponB) | K05365 | Penicillin‐binding protein 1B/PBP1B (glycosyltransferase/DD‐transpeptidase) |
| mrdA (pbpA) | K05515 | Penicillin‐binding protein 2/PBP2 (DD‐transpeptidase) |
| mreB (mbl) | K03569 | Rod shape‐determining protein MreB and related proteins (actin homolog) |
| mreC | K03570 | Rod shape‐determining protein MreC |
| mreD | K03571 | Rod shape‐determining protein MreD |
| mtgA | K03814 | Monofunctional glycosyltransferase |
| murA (murZ) | K00790 | UDP‐N‐acetylglucosamine 1‐carboxyvinyltransferase |
| murB | K00075 | UDP‐N‐acetylpyrovoylglucosamine dehydrogenase |
| murC | K01924 | UDP‐N‐acetylmuramate‐L‐alanine ligase |
| murD | K01925 | UDP‐N‐acetylmuramoylalanine‐‐D‐glutamate ligase |
| murE | K01928 | UDP‐N‐acetylmuramoyl‐L‐alanyl‐D‐glutamate‐‐2,6‐diaminopimelate ligase |
| murF | K01929 | UDP‐N‐acetylmuramoyl‐tripeptide‐‐D‐alanyl‐D‐alanine ligase |
| murG | K02563 | UDP‐N‐acetylglucosamine‐‐N‐acetylmuramyl‐(pentapeptide) pyrophosphoryl‐undecaprenol N‐acetylglucosamine transferase |
| murI (glr) | K01776 | Glutamate racemase |
| murJ (mviN) | K03980 | Putative peptidoglycan lipid II flippase |
| nagK | K00884 | N‐acetylglucosamine kinase |
| nagZ | K01207 | Beta‐N‐acetylhexosaminidase |
| pbpC | K05367 | Penicillin‐binding protein 1C (glycosyltransferase/DD‐transpeptidase) |
| pbpG | K07262 | Serine‐type D‐Ala‐D‐Ala endopeptidase PBP7 |
| pgpB | K01096 | PAP2‐type phosphatidylglycerophosphatase / undecaprenyl‐diphosphate diphosphatase |
| rodA (mrdB) | K05837 | Rod shape determining protein RodA, hypothetical lipid II flippase and/or glycosyltransferase |
| rodZ (yfqA) | K15539 | Cytoskeleton‐associated protein |
| slt (mltE) | K08309 | Soluble lytic murein transglycosylase |
| uppS (yaeS) | K00806 | Undecaprenyl diphosphate synthase |
| ybjG (bcrC) | K19302 | PAP2‐type undecaprenyl‐diphosphate diphosphatase |
Pathways for peptidoglycan biosynthesis and remodelling
Overview of peptidoglycan biosynthesis
The peptidoglycan synthesis pathway starts in the cytoplasm with the formation of UDP‐MurNAc (Barreteau et al., 2008). Several further steps involving cytoplasmic amino acid racemaces, and D‐ and L‐amino acid ligases result in the synthesis of UDP‐MurNAc pentapeptide, which is the substrate for subsequent steps at the cytoplasmic membrane. MraY transfers MurNAc (pentapeptide) phosphate to the transport lipid undecaprenol phosphate (C55‐P) to form lipid I, and MurG catalyses the transfer of GlcNAc from UDP‐GlcNAc to lipid I to synthesize lipid II, the ultimate precursor for peptidoglycan synthesis (Bouhss et al., 2008). C55‐P is generated by UppS and a C55‐PP pyrophosphatase (BacA or PAP2 type). Lipid II is transported from the inner to the outer leaflet of the cytoplasmic membrane by a flippase, a process that remains an active area of research, with either SEDS proteins or MurJ being proposed as the lipid II flippase (Mohammadi et al., 2011; Sham et al., 2014). Lipid II is polymerized and the resulting nascent chains incorporated into the existing peptidoglycan layer by glycosyltransfer and transpeptidation reactions (Egan et al., 2015). These are catalysed by a group of membrane‐bound enzymes including penicillin‐binding proteins (PBPs), monofunctional glycosyltransferases and perhaps SEDS proteins (Meeske et al., 2016), and take place in the periplasm (Gram‐negative/diderm bacteria) or on the extracellular surface (Gram‐positive/monoderm bacteria) (Egan and Vollmer, 2013; Egan et al., 2017). The newly inserted peptidoglycan matures by the activities of synthetic and hydrolytic enzymes, such as LD‐transpeptidases and carboxypeptidases, and is eventually turned over by peptidoglycan hydrolases (amidases, endopeptidases, muramidases) during cell growth and division (Figs 3 and 7) (Höltje, 1998; Vollmer et al., 2008b). Some of the released peptidoglycan fragments are transported into the cytoplasm and trimmed for use in lipid II biosynthesis in a process called peptidoglycan recycling (Park and Uehara, 2008).
Figure 3.
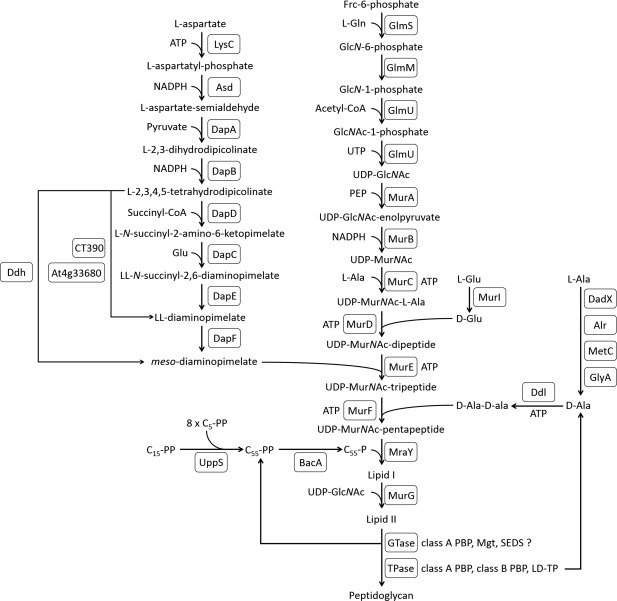
Peptidoglycan biosynthesis pathway. An overview of proteins involved in peptidoglycan biosynthesis. GTase, glycosyltransferase; TPase, transpeptidase; Mgt, monofunctional glycosyltransferase; LD‐TP, LD‐transpeptidase; SEDS, shape, elongation, division and sporulation; CT390, LL‐diaminopimelate aminotransferase from Chlamydia trachomatis; At4g33680, LL‐diaminopimelate aminotransferase from Arabidopsis thaliana; C5‐PP, isopentenyl‐pyrophosphate; C15‐PP, farnesyl‐pyrophosphate; C55‐PP, undecaprenyl‐pyrophosphate.
Figure 7.
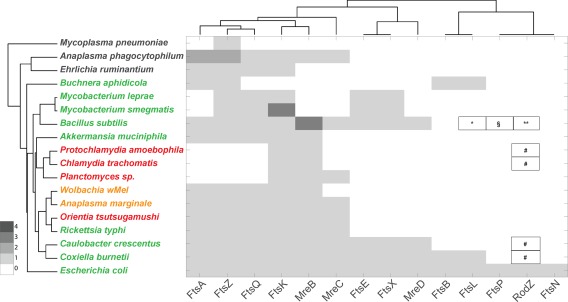
Dendogram showing the presence of cell morphogenesis proteins encoded by selected bacterial genomes. The organism name is coloured according to peptidoglycan status: black, no peptidoglycan; red, intermediate or low‐level peptidoglycan; green, classical peptidoglycan sacculus; orange, peptidoglycan status unknown. The organisms are grouped according to the similarities in the presence/absence profiles for all proteins considered in this study (full list shown in Supporting Information Table S1) and the proteins are grouped according to their similarities across all organisms considered in this work. The presence of multiple orthologs is indicated by colouring according to the key. # Proteins annotated to this function are present in the genomes of these species, Coxiella burnetii NP_820244.1; C. crescentus ADW96154.1; Chlamydia trachomatis NP_219511.1; Protochlamydia amoebophila CAF23404. *I33_1701 is annotated as FtsL but it is not included in any ortholog group. **Orthologs of I33_1877 in other Bacillus species are annotated as RodZ, but these sequences are not included in the RodZ ortholog group in Kegg. § A Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) is present in strain 168 (BSU06300) but not in RO‐NN‐1
Biosynthesis of peptidoglycan precursors
The classical peptidoglycan precursor contains D‐Glu at position 2 and D‐Ala at positions 4 and 5 of the pentapeptide side chain (Vollmer et al., 2008a). Incorporation of D‐Glu and the D‐Ala‐D‐Ala dipeptide, which is synthesized by Ddl proteins, are catalysed by MurD and MurF respectively (Barreteau et al., 2008). The genes encoding these proteins are present across all organisms thought to possess peptidoglycan (Fig. 4A), although Ddl is present as a fusion protein with MurC in many Chlamydiae. The distribution of enzymes that catalyse amino acid racemisation reactions is more complicated. The classical amino acid racemases for these reactions are MurI, Alr and DadX. Whilst these are present in most of the organisms possessing classical peptidoglycan, their presence in intermediate peptidoglycan species is inconsistent. MetC has been shown to possess alternative alanine racemase activity in E. coli (Kang et al., 2011), and GlyA has been shown to be the alanine racemase in Chlamydia pneumoniae (De Benedetti et al., 2014), suggesting that the proteins used for racemase activity need not be strictly conserved in these pathways. It is also possible that Ddl, MurF and MurD may have different amino acid specificities in different organisms, and this can ultimately only be resolved by determining the structure of purified peptidoglycan from specific bacterial species.
Figure 4.
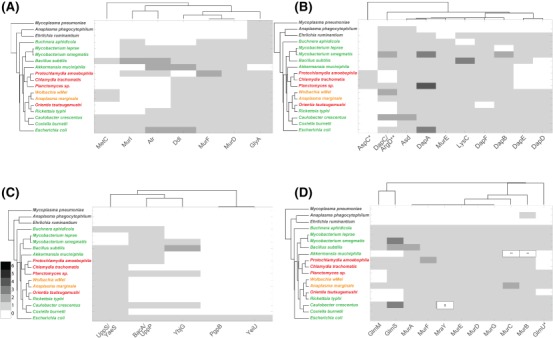
Peptidoglycan precursor enzymes. Dendograms showing the presence of proteins involved in different stages of PG biosynthesis in selected bacterial genomes.
A. D‐amino acid generation and incorporation. B. meso‐DAP generation and incorporation. C. Generation and recycling of undecaprenyl phosphate. D. lipid II biosynthesis. The presence of multiple orthologs is indicated by colouring according to the key. The organism name is coloured according to peptidoglycan status: black, no peptidoglycan; red, intermediate or low‐level peptidoglycan; green, classical peptidoglycan sacculus; orange, peptidoglycan status unknown. The organisms are grouped according to the similarities in the presence/absence profiles for all proteins considered in this study (full list shown in Supporting Information Table S1) and the proteins are grouped according to their similarities across all organisms considered in this work. (B) *Including CT390 AT4G33680; **Merged profiles for K00821 and K14267. (D) *Merged profile for GlmU (K04042 and K11528); **Present as a fusion protein MurC‐MurB (ncbi id: ASB36453.1); § Genuine frameshift in C. crescentus CB15 but present and functional in NA1000.
Peptidoglycan from diderm bacteria typically contains meso‐DAP in position 3, which is linked to D‐Ala in position 4 of adjacent strands in mature peptidoglycan. MurE is the enzyme that mediates the incorporation of meso‐DAP into the peptidoglycan precursor (Barreteau et al., 2008) and this gene is present in all organisms that possess complete or intermediate peptidoglycan in this study (Fig. 4B). The distribution of meso‐DAP biosynthetic proteins is more complicated. DapD and DapE are missing in P. amoebophila, C. trachomatis, Planctomycetes sp. and A. muciniphila, but it has been shown that the LL‐diaminopimelate transferase (CT390) can perform the same reaction in C. trachomatis (McCoy et al., 2006). O. tsutsugamushi lacks the enzyme catalysing the final step of meso‐DAP biosynthesis, DapF. Whilst no direct alternative to this enzyme has been described, a recent study showed the presence of meso‐DAP in purified O. tsutsugamushi by mass spectrometry, suggesting the presence of an unidentified alternative gene or pathway (Atwal et al., 2017). Surprisingly, E. ruminantium possesses a complete set of genes required for meso‐DAP biosynthesis, although it lacks MurE that would be required to incorporate it into the peptidoglycan precursor. This pathway is conserved in E. chaffeensis and E. muris (Supporting Information Table S1) and may have been retained during reductive evolution due to requirement of this metabolite in a different cellular pathway.
The MurNAc(‐pentapeptide) phosphate moiety from UDP‐MurNAc‐pentapeptide is transferred to the lipid anchor C55‐P to form lipid I. This bacterial polyisoprenoid anchors the building blocks for peptidoglycan and LPS synthesis to the inner leaflet of the membrane and enables flippases to shuttle them across the membrane. The MraY transferase synthesizing lipid I (Al‐Dabbagh et al., 2016) is present in all species that have complete or intermediate peptidoglycan, and absent in all those lacking peptidoglycan (Fig. 4D). The assembly of C55‐PP requires UppS (Manat et al., 2014), and its corresponding gene is encoded in almost all genomes of the peptidoglycan‐positive species in our study with the exception of M. leprae and M. smegmatis (Fig. 4C). In fact, uppS is absent in all mycobacteria (Supporting Information Table S1) and it is known that alternative, shorter lipid carriers can perform glycan transport in these organisms. The pyrophosphatases BacA (also called UppP), PgpB and YbjG (E. coli), required for the dephosphorylation of C55‐PP to C55‐P (Manat et al., 2014), were absent in many organisms in our study and this may reflect poor conservation of genes in the pathway.
MurG transfers GlcNAc from UDP‐GlcNAc to lipid I to form lipid II (Chen et al., 2002), which is transported across the membrane for polymerization (Fig. 3). Genes encoding MurA‐G and MraY are present in all organisms with complete and intermediate‐peptidoglycan, and are almost completely absent in those organisms lacking peptidoglycan (Fig. 4D). As expected, the mur genes are therefore a strong predictor of peptidoglycan status.
The generation of UDP‐GlcNAc is more problematic, with GlmM absent from some peptidoglycan‐intermediate species and GlmM and GlmS absent from all Rickettsiaceae and A. muciniphila (Fig. 4D and Supporting Information Table S1). It is possible that an alternative pathway is used for the generation of UDP‐GlcNAc, potentially employing novel importers.
Growth of a peptidoglycan sacculus
Here, we discuss the membrane steps in peptidoglycan synthesis, including the flipping of lipid II across the cytoplasmic membrane, and the synthesis of cross‐linked peptidoglycan strands.
First, lipid II is transported from the inner to the outer leaflet of the cytosolic membrane to make it accessible for incorporation into existing peptidoglycan. The identity of lipid II flippase is an area of active research, with MurJ and SEDS proteins (FtsW, RodA) being primary candidates (Ruiz, 2008; Mohammadi et al., 2014, 2011; Sham et al., 2014). At least one copy of MurJ, FtsW and RodA‐like proteins were present in all peptidoglycan‐positive and peptidoglycan‐intermediate species in our analysis, with the exception of Buchnera and A. muciniphila which lack RodA, and whilst FtsW was absent from all peptidoglycan‐negative species MurJ was found in A. phagocytophilum (Fig. 5).
Figure 5.
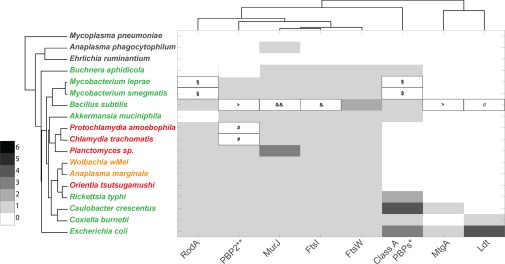
Dendogram showing the presence of peptidoglycan synthases, putative flippases, and SEDS proteins encoded by selected bacterial genomes. The organism name is coloured according to peptidoglycan status: black, no peptidoglycan; red, intermediate or low‐level peptidoglycan; green, classical peptidoglycan sacculus; orange, peptidoglycan status unknown. The organisms are grouped according to the similarities in the presence/absence profiles for all proteins considered in this study (full list shown in Supporting Information Table S1) and the proteins are grouped according to their similarities across all organisms considered in this work. The presence of multiple orthologs is indicated by colouring according to the key. *Class A PBPs contain merged profiles of MrcA (Kegg Accession Number KO5366, see also Supporting Information Table S1), MrcA2/PbpC (KO5367) and MrcB (KO5365). **PBP2 contains merged profiles of MrdA (KO5515) and PBPA (K05364). § Proteins annotated as RodA‐like are present (YP_884452 and NP_301145). # Protein NP_220201 is not present in Kegg as belonging to the PBP2 family, but it has been identified as such in Ouellette et al. (2012). The most similar sequence in Protochlamydia amoebophila is WP_011174685.1 (36% identical). $MSMEG_0031 and MLBr00018 are annotated as PbpA/PBP2 (K05364) they share < 30% identity with MrdA from E. coli. This ortholog group appears to be present in Mycobacterium spp. only (among the organisms considered in this work). & Protein annotated as FtsI‐PbpB is present (I33_1702), but it belongs to a different ortholog group in Kegg (K08724) && Protein similar to MurJ is present (I33_3060) assigned to no orthologous group.//Protein similar to Ldt proteins from E. coli, but not assigned to a K number in Kegg: I33_1583 (YkuD). > A protein belonging to ortholog group K21464 (PbpG) is present (I33_3896). This is also similar to MtgA from E. coli.
Following flippase activity, two specific enzymatic activities are required to incorporate lipid II into an extended peptidoglycan structure. Glycosyltransferase activity polymerizes the glycan strands and transpeptidase activity cross‐links peptides from adjacent glycan strands. The most common cross‐links are DD‐type, between D‐Ala in position 4 of one peptide and meso‐DAP in position 3 of another, but they can also be of the LD‐type which forms between two meso‐DAP residues (Höltje, 1998; Vollmer et al., 2008a). Glycosyltransferase activity in E. coli is largely performed by class A bifunctional PBPs, which possess both glycosyltransferase and transpeptidase activity, and a monofunctional glycosyltransferase (MtgA) (Egan et al., 2015). It was recently suggested that RodA from Bacillus subtilis also possesses glycosyltransferase activity, and it was hypothesised that this may be a general property of SEDS proteins and a possible source of glycosyltransferase activity in organisms lacking class A PBPs and monofunctional glycosyltransferases (Meeske et al., 2016). However, the E. coli SEDS protein FtsW lacked glycosyltransferase activity and instead controlled the glycosyltransferase activity of PBP1B in the presence of the class B PBP3 (Leclercq et al., 2017). DD‐transpeptidase activity is performed by class A PBPs (E. coli PBP1A, PBP1B, PBP1C) and class B PBPs (E. coli PBP2, PBP3), LD‐transpeptidase activity for the formation of 33‐cross‐links is performed by LD‐transpeptidases (E. coli YcbB, YnhG) (Magnet et al., 2008).
In our bioinformatics analysis of transpeptidase and glycosyltransferase activity (Fig. 5), we found that class A PBPs are absent from all the bacteria with intermediate peptidoglycan, but that representatives of both class B PBPs and SEDS proteins are present. Assuming that the transient peptidoglycan of these organisms contains glycan strands as for members of the chlamydia group (Pilhofer et al., 2013; Packiam et al., 2015), then the required glycosyltransferase activity comes either from the SEDS protein(s) or from a yet unknown glycosyltransferase. LD‐transpeptidases are present in E. coli, B. subtilis and C. burnettii but could not be identified in any of the other obligate intracellular organisms that we analysed, based on classifications provided by the Kegg database.
Peptidoglycan trimming, degradation and recycling
Nascent or polymerised peptidoglycan is often subject to secondary modifications, such as N‐deacetylation of residues in glycan chains, and trimming of the peptides by carboxypeptidases (Vollmer, 2008; Peters et al., 2016). Other peptidoglycan hydrolases cleave at various sites in the glycan and peptide components of peptidoglycan and these activities cause a release of peptidoglycan fragments from the sacculus (peptidoglycan turnover) (Vollmer et al., 2008b). Peptidoglycan hydrolases are a large and diverse group of enzymes, whose cleavage sites are shown in Fig. 6A. The released fragments are partly released into the extracellular space, and partly taken up and recycled into new peptidoglycan precursors (Park and Uehara, 2008; Borisova et al., 2016). The release of extracellular peptidoglycan is immunogenic and it is likely that obligate intracellular bacteria would limit the release of these molecules. Our bioinformatics analysis of peptidoglycan degradation and recycling genes shows a diverse pattern of genes in the selected organisms (Fig. 6B). The DD‐carboxypeptidase proteins DacA, DacC, DacD and DacF are present in many peptidoglycan‐positive and peptidoglycan‐negative organisms and may have another role in addition to peptidoglycan degradation. There was no consistent pattern in the distribution of hydrolase activities in the intermediate‐peptidoglycan group. Orientia and A. marginale genomes do not encode an AmiA/B/C‐like amidase, in common with all Rickettsiales, but possess a lytic transglycosylase (MltE) homologue. In contrast, lytic transglycosylase genes are absent in peptidoglycan‐negative A. phagocytophilum and E. ruminantium and obvious lytic transglycosylase‐like genes were not detected in the chlamydial genomes either, raising the possibility that additional enzymes can cleave glycan strands. Interestingly, AmiA from C. pneumoniae showed not only amidase but also DD‐carboxypeptidase activity (Klöckner et al., 2014). The latter, unexpected activity could be attributed to two motifs usually found in PBPs and was inhibited by β‐lactams.
Figure 6.
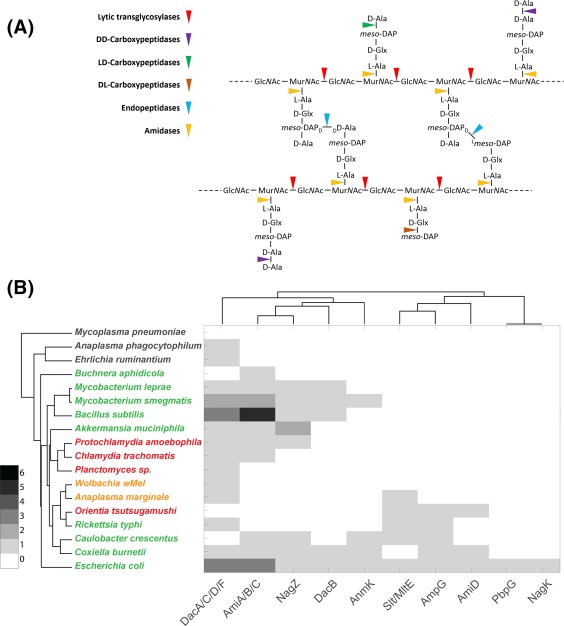
Peptidoglycan degradation and recycling.
A. Overview of enzymes involved in peptidoglycan degradation.
B. Dendogram showing the presence of peptidoglycan degradation and recycling proteins encoded by selected bacterial genomes. The organism name is coloured according to peptidoglycan status: black, no peptidoglycan; red, intermediate or low‐level peptidoglycan; green, classical peptidoglycan sacculus; orange, peptidoglycan status unknown. The organisms are grouped according to the similarities in the presence/absence profiles for all proteins considered in this study (full list shown in Supporting Information Table S1) and the proteins are grouped according to their similarities across all organisms considered in this work. The presence of multiple orthologs is indicated by colouring according to the key.
Cell morphogenesis proteins
The peptidoglycan biosynthesis machinery is positioned and/or regulated through association with a network of cytosolic and membrane‐bound proteins involved in bacterial growth and morphogenesis (Egan et al., 2017). Two major components of the bacterial cytoskeleton are the tubulin homolog FtsZ and the actin homolog MreB (Ouellette et al., 2012; Celler et al., 2013). Both have been shown to generate dynamic filaments that are associated with PBPs and to guide the incorporation of nascent peptidoglycan (Dominguez‐Escobar et al., 2011; Garner et al., 2011). Whilst the Chlamydiae lack FtsZ (Frandi et al., 2014), MreB is absent in Buchnera and mycobacteria (Letek et al., 2008) (as well as the peptidoglycan‐negative mycoplasma and ehrlichiae). This underscores the fact that cytoskeleton‐guided peptidoglycan incorporation may be conserved in bacteria, but that there are different combinations of cytoskeletal elements and peptidoglycan synthesis enzymes across the kingdom, presumably reflecting unique aspects of the cell division apparatus and modes of peptidoglycan synthesis in different organisms.
Predicting the peptidoglycan status of obligate intracellular bacteria
A small number of genes were highly correlated with the predicted peptidoglycan status, and these are summarised in Fig. 8. We observed that those bacteria with demonstrated low level of peptidoglycan or an incomplete sacculus were associated with a signature gene set: the presence of lipid II biosynthesis genes murA‐murG and mraY; the presence of at least one SEDS gene ftsW or rodA; the presence of at least one gene encoding a class B monofunctional PBP, but the notable absence of any detectable genes for class A bifunctional PBPs. Based on this classification we would predict that Wolbachia and Anaplasma marginale both possess some sort of peptidoglycan‐like structure, and we term this group ‘peptidoglycan‐intermediate’ organisms.
Figure 8.
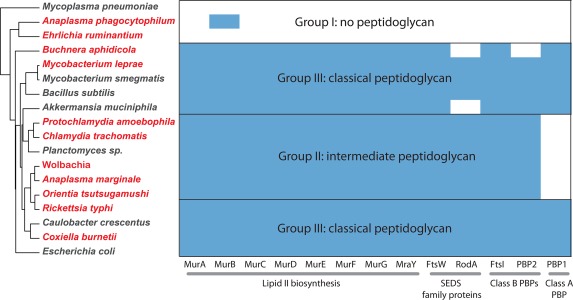
Summary of the presence/absence of key peptidoglycan biosynthesis gene homologs together with predictions about corresponding PG status. Obligate intracellular bacteria are shown in red, and free‐living bacteria are shown in black.
Discussion
In this review, we explored the relationship between the distribution of genes involved in various aspects of cell wall biology, and the intracellular replicative niche adopted by obligate intracellular bacteria. We hypothesise that the selective pressures of an osmoprotective environment, combined with proximity to cellular host immune responses, would lead organisms to reduce their levels of peptidoglycan. Indeed, we observed common patterns of gene loss and retention in groups of unrelated obligate intracellular bacteria (Fig. 8). We termed this group peptidoglycan‐intermediate. This group was characterised by the presence of genes encoding orthologs of MurA‐MurG, MraY, SEDS proteins RodA/FtsW and the class B PBPs PBP2/PBP3(FtsI), but the notable absence of any class A PBPs. This group of organisms included pathogenic and environmental Chlamydiae as well as Orientia tsutsugamushi, Wolbachia strain wMel and Anaplasma marginale. Within this group the detailed structure of peptidoglycan determined by mass spectrometry is only known for Chlamydia trachomatis and it will be interesting to see how conserved patterns of gene retention translates to commonalities and differences in peptidoglycan structure across this group. It is expected that there will remain substantial differences in structure and arrangement, since it is already known that the peptidoglycan of Protochlamydia amoebophila forms a complete sacculus but peptidoglycan of Chlamydia trachomatis has only been detected as a discrete ring located at the septum and no peptidoglycan sacculus could be isolated from the chlamydia‐like bacterium Simkania negevensis. There are also likely to be lineage‐specific differences in the composition of peptidoglycan, for example unidentified chemical modifications have been shown for Protochlamydia amoebophila (Pilhofer et al., 2013) and there is some circumstantial evidence consistent with a modification of muropeptides after antibiotic treatment in Chlamydia trachomatis (Packiam et al., 2015). Species‐specific variations in peptidoglycan structure and composition might therefore reflect unique aspects of the individual cell biology and the host‐pathogen interactions of each obligate intracellular lineage.
The classification of an intermediate peptidoglycan group raises questions about why different closely related obligate intracellular bacteria would adopt different peptidoglycan statuses. For example, whilst all Rickettsia encode genes for a classical peptidoglycan sacculus, the sister genus Orientia lacks class A PBPs and was classified as peptidoglycan‐intermediate. The pressure for a reduced peptidoglycan structure in Orientia is unlikely to result solely from vector difference (many Rickettsia are tick‐borne whilst Orientia is mite‐borne) because Rickettsia akari is also mite‐borne and possesses complete peptidoglycan genes in common with other Rickettsia. Orientia is associated with dendritic cells and monocytes/macrophages in addition to endothelial cells in human patient eschar tissue samples, in contrast to most Rickettsia, which are predominantly endothelial‐cell localised. It is possible that this has led to a specific selective pressure on Orientia to reduce peptidoglycan, although it is worth noting that Rickettsia akari also localises in monocyte/macrophage cells. In common with this line of reasoning, it is notable that A. marginale possesses a complement of genes supporting the production of intermediate peptidoglycan, whilst the closely related A. phagocytophilum completely lacks the ability to produce peptidoglycan. This may also reflect differences in cell tropism since A. marginale is localised in erythrocytes whilst A. phagocytophilum is localised in monocytes/macrophages, potentially resulting in a difference in immune‐driven selective pressure on peptidoglycan status. Lastly, it is also possible that species‐specific chemical modifications exist that influence the magnitude of the signalling response of peptidoglycan innate immune surveillance systems.
The total lack of peptidoglycan in some organisms, as well as the presence of only intermediate peptidoglycan in others raises the question of how bacterial cells are structured in the absence of a complete classical peptidoglycan sacculus. Mycoplasma is unusual in lacking peptidoglycan but not being an obligate intracellular bacterium. It can replicate both in extracellular tissue fluid as well as within eukaryotic cells, and this lifestyle confers osmotic protection as well as a source of sterol lipids that are essential for their growth and likely protects the cell from rupture in the absence of a rigid cell wall. Host‐derived lipids are also important features of the membranes of Anaplasmataceae, conferring structural rigidity in the absence of a cell wall. Both Chlamydiae and Orientia possess disulphide cross‐linked proteins on their outer membranes and these confer additional structural rigidity in the absence of a full peptidoglycan cell wall sacculus. It is also conceivable that the proliferation mode can shape the genomic repertoire of peptidoglycan biosynthesis and remodelling genes. For example although the peptidoglycan‐intermediate Planctomycetes are free‐living bacteria, they reproduce by budding rather than binary fission typically executed by most other bacteria.
In conclusion, our analyses suggest that a group of diverse obligate intracellular bacteria have responded to selective pressures imposed by this lifestyle by selectively retaining a subset of genes in the peptidoglycan‐biosynthesis pathway. We hypothesise that this will result in some commonalities in peptidoglycan structure, which may include reduced overall abundance, differences in chain length and peptide cross linking, and regulated or limited biosynthesis in time and space. Future structural and biochemical analyses of the peptidoglycan from this group will lead to a greater understanding of the relationship between bacterial cell growth and host immune recognition, as well as commonalities of a minimal peptidoglycan cell wall, in this unique group of bacterial organisms.
Supporting information
Supporting Table
Supporting Information
Acknowledgements
WV was supported by the UK Medical Research Council (MRC) within the Joint Programming Initiative on Antimicrobial Resistance ANR‐14‐JAMR‐0003 (NAPCLI) and a Senior Investigator Award from the Wellcome Trust (101824/Z/13/Z). JS was supported by a Dorothy Hodgkin Research Fellowship from the Royal Society.
References
- Al‐Dabbagh, B. , Olatunji, S. , Crouvoisier, M. , El Ghachi, M. , Blanot, D. , Mengin‐Lecreulx, D. , and Bouhss, A. (2016) Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl‐phosphate N‐acetylhexosamine 1‐phosphate transferase superfamily. Biochimie 127: 249–257. [DOI] [PubMed] [Google Scholar]
- Amano, K. , Tamura, A. , Ohashi, N. , Urakami, H. , Kaya, S. , and Fukushi, K. (1987) Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi . Infect Immun 55: 2290–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerdorffer, A. , Stojanov, M. , Greub, G. , and Baud, D. (2017) Chlamydia trachomatis and chlamydia‐like bacteria: new enemies of human pregnancies. Curr Opin Infect Dis 30: 289–296. [DOI] [PubMed] [Google Scholar]
- Atwal, S. , Giengkam, S. , Chaemchuen, S. , Dorling, J. , Kosaisawe, N. , VanNieuwenhze, M. , et al (2017) Evidence for a peptidoglycan‐like structure in Orientia tsutsugamushi . Mol Microbiol 105: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, A.F. (2007) Pathogenic rickettsiae as bioterrorism agents. Clin Infect Dis 45 (Suppl. 1): S52–S55. [DOI] [PubMed] [Google Scholar]
- Barreteau, H. , Kovac, A. , Boniface, A. , Sova, M. , Gobec, S. , and Blanot, D. (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 32: 168–207. [DOI] [PubMed] [Google Scholar]
- Boneca, I.G. (2005) The role of peptidoglycan in pathogenesis. Curr Opin Microbiol 8: 46–53. [DOI] [PubMed] [Google Scholar]
- Bonen, L. , Cunningham, R.S. , Gray, M.W. , and Doolittle, W.F. (1977) Wheat embryo mitochondrial 18S ribosomal RNA: evidence for its prokaryotic nature. Nucleic Acids Res 4: 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisova, M. , Gaupp, R. , Duckworth, A. , Schneider, A. , Dalugge, D. , Muhleck, M. , et al (2016) Peptidoglycan recycling in Gram‐positive bacteria is crucial for survival in stationary phase. mBio 7: pii e00923‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhss, A. , Trunkfield, A.E. , Bugg, T.D. , and Mengin‐Lecreulx, D. (2008) The biosynthesis of peptidoglycan lipid‐linked intermediates. FEMS Microbiol Rev 32: 208–233. [DOI] [PubMed] [Google Scholar]
- Busiek, K.K. , and Margolin, W. (2015) Bacterial actin and tubulin homologs in cell growth and division. Curr Biol 25: R243–R254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra, T. (2001) Pathogenesis of septic shock: implications for prevention and treatment. J Chemother 13 (Spec No. 1): 173–180. [DOI] [PubMed] [Google Scholar]
- Carlyon, J. , and Fikrig, E. (2003) Invasion and survival strategies of Anaplasma phagocytophilum . Cell Microbiol 5: 743–754. [DOI] [PubMed] [Google Scholar]
- Carvalho, D. , Andrade, R. , Pinho, S. , Goes‐Neto, A. , Lobao, T. , Bomfim, G. , and El‐Hani, C. (2015) What are the evolutionary origins of mitochondria? A complex network approach. PLoS One 10: e0134988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celler, K. , Koning, R.I. , Koster, A.J. , and van Wezel, G.P. (2013) Multidimensional view of the bacterial cytoskeleton. J Bacteriol 195: 1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput, C. , and Boneca, I.G. (2007) Peptidoglycan detection by mammals and flies. Microbes Infect 9: 637–647. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Men, H. , Ha, S. , Ye, X.Y. , Brunner, L. , Hu, Y. , and Walker, S. (2002) Intrinsic lipid preferences and kinetic mechanism of Escherichia coli MurG. Biochemistry 41: 6824–6833. [DOI] [PubMed] [Google Scholar]
- Cho, B. , Cho, N. , Seong, S. , Choi, M. , and Kim, I. (2010a) Intracellular invasion by Orientia tsutsugamushi is mediated by integrin signaling and actin cytoskeleton rearrangements. Infect Immun 78: 1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Uehara, T. , and Bernhardt, T.G. (2014) Beta‐lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159: 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K. , Jun, Y. , Suh, J. , Kang, J. , Choi, H. , and Woo, S. (2010b) Orientia tsutsugamushi induced endothelial cell activation via the NOD1‐IL‐32 pathway. Microb Pathog 49: 95–104. [DOI] [PubMed] [Google Scholar]
- Cho, N.‐H. , Kim, H.‐R. , Lee, J.‐H. , Kim, S.‐Y. , Kim, J. , Cha, S. , et al (2007) The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host– cell interaction genes. Proc Natl Acad Sci U S A 104: 7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Cheong, T. , Ha, N. , Ko, Y. , Cho, C. , Jeon, J. , et al (2013) Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration. PLoS Negl Trop Dis 7: e1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Lee, J. , Han, S. , Kim, S. , Cho, N. , Kim, I. , and Choi, M. (2006) Exploitation of the endocytic pathway by Orientia tsutsugamushi in nonprofessional phagocytes. Infect Immun 74: 4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S.T. , Eiglmeier, K. , Parkhill, J. , James, K.D. , Thomson, N.R. , Wheeler, P.R. , et al (2001) Massive gene decay in the leprosy bacillus. Nature 409: 1007–1011. [DOI] [PubMed] [Google Scholar]
- De Benedetti, S. , Buhl, H. , Gaballah, A. , Klöckner, A. , Otten, C. , Schneider, T. , et al (2014) Characterization of serine hydroxymethyltransferase GlyA as a potential source of D‐alanine in Chlamydia pneumoniae . Front iCell Infect Microbiol 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich, S. , Phuklia, W. , Turner, G. , Rattanavong, S. , Chansamouth, V. , Dumler, S. , et al (2015) Neorickettsia sennetsu as a neglected cause of fever in south‐east asia. PLoS Negl Trop Dis 9: e0003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez‐Escobar, J. , Chastanet, A. , Crevenna, A.H. , Fromion, V. , Wedlich‐Soldner, R. , and Carballido‐Lopez, R. (2011) Processive movement of MreB‐associated cell wall biosynthetic complexes in bacteria. Science 333: 225–228. [DOI] [PubMed] [Google Scholar]
- Douglas, A.E. , Bouvaine, S. , and Russell, R.R. (2011) How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci 278: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski, R. , and Gupta, D. (2010) Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun 16: 168–174. [DOI] [PubMed] [Google Scholar]
- Egan, A.J. , Biboy, J. , Van't Veer, I. , Breukink, E. , and Vollmer, W. (2015) Activities and regulation of peptidoglycan synthases. Phil Trans Roy Soc B, Biol Sci 370: pii 20150031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, A.J. , Cleverley, R.M. , Peters, K. , Lewis, R.J. , and Vollmer, W. (2017) Regulation of bacterial cell wall growth. Febs J 284: 851–867. [DOI] [PubMed] [Google Scholar]
- Egan, A.J. , and Vollmer, W. (2013) The physiology of bacterial cell division. Ann N Y Acad Sci 1277: 8–28. [DOI] [PubMed] [Google Scholar]
- Emelyanov, V. (2001) Evolutionary relationship of Rickettsiae and mitochondria. FEBS Lett 501: 11–18. [DOI] [PubMed] [Google Scholar]
- Eremeeva, M. , and Dasch, G. (2015) Challenges posed by tick‐borne rickettsiae: eco‐epidemiology and public health implications. Front Public Health 3: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremeeva, M. , Madan, A. , Shaw, C. , Tang, K. , and Dasch, G. (2005) New perspectives on rickettsial evolution from new genome sequences of rickettsia, particularly R. canadensis, and Orientia tsutsugamushi . Ann N Y Acad Sci 1063: 47–63. [DOI] [PubMed] [Google Scholar]
- Figueiredo, T.A. , Sobral, R.G. , Ludovice, A.M. , Almeida, J.M. , Bui, N.K. , Vollmer, W. , et al (2012) Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus . PLoS Pathogens 8: e1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandi, A. , Jacquier, N. , Théraulaz, L. , Greub, G. , and Viollier, P. (2014) FtsZ‐independent septal recruitment and function of cell wall remodelling enzymes in chlamydial pathogens. Nat Commun 5: 4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, E.C. , Bernard, R. , Wang, W. , Zhuang, X. , Rudner, D.Z. , and Mitchison, T. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis . Science 333: 222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen, J.M. , and Goffin, C. (1999) Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother 43: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin, S. , and Philpott, D. (2004) Mini‐review: the role of peptidoglycan recognition in innate immunity. Eur J Immunol 34: 1777–1782. [DOI] [PubMed] [Google Scholar]
- Goodell, E.W. , and Schwarz, U. (1985) Release of cell wall peptides into culture medium by exponentially growing Escherichia coli . J Bacteriol 162: 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, G.W. , and Beck, S.D. (1974) Effects of antibiotics on intracellular symbiotes in the pea aphid, Acyrthosiphon pisum . Cell Tissue Res 148: 287–300. [DOI] [PubMed] [Google Scholar]
- Gutierrez, M.C. , Supply, P. , and Brosch, R. (2009) Pathogenomics of mycobacteria. Genome Dyn 6: 198–210. [DOI] [PubMed] [Google Scholar]
- Hatch, T.P. , Miceli, M. , and Sublett, J.E. (1986) Synthesis of disulfide‐bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis . J Bacteriol 165: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichfreise, B. , Schiefer, A. , Schneider, T. , Nzukou, E. , Poellinger, C. , Hoffmann, T.J. , et al (2009) Functional conservation of the lipid II biosynthesis pathway in the cell wall‐less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol Microbiol 73: 913–923. [DOI] [PubMed] [Google Scholar]
- Höltje, J.V. (1998) Growth of the stress‐bearing and shape‐maintaining murein sacculus of Escherichia coli . Microbiol Mol Biol Rev 62: 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk, E.J. , Griffiths, G.W. , Hadjokas, N.E. , and Beck, S.D. (1977) Peptidoglycan in the cell wall of the primary intracellular symbiote of the pea aphid. Science 198: 401–403. [DOI] [PubMed] [Google Scholar]
- Ihn, K. , Han, S. , Kim, H. , Huh, M. , Seong, S. , Kang, J. , et al (2000) Cellular invasion of Orientia tsutsugamushi requires initial interaction with cell surface heparan sulfate. Microb Pathog 28: 227–233. [DOI] [PubMed] [Google Scholar]
- Jacquier, N. , Frandi, A. , Pillonel, T. , Viollier, P.H. , Viollier, P. , and Greub, G. (2014) Cell wall precursors are required to organize the chlamydial division septum. Nat Commun 5: 3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, N. , Viollier, P.H. , and Greub, G. (2015) The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly. FEMS Microbiol Rev 39: 262–275. [DOI] [PubMed] [Google Scholar]
- Jankute, M. , Cox, J.A. , Harrison, J. , and Besra, G.S. (2015) Assembly of the mycobacterial cell wall. Annu Rev Microbiol 69: 405–423. [DOI] [PubMed] [Google Scholar]
- Jeske, O. , Schuler, M. , Schumann, P. , Schneider, A. , Boedeker, C. , Jogler, M. , et al (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6: 7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, L. , Shaw, A.C. , Xu, D. , Xia, W. , Zhang, J. , Deng, J. , et al (2011) Upregulation of MetC is essential for D‐alanine‐independent growth of an alr/dadX‐deficient Escherichia coli strain. J Bacteriol 193: 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebbi‐Beghdadi, C. , and Greub, G. (2014) Importance of amoebae as a tool to isolate amoeba‐resisting microorganisms and for their ecology and evolution: the Chlamydia paradigm. Environ Microbiol Rep 6: 309–324. [DOI] [PubMed] [Google Scholar]
- Kemege, K.E. , Hickey, J.M. , Barta, M.L. , Wickstrum, J. , Balwalli, N. , Lovell, S. , Battaile, K.P. , and Hefty, P.S. (2015) Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Mol Microbiol 95: 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöckner, A. , Otten, C. , Derouaux, A. , Vollmer, W. , Buhl, H. , De Benedetti, S. , et al (2014) AmiA is a penicillin target enzyme with dual activity in the intracellular pathogen Chlamydia pneumoniae . Nat Commun 5: 4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, Y. , Choi, J. , Ha, N. , Kim, I. , Cho, N. , and Choi, M. (2013) Active escape of Orientia tsutsugamushi from cellular autophagy. Infect Immun 81: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, L. , and Roy, C. (2015) Biogenesis of the lysosome‐derived vacuole containing Coxiella burnetii . Microbes Infect 17: 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlrausch, U. , and Höltje, J.V. (1991) Analysis of murein and murein precursors during antibiotic‐induced lysis of Escherichia coli . J Bacteriol 173: 3425–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzel, H. , Heidrich, M. , and Piechulla, B. (1981) Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res 9: 1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier, J.C. , Edouard, S. , Pagnier, I. , Mediannikov, O. , Drancourt, M. , and Raoult, D. (2015) Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev 28: 208–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq, S. , Derouaux, A. , Olatunji, S. , Fraipont, C. , Egan, A.J. , Vollmer, W. , et al (2017) Interplay between Penicillin‐binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep 7: 43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Cho, N. , Kim, S. , Bang, S. , Chu, H. , Choi, M. , and Kim, I. (2008) Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56‐kDa type‐specific antigen. J Infect Dis 198: 250–257. [DOI] [PubMed] [Google Scholar]
- Letek, M. , Ordonez, E. , Vaquera, J. , Margolin, W. , Flardh, K. , Mateos, L.M. , and Gil, J.A. (2008) DivIVA is required for polar growth in the MreB‐lacking rod‐shaped actinomycete Corynebacterium glutamicum . J Bacteriol 190: 3283–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti, G. , Kuru, E. , Packiam, M. , Hsu, Y.P. , Tekkam, S. , Hall, E. , et al (2016) Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid‐cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathogens 12: e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti, G.W. , Kuru, E. , Hall, E. , Kalinda, A. , Brun, Y.V. , VanNieuwenhze, M. , and Maurelli, A.T. (2013) A new metabolic cell‐wall labelling method reveals peptidoglycan in Chlamydia trachomatis . Nature 506: 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M. , and Rikihisa, Y. (2003) Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun 71: 5324–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet, S. , Dubost, L. , Marie, A. , Arthur, M. , and Gutmann, L. (2008) Identification of the L,D‐transpeptidases for peptidoglycan cross‐linking in Escherichia coli . J Bacteriol 190: 4782–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra, S. , Crick, D. , McNeil, M. , and Brennan, P. (2008) Unique structural features of the peptidoglycan of Mycobacterium leprae . J Bacteriol 190: 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manat, G. , Roure, S. , Auger, R. , Bouhss, A. , Barreteau, H. , Mengin‐Lecreulx, D. , and Touze, T. (2014) Deciphering the metabolism of undecaprenyl‐phosphate: the bacterial cell‐wall unit carrier at the membrane frontier. Microb Drug Res 20: 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J. , Adams, N.E. , Hudson, A.O. , Gilvarg, C. , Leustek, T. , and Maurelli, A.T. (2006) L,L‐diaminopimelate aminotransferase, a trans‐kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci U S A 103: 17909–17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske, A.J. , Riley, E.P. , Robins, W.P. , Uehara, T. , Mekalanos, J.J. , Kahne, D. , et al (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, C. , Yang, J. , Kim, S. , Choi, M. , Kim, I. , and Cho, N. (2008) Genome‐based construction of the metabolic pathways of Orientia tsutsugamushi and comparative analysis within the Rickettsiales order. Comp Funct Genomics 2008: 623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, T. , Sijbrandi, R. , Lutters, M. , Verheul, J. , Martin, N.I. , den Blaauwen, T. , et al (2014) Specificity of the transport of lipid II by FtsW in Escherichia coli . J Biol Chem 289: 14707–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, T. , van Dam, V. , Sijbrandi, R. , Vernet, T. , Zapun, A. , Bouhss, A. , et al (2011) Identification of FtsW as a transporter of lipid‐linked cell wall precursors across the membrane. Embo J 30: 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moron, C. , Popov, V. , Feng, H. , Wear, D. , and Walker, D. (2001) Identification of the target cells of Orientia tsutsugamushi in human cases of scrub typhus. Mod Pathol 14: 752–759. [DOI] [PubMed] [Google Scholar]
- Munderloh, U. , Jauron, S. , Fingerle, V. , Leitritz, L. , Hayes, S. , Hautman, J. , et al (1999) Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 37: 2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, K. , Yamashita, A. , Kurokawa, K. , Morimoto, T. , Ogawa, M. , Fukuhara, M. , et al (2008) The Whole‐genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res 15: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nans, A. , Ford, C. , and Hayward, R.D. (2015) Host‐pathogen reorganisation during host cell entry by Chlamydia trachomatis . Microbes Infect 17: 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataraj, V. , Varela, C. , Javid, A. , Singh, A. , Besra, G.S. , and Bhatt, A. (2015) Mycolic acids: deciphering and targeting the Achilles' heel of the tubercle bacillus. Mol Microbiol 98: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen, C. , and Lemaitre, B. (2016) Sensing Gram‐negative bacteria: a phylogenetic perspective. Curr Opin Immunol 38: 8–17. [DOI] [PubMed] [Google Scholar]
- Omsland, A. , Cockrell, D.C. , Howe, D. , Fischer, E.R. , Virtaneva, K. , Sturdevant, D.E. , et al (2009) Host cell‐free growth of the Q fever bacterium Coxiella burnetii . Proc Natl Acad Sci U S A 106: 4430–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette, S. , Karimova, G. , Subtil, A. , and Ladant, D. (2012) Chlamydia co‐opts the rod shape‐determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85: 164–178. [DOI] [PubMed] [Google Scholar]
- Oyston, P.C. , and Davies, C. (2011) Q fever: the neglected biothreat agent. J Med Microbiol 60: 9–21. [DOI] [PubMed] [Google Scholar]
- Packiam, M. , Weinrick, B. , Jacobs, W.R., Jr ., and Maurelli, A.T. (2015) Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly.” Proc Natl Acad Sci U S A 112: 11660–11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, D. , Phetsouvanh, R. , Tanganuchitcharnchai, A. , Jones, M. , Jenjaroen, K. , Vongsouvath, M. , et al (2012) Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl Trop Dis 6: e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.T. , and Uehara, T. (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). MIcrobiol Mol Biol Rev 72: 211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola, P. , and Raoult, D. (2006) Tropical rickettsioses. Clin Dermatol 24: 191–200. [DOI] [PubMed] [Google Scholar]
- Pazos, M. , Peters, K. , and Vollmer, W. (2017) Robust peptidoglycan growth by dynamic and variable multi‐protein complexes. Curr Opin Microbiol 36: 55–61. [DOI] [PubMed] [Google Scholar]
- Peters, K. , Kannan, S. , Rao, V.A. , Biboy, J. , Vollmer, D. , Erickson, S.W. , et al (2016) The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli . mBio 7: e00819–e00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer, M. , Aistleitner, K. , Biboy, J. , Gray, J. , Kuru, E. , Hall, E. , et al (2013) Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4: 2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic, S. , Price, P. , Beier, M. , Gaywee, J. , Macaluso, J. , and Azad, A. (2002) Rickettsia‐macrophage interactions: host cell responses to Rickettsia akari and Rickettsia typhi . Infect Immun 70: 2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa, Y. (2003) Mechanisms to create a safe Haven by members of the family anaplasmataceae. Ann N Y Acad Sci 990: 548–555. [DOI] [PubMed] [Google Scholar]
- Rodrigues, L.C. , and Lockwood, D. (2011) Leprosy now: epidemiology, progress, challenges, and research gaps. Lancet Infect Dis 11: 464–470. [DOI] [PubMed] [Google Scholar]
- Royet, J. , and Dziarski, R. (2007) Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol 5: 264–277. [DOI] [PubMed] [Google Scholar]
- Ruiz, N. (2008) Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A 105: 15553–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscitto, A. , Hottmann, I. , Stafford, G.P. , Schaffer, C. , Mayer, C. , and Sharma, A. (2016) Identification of a novel N‐acetylmuramic acid transporter in Tannerella forsythia . J Bacteriol 198: 3119–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz, K.M. , Popham, D.L. , Beare, P.A. , Sturdevant, D.E. , Hansen, B. , Nair, V. , et al (2016) Transcriptional profiling of Coxiella burnetii reveals extensive cell wall remodeling in the small cell variant developmental form. PLoS One 11: e0149957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. , Sullivan, C.J. , and Kuehn, M.J. (2013) Envelope control of outer membrane vesicle production in Gram‐negative bacteria. Biochemistry 52: 3031–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, L.T. , Butler, E.K. , Lebar, M.D. , Kahne, D. , Bernhardt, T.G. , and Ruiz, N. (2014) MurJ is the flippase of lipid‐linked precursors for peptidoglycan biogenesis. Science 345: 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, E. , Roh, J. , Park, W. , Song, B. , Chang, K. , Lee, W. , et al (2014) Transovarial transmission of Orientia tsutsugamushi in Leptotrombidium palpale (Acari: Trombiculidae). PLoS One 9: e88453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard, M. , Dittmer, J. , Greve, P. , Bouchon, D. , and Braquart‐Varnier, C. (2014) A host as an ecosystem: Wolbachia coping with environmental constraints. Environ Microbiol 16: 3583–3607. [DOI] [PubMed] [Google Scholar]
- Silhavy, T.J. , Kahne, D. , and Walker, S. (2010) The bacterial cell envelope. Cold Spring Harbor Perspect Biol 2: a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, F.J. , Latorre, A. , and Moya, A. (2001) Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet 17: 615–618. [DOI] [PubMed] [Google Scholar]
- Silver, L.L. (2013) Viable screening targets related to the bacterial cell wall. Ann N Y Acad Sci 1277: 29–53. [DOI] [PubMed] [Google Scholar]
- Silverman, D. , and Wisseman, C. (1978) Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi . Infect Immun 21: 1020–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skilton, R.J. , Cutcliffe, L.T. , Barlow, D. , Wang, Y. , Salim, O. , Lambden, P.R. , et al (2009) Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS One 4: e7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhithasri, V. , Nisha, N. , Biswas, L. , Anil Kumar, V. , and Biswas, R. (2013) Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol Res 168: 396–406. [DOI] [PubMed] [Google Scholar]
- Takhampunya, R. , Tippayachai, B. , Korkusol, A. , Promsathaporn, S. , Leepitakrat, S. , Sinwat, W. , et al (2016) Transovarial transmission of co‐existing Orientia tsutsugamushi genotypes in laboratory‐reared Leptotrombidium imphalum . Vector Borne Zoonotic Dis 16: 33–41. [DOI] [PubMed] [Google Scholar]
- Tomasz, A. , and Waks, S. (1975) Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc Natl Acad Sci U S A 72: 4162–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese, M. , and Carlyon, J. (2009) Anaplasma phagocytophilum dense‐cored organisms mediate cellular adherence through recognition of human P‐selectin glycoprotein ligand 1. Infect Immun 77: 4018–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas, A. , Banzhaf, M. , Gross, C.A. , and Vollmer, W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, T. , and Park, J.T. (2008) Peptidoglycan recycling. EcoSal Plus 3(1). [DOI] [PubMed] [Google Scholar]
- van Schaik, E. , Chen, C. , Mertens, K. , Weber, M. , and Samuel, J. (2013) Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii . Nat Rev Microbiol 11: 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling, M.C. , Mesman, R.J. , Kuru, E. , Espaillat, A. , Cava, F. , Brun, Y.V. , et al (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6: 6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, J. , Schiefer, A. , Schneider, T. , Julicher, K. , Johnston, K.L. , Taylor, M.J. , et al (2013) Requirement of lipid II biosynthesis for cell division in cell wall‐less Wolbachia, endobacteria of arthropods and filarial nematodes. Int J Med Microbiol 303: 140–149. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev 32: 287–306. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , Blanot, D. , and de Pedro, M.A. (2008a) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , and Höltje, J.V. (2004) The architecture of the murein (peptidoglycan) in gram‐negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol 186: 5978–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, W. , Joris, B. , Charlier, P. , and Foster, S. (2008b) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32: 259–286. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , and Seligman, S.J. (2010) Architecture of peptidoglycan: more data and more models. Trends Microbiol 18: 59–66. [DOI] [PubMed] [Google Scholar]
- Vollmer, W. , and Tomasz, A. (2002) Peptidoglycan N‐acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae . Infect Immun 70: 7176–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouga, M. , Baud, D. , and Greub, G. (2017) Simkania negevensis may produce long‐lasting infections in human pneumocytes and endometrial cells. Pathog Dis 75(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D. (2007) Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis 45 (Suppl. 1): S39–S44. [DOI] [PubMed] [Google Scholar]
- Weidel, W. , and Pelzer, H. (1964) Bagshaped macromolecules–a new outlook on bacterial cell walls. Adv Enzym Biochem 26: 193–232. [DOI] [PubMed] [Google Scholar]
- Weidenmaier, C. , and Peschel, A. (2008) Teichoic acids and related cell‐wall glycopolymers in Gram‐positive physiology and host interactions. Nat Rev Microbiol 6: 276–287. [DOI] [PubMed] [Google Scholar]
- Wilmes, M. , Meier, K. , Schiefer, A. , Josten, M. , Otten, C.F. , Klöckner, A. , et al (2017) AmiD is a novel peptidoglycan amidase in Wolbachia endosymbionts of Drosophila melanogaster . Front Cell Infect Microbiol 7: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisseman, C.J. , Silverman, D. , Waddell, A. , and Brown, D. (1982) Penicillin‐induced unstable intracellular formation of spheroplasts by rickettsiae. J Infect Dis 146: 147–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table
Supporting Information


