Abstract
Genome editing with the CRISPR–Cas9 system has enabled unprecedented efficacy for reverse genetics and gene correction approaches. While off-target effects have been successfully tackled, the effort to eliminate variability in sgRNA efficacies—which affect experimental sensitivity—is in its infancy. To address this issue, studies have analyzed the molecular features of highly active sgRNAs, but independent cross-validation is lacking. Utilizing fluorescent reporter knock-out assays with verification at selected endogenous loci, we experimentally quantified the target efficacies of 430 sgRNAs. Based on this dataset we tested the predictive value of five recently-established prediction algorithms. Our analysis revealed a moderate correlation (r = 0.04 to r = 0.20) between the predicted and measured activity of the sgRNAs, and modest concordance between the different algorithms. We uncovered a strong PAM-distal GC-content-dependent activity, which enabled the exclusion of inactive sgRNAs. By deriving nine additional predictive features we generated a linear model-based discrete system for the efficient selection (r = 0.4) of effective sgRNAs (CRISPRater). We proved our algorithms’ efficacy on small and large external datasets, and provide a versatile combined on- and off-target sgRNA scanning platform. Altogether, our study highlights current issues and efforts in sgRNA efficacy prediction, and provides an easily-applicable discrete system for selecting efficient sgRNAs.
INTRODUCTION
Genome editing promises the ability to probe genetic interactions at their origin and the opportunity to cure severe inherited diseases. With the development of the clustered regularly interspaced short palindromic repeats (CRISPR)—CRISPR-associated-9 (Cas9) technology, new avenues for versatile genome editing have opened due to the minimal selection criteria of its target site—the protospacer adjacent motif (PAM)—and its high overall activity (1–3). After initial doubts about off-target activity-induced safety issues (4–6), recent advances in understanding sgRNA/Cas9 function –based on crystal structures of Cas9-DNA-RNA interaction (7,8)—guided protein engineering of the S. pyogenes Cas9 towards an expanded PAM repertoire (9) and increased safety (10). This, as well as the availability of Cas9 proteins from other bacterial strains (11–13), and a plethora of approaches to decrease off-target activity (14–16), have since eliminated nearly all restrictions of target site selection. However, variance in genome editing efficacy remains a limitation for translational approaches and lowers the signal-to-noise ratio of large-scale genetic screens. While this constraint may have its basis in Cas9 functions, like efficient DNA-scanning (17) and PAM detection (7,8), or nuclease domain activation (8,18), a distinct requirement for nucleotide composition of the sgRNA may cause reasonable variances in genome editing efficacy.
When short hairpin RNAs (shRNAs) were first developed they similarly showed highly variable knock-down results, which were overcome by tiling experiments that determined sequence requirements for highly active shRNAs (19,20). In line with this approach, several recent studies have aimed to decipher the molecular nature of active versus inactive sgRNAs (21–29). Wang et al. detected an increased loading of efficient sgRNAs onto the Cas9 protein by Cas9-RIP, and derived sequence properties for these sgRNAs (21). This theme was further addressed by Moreno-Mateos et al., and connected to guanine-dependent sgRNA stability (26). Tiling approaches similar to the successful screenings for efficient shRNAs (19) have focused more on the genomic target site of the sgRNA/Cas9 complex, including the influence of the precise PAM sequence and both upstream and downstream sequences (22,24,26,29). In addition, Xu et al. performed RNA-seq and incorporated sgRNA folding energies into the mix (27), while Malina et al. discovered that PAM sequences within the sgRNA inhibit its activity (25). The idea that sgRNA folding may be a confounding factor for sgRNA-dependent genome editing activity was further addressed by Chu et al., who evaluated spacer sequence-determined sgRNA folding as a potential modifier of genome editing activity which may cause improper recognition by Cas9 and thus reduced genome editing (30). These studies have all generated predictive algorithms and provided scoring systems to identify highly active sgRNAs compared to less active ones. However, an independent validation and comparison between these prediction algorithms using an experimentally-established, quantitative dataset of sgRNAs was lacking until now.
We present findings from a dataset of 430 lentivirally-delivered sgRNAs, tested via surrogate fluorescent reporter knock-out assays that allow the assessment of sgRNAs at the genomic level with single-cell resolution. Meta-analysis of existing algorithms and features for sgRNA prediction suggested only a moderate capacity to recapitulate the observed activities of lentivirally-delivered sgRNAs from our dataset. This was further underlined by modest concordance between prediction algorithms. By probing our dataset for potential predictive features, we developed an experimentally-defined discrete model for the prediction of sgRNA efficacy, which enables the exclusion of low-efficacy candidates and is publicly available as an additional tool within the CCTop online platform (http://crispr.cos.uni-heidelberg.de/). Our study thereby highlights current obstacles in the prediction of sgRNA activity, outlines potential ways to overcome these hurdles, and provides efficient methodologies to select highly active sgRNAs for focused approaches.
MATERIALS AND METHODS
Vectors and sgRNA design
The lentiviral CRISPR–Cas9 plasmids, and the tRFP657- and dTomato-labeled CRISPR–Cas9 efficacy reporter plasmids have been described before (31,32). The sgRNA expression vector SGL40C.EFS.E2Crimson (#100894) and the SIN40C.SFFV.sfGFP-Rep.iPAC (#100893) CRISPR–Cas9 efficacy reporter plasmids have been deposited at Addgene.
CRISPR–Cas9 target sites and sgRNAs were selected and designed using the CCTop online target prediction tool with maximal avoidance of off-target effects (33).
Viral particle production
Lentiviral particles were generated by transient transfection of 293T cells using polyethylenimine (PEI, Polysciences). Lentiviral vectors (described above) were co-transfected with the psPAX2 packaging plasmid (Addgene, Plasmid #12260) and the pMD2.G envelope plasmid (Addgene, Plasmid #12259). Particles were either used directly by applying the supernatant of transfected 293T cells, or they were concentrated by ultracentrifugation.
Cell culture and cell transduction
293T cells (German National Resource Center for Biological Material (DSMZ)) were cultured in DMEM (Biochrom) supplemented with 10% FCS, 1% penicillin/streptomycin, 1% sodium pyruvate, 1% L-glutamine, and 1% non-essential amino acids (all Life Technologies). HEL cells (DSMZ) were cultured in equally supplemented RPMI 1640 (Lonza). HEL cells were transduced in the presence of 5 μg/ml hexadimethrine bromide (Polybrene, Life Technologies).
Fluorescent reporter assay
HEL cells were transduced with the CRISPR–Cas9 reporter vector containing the respective target sites in-frame with a sfGFP cDNA. Fluorescence was checked 48 h after transduction by flow cytometry and cultures with 15–25% transduction rate were subjected to selection with 2 μg/ml puromycin (Life Technologies) for 72 h. The selection step was verified by flow cytometry.
Subsequently, cells expressing the reporter construct were super-transduced with lentiviral vectors encoding SpCas9, the sgRNA and a fluorescent protein. Six days after super-transduction, cells were analyzed by flow cytometry (BD FACSCanto™ [BD Biosciences]). Cleavage efficacies of sgRNAs were calculated by the loss of reporter fluorescence compared to a non-targeting sgRNA.
T7-endonuclease I assay
HEL cells were transduced with CRISPR–Cas9 vectors containing the respective sgRNA, the SpCas9 cDNA, a puromycin resistance cassette and the eGFP cDNA. Starting at 48h post-transduction, cells were selected with 2 μg/ml puromycin (Life Technologies) for 72–96 h, followed by flow cytometric verification. Genomic DNA was isolated using the DNA Mini Kit (Qiagen) according to the manufacturer's instructions. PCR amplicons asymmetrically spanning the genomic target site of the individual sgRNAs (Supplementary Table S2) were produced using standard protocols with the Extensor 2x Master Mix (Thermo Scientific). Fragments were purified (Gel Extraction Kit [Thermo Scientific]), then melted and re-annealed by heating to 96°C for 5 min and slowly cooling down to room temperature. 350 ng of PCR products were incubated with 10 U T7-endonuclease I (NEB) in a total volume of 20 μl at 37°C for 22 min, and loaded onto a 1.5% agarose gel for analysis. For absolute quantification of cleaved products, the signal intensities of DNA bands were determined and converted by reference to defined amounts of DNA ladder input (Thermo Scientific) using the GelDoc XR system and ImageLab 3.0 software (both BioRad).
Statistical analyses
Statistical analyses were performed using GraphPad Prism® version 6.07. Significance levels of normally and non-normally distributed data were calculated using the Student's t-test and the Mann–Whitney test, respectively. All boxplot graphs show the median and interquartile ranges, unless otherwise stated. Significance levels of linear correlations were calculated using a two-tailed f-test.
Prediction model creation
Feature extraction and modeling of sgRNA efficacy was performed as previously described (34). In brief, adopting a similar approach as Vert et al. (35), the efficacy prediction model was built by applying the R package lars (36) to the knock-out efficacies of 426 sgRNAs assessed by the surrogate fluorescent reporter assay (flow cytometry-based). The predictive capacity of each positional sequence feature was assessed via linear modelling and calculation of the respective root-mean-square-error (RMSE) as the mean over all sgRNAs. After subsequent ranking on RMSE, the 10 best features available for all sgRNAs were selected. To obtain a feature combination capable of rating sgRNA efficacies (CRISPRater), the mean RMSE for all possible feature combinations was calculated and the best model with lowest RMSE was selected. Validation of the model was performed on the 426 sgRNAs, one internal dataset, and three published external datasets: (i) an independent set of 45 sgRNAs not used to train the algorithm (Supplementary Table S1), (ii) a dataset by (21,27,37) consisting of 3141 sgRNAs assessed by their effects on proliferation (27), (iii) 20 sgRNAs assessed by cleavage efficacy (27) and (iv) 15 sgRNAs assessed by protein level expression (27) (all Supplementary Table S3).
RESULTS
Surrogate fluorescent reporter knock out assays are predictive for targeted DNA cleavage at genomic loci
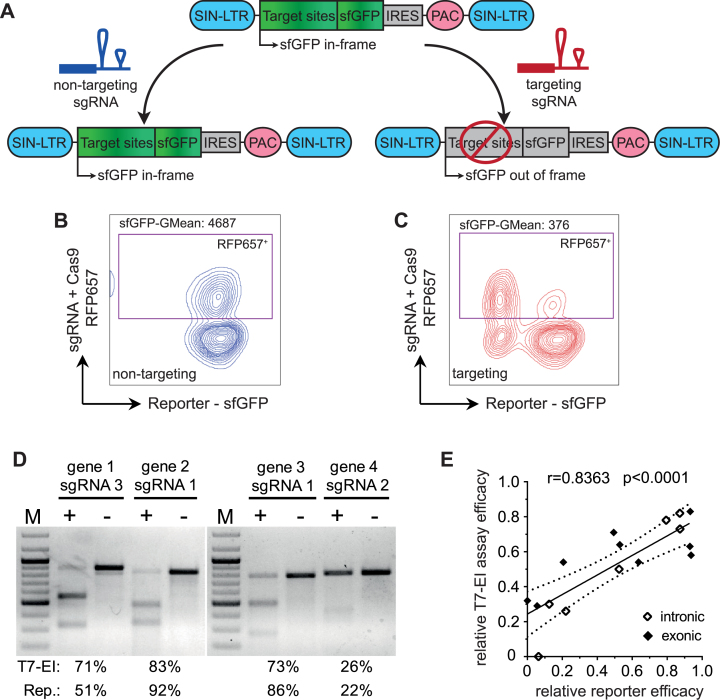
Aiming to establish highly efficient sgRNAs for genome editing applications, we developed a lentiviral-based reporter system that allows quantitative testing of the genome editing efficacies of 20–40 sgRNAs simultaneously at the genomic level (31) (Figure 1A). The vector was optimized to provide a robust fluorescence signal despite insertion of random amino acids into the fluorescent protein. This was tested by inserting the same sgRNA target sequence into different fluorescent proteins (n = 5, 126–276 amino acids in length). Among the tested fluorescent proteins, the fast-folding version of eGFP (sfGFP) (38) showed the highest and most stable fluorescence (Supplementary Figure S1A-B). Coupled to flow cytometry, this reporter system allows us to assess genome editing efficacy at the single-cell level, and to distinguish highly active sgRNAs from less active or inactive ones in a highly consistent manner (Figure 1B–C; Supplementary Figure S1C). Incorporation of the highly efficient target site of murine Tet2 yielded an average cleavage rate of 89.44% (n = 16, SD: ±2.64%), thus assuring the reproducibility and cross-experiment comparability of these assays. This result is in line with recent findings that CRISPR–Cas9 efficacy is independent from target site copy number variations (39).
Figure 1.
Evaluation of CRISPR–Cas9 cleavage utilizing a fluorescent based reporter-assay. (A) Schematic presentation of the reporter construct. CRISPR–Cas9 target sites are integrated into the ORF of a sfGFP (super-folder GFP) cDNA. Puromycin resistance enables selection of HEL cells harboring the reporter construct. Genome editing generates frame-shift mutations leading to quantifiable loss of fluorescence. (B, C) Representative flow cytometry analyses of a reporter assay. X-axis: sfGFP reporter fluorescence; Y-axis: fluorescence of the provided CRISPR–Cas9 components, namely, a non-targeting sgRNA (B) and a targeting sgRNA (C). (D) An example of T7-endonuclease I assay results, and correlation of genomic modification efficacies at endogenous loci with results from the reporter assay. [M] DNA marker; [+] targeting sgRNA; [–] non-targeting sgRNA. (E) Correlation of reporter assay efficacies (x-axis) and T7-endonuclease I assay efficacies (y-axis). Pearson correlation (r) and P value (p) are indicated.
Chromatin accessibility has been reported to alter CRISPR–Cas9 efficacies (40). In utilizing a non-randomly integrating lentiviral reporter system that preferentially integrates in open chromatin (41), the native chromatin structure of the target site is eliminated, which may limit the predictive value of our assay. To address this potential issue, we selected a total of 17 sgRNAs with different activities in our reporter assay: six sgRNAs with low activity (∼0–25% cleavage efficacy), four sgRNAs with intermediate activity (∼50–65%), and seven sgRNAs with high activity (∼80–95%). T7-EI assays were performed to test DNA modification at the designated genomic loci (Figure 1D–E). These results verified a high correlation (r = 0.84) between sgRNA activity in the reporter assay and at the genomic locus. Thus, high predictivity for genome editing activity at the endogenous genomic site was retained despite the use of a non-randomly integrating reporter system.
Lentivirally-delivered CRISPR–Cas9 has an inherent high efficacy
Having experimentally established a biologically neutral readout-based reporter system capable of single-cell resolution, we generated a dataset of 430 sgRNAs (Supplementary Table S1) and first analyzed their individual cutting efficacies (Figure 2A). Of note, sgRNA design was restricted to the avoidance of off-target sites as the only selection criterion for spacers/protospacers (5,33). Following these guidelines, we have shown before that high-scoring sgRNAs in our reporter assays have no detectable off-target activity (31,32). Without any further consideration for target sequence features or genomic context, our dataset had a median cleavage activity of 76.1% (mean: 67.2%; range: 0% to 99.7%). The likelihood of retrieving highly efficient (efficacy >80%) or inefficient sgRNAs (efficacy <40%) after random selection of target sites was 40.7% and 16.3%, respectively. Based on these observations, evaluating three sgRNAs per target gene should be sufficient to yield a highly efficient one.
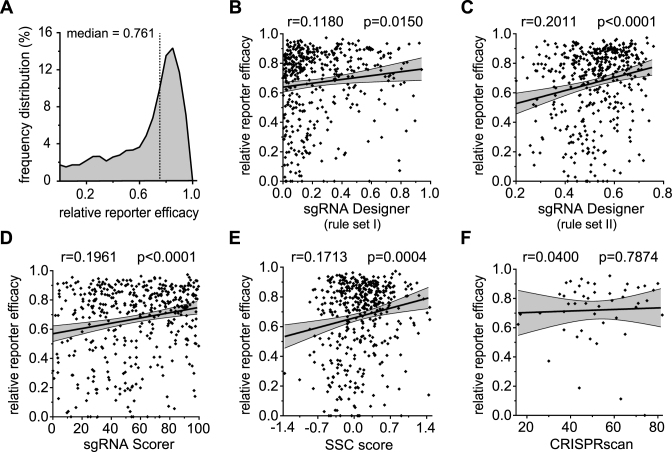
Figure 2.
Experimentally-ascertained cutting efficacies of 430 sgRNAs can be partially predicted by five up-to-date online prediction tools. (A) Distribution of the individually-assessed cleavage efficacies of 430 sgRNAs targeting a total of 92 genes (54 human and 38 murine), calculated on the basis of reporter assay analyses. The dataset shows median cleavage efficacy of 76.1% (interquartile range: 52.9 to 85.8%). (B–F) Scatter plots showing the correlation between sgRNA-specific cleavage efficacies obtained from reporter assays (y-axes) and predicted scores obtained from sgRNA sequence analysis using the online tools sgRNA Designer (rule set I) (22) (B), sgRNA Designer (rule set II) (29) (C), sgRNA Scorer (24) (D), SSC score (27) (E) and CRISPRscan (26) (F) (x-axes). The Pearson correlations (r) and P values (p) are indicated.
Established prediction algorithms for sgRNA efficacy have limited value
Next, we investigated whether the cleavage efficiencies of sgRNAs in our dataset could be correctly predicted by existing algorithms, all of which were established with large-scale library approaches (22,26,27,29). To this end, we retrieved efficiency scorings for our tested sgRNAs using sgRNA Designer, the first openly available platform generated by Doench et al. (22). By targeting twelve cell surface receptors on murine and human cell lines with a total of 1,841 sgRNAs, Doench et al. found that only ∼5% of all sgRNAs are highly effective. In line with this result, their algorithm (rule set I) predicted the majority of sgRNAs in our dataset to have low cleavage efficiency (score <0.25: 62.1%). This resulted in a poor but statistically significant correlation between the prediction score of sgRNA Designer and our measured cleavage efficiency (r = 0.118; p = 0.015; 1.04-fold activity at rule set I score >0.7 versus <0.2 (Figure 2B)). With a more advanced algorithm by Doench et al. (rule set II) (29), the predictive value increased and higher activity scores were obtained in our dataset (Figure 2C). Cross-comparison between the two algorithms (rule set I and II) illustrated the different predicted activities they assigned to our sgRNAs (Supplementary Figure S2A).
We next tested the alternative library-on-library approach, sgRNA Scorer by Chari et al. (24), on our dataset. The correlation between our data and the predicted activities from this algorithm did not improve compared to rule set II (29), although a trend of high scores matching to higher measured efficacy was still visible (Figure 2D). To further compare the predictive capacity of these algorithms, we performed a cross-comparison of rule set I (Supplementary Figure S2B) and rule set II (Supplementary Figure S2C) with Chari et al.’s approach, yielding positive correlation of the incorporated features but with reasonable variability.
Contrasting these approaches, efforts have also been dedicated towards distinguishing efficient from non-efficient sgRNAs. Xu et al. utilized published CRISPR–Cas9 drop-out screening data (21,37) to derive 28 characteristics from sgRNAs showing twofold higher sgRNA efficacy than their less active counterparts for the same genes (SSC score). Analysis of our sgRNAs based on these characteristics resulted in a moderate correlation between the provided scores and measured efficacies, while cross-comparison with formerly tested algorithms also showed intermediate-level correlation (Figure 2E; Supplementary Figure S2D-F).
We further proceeded to compare our data to the sgRNA prediction algorithm produced by Moreno-Mateos et al., CRISPRscan, which is based on in vivo knock-out data from zebrafish, and which indicated strong G-richness and higher stability within efficient sgRNAs (26). Hampered by the algorithm's immediate exclusion of sgRNAs labeled to be inefficient, we only retrieved scorings for 11.2% of our sgRNAs. Since this result may indicate inferior performance of the excluded sgRNAs, we compared efficiencies but did not observe any significant differences between the included and excluded sgRNAs (76.1% versus 75.7%, p = 0.4380, Supplementary Figure S2G). In line with this result, the characteristics derived by Moreno-Mateos et al. had no predictive capacity in our dataset and poor or no correlation with other tested algorithms (Figure 2F; Supplementary Figure S2H-K).
In conclusion, we retrospectively tested five sgRNA prediction algorithms on our experimentally-established dataset of 430 CRISPR–Cas9 targets. We observed moderate predictive capacity (r = 0.17 to r = 0.2) for three (24,27,29) out of five algorithms and moderate cross-comparability of the tested tools, indicating their benefit for large-scale library generation but modest performance for individual sgRNA/target design.
Purine bases at PAM-proximal sgRNA position 20 enhance genome editing efficacy
Next, we thought to interrogate our dataset for existing and novel features that may help in the design of efficient sgRNAs. Previously-identified features involving a nucleotide-dependent influence on CRISPR–Cas9 cutting efficacy are the nucleotide at the PAM-proximal position 20 (22) and the variable nucleotide of the PAM (22,42). Analysis of the variable nucleotide of the S.pyogenes PAM showed no impact on sgRNA activity in our dataset (Supplementary Figure S3A-D).
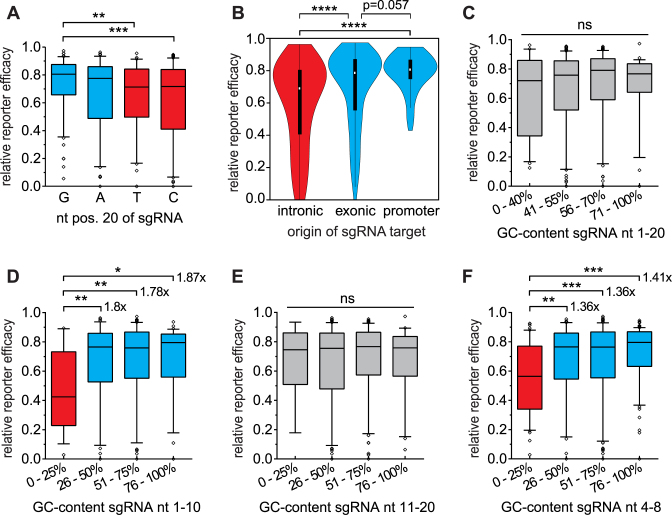
In contrast, sgRNA position 20 exhibited a guanine (G20)-dependent increase of genome editing activity in our dataset, as has been previously shown (21). Additional assessment of all four nucleotide options emphasized the impact of G20 on sgRNA activity, with a 9.2% and 8.8% increase compared to thymine and cytosine at position 20, respectively. However, no significant increase was observed between G20 versus an adenine at this position (Figure 3A). Thus, both purine bases (G and A) appear to have a favorable impact on Cas9 performance compared to pyrimidine bases (C and T) (Supplementary Figure S3E–G). Inclusion of A20 into the criteria for sgRNA selection could thus increase the likelihood of retrieving efficient sgRNAs compared to selection based on G20 only.
Figure 3.
Investigation of sgRNA sequence features capable of increasing genome editing capacity. (A) Efficacies of sgRNAs subdivided based on nucleotide usage at position 20 (adjacent to the PAM). Efficacy increased by 9.2% and 8.8% for G versus T and G versus C, respectively, and both are statistically significant by the Mann–Whitney test. (B) Comparison of sgRNA efficacy based on genomic origin of the target site. The sgRNAs target intronic regions (median 69.0%, interquartile range: 40.5–80.6%, n = 154), exonic regions (median 78.7%, interquartile range: 55.1–87.1%, n = 222) and promoter regions (median 80.7%, interquartile range: 74.8–87.2%, n = 54). Mann-Whitney test results are as indicated. (C–F) Depiction of GC-content dependent sgRNA activity. Overall GC: no significant differences (C). 1.78-fold to 1.87-fold reduction of efficacy in sgRNAs with <25% GC within nt 1–10 (n = 14) (D). GC-content has no effect on activity within nt 11–20 (E). Narrowed-down GC window size (5 nt) of PAM-distal GC-content: 1.36–1.41-fold reduction of efficacy with <25% GC within nt 4–8 (n = 50) (F). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; Mann–Whitney test used in all cases.
Genomic origin of target sites influences sgRNA efficacy in a chromatin-independent manner
Since our dataset contained not only sgRNAs targeting coding regions, but also included targets in non-coding genome elements, we next asked if the genomic origin of the target sites may help identify molecular features that guide the activity of the CRISPR–Cas9 system. We therefore first subdivided our dataset into sgRNAs targeting sites derived from coding (n = 222) or non-coding (n = 208) regions of the genome (Supplementary Figure S4A-B). Indeed, sgRNAs targeting non-coding regions of the genome performed significantly worse compared to coding regions (median: 73.6% versus 78.7%; p = 0.0082) (Supplementary Figure S4C). The non-coding part of the genome is versatile in its function and nucleotide composition, and this initial observation led us to interrogate sgRNA efficacies at non-coding loci in more detail. Since we targeted promoter regions (defined as TSS –200 to +50) and intronic regions, we again subdivided the non-coding sgRNAs into these categories and compared them to sgRNAs targeting exonic regions (Supplementary Figure S4D-E). Interestingly, sgRNAs targeting promoter regions yielded the highest median efficacy (80.7%, n = 54), while sgRNAs targeting intronic regions performed worst overall (69.0%, n = 154). Those sgRNAs targeting exonic regions showed an intermediate level of efficacy (78.7%, n = 222) (Figure 3B). Of note, since these results were obtained by lentiviral reporter assays, all protospacers were non-randomly integrated into the genome (41) at different loci without their native neighboring sequence or native epigenetic state. Due to the absence of these potential modifiers of genome editing activity in the lentiviral reporter assay, it is more likely that the detected differences between sgRNAs derived from different genomic regions are caused by particular sequence features.
GC-content discriminates low efficacy sgRNAs but is no identifier for maximum efficacy
The significant differences in sgRNA activity at sequences derived from different regions of the genome prompted us to investigate the molecular features of the sgRNAs in terms of nucleotide composition. First, the overall GC-content of the sgRNA spacers were tested, as GC-content differs between non-coding and coding genome regions (43,44) and as it was previously shown to influence sgRNA activity (21,22). Only a minor and non-significant increase in performance was found for sgRNAs with a GC-content of 41–55% (p = 0.3713) or 56–70% (p = 0.1346), compared to sgRNAs with <40% or >70% GC-content (Figure 3C). Our dataset thus does not reproduce former findings that indicate an optimal intermediate GC-content, with decreasing sgRNA performance towards low and high GC-content (22,23).
Nevertheless, we saw variations in sgRNA efficacy with changes in GC-content and we hypothesized that only some regions of the sgRNA are affected by this feature, as has been previously observed (23). To this end we split the sgRNA sequence into sub-intervals. Due to the controversy regarding a functional seed region for logical splitting (1,40,45,46), we split the sgRNA sequence into 50% intervals starting with a PAM-proximal and a PAM-distal half and analyzed their GC-content independently (Figure 3D–E). Indeed, medium and high GC-content within the first 10 nt of the sgRNA (distal to the PAM) significantly increased genome editing activity. In comparison to a GC-content of <25%, a GC-content of 26–50% increased sgRNA efficacy by 1.80-fold (p = 0.0077). The same result was seen when comparing <25% to even higher GC-content (GC 51–75%: 1.78-fold; p = 0.0088 and 76–100%: 1.87-fold; p = 0.0281). Notably, GC-content in the PAM-proximal half showed no significant impact on sgRNA efficacy.
Having verified our hypothesis that sub-regions of the sgRNA are sensitive towards varying GC-content, we next performed complete tiling with a 10 nt window size (Supplementary Figure S5A), and found a positive impact of high GC up to position 13 (Supplementary Figure S6). However, the strongest resolution of sgRNA efficacy could be obtained by analysis of nucleotides 1–10 (GC >25% versus <25%: 1.78-fold to 1.87-fold efficacy), thereby marking 3.3% of all sgRNAs as very ineffective. Of note, our analyses were performed on native sequences before the addition/substitution of a guanine at position one for U6 promoter initiation. Tiling for GC-content-dependent efficacy with a 5 nt window (Supplementary Figure S5B) yielded similar results for the PAM-distal half of the sgRNA (Supplementary Figure S7). The highest resolution of sgRNA activity was obtained in the window from position 4–8 (GC >25% versus <25%: 1.36-fold to 1.41-fold efficacy, Figure 3F). Application of this narrowed window size elevated the proportion of excludable low-efficacy sgRNAs to 11.2%.
Given the initial observation that genome editing efficacy varied based on which genomic origin (exon, intron, promoter) the sgRNA target sites were derived from, and considering known differences in GC-content in the genome (43,44), we wondered if the likelihood of retrieving sgRNAs with low GC1–10 or GC4–8 differs based on genomic origin. While the average GC1–10 and GC4–8 was within the optimal range for all genomic origins, we could observe a gradual and significant increase of GC1–10 and GC4–8 from intron- to exon- to promoter-derived sgRNAs, and a concomitantly decreased likelihood of retrieving sgRNAs with critically low (<25%) GC1–10 and GC4–8 (Supplementary Figure S8). In detail, the likelihood of retrieving a sgRNA with critically low GC1–10 was 6.5%, 2.3%, and 0% for intron-, exon- and promoter-derived sgRNAs, respectively. The likelihood of retrieving a sgRNA with critically low GC4–8 was 17.5%, 9.3%, and 1.9% for intron-, exon-, and promoter-derived sgRNAs, respectively. Thus, intron-derived sgRNAs are more likely to have a critically low GC1–10 and GC4–8, which may be caused by known differences in GC content within the genome (43,44).
Next, we sought to perform a biased search for genome editing activity-determining sequence features within defined sgRNA efficacy windows (Figure 4A). A similar approach has recently been successfully used to refine shRNA prediction (34). We tested different sgRNA efficacy windows to achieve the best separation of poorly- and well-performing sgRNAs, resulting in grouped sgRNA fractions with activities of <40% (sgRNAL) and >70% (sgRNAH) (Figure 4A), respectively. Comparison between the sgRNAH and sgRNAL groups again displayed a marked difference in GC-content spanning the PAM-distal nucleotides 4–13 (GC4–13) (Figure 4B).
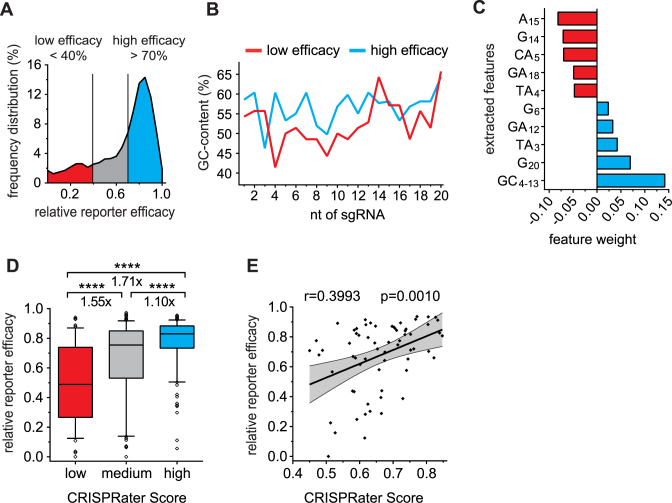
Figure 4.
CRISPRater: a 10-feature-based algorithm capable of predicting sgRNA activity via a discrete model. (A) Distribution of sgRNA efficacies within the reporter assay dataset. sgRNAs defined as low-efficiency (<40% efficacy, n = 70) and high-efficiency (>70%, n = 265) were segregated. (B) Average GC-content compared between high-efficiency and low-efficiency sgRNAs. The high-efficiency group displayed a higher overall GC-content from nt 4 to nt 13. (C) Most potent sequence features modulating sgRNA efficacy (five positively- and five negatively-modulating) extracted from 1024 features by individual feature weight. (D) Boxplot showing separation of the sgRNA dataset into low-efficiency (score <0.56, n = 52), medium-efficiency (score 0.56–0.74, n = 274) and high-efficiency (score >0.74, n = 100) groups according to discrete CRISPRater modeling, thereby excluding 12.2% of sgRNAs as low-efficiency (****P ≤ 0.0001 by use of Mann–Whitney test). (E) Validation of CRISPRater on 65 subsequently-designed sgRNAs not used to train the algorithm. The scatter plot shows a positive correlation between CRISPRater scores and measured sgRNA efficacies. Pearson correlation (r) and P value (p) are indicated.
In summary, our analyses strongly suggest that monitoring the GC-content in the PAM-distal 4–13 nt allows for the exclusion of the most inefficient sgRNAs. Together with an expanded repertoire for position 20 of the sgRNA (G/A versus G only), these features could be leveraged to enhance sgRNA library construction in the future.
Sequence logo examination outlines a PAM-distal GC-content led prediction model
To obtain a more complete picture of the features that shape genome editing efficacy, and to utilize them for rating prospective sgRNAs, we performed an expanded feature analysis that further tested single-base features including single nucleotide exclusion, and dual-base features. The predictive value of each potential feature was assessed by calculating a linear model and the respective root-mean-square-error (RMSE) as the mean over all sgRNAs, as has been performed before for shRNAs (34). After ranking on RMSE, we derived a set of ten potent predictors of sgRNA efficacy (Figure 4C): five features positively affecting sgRNA activity (GC4–13, weight = 0.14; G20, weight = 0.07; TA3, weight = 0.04; GA12, weight = 0.03; G6, weight = 0.02) and five features negatively affecting sgRNA efficacy (TA4, weight = –0.05; GA18, weight = –0.05; CA5, weight = –0.07; G14, weight = –0.07; A15, weight = –0.08). Notably, GC4–13 had a larger positive impact on sgRNA efficacy than the commonly-detected G20 (Figure 4C).
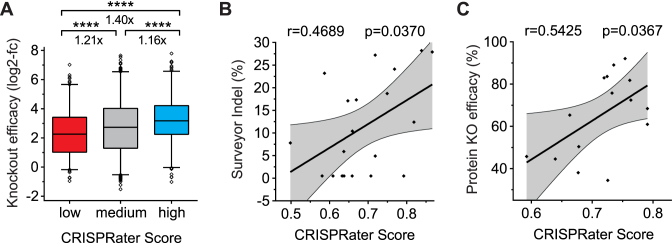
To obtain a model capable of rating sgRNAs based on efficacy (CRISPRater), the mean RMSE for all possible feature combinations was calculated and the best model with the lowest RMSE was selected. The overall difficulties with presenting sgRNA efficacy as a continuous model that we observed in our study prompted us to generate a discrete exclusion approach for the separation of low (<0.56), medium (0.56–0.74) and high (>0.74) efficacy sgRNAs (Figure 4D). CRISPRater proved to be superior at predicting sgRNA efficacy compared to the single feature of PAM-distal GC. Moreover, CRISPRater selected substantially more efficient sgRNAs than random selection (low versus medium: 1.55-fold; low versus high: 1.71-fold), while maintaining high dataset coverage (low-efficacy group: 12.2% of total). This we validated on an independently-tested set of 65 sgRNAs (Figure 4E). Notably, the prediction accuracy obtained by CRISPRater exceeds what we were able to achieve with established algorithms by 1.5–1.6-fold (Supplementary Figure S9; Figure 2B–F).
CRISPRater efficiently predicts sgRNA activity in small- and large-scale sgRNA datasets
For in-depth analysis of CRISPRater, we first selected independent large-scale datasets from various experimental sources generated by Wang et al. (21) and Koike-Yusa et al. (37), as derived by Xu et al. (27). Using the CRISPRater algorithm to predict efficient sgRNAs in a discrete manner yielded a highly significant selection of efficiently-acting sgRNAs—improving library design by 1.27-fold while retaining 91.2% of the libraries’ sgRNAs (combined medium and high efficiency groups), or by 1.19-fold while retaining 24.0% of the sgRNAs (high efficiency group only) (Figure 5A). Given that established sgRNA prediction algorithms are mainly limited to improving large-scale libraries, we aimed to test CRISPRater on independently established small datasets. We therefore retrieved the genome editing efficacy data of 20 sgRNAs tested for DNA modification via T7-EI (27), and of 15 sgRNAs tested for protein reduction (27), and we were able to show significant predictive value in both cases (Figure 5B–C).
Figure 5.
Predictivity validation of the CRISPRater score based on external experimentally-assessed cutting efficacies. (A) Validation of CRISPRater on a combined sgRNA dataset (n = 3141 sgRNAs) derived by Xu et al., proving its predictive capacity (****P ≤ 0.0001 by use of Mann–Whitney test). (B and C) Scatter plots showing correlations of CRISPRater predicted cutting efficacies (x-axes) and experimentally-tested efficacies from Xu et al. (27) (y-axes). Positive correlations can be seen with n = 20 efficacies tested on the genomic level via the Surveyor assay (B), and with n = 15 efficacies tested on the protein expression level (C). Pearson correlations (r) and P values (p) are indicated.
In summary, our studies resulted in a feature-based discrete model of sgRNA efficacy, providing an efficient and verified tool for future studies and enabling sgRNA selection processes for small and large-scale projects. Combined with established, powerful off-target prediction (33) our novel selection system is publicly available (http://crispr.cos.uni-heidelberg.de/) to help improving future genome editing efforts.
DISCUSSION
Despite increased application of the CRISPR–Cas9 system in basic research and translational approaches, guidelines for efficacy-orientated design of sgRNAs are still controversial. To investigate this issue, we utilized an optimized reporter system and individually analyzed the genome editing efficacies of 430 sgRNAs derived from various genomic origins. Based on this dataset we evaluated the performance of established sgRNA efficacy prediction algorithms, outlined novel molecular aspects that enable the discrete rating of sgRNAs efficacies, and provide an advanced online tool that predicts both on- and off-target sgRNA efficacies.
The biologically neutral readout of our reporter assay, its high concordance with the actual genomic cleavage efficacy, its reproducibility and its throughput demonstrate that it is a practical and verified method for focused evaluation of sgRNA efficacy. Meanwhile, the inherently high efficacy of the CRISPR–Cas9 system resulted in a low degree of separation of measured sgRNA activities, illustrating a major hurdle on the road towards accurate prediction of genome editing efficacy. In line with this challenge, retrospective analysis of established efficacy guidelines (22,24,26,27,29) on our experimentally-tested sgRNAs revealed that these prediction algorithms achieved only moderate benefit and reliability in the design of individual sgRNAs, despite having a significant impact on large-scale datasets. The difficulties of predicting sgRNA-mediated genome editing efficacy were further highlighted in cross-comparisons between the different algorithms, which again showed only moderate concordance. These results indicate a high degree of variability in the sgRNA features that were detected and incorporated into the different prediction models.
Aside from the high overall activity of the CRISPR–Cas9 system hampering a good separation of low and high activity sgRNAs, low cutting efficacies (24) or failure to avoid off-target effects (22) may have introduced biases in the datasets utilized to establish prediction rules, thus preventing unimpaired transfer of the underlying features to other datasets. These potentially confounding factors differ between the studies, and may have been amplified by the use of dissimilar Cas9/sgRNA delivery methods, possibly resulting in differences in genome editing kinetics or efficacy. In line with this, the use of different sgRNA backbones (47) may impact the ability to utilize features like sgRNA folding (30) for efficacy prediction, which we could not verify in our study (Supplementary Figure S10). Furthermore, the use of biological readouts like cell depletion (21,26) or drug resistance (29)—which can greatly vary between the chosen targets—may have added to the difficulties preventing more precise predictions of sgRNA efficacy. Notably, although genomic accessibility has been controversially discussed with regard to its impact on genome editing activity (24,26), it is unlikely to explain any variation in our study, given our use of the non-randomly integrating lentiviral reporter assay and its high correlation with cleavage at the respective endogenous locus. Further contrasting former studies, our data is characterized by high cleavage efficacy, prior scanning for off-target activity, and a biologically neutral readout on top of constitutive genomic accessibility, and may thus serve as a solid basis to establish more reliable prediction guidelines.
Since not only epigenetic state but also nucleotide content differs between functionally diverse regions of the genome (43,44), we additionally separated our sgRNA dataset by genomic origin of the target sites. Indeed, we could show that genomic origin is a major confounding factor for sgRNA activity in our dataset (efficacy: promoter > exonic > intronic). Upon interrogating sequence features that may explain these differences, we discovered that the PAM-distal—but not overall—GC-content of the sgRNA correlates with its efficacy, which we could track to an increased likelihood of retrieving sgRNAs with critically low PAM-distal GC content (<25%) when scanning intronic DNA regions. Monitoring of PAM-distal GC-content thus represents an easily-applicable method for excluding sgRNAs with the lowest activity level (11.3%, 1.36-fold activity; 3.3%, 1.8-fold activity), which may have been missed by studies testing for overall GC-content (21,22). In contrast, high overall GC-content (26) and PAM-proximal high GC-content (23) have both been found to positively affect sgRNA activity in non-mammalian systems, indicating species-specific requirements for optimal sgRNA activity. Of note, the proposed negative impact of PAM motifs within the sgRNA target region (25), which occur with increased likelihood in sgRNAs with higher GC-content, could not be verified in our dataset. This feature was only marked as a negative one by the sgRNA Designer rule set I algorithm, which also considers high GC-content as a negative marker of sgRNA efficacy (Supplementary Figure S11).
Finally, we performed linear model-based feature analysis of our sgRNA dataset, in order to extend the strength of our sgRNA efficacy prediction beyond PAM-distal GC. This resulted in a ten-feature model (CRISPRater). Given the existing issues with modeling sgRNA efficacy in a continuous manner, CRISPRater is primarily designed as a discrete selection tool. It is capable of rating sgRNA efficacy on small sets of sgRNAs -as well as large ones-, and thus has broad utility for a wide range of projects. Validation of CRISPRater on small- and large-scale external datasets resulted in an increase in efficacy of up to 1.4-fold, promising significantly improved signal-to-noise rates in future studies based on our work. These findings strongly support transferability to multiple experimental settings.
Despite these advances towards more uniform, highly active genome editing with the CRISPR–Cas9 system, our work also highlights the limitations of current genome editing predictions, as seen by the moderate predictivity of intermediate genome editing activity of all current algorithms. Studies like ours and others before (22,24,27–29) have mainly been focused on correlating sgRNA nucleotide composition to genome editing activity at the single nucleotide or small motif level. Larger studies with uniform, highly quantifiable detection systems will be needed to successfully evaluate the effects of larger sequence motifs or sgRNA secondary structures. Beyond the sgRNA component, Cas9 target finding, binding, and nuclease domain activation independent of a matching sgRNA may be factors not yet properly evaluated by current studies (7,8,17,18). At last, the understanding of factors that potentially affect genome editing activity at different loci and in different experimental settings may be challenged by new findings on sgRNA or Cas9 function, as well as on sgRNA–Cas9 interaction, all of which could expand the repertoire of variables to be included in future in silico predictions.
Apart from the benefits and limitations in silico prediction of genome editing activity provides for diverse applications, increased monitoring for off-target activities may be warranted with increased on-target activity—which we have partially addressed by implementing CRISPRater into the off-target evaluation platform CCTop (33). Furthermore, future studies may incorporate on-target activity predictions like CRISPRater into the evaluation of potential off-target sites, and thus their risk stratification.
As of yet, high-confidence prediction of genome editing efficacy remains a challenge. With what is currently an unique approach involving large-scale assessment of individual sgRNA efficacies at the single-cell level, and utilizing widely-applied delivery tools (31,32,48) in mammalian cells, we were able to develop and provide CRISPRater—(http://crispr.cos.uni-heidelberg.de/)—an efficient way to prospectively increase genome editing performance with concurrent prediction of potential off-target sites.
ACCESSION NUMBERS
Lentiviral vector constructs deposited at Addgene (#100893, #100894).
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Trono of EPFL, Lausanne, Switzerland, for kindly providing both pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Cancer Aid [111743 to D.H.]; European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program [714226 to J.H.K.]; DFG [HE-7482/1-1 to D.H.]; DFG [KL-2374/2-1 to J.H.K.]; Cluster of Excellence REBIRTH and SFB738 (DFG) (to F.F.A., A.Scha. and A.Schw.); Hannover Biomedical Research School (to M.L., M.N.). Funding for open access charge: DFG [HE-7482/1-1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R.. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013; 31:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M.. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013; 10:1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. In vivo genome editing using staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K.. Broadening the targeting range of staphylococcus aureus CRISPR–Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015; 33:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K.. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014; 32:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A.. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature. 2015; 527:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fellmann C., Zuber J., McJunkin K., Chang K., Malone C.D., Dickins R.A., Xu Q., Hengartner M.O., Elledge S.J., Hannon G.J. et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 2011; 41:733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pelossof R., Fairchild L., Huang C.H., Widmer C., Sreedharan V.T., Sinha N., Lai D.Y., Guan Y., Premsrirut P.K., Tschaharganeh D.F. et al. Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 2017; 35:350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR–Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q. et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in drosophila. Cell. Rep. 2014; 9:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chari R., Mali P., Moosburner M., Church G.M.. Unraveling CRISPR–Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015; 12:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malina A., Cameron C.J., Robert F., Blanchette M., Dostie J., Pelletier J.. PAM multiplicity marks genomic target sites as inhibitory to CRISPR–Cas9 editing. Nat. Commun. 2015; 6:10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J.. CRISPRscan: designing highly efficient sgRNAs for CRISPR–Cas9 targeting in vivo. Nat. Methods. 2015; 12:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015; 25:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong N., Liu W., Wang X.. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015; 16:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu V.T., Graf R., Wirtz T., Weber T., Favret J., Li X., Petsch K., Tran N.T., Sieweke M.H., Berek C. et al. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12514–12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L.. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR–Cas9 genome editing. Nat. Biotechnol. 2014; 32:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reimer J., Knoess S., Labuhn M., Charpentier E.M., Gohring G., Schlegelberger B., Klusmann J.H., Heckl D.. CRISPR–Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017; 102:1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L.. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015; 10:e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams F.F., Heckl D., Hoffmann T., Talbot S.R., Kloos A., Thol F., Heuser M., Zuber J., Schambach A., Schwarzer A.. An optimized lentiviral vector system for conditional RNAi and efficient cloning of microRNA embedded short hairpin RNA libraries. Biomaterials. 2017; 139:102–115. [DOI] [PubMed] [Google Scholar]
- 35. Vert J.P., Foveau N., Lajaunie C., Vandenbrouck Y.. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006; 7:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Efron B., Hastie T., Johnstone I., Tibshirani R.. Least angle regression. The Annals of Statistics. 2004; 32:407–499. [Google Scholar]
- 37. Koike-Yusa H., Li Y., Tan E.P., Velasco-Herrera Mdel C., Yusa K.. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014; 32:267–273. [DOI] [PubMed] [Google Scholar]
- 38. Pedelacq J.D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S.. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006; 24:79–88. [DOI] [PubMed] [Google Scholar]
- 39. Yuen G., Khan F.J., Gao S., Stommel J.M., Batchelor E., Wu X., Luo J.. CRISPR/Cas9-mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res. 2017; 45:12039–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014; 32:670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang G.P., Ciuffi A., Leipzig J., Berry C.C., Bushman F.D.. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007; 17:1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F.. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014; 9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pozzoli U., Menozzi G., Fumagalli M., Cereda M., Comi G.P., Cagliani R., Bresolin N., Sironi M.. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evol. Biol. 2008; 8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amit M., Donyo M., Hollander D., Goren A., Kim E., Gelfman S., Lev-Maor G., Burstein D., Schwartz S., Postolsky B. et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell. Rep. 2012; 1:543–556. [DOI] [PubMed] [Google Scholar]
- 45. O’Geen H., Henry I.M., Bhakta M.S., Meckler J.F., Segal D.J.. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015; 43:3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuscu C., Arslan S., Singh R., Thorpe J., Adli M.. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014; 32:677–683. [DOI] [PubMed] [Google Scholar]
- 47. Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/cas system. Cell. 2013; 155:1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.