Abstract
Post-transcriptional base modifications are important to the maturation process of transfer RNAs (tRNAs). Certain modifications are abundant and present at several positions in tRNA as for example the dihydrouridine, a modified base found in the three domains of life. Even though the function of dihydrourine is not well understood, its high content in tRNAs from psychrophilic bacteria or cancer cells obviously emphasizes a central role in cell adaptation. The reduction of uridine to dihydrouridine is catalyzed by a large family of flavoenzymes named dihydrouridine synthases (Dus). Prokaryotes have three Dus (A, B and C) wherein DusB is considered as an ancestral protein from which the two others derived via gene duplications. Here, we unequivocally established the complete substrate specificities of the three Escherichia coli Dus and solved the crystal structure of DusB, enabling for the first time an exhaustive structural comparison between these bacterial flavoenzymes. Based on our results, we propose an evolutionary scenario explaining how substrate specificities has been diversified from a single structural fold.
INTRODUCTION
Transfer RNAs (tRNAs) are key actors of the translational machinery. Transcribed into an inactive form, these molecules gain their functional state after numerous steps of maturation, including chemical modifications of their canonical bases (1,2). To date, more than a hundred different chemically altered bases have been identified, making the biology of tRNA the most diverse biochemical process (3). These modifications stabilize the peculiar tRNA L-shaped structure promoting efficient and specific interactions with its cellular partners such as aminoacyl-tRNA-synthetases, translation factors or mRNA (4–7). Beyond these classical functions, recent evidences have shown their implications in regulation of genetic expression and in cell adaptation to environmental changes (8).
Generated from uridine by a simple reduction of the C5 = C6 double bond, the dihydrouridine (D) is a quite unique base owing to its non-aromatic character, which makes it refractory to stacking interactions (9–13). As a result, D-containing nucleoside is more flexible promoting tertiary interactions essential for stabilizing the functional tRNA structure. The flexibility introduced by such a base appears to be important for the adaptability of organisms to their environment, as suggested by the fact that psychrophilic organisms contain on average more dihydrouridine per tRNA than thermophilic organisms (14,15). Similarly, cancer cells have a higher D-level than healthy cells probably to adapt to fast metabolism (16,17).
A broad family of flavin mononucleotide (FMN)-dependent enzymes, classified in eight subfamilies, and called tRNA-dihydrouridine synthases (Dus) catalyzes the biosynthesis of D (18). DusA, B and C are prokaryotic enzymes; Dus1, 2, 3 and 4 are eukaryotic and the last subgroup is the archaeal Dus. Considered as the oldest prokaryotic member, DusB is present in most bacteria whereas DusA and DusC, observed mainly in proteobacteria, originate from dusB duplications (18).
The D content and position vary according to the organism and tRNA species. Commonly observed in the so-called D-Loop at positions 16, 17, 20, 20a and 20b, it can also be found occasionally in the V-loop at position 47 (19,20), with positions 20b and 47 being specific to eukaryotes. While the substrate specificity of eukaryotic enzymes is established for Saccharomyces cerevisiae (D16-D17 synthesized by Dus1p, D20 by Dus2p, D20a/D20b by Dus4p and D47 by Dus3p) (20–22), the enzymatic specificity remains partially unsolved for prokaryotic proteins (19,23). In Escherichia coli, which carries the three Dus members, D16 and D20 are synthesized by DusC and A, respectively (19,24), while the enzymes responsible for the formation of D17 and D20a as well as the specificity of DusB remain to be identified. In contrast, Thermus thermophilus has a single Dus, belonging to the A-type, and is a bi-site-specific enzyme catalyzing D20 and 20a synthesis (25,26).
Recent crystallographic structures of T. thermophilus DusA and E. coli DusC in complex with a tRNA transcript have considerably improved our understanding on the exquisite substrate specificity of these RNA-modifying enzymes (24,26). These structures show that despite an overall conservation of the Dus fold (a N-terminal TIM-Barrel catalytic domain followed by a C-terminal helical domain, HD), DusA and DusC gain access to their respective uridine substrates by using two drastically distinct tRNA orientations (24,26). However, the absence of a DusB structure as well as the incomplete assignment of bacterial Dus substrate specificities prevent us from fully understanding the functional and structural evolution of these flavoenzymes. Here, we successfully address these issues leading us to propose a new tRNA binding mode in this RNA-modifying enzymes family.
MATERIALS AND METHODS
MS analysis of purified E. coli tRNAs
Bulk tRNA was extracted from E. coli strains BW25113 (F−, Δ(araD-araB)567, ΔlacZ4787::rrnB-3, λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514) and its derivatives ΔdusA743::kan, ΔdusB778::kan or ΔdusC767::kan. Cells were grown in 500 ml of LB (tryptone 10%, yeast extract 5%, NaCl 10 g l−1) to an OD600 of 0.8 and treated as previously described (21). Purification of ArgtRNAICG, IletRNAGAU and LeuRNACAG, was made with 5′ biotinylated complementary oligonucleotide (TGGTGCATCCGGGAGGATTCGAACCTCCGACCG, TGGTAGGCCTGAGTGGACTTGAACCACCG and GGACTTGAACCCCCACGTCCGTAAGGACACTAACACCTGAAGCTAGCG, respectively) coupled to Streptavidin Magnesphere Paramagnetic particles (Promega). Annealing of specific tRNA was made in 1 × TMA buffer (Tris–HCl pH 7.5 10 mM, ethylenediaminetetraacetic acid (EDTA) 0.1 mM, tetramethylammonium chloride 0.9 M) by heating the mixture at 95°C for 3 min and 60°C for 30 min. Paramagnetic particles were then washed three times with 1 × TMA buffer and specific tRNA was recovered by heating the final suspension at 95°C for 3 min. Specific tRNAs were desalted and concentrated four times to 50 μl in Vivaspin 500 devices (Sartorius; 3000 MWCO) using 100 mM ammonium acetate (pH 5.3) as a final buffer.
For mass spectrometry analysis, about 50 μg of tRNAs were digested with 10 μg of RNAse A (Euromedex), which cleaves after C and U and generates 3′-phosphate nucleosides, in a final volume of 10 μl at 37°C for 4 h. One microliter of digest was mixed with 9 μl HPA (40 mg/ml in water: acetonitrile 50:50) and 1 μl of the mixture was spotted on the MALDI plate and air-dried (‘dried droplet’ method) as described previously. MALDI-TOF MS analyses were performed directly on the digestion products using an UltrafleXtreme spectrometer (Bruker Daltonique, France). Acquisitions were performed in positive ion mode. An identical strategy was applied for RNase T1 digests (cleavage after G generating 3′-phosphate nucleosides).
Expression and purification of E. coli Dus
Escherichia coli DusA, DusB and DusC genes were cloned between BamHI and NcoI in a pET15b vector and expressed in BL21DE3. Cells were grown in LB medium with the addition of 200 μM riboflavin and induction was carried out with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at OD600 of 0.6 and left overnight at 29°C. Cells were centrifuged and lysis was carried out by sonication in 50 mM sodium phosphate pH 8 (NaP8), 2 M sodium chloride, 25 mM imidazole and 10% v/v glycerol with the addition of 10 mM β-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride (PMSF). Lysate was centrifuged for 40 min at 35 000 rpm and loaded on a NiNTA column (Qiagen) previously equilibrated with 50 mM NaP8, 2 M NaCl, 25 mM imidazole and 10% glycerol. Extensive washing with the same buffer was performed prior to elution in 50 mM NaP8, 200 mM NaCl, 10% glycerol and 250 mM imidazole. Concentrated protein was further purified by size exclusion chromatography on a HiLoad 16/600 Superdex 200 (GE Healthcare) equilibrated in 50 mM tris(hydroxymethyl)aminomethane pH 8 and 150 mM NaCl at 4°C. Purity of the proteins was assessed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Crystallization and structure determination
Initial crystals of DusB were obtained by mixing 1 μl of protein at 10 mg/ml in 50 mM Tris pH 8 and 150 mM NaCl with 1 μl of 1.6 M lithium sulfate and 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5 by vapor diffusion at 292 K. Due to poor diffraction quality, crystallization assays at 277 K were carried out and led to higher diffraction properties. Crystals were cryoprotected with a solution consisting of the reservoir supplemented with 15% v/v glycerol and flash frozen. X-ray diffraction data were collected at 100 K on a single crystal at PROXIMA-2a beamline at the SOLEIL synchrotron (Saint-Aubin, France) using an Eiger X-9M. Data were indexed and processed using XDS and corrected for anisotropy with the STARANISO server (staraniso.globalphasing.org). Structure solution was obtained by molecular replacement with Phaser using as a search template 2 monomers of E. coli DusC (PDB code: 4BFA) that shares 29.4% sequence identity with DusB. A clear solution was obtained (TFZ = 12.8; LLG = 154 723). Refinement was done with autoBUSTER using NCS and alternating with manual building in Coot. The entire HD was rebuilt manually into the density. One TLS group per chain was used at the end of the refinement.
Activity assay and dihydrouridine quantification
In vitro activity was assayed for 1 h at 30°C in 50 mM Tris pH 8, 150 mM ammonium acetate, 1 mM dithiothreitol (DTT), 2 mM MgCl2, 10% v/v glycerol. Bulk tRNAs (100 μM) were incubated with 5 μM of protein in a total volume of 50 μl and reaction was started with addition of 2 mM NADPH. Quenching was performed by adding 50 μl of acidic phenol (Sigma Aldrich) followed by centrifugation at 13 000 rpm for 10 min. tRNA in the aqueous phase were ethanol precipitated and further purified using a MicroSpin G-25 column (GE-healthcare). Dihydrouridine quantification was carried out by means of a colorimetric method as described previously (18). Briefly, samples were incubated at 40°C for 30 min after addition of 5 μl of 1 M KOH. The solutions were neutralized with 25 μl of 96% H2SO4 followed by 25 μl of a 3% solution 2,3-butanedione monoxime (Sigma Aldrich) and 25 μl of a saturated solution in N-Phenyl-p-phenylenediamine (Sigma-Aldrich). Samples were then heated at 95°C for 10 min and cooled to 55°C. Following addition of 50 μl of 1 mM FeCl3, a violet-red coloration appeared allowing quantification via absorption at 550 nm. Concentration of D in tRNA was determined by using a standard curve obtained with variable amounts of dihydrouracil.
Binding assay
Electrophoretic mobility shift assays were carried out for DusB using a 6% native (19:1) PAGE at 4°C with 100 V. Increased concentrations of proteins were added to a fixed concentration of bulk tRNA lacking D17 (1μM) and incubated at room temperature for 20 min in 50 mM Tris pH 8, 10% glycerol, 5 mM DTT, 150 mM ammonium acetate and 1 mM EDTA prior to migration. RNA was visualized by toluidine coloration.
SEC-MALLS
Analysis were done by size-exclusion chromatography (SEC) on a Superdex 200 10/300 GL (GE Healthcare) using a Shimadzu Prominence high-performance liquid chromatography (HPLC). Multi-angle laser light scattering (MALLS) was measured with a MiniDAWN TREOS equipped with a quasi-elastic light scattering module and a refractometer Optilab T-rEX (Wyatt Technology). Protein concentration was determined using a specific refractive index (dn/dc) of 0.183 at 658 nm.
SAXS data collection, analysis and model generation
Small-angle X-ray scattering (SAXS) experiments were performed at the SWING beamline at the SOLEIL synchrotron (Saint-Aubin, France) using an online HPLC. All scattering intensities were collected on the elution peaks after injection on a BioSEC-3 column (Agilent) equilibrated in 25 mM HEPES pH 8, 500 mM NaCl and 5% glycerol. Data were processed using FOXTROT (27). The radius of gyration (Rg) was calculated by the Guinier approximation.
Coral or bunch were used to construct the 19 missing residues (44–62) not observed in the X-ray structure (28,29). Ten independent runs in each case were averaged and compared to the experimental curve with crysol. Both approaches led to similar models.
Circular dichroism
Circular dichroism (CD) spectra and thermal denaturation of Dus proteins were recorded on a Chirascan™-plus CD Spectrometer (Applied Photophysics). The far ultraviolet spectra (190–260 nm) were measured at 20 and 90°C in quartz cells of 0.4 cm optical path length. The final concentration of Dus protein was 0.05 mg/ml in 25 mM potassium phosphate pH 8. Spectra were acquired at a resolution of 1 nm, with time per points set at 0.7 s and a bandwidth of 1 nm. All spectra were corrected from the contribution of the buffer and are an average of four accumulations. The thermal stability of the proteins was monitored by following the changes of CD at 222 nm from 25 to 90°C. Thermal unfolding curves were continuously scanned at a temperature ramping rates of 1.5 K min−1.
RESULTS
Unraveling substrate specificities of bacterial Dus
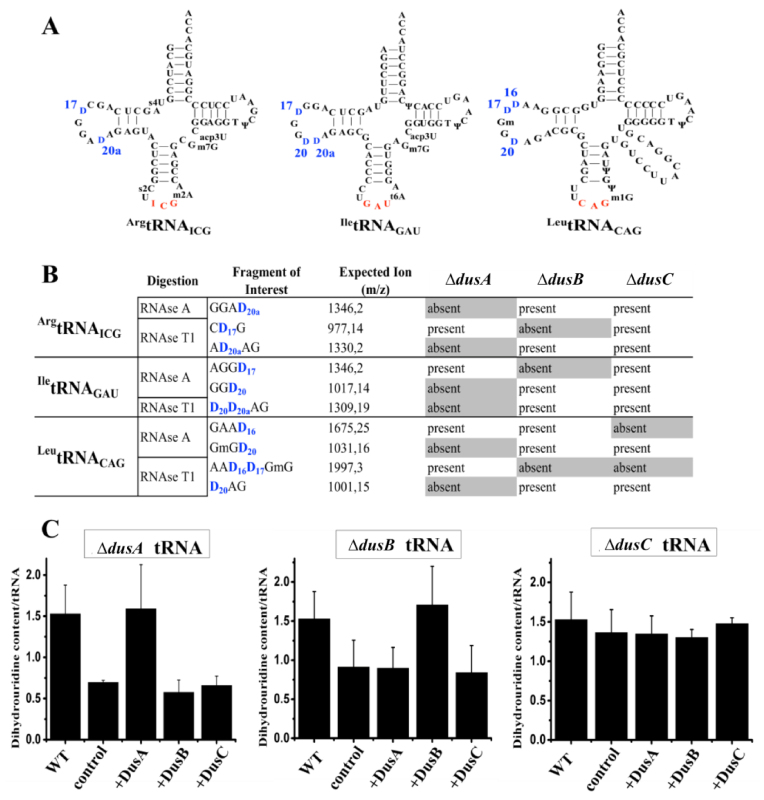
The incomplete determination of Dus specificities is mainly due to the absence of a reliable test for the detection of dihydrouridine. Although a classical methodology based on reverse transcriptase stop at hydrolyzed dihydrouridine site provides some information, it does not detect consecutive D. To circumvent this issue, we applied a new methodology combining (i) isolation and purification of specific tRNAs harboring various D by hybridization with a complementary biotinylated oligonucleotide from single E. coli dus knockout mutants, (ii) determination of D positions by MALDI-MS via analysis of RNA fragments resulting from RNAses (A or T1) digestion of the corresponding tRNA. Three tRNAs were specifically chosen to screen all the D sites (Figure 1A and Supplementary Figure S1).
Figure 1.
Identification of Escherichia coli Dus specificity from single knockout strains. (A) Secondary cloverleaf structure and sequence of E. coliArgtRNAICG, IletRNAGAU and LeutRNACAG used in this study. The anticodon is shown in red while the positions of D are labeled in blue. (B) Table showing the D-containing fragment and their sizes (m/z) after RNAse A or T1 digestion of ArgtRNAICG, IletRNAGAU or LeutRNACAG extracted from a wild-type E. coli strain. Presence or absence of these fragments is indicated for each tRNAs originated from ΔdusA, ΔdusB or ΔdusC E. coli strains. (C) In vitro quantification assay of D in E. coli tRNA. For each panel, WT corresponds to the amount of D per tRNA in wild-type bulk E. coli tRNAs. Left, middle and right panels show the amount of D per tRNA in ΔdusA, ΔdusB and ΔdusC bulk E. coli tRNAs, respectively, in the absence of added enzyme (control), or after incubation with the indicated enzymes. Error bars represent the standard error to the mean of three independent experiments.
As shown in Supplementary Figure S2, RNAse A digest of ArgtRNAICG generates the protonated GGAD20a fragment (m/z 1346), while treatment with RNAse T1 produces the protonated CD17G and AD20aAG fragments (m/z of 977 and 1330, respectively). Absence of D should induce a −2 Da shift in m/z value. RNAse A digest of ArgtRNAICG from ΔdusA strain leads to (i) a peak at m/z 1344 corresponding to both GGAU70 and GGAU20a fragments and (ii) to a loss of the peak at m/z 1346. With RNAse T1 two peaks are observed at m/z 977 and 1328 attributed to CD17G and AU20aAG, respectively (Supplementary Figure S2). In contrast, in ΔdusB, only the fragment m/z 977 undergoes a −2 Da due to the loss of D17 (Supplementary Figure S2C) while in ΔdusC, the three wild-type fragments are conserved (Supplementary Figure S2). Hence, these results suggest that DusA and DusB are involved in the biosynthesis of D20a and D17, respectively.
IletRNAGAU treated with RNAse A generates the protonated AGGD17 (m/z 1346) and GGD20 (m/z 1017) fragments whereas RNAse T1 generates a single fragment, D20D20aAG (m/z 1309) (Supplementary Figure S3). In ΔdusA, the m/z 1346 remains unaffected while the two other peaks are converted to a −2 and a −4 Da species attributed to GGU20 and U20U20aAG, respectively. In ΔdusB, the peak at m/z 1346 disappears and gives rise to an m/z 1344 fragment confirming the presence of AGGU17 (Supplementary Figure S3C). Furthermore, these three D-containing fragments (m/z 1346, 1017 and 1309) are unaffected in ΔdusC (Supplementary Figure S3). These results suggest that DusA is also involved in the D20 biosynthesis.
Finally, LeutRNACAG treated with RNAse A generates GAAD16 (m/z 1675) and GmGD20 (m/z 1031) while RNAse T1 generates AAD16D17GmG (m/z 1997) and D20AG (m/z 1001) (Supplementary Figure S4). All Δdus single mutants show an altered pattern. As expected, ΔdusA mutant does not convert U20 into D20 containing fragments (Supplementary Figure S4). In ΔdusB, only AAD16D17GmG loses a −2 Da and in ΔdusC, both m/z 1675 and 1997 loses −2 Da likely arising from the lack of D16 (Supplementary Figure S4).
Altogether these results summarized in Figure 1B show that DusC and DusB are mono-site specific flavoenzymes involved in D16 and D17 biosynthesis, respectively, while DusA is a U20/U20a bi-site specific enzyme.
Biochemical characterization of E. coli DusB and in vitro dihydrouridine synthase activity of bacterial Dus
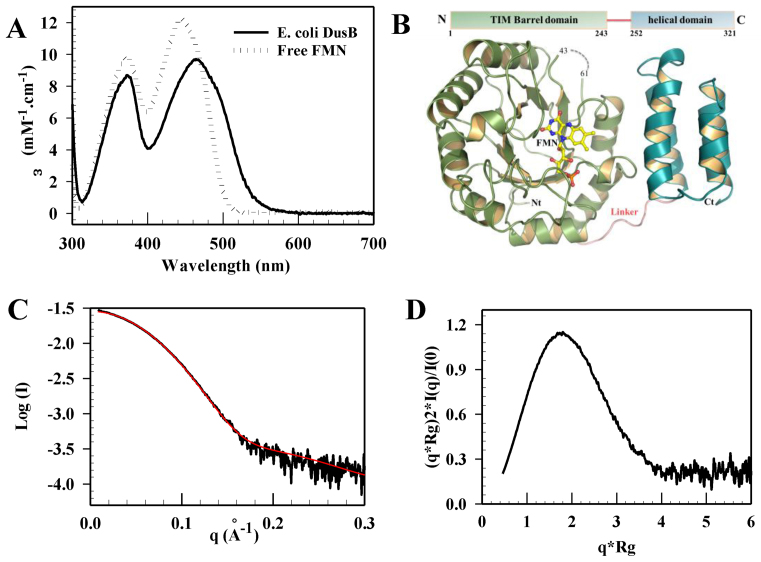
The absence of a DusB structure prevented us from understanding how this Dus subfamily specifically targets U17 position. We thus expressed, purified to homogeneity (Supplementary Figure S5A) and characterize E. coli DusB. Like its orthologs, the enzyme is a monomer with an experimental molecular weight of ∼34.7 kDa measured by SEC-MALLS (Supplementary Figure S5B). The absorption spectrum of DusB shows that the protein is isolated with a flavin derivative (Figure 2A). This spectrum presents peaks at 373 and 464 nm, characteristic of a protein-bound oxidized flavin. Addition of 0.1% SDS to the protein sample releases the flavin as evidenced by an absorption spectrum typical of free FMN. Therefore, DusB is a flavoprotein that binds FMN non-covalently.
Figure 2.
Spectroscopic and structural characterizations of Escherichia coli DusB. (A) UV-visible absorbance spectra of folded and SDS-induced unfolded E. coli DusB. (B) X-ray crystal structure of E. coli DusB. The protein is organized into two domains, a typical (α/β)8 Tim-barrel (in green cartoon) harboring the FMN cofactor at its center (yellow sticks) and an helical domain named HD (blue helix). The missing residues (44–62), not observed in the X-ray structure, are indicated as a dashed line. (C) Superposition of the experimental (black) and theoretical scattering curves (red) computed by crysol using a DusB model obtained from coral or bunch. (D) Normalized Kratky plot of E. coli DusB.
We also tested in vitro activities of the three recombinant E. coli DusA, B and C. Each flavoenzyme restores the native dihydrouridine level only when mixed with bulk tRNAs originating from the corresponding Δdus strain. In agreement with the in vivo results, this demonstrates that all Dus enzymes carry out non redundant enzymatic activity (Figure 1C).
Crystal structure and conformational dynamic characterization of E. coli DusB
The crystal structure of E. coli DusB was solved by molecular replacement at 2.5 Å using E. coli DusC as search template (Supplementary Table S1). The asymmetric unit contains two identical DusB molecules (RMSD = 0.18 over 242 Cα) (Supplementary Figure S6A) resulting from the crystal packing since the protein is monomeric in solution (Supplementary Figure S5B). DusB is organized into two domains (Figure 2B). The N-terminal domain comprising residues 1–243 adopts a classical (α/β)8-Tim barrel fold. A clear density attributed to non-covalently bound FMN lies in the center of the β-barrel, which is consistent with the catalytic function of this domain (Supplementary Figure S6B). The residues S43 to I61 located between β3–β4 are disordered. A linker of eight amino acids directly connects the last helix of the catalytic domain to helix 1 of the C-terminal HD formed by four parallel helix bundle.
Spatial orientation of both domains is maintained throughout extensive buried interfaces of 685 Å2. This interface involves mainly hydrophobic interactions with residues located in helix α1 and α8–β8 loop of the catalytic domain and helix 1 and 2 of HD (Supplementary Figure S7). Curiously, the relevance of such orientation in solution has never been investigated so far. We therefore performed small-angle X-ray scattering coupled to SEC-SAXS experiments on E. coli DusB. The resulting scattering curve is shown in Figure 2C. Curve analysis using Guinier approximation leads to a gyration radius ∼23.3 Å (Supplementary Figure S8A). The normalized Kratky plot indicates that DusB is globular and carries a compact conformation (Figure 2D and Supplementary Figure S8B). The crystal structure being incomplete, we generated the 19 missing amino acids between S43 and I61 using bunch or coral (both gave similar results). As shown in Figure 2C, the theoretical SAXS curve of the resulting model fits well the experimental data with a χ2 ∼1.8. This indicates that the crystal structure of monomeric DusB is a relevant form in solution.
Structural similarities and divergences between the three bacterial Dus paralogs
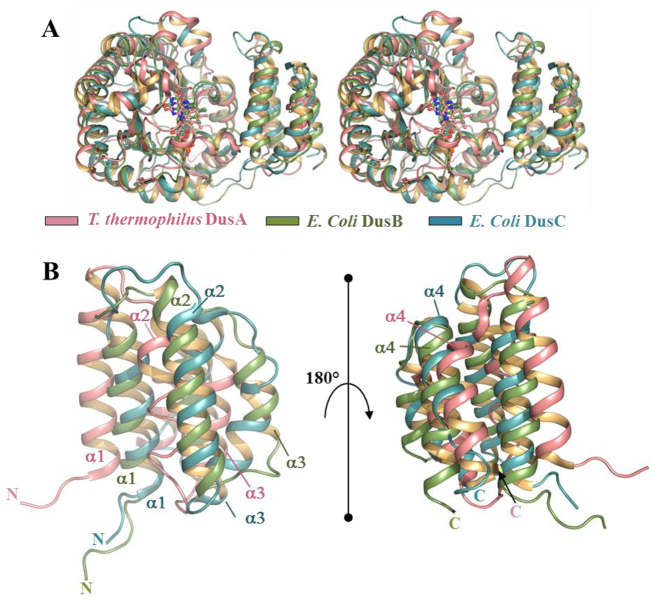
We generated a structural alignment between E. coli DusB, E coli DusC and T. thermophilus DusA (Figure 3A). The catalytic domains align globally well between the three enzymes (RMSD A versus B = 1.15 for 162 Cα and RMSD B versus C = 0.88 for 153 Cα). A significant divergence between DusA on the one hand and DusB and DusC on the other hand appears at the C-terminus of the Tim barrel, resulting in different linker orientations. The structural divergence is even more prominent between the HD of DusB and DusA (Figure 3B) (RMSD 7.09 over 50 Cα). In contrast, the HD of DusC and DusB are structurally similar (RMSD 3.89 over 62 Cα). Given that this domain is a major tRNA recognition module in this enzyme family (24,26), the tRNA binding mode of DusB should be close to that of DusC (see below).
Figure 3.
Structural alignment between bacterial Dus paralogs. (A) Stereo view of a structural alignment between Thermus thermophilus DusA (3B0P), Escherichia coli DusB (this study) and E. coli DusC (4BFA) in pink, green and blue, respectively. (B) Zoom on two different orientations of the structural alignment of T. thermophilus DusA, E. coli DusB and E. coli DusC HD.
The thermal stability of E. coli Dus followed by the CD at 222 nm corresponding to the α-helix signal versus temperature also offers additional evidence that DusB and C are closely related in terms of structural stability. As shown in Supplementary Figure S9, the three Dus are thermostable proteins since at 90°C they retain a significant amount of secondary structures. One can note that the thermal stability can be classified from the least stable to the most stable as follows: DusB < DusC < DusA, reminiscent of the evolutionary history of prokaryotic Dus enzymes.
Analysis of the active site of bacterial Dus enzymes
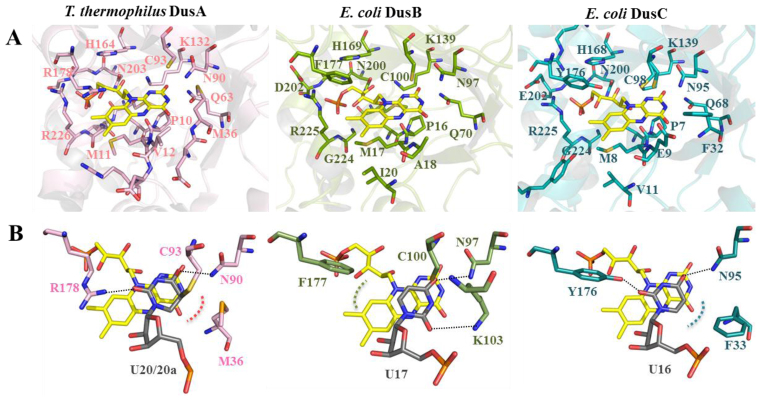
Most of the strictly conserved active site residues in Dus enzymes are in interaction with FMN (Figure 4A). For instance in E. coli DusB, OE1 and NE2 of Q70 (Q68 in E. coli DusC and Q63 in T. thermophilus DusA) make hydrogen bond with N3 and O2’ of the isoalloxazine ring, respectively. In a similar way, NZ of the conserved K139 (K139 in E. coli DusC and K132 in T. thermophilus DusA) interacts with O2’ of the isoalloxazine. This conserved network of hydrogen bonds around the pyrimidine moiety likely contributes to the stabilization of the anionic hydroquinone, FMNH−, formed during the catalytic cycle. Hydrophobic residues (P16 and M17 in DusB, P8 and M8 in DusC, P10 and M11 in DusA) and a portion of the amide bond chain interact with the re-face of the isoalloxazine. The ribityl-phosphate moiety is stabilized by several ionic and hydrophobic conserved interactions involving H169, residues N200 to D202, G224 and R225 in E. coli DusB. These interactions involving similar residues are also present in DusA and C (Figure 4A). Interestingly, Sγ of the catalytic cysteine, considered as a general acid, is located above the si-face of FMN at 6.4, 4.4 and 4 Å from the N5 of the isoalloxazine ring of DusA, B and C, respectively. This cysteine has been proposed to protonate the reduced nucleobase enolate intermediate generated during the modification reaction (9).
Figure 4.
Structure of Dus active site and uridine binding mode. (A) Structure of the FMN binding sites of Thermus thermophilus DusA (3B0P), Escherichia coli DusB (this study) and E. coli DusC (4BFA) in pink, green and blue, respectively. Residues involved in polar contacts with FMN (yellow stick) are shown as sticks. (B) Uridine binding mode in Dus active site as seen in the crystal structures of T. thermophilus DusA and E. coli DusC in complex with tRNA. In the case of E. coli DusB, the uridine was placed manually.
Evidence for a new uridine binding mechanism in Dus family
Interestingly, in the structure of Dus/tRNA complexes, several conserved interactions stabilize the C6-uracil in a competent orientation for receiving a hydride anion from N5-FMNH− (24,26) (Figure 4B). The first one involves an ionic interaction between a strictly conserved asparagine side chain and the uracil C = O4’ (N90, 97 and 95 in DusA, B and C, respectively). The second one involves an ionic interaction between the side chain of a polar residue and the uracil C = O2’ carbonyl group (R178 in DusA and Y176 in DusC). In addition to these two ionic interactions, we have noticed that in DusA and DusC, the C5 = C6 edge of the pyrimidine is in hydrophobic interaction with M36 and F33, respectively. Remarkably, in DusB, the residue corresponding to R178 in DusA and Y176 in DusC is the hydrophobic F177 residue. In addition, a positively charged residue K103 occupies the space corresponding to M36 in DusA and F33 in DusC. Thus, in E. coli DusB the active site polarity is inverted and appears to be a feature specific to this bacterial Dus subfamily (see alignments Supplementary Figures S10–12). This prompted us to verify how a uridine could be placed within DusB active site. Interestingly, when we manually place a uridine adopting the same orientation as in DusA or DusC, the phenyl group of F177 is in the vicinity of C = O2’ carbonyl group of uracil, an energetically unfavorable configuration. However, when we rotate the base by 180° around the N1-C1 glycosidic bond, C = O2’ becomes perfectly poised to make a hydrogen bond with K103 side chain and furthermore, the uracil C5 = C6 edge is in hydrophobic interaction with F177 (Figure 4B). This orientation is more favorable as it allows to recover the three peculiar interactions stabilizing the nucleobase (namely, two ionic interactions, N97:C = O4’, K103: C = O2’, and one hydrophobic interaction, F177:C5 = C6). Moreover, such orientation would not affect the hydride transfer because the distance between N5-FMN and C6-uracil is similar to that observed in DusA and DusC.
tRNA binding mode in Dus family
Electrophoretic mobility shift assay shows that DusB is able to bind tRNA by itself (Supplementary Figure S13A) but in contrast to DusA (26), it does not form a covalent complex between the strictly conserved catalytic cysteine and the uridine substrate.
As shown in Supplementary Figure S13B, DusC-oriented tRNA is well adapted to the positive patches on the surface of DusB that appears complementary in shape and in charge to the tRNA. Such an arrangement does not produce clashes between the protein and the tRNA, except for residues K104 and R107, and most importantly U17 is located not far from the N5-FMN (at ∼8 Å). In drastic contrast, DusA-oriented tRNA produces numerous clashes mainly with the HD (from Y286 to N301) (Supplementary Figure S13C) and U17 is found outside the active site, with N5-FMN located at more than 18 Å away from C6-uracil.
Thus, with slight changes in conformation of the protein and/or DusC-oriented tRNA to eliminate clashes, the overall orientation of tRNA on DusB should likely be close to that of DusC.
DISCUSSION
Our structural and functional characterization of bacterial Dus explain how these flavoenzymes achieve synthesis of D at spatially distinct positions with an overall conserved protein architecture but with, nonetheless, specific peculiarities. This was possible because (i) we unveiled the ambiguity on their substrate specificity and (ii) we obtained the first crystal structure of a DusB member.
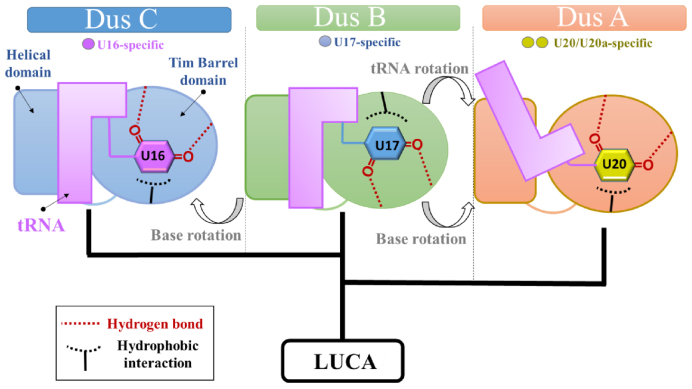
Indeed, we established that DusB and DusC are functionally and structurally more related to each other than to DusA. Firstly, DusB and DusC are mono-site specific enzymes catalyzing formation of two adjacent dihydrouridines, D17 and D16, respectively while DusA is a U20/U20a bispecific enzyme (Figure 1). This contrasts with the specificity pattern of S. cerevisiae enzymes where D16/D17 are both synthesized by Dus1p, D20 by Dus2p and D20a/D20b by Dus4p (20). As Dus1 and Dus4 derived from Dus2, it has been proposed that the mono-site specificity is an evolutionary older feature than the bi-site specificity (18). Our results strengthen this hypothesis as DusA derives from DusB. Second, DusB and DusC likely share a similar tRNA binding mechanism as inferred from structural similarities between (i) their HD (Figure 3) and (ii) their positive electrostatic surface delimiting the RNA binding site (Supplementary Figure S12).
The crystal structures of DusA and DusC in complex with a tRNA substrate show that despite their different tRNA recognition modes, the catalytic sites of both enzymes have a similar polarity and bind the target uridine in the same orientation (24,26). Additionally, the N + 1 base (N being the target base) is often flipped from its initial position to facilitate stacking of uridine against FMN without major tRNA conformational changes (24,26). It is unlikely that such base flipping occurs with DusB since its U17 substrate is invariably followed by G18 involved in an interaction network between the D- and T-loops (G18-Ψ55, T54-A58 reverse-Hoogsteen, G19-C56) to maintain the L-shaped conformation. In addition, G18 also participates in the complex purine base intercalation comprising of A58-G18/G57-G19. Therefore, a flipping of any of these bases, including G18, would have major consequences on tRNA structure. Such conformational changes have been recently observed in the crystal structure of human m1A58 methyltransferase (m1A58 MTase) in complex with tRNA. To gain access and methylate N1-A58 buried within the tRNA TψC-loop, m1A58 MTase dramatically distorts the tRNA structure splaying apart the D- and T-loops (30). As m1A58 MTase is a tetramer, the resulting large tRNA binding interface can compensate for the energy cost caused by the rupture of tRNA elbow structure. DusB is a monomer and its tRNA binding interface is not larger than that of DusC, making such drastic conformational change of tRNA unlikely to occur. However, a 180° rotation of the uracil around U17glycosidic bond could avoid flipping of G18, eventually minimizing tRNA structural changes induced by DusB. Such orientation of the uracil could be stable enough in DusB active site as the latter carries an inverted polarity compared to that of both DusA and DusC (Figure 4B).
Altogether, our results lead us to propose an evolutionary scenario for adapting various Dus functional specificities to a similar protein structure (Figure 5). DusB is considered as the common ancestor of all bacterial Dus. Thus, the transition from DusB to DusC did not require a major tRNA reorientation since the target base is adjacent to that of DusB. Additionally, the N + 1 not being involved in tertiary interactions, recognition of U16 by DusC could be relaxed allowing a change of the orientation of the base via a reversal of the active site polarity. On the other hand, the bi-specificity of DusA was generated by reorientation not only of the base but also of the tRNA enabling the modification of uridines on opposite sides of the D-loop. To our knowledge, such evolutionary strategy remains unprecedented in RNA enzymology.
Figure 5.
Proposed evolutionary scenario for the structural and functional differences between bacterial Dus enzymes.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure of DusB from E. coli has been deposited with the Protein Data bank under accession number 6EI9.
Supplementary Material
ACKNOWLEDGEMENTS
Use of the crystallography laboratory at IBPC is acknowledged. This work has benefited from the facilities and expertise of the Macromolecular Interaction Platform of I2BC. We acknowledge SOLEIL for provision of synchrotron radiation facilities (proposals ID20160782) in using proxima beamlines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; University Pierre et Marie Curie (Emergence Program); French State Program ‘Investissements d’Avenir’ [ANR-15-CE11-0004-01 to D.H.]. Funding for open access charge: French Government (ANR funding).
Conflict of interest statement. None declared.
REFERENCES
- 1. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El Yacoubi B., Bailly M., de Crecy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 3. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. . MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gustilo E.M., Vendeix F.A., Agris P.F.. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008; 11:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 6. Duechler M., Leszczynska G., Sochacka E., Nawrot B.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life. Sci. 2016; 73:3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lorenz C., Lunse C.E., Morl M.. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017; 7:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu C., Begley T.J., Dedon P.C.. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014; 588:4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rider L.W., Ottosen M.B., Gattis S.G., Palfey B.A.. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J. Biol. Chem. 2009; 284:10324–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lombard M., Hamdane D.. Flavin-dependent epitranscriptomic world. Arch. Biochem. Biophys. 2017; 632:28–40. [DOI] [PubMed] [Google Scholar]
- 11. Dalluge J.J., Hashizume T., Sopchik A.E., McCloskey J.A., Davis D.R.. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996; 24:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyubankova N., Sochacka E., Kraszewska K., Nawrot B., Herdewijn P., Lescrinier E.. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem. 2015; 13:4960–4966. [DOI] [PubMed] [Google Scholar]
- 13. Suck D., Saenger W., Zechmeister K.. Conformation of the tRNA minor constituent dihydrouridine. FEBS Lett. 1971; 12:257–259. [DOI] [PubMed] [Google Scholar]
- 14. Edmonds C.G., Crain P.F., Gupta R., Hashizume T., Hocart C.H., Kowalak J.A., Pomerantz S.C., Stetter K.O., McCloskey J.A.. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 1991; 173:3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dalluge J.J., Hamamoto T., Horikoshi K., Morita R.Y., Stetter K.O., McCloskey J.A.. Posttranscriptional modification of tRNA in psychrophilic bacteria. J. Bacteriol. 1997; 179:1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuchino Y., Borek E.. Tumour-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature. 1978; 271:126–129. [DOI] [PubMed] [Google Scholar]
- 17. Kato T., Daigo Y., Hayama S., Ishikawa N., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y.. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005; 65:5638–5646. [DOI] [PubMed] [Google Scholar]
- 18. Kasprzak J.M., Czerwoniec A., Bujnicki J.M.. Molecular evolution of dihydrouridine synthases. BMC Bioinformatics. 2012; 13:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bishop A.C., Xu J., Johnson R.C., Schimmel P., de Crecy-Lagard V.. Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 2002; 277:25090–25095. [DOI] [PubMed] [Google Scholar]
- 20. Xing F., Hiley S.L., Hughes T.R., Phizicky E.M.. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004; 279:17850–17860. [DOI] [PubMed] [Google Scholar]
- 21. Bou-Nader C., Pecqueur L., Bregeon D., Kamah A., Guerineau V., Golinelli-Pimpaneau B., Guimaraes B.G., Fontecave M., Hamdane D.. An extended dsRBD is required for post-transcriptional modification in human tRNAs. Nucleic Acids Res. 2015; 43:9446–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Betteridge T., Liu H., Gamper H., Kirillov S., Cooperman B.S., Hou Y.M.. Fluorescent labeling of tRNAs for dynamics experiments. RNA. 2007; 13:1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savage D.F., de Crecy-Lagard V., Bishop A.C.. Molecular determinants of dihydrouridine synthase activity. FEBS Lett. 2006; 580:5198–5202. [DOI] [PubMed] [Google Scholar]
- 24. Byrne R.T., Jenkins H.T., Peters D.T., Whelan F., Stowell J., Aziz N., Kasatsky P., Rodnina M.V., Koonin E.V., Konevega A.L. et al. . Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6033–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kusuba H., Yoshida T., Iwasaki E., Awai T., Kazayama A., Hirata A., Tomikawa C., Yamagami R., Hori H.. In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium. J. Biochem. 2015; 158:513–521. [DOI] [PubMed] [Google Scholar]
- 26. Yu F., Tanaka Y., Yamashita K., Suzuki T., Nakamura A., Hirano N., Yao M., Tanaka I.. Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:19593–19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. David G., Perez J.. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009; 42:892–900. [Google Scholar]
- 28. Petoukhov M.V., Svergun D.I.. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005; 89:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D., Konarev P.V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finer-Moore J., Czudnochowski N., O’Connell J.D. 3rd, Wang A.L., Stroud R.M.. Crystal structure of the human tRNA m(1)A58 Methyltransferase-tRNA(3)(Lys) complex: refolding of substrate tRNA allows access to the methylation target. J. Mol. Biol. 2015; 427:3862–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure of DusB from E. coli has been deposited with the Protein Data bank under accession number 6EI9.