Abstract
Rpc82 is a TFIIE-related subunit of the eukaryotic RNA polymerase III (pol III) complex. Rpc82 contains four winged-helix (WH) domains and a C-terminal coiled-coil domain. Structural resolution of the pol III complex indicated that Rpc82 anchors on the clamp domain of the pol III cleft to interact with the duplex DNA downstream of the transcription bubble. However, whether Rpc82 interacts with a transcription factor is still not known. Here, we report that a structurally disordered insertion in the third WH domain of Rpc82 is important for cell growth and in vitro transcription activity. Site-specific photo-crosslinking analysis indicated that the WH3 insertion interacts with the TFIIB-related transcription factor Brf1 within the pre-initiation complex (PIC). Moreover, crosslinking and hydroxyl radical probing analyses revealed Rpc82 interactions with the upstream DNA and the protrusion and wall domains of the pol III cleft. Our genetic and biochemical analyses thus provide new molecular insights into the function of Rpc82 in pol III transcription.
INTRODUCTION
Eukaryotic RNA polymerase III (pol III) is responsible for transcribing transfer RNAs (tRNA), 5S ribosomal RNA, small nuclear RNAs such as U6 and 7SK RNAs, and a number of small nucleolar and microRNAs (1,2). The pol III complex contains a 12-subunit core structure. Its two largest subunits, Rpc160 and Rpc128, form the DNA-binding cleft, and the remaining 10 smaller subunits of the core are located at its periphery (3). The overall structural arrangement of the core is similar to the structures of pol II and the core structure of pol I (4–7). In addition to its core, pol III contains two specific subcomplexes: the Rpc82/34/31 trimer and the Rpc53/37 dimer. A previous analysis suggested that the pol III-specific subunits act as basal transcription factors that are permanently recruited to the polymerase core (8).
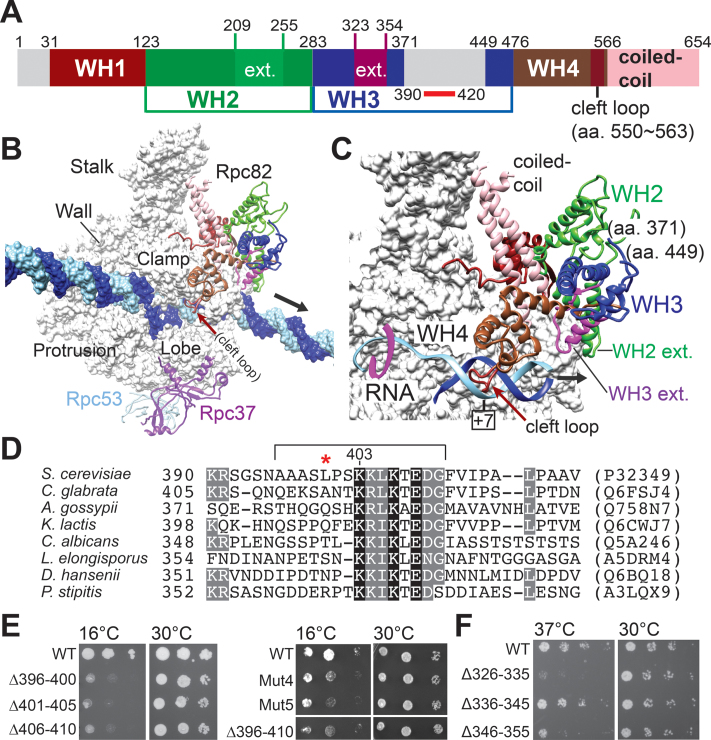
Both Rpc34 and Rpc82 contain multiple copies of a significant structural fold, i.e. the winged-helix (WH) domain. WH domains are also present in the transcription factors of eukaryotic and archaeal transcription machineries, such as TFIIE, TFIIF and TFE (5,8–10). The WH domain is essential for protein-DNA and protein-protein interactions during the transcription process (11). Rpc34 contains three WHs, with the first two WHs (WH1&2) generally being compared to the tandem WH domain of the Rpa49 subunit of pol I and to the TFIIEβ subunit of TFIIE (12–14). Rpc82 (and its human counterpart hRPC62) contains four copies of the WH domain (WH1–4) from the N-terminus end and a coiled-coil domain at the C-terminus (Figure 1A) (3,10). Rpc82 WH domains are related to the extended WH (eWH) fold first described for the archaeal transcription factor, TFE (15). The eWH fold is also present as a single copy in the TFIIEα subunit of TFIIE. In recent cryo-electron microscopy (EM) structures of the free and elongating pol III complexes (Figure 1B), Rpc82 anchors on the clamp domain of Rpc160 mainly through its WH1 and WH4 domains, which is consistent with a previous protein crosslinking analysis (3,16,17). This localization also allows the WH4 domain to contact with the duplex DNA downstream of the DNA bubble in the pol III cleft (Figure 1C). In addition, both WH2 and WH3 domains contain helical extensions oriented towards the duplex DNA entry face of the pol III active site cleft (Figure 1A and C).
Figure 1.
A functionally important insertion in the WH3 domain of Rpc82. (A) Structural domains of Rpc82 are colored differently. WH: winged-helix domain. Gray colored regions are structurally flexible. Approximate position of a conserved sequence block within the insertion of WH3 is marked with a red bar under the schematic. ext.: helical extensions within the WH2 and WH3 domains. (B) The elongating pol III complex. The structure model is derived from the coordinate file 5FJ8 (PDB) published by the Müller group (3). The molecular surface of the 12-subunit core is colored in gray. Template and non-template DNA strands are colored in light blue and dark blue, respectively. Rpc82, Rpc37 and Rpc53 are shown, with ribbons representing their backbone traces. Rpc82 is colored based on the structural domains in the schematic in (A). An arrow points to the downstream DNA. (C) An enhanced view of Rpc82 in the pol III structure. As indicated, an insertion loop between amino acid residues 371 and 449 of the WH3 domain is not resolved. In order to visualize the downstream duplex DNA (displayed as the phosphate backbone trace model), the molecular surface of pol III has been partially removed. Helical extensions (ext.) of WH2 and WH3 domains are indicated. (D) Multiple sequence alignment of the conserved sequence block. The lysine-rich sequence block of amino acids 396–410 (S. cerevisiae) subjected to mutational analysis is bracketed. Uniprot accession numbers are listed on the right (in parentheses). A red asterisk indicates the amino acid position Leu400 involved in BPA crosslinking with Brf1 (see below). (E) Cell growth of the yeast strains with mutations in the WH3 insertion. A series of deletion and alanine replacement mutants were analyzed by serial dilution spot assays. Mut4 mutant: alanine replacement in residues K403K404L405K406; Mut5 mutant: alanine replacement in residues K403K404L405K406E408D409. (F) Cell growth of the yeast strains with mutations in the WH3 helical extension.
Functional roles of the Rpc82/34/31 and Rpc53/37 subcomplexes in pol III transcription have been derived from their respective locations on the pol III DNA-binding cleft. For example, the Rpc53/37 dimerization domain is positioned on the Rpc128 lobe/external surface above the active site cleft, and previous functional analyses have revealed the importance of this subcomplex in transcription initiation, termination and reinitiation (18–20). In particular, an Rpc37 C-terminal loop regulates transcription termination, possibly through an interaction with the non-template strand of the DNA bubble (18,21). Functional analyses have also revealed the importance of the Rpc82/34/31 subcomplex in transcription initiation (22). The Rpc34 subunit of this subcomplex contributes to promoter DNA melting, as well as interacts with the TFIIB-related Brf1 subunit of the transcription factor TFIIIB within the pre-initiation complex (PIC) (23–25). The functional role of Rpc34 in DNA interaction has been explained based on the localization of its tandem WH domain above the polymerase active site cleft (17,26,27). Although cryo-EM structures have revealed the structural localization of Rpc82 in the elongating pol III structure, it remains unclear how exactly Rpc82 functions in the transcription initiation process.
To provide functional insights, we conducted a mutational analysis on yeast Rpc82. Surprisingly, we found that a structurally disordered insertion in the WH3 domain is indispensable for cell viability. Small internal deletions and point mutations in the insertion also impaired cell growth, which was attributable to defective pol III transcription activity. We site-specifically incorporated a non-natural amino acid photo-crosslinker into Rpc82 and identified a site within the WH3 insertion that is involved in binding to Brf1 in the PIC. Additional biochemical analyses also indicated that Rpc82 interacts with the Rpc128 protrusion and wall domains of the pol III cleft and the DNA region upstream of the transcription start site. Our study thus provides an Rpc82 interaction network within the pol III cleft crucial to transcription initiation.
MATERIALS AND METHODS
Yeast strain and plasmids
The yeast (Saccharomyces cerevisiae) strains used in this study were derived from BY4705 (28), with chromosomal disruptions of individual genes introduced by the KanMX4 cassette, yielding the following yeast strains: RPC82 shuffle strain YLy2 (MATα ade2::his3G his3Δ200 leu2Δ0 met15Δ0 lys2Δ0 trp1Δ63 ura3Δ0 [C82::KanMX4] RPC82-pRS316(URA3+)) and RPC128 shuffle strain YLy1 (MATα ade2::his3G his3Δ200 leu2Δmet15Δ lys2Δ trp1Δ63 ura3Δ [rpc2::KanMX4] RPC128-pRS316(URA3+)).
For our Rpc82 and Rpc128 mutagenesis study, the genes along with their respective endogenous promoters were separately cloned into the vector pRS315 with a single HA-epitope tag at the C-terminus, yielding pYL4 (RPC82-HA, ars cen, LEU2) and pYL2 (RPC128, ars cen, LEU2). All Rpc82 and Rpc128 mutant plasmids were based on pYL4 and pYL2, respectively. Rpc82 wild-type (WT) and mutant plasmids were transformed into the RPC82 shuffle strain (YLy2). Mutant strains additionally hosted a Flag epitope at the C-terminus of RPC128 and a V5 epitope at the C-terminus of Rpc31. Rpc128 WT and mutant plasmids were transformed into the RPC128 shuffle strain (YLy1). Rpc128-associated strains also hosted a Flag epitope at the C-terminus of RPC82. Rpc82 and Rpc128 mutant strains were separately generated using the 5-FOA drop-out method to replace the URA3-marked plasmid with LEU2-marked plasmids containing mutant gene copies. For cell growth assays, both the WT and mutant strains were grown in YPD to an optical density (600 nm) of 1.0 and the cell cultures were subsequently diluted 10−1 to 10−4. The diluted cells were spotted onto synthetic complete glucose plates lacking leucine, and their growth phenotypes were monitored at 16, 25, 30 and 37°C. The incubation time for cell growth at 25, 30, and 37°C was 3 days and at 16°C it was 7 days.
The non-natural amino acid p-benzoyl-l-phenylalanine (BPA; Bachem) was used for photo-crosslinking studies. Rpc82 and Rpc128 were separately cloned into the yeast 2-micron vector pRS425 with the LEU2 selection marker (29). Both genes were driven by the yeast ADH1 promoter. The C-termini of Rpc82 and Rpc128 respectively contained a V5-epitope and 13 copies of Myc epitopes. Plasmids are referred to as pYL3 (Adh1-Rpc82 C-ter V5-pRS425) and pYL1 (Adh1-Rpc128 C-ter 13Myc-pRS425), respectively. To generate individual plasmids for subsequent BPA incorporations into Rpc82 and Rpc128, the ‘TAG’ (amber) nonsense codon was introduced into the above plasmids at intended amino acid positions. The resulting plasmids, referred to as ‘amber plasmids’, were transfected into respective shuffle strains to generate mutant strains. To allow site-specific BPA substitutions in proteins through non-sense suppression, the plasmid pLH157 containing coding sequences of both the suppressor tRNACUA and the BPA-tRNA synthetase was also co-transfected. Procedures of BPA incorporation into yeast and the preparation of yeast whole-cell extracts for photo-crosslinking analyses have been described previously in more detail (17,18,25).
Immunoprecipitation (IP)
To prepare the yeast whole cell extract (WCE) for IP assays, 1 L yeast cell culture was grown in YPD medium to an O.D. of 1.5. The harvested cells were lysed and processed for WCE preparation following a previously described protocol (18,25). WCE of 1 mg was mixed with 50 μl of anti-flag antibody agarose beads (M2; Sigma-Aldrich) and incubated at 4°C for 2 h in WCE buffer [20 mM HEPES (pH 7.9), 100 mM KCl, 5 mM MgCl2, 1 mM EDTA and 20% glycerol]. The bound proteins were washed three times with 500 μl of WCE buffer, and the proteins were extracted by boiling with NuPAGE sample buffer (Invitrogen) for subsequent SDS-PAGE and Western blot analysis. For Western analysis, immuno-stained protein bands were visualized using the Odyssey infrared imaging system (LI-COR Biosciences).
Immobilized template assay and BPA cross-linking
To isolate the pol III PIC by immobilized template (IMT) assay, yeast WCEs from 1 L cell culture were incubated with the 5′-end biotin-conjugated DNA fragment containing the Sup4 tRNA gene. Briefly, the biotinylated DNA of 603-bp was amplified by PCR and subsequently immobilized on Streptavidin magnetic beads (Dynal) in a transcription buffer containing 20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol and 0.01% Tween 20. Each WCE of 800 μg was mixed with 2 μg of immobilized DNA in a final volume of 100 μl transcription buffer for 30 mins incubation at 30°C. The isolated PICs were washed three times with the transcription buffer prior to Western blot analysis. For the BPA photo-crosslinking experiment, the isolated PICs from the IMT assay were resuspended in a volume of 200 μl transcription buffer and treated with a total energy of 8000 μJ cm−2 UV (UVB) irradiation in a Spectrolinker XL-1500 UV oven (Spectronics). After UV treatment, the reaction mixture was washed three times, separated by SDS-PAGE, and analyzed by western blot analysis. A detailed protocol was previously described (18).
In vitro transcription
To analyze in vitro transcription activity with the IMT assay, the isolated PICs from above were resuspended in 17 μl of transcription buffer containing 200 ng α-amanitin, 4 units of RNase inhibitor (Promega), and 1 mM DTT. A mixture of NTPs (3 μL) was subsequently added, and the resulting reaction mixture contained 500 μM each of ATP, UTP, CTP, 50 μM GTP and 0.16 μM [α-32P] GTP (3000 Ci/mmol). After allowing the reaction to proceed at 30°C for 30 min, transcription was quenched by adding 180 μl of 0.1 M sodium acetate, 10 mM EDTA, 0.5% SDS and 200 μg/ml glycogen. The transcripts were extracted by phenol/chloroform and ethanol precipitated, separated on 6% (w/v) denaturing urea polyacrylamide gel and visualized by autoradiogram.
In vitro transcription using circular DNA plasmid and yeast WCE was conducted according to a protocol described previously (18,25). Briefly, WCE of 40 μg was pre-incubated with 100 ng plasmid DNA containing the SUP4 (tRNA-tyr) gene in a final volume of 17 μl transcription buffer (20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol and 0.01% Tween 20) together with 200 ng α-amanitin, 4 units of RNase inhibitor (Roche), and 1 mM dithiothreitol (DTT). Transcription was started with a further addition of 3 μl NTPs [ATP (500 μM), UTP (500 μM), CTP (500 μM), GTP (50 μM) and [α-32P] GTP (0.08 μM [3,000 Ci/mmol])] to a final volume of 20 μl. After reaction at 30°C for 3 or 30 min for single- or multiple-round transcription, respectively, RNA synthesis was terminated by adding the quenching buffer as described above. RNA products were extracted and analyzed by denaturing polyacrylamide (6%) gel electrophoresis. The SUP4 pre-tRNA transcripts were visualized by autoradiography and quantified with the ImageQuant TL program (GE). The amount of recombinant protein used in the Brf1-supplemented transcription assay was 200 ng.
FeBABE conjugation to phosphorothioate-containing SUP4 DNA and hydroxyl radical protein cleavage assay
FeBABE was conjugated at the phosphorothioate sites of the DNA template containing the SUP4 tRNA gene sequence. Two DNA fragments, referred to as the upstream and downstream DNAs, were separately generated and ligated. First, two synthetic oligonucleotides were designed with the following sequences: (A) 5′-gataattatttgaaatctctttttcaattgtatatgtgttatgtagtatactctttcttcAACAATTAAATACTCTCGGTAGCCAAGTTGGcacaag-3′; (B) 5′-gtgCCAACTTGGCTACCGAGAGTATTTAATTGTTgaagaaagagtatactacataacacatatacaattgaaaaagagatttcaaataattatc-3′. The sequence of oligonucleotide-A (top strand) covers the nucleotide positions from –60 to +31 of the SUP4 tRNAtyr gene with position +1 representing the transcription start site (+1 to +31 are in upper case), whereas oligonucleotide-B (bottom strand) contains the complementary sequence (upper case text: sequence complementary to the +1 to +31 coding sequence of the SUP4 gene). Note that the nucleotide position +30 of SUP4 marks the last position of the internal promoter element A box (ranging from position +20 to +30). A total of five different synthetic oligonucleotides were designed for oligonucleotide-A, with each containing six phosphorothioate moieties from –16 to +14 (the underlined sequence block) as shown in Figure 4B. Oligonucleotides-A and -B were annealed to generate the upstream DNA fragment that contains a 94-bp duplex DNA region and a triple nucleotide (aag) overhanging sequence at the 3′-end of the top strand. The annealed DNA product was further purified by agarose-gel separation and DNA extraction. The downstream DNA fragment was generated by PCR using primers 5′-GCCAAGTTGGCACAAGGTGCAA-3′ (top strand primer; underlined text: DraIII restriction sequence) and 5′-biotin-CAGGAAACAGCTATGACCATG-3′ (bottom strand primer) to amplify a 272-bp DNA-containing sequence starting from the box A element and extending to the transcription terminator of the SUP4 tDNA gene as well as downstream DNA. Subsequently, the PCR-generated DNA fragment was subjected to restriction enzyme digestion by DraIII, agarose-gel separation, and DNA extraction. The resulting DNA fragment contained a duplex DNA region of 269-bp, as well as a triple nucleotide (ctt) overhanging sequence at the 3′ end and a biotin moiety at the 5′-end of the bottom strand. Finally, the phosphorothioate-containing upstream DNA and the biotinylated downstream DNA fragments were ligated and further purified by agarose-gel separation and extraction to generate a 366-bp DNA for subsequent incorporation of the thio-reactive FeBABE reagent. FeBABE conjugation was conducted in a reaction containing 7 mM FeBABE (Dojindo), 1 μg phosphorothioate-containing DNA, and 20 mM MOPS, pH7.9. After 16 h of incubation at 50°C, the FeBABE-tethered and biotinylated SUP4 tDNAs were immobilized on Streptavidin magnetic beads (Dynal) and excess FeBABE was removed by washing three times with 200 μl of transcription buffer (20 mM HEPES, pH 7.9, 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol) additionally containing 0.05% (w/v) NP-40. Subsequent pol III PIC isolation and hydroxyl radical cleavage were performed as described previously (17,18,25).
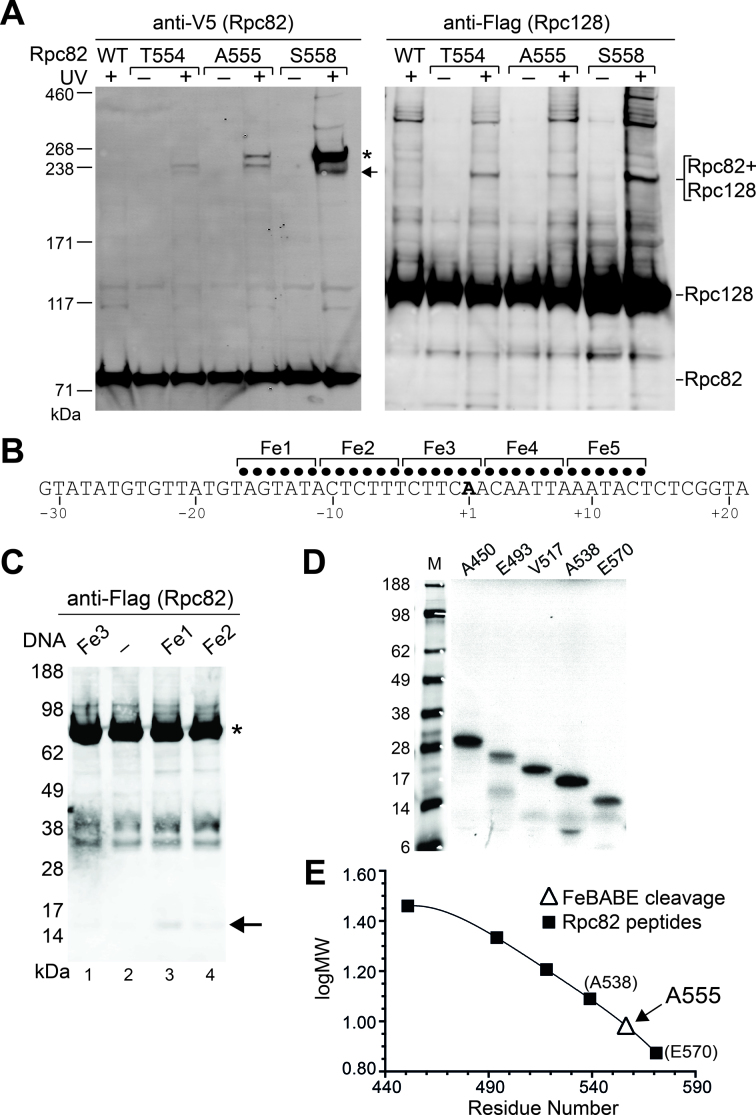
Figure 4.
The cleft loop of the Rpc82 WH4 domain interacts with Rpc128 and the DNA bubble. (A) BPA crosslinking of the cleft loop. In the left panel, BPA crosslinking for the three amino acid positions Thr554, Ala555 and Ser558 was probed with anti-V5 antibody for V5 epitope-tagged Rpc82. The slower (asterisk) and faster (arrow) migrating crosslinking bands were identified as the Rpc82-Rpc160 and Rpc82-Rpc128 crosslinks, respectively. In the right panel, the Rpc82–Rpc128 crosslink was confirmed by probing the Flag epitope-tagged Rpc128. (B) Phosphorothioate moieties in the SUP4 tDNA are indicated with dots above the transcribed sequence (non-template strand). A total of five phosphorothioate-containing DNA templates (Fe1∼5) were used for FeBABE conjugation. Numbers below the sequence denote the nucleotide positions relative to the transcription start site +1. (C) Western analysis showing hydroxyl radical cleavage of Rpc82 by the FeBABE-conjugated DNA probes. Anti-Flag antibody was used to probe the Flag epitope-tagged Rpc82. A control (–) SUP4 tDNA template does not contain any phosphorothioate. An arrow points to the Rpc82 peptide fragment cleaved by the probes. An asterisk indicates the uncleaved full-length Rpc82. (D) In vitro-translated Rpc82 peptide fragments. The residue number above each lane indicates the second amino acid residue (after the first methionine residue) of the Rpc82 fragment. Each in vitro-translated Rpc82 C-terminal peptide fragment contains a Flag epitope at the C-terminus. The first lane (M) contains molecular weight makers. (E) Determination of the hydroxyl radical cleavage site in Rpc82. The calibration curve is derived from the gel migration (log MW) of in vitro-translated Rpc82 peptide fragments (filled squares) and the corresponding starting residue numbers in (D). The size of the FeBABE cleavage fragment is mapped to the calibration curve (open triangle), and the corresponding cleavage site (A555) in Rpc82 is indicated.
RESULTS
A structurally disordered insertion in the WH3 domain is indispensable for cell growth
Although cryo-EM analysis of the pol III complex has revealed the structural arrangement of Rpc82, disordered regions remain (Figure 1A; gray colored regions). A disordered region lies within the WH3 domain of Rpc82, which spans amino acid residues 371–449 (Figure 1A–C). In an initial study, we deleted this 79-residue sequence block (referred to as the WH3 insertion). We found that yeast cells lacking the entire insertion were not viable. Based on multiple sequence alignment of the WH3 insertion, we found a lysine-rich sequence conserved among certain yeast species (Figure 1D). Internal deletions and multiple alanine substitutions were subsequently introduced to the sequence. As shown in Figure 1E, these mutations conferred a cold-sensitive growth phenotype at 16°C. In addition to the disordered insertion, the WH3 domain contains an extended structural motif, termed the helical extension (Figure 1A and C; WH3 ext.). Further mutational analysis in the extension region yielded a temperature-sensitive growth phenotype at 37°C (Figure 1F), consistent with a role to interface the Rpc160 clamp domain for pol III structural integrity (Figure 1C and Supplementary Figure S1). In contrast, the WH3 insertion is disordered and unlikely to assume a structural role within the pol III complex. We surmise that this insertion loop is involved in higher-order complex assembly in pol III transcription, as evidenced by the cold-sensitive cell growth phenotype.
The WH3 insertion is important for transcription initiation
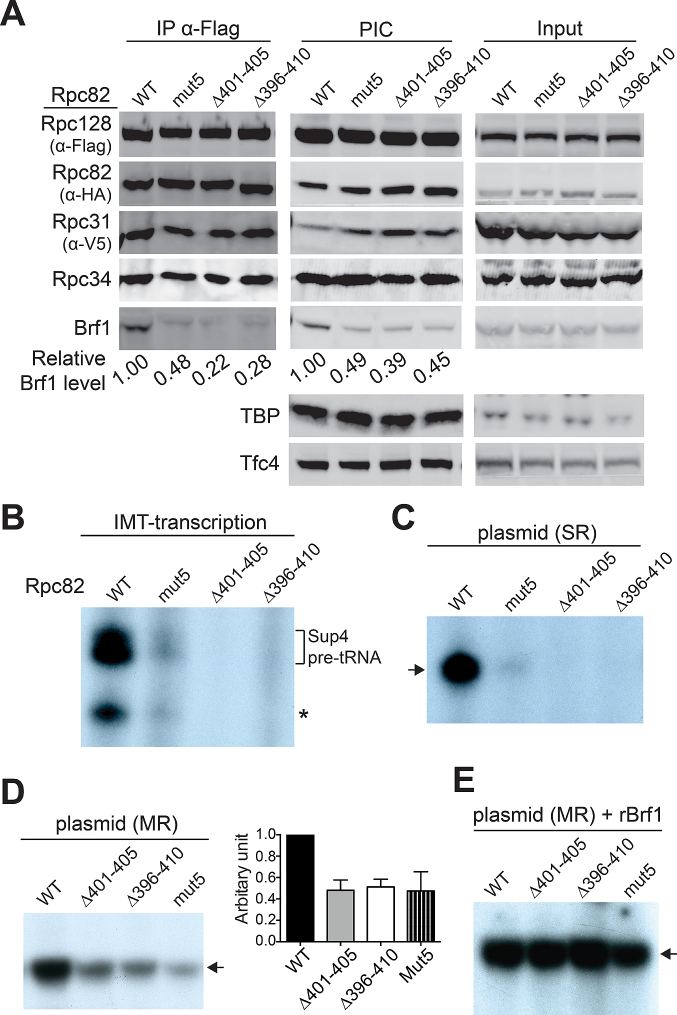
To characterize the functional importance of the WH3 insertion, we utilized whole cell extracts (WCE) from the cold-sensitive mutant strains to conduct in vitro assays. We first used immunoprecipitation (IP) to isolate the pol III complex from the WCEs. As shown in Figure 2A (left panel), when the pol III complex was precipitated by immobilizing the Flag epitope-tagged Rpc128, the co-IP protein levels were approximately equal for the WT and mutant Rpc82. Similar results were observed for the levels of Rpc34 and Rpc31, indicating that the WH3 insertion motif is not involved in stable association of the Rpc82/34/31 subcomplex to the pol III complex. Strikingly, whereas no co-IP of TFIIIB subunit TBP or TFIIIC subunit Tfc4 was found, levels of co-precipitated Brf1 were strongly compromised by the WH3 mutations.
Figure 2.
In vitro functional analysis. (A) Immunoprecipitation (IP) and IMT assays. Mutations in the WH3 insertion are indicated above lanes. The Flag-epitope-tagged Rpc128 was precipitated with anti-Flag antibody beads and co-IP polypeptides were probed with respective antibodies. Proteins from the IMT assay were revealed with respective antibodies. Input: proteins in the WCEs. The relative protein levels for Brf1 are listed below the gel bands. (B) In vitro transcription activity from the IMT assay. The autoradiogram shows tRNA transcripts generated from the isolated PICs of the IMT assays. The gel bands corresponding to the preliminary tRNA transcripts from the Sup4 promoter are indicated. An asterisk points to possible processed tRNAs. (C) Single-round transcription activity. The autoradiogram shows tRNA transcripts generated from the transcription assays with plasmid DNA and WCEs. An arrow points to the gel band corresponding to the Sup4 pre-tRNA. (D) Multiple-round transcription activity from the plasmid transcription assay. Quantitation of transcription activity is shown in the bar graph on right. Error bars indicate SD from three independent experiments using separately prepared WCEs. (E) Supplementation of Brf1 to the plasmid transcription assay.
We subsequently utilized the WCEs to analyze PIC formation by immobilized template (IMT) assay using DNA containing the Sup4 tRNA gene. As demonstrated in Figure 2A (middle panel), the mutants exhibited reduced Brf1 in the isolated PICs, whereas the levels of pol III, TBP, and Tfc4 were unaffected. Although TFIIIC and TFIIIB are known to stably bind to the promoter for subsequent recruitment of pol III (30), our data indicate that association of Brf1 in the isolated PICs is destabilized by the WH3 mutations. We speculate that a conformational change likely occurs within the PIC, and that the Rpc82 WH insertion contributes to the post-recruitment protein network for Brf1 association. Further in vitro RNA transcription analysis was conducted with addition of nucleoside triphosphates to the isolated PICs from IMT assays. In correlation with the observed Brf1 levels in the IMT assays, the WH3 mutations severely compromised Sup4 tRNA synthesis (Figure 2B). Similarly, the mutants exhibited reduced pol III transcription activity in the single-round or multiple-round transcription assays with plasmid DNA (Figure 2C and D, respectively). However, by supplementing mutant WCEs with recombinant Brf1 protein, transcriptional activity was restored to WT levels (Figure 2E). Based on these in vitro analyses, we have established an important functional link between the WH3 insertion and Brf1.
The WH3 insertion crosslinks with Brf1 in the PIC
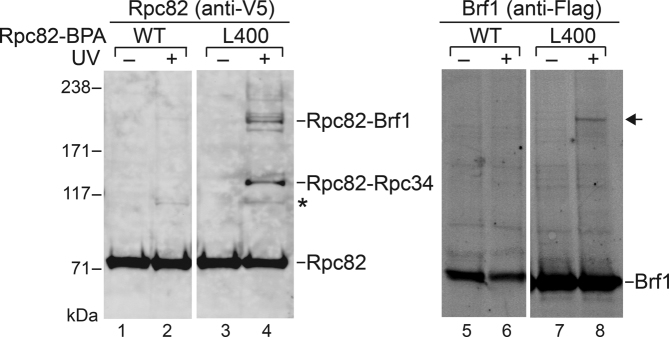
We previously incorporated a photo-crosslinking reagent, p-Benzoyl-L-Phenylalanine (BPA), as a non-natural amino acid into proteins in yeast (18). BPA substitution at a specific amino acid site was achieved by utilizing the suppressor tRNA/tRNA synthetase plasmid system targeted to the non-sense TAG codon inserted in the protein coding sequence (31). We reported our BPA crosslinking analysis for pol III subunits including Rpc160, Rpc128, Rpc82, Rpc53 and Rpc37 (17,18). In particular, our results generally agreed with the recently published cryo-EM structure supporting protein interactions for the Rpc82/34/31 and Rpc53/37 subcomplexes within the pol III cleft (Supplementary Figure S1A and B). Given our new functional analyses revealing a connection between the Rpc82 WH3 insertion and the transcription factor Brf1, we re-inspected the Rpc82 crosslinking results reported in our previous publication (17). A series of WCEs containing BPA substitutions within the WH3 insertion were re-generated for crosslinking analysis within the PIC by means of IMT assays. BPA incorporation in the WH3 insertion and crosslinking results are summarized in Supplementary Table S1. As shown in Figure 3 (left panel), a BPA substitution at Leu400 (i.e. close to the conserved sequence motif of the insertion, Figure 1D) yielded two crosslinking gel bands of approximately 200 kDa and 120 kDa. By subtracting the molecular mass of 85 kDa for Rpc82, the two crosslinked polypeptides were estimated to have molecular masses of 115 kDa and 35 kDa. Using a previously published epitope tagging approach, we inserted a Flag epitope at the C-terminus of Brf1 and repeated the crosslinking experiment (25), which confirmed Brf1 as the crosslinked protein target of 200 kDa (Figure 3; right panel, anti-Flag antibody staining). The same approach also allowed us to re-confirm Rpc34 as the crosslinked protein of 120 kDa reported in the previous publication (17). The evidence of Rpc34 crosslinking is also consistent with a previously reported chemical crosslinking/mass spectrometric analysis linking Lys403 of Rpc82 and Lys204 of Rpc34 (17). However, that previous chemical crosslinking analysis was conducted only with a purified pol III sample. Our BPA crosslinking reveals a PIC-specific Brf1 interaction for the conserved sequence motif of the WH3 insertion.
Figure 3.
The Rpc82 WH3 insertion interacts with Brf1. The photo-crosslinker BPA was incorporated at Leu400 near the conserved sequence block within the WH3 insertion. Crosslinking analysis was conducted within the PIC by means of IMT assays. Rpc82 and crosslinking gel bands were revealed with anti-V5 antibody against V5-epitope-tagged Rpc82 (left panel; lanes 1–4) in the Western analysis. Anti-Flag antibody was used to confirm the Rpc82-Brf1 crosslink (right panel; lanes 5–8). An arrow highlights the Rpc82-Brf1 crosslinking gel band (lane 8) revealed by the anti-Flag antibody. In addition to the Rpc82-Brf1 crosslink, BPA incorporation at Leu400 also yielded another crosslink with Rpc34 (lane 4). UV + or –, with or without UV irradiation. WT, wild-type Rpc82 without BPA incorporation. *, a background gel band due to UV irradiation. Protein molecular weight marker positions are indicated at left (kDa).
The Rpc82 cleft loop interacts with Rpc128 and the transcription bubble
In the pol III cryo-EM structure, the WH4 domain of Rpc82 contains a notable structural motif termed the cleft loop that spans amino acid residues 550–563 (Figure 1A–C) (3). This cleft loop is positioned within a canyon in the clamp head region of Rpc160 and close to the +7 bp position downstream of the DNA bubble (Figure 1C). Consistent with the pol III structure, our previous study reported that when BPA was incorporated into the cleft loop, there was a crosslink between Rpc82 and Rpc160 (Supplementary Figure S1B) (17). However, we also observed a faster migrating crosslinking band for BPA substitutions at Thr554, Ala555 and Ser558 around the turn of the cleft loop (Figure 4A; left panel). We re-conducted the crosslinking analyses and found that the lower molecular-weight band represented Rpc82–Rpc128 crosslinking (Figure 4A; right panel; Supplementary Table S1). Therefore, the cleft loop is structurally flexible, stretching from the clamp domain to an Rpc128 region such as the lobe or protrusion domain in the pol III cleft (Supplementary Figure S1A). Supporting structural flexibility of the cleft loop, the x-ray crystal structure of the human ortholog of Rpc82 (hRpc62) indicates that this conserved sequence region (aa 408–421 of hRpc62) is disordered (10).
To further investigate the DNA interactions of Rpc82, we conducted a directed hydroxyl radical protein cleavage analysis. Site-directed hydroxyl radical analyses have been applied to map protein-protein and protein–DNA interactions in the pol II and pol III PICs (18,32). To perform this analysis, we first conjugated the iron-EDTA-containing reagent FeBABE to the non-template strand of SUP4 tDNA (Figure 4B). A series of SUP4 DNA templates with phosphorothioate substitutions from nucleotide positions -16 to +14 was synthesized for FeBABE conjugation. The FeBABE-conjugated DNA templates were utilized in IMT assays for site-directed hydroxyl radical protein cleavage analysis on the PIC. On the basis of a previous KMnO4 hypersensitivity analysis (33), the FeBABE probe was tethered at the nucleotide positions of the DNA bubble (nucleotides –10 to +6) and the flanking duplex DNA of the SUP4 tDNA. As shown in Figure 4C, a DNA probe (termed Fe1) with FeBABE tethered at nucleotide positions ranging from –16 to –11 yielded cleaved Rpc82 peptides. In addition, weaker cleavages were observed with FeBABE at downstream nucleotides –10 to +2 (probes Fe2 and 3). No Rpc82 cleavage was found with probes (Fe4 and 5) containing FeBABE at further downstream nucleotide positions (data not shown). Based on a previously established method using a series of in vitro-translated Rpc82 peptides as the molecular weight standards (18), we approximated the cleavage site in Rpc82 to be at Ala555 in the cleft loop of WH4 (Figure 4D and E). Our directed hydroxyl radical cleavage analysis suggests that the cleft loop lies in close proximity to the upstream region of the DNA bubble, in addition to its resolved position near the downstream duplex DNA (Figure 1B and C). Strikingly, this FeBABE cleavage site at Ala555 coincides with BPA crosslinks identified for Rpc160 and Rpc128 (see above; Figure 4A). Therefore, we propose that the cleft loop participates in a dynamic molecular network with the pol III cleft and the DNA bubble.
Rpc82 interacts with the wall and protrusion domains of Rpc128
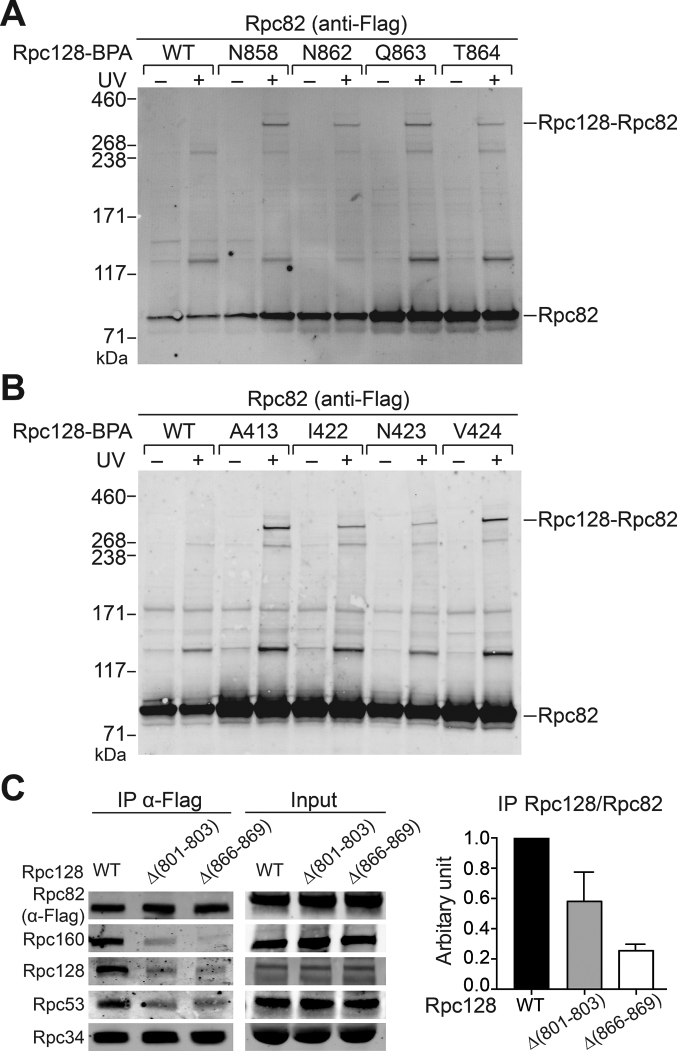
Since our crosslinking analysis revealed Rpc128 as a binding target for the WH4 cleft loop, we extended our BPA crosslinking analysis to Rpc128. We incorporated the BPA crosslinker in Rpc128 structural regions, such as the wall and protrusion domains within the pol III cleft. We then conducted photo-crosslinking analyses via IMT assays to probe PIC-specific interactions. We observed a number of Rpc128–Rpc82 crosslinks, including 15 and 12 positions in the wall and protrusion domains, respectively (Figure 5A and B; Supplementary Table S2). Interestingly, the Rpc82 crosslinking sites are scattered over the wall and protrusion surfaces and are distant from the resolved Rpc82 structure in the elongating pol III complex (Figure 6A). To further analyze the Rpc128–Rpc82 interaction, we mutagenized the Rpc128 structural regions that crosslinked with Rpc82. We obtained two Rpc128 wall domain mutations (Δ(801–803) and Δ(866–869)) with a slow growth phenotype at non-permissive temperatures of 37 and 16°C (data not shown). We isolated WCEs from these Rpc128 mutants to analyze protein interactions by IP assay. As illustrated in Figure 5C, Rpc128 wall mutations severely compromised the association of Rpc82 with the pol III core subunits Rpc128 and Rpc160, as well as the TFIIF-related subunit Rpc53. In summary, our BPA crosslinking analysis suggests an interaction of Rpc82 with the wall and protrusion domains of the pol III cleft, and our co-IP analysis further supports the Rpc82–Rpc128 wall domain interaction.
Figure 5.
Rpc82 interacts with the wall and protrusion domains of the pol III cleft. (A) Representative Rpc128-Rpc82 crosslinking assay with BPA incorporated in the wall domain. Anti-Flag antibody was used to probe the Flag epitope-tagged Rpc82 in the crosslinking gel bands. The Rpc128 protein is C-terminally 13-Myc epitope-tagged. (B) Representative Rpc128–Rpc82 crosslinking assay with BPA incorporated in the protrusion domain. As in (A), Rpc128–Rpc82 crosslinking is revealed with anti-Flag antibody. (C) Immunoprecipitation (IP) analysis. The IP assay was conducted by immobilizing the Flag epitope-tagged Rpc82 proteins. Mutations of the Rpc128 wall domain are indicated above the respective gel lanes. Proteins were revealed with the antibodies indicated at left. Input, proteins in the WCEs used in the IP assay. Right panel: quantitation of co-IP Rpc128 normalized by the level of Rpc82; error bars indicate SD from three independent IP experiments using separately prepared WCEs.
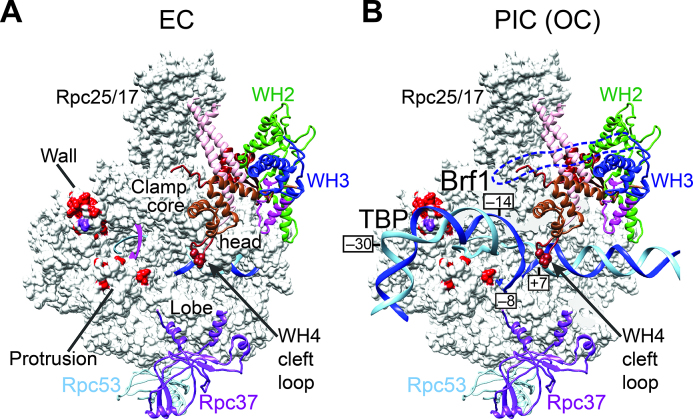
Figure 6.
Network of Rpc82 interactions in the pol III structure. (A) Sites of BPA crosslinking with Rpc82 in the pol III cleft. The pol III elongating complex (EC) is illustrated as in Figure 1B and C. The core and head regions of the clamp domain are indicated. Positions of BPA crosslinking with Rpc82 in the wall and protrusion domains are colored in red. The BPA substitution in His801 of the wall domain (colored purple) crosslinks to both Rpc82 and Brf1. Three amino acid positions in the Rpc82 WH4 cleft loop—Thr554, Ala555, and Ser558—are highlighted with spheres to indicate BPA crosslinking with Rpc128. (B) A model of the pol III open complex (OC). The pol III structure is shown as in (A). The molecular surface of Rpc160 has been partially removed to reveal the open promoter DNA. The open promoter DNA model is derived from PDB coordinate file 5iy7 of the pol II open complex structure (12). Template and non-template DNA strands are illustrated with the phosphate backbone traces (ribbons) colored in light and dark blue, respectively. Nucleotide positions relative to the transcription start site are labeled. Approximate locations of TBP and Brf1 on the DNA are indicated based on earlier DNase I protection and DNA photo-affinity studies (40,41). A blue dashed line denotes a speculative position of the WH3 insertion.
DISCUSSION
The cryo-EM structures of free and elongating pol III complexes have provided the atomic detail necessary to understand the roles of individual subunits in its structural integrity and transcriptional activity (3). Structural analyses have also revealed flexible regions in the TFIIE- and TFIIF-related subunits (3,26,27). In this paper, we reveal that a structurally flexible insertion in the WH3 domain of the TFIIE-related Rpc82 subunit interacts with the TFIIB-related transcription initiation factor Brf1. This WH3 insertion is one of the functionally important SUMO modification regions of the pol III complex (34). Our site-specific photo-crosslinking and hydroxyl radical probing analyses further indicated that the cleft loop of the Rpc82 WH4 domain interacts with both the Rpc128 and Rpc160 subunits of the pol III cleft, as well as the DNA bubble. Additional Rpc82 binding sites were also mapped to the wall and protrusion domains of Rpc128 within the PIC. Based on our biochemical analysis, we provide a model for the pol III open promoter complex in Figure 6B to indicate the more extensive Rpc82 interactions with the pol III cleft and the DNA bubble. Our study thus completes the protein/DNA network for Rpc82 and provides new insights into the functional roles of Rpc82 in transcription initiation.
Since the conserved sequence block in the WH3 insertion is only found in yeast species (Figure 1D) (10), its interaction with highly conserved transcription factor Brf1 is surprising. We speculate that a possible Rpc82-binding region in Brf1 resides in the C-terminal domain (CTD). Unlike the evolutionarily conserved N-terminal zinc ribbon and cyclin repeats, the Brf1 CTD contains several conserved sequence blocks separated by linker regions (24,35). Therefore, it is likely that a CTD linker sequence acts as the yeast-specific Brf1–Rpc82 interface. In addition, as indicated by multiple sequence alignment (Supplementary Figure S1C), the WH3 helical extension is also specific in yeast species. However, this helical extension is structurally important for the pol III complex, as validated by our mutational and crosslinking studies (Figure 1F and Supplementary Figure S1B/C) as well as the cryo-EM structural analysis (3,17). We suggest that the WH3 helical extension and insertion were incorporated specifically into yeast Rpc82 and evolved to become essential structural regions of the pol III machinery. Specific functional regions are present in other pol III subunits in higher eukaryotic systems. For example, the human ortholog of yeast Rpc37, HsRpc5, is a much larger protein (708 aa. vs. 282 aa.), and the expanded sequence of HsRpc5 is conserved among higher eukaryotic species (36).
Our study reveals a more flexible molecular network for the WH4 cleft loop to include the Rpc128 and Rpc160 subunits, as well as the DNA bubble (Figure 6B). Based on topological similarity, the cleft loop could be compared to the anti-parallel β-sheet of the TFIIEα WH domain. In recent cryo-EM analyses of the pol II closed and open promoter complexes, the anti-parallel β-sheet of TFIIEα, which is also referred to as the E-wing, was proposed to modulate the DNA melting process through an interaction with the –7 nucleotide position of the DNA bubble (12,13). Those pol II structural analyses and our biochemical study of the pol III PIC suggest a common role of the WH β-sheet in interacting with the DNA bubble. Supporting this hypothesis, previous DNA-protein crosslinking analyses also mapped Rpc82 to nucleotide positions from –8/–7 to +11 (37,38).
Our BPA crosslinking analysis of the pol III PIC mapped a number of Rpc82 binding sites to the Rpc128 protrusion and wall domains of the pol III cleft (Figures 5 and 6). These Rpc82 interactions are unexpected as the cryo-EM structures of free and elongating pol III revealed that Rpc82 is tightly bound to the Rpc160 clamp (Supplementary Figure S1A and B). Although we identified the Rpc128 interaction for the WH4 cleft loop, this structural motif is ∼30 Å and ∼60 Å from the protrusion and wall domains of Rpc128, respectively. Another Rpc128 interacting region for Rpc82 likely lies within the disordered WH3 insertion. Assuming a random coil conformation with the peptide backbone configuration of ∼3.8 Å between consecutive Cα atoms (39), the 79-residue WH3 insertion could form a loop of ∼150 Å. As both of the wall and protrusion domains of Rpc128 are located ∼100 Å from the WH3 domain, the insertion loop could thus reach over the two Rpc128 domains (Figure 6B; dashed blue line). This proposed positioning is further supported by the newly identified interaction between the WH3 insertion and the transcription factor Brf1 (Figure 6B), which is in contact with the wall domain (25). The structural extension likely occurs transiently upon PIC formation, as the WH3 insertion is not resolved in the cryo-EM structural analyses of free and elongating pol III complexes. This speculative WH3 interaction mode warrants further structural analysis of the pol III PIC.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yue-Chang Chou and Chih-Syuan Liu for technical assistance throughout the study. We thank Dr John O’Brien for English editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan, R.O.C. [103-2311-B-001-021-MY3, 105-2627-M-001-009]; Career Development Award to (H.-T.C.) from Academia Sinica, Taiwan, R.O.C. Funding for open access charge: Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, R.O.C.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A.. The expanding RNA polymerase III transcriptome. Trends Genet. 2007; 23:614–622. [DOI] [PubMed] [Google Scholar]
- 2. Roeder R.G., Rutter W.J.. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969; 224:234–237. [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann N.A., Jakobi A.J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W.J., Sachse C., Muller C.W.. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015; 528:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cramer P., Armache K.J., Baumli S., Benkert S., Brueckner F., Buchen C., Damsma G.E., Dengl S., Geiger S.R., Jasiak A.J. et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 2008; 37:337–352. [DOI] [PubMed] [Google Scholar]
- 5. Vannini A., Cramer P.. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012; 45:439–446. [DOI] [PubMed] [Google Scholar]
- 6. Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P.. RNA polymerase I structure and transcription regulation. Nature. 2013; 502:650–655. [DOI] [PubMed] [Google Scholar]
- 7. Fernandez-Tornero C., Moreno-Morcillo M., Rashid U.J., Taylor N.M., Ruiz F.M., Gruene T., Legrand P., Steuerwald U., Muller C.W.. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013; 502:644–649. [DOI] [PubMed] [Google Scholar]
- 8. Carter R., Drouin G.. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 2010; 27:1035–1043. [DOI] [PubMed] [Google Scholar]
- 9. Blombach F., Salvadori E., Fouqueau T., Yan J., Reimann J., Sheppard C., Smollett K.L., Albers S.V., Kay C.W., Thalassinos K. et al. Archaeal TFEalpha/beta is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. eLife. 2015; 4:e08378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lefevre S., Dumay-Odelot H., El-Ayoubi L., Budd A., Legrand P., Pinaud N., Teichmann M., Fribourg S.. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat. Struct. Mol. Biol. 2011; 18:352–358. [DOI] [PubMed] [Google Scholar]
- 11. Teichmann M., Dumay-Odelot H., Fribourg S.. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription. 2012; 3:2–7. [DOI] [PubMed] [Google Scholar]
- 12. He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., Nogales E.. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016; 533:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P.. Transcription initiation complex structures elucidate DNA opening. Nature. 2016; 533:353–358. [DOI] [PubMed] [Google Scholar]
- 14. Geiger S.R., Lorenzen K., Schreieck A., Hanecker P., Kostrewa D., Heck A.J., Cramer P.. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell. 2010; 39:583–594. [DOI] [PubMed] [Google Scholar]
- 15. Meinhart A., Blobel J., Cramer P.. An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 2003; 278:48267–48274. [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann N.A., Jakobi A.J., Vorlander M.K., Sachse C., Muller C.W.. Transcribing RNA polymerase III observed by electron cryo-microscopy. FEBS J. 2016; 283:2811–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu C.C., Herzog F., Jennebach S., Lin Y.C., Pai C.Y., Aebersold R., Cramer P., Chen H.T.. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19232–19237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C.C., Lin Y.C., Chen H.T.. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell. Biol. 2011; 31:2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landrieux E., Alic N., Ducrot C., Acker J., Riva M., Carles C.. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006; 25:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kassavetis G.A., Prakash P., Shim E.. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 2010; 285:2695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arimbasseri A.G., Maraia R.J.. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol. Cell. 2015; 58:1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z., Roeder R.G.. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997; 11:1315–1326. [DOI] [PubMed] [Google Scholar]
- 23. Brun I., Sentenac A., Werner M.. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997; 16:5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khoo B., Brophy B., Jackson S.P.. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994; 8:2879–2890. [DOI] [PubMed] [Google Scholar]
- 25. Khoo S.K., Wu C.C., Lin Y.C., Lee J.C., Chen H.T.. Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol. Cell. Biol. 2014; 34:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vannini A., Ringel R., Kusser A.G., Berninghausen O., Kassavetis G.A., Cramer P.. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010; 143:59–70. [DOI] [PubMed] [Google Scholar]
- 27. Fernandez-Tornero C., Bottcher B., Rashid U.J., Steuerwald U., Florchinger B., Devos D.P., Lindner D., Muller C.W.. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010; 29:3762–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D.. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998; 14:115–132. [DOI] [PubMed] [Google Scholar]
- 29. Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P.. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992; 110:119–122. [DOI] [PubMed] [Google Scholar]
- 30. Kassavetis G.A., Joazeiro C.A., Pisano M., Geiduschek E.P., Colbert T., Hahn S., Blanco J.A.. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992; 71:1055–1064. [DOI] [PubMed] [Google Scholar]
- 31. Chen H.T., Warfield L., Hahn S.. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007; 14:696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller G., Hahn S.. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 2006; 13:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kassavetis G.A., Blanco J.A., Johnson T.E., Geiduschek E.P.. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol. 1992; 226:47–58. [DOI] [PubMed] [Google Scholar]
- 34. Chymkowitch P., Nguea P.A., Aanes H., Robertson J., Klungland A., Enserink J.M.. TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase III assembly and activity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schramm L., Hernandez N.. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002; 16:2593–2620. [DOI] [PubMed] [Google Scholar]
- 36. Hu P., Wu S., Sun Y., Yuan C.C., Kobayashi R., Myers M.P., Hernandez N.. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol. Cell. Biol. 2002; 22:8044–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartholomew B., Durkovich D., Kassavetis G.A., Geiduschek E.P.. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 1993; 13:942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kassavetis G.A., Han S., Naji S., Geiduschek E.P.. The role of transcription initiation factor IIIB subunits in promoter opening probed by photochemical cross-linking. J. Biol. Chem. 2003; 278:17912–17917. [DOI] [PubMed] [Google Scholar]
- 39. Smith L.J., Fiebig K.M., Schwalbe H., Dobson C.M.. The concept of a random coil: Residual structure in peptides and denatured proteins. Folding Des. 1996; 1:R95–R106. [DOI] [PubMed] [Google Scholar]
- 40. Kassavetis G.A., Riggs D.L., Negri R., Nguyen L.H., Geiduschek E.P.. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 1989; 9:2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lannutti B.J., Persinger J., Bartholomew B.. Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry. 1996; 35:9821–9831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.