Abstract
The soil bacterium Pseudomonas putida KT2440 has gained increasing biotechnological interest due to its ability to tolerate different types of stress. Here, the tolerance of P. putida KT2440 toward eleven toxic chemical compounds was investigated. P. putida was found to be significantly more tolerant toward three of the eleven compounds when compared to Escherichia coli. Increased tolerance was for example found toward p‐coumaric acid, an interesting precursor for polymerization with a significant industrial relevance. The tolerance mechanism was therefore investigated using the genome‐wide approach, Tn‐seq. Libraries containing a large number of miniTn5‐Km transposon insertion mutants were grown in the presence and absence of p‐coumaric acid, and the enrichment or depletion of mutants was quantified by high‐throughput sequencing. Several genes, including the ABC transporter Ttg2ABC and the cytochrome c maturation system (ccm), were identified to play an important role in the tolerance toward p‐coumaric acid of this bacterium. Most of the identified genes were involved in membrane stability, suggesting that tolerance toward p‐coumaric acid is related to transport and membrane integrity.
Keywords: p‐coumaric acid, Pseudomonas putida, Tn‐seq, tolerance
1. INTRODUCTION
Pseudomonas putida is one of the soil organisms that is gaining interest due to its natural resistance to a number of hydrophobic solvents, such as xylenes, toluene, or styrene (Cruden, Wolfram, Rogers, & Gibson, 1992; Inoue, Yamamoto, & Horikoshi, 1991; Weber, Ooijkaas, Schemen, Hartmans, & De Bont, 1993). Some of the mechanisms leading to increased tolerance in these solvent‐tolerant strains have been identified and described, and efflux pumps (Kieboom & de Bont, 2001; Mosqueda & Ramos, 2000; Ramos, Duque, Godoy, & Segura, 1998) as well as changes in the membrane composition (Heipieper & De Bont, 1994), have been shown to be important for tolerance in these strains. Some of the changes that can occur in the membrane, when bacteria are exposed to hydrophobic solvents, include vesicle formation, alteration of phospholipid composition, and reduced permeability of the cell membrane (Heipieper & De Bont, 1994; Nicolaou, Gaida, & Papoutsakis, 2010; Ramos et al., 2002). The mechanisms behind tolerance in the model strain P. putida KT2440 has also been studied for different compounds (Benndorf, Thiersch, Loffhagen, Kunath, & Harms, 2006; Domínguez‐Cuevas, González‐Pastor, Marqués, Ramos, & de Lorenzo, 2006; Fernandez, Conde et al., 2012; Fernandez, Niqui‐Arroyo, Conde, Ramos, & Duque, 2012; Roca, Rodríguez‐Herva, Duque, & Ramos, 2008; Santos, Benndorf, & Sá‐Correia, 2004).
Traditionally, transposon insertion mutagenesis has been used to identify gene functions in many different bacteria (Glass et al., 2006; Hensel et al., 1995; Jacobs et al., 2003; Mack et al., 1994), including P. putida KT2440 (de Lorenzo & Timmis, 1994; Herrero, Lorenzo, & Timmis, 1990; Molina‐Henares et al., 2010). However, genome wide screening and identification of all genes involved in tolerance has been a limiting factor, which in the last years have been overcome through the use of next generation sequencing (NGS) techniques. Transposon insertion sequencing (Tn‐seq) (Figure 1a), enables high‐throughput identification of genomic regions involved in the survival of cells when exposed to various conditions (Barquist, Boinett, & Cain, 2013; van Opijnen & Camilli, 2013). A number of organisms have been investigated using this technique, thereby successfully identifying genes important for growth under different stress conditions, essentially by simultaneously investigating the fitness of all single mutants (Gawronski, Wong, Giannoukos, Ward, & Akerley, 2009; Langridge et al., 2009; Lennen & Herrgård, 2014; Santiago et al., 2015). So far, a Tn‐seq method has, however, not been developed for P. putida.
Figure 1.
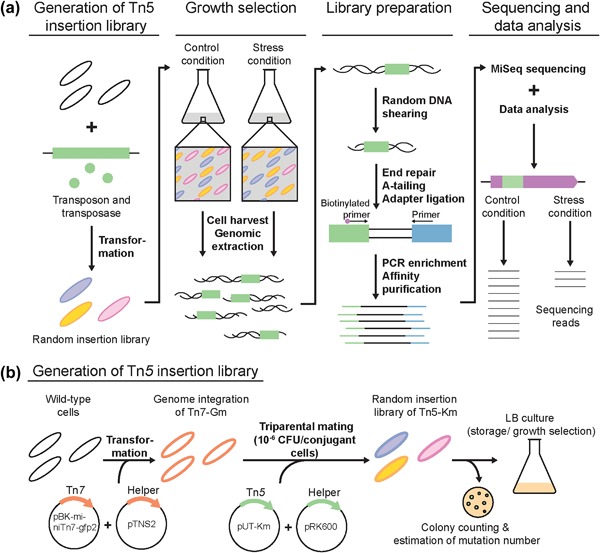
(a) Diagram of the experimental procedure used in a Tn‐seq approach. A Tn5 insertion library (transposons depicted as green boxes) is constructed by transformation and used in growth assays in different conditions, in order to assess differential growth of each insertion mutant. After harvesting the cells, the extracted genomic DNA is processed by DNA shearing, A‐tailing, and ligation of adapters (blue boxes), followed by an enrichment using PCR. This enriched library is subsequently sequenced by NGS and the results of the number of readings in the different conditions are compared in order to identify the differential abundance of insertions. (b) The method used in this study for generating the Tn5 library generating a combination of miniTn7‐Gm and miniTn5‐Km. The efficiency of the method to generate random insertion libraries was assessed by counting colonies of a sample
Metabolic engineering of microorganisms for production of chemical compounds from renewable resources aims at providing an alternative to petroleum‐based chemical processes. However, it still faces some challenges, such as ameliorating the toxic effects of intermediates and products of interest (Keasling, 2010). Numerous studies have focused on increasing the tolerance of different production strains toward various biochemicals using different strategies (Alper, Moxley, Nevoigt, Fink, & Stephanopoulos, 2006; Atsumi et al., 2010; Dunlop et al., 2011; Goodarzi et al., 2010). An alternative to this approach is to identify alternative production hosts that are naturally tolerant to higher concentrations of the chemicals of interest. P. putida is one such strain that is often utilized for bioremediation and production of more toxic chemicals (Loeschcke & Thies, 2015; Nikel, Martínez‐García, & de Lorenzo, 2014; Poblete‐Castro, Becker, Dohnt, dos Santos, & Wittmann, 2012; Verhoef, Ruijssenaars, de Bont, & Wery, 2007; Verhoef, Wierckx, Westerhof, De Winde, & Ruijssenaars, 2009).
In this study, we have investigated the tolerance of P. putida KT2440 toward a range of different biochemicals and found that it exhibits significantly increased tolerance toward the aromatic organic compound p‐coumaric acid, a low molecular weight phenolic acid present in soils and plants (Mussatto, Dragone, & Roberto, 2007; Strobel, 2001). p‐Coumaric acid is released as an inhibitory by‐product during the pretreatment of the lignocellulosic feedstocks (Jönsson & Martín, 2016). There is furthermore significant industrial interest in producing p‐coumaric acid since it can be used for making high performance polymers. Aditionally, it is a precursor for a number of interesting high value compounds (Santos, Koffas, & Stephanopoulos, 2011).
To study the mechanism of tolerance toward p‐coumaric acid, we developed a novel method for generating random transposon insertion mutant libraries in P. putida KT2440. In combination with transposon insertion sequencing, the most important genes required for growth in the presence of p‐coumaric acid were identified. A number of genes involved in maintenance of the membrane structure and efflux of solvent compounds were found to be important for growth, when P. putida KT2440 was exposed to this compound.
2. EXPERIMENTAL PROCEDURES
2.1. Bacterial strains and chemicals
P. putida KT2440 (DSM‐6125) and P. putida KT2440 Δfcs (Calero, Jensen, & Nielsen, 2016) were grown at 30°C and E. coli K‐12 MG1655 was grown at 37°C. Cells were routinely cultured in LB medium or on LB agar plates according to standard protocols (Sambrook & Russel, 2000). Modified M9 minimal medium containing 5 g L−1 of glucose as carbon source (Abril, Michan, Timmis, & Ramos, 1989) was used for all assays. E. coli DH5α and E. coli DH5α‐Pir were used for plasmid maintenance and were grown in LB medium at 37°C.
Antibiotics and other supplements were used at the following concentrations: chloramphenicol (Cm) 30 µg ml−1, kanamycin (Km) 50 µg ml−1, and gentamicin (Gm) 10 µg ml−1. A total of 300 mM sucrose was used for preparing electrocompetent P. putida cells. The chemicals used for toxicity screening included sodium acetate (Sigma, S8750), n‐butanol (Sigma, 281549), 3‐hydroxy‐γ‐butyrolactone (TCI Chemicals, H0939), 1,4‐butanediol (Merck, 801534), furfural (Sigma, 185914), itaconic acid (Sigma, I29204), levulinic acid (Sigma, L2009), succinic acid (Sigma, S9512), L‐threonine (Sigma, T8441), p‐coumaric acid (TCI Chemicals, C0393), and octanoic acid (Sigma, O3907). Stocks of these chemicals were prepared in modified minimal medium, and neutralized with NaOH to pH 7 when acidic chemicals were used.
2.2. Plasmids and strains constructions
P. putida KT2440::miniTn7‐Gm:GFP and P. putida KT2440 Δfcs::miniTn7‐Gm:GFP (Table 1) were constructed by transformation of plasmid pBK‐miniTn7‐gfp2 (Koch, Jensen, & Nybroe, 2001) as previously described (Choi, Kumar, & Schweizer, 2006), using plasmid pTNS2 (Choi et al., 2005) as a helper. The correct integration of the transposon was checked by colony PCR using primers Tn7‐GlmS and Tn7R109 (Table 2).
Table 1.
Plasmids and strains used in this study
| Genotype | Source | |
|---|---|---|
| Plasmid | ||
| pBK‐miniTn7‐gfp2 | pUC19‐based delivery plasmid for miniTn7‐gfp2. GmR, CmR, ApR, mobq | Koch et al. (2001) |
| pTNS2 | R6K replicon‐based helper plasmid, providing the Tn7 transposition functions in trans. ApR Mob+ | Choi et al. (2005) |
| pUT‐Km | R6K replication origin‐based suicide delivery plasmid for miniTn5‐Km. ApR KmR | Herrero et al. (1990) |
| pRK600 | ColE1 oriV; RP4tra+ RP4oriT; CmR; helper in triparental matings | Kessler, de Lorenzo, and Timmis (1992) |
| pEMG | oriV(R6K), lacZα fragment with I‐SceI sites; KmR | Martínez‐García and de Lorenzo (2011) |
| pEMG‐vacJ | oriV(R6K), lacZα fragment with I‐SceI sites, with 500 bp of gene vacJ; KmR | This study |
| pEMG‐tyrB | oriV(R6K), lacZα fragment with I‐SceI sites, with 500 bp of gene tyrB; KmR | This study |
| pEMG‐fleN | oriV(R6K), lacZα fragment with I‐SceI sites, with 500 bp of gene fleN; KmR | This study |
| Strains | ||
| P. putida KT2440 | Mt‐2 hsdR1 (r‐ m+) | Bagdasarian et al. (1981) |
| P. putida KT2440 Δfcs | P. putida KT2440 derivative with gene fcs deleted | Calero et al. (2016) |
| P. putida KT2440::miniTn7‐Gm:GFP | P. putida KT2440 with Gentamycin resistance gene in a miniTn7 | This study |
| P. putida KT2440 Δfcs::miniTn7‐Gm:GFP | P. putida KT2440 Δfcs with Gentamycin resistance gene in a miniTn7 | This study |
| P. putida KT2440 ttg2A‐ | P. putida KT2440 derivative with a miniTn5‐Km insertion gene ttg2A | Duque et al. (2007) |
| P. putida KT2440 ttg2B‐ | P. putida KT2440 derivative with a miniTn5‐Km insertion gene ttg2B | Duque et al. (2007) |
| P. putida KT2440 ccmC‐ | P. putida KT2440 derivative with a miniTn5‐Km insertion gene ccmC | Duque et al. (2007) |
| P. putida KT2440 ccmF‐ | P. putida KT2440 derivative with a miniTn5‐Km insertion gene ccmF | Duque et al. (2007) |
| P. putida KT2440 vacJ‐ | P. putida KT2440 derivative with an insertion in gene vacJ | This study Duque et al. (2007) |
| P. putida KT2440 tyrB‐ | P. putida KT2440 derivative with an insertion in gene tyrB | This study Duque et al. (2007) |
| P. putida KT2440 fleN‐ | P. putida KT2440 derivative with an insertion in gene fleN | This study Duque et al. (2007) |
| E. coli K‐12 MG1655 | F‐ hsdS gal | |
| E. coli DH5α | ϕ80dlacZΔM15 Δ(lacZYA‐argF)U169 recA1 endA1 hsdR17 (rk ‐ mk+) supE44 thi‐1 gyrA relA1 | Hanahan (1985) |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Purpose |
|---|---|---|
| Tn7‐GlmS | AATCTGGCCAAGTCGGTGAC | Check miniTn7 integration after glmS |
| Tn7R109 | CAGCATAACTGGACTGATTTCAG | |
| BioTEG‐Tn5‐fw | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAGCCGGATCCTCTAGAGTCGACC | Biotinylated primer with TEG in 5′ for enrichment of the Tn5 fragments. Bold sequence corresponds to the sequence that anneals with the transposon. |
| Tn5seq_rev | CAAGCAGAAGACGGCATACGAGAT | Enrichment of the Tn5 fragments. |
| VacJ‐ins‐U‐fw | ATCTGAGTU TAATTAATTAATTAGGGAAGCGGTCAACCGCCC | USER‐cloning of the homologous region of gene vacJ in pEMG. In bold STOP codons. In italics overhangs homologous to pEMG_phuser_rv. |
| vacJ‐ins‐U‐rv | ATCCCTAGAAAU TAATTAATTAATTACTTTTCGGCCGAGAGCAGGCTG | Same as the previous one. In italics overhangs homologous to pEMG_phuser_fw. |
| TyrB‐ins‐U‐fw | ATCTGAGTU TAATTAATTAATTAGATACCATCTGCTTGCGCATG | Same as the previous pair fot gene tyrB. |
| TyrB‐ins‐U‐rv | ATCCCTAGAAAU TAATTAATTAATTAGGCATGCTCGAAGACCTCAAC | Same as the previous pair fot gene tyrB. |
| FleN‐ins‐U‐fw | ATCTGAGTU TAATTAATTAATTATCGTAGACGGCACGCTGCTTC | Same as the previous pair fot gene fleN. |
| FleN‐ins‐U‐rv | ATCCCTAGAAAU TAATTAATTAATTACTCGCCGATGTGATTGAAGGGC | Same as the previous pair fot gene fleN. |
| pEMG_phuser_fw | ATTTCTAGGGAUAACAGGGTAATCCGGCGTAATCAT | Cloning in pEMG |
| pEMG_phuser_rv | AACTCAGAUTACCCTGTTATCCCTATACTGGCC | Cloning in pEMG |
Mutants in genes ttg2A, ttg2B, ccmC, and ccmF were obtained from the Pseudomonas Reference Culture Collection (PRCC), consisting of insertion mutants generated by disruption of the genes using a miniTn5‐Km (Duque et al., 2007).
Mutants in genes vacJ, tyrB, and fleN were constructed by introducing, by homologous recombination, a 3 kb fragment of DNA with a kanamycin resistance gene for its selection. For this purpose, plasmids pEMG‐vacJ, pEMG‐tyrB and pEMG‐fleN were constructed by cloning a 500 bp homologous fragment of the gene to be disrupted, using USER‐cloning (Nour‐Eldin, Geu‐Flores, & Halkier, 2010). The correct plasmids, which were checked by colony PCR and sequencing, were electroporated into P. putida KT2440 strains and the resistance to kanamycin was selected. The correct insertion was checked by PCR.
2.3. Chemical tolerance assays
Overnight cultures of P. putida KT2440 were diluted 20‐fold in 10 ml of modified M9 minimal medium. Cells were grown in 96‐well microtiter plates (flat bottom, Greiner Bio‐one, Frickenhausen, Germany) at 30°C and 200 rpm in an ELx808 microtiter plate reader (BioTek, Winooski, VT). After 3 hr of incubation, the cultures had reached an OD630 of approximately 0.1, and the stressors were added at different concentrations. Cells exposed to the different chemicals at various concentrations were incubated in a microtiter plate reader for 24 hr, and growth was followed by measuring OD630 every 30 min. The concentrations used for each compound were: 0, 2.25, 9, 22.5, and 45 g L−1 of threonine; 0, 8, 20, 30, and 40 g L−1 of itaconic acid; 0, 7.3, 18.18, 27.27, and 36.36 g L−1 of succinic acid; 0, 0.25, 0.5, 2.5, and 5 % (v/v) of 3‐hydroxy‐butyrolactone; 0, 5, 10, 50, and 100 g L−1 of sodium acetate; 0, 0.1, 0.5, 2.5, and 5 % (v/v) of levulinic acid; 0, 0.075, 0.15, 0.75, and 1.5 % (v/v) of furfural; 0, 0.25, 0.5, 2.5, and 5 % (v/v) of 1,4‐butanediol; 0, 5, 10, 50, and 100 g L−1 of butanol; and 0, 15, 30, and 60 mM of p‐coumaric acid.
Due to absorbance interference of the compound in the readings in 96‐well microtiter plates, octanoic acid toxicity assays in P. putida KT2440 and E. coli K‐12 MG1655 were performed in 50 ml of modified M9 minimal medium in 250 ml shake flasks at 30 and 37°C, respectively, and 250 rpm of shaking. Overnight cultures grown in modified M9 minimal medium were diluted to obtain an initial OD600 of 0.1 and incubated until OD600 reached 0.5, after which octanoic acid was added to the culture. The concentrations used were 0, 30, 40, 60, and 100 mM. Growth was followed by measuring OD600 every hour for 5 hr.
p‐Coumaric acid tolerance in strains P. putida KT2440 and P. putida KT2440 Δfcs, ttg2A‐, ttg2B‐, ccmC‐, ccmF‐, vacJ‐, tyrB‐, or fleN‐, as well as in E. coli K‐12 MG1655 was tested following the same protocol described above using 0, 15, and 30 mM of p‐coumaric acid.
2.4. Generation of miniTn5 insertions libraries
Introduction of plasmid pUT‐Km into P. putida KT2440::miniTn7‐Gm and P. putida KT2440 Δfcs::miniTn7‐Gm was performed by triparental mating. A total of 20 ml of overnight LB pre‐cultures of the recipient strains, the donor strain containing pUT‐Km, and the strain containing the helper plasmid pRK600, were harvested and washed twice in a phosphate buffer, after which they were mixed in a 1:1:1 ratio and adjusted to a volume of 1 ml in phosphate buffer. A volume of 100 µl was placed on an autoclaved 0.45 µm filter (Durapore membrane filters, Merck Millipore, Hellerup, Denmark), using a total of 10 filters, and incubated overnight at 30°C on LB plates. Subsequently, cells on all filters were re‐suspended in 8 ml of buffer. A total of 100 µl of this mix was diluted and plated on LB agar plates containing kanamycin and gentamycin for counting the number of mutants in the library, and the rest was used to inoculate 400 ml of LB with kanamycin and gentamycin in a 2 L shake flask, which was incubated at 30°C with 250 rpm shaking overnight. Aliquots of this culture were used to prepare cryo stocks in glycerol and stored at −80°C.
2.5. Transposon mutant library growth selection
Tn5 mutant libraries of P. putida KT2440::miniTn7‐Gm and P. putida KT2440 Δfcs::miniTn7‐Gm were used for growth selection under p‐coumaric acid stress conditions. The cryo stocks, containing approximately 100,000 mutants, determined by colony counting on agar plates, were thawed on ice, washed twice with LB and grown overnight in 50 ml of LB with proper antibiotics at 30°C in 250 ml shake flasks. A total of 10 ml of the overnight LB culture was harvested for genomic DNA extraction. Part of the overnight LB culture was used to inoculate 50 ml of modified M9 minimal medium with antibiotics to a starting OD600 of 0.1 after washing the cells with modified M9 minimal medium. Cells were incubated at 30°C and 250 rpm and growth was followed by measuring OD600 every 2 hr. When cultures reached an OD600 of 0.5, they were divided into 25 ml cultures and diluted two fold in preheated minimal medium or in minimal medium with 50 mM of p‐coumaric acid. Growth was followed until cultures reached late exponential phase at an OD600 of approximately 2, after which the cells were harvested and stored at −20°C prior to gDNA extraction. Three independent biological replicates were carried out.
2.6. Library preparation for Tn‐seq
gDNA libraries were prepared as previously described (Lennen & Herrgård, 2014). P. putida gDNA was extracted using the PureLink genomic DNA kit (Invitrogen, Thermo Fisher, Waltham, MA) and 3 µg of total gDNA was sheared in 300‐bp fragment size using a Covaris E220 ultrasonicator. End repair (NEBNext End repair module, New England Biolabs, Ipswich, MA) and A‐tailing using Klenow Fragment (Thermo Scientific, Waltham, MA) was performed on the fragments prior to ligation of adapters. Illumina TruSeq adapters were ligated using the NEBNext Quick Ligation Module (New England Biolab) and the transposon‐chromosome junctions were amplified by PCR with a primer specific for the adapter and a biotinylated primer specific for the miniTn5 inserts. Biotinylated PCR products were subsequently purified by affinity capture using the Dynal MyOne Streptavidin C1 beads (Invitrogen). Library size distribution was validated by qPCR, using the KAPA qPCR quantification kit, the Agilent RNA 6000 Pico kit, and the Agilent 2100 Bioanalyzer. Finally, the library was sequenced on an Illumina MiSeq.
2.7. Data analysis
Reads obtained from sequencing were initially processed in order to discard the unspecific sequences and trim the length of the correct ones to 120 bp, leaving 25 bp of genomic sequence that can be used for comparison to the P. putida KT2440 genome. Genes that were conditionally more abundant were identified using the ESSENTIALS pipeline (Zomer, Burghout, Bootsma, Hermans, & van Hijum, 2012). Gene‐level insertion counts were normalized using TMM normalization and p‐values adjusted by Benjamini‐Hochberg correction. Genbank version NC_002947.3 of P. putida KT2440 was used as the reference genome.
3. RESULTS
3.1. Tolerance of P. putida KT2440 toward industrially relevant biochemicals
P. putida has been widely used for bioremediation because of its high degree of tolerance toward many organic compounds, and it is furthermore a promising host for metabolic engineering. We therefore decided to screen P. putida KT2440 for its tolerance toward a number of chemical compounds that could be relevant for biobased production, and that are known to be relatively toxic to E. coli (Rau, Calero, Lennen, Long, & Nielsen, 2016). Six of the compounds, hydroxy‐γ‐butyrolactone, furfural, itaconic acid, levulinic acid, succinic acid, and L‐threonine, have previously been identified as being among the top 30 predicted chemical building blocks in a report from the U.S Department of Energy (Werpy & Petersen, 2004). Five other compounds were also selected, including acetic acid, which is a common inhibitor present in biomass hydrolysates; n‐butanol, a potential biofuel and also widely used in the chemical industry; octanoic acid and 1,4‐butanediol, which are polyester precursors; and p‐coumaric acid, an aromatic polymer precursor, for which there is currently no efficient industrial production method.
P. putida KT2440 tolerance to these chemical compounds was tested by assaying their effect on the growth rate using different concentrations, using conditions identical to those used previously for E. coli K‐12 MG1655 (Rau et al., 2016). In order to compare the effect the different compounds have on P. putida KT2440 and E. coli K‐12 MG1655 growth, we calculated the concentration of each compound required to reduce the growth rate by 33%, as described previously (Rau et al., 2016). The concentrations of compounds needed to reduce the growth rate in P. putida KT2440 varied significantly. Some of the chemicals were found to effect growth at very low concentrations, including furfural, which was found to be toxic at a concentration of approximately 15 mM; levulinic acid and n‐butanol at 40 mM; and hydroxy‐γ‐butyrolactone at 58 mM (Table 3). Others were found to require higher concentrations to achieve the same effect on the growth, such as threonine or 1,4‐butanediol, for which concentrations of 412 and 399 mM were required, respectively.
Table 3.
Concentrations of biochemicals leading to a 33% reduction in the growth rate (mM)
| P. putida KT2440 | E. coli MG1655 | |
|---|---|---|
| Itaconic acid | 189.5 | 215.2 * |
| Succinic acid | 148.3 | 262.5 * |
| Threonine | 412.0 | 58.8 * |
| Sodium acetate | 65.6 | 91.4 * |
| Butanol | 40.2 | 82.0 * |
| Levulinic acid | 40.0 | 67.0 * |
| 1,4‐butanediol | 398.7 | 452.7 * |
| Furfural | 14.9 | 12.0 * |
| 3‐hydroxy‐γ‐butyrolactone | 58.0 | 133.7 * |
| Octanoic acid | 39.5 | 15.0 |
| p‐Coumaric acid | 61.0 | 30.4 |
Data obtained from (Rau et al., 2016).
When comparing to E. coli K‐12 MG1655, most of the chemical compounds tested were found to have a similar or even higher inhibitory effect on P. putida KT2440 in the conditions used. However, higher tolerance toward L‐threonine, octanoic acid, p‐coumaric acid, and furfural was found in P. putida KT2440. The highest difference in tolerance was found for L‐threonine, where P. putida KT2440 was found to be sevenfold more tolerant when compared to E. coli K‐12 MG1655. P. putida KT2440 was also found to be twofold more tolerant to octanoic acid and p‐coumaric acid, whereas a lower difference of 1.25‐fold increased tolerance was found for furfural (Table 3). Since p‐coumaric acid is a known toxic compound in biomass hydrolysate, and since its toxicity has been shown to affect production by fermentation (Sariaslani, 2007), we decided to study the mechanism of tolerance toward this compound in further detail.
3.2. The deletion of fcs does not affect the tolerance toward p‐coumaric acid
P. putida KT2440 is able to degrade p‐coumaric acid and use it as carbon source (Figure 2a) (Jiménez, Miñambres, Luis, & Díaz, 2002). In order to investigate the effect of the degradation of p‐coumaric acid on tolerance, a strain with a deletion of the first gene in the p‐coumaric acid degradation pathway, fcs, which abolishes p‐coumaric acid degradation, or conversion to another compound (Calero et al., 2016), was tested in different concentrations of the compound. Its tolerance to p‐coumaric acid was compared to the wild‐type strain, which is able to degrade p‐coumaric acid, and to E. coli K‐12 MG1655. Whereas a p‐coumaric acid concentration of 30 mM was enough to reduce the growth rate of E. coli to 68% of the control grown without the compound, no effect on growth was observed when this concentration was added to the P. putida KT2440 Δfcs strain (Figure 2b).
Figure 2.
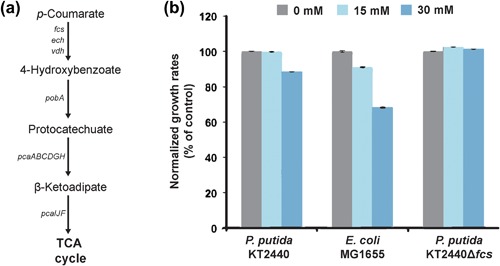
(a) Degradation pathway of p‐coumaric acid in P. putida KT2440 ending in the tricarboxylic acid (TCA) cycle. (b) Growth rates of P. putida KT2440, E. coli K‐12 MG1655, and P. putida KT2440 Δfcs when exposed to different concentrations of p‐coumaric acid. Growth rates were normalized to the growth rate in control conditions without p‐coumaric acid. Error bars indicate the standard deviation of four different biological replicates
3.3. Tn5 library generation in P. putida KT2440 and putida KT2440 Δfcs
Since degradation of p‐coumaric acid was shown not to be involved in the increased tolerance of P. putida KT2440 toward p‐coumaric acid, a random library of mini‐Tn5 mutants was generated in order to further investigate the possible mechanisms of tolerance (Figure 1b). To obtain a mutant library with a high genomic coverage, a triparental mating of two plasmids, pUT‐Km, containing the transposon, and pRK600, a conjugation helper plasmid was performed (Herrero et al., 1990). To have a heterologous antibiotic resistance cassette for selection of the recipients with the mini‐Tn5 insertions and to avoid growth of other strains used in the triparental mating, a gentamycin resistance gene was introduced into the genome of P. putida KT2440 and P. putida KT2440 Δfcs using a mini‐Tn7‐Gm transposon (Figure 1b). This approach inserts the gene in a fixed position on the genome, after the gene glmS, which has previously been shown to be innocuous to the cell physiology (Lambertsen, Sternberg, & Molin, 2004). Using this method we avoided the natural chloramphenicol resistance of P. putida KT2440 as a selection marker, which would introduce a bias in the transposon insertion sequencing results for mutants involved in the chloramphenicol resistance, such as the efflux pump TtgABC (Fernandez, Conde et al., 2012). The use of alternative carbon sources, like citrate, that only the receptor strain could degrade was also avoided, and the developed method thereby enabled the use of LB in the creation of the library.
An efficiency of 10−6 CFU/conjugant cells when using triparental mating was achieved (Figure 1b). Using this method, a pooled library of ∼125.000 mutants in P. putida KT2440 and ∼95.000 mutants in the strain P. putida KT2440 Δfcs was generated. The number of transposon insertions corresponds to a theoretical insertion average of one insertion every 52 bp or one insertion every 65 bp, respectively.
The two independent P. putida KT2440 and P. putida KT2440 Δfcs libraries of random transposon insertion mutants were grown in modified M9 minimal media with or without supplementation of 50 mM p‐coumaric acid as stressor. A comparison of the resulting populations of random insertions in each condition can be used to identify genes that are related to the growth under the stressful conditions. Cells were harvested for further analysis during late exponential growth, after approximately five generations in the presence of p‐coumaric acid (Figure 3a). The growth rate of the transposon insertion libraries when exposed to p‐coumaric acid stress was 77% and 83% for P. putida KT2440 and P. putida KT2440 Δfcs, respectively, compared to the libraries that were grown in modified M9 minimal medium, corresponding to increased doubling times from 82 to 107 min in the P. putida KT2440 strain, and from 81 to 98 min in the P. putida KT2440 Δfcs strain. The populations furthermore had an approximately 6 hr lag phase compared to the cultures without p‐coumaric acid.
Figure 3.
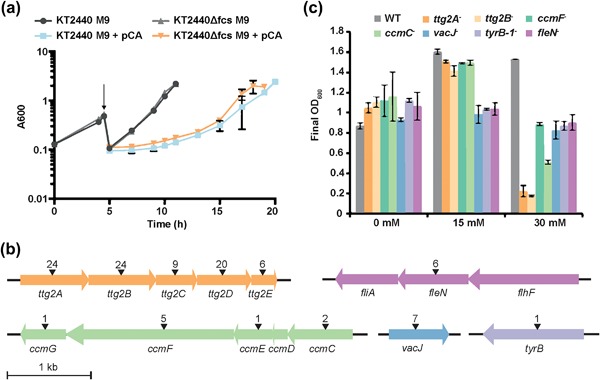
(a) Growth of the Tn5 libraries in P. putida KT2440 and P. putida KT2440 Δfcs in M9 and M9 supplemented with 50 mM of p‐coumaric acid (pCA) is shown. The arrow indicates when p‐coumaric acid was added to the cultures. The growth experiments ended when cells were harvested for DNA sequencing. (b) Some of the most important genes identified to be involved in the tolerance of P. putida KT2440 toward p‐coumaric acid in their genetic landscape. The number of different insertions found in the Tn‐seq assay is shown on top of each gene with arrows. (c) Final OD600 of strains of P. putida KT2440 (WT), ttg2A‐, ttg2B‐, ccmF‐, ccmC‐, vacJ‐, tyrB‐, and fleN‐ at three different concentrations of p‐coumaric acid: 0, 15, and 30 mM. Error bars indicate standard deviation of three independent biological replicates
3.4. Tn‐seq results demonstrate that the structure of the membrane has an important role in the tolerance of putida KT2440 to p‐coumaric acid
Sequencing of the junctions between the transposon and the genome was achieved by generating an enriched library as previously described (Lennen & Herrgård, 2014). A pUT‐Km transposon specific primer, BioTEG‐Tn5‐fw, was designed for the enrichment of the junctions (Table 2). Sequencing of the enriched library resulted in the identification of approximately 12.900 different insertions with 20 reads or more in the library derived from P. putida KT2440 Δfcs, whereas approximately 6,000 insertions with 20 reads or more were found in the P. putida KT2440 library. Using the generated libraries, we sought to identify genes involved in tolerance toward p‐coumaric acid. A number of genes were found to be important for the growth of both P. putida KT2440 and P. putida KT2440 Δfcs when exposed to high concentrations of p‐coumaric acid. Genes showing a very high impact on tolerance included the genes in the ttg2ABCDE operon, encoding an ABC (ATP‐binding cassette) transporter. Transposon insertions in all of the genes in this operon had a strong effect on growth in the presence of p‐coumaric acid, and these insertions were represented between 11‐ and 15‐fold less in the presence of p‐coumaric acid. Moreover, all the genes were found to have more than one insertion in different positions of the ORFs. The longest genes in the operon, ttg2A, ttg2B, and ttg2D had 16, 17, and 13 different insertion sites, respectively, in P. putida KT2440, and 8, 7, and 7 in strain P. putida KT2440 Δfcs. On the other hand, ttg2C and ttg2E, which are shorter genes, were found to have 6 and 5 different insertion sites in strain P. putida KT2440 and 3 and 1 in strain P. putida KT2440 Δfcs. The loss of function of two other genes, vacJ and fleN, encoding an outer membrane protein and a flagellar number regulator, respectively, was also shown to impair growth in both strains in the presence of p‐coumaric acid. Transposon insertions in vacJ were found to be approximately 15‐fold less abundant when p‐coumaric acid was present in the media, and mutations in fleN were shown to be around 15‐fold less represented when p‐coumaric acid was present. The gene fleN forms an operon together with fliA, flhN, and flhA, but none of the other operon members appeared to play a role in the tolerance to p‐coumaric acid (Table 4).
Table 4.
Genes involved in tolerance toward p‐coumaric acid
| Gene | Number of insertions | Fold change | Gene name | Gene function |
|---|---|---|---|---|
| a | ||||
| PP_0958 | 24 | 11.3 | ttg2A | Toluene tolerance ABC efflux transporter ATP‐binding protein |
| PP_0959 | 24 | 10.4 | ttg2B | Hypothetical protein |
| PP_0960 | 9 | 15.2 | ttg2C | Hypothetical protein |
| PP_0961 | 20 | 11.7 | ttg2D | Toluene tolerance family protein |
| PP_0962 | 6 | 11.1 | ttg2E | Toluene‐tolerance protein |
| PP_2163 | 7 | 13.4 | vacJ | VacJ family lipoprotein |
| PP_4342 | 6 | 8.16 | fleN | Flagellar number regulator FleN |
| b | ||||
| PP_3286 | 1 | 9.3 | phaN | PaaX family transcriptional regulator |
| PP_4320 | 1 | 8.1 | ccmH | Hypothetical protein |
| PP_4321 | 1 | 7.7 | ccmG | Thiol‐disulfide oxidoreductase |
| PP_4322 | 5 | 7.4 | ccmF | Cytochrome C biogenesis protein CcmF |
| PP_4323 | 1 | 8.2 | ccmE | Cytochrome C biogenesis protein CcmE |
| PP_4325 | 2 | 7.2 | ccmC | Heme exporter protein CcmC |
| PP_4937 | 1 | 7.1 | – | Toluene tolerance protein |
| c | ||||
| PP_0168 | 1 | 18.8 | – | Surface adhesion protein |
| PP_0674 | 1 | 6.1 | – | ABC transporter ATP‐binding protein |
| PP_1185 | 1 | 7.5 | oprH | Outer membrane protein H1 |
| PP_1972 | 1 | 6.3 | tyrB‐1 | Aromatic amino acid aminotransferase |
| PP_2889 | 2 | 10.2 | – | Transmembrane anti‐sigma factor |
a, Important genes found in both strains; b, Important genes found in the strain P. putida KT2440 fcs; c, Important genes found in the strain P. putida KT2440.
A number of transposon insertions were also identified in just one of the two strain backgrounds. One of the most significant was the operon consisting of the genes ccmCDEF and dsbE in the strain P. putida KT2440 Δfcs. All the genes in this operon, which encodes the cytochrome c maturation system, were shown to be involved in tolerance toward p‐coumaric acid, except for ccmD, likely due to the fact that it is the shortest gene in the operon, with approximately 200 bp and therefore may not have been targeted by transposon insertions. The genes in the operon with intermediate lengths (450–550 bp), dsbE and ccmE were targeted by one type of insertion whereas the longest genes, ccmF and ccmC (2,000 and 750 bp, respectively) were found to have five and two different insertions, respectively. In all cases, the mutants in these genes were found to be represented approximately 13‐fold less in the populations when p‐coumaric acid was present in the media. In the P. putida KT2440 library, genes encoding some other membrane proteins such as the outer membrane protein OprH, a surface adhesion protein encoded by gene PP_0168 and an ABC transporter encoded by PP_0674 were found to be related to the tolerance toward p‐coumaric acid (Table 4).
The loss‐of‐function of two genes was found to increase the growth rate of both P. putida KT2440 Δfcs and P. putida KT2440 in the presence of p‐coumaric acid. These genes include the LysR family transcriptional regulator encoded by the gene PP_1262 and the MarR family transcriptional regulator encoded by the gene PP_4515. In both cases, the cells containing transposon insertions in these genes were found to grow better when p‐coumaric acid was present and were approximately 15‐fold more abundant under these conditions.
3.5. Loss of function in genes identified through the Tn‐seq leads to impaired growth in the presence of p‐coumaric acid
Two complete operons, ttg2 and ccm, were identified in both the assayed strain as playing an important role in the tolerance of P. putida KT2440 toward p‐coumaric acid (Figure 3b). To further investigate the role of these genes, independent insertion mutants available from the PRCC (Duque et al., 2007) in genes ttg2A, ttg2B, ccmC, and ccmF were acquired, while insertion mutants in genes vacJ, tyrB, and fleN were constructed. The different strains were grown in medium containing different concentrations of p‐coumaric acid, and a clear impact on the final optical density of the mutant strains was found when compared to the wild‐type strain (Figure 3c), and a similar effect was observed for the growth rate (Figure S1). This result confirms that both the ttg2 and ccm operons as well as other genes identified through the Tn‐seq experiment are strongly involved in the tolerance toward the aromatic compound p‐coumaric acid.
4. DISCUSSION
When compared to E. coli, P. putida KT2440 was found to have enhanced tolerance to only three out of the 11 tested industrially relevant compounds, when these were supplemented to the growth medium. Even though P. putida strains have been extensively studied for their tolerance to certain organic solvents such as toluene, P. putida KT2440 was not found to be more tolerant toward carboxylic acids (levulinic acid, acetic acid, itaconic acid, and succinic acid) and furfural when compared to E. coli. Surprisingly, P. putida KT2440 was not found to be more tolerant toward n‐butanol, even though some P. putida strains, including P. putida DOT‐T1E and S12, have a high tolerance toward organic solvents such as n‐butanol (Rühl, Schmid, & Blank, 2009). It should be noted that the compounds were added to the growth medium in the given experiments. It is possible that the effect and tolerance mechanisms could be different if the compounds were produced intracellularly.
P. putida KT2440 was shown to be significantly more tolerant toward p‐coumaric acid when compared to E. coli. Deletion of the first gene (fcs) of the degradation pathway revealed that degradation of the compound was not the mechanism of detoxification. This observation agrees with previous observations showing that even though metabolism of toxic chemicals can help to alleviate toxicity, it is usually a minor mechanism of tolerance (Ramos et al., 2002). Interestingly, some defect on the growth was observed only for the wild‐type strain when exposed to 30 mM of p‐coumaric acid, and not for the deletion mutant, suggesting that certain intermediates in the degradation pathway may be toxic (Figure 2b).
To further investigate the mechanism of tolerance toward p‐coumaric acid, we developed a new method for generating large transposon libraries compatible with Tn‐seq. By comparing sequences of libraries grown with or without inhibitory concentrations of p‐coumaric acid, we identified 19 genes involved in the tolerance to p‐coumaric acid, many of them involved in membrane processes. These results are in accordance to a previous study that investigated the mechanism of action of p‐coumaric acid in different bacteria, and pointed toward membrane disruption as a main effect (Lou et al., 2012). The genes in the ABC (ATP‐binding cassette) transporter, Ttg2ABC, were part of the seven genes for which loss‐of‐function indicated impaired growth in both strain libraries when exposed to high concentrations of p‐coumaric acid. This transporter has previously been identified to play a role in the tolerance toward toluene in P. putida (Garcia et al., 2010; Kim, Lee, Lee, & Lim, 1998), and it has been described to be involved in resistance to a number of other stress agents (Garcia et al., 2010).
Another gene with decreased abundance in both libraries when exposed to p‐coumaric acid, vacJ (PP_2163), contains the domain of the VacJ superfamilies. VacJ has been characterized as an outer membrane protein associated to an ABC transporter system in other organisms, such as E. coli and Shigella flexneri, in which it has been proposed to be involved in the maintenance of outer membrane stability in the presence of membrane disruptors such as SDS (Carpenter et al., 2014; Malinverni & Silhavy, 2009). Its deletion has been shown to increase permeability of the outer membrane (Malinverni & Silhavy, 2009) and increase the formation of vesicles in E. coli and other gram‐negative bacteria (Roier et al., 2016). In P. aeruginosa, a VacJ homolog has been described to play a role in antibiotic resistance. VacJ has also been shown to be more abundant in cells growing in the presence of phenol in P. putida KT2440 (Santos et al., 2004).
A transposon insertion in the flagellar number regulator fleN was shown to be important for tolerance toward p‐coumaric acid. The deletion of this gene in P. aeruginosa has been shown to alter the single polar flagella localization, affecting the motility of the cells (Dasgupta, Arora, & Ramphal, 2000). Even though some genes of the flagella systems have been identified in the tolerance toward toluene in the strain P. putida DOT‐T1E, their function in the resistance is not clear, since it does not seem to be related to the cells motility (Ramos et al., 2002).
In one of the libraries, the complete ccmCDEF‐dsbE operon, containing the genes PP_4321‐PP_4325, was found to be of especial importance for tolerance toward p‐coumaric acid. These genes encode the cytochrome c maturation system, which is known to be present in Proteobacteria, where it promotes the attachment of the heme group to the apocytochrome c in the periplasm, allowing the correct function of the type c‐cytochrome in cell respiration and ATP synthesis (Thöny‐Meyer, 2000). The effect of the lack of a functional operon in tolerance toward p‐coumaric acid may indicate a high energy requirement or disruption of the membrane potential, causing the release of cytochrome c, as it has previously been shown in mitochondria (Shailasree, Venkataramana, Niranjana, & Prakash, 2014). Moreover, an increased relative abundance of genes involved in energy metabolism in E. coli has also been shown to play a role in free fatty acids stress conditions (Lennen et al., 2011). Furthermore, the cytochrome c maturation system has been described to play a role in other cellular processes in other organisms (Cianciotto, Cornelis, & Baysse, 2005), such as P. aeruginosa (Baert, Baysse, Matthijs, & Cornelis, 2008) or P. fluorescens (Yang, Azad, & Cooksey, 1996). A transposon insertion mutant in this operon in the strain P. putida P8 was found to have reduced cis‐trans isomerization of unsaturated fatty acids, since a cytochrome c‐type heme‐binding motif was found to catalyze the activity of this enzyme.
In addition to the genes discussed above, other genes such as the transcriptional repressor phaN, a number of outer membrane proteins and surface adhesion proteins as well as other ABC transporters (PP_0168, PP_0674, and PP_1185) were identified to be important for tolerance, most of them in the P. putida KT2440 background. It is possible that these genes may be related to the detoxification of the intermediates of the p‐coumaric acid degradation pathway. The identification of these efflux pumps supports the observation that the role of transporters are of major importance when P. putida KT2440 is exposed to chemical stress, as it has also been observed in other organisms such as Clostridium acetobutylicum in the presence of butanol.
The importance of some of these genes was validated by assessing the effect of p‐coumaric acid on the growth of a number of independent insertion mutants in the genes ttg2A and ttg2B, ccmC and ccmF, vacJ, tyrB, and fleN, which displayed clear growth impairment at high concentrations of p‐coumaric acid when compared to the wild‐type (Figure 3b).
One of the beneficial insertions found, in gene PP_4515, is annotated in P. putida KT2440 as belonging to the MarR transcriptional regulator family. However, a BLASTp analysis of the protein sequence showed that this transcriptional regulator seems to be more closely related to the transcriptional regulator SlyA, with a 38% of identity in the amino acid sequence. SlyA belongs to a large family of regulators involved in different processes, and it has been shown to regulate chaperones and other stress proteins in one strain of E. coli (Spory et al., 2002). However, the activity of various homologs differs widely among species, thus further work is required in order to understand the cellular mechanism in P. putida KT2440 of this regulator. The other gene, PP_1262, belonging to the LysR‐type transcriptional regulator family has no known function. The LysR‐type transcriptional regulator family has been described to regulate a number of functions, such as aromatic acid metabolism, and both activators and repressors have been identified. Most of them are described to be transcribed divergently of the genes they regulate. Here, the operon PP_1263 to PP_1266, which is divergently transcribed, has been annotated as an efflux system for the export of fusaric acid, a fungal toxin released by phytopathogenes (Martins Dos Santos, Heim, Moore, Strätz, & Timmis, 2004). Further studies are required to elucidate the effect of this regulator and the genes that it possibly regulates for the tolerance.
In conclusion, we have developed a method for generating large libraries of random insertion mutants in P. putida KT2440 with a minimum amount of bias by avoiding natural P. putida selection methods. P. putida KT2440 was found to have enhanced tolerance toward p‐coumaric acid when compared to E. coli. Using Tn‐seq analysis, the mechanisms of tolerance were found to involve membrane stability, exclusion, and secretion of the toxic compound to the outside of the cell. Due to its significant tolerance, P. putida could be a promising production host for p‐coumaric acid. The developed Tn‐seq method will be generally useful for investigating a wide range of conditions in this organism.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Growth curves of the P. putida KT2440 (WT) in M9 (blue) and M9 supplemented with p‐coumaric acid at a concentration of 15 (red) and 30 (green) mM in 96‐well microtiter plates.
ACKNOWLEDGMENTS
This work was supported by The Novo Nordisk Foundation, and a PhD grant from the People Programme (Marie Curie Actions) of the European Union Seventh Framework Programme FP7‐People‐2012‐ITN, under grant agreement No. 317058, “BACTORY.” We thank Juan Luis Ramos for providing strains from the PRCC and plasmids for the constructions of the libraries.
Calero P, Jensen SI, Bojanovič K, Lennen RM, Koza A, Nielsen AT. Genome‐wide identification of tolerance mechanisms toward p‐coumaric acid in Pseudomonas putida . Biotechnology and Bioengineering. 2018;115: 762–774. https://doi.org/10.1002/bit.26495
REFERENCES
- Abril, M. A. , Michan, C. , Timmis, K. N. , & Ramos, J. L. (1989). Regulator and enzyme specificities of the TOL plasmid‐encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. Journal of Bacteriology, 171, 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper, H. , Moxley, J. , Nevoigt, E. , Fink, G. R. , & Stephanopoulos, G. (2006). Tolerance and Production. Science, 314, 1565–1568. [DOI] [PubMed] [Google Scholar]
- Atsumi, S. , Wu, T.‐Y. , Machado, I. M. P. , Huang, W.‐C. , Chen, P.‐Y. , Pellegrini, M. , & Liao, J. C. (2010). Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli . Molecular Systems Biology, 6, 449 https://doi.org/10.1038/msb.2010.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert, B. , Baysse, C. , Matthijs, S. , & Cornelis, P. (2008). Multiple phenotypic alterations caused by a c‐type cytochrome maturation ccmC gene mutation in Pseudomonas aeruginosa . Microbiology, 154, 127–138. [DOI] [PubMed] [Google Scholar]
- Bagdasarian, M. , Lurz, R. , Rückert, B. , Franklin, F. C. H. , Bagdasarian, M. M. , Frey, J. , & Timmis, K. N. (1981). Specific‐purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010‐derived vectors, and a host‐vector system for gene cloning in Pseudomonas . Gene, 16, 237–247. [DOI] [PubMed] [Google Scholar]
- Barquist, L. , Boinett, C. J. , & Cain, A. K. (2013). Approaches to querying bacterial genomes with transposon‐insertion sequencing. RNA Biology, 10, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf, D. , Thiersch, M. , Loffhagen, N. , Kunath, C. , & Harms, H. (2006). Pseudomonas putida KT2440 responds specifically to chlorophenoxy herbicides and their initial metabolites. Proteomics, 6, 3319–3329. [DOI] [PubMed] [Google Scholar]
- Calero, P. , Jensen, S. I. , & Nielsen, A. T. (2016). Broad‐Host‐Range ProUSER vectors enable fast characterization of inducible promoters and optimization of p‐Coumaric acid production in Pseudomonas putida KT2440. ACS Synthetic Biology, 5, 741–753. [DOI] [PubMed] [Google Scholar]
- Carpenter, C. D. , Cooley, B. J. , Needham, B. D. , Fisher, C. R. , Trent, M. S. , Gordon, V. , & Paynea, S. M. (2014). The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri . Infection and Immunity, 82, 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. H. , Kumar, A. , & Schweizer, H. P. (2006). A 10‐min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. Journal of Microbiological Methods, 64, 391–397. [DOI] [PubMed] [Google Scholar]
- Choi, K.‐H. , Gaynor, J. B. , White, K. G. , Lopez, C. , Bosio, C. M. , Karkhoff‐Schweizer, R. R. , & Schweizer, H. P. (2005). A Tn7‐based broad‐range bacterial cloning and expression system. Nature Methods, 2, 443–448. [DOI] [PubMed] [Google Scholar]
- Cianciotto, N. P. , Cornelis, P. , & Baysse, C. (2005). Impact of the bacterial type I cytochrome c maturation system on different biological processes. Molecular Microbiology, 56, 1408–1415. [DOI] [PubMed] [Google Scholar]
- Cruden, D. L. , Wolfram, J. H. , Rogers, R. D. , & Gibson, D. T. (1992). Physiological properties of a Pseudomonas strain which grows with p‐xylene in a two‐phase (organic‐aqueous) medium. Applied and Environmental Microbiology, 58, 2723–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, N. , Arora, S. K. , & Ramphal, R. (2000). fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa . Journal of Bacteriology, 182, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V. , & Timmis, K. N. (1994). Analysis and construction of stable phenotypes in gram‐negative bacteria with Tn5‐ and Tn10‐derived minitransposons. Methods in Enzymology, 235, 386–405. [DOI] [PubMed] [Google Scholar]
- Domínguez‐Cuevas, P. , González‐Pastor, J.‐E. , Marqués, S. , Ramos, J.‐L. , & de Lorenzo, V. (2006). Transcriptional tradeoff between metabolic and stress‐response programs in Pseudomonas putida KT2440 cells exposed to toluene. The Journal of Biological Chemistry, 281, 11981–11991. [DOI] [PubMed] [Google Scholar]
- Dunlop, M. J. , Dossani, Z. Y. , Szmidt, H. L. , Chu, H. C. , Lee, T. S. , Keasling, J. D. , … Mukhopadhyay, A. (2011). Engineering microbial biofuel tolerance and export using efflux pumps. Molecular Systems Biology, 7, 487 https://doi.org/10.1038/msb.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque, E. , Molina‐Henares, A. J. , De Torre, J. , Molina‐Henares, M. A. , Castillo, T. , Lam, J. , & Ramos, J. L. (2007). Towards a genome‐wide mutant library of Pseudomonas puida strain KT2440. In: Ramos, J. L., & Filloux, A. (Eds.), Pseudomonas, (pp. 227–251). Dordrecht: Springer. [Google Scholar]
- Fernandez, M. , Conde, S. , De La Torre, J. , Molina‐Santiago, C. , Ramos, J. L. , & Duque, E. (2012). Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrobial Agents and Chemotherapy, 56, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, M. , Niqui‐Arroyo, J. L. , Conde, S. , Ramos, J. L. , & Duque, E. (2012). Enhanced tolerance to naphthalene and enhanced rhizoremediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid. Applied and Environmental Microbiology, 78, 5104–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V. , Godoy, P. , Daniels, C. , Hurtado, A. , Ramos, J. L. , & Segura, A. (2010). Functional analysis of new transporters involved in stress tolerance in Pseudomonas putida DOT‐T1E. Environmental Microbiology Reports, 2, 389–395. [DOI] [PubMed] [Google Scholar]
- Gawronski, J. D. , Wong, S. M. S. , Giannoukos, G. , Ward, D. V. , & Akerley, B. J. (2009). Tracking insertion mutants within libraries by deep sequencing and a genome‐wide screen for Haemophilus genes required in the lung. Proceedings of the National Academy of Sciences of the United States of America, 106, 16422–16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, J. I. , Assad‐Garcia, N. , Alperovich, N. , Yooseph, S. , Lewis, M. R. , Maruf, M. , … Venter, J. C. (2006). Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences of the United States of America, 103, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi, H. , Bennett, B. D. , Amini, S. , Reaves, M. L. , Hottes, A. K. , Rabinowitz, J. D. , & Tavazoie, S. (2010). Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Molecular Systems Biology, 6, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1985). Techniques for transformation of E. coli In Glover D. M. (Ed.), DNA cloning a Pract. approach (pp. 109–135). Oxford: United Kingdom IRL Press. [Google Scholar]
- Heipieper, H. J. , & De Bont, J. A. M. (1994). Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Applied and Environmental Microbiology, 60, 4440–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel, M. , Shea, J. E. , Gleeson, C. , Jones, M. D. , Dalton, E. , & Holden, D. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science, 269, 400–403. [DOI] [PubMed] [Google Scholar]
- Herrero, M. , Lorenzo, V. D. E. , & Timmis, K. N. (1990). Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram‐negative bacteria. Journal of Bacteriology, 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, A. , Yamamoto, M. , & Horikoshi, K. (1991). Pseudomonas putida which can grow in the presence of toluene. Applied and Environmental Microbiology, 57, 1560–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, M. , Alwood, A. , Thaipisuttikul, I. , Spencer, D. , Haugen, E. , Ernst, S. , … Manoil, C. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America, 100, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, J. I. , Miñambres, B. , Luis, J. , & Díaz, E. (2002). Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environmental Microbiology, 4, 824–841. [DOI] [PubMed] [Google Scholar]
- Jönsson, L. J. , & Martín, C. (2016). Pretreatment of lignocellulose: Formation of inhibitory by‐products and strategies for minimizing their effects. Bioresource Technology, 199, 103–112. [DOI] [PubMed] [Google Scholar]
- Keasling, J. D. (2010). Manufacturing molecules through metabolic enginering. Science, 330, 1355–1358. [DOI] [PubMed] [Google Scholar]
- Kessler, B. , de Lorenzo, V. , & Timmis, K. (1992). A general system to integrate lacZ fusions into the chromosomes of gram‐negative eubacteria: Regulation of the Pm promoter of the TOL plasmid. Molecular and General Genetics, 233, 293–301. [DOI] [PubMed] [Google Scholar]
- Kieboom, J. , & de Bont, J. (2001). Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology, 147, 43–51. [DOI] [PubMed] [Google Scholar]
- Kim, K. , Lee, S. , Lee, K. , & Lim, D. (1998). Isolation and characterization of toluene‐sensitive mutants from the toluene‐resistant bacterium Pseudomonas putida GM73. Journal of Bacteriology, 180, 3692–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, B. , Jensen, L. E. , & Nybroe, O. (2001). A panel of Tn7‐based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into gram‐negative bacteria at a neutral chromosomal site. Journal of Microbiological Methods, 45, 187–195. [DOI] [PubMed] [Google Scholar]
- Lambertsen, L. , Sternberg, C. , & Molin, S. (2004). Mini‐Tn7 transposons for site‐specific tagging of bacteria with fluorescent proteins. Environmental Microbiology, 6, 726–732. [DOI] [PubMed] [Google Scholar]
- Langridge, G. C. , Phan, M. D. , Turner, D. J. , Perkins, T. T. , Parts, L. , Haase, J. , … Turner, A. K. (2009). Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research, 19, 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen, R. , & Herrgård, M. J. (2014). Combinatorial strategies for improving multiple‐stress resistance in industrially relevant Escherichia coli strains. Applied and Environmental Microbiology, 80, 6223–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen, R. M. , Kruziki, M. A. , Kumar, K. , Zinkel, R. A. , Burnum, K. E. , Lipton, M. S. , … Pfleger, B. F. (2011). Membrane stresses induced by overproduction of free fatty acids in Escherichia coli . Applied and Environmental Microbiology, 77, 8114–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeschcke, A. , & Thies, S. (2015). Pseudomonas putida—A versatile host for the production of natural products. Applied Microbiology and Biotechnology, 99, 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Z. , Wang, H. , Rao, S. , Sun, J. , Ma, C. , & Li, J. (2012). P‐Coumaric acid kills bacteria through dual damage mechanisms. Food Control, 25, 550–554. [Google Scholar]
- Mack, D. , Nedelmann, M. , Krokotsch, A. , Schwarzkopf, A. , Heesemann, J. , & Laufs, R. (1994). Characterization of transposon mutants of biofilm‐producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: Genetic identification of a hexosamine‐containing polysaccharide intercellular adhesin. Infection and Immunity, 62, 3244–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni, J. C. , & Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the gram‐negative outer membrane. Proceedings of the National Academy of Sciences of the United States of America, 106, 8009–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, E. , & de Lorenzo, V. (2011). Engineering multiple genomic deletions in gram‐negative bacteria: Analysis of the multi‐resistant antibiotic profile of Pseudomonas putida KT2440. Environmental Microbiology, 13, 2702–2716. [DOI] [PubMed] [Google Scholar]
- Martins Dos Santos, V. A. P. , Heim, S. , Moore, E. R. B. , Strätz, M. , & Timmis, K. N. (2004). Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environmental Microbiology, 6, 1264–1286. [DOI] [PubMed] [Google Scholar]
- Molina‐Henares, M. A. , de la Torre, J. , Garcia‐Salamanca, A. , Molina‐Henares, A. J. , Herrera, M. C. , Ramos, J. L. , & Duque, E. (2010). Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome‐wide mutant library. Environmental Microbiology, 12, 1468–1485. [DOI] [PubMed] [Google Scholar]
- Mosqueda, G. , & Ramos, J. L. (2000). A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT‐T1E is linked to the tod genes for toluene metabolism. Journal of Bacteriology, 182, 937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussatto, S. I. , Dragone, G. , & Roberto, I. C. (2007). Ferulic and p‐coumaric acids extraction by alkaline hydrolysis of brewer's spent grain. Industrial Crops and Products, 25, 231–237. [Google Scholar]
- Nicolaou, S. A. , Gaida, S. M. , & Papoutsakis, E. T. (2010). A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metabolic Engineering, 12, 307–331. [DOI] [PubMed] [Google Scholar]
- Nikel, P. I. , Martínez‐García, E. , & de Lorenzo, V. (2014). Biotechnological domestication of pseudomonads using synthetic biology. Nature Reviews Microbiology, 12, 368–379. [DOI] [PubMed] [Google Scholar]
- Nour‐Eldin, H. H. , Geu‐Flores, F. , & Halkier, B. (2010). USER cloning and USER fusion: The ideal cloning techniques for small and big laboratories. Methods in Molecular Biology, 643, 185–200. [DOI] [PubMed] [Google Scholar]
- Poblete‐Castro, I. , Becker, J. , Dohnt, K. , dos Santos, V. M. , & Wittmann, C. (2012). Industrial biotechnology of Pseudomonas putida and related species. Applied Microbiology and Biotechnology, 93, 2279–2290. [DOI] [PubMed] [Google Scholar]
- Ramos, J. L. , Duque, E. , Gallegos, M. T. , Godoy, P. , Ramos‐González, M. I. , Rojas, A. , … Segura, A. (2002). Mechanisms of solvent tolerance in gram‐negative bacteria. Annual Review of Microbiology, 56, 743–768. [DOI] [PubMed] [Google Scholar]
- Ramos, J. L. , Duque, E. , Godoy, P. , & Segura, A. (1998). Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT‐T1E. Journal of Bacteriology, 180, 3323–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, M. H. , Calero, P. , Lennen, R. M. , Long, K. S. , & Nielsen, A. T. (2016). Genome‐wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microbial Cell Factories, 15, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca, A. , Rodríguez‐Herva, J. J. , Duque, E. , & Ramos, J. L. (2008). Physiological responses of Pseudomonas putida to formaldehyde during detoxification. Microbial Biotechnology, 1, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier, S. , Zingl, F. G. , Cakar, F. , Durakovic, S. , Kohl, P. , Eichmann, T. O. , … Schild, S. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in gram‐negative bacteria. Nature Communications, 7, 10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl, J. , Schmid, A. , & Blank, L. M. (2009). Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations. Applied and Environmental Microbiology, 75, 4653–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , & Russel, D. W. (2000). Molecular cloning: A laboratory manual. 3rd ed. New York: Cold Spring Harboc Lab. Press. [Google Scholar]
- Santiago, M. , Matano, L. M. , Moussa, S. H. , Gilmore, M. S. , Walker, S. , & Meredith, T. C. (2015). A new platform for ultra‐high density Staphylococcus aureus transposon libraries. BMC Genomics, 16, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, C. N. S. , Koffas, M. , & Stephanopoulos, G. (2011). Optimization of a heterologous pathway for the production of flavonoids from glucose. Metabolic Engineering, 13, 392–400. [DOI] [PubMed] [Google Scholar]
- Santos, P. M. , Benndorf, D. , & Sá‐Correia, I. (2004). Insights into Pseudomonas putida KT2440 response to phenol‐induced stress by quantitative proteomics. Proteomics, 4, 2640–2652. [DOI] [PubMed] [Google Scholar]
- Sariaslani, F. S. (2007). Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annual Review of Microbiology, 61, 51–69. [DOI] [PubMed] [Google Scholar]
- Shailasree, S. , Venkataramana, M. , Niranjana, S. R. , & Prakash, H. S. (2014). Cytotoxic effect of p‐Coumaric acid on neuroblastoma, N2a cell via generation of reactive oxygen species leading to dysfunction of mitochondria inducing apoptosis and autophagy. Molecular Neurobiology, 51, 119–130. [DOI] [PubMed] [Google Scholar]
- Spory, A. , Spory, A. , Bosserhoff, A. , Bosserhoff, A. , von Rhein, C. , von Rhein, C. , … Ludwig, A. (2002). Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. Journal of Bacteriology, 184, 3549–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, B. W. (2001). Influence of vegetation on low‐molecular‐weight carboxylic acids in soil solution—A review. Geoderma, 99, 169–198. [Google Scholar]
- Thöny‐Meyer, L. (2000). Haem‐polypeptide interactions during cytochrome c maturation. Biochimica et Biophysica Acta (BBA) ‐ Bioenergetics, 1459, 316–324. [DOI] [PubMed] [Google Scholar]
- van Opijnen, T. , & Camilli, A. (2013). Transposon insertion sequencing: A new tool for systems‐level analysis of microorganisms. Nature Reviews Microbiology, 11, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef, S. , Ruijssenaars, H. J. , de Bont, J. A. M. , & Wery, J. (2007). Bioproduction of p‐hydroxybenzoate from renewable feedstock by solvent‐tolerant Pseudomonas putida S12. Journal of Biotechnology, 132, 49–56. [DOI] [PubMed] [Google Scholar]
- Verhoef, S. , Wierckx, N. , Westerhof, R. G. M. , De Winde, J. H. , & Ruijssenaars, H. J. (2009). Bioproduction of p‐hydroxystyrene from glucose by the solvent‐tolerant bacterium Pseudomonas putida S12 in a two‐phase water‐decanol fermentation. Applied and Environmental Microbiology, 75, 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, F. J. , Ooijkaas, L. P. , Schemen, R. M. W. , Hartmans, S. , & De Bont, J. A. M. (1993). Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Applied and Environmental Microbiology, 59, 3502–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werpy, T. , & Petersen, G. (2004). Top Value Added Chemicals from Biomass Volume I — Results of Screening for Potential Candidates from Sugars and Synthesis Gas Top Value Added Chemicals From Biomass Volume I: Results of Screening for Potential Candidates. Oak Ridge, TN: US Department of Energy.
- Yang, C. H. , Azad, H. R. , & Cooksey, D. A. (1996). A chromosomal locus required for copper resistance, competitive fitness, and cytochrome c biogenesis in Pseudomonas fluorescens . Proceedings of the National Academy of Sciences of the United States of America, 93, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer, A. , Burghout, P. , Bootsma, H. J. , Hermans, P. W. M. , & van Hijum, S. A. F. T. (2012). Essentials: Software for rapid analysis of high throughput transposon insertion sequencing data. PLoS ONE, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Growth curves of the P. putida KT2440 (WT) in M9 (blue) and M9 supplemented with p‐coumaric acid at a concentration of 15 (red) and 30 (green) mM in 96‐well microtiter plates.


