Key Points
The binding of mutant calreticulin to MPL can be uncoupled from MPL activation.
The lectin activity but not the chaperone functionality of mutant CALR is required for cytokine-independent growth.
Abstract
Mutations in calreticulin (CALR) are phenotypic drivers in the pathogenesis of myeloproliferative neoplasms. Mechanistic studies have demonstrated that mutant CALR binds to the thrombopoietin receptor MPL, and that the positive electrostatic charge of the mutant CALR C terminus is required for mutant CALR-mediated activation of JAK-STAT signaling. Here we demonstrate that although binding between mutant CALR and MPL is required for mutant CALR to transform hematopoietic cells; binding alone is insufficient for cytokine independent growth. We further show that the threshold of positive charge in the mutant CALR C terminus influences both binding of mutant CALR to MPL and activation of MPL signaling. We find that mutant CALR binds to the extracellular domain of MPL and that 3 tyrosine residues within the intracellular domain of MPL are required to activate signaling. With respect to mutant CALR function, we show that its lectin-dependent function is required for binding to MPL and for cytokine independent growth, whereas its chaperone and polypeptide-binding functionalities are dispensable. Together, our findings provide additional insights into the mechanism of the pathogenic mutant CALR-MPL interaction in myeloproliferative neoplasms.
Introduction
Recurrent mutations in calreticulin (CALR), an endoplasmic reticulum (ER) resident chaperone protein, represent the second most common mutation in patients with myeloproliferative neoplasms (MPNs) after JAK2V617F.1-4 CALR mutations in MPN occur as a heterogeneous set of indel mutations in exon 9 of CALR that all result in a +1 bp frameshift in the CALR reading frame.5,6 Although the mutant CALR C-terminal alterations vary, all CALR mutations lead to a loss of most of the C-terminal acidic domain and a concomitant gain of a novel C terminus consisting of 36 amino acids that are enriched for positively charged residues. Mechanistic studies have demonstrated that mutant CALR (CALRMUT) binds to the thrombopoietin receptor, MPL, to activate the MPL-JAK-STAT signaling axis,7-10 and that the positive charge of the CALRMUT C terminus is required to mediate this interaction.9
Several unanswered questions remain regarding the molecular and functional basis of CALRMUT oncogenic activity. In this report, we use a series of mutagenesis experiments to address some of these questions, focusing on the structural determinants of CALRMUT and MPL that are necessary for hematopoietic cell transformation.
Study design
Ba/F3 cell growth assays
Ba/F3 cells expressing CALRMUT or MPL variants were generated by retroviral transduction and assayed for cytokine-independent growth as previously described.9
In vitro binding assay
V5-tagged purified recombinant wild-type CALR (CALRWT), CALRMUT, CALRMUT-D135L, and CALRMUT-D317A proteins were incubated with purified recombinant ERp57 or MPL for 30 minutes at 4°C, and washed with N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid acetate buffer. Bound proteins were eluted from beads and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Results and discussion
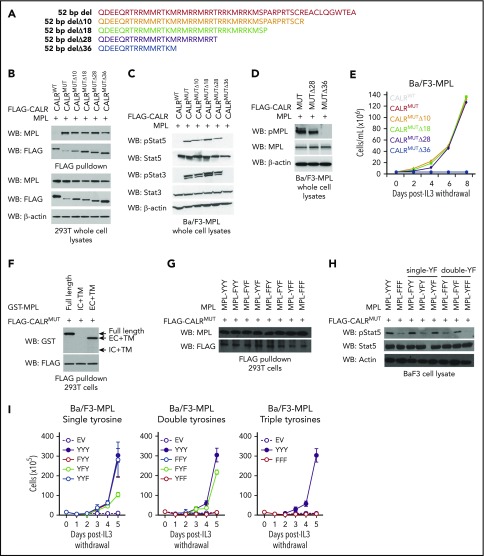
To resolve which amino acids within the CALRMUT C terminus are required for CALRMUT activity, we generated CALRMUT variants harboring serial truncations of the mutant C-terminal tail in blocks of 8 to 10 amino acids (Figure 1A) and tested their ability to associate with MPL in pull-down assays. All CALRMUT variants examined retained the ability to bind MPL (Figure 1B). However, the most severely truncated form (CALRMUTΔ36) failed to activate JAK-STAT signaling (Figure 1C), stimulate phosphorylation of MPL (Figure 1D), and transform Ba/F3-MPL cells to interleukin-3 independence (Figure 1E). Further truncation of an 11 amino acid positively charged stretch (QRTRRMMRTKM) that is also present in CALRMUT protein species generated by the 52-bp deletion (CALRMUTΔ47; see supplemental Figure 1A , available on the Blood Web site) led to loss of MPL binding (supplemental Figure 1B) and inability to transform Ba/F3-MPL cells (supplemental Figure 1C). A CALRMUT variant in which these 11 residues are deleted but the distal 36 mutant-specific amino acids are retained (CALRMUTΔ37-47) was still able to bind to MPL and transform Ba/F3-MPL cells (supplemental Figure 1A-C), suggesting that the QRTRRMMRTKM stretch is sufficient but not strictly required for MPL binding. Our data also suggest that although CALRMUTΔ36 can bind to MPL, this binding is insufficient to activate MPL signaling. To our knowledge, these data provide the first evidence that physical interaction between CALRMUT and MPL is not ipso facto sufficient to activate MPL. Rather, our data argue for a model whereby different thresholds of positive charge in the CALRMUT C terminus are required to enable binding of CALRMUT to MPL and to activate MPL signaling. Because MPL phosphorylation is dependent on homodimerization of single MPL chains, these data may suggest that CALRMUTΔ36 retains the ability to interact with single MPL chains but is unable to induce homodimerization, which is required for receptor activation. Further studies are warranted to fully resolve the 3-dimensional structure of the CALRMUT-MPL interaction.
Figure 1.
Binding of mutant calreticulin to MPL is required to transform hematopoietic cells but binding alone is insufficient for cytokine-independent growth. (A) Schema depicting serial C-terminal truncation mutants of mutant CALR. (B) Immunoblotting of FLAG-immunoprecipitated proteins and whole cell lysates from 293T cells cotransfected with wild-type FLAG-CALR (CALRWT), FLAG-CALR 52-bp deletion (CALRMUT), or FLAG-CALR 52-bp deletion serial C-terminal truncation mutants (CALRMUT Δ10-Δ36) demonstrates that mutant CALR truncated up to Δ36 still binds to MPL. (C) Immunoblotting demonstrates phosphorylation of Stat5 and Stat3 in Ba/F3-MPL cells expressing CALRMUT and truncation variants Δ10, Δ18, and Δ28, but not Δ36. (D) Immunoblotting demonstrates phosphorylation of MPL in Ba/F3-MPL cells expressing CALRMUT and Δ28, but not Δ36. (E) Growth curves in Ba/F3-MPL cells expressing CALRWT, CALRMUT, or CALRMUT C-terminal truncation variants demonstrates that only severe truncation of the mutant CALR C terminus (Δ36) abolishes the transforming capacity of mutant CALR. (F) Immunoblotting of FLAG immunoprecipitated proteins from 293T cells co-transfected with FLAG-CALR 52 bp deletion (CALRMUT) and glutathione S-transferase (GST)–tagged full-length MPL, GST-tagged MPL intracellular + transmembrane domains, or GST-tagged MPL extracellular + transmembrane domains demonstrates that mutant CALR binds to the extracellular domain of MPL. (G) Immunoblotting of FLAG immunoprecipitated proteins from 293T cells coexpressing FLAG-tagged mutant CALR and MPL YF variants demonstrates that mutations of intracellular tyrosine residues on MPL does not affect the ability of mutant CALR to bind to MPL. (H) Immunoblotting demonstrates that phosphorylation of Stat5 is abrogated in Ba/F3 cells expressing MPL YF variants harboring loss of Y626. (I) Growth curves in Ba/F3 cells stably expressing MPL-YF variants demonstrate that all 3 intracellular tyrosines play a role in supporting cytokine independent growth in Ba/F3 cells mediated by mutant CALR. WB, western blot.
We next sought to elucidate the regions of MPL that are essential to support CALRMUT activity. We observed that CALRMUT binds to full-length MPL and to the extracellular and transmembrane fragment of MPL, but not to the intracellular and transmembrane fragment (Figure 1F). Furthermore, an MPL variant in which the thrombopoietin (TPO) binding site is mutated (D235A/L239A)11 was still able to bind to CALRMUT (supplemental Figure 1D) and could support CALRMUT-mediated cytokine-independent growth in Ba/F3 cells (supplemental Figure 1E). This suggests that CALRMUT does not occupy the same binding pocket as TPO and is consistent with CALRMUT-driven hematopoietic transformation being a TPO-independent process.7
Analysis of the intracellular portion of MPL reveals 3 tyrosine residues (Y591, Y626, and Y631) that may also be important for CALRMUT-MPL signaling.12 MPL variants were therefore generated where these residues were systematically mutated to phenylalanine individually (FYY, YFY, YYF), in tandem (FFY, FYF, YFF), or altogether (FFF). As expected, all MPL variants were able to physically interact with CALRMUT in FLAG pull-down assays (Figure 1G). However, we found differences in their ability to support CALRMUT signaling. MPL variants harboring intact Y626 (MPL-FYY, MPL-YYF, MPL-FYF) supported robust Stat5 phosphorylation (Figure 1H) in association with cytokine-independent growth (Figure 1I), but not MPL variants in which the Y626 is mutated (MPL-YFY, MPL-YFF, MPL-FFY, MPL-FFF). These data indicate that Y626 plays a more prominent role than Y591 and Y631 in MPL signaling downstream of CALRMUT, which is consistent with a previous report identifying Y626 as the major signaling tyrosine in canonical TPO-MPL signaling.12 Our data therefore highlight the importance of tyrosine-mediated MPL signaling as a pathway coopted by CALRMUT to effect cytokine-independent proliferation. Our studies do not rule out a role for nontyrosine residues of MPL, which may be required for CALRMUT-mediated activation of other downstream signaling pathways (eg, extracellular signal-regulated kinase)13; future studies to explore this question are warranted.
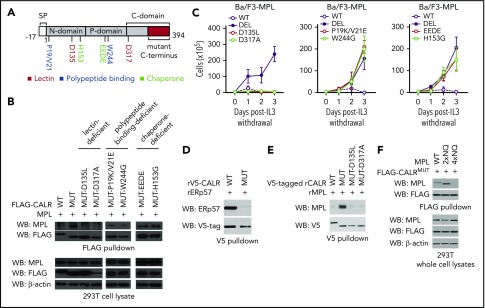
Finally, we sought to gain functional insights into how CALRMUT interacts with MPL to confer cytokine-independent growth. Wild-type calreticulin is an ER-resident chaperone that interacts with glycoproteins by binding to Glc1Man9GlcNAc2 oligosaccharides and the polypeptide backbone to facilitate proper protein folding. We therefore created variants of CALRMUT harboring mutations in critical residues implicated in 3 key functionalities of wild-type CALR: (1) polypeptide binding, (2) chaperone activity, and (3) lectin activity. We then tested their capacity to bind to MPL and confer cytokine independence (Figure 2A).
Figure 2.
The lectin-dependent function of mutant CALR is required for cytokine-independent growth, whereas its chaperone and polypeptide binding functionalities are dispensable. (A) Schema depicting mutations introduced into the lectin (dark red), polypeptide binding (blue), and chaperone (light green) domains of mutant CALR. (B) Immunoblotting of FLAG immunoprecipitated proteins from 293T cells cotransfected with wild-type CALR, mutant CALR, or mutant CALR lectin-, chaperone-, and polypeptide binding-deficient variants demonstrates that binding between mutant CALR and MPL is lost when residues required for lectin binding are mutated, but binding is retained when residues critical for CALR chaperone and polypeptide binding functionality are mutated. (C) Growth curves in Ba/F3-MPL cells expressing wild-type CALR, mutant CALR, or mutant CALR lectin- (left), chaperone- (center), and polypeptide binding-deficient (right) variants demonstrates that mutant CALR loses its ability to drive cytokine-independent growth when residues required for lectin binding are mutated but retains its ability to drive cytokine-independent growth in when residues critical for CALR chaperone and polypeptide-binding functionality are mutated. (D) In vitro binding assay between purified recombinant CALRWT or CALRMUT and purified recombinant ERp57 demonstrates that CALRWT binds directly to ERp57 but CALRMUT does not. (E) In vitro binding assay between purified recombinant CALRWT, CALRMUT, CALRMUT-D135L, or CALRMUT-D317A demonstrates that CALRMUT binds directly to MPL but not CALRWT or altered CALRMUT variants. (F) FLAG pulldown in 293T cells coexpressing CALRMUT and MPL glycosylation mutants (2xNQ = N117/178Q; 4xNQ = N117/178/298/358Q), shows CALRMUT binding to MPL-2xNQ but not to MPL-4xNQ.
We observed that both polypeptide binding-deficient variants of CALRMUT (CALRMUT-P19K/V21E and CALRMUT-W244G) and chaperone-deficient variants of CALRMUT (CALRMUT-H153G and CALRMUT-EEDE)14,15 retained MPL binding ability (Figure 2B) and conferred cytokine-independent growth (Figure 2C). Consistent with the nonessentiality of chaperone functionality in CALRMUT oncogenic activity, CALRWT exhibits strong, direct binding to the ERp57 cochaperone, whereas CALRMUT does not (Figure 2D). In contrast, lectin-deficient CALRMUT variants harboring mutations in Asp-135 and Asp-317 (CALRMUT-D135L and CALRMUT-D317A)16,17 were both unable to bind to MPL or confer cytokine independence (Figure 2B-C). In accordance, we also found that only recombinant CALRMUT protein directly binds to recombinant MPL in an in vitro binding assay, whereas neither CALRWT nor lectin-deficient CALRMUT do (Figure 2E). These data explain the previously reported essential role for Asp-135 in mediating CALRMUT-driven STAT5 activation8 as being from a requirement for Asp-135 in mediating binding between CALRMUT to MPL. Finally, to determine the requisite glycosylation status of MPL that enables CALRMUT binding, we tested CALRMUT binding to MPL mutants where either 2 (2xNQ = N117/178Q) or all 4 (4xNQ = N117/178/298/358Q) glycosylation sites in the extracellular domain of MPL were abolished. We found that CALRMUT can still bind to MPL-2xNQ but not to MPL-4xNQ (Figure 2F). These data are consistent with a previous report that demonstrated that MPL variants devoid of the same 4 N-glycosylation sites failed to support STAT5 activation by CALRMUT8 and suggests that this defect is due to an inability of unglycosylated MPL to bind CALRMUT.
In conclusion, our data provide additional insights into the molecular mechanism by which CALRMUT interacts with MPL to induce MPN (supplemental Table 1). Specifically, we (1) uncouple the binding of CALRMUT to MPL from MPL activation, (2) define the key properties of MPL required for CALRMUT binding and for its activation, and (3) decipher the key functionalities of CALRMUT required for its oncogenic activity.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI; grant R01HL131835) (A.M.), Damon Runyon clinical investigator award (A.M.), the Starr Cancer Consortium (A.M.), Leukemia and Lymphoma Society (LLS) (A.M.), Wellcome Trust (E.C.), Academy of Medical Science Springboard Award (E.C.), Leuka John Goldman Fellowship (E.C.), and Gabrielle’s Angel Foundation for Cancer Research (G.B.). S.E. is a recipient of a T32 molecular hematology training award (NHLBI) and an LLS Special Fellow Award.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.E., N.S.A., E.C., and A.M. designed the study; G.B. provided guidance on experimental design; S.E., N.S.A., A.J.B., D.B., G.B., J.F.R., A.K., and N.F. performed experiments and collected the data; S.E., N.S.A., A.J.B., and D.B. analyzed the data; and S.E., N.S.A., E.C., and A.M. wrote the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Ann Mullally, Harvard Institutes of Medicine Building, Room 738, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail: amullally@partners.org; or Edwin Chen, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Woodhouse Ln, Leeds LS2 9JT, United Kingdom; e-mail: e.chen@leeds.ac.uk.
References
- 1.Baxter EJ, Scott LM, Campbell PJ, et al. ; Cancer Genome Project. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054-1061. [DOI] [PubMed] [Google Scholar]
- 2.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387-397. [DOI] [PubMed]
- 3.James C, Ugo V, Le Couédic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144-1148. [DOI] [PubMed] [Google Scholar]
- 4.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779-1790. [DOI] [PubMed] [Google Scholar]
- 5.Klampfl T, Gisslinger H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369(25):2379-2390. [DOI] [PubMed] [Google Scholar]
- 6.Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369(25):2391-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marty C, Pecquet C, Nivarthi H, et al. Calreticulin mutants in mice induce an MPL-dependent thrombocytosis with frequent progression to myelofibrosis. Blood. 2016;127(10):1317-1324. [DOI] [PubMed] [Google Scholar]
- 8.Chachoua I, Pecquet C, El-Khoury M, et al. Thrombopoietin receptor activation by myeloproliferative neoplasm associated calreticulin mutants. Blood. 2016;127(10):1325-1335. [DOI] [PubMed] [Google Scholar]
- 9.Elf S, Abdelfattah NS, Chen E, et al. Mutant calreticulin requires both its mutant C-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov. 2016;6(4):368-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araki M, Yang Y, Masubuchi N, et al. Activation of the thrombopoietin receptor by mutant calreticulin in CALR-mutant myeloproliferative neoplasms. Blood. 2016;127(10):1307-1316. [DOI] [PubMed] [Google Scholar]
- 11.Chen WM, Yu B, Zhang Q, Xu P. Identification of the residues in the extracellular domain of thrombopoietin receptor involved in the binding of thrombopoietin and a nuclear distribution protein (human NUDC). J Biol Chem. 2010;285(34):26697-26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drachman JG, Kaushansky K. Dissecting the thrombopoietin receptor: functional elements of the Mpl cytoplasmic domain. Proc Natl Acad Sci U S A. 1997;94(6):2350-2355. [DOI] [PMC free article] [PubMed]
- 13.Han L, Schubert C, Köhler J, et al. Calreticulin-mutant proteins induce megakaryocytic signaling to transform hematopoietic cells and undergo accelerated degradation and Golgi-mediated secretion. J Hematol Oncol. 2016;9(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L, Groenendyk J, Papp S, et al. Identification of an N-domain histidine essential for chaperone function in calreticulin. J Biol Chem. 2003;278(50):50645-50653. [DOI] [PubMed] [Google Scholar]
- 15.Martin V, Groenendyk J, Steiner SS, et al. Identification by mutational analysis of amino acid residues essential in the chaperone function of calreticulin. J Biol Chem. 2006;281(4):2338-2346. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor M, Ellgaard L, Gopalakrishnapai J, et al. Mutational analysis provides molecular insight into the carbohydrate-binding region of calreticulin: pivotal roles of tyrosine-109 and aspartate-135 in carbohydrate recognition. Biochemistry. 2004;43(1):97-106. [DOI] [PubMed] [Google Scholar]
- 17.Thomson SP, Williams DB. Delineation of the lectin site of the molecular chaperone calreticulin. Cell Stress Chaperones. 2005;10(3):242-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.