Abstract
Prostate cancer (PC) is a very important kind of male malignancies. When PC evolves into a stage of hormone resistance or metastasis, the fatality rate is very high. Currently, discoveries and advances in miRNAs as biomarkers have opened the potential for the diagnosis of PC, especially early diagnosis. miRNAs not only can noninvasively or minimally invasively identify PC, but also can provide the data for optimization and personalization of therapy. Moreover, miRNAs have been shown to play an important role to predict prognosis of PC. The purpose of this meta‐analysis is to integrate the currently published expression profile data of miRNAs in PC, and evaluate the value of miRNAs as biomarkers for PC. All of relevant records were selected via electronic databases: Pubmed, Embase, Cochrane, and CNKI based on the assessment of title, abstract, and full text. we extracted mean ± SD or fold change of miRNAs expression levels in PC versus BPH or normal controls. Pooled hazard ratios (HRs) with 95% confidence intervals (CI) for overall survival (OS) and recurrence‐free survival (RFS), were also calculated to detect the relationship between high miRNAs expression and PC prognosis. Selected 104 articles were published in 2007‐2017. According to the inclusion criteria, 104 records were included for this meta‐analysis. The pooled or stratified analyze showed 10 up‐regulated miRNAs (miR‐18a, miR‐34a, miR‐106b, miR‐141, miR‐182, miR‐183, miR‐200a/b, miR‐301a, and miR‐375) and 14 down‐regulated miRNAs (miR‐1, miR‐23b/27b, miR‐30c, miR‐99b, miR‐139‐5p, miR‐152, miR‐187, miR‐204, miR‐205, miR‐224, miR‐452, miR‐505, and let‐7c) had relatively good diagnostic and predictive potential to discriminate PC from BPH/normal controls. Furthermore, high expression of miR‐32 and low expression of let‐7c could be used to differentiate metastatic PC from local/primary PC. Additional interesting findings were that the expression profiles of five miRNAs (miR‐21, miR‐30c, miR‐129, miR‐145, and let‐7c) could predict poor RFS of PC, while the evaluation of miR‐375 was associated with worse OS. miRNAs are important regulators in PC progression. Our results indicate that miRNAs are suitable for predicting the different stages of PC. The detection of miRNAs is an effective way to control patient's prognosis and evaluate therapeutic efficacy. However, large‐scale detections based on common clinical guidelines are still necessary to further validate our conclusions, due to the bias induced by molecular heterogeneity and differences in study design and detection methods.
Keywords: biomarker, meta‐analysis, microRNA, prostate cancer
1. INTRODUCTION
Prostate cancer (PC) is the leading male cancer worldwide. In 2016, PC is estimated to be responsible for 26 120 deaths in the United States.1 Early PC is localized which can be curable by a variety of therapies: chemotherapy, radiation therapy, radical prostatectomy, and cryotherapy, etc. Unfortunately, approximately 23‐40% of these patients would go on to develop metastatic tumors after initial therapy.2 Prostate tumors often metastasize to bone and other organs to cause patients death.3 At present, metastatic cases are treated with androgen‐deprivation therapy to induce apoptosis of tumor cells or to inhibit cells growth. This will further induce PC to be insensitive to hormone and progress to CRPC, which is essentially untreatable.
In spite of the prevalence of PC, there are no diagnostic or prognostic biomarkers to specifically and precisely distinguish its aggressiveness. In the early 1990s, the detection of PC dramatically increased due to the introduction of the prostate‐specific antigen (PSA) test, which had been used as a routine assay in clinic. PSA levels are not specific for PC, and may fluctuate to induce false‐positive due to infections, inflammation, or hyperplasia, etc. Due to the poor correlation between PSA levels and PC which leads to overdiagnosis and overtreatment,4, 5 the US Preventive Services Task Force recommends physicians not to routinely perform PSA‐based screening.6, 7, 8 Moreover, the prostate needle biopsy has also obvious defects because only 2% of the prostate tumor samples can be sampled by puncture.9 Therefore, we still need to seek unique biomarkers discriminating different stages of PC.
miRNAs are small, single‐stranded, non‐coding, 21‐23 nucleotides RNAs that are conserved and endogenous, and have been shown to regulate the expression of approximately 60% of human genes.10 miRNAs post‐transcriptionally regulate gene expression via base‐pairing with 3′‐untranslated regions (UTRs) of mRNA, and are found to be located in fragile regions involved in various cancers.11 miRNAs may regulate a wide range of biological processes: proliferation, apoptosis, development, and differentiation, etc, and are discovered to be aberrantly expressed in various carcinomas. Thus, more and more researchers are willing to consider miRNAs as diagnostic or prognostic biomarkers. Recently, miRNAs have attracted the attention of urologists and oncologists, because of their potential uses for the urologic cancers diagnosis, monitoring, and treatment. Specific miRNA may be used as marker to detect PC, predict prognosis, and monitor therapy. miRNAs are attractive biomarkers because they can be easily extracted from a wide range of biologic samples, and are stable in various storage conditions. Furthermore, miRNAs can be accurately detected by a variety of techniques, for example, qRT‐PCR, microarray, and next‐generation sequencing, etc. However, there are some controversies on miRNAs as biomarkers, because some studies obtain statistically insignificant results, and some draw inconsistent conclusions. In view of these results from different patient cohorts or various detection methods or different data analysis platforms, miRNAs are still considered an attractive biomarkers to assess recurrence and therapeutic effect. Therefore, we conducted a meta‐analysis to clarify the role of miRNAs for tumor progression and RFS and OS in PC clinical specimens.
2. MATERIALS AND METHODS
2.1. Search strategy
We performed a detailed literature search in PubMed, Embase, Cochrane, and Chinese National Knowledge Infrastructure databases to obtain relevant articles for this meta‐analysis. Relevant studies were selected according to a combination of keywords and Medical Subject Headings (MeSH): (“prostate cancer” or “prostate neoplasm” or “prostate tumor”) and (“microRNAs” or “miRNAs” or “miR‐”) and (“marker or “biomarker”). All selected studies in English or Chinese were viewed, and their reference lists were also examined for other eligible publications. Most studies were published between 2007and 2017. The last search update was finished on July 8, 2017. These studies regarding miRNAs and PC are performed in clinical samples or PC cell lines. Published data are subject to the limitation of small sample size and selection bias.
2.2. Inclusion and exclusion criteria
More than 1300 articles were retrieved, and 104 publications were included and reviewed in the meta‐analysis (Figure 1). Eligible studies had to fit the following inclusion criteria: (i) a kind of miRNA was involved in the studies; (ii) patients with PC were studied, and gold standard test (eg, histological examination) was used for the PC diagnosis; (iii) prostate tissue or serum or urine samples were used from PC patients or non‐PC patients for miRNA expression comparison; and (iv) validation method and enough patients' information were reported. Eligible studies that met above mentioned criteria were further evaluated and excluded according to a selection process showed in Figure 1. Exclusion criteria were as follows: (i) reviews, letters, commentary, or erratum; (ii) non‐English or non‐Chinese studies; (iii) data was obtained from PC cell lines; (iv) no sufficient data to extract; and (v) duplicate records.
Figure 1.
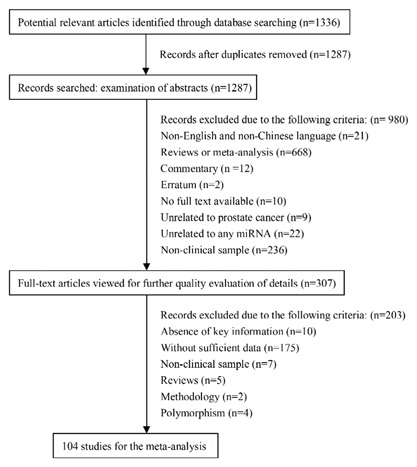
Flowchart of study selection process in this meta‐analysis
2.3. Data extraction
We assessed the data quality of each publication and extracted the following information: (i) basic features, such as first author, publication year, case region, study design, sample number, validation method, and detected miRNAs, as showed in Table 1; (ii) expression levels or fold‐change of detected miRNAs and predictive data, including OS and RFS; and (iii) information needed for quality assessment. If there were no data that could be extracted directly, we used the computer of revman 5.3 software to calculate and generate the relevant data.
Table 1.
The main characteristics of included studies
| First author & publishing year | Region | Study design | Detected samples | Validation | miRNA | Refs. |
|---|---|---|---|---|---|---|
| Robert S. Hudson 2012 | USA | PR | A large publicly available data set consisting of 99 primary tumors and 14 distant metastasis and patient data for disease recurrence | qRT‐PCR | miR‐1 | 32 |
| Yun‐Li Chang 2015 | China | P | 20 paired of PC tumors and adjacent normal tissues | qRT‐PCR | miR‐7 | 33 |
| Annika Fendler 2011 | Canada Germany | P | 52 primary prostate cancers and normal adjacent tissues | qRT‐PCR | miR‐10b | 34 |
| Bing Yang 2016 | China | P | 92 PC 85 BPH 97 controls | qRT‐PCR | miR‐21 | 16 |
| Christian Melbø‐Jørgensen 2014 | Norway | P | 535 PC patients 30 patients (14 patients with rapid biochemical failure (BF) and 16 patients without BF) with Gleason score 7 | microarray qRT‐PCR ISH | miR‐21 | 35 |
| Ernest K Amankwah 2013 | USA | P | 28 recurrent and 37 non‐recurrent prostate cancer cases | qRT‐PCR | miR‐21 | 28 |
| Judit Ribas 2009 | USA | P | 10 pairs | Northern blot | miR‐21 | 36 |
| Marco Folini 2010 | Italy | P | 36 pairs of PC and N tissues | qRT‐PCR | miR‐21 | 37 |
| Sabrina Thalita Reis 2012 | Brazil | P | 53 PC 11 benign prostatic hyperplasia (BPH) | qRT‐PCR | miR‐21 | 38 |
| Sarvesh Jajoo 2013 | USA | P | 18 PC | qRT‐PCR | miR‐21 | 39 |
| Tao Li 2012 | China | P | 169 radical prostatectomy tissue samples | ISH microarray | miR‐21 | 40 |
| Wei Huang 2015 | China | P | 75 localized PC 75 healthy volunteers | qRT‐PCR | miR‐21 | 17 |
| Yangbo Guan 2016 | China | P | 85 PC patients and 40 adjacent noncancerous biospy specimens | qRT‐PCR ISH | miR‐21 | 19 |
| Songwang Cai 2015 | China | P | 3 pairs of primary human prostate cancer and adjacent non‐tumor tissues 20 pairs of human prostate cancer and adjacent non‐tumor tissues. 123 prostate cancer tissues | Sequencing qRT‐PCR | miR‐23a | 41 |
| Hui‐chan He 2012 | China | P | 4 pairs 20 pairs 26 PC 20N | microarray qRT‐PCR ISH | miR‐23b | 42 |
| Shahana Majid 2012 | USA | P | 118 pairs of laser captured microdisected tissue samples an unmatched group of 27 benign prostatic hyperplasia (BPH) and 20 tumor samples another cohort of 48 samples | qRT‐PCR qMSP ISH | miR‐23b | 43 |
| Yusuke Goto 2014 | Japan | P | 41 noncancerous tissues 49 PC tissues | qRT‐PCR | miR‐23b/27b/24‐1 | 44 |
| Kai Guo 2016 | China | P | 140 pairs of fresh PC tissues and normal control tissues | qRT‐PCR | miR‐26a‐5p | 45 |
| Xiao‐hui Ling 2014 | China | P | 103 pairs of prostate tumor tissues and adjacent benign tissues and 28 benign prostate tissues gene expression omnibus (GEO) repository database (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE34932). | qRT‐PCR | miR‐30c | 46 |
| Xiao‐Hui Ling 2016 | China | P | 98 tumor tissue 20 benign prostate hyperplasia (BPH) specimens | qRT‐PCR | miR‐30c | 47 |
| Naohito Kobayashi 2012 | Japan | P | 56 pairs of primary PC and controls | Oligo chips qRT‐PCR | miR‐30d | 48 |
| SE Jalava 2012 | Finland | P | 5 benign prostatic hyperplasia (BPH) and 28 primary PCs 7 BPH and 14 CRPCs | microarray | miR‐32 | 49 |
| Q. Li 2017 | China | P | paired prostate cancer tissue and adjacent normal tissue | qRT‐PCR | miR‐33a | 50 |
| Shahana Majid 2013 | USA | P | 148 matched human tissue samples an unmatched group of 27 benign prostatic hyperplasia (BPH) and 20 tumor samples | qRT‐PCR ISH | miR‐34b | 51 |
| Zandra Hagman 2010 | Sweden | P | 49 PC patients and 25 benign prostatic hyperplasia | qRT‐PCR | miR‐34c | 52 |
| Robert S. Hudson 2013 | USA | PR | dataset for 28 non‐cancerous tissues, 99 primary tumors and 14 distant metastases with patient data for disease recurrence. | qRT‐PCR | miR‐106b‐25 | 53 |
| Xu‐Bao Shi 2013 | USA | P | 19 BPHs, 44 primary CaPs, 6 lymph node metastases, and 10 CR tumors | qRT‐PCR | miR‐124 | 54 |
| Xiaoke Sun 2013 | China | P | A series of 128 cases with PCa | qRT‐PCR | miR‐126 | 55 |
| Xiaoke Sun 2015 | China | P | 128 PC tissue and serum and matched controls | qRT‐PCR | miR‐128 | 56 |
| Song Xu 2015 | China | P | 98 PC and 56 health controls | qRT‐PCR | miR‐129 | 57 |
| Song Xu 2016 | China | P | 118 pairs of PC and noncancerous tissue | qRT‐PCR | miR‐129 | 58 |
| Xia Li 2014 | China | P | 135 specimens of patients with prostate cancer, 18 patients with prostatic intraepithelial neoplasia (PIN), and 25 normal prostate tissue samples) | qRT‐PCR ISH | miR‐133b | 59 |
| Cheng Pang 2016 | China | P | 45 PC patients, 45 benign prostatic hyperplasia (BPH) patients and 50 healthy controls serum peripheral whole blood samples | qRT‐PCR | miR‐139‐5p | 24 |
| Jason C. Gonzales 2011 | USA | P | 21 PC | qRT‐PCR | miR‐141 | 60 |
| Zhuo Li 2015 | China | P | 20 PCa, 20 BPH, and 20 control volunteers 51 PC and 40 control volunteers | qRT‐PCR | miR‐141 | 61 |
| M Avgeris 2013 | Greece | P | 73 radical prostatectomy‐treated PC patients and 64 benign prostate hyperplasia (BPH) patients | qRT‐PCR | miR‐145 | 62 |
| Bin Xu 2015 | China | PR | 13 ADPC 9 AIPC MSKCC prostate cancer database (GSE21032) | microarray qRT‐PCR | miR‐146a‐5p | 25 |
| Liu Dezhong 2015 | China | P | 167 PC 4 pairs of PC and adjacent to tumor healthy tissues to tumor | qRT‐PCR | miR‐150 | 63 |
| Shaniece C. Theodore 2014 | USA UK | P | 39 pairs of prostate cancer tissues and controls (20 AA and 19 CA) 97 primary tumors and 13 metastases | qRT‐PCR | miR‐152 | 64 |
| Zsuzsanna Lichner 2013 | Canada Germany | P | 41 prostatectomy samples were dichotomized to 27 high‐risk and 14 low‐risk The validation set: 35 high‐risk patients and 29 low‐risk patients | microarray qRT‐PCR | miR‐152 | 65 |
| Ranlu Liu 2013 | China | P | 5 PC 3 BPH | array | miR‐182 | 66 |
| Katsuki Tsuchiyama 2013 | Japan | P | patient set 1: 22 GP 3, 35 GP 4, and 12 GP 5 patient set 2: 10 GP 4 Cancer tissues from each GP and adjacent normal counterparts were separately collected using LCM | qRT‐PCR | miR‐182‐5p | 67 |
| Hongtuan Zhang 2015 | China | P | 180 pairs of PC and adjacent noncancerous tissues | qRT‐PCR | miR‐188‐5p | 68 |
| Amelie Hailer 2014 | Germany | P | 15 BPH 161 PC 17 LNM | qRT‐PCR | miR‐203 | 69 |
| Berlinda Verdoodt 2013 | Germany | P | 111 pairs of formalin‐fixed paraffin‐embedded (FFPE) prostatectomy specimens with primary prostate adenocarcinoma (PCa) and control | qRT‐PCR | miR‐205 | 70 |
| Charis Kalogirou 2013 | Germany Belgium | P | 105 HRPCa for study collective and 10 BHP 78 HRPCa for validation | qRT‐PCR | miR‐205 | 71 |
| Sigve Andersen 2016 | Norway | P | 535 prostatectomy patients | microarray ISH | miR‐210 | 72 |
| Aida Gordanpour 2011 | Canada | P | 153 radical prostatectomy samples | microarray qRT‐PCR | miR‐221 | 73 |
| Burkhard Kneitz 2014 | Germany Belgium | P | cohort 1, N = 134; cohort 2, n = 89 | qRT‐PCR | miR‐221 | 74 |
| Yongbao Wei 2014 | China | P | 10 pairs of PC tissues and adjacent non‐cancerous tissues | qRT‐PCR | miR‐223‐3p | 75 |
| Hao Fu 2015 | China | PR | A 4 and 20 pairs of primary PC and adjacent non‐tumor frozen samples the Taylor dataset (149 primary PC tissues and 29 adjacent non‐cancerous prostate tissues) | array qRT‐PCR | miR‐224 | 76 |
| Konstantinos Mavridis 2013 | Greece | P | 66 BPH or 73 CaP | qRT‐PCR | miR‐224 | 77 |
| Zhuo‐Yuan Lin 2014 | China USA | P | 4 and 20 pairs of primary PC and adjacent non‐tumor frozen samples Human PC tissue microarrays (TMA) consisting 114 PC tissues respectively from Caucasian and African‐American PC patients | array qRT‐PCR ISH | miR‐224 | 78 |
| Jian‐Jun Wei 2011 | USA | P | TMA100 contained 100 PC cases (from the Cooperative PC Tissue Resource at New York University) and TMA96 contained 96 cases (from Northwestern University). | microarray qRT‐PCR ISH | miR‐296 | 79 |
| Chendil Damodaran 2016 | USA | P | 58 FFPE 4 metastatic tumors, 6 fresh tumor tissues and 13 BPH | qRT‐PCR | miR‐301a | 80 |
| Robert K. Nam 2016 | Canada | P | 585 prostate cancer | qRT‐PCR | miR‐301a | 81 |
| Si‐wei Xiong 2013 | China | P | 20 clinical PC tissues 104 clinical PC tissues | qRT‐PCR ISH | miR‐335 | 82 |
| Sven Wach 2015 | Germany | P | 146 PC patients, 35 benign prostate hyperplasia (BPH) patients and 18 healthy controls serum | qRT‐PCR | miR‐375 | 31 |
| Yuan Wang 2016 | USA | R | 495 tumor tissues and 52 normal tissues from TCGA data | qRT‐PCR | miR‐375 | 83 |
| N Bucay 2017 | USA | PR | TCGA(187 primary prostate adenocarcinoma cases) validation cohort: 112 PC FFPE tissues and matched adjacent normals | qRT‐PCR | miR‐383 | 84 |
| Martin Mørck Mortensen 2014 | Denmark | P | 36 prostate cancer Samples 163 radical prostatectomy patients 40 patients (20 recurrent and 20 non‐recurrent patients) | qRT‐PCR | miR‐449b | 85 |
| Melissa Colden 2017 | USA | PR | 48 pairs of LCM tissue samples validation cohort: 56 prostate adenocarcinoma (TCGA database) | qRT‐PCR | miR‐466 | 86 |
| X. M. Tian 2017 | China | P | 20 prostate cancer tumor tissues 20 tumor‐adjacent tissues and 20 normal prostate tissues | qRT‐PCR | miR‐509‐5p | 87 |
| Jayant K. Rane 2015 | UK | P | 5 benign prostatic hyperplasia 5 G7 prostate cancer, and 3 castration‐resistant PC (CRPC) | microarray | miR‐548c‐3p | 88 |
| Takeshi Chiyomaru 2013 | USA | P | 48 pairs of PC tissues and adjacent non‐cancerous tissues | microarray qRT‐PCR | miR‐574‐3p | 89 |
| Ze‐Hua Zuo 2015 | USA | P | 77 organ donor (OD) prostates, 324 benign prostate tissues adjacent to cancer, and 216 PCs | qRT‐PCR ISH | miR‐650 | 90 |
| Li Jiao 2014 | China | P | 127 patients with prostate cancer and 10 patients with benign prostatic hyperplasia (BPH) | qRT‐PCR microarray ISH | miR‐663 | 91 |
| Sharanjot Saini 2012 | USA | P | 40 PC and 8 normal 96 paired | ISH qRT‐PCR | miR‐708 | 92 |
| Dibash K. Das 2016 | USA | P | 404 PC (389 CA and 15 AA) | qRT‐PCR | miR‐1207‐3p | 93 |
| Nathan Bucay 2016 | USA | P | 100 pairs of PC and adjacent normals | qRT‐PCR | miR‐3622b | 94 |
| Yang Wang 2016 | China | P | 3 CRPC and 3 ADPC samples 30 ADPC tissues and 18 CRPC tissues | microarray qRT‐PCR | miR‐4638‐5p | 95 |
| Albertoivan S. Guadarrama 2016 | Mexico | P | 73 PC urine and 70 BPH urine | qRT‐PCR | miR‐100 miR‐200b | 96 |
| Betina Katz 2014 | Brazil | P | 51 localized prostate cancer (PCa) | qRT‐PCR | miR‐30a miR‐200b | 15 |
| Chunjiao Song 2015 | China | P | 7 G>7 8 G7 9 Non‐cancerous Samples 7 8 9 12 G>7 12 G7 12 BPH | sequencing qRT‐PCR | miR‐125b‐5p miR‐126‐5p miR‐151a‐5p miR‐221‐3p miR‐222‐3p miR‐486‐5p | 97 |
| Darina Kachakova 2015 | Bulgaria | P | 59 prostate cancer (PC) patients and two groups of controls: 16 benign prostatic hyperplasia (BPH) samples and 11 young asymptomatic men | qRT‐PCR | miR‐30c miR‐141 miR‐375 let‐7c | 98 |
| D Lin 2011 | China | P | 35 PC (17 aggressive and 18 non‐aggressive) | qRT‐PCR | miR‐221 miR‐222 | 99 |
| Fulya Yaman Agaoglu 2011 | Turkey | P | 51 PC (26 local/local advanced or 25 metastatic PCa) 20 healthy individuals | qRT‐PCR | miR‐21 miR‐141 miR‐221 | 100 |
| Heather H. Cheng 2013 | USA | P | 25 mCRPC and 25 healthy donor serum pools the sera of an additional 21 mCRPC patients and 20 age‐matched healthy Controls for validation | array qRT‐PCR | miR‐141 miR‐200a/c miR‐210 miR‐375 | 20 |
| Hui‐Ming Lin 2017 | Australia | P | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | qRT‐PCR | miR‐20a/20b miR‐21 miR‐25 miR‐132 miR‐145a miR‐200a/b/c miR‐222 miR‐301b miR‐375 miR‐429d miR‐590‐5p | 29 |
| Irene Casanova‐Salas 2014 | Spain | PR | 10 normal prostate and 50 prostate cancer samples an independent cohort of 273 paraffin embedded prostate cancer samples Another 92 urine samples GEO (Gene Expression Omnibus) database Accession No. GSE45604 (http://www.ncbi.nlm.nih.gov/geo/) | qRT‐PCR | miR‐182 miR‐187 | 101 |
| Ivan D. Osipov 2016 | Russia | P | Blood samples from 47 healthy donors and 48 prostate cancer (PC) patients | qRT‐PCR | miR‐141 miR‐205 | 102 |
| Jorge Torres‐Ferreira 2017 | Portugal | P | 180 localized PC and 15 control 95 urine sediments and 46 controls 74 prostate biopsies | Human Methylation 450 Bead Chip qMSP | miR‐34b/c miR‐129‐2 miR‐152 miR‐193b miR‐663a miR‐1258 | 103 |
| Katia R. M. Leite 2011‐1 | Brazil | P | 18 localized high grade prostate carcinoma (PC) with mean Gleason score 8.6, all staged pT3 4 patients with metastatic, androgen‐independent prostate carcinoma 6 nonneoplastic tissue (benign prostate hyperplasia) | qRT‐PCR | miR‐100 miR‐218 Let‐7c | 26 |
| Katia R. M. Leite 2011‐3 | Brazil | P | 49 prostate cancer (28 men without and 21 with biochemical recurrence) | qRT‐PCR | miR‐100 miR‐145 miR‐191 | 27 |
| Katia R. M. Leite 2013 | Brazil | P | 63 localized prostate carcinoma 15 high grade prostate intraepithelial neoplasia (HGPIN) 14 localized favorable CaP and 34 unfavorable, mostly non‐organ‐confined disease. | qRT‐PCR | miR‐21 miR‐206 | 22 |
| Kristina Stuopelytė 2016 | Lithuania | P | 13 PC 143 urine PC and 23 urine BPH 52 PC and 12 N | microarray qRT‐PCR | miR‐19a/b miR‐21 miR‐95 | 104 |
| Kristina Stuopelyte 2016 | Lithuania | P | 56 Cancerous and 16 non‐cancerous 215 PC 23 benign prostatic hyperplasia and 62 asymptomatic controls | array qRT‐PCR | miR‐148a miR‐375 | 105 |
| Maria Giulia Egidi 2015 | Italy | P | 35 urine sediments of PC and 26 benign prostatic hyperplasia (BPH). | qRT‐PCR | miR‐25 miR‐191 miR‐200b miR‐452 | 106 |
| Maria Schubert 2013 | Germany Belgium | P | cohort A: 98 high‐risk PC Cohort B: 92 FFPE samples from RP Cohort C: 21 pairs of PC tissues and adjacent benign tissues | microarray qRT‐PCR | miR‐146b miR‐181b miR‐361 miR‐515‐3p/5p let‐7a/b/c | 107 |
| Matthew J. Roberts 2015 | Australia Germany | 20 specimens 54 non‐cancerous histology and 98 cancer tissues | qRT‐PCR | miR‐125b miR‐200b/c miR‐375 | 108 | |
| Robert Mahn 2011 | Germany | P | 37 localized PC 18 BPH 8 metastatic PC 20 healthy volunteers 10 PC and adjacent tissues and pre/post prostatectomy serum | qRT‐PCR | miR‐16 miR‐26a miR‐32 miR‐195 Let‐7i | 109 |
| Stefan Ambs 2008 | USA | P | 60 primary prostate tumors and 16 non‐tumor prostate tissues | qRT‐PCR microarray | miR‐1 miR‐32 miR‐106a/106b | 110 |
| Taha A Haj‐Ahmad 2014 | Egypt | PR | 8 PC patients, 12 BPH patients and 10 healthy males urine samples | microarray qRT‐PCR | miR‐484 miR‐1825 | 111 |
| Tong Sun 2012 | USA | P | 86 individuals, prostate tumor tissues from 34 individuals with localized hormone naïve disease, and bone‐derived metastatic CRPC tissues from 17 individuals. | qRT‐PCR | miR‐23b/27b miR‐221/222 | 112 |
| William T. Budd 2015 | USA | P | 4 pairs of frozen PC and BPH tissue samples 1 FFPE prostate sample | qRT‐PCR | miR‐22 miR‐125b | 113 |
| Xiaoyi Huang 2015 | USA | P | 23 CRPC patients 100 CRPC | sequencing qRT‐PCR | miR‐375 miR‐1290 | 30 |
| Yubin Hao 2011 | China | P | 20 human prostate specimens (8 prostate cancer tissues and 12 benign prostatic hyperplasia tissues | qRT‐PCR | miR‐16 miR‐21 miR‐34c miR‐101 miR‐125b miR‐141 | 18 |
| Beatriz A. Walter 2013 | USA | P | 37 matched prostate tumors, normal epithelium and adjacent stroma. 40 PC 10 N 10stroma | microarray qRT‐PCR | 34 deregulated | 23 |
| Fan Feng 2017 | Spain | R | Dataset (GSE45604) 50 PC and 10 normal specimens urine | data analysis | 7 up 59 down | 21 |
| Rihan El Bezawy 2017 | Italy | P | 44 pairs of PC specimens and normal tissues (GSE76260) | qRT‐PCR | 5 up 13 down | 114 |
| Robert K. Nam 2015 | Canada. | P | 546 prostate cancer | qRT‐PCR | 29 up 4 down | 115 |
| Yanan Sun 2016 | Nonchina | R | 3 microarray studies: 197 samples of PC and 43 samples of normal control | data analysis | 10 up 19 down | 116 |
P, prospective study; R, retrospective study; ISH, in situ hybridization; Refs, references; PC, prostate cancer; BPH, benign prostate hyperplasia; NM, not mentioned; LCM, laser‐captured microdissection; CRPC, castration resistant prostate cancer.
2.4. Statistical analysis
We drew forest plots to estimate miRNAs expression levels in PC and control patients' samples, and their effects on PC patients' OS and RFS. Publication bias was explored by funnel plots.12, 13 The fixed‐effects model was used to calculate HR and 95% CI in all enrolled studies.14 We used Chisquared and the inconsistency index (I 2) tests to assess the heterogeneities (P value ≤0.1 and I 2 value ≥50 %). To avoid the influence of heterogeneity, subgroup analyses were performed based on the characteristics of included studies, such as patients' ethnicities, pathological types, and detected sample types, etc. All P values were two‐tailed and a P value <0.05 was considered to be statistically significant.
3. RESULTS
3.1. Summary of included studies
A total of 1336 primary literatures were searched in PubMed, Embase, Cochrane, and CNKI. As shown in the selection process (Figure 1), we firstly removed 49 studies due to duplication. Then, we excluded 980 and 202 studies, respectively, after abstracts and full texts were reviewed. Ultimately, only 104 articles were considered eligible for the meta‐analysis. The characteristics of 104 included studies were summarized in Table 1 in alphabetical order of the miRNAs. The publication years of these records ranged from 2007 to 2017. In these 104 studies, some were divided into several parts because of multiple miRNAs. Data of enrolled records were collected from the United States, China, Germany, Greece, Italy, Austria, Korea, and Brazil, etc. The dominant ethnicity was Caucasian in more than half of studies, while 38‐2 studies were executed in Asians. Most studies were prospective in design. The expression level of miRNA was usually detected by quantitative real‐time polymerase chain reaction (qRT‐PCR) and microarray in tissue samples, while 6 + 2 studies were in serum or plasma samples, 6 + 2 studies were in urine (Table 1). Among these studies, 71 records were associated with Mean ± SD and fold‐change of miRNA expression level in tumor or control samples (Table 2 and Figures 2, 3, 4, 5). A 29 focused on RFS (Table 3 and Figure 6A‐E), and 11 focused on OS (Table 4 and Figure 6F). In the analysis of RFS and OS, 26, and 9 records directly reported HRs and 95% CIs, respectively, while in other studies we extrapolated these necessary variables by available original data (Tables 3, 4 and Figure 6).
Table 2.
The expression levels of miRNAs
| miRNA | Samples | Mean ± SD (PC vs control) | Fold change (PC/control) | P value | Refs |
|---|---|---|---|---|---|
| miR‐7 | 20 pairs of tumors and adjacent normal tissues | 1.7 ± 1.04 vs 1.21 ± 0.55 | 0.6569 | 33 | |
| miR‐7‐2* | 44 pairs of PC and normal | 0.806642 | 2.19E‐02 | 114 | |
| miR‐7c | 50 PC and 10 normal | 0.001272 | 1.56E‐02 | 21 | |
| miR‐9 | 51 localized PC | 0.96 ± 0.89 vs 1.34 ± 2.47 | 0.637 | 15 | |
| miR‐9‐1 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.9 vs 4.98 | 0.04723 | 115 | |
| miR‐9‐2 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.89 vs 4.97 | 0.04892 | 115 | |
| miR‐9‐3 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 6.12 vs 4.96 | 0.01907 | 115 | |
| miR‐15b | 40 PC and 10 normal | 3.4761 | 0.0418 | 23 | |
| miR‐18b | 40 PC and 10 normal | 6.8061 | 0.0133 | 23 | |
| miR‐20b | 40 PC and 10 normal | 3.1928 | 0.0501 | 23 | |
| miR‐22 | 4 frozen tissue samples 1 FFPE prostate sample | 3.2 | NM | 113 | |
| miR‐24 | 50 PC and 10 normal | 0.27 | 3.68E‐03 | 21 | |
| miR‐24‐2 | 50 PC and 10 normal | 0.164459 | 9.76E‐03 | 21 | |
| miR‐26a‐5p | 140 pairs of fresh PC tissues and normal tissues | 0.058 ± 0.016 vs 0.115 ± 0.043 | <0.001 | 45 | |
| miR‐28‐3p | 50 PC and 10 normal | 0.00668 | 1.08E‐02 | 21 | |
| miR‐28‐5p | 50 PC and 10 normal | 0.003839 | 3.28E‐03 | 21 | |
| miR‐29b | 51 localized PC | 0.51 ± 0.64 vs 0.56 ± 0.77 | 0.852 | 15 | |
| miR‐30a | 51 localized PC | 6.37 ± 7.91 vs 1.7 ± 2.77 | 0.039 | 15 | |
| miR‐30c‐1 | 50 PC and 10 normal | 0.257951 | 3.18E‐02 | 21 | |
| miR‐30d | 56 pairs of primary PC and control | 7.95 ± 7.03 vs 6.23 ± 6.06 | 0.03 | 48 | |
| miR‐30e* | 44 pairs of PC and normal | 0.840896 | 4.10E‐03 | 114 | |
| miR‐33a | Paired prostate cancer tissue and adjacent normal tissue | 0.1389 | <0.01 | 50 | |
| miR‐34c‐3p | 50 PC and 10 normal | 0.17691 | 7.42E‐03 | 21 | |
| miR‐34c‐5p | 40 PC and 10 normal | 8.0395 | 0.0283 | 23 | |
| miR‐92a | 40 PC and 10 normal | 3.0015 | 0.0177 | 23 | |
| miR‐93 | 197 PC and 43 normal | 2.14 | 1.69E‐09 | 116 | |
| miR‐96 | 197 PC and 43 normal | 2.35 | 2.33E‐12 | 116 | |
| miR‐101 | 8 PC and 12 BPH | 0.91 | >0.05 | 18 | |
| miR‐122 | 40 PC and 10 normal | 5.5663 | 0.0054 | 23 | |
| miR‐126 | 128 PCa | 1.05 ± 0.63 vs 2.92 ± 0.98 | <0.001 | 55 | |
| miR‐126‐5p | 12 G > 7, 12 G7, and 12 non‐cancerous samples | 2.22 | <0.05 | 97 | |
| miR‐128 | 128 PC tissue and serum and matched normal | 1.05 ± 0.63 vs 2.92 ± 0.98 | <0.001 | 56 | |
| miR‐128a | 40 PC and 10 normal | 4.5004 | 0.0143 | 23 | |
| miR‐130b | 197 PC and 43 normal | 1.974463 | 3.52E‐07 | 116 | |
| miR‐134 | 40 PC and 10 normal | 23.1323 | 0.0125 | 23 | |
| miR‐135b | 40 PC and 10 normal | 4.0019 | 0.0141 | 23 | |
| miR‐138‐2 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.23 vs 4.25 | 0.03941 | 115 | |
| miR‐139 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 7.24 vs 8.08 | 0.03061 | 115 | |
| miR‐146b‐5p | 40 PC and 10 normal | 3.5577 | 0.0019 | 23 | |
| miR‐148b | 40 PC and 10 normal | 2.8135 | 0.0358 | 23 | |
| miR‐149 | 44 pairs of PC and normal | 0.796 | 0.416 | 114 | |
| miR‐151a‐5p | 12 G > 7, 12 G7, and 12 non‐cancerous samples | 2.02 | <0.05 | 97 | |
| miR‐153 | 197 PC and 43 normal | 3.1425 | 2.74E‐13 | 116 | |
| miR‐155 | 51 localized PC | 3.12 ± 4.56 vs 2.09 ± 3.8 | 0.463 | 15 | |
| miR‐181d | 50 PC and 10 normal | 0.062341 | 9.34E‐03 | 21 | |
| miR‐182‐5p | patient set 1:69 PC patient set 2:10 PC | Patient set 1: 1.745 ± 0.278 vs 0.864 ± 0.136 Patient set 2: 1.863 ± 0.381 vs 0.761 ± 0.158 | 0.021 | 66 | |
| miR‐183* | 44 pairs of PC and normal | 1.505247 | 7.68E‐03 | 114 | |
| miR‐184 | 40 PC and 10 normal | 4.0633 | 0.0086 | 23 | |
| miR‐188 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 8.48 vs 7.5 | 0.01878 | 115 | |
| miR‐188‐5p | 180 pairs of PC and normal | 0.0956 | NM | 68 | |
| miR‐193a‐5p | 40 PC and 10 normal | 4.5984 | 0.0094 | 23 | |
| miR‐193b | 40 PC and 10 normal | 12.649 | 0.0021 | 23 | |
| miR‐199a‐1 | 50 PC and 10 normal | 0.451942 | 2.06E‐02 | 21 | |
| miR‐199a‐3p | 50 PC and 10 normal | 0.000759 | 1.08E‐02 | 21 | |
| miR‐214 | 40 PC and 10 normal | 9.9075 | 0.0055 | 23 | |
| miR‐215 | 40 PC and 10 normal | 8.4863 | 0.038 | 23 | |
| miR‐220a | 44 pairs of PC and normal | 0.907519 | 0.355 | 114 | |
| miR‐221‐3p | 12 G > 7, 12 G7, and 12 non‐cancerous samples | 5.47 | <0.05 | 97 | |
| miR‐222‐3p | 12 G > 7, 12 G7, and 12 non‐cancerous samples | 3.88 | <0.05 | 97 | |
| miR‐223 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 10.66 vs 11.9 | 0.00179 | 115 | |
| miR‐223‐3p | 10 pairs of PC and adjacent non‐cancerous tissues | 2.98 ± 1.45 vs 1.55 ± 0.38 | <0.01 | 75 | |
| miR‐296 | TMA100: 100 PC cases TMA96: 96 cases | 1.79 ± 0.19 vs 2.71 ± 0.16 | <0.05 | 79 | |
| miR‐296‐5p | 44 pairs of PC and normal | 0.646176 | 1.49E‐02 | 114 | |
| miR‐301b | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.61 vs 3.65 | 0.02116 | 115 | |
| miR‐320c‐2 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 3.54 vs 2.39 | 0.0393 | 115 | |
| miR‐324‐5p | 197 PC and 43 normal | 0.565156 | 2.06E‐05 | 116 | |
| miR‐328 | 197 PC and 43 normal | 0.511 | 7.85E‐07 | 116 | |
| miR‐335 | 20 pairs of primary PC and adjacent 104 PC and 20 benign | 3.27 ± 0.99 vs. 4.55 ± 1.34 | <0.05 | 82 | |
| miR‐338‐5p | 50 PC and 10 normal | 14.70974 | 9.88E‐03 | 21 | |
| miR‐362‐3p | 50 PC and 10 normal | 0.265027 | 3.18E‐02 | 21 | |
| miR‐372 | 40 PC and 10 normal | 6.8639 | 0.0184 | 23 | |
| miR‐373 | 51 localized PC | 0.26 ± 0.37 vs. 0.29 ± 0.32 | 0.186 | 15 | |
| miR‐376a | 50 PC and 10 normal | 0.457502 | 1.41E‐02 | 21 | |
| miR‐378* | 197 PC and 43 normal | 0.476022 | 1.64E‐08 | 116 | |
| miR‐378c | 50 PC and 10 normal | 0.011878 | 1.40E‐03 | 21 | |
| miR‐381 | 50 PC and 10 normal | 0.20897 | 2.30E‐02 | 21 | |
| miR‐383 | TCGA:187 primary PC validation cohort: 112 pairs of PC and adjacent normals | 0.25 | 0.05 | 84 | |
| miR‐411 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 3.83 vs 2.73 | 0.02673 | 115 | |
| miR‐421 | 50 PC and 10 normal | 0.03487 | 6.02E‐04 | 21 | |
| miR‐422a | 50 PC and 10 normal | 0.014149 | 8.65E‐05 | 21 | |
| miR‐424 | 50 PC and 10 normal | 0.088399 | 2.94E‐02 | 21 | |
| miR‐429 | 51 localized PC | 7.74 ± 7.34 vs 7.75 ± 17.18 | 0.998 | 15 | |
| miR‐433 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.21 vs 3.15 | 0.030601 | 115 | |
| miR‐455‐3p | 50 PC and 10 normal | 0.001986 | 2.07E‐02 | 21 | |
| miR‐455‐5p | 50 PC and 10 normal | 0.093956 | 9.76E‐03 | 21 | |
| miR‐485‐3p | 50 PC and 10 normal | 0.2564 | 1.82E‐02 | 21 | |
| miR‐486 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.49 vs 5.6 | 0.03746 | 115 | |
| miR‐486‐5p | 12 G > 7, 12 G7, and 12 non‐cancerous samples | 0.3937 | <0.05 | 97 | |
| miR‐487b | 197 PC and 43 normal | 0.565379 | 3.69E‐05 | 116 | |
| miR‐489 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 3.66 vs 2.67 | 0.02183 | 115 | |
| miR‐490‐5p | 50 PC and 10 normal | 0.184615 | 1.56E‐02 | 21 | |
| miR‐495 | 51 localized PC | 0.77 ± 0.39 vs 0.93 ± 0.32 | 0.78 | 15 | |
| miR‐497 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 11.19 vs 10.28 | 0.01111 | 115 | |
| miR‐501 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.74 vs 4.91 | 0.00525 | 115 | |
| miR‐502‐5p | 197 PC and 43 normal | 0.573804 | 3.86E‐05 | 116 | |
| miR‐503 | 50 PC and 10 normal | 0.376508 | 1.41E‐02 | 21 | |
| miR‐507 | PC and matched Normal | 0.858565 | 4.88E‐03 | 114 | |
| miR‐509‐3‐5p | 50 PC and 10 normal | 0.318223 | 3.62E‐02 | 21 | |
| miR‐509‐5p | 20 PC, 20 tumor‐adjacent tissues, and 20 normal prostate tissues | 0.314 ± 0.048 vs 1.532 ± 0.015 | <0.05 | 87 | |
| miR‐518b | 44 pairs of PC and normal | 0.779165 | 4.23E‐02 | 114 | |
| miR‐543 | 50 PC and 10 normal | 0.270522 | 1.08E‐02 | 21 | |
| miR‐545 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.1 vs 4.04 | 0.00524 | 115 | |
| miR‐574‐3p | 48 pairs of PC and adjacent non‐cancerous tissues | 0.5 | <0.0001 | 89 | |
| miR‐612 | 44 pairs of PC and normal | 1.658639 | 5.31E‐03 | 114 | |
| miR‐624 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.66 vs 4.07 | 0.030601 | 115 | |
| miR‐628‐3p | 50 PC and 10 normal | 0.04014 | 1.80E‐03 | 21 | |
| miR‐650 | 216 PC, 324 benign, and 77 control 22 PC, 20 benign, and 11 control | 1.29 ± 0.08 vs 1.07 ± 0.05 | 0.012 | 90 | |
| miR‐652 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 8.3 vs 6.73 | 0.00124 | 115 | |
| miR‐659 | 44 pairs of PC and normal | 0.795536 | 4.10E‐03 | 114 | |
| miR‐663 | 197 PC and 43 normal | 0.545382 | 2.62E‐09 | 116 | |
| miR‐671 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 7.75 vs 6.94 | 0.00072 | 115 | |
| miR‐708 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 6.67 vs 5.45 | 0.01206 | 115 | |
| miR‐875‐3p | 50 PC and 10 normal | 0.383402 | 3.12E‐02 | 21 | |
| miR‐875‐5p | 44 pairs of PC and normal | 0.632878 | 1.31E‐02 | 114 | |
| miR‐887 | 50 PC and 10 normal | 0.211747 | 2.33E‐02 | 21 | |
| miR‐1184 | 50 PC and 10 normal | 3.450542 | 1.56E‐02 | 21 | |
| miR‐1206 | 44 pairs of PC and normal | 0.907519 | 2.68E‐02 | 114 | |
| miR‐1207‐3p | PC patients of 389 CA and 15 AA | Black: 3.00 ± 2.65 White 5.36 ± 3.76 | 0.062 | 93 | |
| miR‐1207‐5p | 50 PC and 10 normal | 180.2841 | 4.72E‐02 | 21 | |
| miR‐1228 | 44 pairs of PC and normal | 1.086735 | 4.90E‐02 | 114 | |
| miR‐1238 | 50 PC and 10 normal | 0.883057 | 2.36E‐02 | 21 | |
| miR‐1244 | 44 pairs of PC and normal | 1.484524 | 5.20E‐03 | 114 | |
| miR‐1245 | 44 pairs of PC and normal | 1.265757 | 3.01E‐02 | 114 | |
| miR‐1248 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 8.91 vs 7.77 | 0.01907 | 115 | |
| miR‐1249 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 5.37 vs 4.47 | 0.00622 | 115 | |
| miR‐1271 | 50 PC and 10 normal | 0.02573 | 3.28E‐03 | 21 | |
| miR‐1302‐1 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.53 vs 2.75 | 0.01529 | 115 | |
| miR‐1302‐3 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.42 vs 2.75 | 0.01529 | 115 | |
| miR‐1302‐7 | 18 PC with recurrence and 13 PC no metastasis no recurrence | 4.19 vs 2.55 | 0.02113 | 115 | |
| miR‐3200‐3p | 50 PC and 10 normal | 0.40029 | 4.48E‐02 | 21 | |
| miR‐4288 | 50 PC and 10 normal | 0.219167 | 3.76E‐03 | 21 | |
| miR‐4328 | 50 PC and 10 normal | 0.470068 | 4.96E‐02 | 21 | |
| miR‐4638‐5p | 3 CRPC and 3 ADPC 18 CRPC and 30 ADPC | 0.4167 0.2128 | 1.44E‐08 | 95 | |
| let‐7b | Cohort A: 6 BPH tissues and 13 high‐risk PC specimens Cohort B: 92 FFPE PC samples Cohort C: 21 pairs of fresh frozen PC tissue and adjacent benign tissue | 3.16 ± 0.76 vs 3.8 ± 0.37 | <0.01 | 107 |
Refs, reference; PC, prostate cancer; BPH, benign prostate hyperplasia; CRPC, castration resistant prostate cancer.
Figure 2.
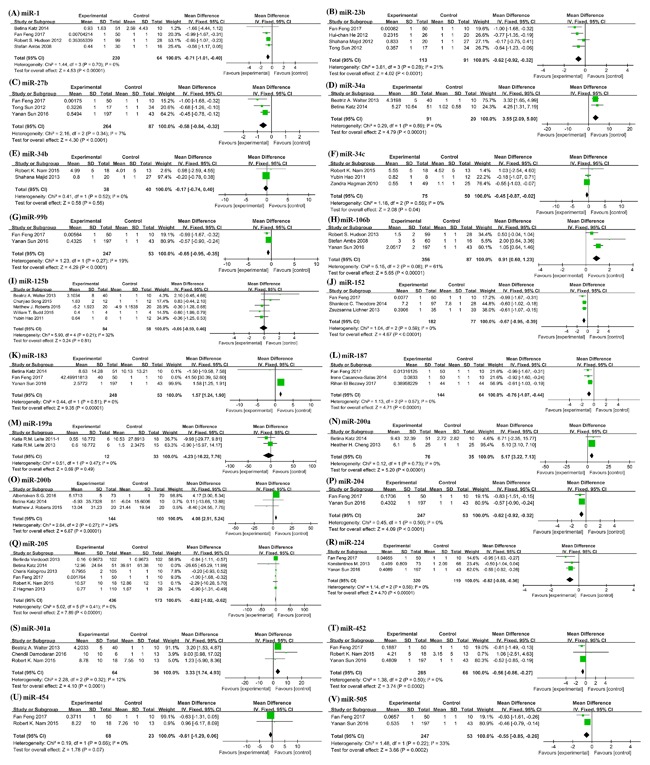
Forest plots showing mean expression levels of different miRNAs with corresponding heterogeneity statistics. (A) miR‐1; (B) miR‐23b; (C) miR‐27b; (D) miR‐34a; (E) miR‐34b; (F) miR‐34c; (G) miR‐99b; (H) miR‐106b; (I) miR‐125b; (J) miR‐152; (K) miR‐183; (L) miR‐187; (M) miR‐199a; (N) miR‐200a; (O) miR‐200b; (P) miR‐204; (Q) miR‐205; (R) miR‐224; (S) miR‐301a; (T) miR‐452; (U) miR‐454; (V) miR‐505. Squares and horizontal lines correspond to study‐specific HRs and 95% CIs; respectively. The area of the squares correlates the weight of each enrolled study and the diamonds represent the summary HRs and 95% CIs
Figure 3.
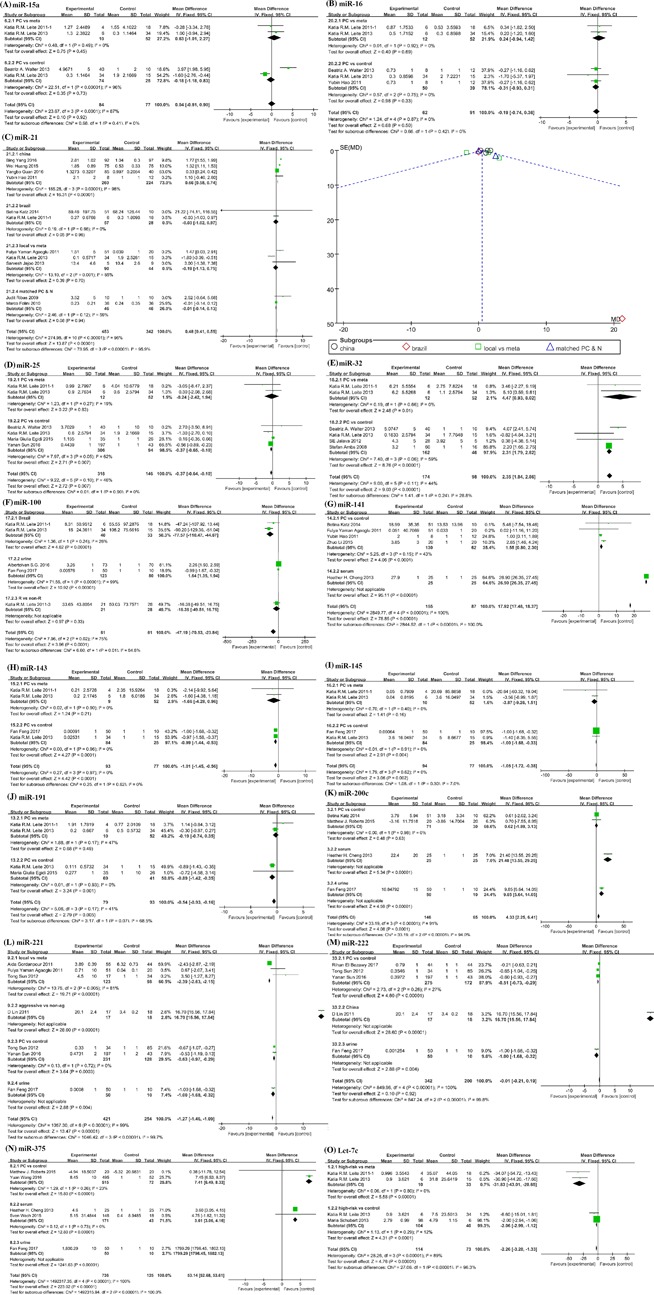
Forest plots of subgroup analyses stratified by ethnicities; main pathologic types and detected samples; showing mean expression levels or fold change with corresponding heterogeneity statistics. (A) miR‐15a; (B) miR‐16; (C) forest plot and funnel plot of miR‐21; each point represents a separate study for publication bias test in funnel plot; (D) miR‐25; (E) miR‐32; (F) miR‐100; (G) miR‐141; (H) miR‐143; (I) miR‐145; (J) miR‐191; (K) miR‐200c; (L) miR‐221; (M) miR‐222; (N) miR‐375; (O) let‐7c. Squares and horizontal lines correspond to study‐specific HRs and 95% CIs; respectively. The area of the squares correlates the weight of each enrolled study and the diamonds represent the summary HRs and 95% CIs
Figure 4.
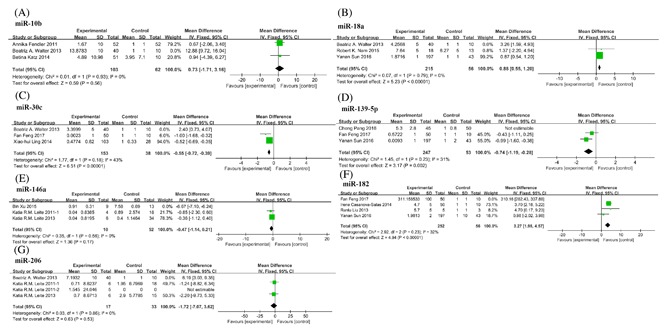
Forest plots showing mean expression levels of miRNAs with corresponding heterogeneity statistics. (A) miR‐10b; (B) miR‐18a; (C) miR‐30c; (D) miR‐139‐5p; (E) miR‐146a; (F) miR‐182; (G) miR‐206. Squares and horizontal lines correspond to study‐specific HRs and 95% CIs; respectively. The area of the squares correlates the weight of each enrolled study and the diamonds represent the summary HRs and 95% CIs
Figure 5.
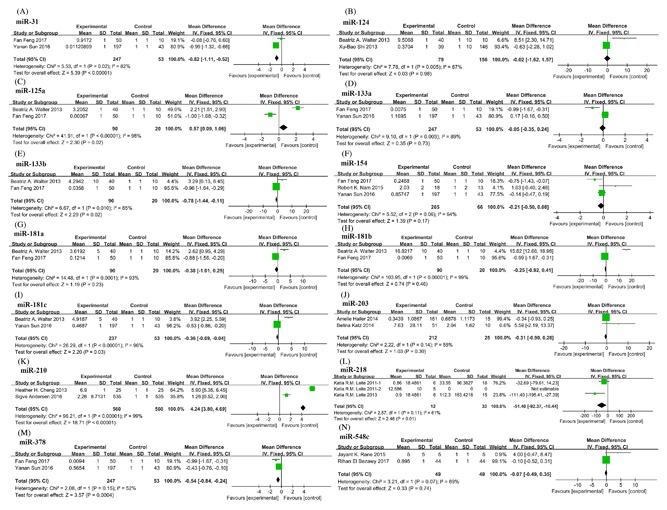
Forest plots showing mean expression levels of miRNAs with significant heterogeneity. (A) miR‐31; (B) miR‐124; (C) miR‐125a; (D) miR‐133a; (E) miR‐133b; (F) miR‐154; (G) miR‐181a; (H) miR‐181b; (I) miR‐181c; (J) miR‐203; (K) miR‐210; (L) miR‐218; (M) miR‐378; (N) miR‐548c. Squares and horizontal lines correspond to study‐specific HRs and 95% CIs; respectively. The area of the squares correlates the weight of each enrolled study and the diamonds represent the summary HRs and 95% CIs
Table 3.
The recurrence‐free survival of miRNAs in enrolled studies
| miRNA | Samples | RFS HR/RR (95%CI) | P value | Refs |
|---|---|---|---|---|
| miR‐10b | 52 primary PC and normal adjacent tissues (24 early biochemical relapse and 22 no/ late biochemical relapse) | 2.15 (1.02‐4.51) | 0.044 | 34 |
| miR‐23a | 3 pairs of primary prostate cancer and adjacent non‐tumor tissues 20 paired of prostate cancer and adjacent non‐tumor tissues. 123 prostate cancer tissues | 0.389 (0.249‐0.608) | <0.0001 | 41 |
| miR‐23b | 118 pairs of PC and control 27 BPH and 20 tumor samples 48 samples | 6 (3‐13) | <0.002 | 43 |
| miR‐27b | 41 noncancerous tissues and 49 PC tissues | 0.255 (0.069‐0.944) | 0.0407 | 44 |
| miR‐34b | 148 LCM matched human tissue samples 27 BPH and 20 tumor samples | 3.3 (1.3‐8.7) | <0.02 | 51 |
| miR‐106b | 28 non‐cancerous tissues, 99 primary tumors, and 14 distant metastases/recurrence. | 2.7 (1.1‐7.3) | 0.014 | 53 |
| miR‐100 | 49 prostate cancer (28 men without and 21 with biochemical recurrence) | 3.045 (1.200‐7.737) | 0.019 | 27 |
| miR‐133b | 135 PC, 18 prostatic intraepithelial neoplasia (PIN), and 25 normals | 1.775 (1.013‐3.108) | 0.045 | 59 |
| miR‐150 | 167 PC 4 pairs of PC and adjacent normal tissues | 1.90 (1.21‐2.98) | 0.005 | 63 |
| miR‐191 | 49 prostate cancer (28 men without and 21 with biochemical recurrence) | 2.642 (1.030‐6.780) | 0.043 | 27 |
| miR‐205 | Study cohort: 105 HRPC, 10 BHP validation cohort:78 HRPCa | Study cohort: 2.01(0.83‐4.85) Validation cohort: 0.82 (0.39‐1.7) | 0.596 | 71 |
| miR‐221 | 28 recurrent and 37 non‐recurrent prostate cancer cases | 0.71 (0.32‐1.61) | 0.42 | 28 |
| miR‐222 | 28 recurrent and 37 non‐recurrent prostate cancer cases | 0.51 (0.22‐1.18) | 0.12 | 28 |
| miR‐224 | 4 and 20 pairs of primary PC and adjacent non‐tumor frozen samples TMA: 114 PC tissues respectively from Caucasian and African‐American PC patients | 0.31 (0.11‐0.86) | 0.017 | 78 |
| miR‐301a | 585 prostate cancer | 1.42 (1.06‐1.90) | 0.002 | 81 |
| miR‐383 | TCGA database: 187 primary PC validation cohort: 112 PC FFPE tissues and matched adjacent normals | TCGA database: 0.661 Validation cohort: 0.897 | 0.0655 | 84 |
| miR‐449b | 36 PC 163 radical prostatectomy patients 40 patients (20 recurrent and 20 non‐recurrent patients) | 1.9 | 0.003 | 85 |
| miR‐466 | 48 pairs of LCM tissue samples validation cohort: 56 PC | 17 (5‐50) | 0.02 | 86 |
| miR‐663 | 127 prostate cancer and 10 benign prostatic hyperplasia (BPH) | 2.924 (1.981‐4.316) | <0.001 | 91 |
| miR‐708 | 40 PC and 8 normal 96 paired of PC and normal | 6 (2.2‐16.4) | 0.0138 | 92 |
| miR‐1207‐3p | PC patients of 389 CA and 15 moAA | 1.8 (0.8‐4.3) | <0.001 | 93 |
| miR‐3622b | 100 pairs of PC and adjacent normals | 0.407 | 0.2 | 94 |
| let‐7b | cohort A: 98 high‐risk PC Cohort B: 92 FFPE samples cohort C: 21 pairs of PC and adjacent benign tissue | 0.36 (0.161‐0.823) | 0.02 | 107 |
RFS, recurrence free survival; HR, hazard ratio; CI, confidence interval; Refs, reference; PC, prostate cancer; BPH, benign prostate hyperplasia; CRPC, castration resistant prostate cancer.
Figure 6.
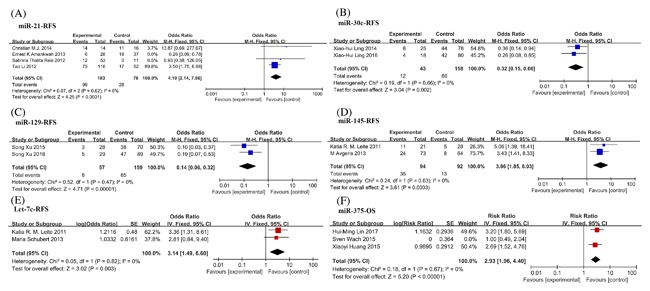
Forest plots for merged analyses of recurrence‐free survival (RFS) and overall survival (OS) associated with different miRNAs expression. Forest plots for RFS analyses of (A) miR‐21 (B) miR‐30c; (C) miR‐129; (D) miR‐145; (E) let‐7c; (F) Forest plots of OS analyses of miR‐375. Squares and horizontal lines correspond to study‐specific HRs and 95% CIs; respectively. The area of the squares correlates the weight of each enrolled study and the diamonds represent the summary HRs and 95% CIs
Table 4.
The overall survival of miRNAs in enrolled studies
| miRNA | Samples | OS HR/RR (95%CI) | P value | Refs |
|---|---|---|---|---|
| miR‐23a | 3 pairs of primary prostate cancer and adjacent non‐tumor tissues 20 paired of prostate cancer and adjacent non‐tumor tissues. 123 prostate cancer tissues | 0.389 (0.249‐0.608) | <0.001 | 41 |
| miR‐23b | 118 pairs of PCs and controls 27 BPH and 20 tumor samples 48 samples | 3.3 (4‐19) | <0.0001 | 43 |
| miR‐132 | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | 1.9 (1.1‐3.4) | 0.02 | 29 |
| miR‐150 | 167 PC 4 pairs of PC and adjacent normal tissues | 1.87 (1.19‐2.94) | 0.006 | 63 |
| miR‐200a | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | 2.1 (1.2‐3.6) | 0.009 | 29 |
| miR‐200b | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | 3.8 (2.0‐6.9) | 0.000006 | 29 |
| miR‐200c | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | 3.8 (2.0‐6.9) | 0.005 | 29 |
| miR‐205 | Study cohort: 105 HRPC, 10 BPH validation cohort: 78 HRPC | Study cohort: 2.04 Validation cohort: 3.1 | 0.0817 | 71 |
| miR‐221 | cohort 1: 134 PC cohort 2: 89 PC | cohort 1: 0 cohort 2: 0.029 | <0.0001 | 74 |
| miR‐224 | 4 and 20 pairs of primary PC and adjacent non‐tumor Taylor dataset: 149 primary PC tissues and 29 adjacent non‐cancerous prostate tissues | 0.73 (0.31‐1.72) | 0.046 | 76 |
| miR‐429 | Phase 1 cohort: 97 patients Phase 2 cohort: 89 patients | 3.3 (1.8‐6.0) | 0.00005 | 29 |
| miR‐708 | 40 PC and 8 normal 96 pairs of PC and normal | 6 (2.2‐16.4) | 0.0223 | 92 |
| miR‐1207‐3p | PC patients of 389 CA and 15 AA | 1.8 (0.8‐4.3) | 0.062 | 93 |
| miR‐1290 | 23 CRPC patients 100 CRPC | 1.79 (1.30‐2.48) | <0.004 | 30 |
OS,overall survival; HR, hazard ratio; CI, confidence interval; Refs, reference; PC, prostate cancer; BPH, benign prostate hyperplasia; CRPC, castration resistant prostate cancer.
3.2. miRNAs and PC diagnosis
miRNAs may regulate the wide range of biologic processes, and their deregulation are associated with PC onset, progression, and metastasis. More and more studies investigated differentially expressed miRNA as PC diagnostic and prognostic markers by comparing the expression levels of miRNAs in tumor tissues to that in BPH or normal controls. But there were the high variability in the data obtained from the different records. These could be caused by several factors as follows: (i) different sample groups; (ii) different detecting and verifying methods; (iii) small sample size. Nonetheless, these studies depicted a starting point, and some of the included records screened the same miRNA which was found with the same trend in multiple studies with different methods, as shown in Table 1. However, a confirmed diagnostic miRNA which could be translated into the clinic was not arised. Further confirmed experiments are needed in additional large patient cohorts.
In Figure 2, 22 miRNAs were reported to be consistently deregulated in different records. Among them, 6 miRNAs (miR‐34a, miR‐106b, miR‐183, miR‐200a/b, and miR‐301a) were up‐regulated in PC, while 16 miRNAs (miR‐1, miR‐23b, miR‐27b, miR‐34b/c, miR‐99b, miR‐125b, miR‐152, miR‐187, miR‐199a, miR‐204, miR‐205, miR‐224, miR‐452, miR‐454, and miR‐505) were down‐regulated. miR‐125b, miR‐205, miR‐1, and miR‐23b were the most commonly detected to evaluate their diagnostic efficacy between PC patients and non‐cancerous individuals. In the studies about the most obviously up‐regulated miR‐200a and miR‐200b, the pooled expression values were 5.17 (95%CI 3.22‐7.13) and 4.08 (95%CI 2.91‐5.24), respectively (Figures 2N and 2O). While miR‐199a was most significantly down‐regulated, which pooled value was −4.23 (95%CI −16.22, 7.76) (Figure 2M). More potential biomarkers were summarized in Table 2, 134 PC related miRNAs were listed which had the diagnostic potential to be aberrantly expressed in PC patients compared with healthy controls. A 60 up‐regulated miRNAs and 63 down‐regulated miRNAs were able to discriminate PC patients from BPH or healthy individuals. The remaining 11 miRNAs were not statistically significant in the studies. Any miRNA‐based clinical screening still lacks a consensus signature to be applied in the routine assay, and needs further validation in an intended use population.
3.3. Publication bias and subgroup analysis
The high heterogeneity between the data from the included records could be associated with several factors: different study design, different races of patients, different methods of sample collection and detection, incomplete information, and small sample size. There were also many difficultly statistical factors: proportion of contaminating cells, limited tumor size and differences in miRNAs stability and processing. In addition, different control samples (BPH or adjacent normal or unmatched normal) and the different characteristics of PC (low/high‐risk or metastasis or recurrence) could explain, at least in part, the different results. Significant heterogeneities (P < 0.05, I 2 > 50%) were found in most miRNAs expression profiles, we performed the subgroup analyses to seek the source of heterogeneity, which include ethnicity, sources of control (BPH or N), and sample types (serum/plasma or urine), etc.
To assess publication bias of 11 studies on miR‐21, the funnel plot was drawed. As shown in the Figure 3C, significant publication bias was found in the pooled analysis of miR‐21 (P < 0.00001, I 2 = 95%), most of the research data was distributed on the edge line. In order to avoid the effect of heterogeneity, we performed four subgroup analyses divided by ethnicity, and sample categories, including: China, Brazil, local versus meta, and PC versus control. Unfortunately, the heterogeneities were significantly reduced in Brazil subgroup and local versus meta subgroup, while the expression of miR‐21 in other subgroups still had obvious heterogeneity. In Brazil subgroup, I2 value was less than 50%, but SD value from the first study by Betina Katz15was too large, and covered the scope of the other two data. Therefore, we believe that the study on miR‐21 still needs to be further expanded. We did not find corresponding increased miR‐21 in Chinese by merging four studies16, 17, 18, 19 (Figure 3C). When stratified by the category of detected samples, increased expression of miR‐21 showed consistency in local versus meta subgroup, but no statistically significant result was observed in PC versus control subgroup.
Five studies on miR‐100 had obvious heterogeneity, as shown in the Figure 3F. After carefully reviewing the five full‐texts, they were divided into three subgroups, including: Brazil, urine and recurrence and non‐recurrence. Among them, heterogeneity in the Brazil subgroup was significantly reduced (P = 0.24, I 2 = 26%). Subgroup analysis of miR‐141 expression showed that miR‐141 expression was consistently up‐regulated in four studies of PC versus control subgroup and more obviously up‐regulated in serum samples data from Heather H. Cheng.20 Four studies on miR‐200c had also obvious heterogeneity, as shown in the Figure 3K. Three subgroups: PC versus control, serum and urine were classified according to different sample characteristics. Fan feng et al21 collected the serum samples from 50 PC patients and 10 normal controls, while Heather H, Cheng et al20 detected the miR‐200c expression levels in patients' urine samples. The heterogeneity of miR‐200c expression in PC versus control subgroup significantly reduced (P = 0.98, I2 = 0%). Six studies on miR‐221 were divided into four subgroups: local versus meta, aggressive versus non‐aggressive, PC versus control and urine, and the heterogeneity in PC versus control subgroup was significantly reduced to 0%. In addition, existing data showed that the expression of miR‐221 in primary PC was less than that in normal tissues, but miR‐221 was significantly increased when PC progressed to more malignant stages (metastasis or recurrence or hormone resistance). Among them, Tong's research data were divided into two parts, which were included in local versus meta subgroup and PC versus control subgroup, respectively. The studies on miR‐15a and miR‐16 were divided into two subgroups: PC versus meta, PC versus control. Results showed that both of miR‐15a and miR‐16 were up‐regulated in metastasis PC, while their expression levels were lower in PC tissues than in non‐cancerous tissues. Inconsistently expression of let‐7c was reported. The three studies on let‐7c were divided into two subgroups: high‐risk versus meta and high‐risk versus control, and the research data from Katia R. M. Leite 201322 were separately counted in the two subgroups because two sets of data were involved. The heterogeneity was significantly reduced to 0% and 12%. The study of miR‐143, 145 191, −25‐32 was divided into two subgroups, PC versus meta, PC versus control. Moreover, miR‐222 and miR‐375 were inconsistently expressed in prostate tumor tissues and matched normal tissues (Figures 3M and 3N). So it was essential to conduct subgroup analyses on miR‐222 and miR‐375 expression. Five studies on miR‐222 could be divided into three subgroups: PC versus control, China, and urine. The study of D Lin was a comparative study on the malignant and non‐malignant PC in China. The heterogeneity in PC versus control subgroup significantly decreased to 27%. Five studies on miR‐375 were divided into three subgroups: PC versus control, serum, and urine. The heterogeneities in PC versus control and serum subgroups were significantly reduced to 23% and 0%, respectively. The analyses of the above‐mentioned subgroups showed that the expression of miR‐375 in the urine samples were widely different, and also deviated from the expression profiles of tissues and plasma samples.
In addition to the above mentioned miRNAs expression data, there were also significant heterogeneities in the studies on seven miRNAs (Figure 4). Among them, studies on miR‐10b, miR‐18a, miR‐30c, and miR‐206, research data from Beatriz A. Walter23 deviated significantly from other research data. The heterogeneity decreased significantly when we rejected the deviant data. Moreover, in several studies on miR‐139‐5p and miR‐182, Cheng Pang24 and Fan Feng21 detected miRNAs expression profiles in whole blood and urine, respectively, which could explain the causes of heterogeneity. Finally, in three studies about miR‐146 a, Bin Xu25 collected ADPC and AIPC patients' samples in China, which were obviously different from the other two studies by Katia R. M. Leite26, 27 in Brazil.
Studies on 14 miRNAs (miR‐31, miR‐124, miR‐125a, miR‐133a/b, miR‐154, miR‐181a/b/c, miR‐203, miR‐210, miR‐218, miR‐378, and miR‐548c) had separately 2‐3 studies with significant heterogeneity (Figure 5). These studies only opened the gateway for the diagnosis and prognosis potential of 14 miRNAs, more researches are needed to confirm their application value in clinic.
3.4. miRNA expression and recurrence‐free survival
Biochemical recurrence (BCR) was considered as the first key point to estimate treatment success after RP. BCR can predate the development of metastases and other signs of clinical progression, or ultimately death. Recently, a lot of studies attempted to find miRNAs to be potential predictors for patients with biochemical failure. We summarized previous data in the meta‐analysis, miR‐30c, miR‐129, miR‐145, and let‐7c were found to have the same trend to predict BCR in eight articles (Figure 6B‐E). While the relationship of miR‐21 and BCR were studied in four articles with significant heterogeneity (Figure 6A). After reviewing the four full texts, we found that Ernest K Amankwah28 examined the effect of the interaction between obesity and miR‐21 expression on PC recurrence. Obese patients were included in the study. Removing the data from Ernest K Amankwah, miR‐21 could distinguish biochemical failure patients from non‐recurrence (Figure 6A).
In remaining 20 articles, we found prostate tumors with high levels of miR‐10b, miR‐100, miR‐106b, miR‐133b, miR‐150, miR‐191, miR‐301a, miR‐449b, miR‐663, or miR‐1207‐3p have significant decrease in RFS, while low levels of miR‐23a/b, miR‐27b, miR‐34b, miR‐224, miR‐466, miR‐709, and let‐7b were significantly correlated with poorer RFS (Table 3). Five miRNAs (miR‐205, miR‐221, miR‐222, miR‐383, and miR‐3622b) were detected no correlation between the expression levels and tumor progression (P > 0.05).
3.5. miRNA expression and overall survival
A total of 11 records comprised OS analysis involving 15 miRNAs (Table 4 and Figure 6F). Among three articles on miR‐375, significant heterogeneity was observed (P = 0.03, I 2 = 70%). After reviewing three full texts, we found plasma samples were used in the studies of Hui‐ming Lin29 and Xiaoyi Huang,30 while serum samples were used in the study of Sven Wach.31 Removing the data from Sven Wach, the heterogeneity was markedly decreased (P = 0.67, I 2 = 0%) (Figure 6F). Hence, a fixed model was applied to calculate a pooled RR and 95%CI, and we found that patients with high miR‐375 expression had significantly poorer OS compared to low miR‐375 expression (RR = 2.93, 95%CI, 1.96‐4.40) (Figure 6F).
In the other eight studies involving 14 miRNAs (Table 4), eight miRNAs (miR‐132, miR‐150, miR‐200a/b/c, miR‐429, miR‐708, and miR‐1290) were showed that increased expression predicted significantly worse OS, and low expression of four miRNAs (miR‐23a, miR‐23b, miR‐221, and miR‐224) were associated with poorer OS. Moreover, in the analyses on miR‐205 and miR‐1207‐3p, no statistically significant results were observed. It was worth noting that miR‐200a, miR‐200b, miR‐200c, and miR‐429 were the members of the same family, their change trends were consistent in different studies, and all of them were associated with poorer OS.
4. DISCUSSION
The major challenges for PC clinical management were its accurate diagnosis and dynamic monitoring after RP, chemotherapy or radiotherapy, etc. Although PSA routinely screening improved the ratio of early detection, its levels was poorly associated with tumor aggressiveness, and had a little help to predict PC patients' prognosis. Moreover, biopsies were not only invasive but also not conclusive, for example, sampling errors could lead to missed diagnosis and wrong therapies in clinic, especially in the cases with multifocal PC.
Recently, miRNAs had been found to be closely associated with a variety of tumors by regulating their target genes to affect carcinogenesis and progression. And a number of researches showed a significant correlation between the expression levels of miRNAs and the diagnosis and prognosis of PC. These study data would be helpful miRNAs as biomarkers to be transfer into the clinical application for diagnosis and prognosis of PC. Moreover, Compared to mRNAs, clinical samples containing miRNAs are more likely to be collected and detested because miRNAs are stable not to be easily degraded. The expression profiles of miRNAs are special in various cancer or normal tissues. And they can be accurately quantified by microarray, qRT‐PCR, and RNA sequencing in not only frozen or fresh or formalin‐fixed paraffin‐embedded tissues, but also serum or plasma samples, even in urine or saliva samples. However, these results on the clinical value of miRNAs were inconsistent and even contradictory due to the clinical complexity of PC. Therefore, it is necessary to conduct stratified and systematic analyses to confirm their expression pattern and application scope.
By meta‐analyses of included studies, we successfully come to some valuable conclusions for future applications in clinic. The most studied miRNA was miR‐21, with 11 articles providing the data of its expression level in clinical PC samples. Secondly, the expression profile data of miR‐221 and miR‐205 were clearly reported in seven and six studies, respectively. And the expression levels of 7 miRNAs (miR‐25, miR‐32, miR‐100, miR‐125b, miR‐141, miR‐222, miR‐375) were reported in five literatures. In addition, the most obviously increased miRNAs were the members of the miR‐200 family: miR‐200a and miR‐200b, their HR and 95%CI were 5.17 (3.22‐7.13) and 4.08 (2.91‐5.24), respectively. The most significantly decreased miRNA was miR‐199a, its pooled HR and 95%CI was −4.23 (−16.22‐7.76).
In order to remove the interference of genetic backgrounds due to patients' ethnic groups, the included studies were classified into China subgroup and Brazil subgroup, etc. We found increased miR‐21 expression could distinguish PC patients from normal controls, and could predict a significantly poor RFS. The expression of miR‐100 in the Brazilian population was significantly reduced, and HR and 95%CI was −77.57 (−110.47, −44.67). The different expression levels and predictive values of miRNAs may be explained by the differences of hereditary backgrounds and environmental exposures.
Second, we conducted subgroup analyses depending on the pathological types of PC to classify the enrolled studies into subgroups of cancer categories: normal controls/BPH, primary/local PC, metastatic PC, high‐risk PC, and recurrence PC/non‐recurrence PC subgroups, etc. In the comparisons of the expression profiles of miRNAs in primary/local PC versus metastatic PC subgroups, we found that miR‐21 and miR‐32 were up‐regulated in metastatic PC tissues, while miR‐25, miR‐143, miR‐145, miR‐191, and let‐7c were down‐regulated. In subgroup analyses of PC versus control, we found that miR‐141, miR‐200c, and miR‐375 were increased, while miR‐30 c, miR‐143, miR‐145, miR‐191, miR‐221, miR‐222, and let‐7c were reduced. Among them, low expression of three miRNAs (miR‐30c, miR‐45, and let‐7c) predicted worse RFS, the HR 95%CI were 0.32 (0.15‐0.66), 3.86 (1.85‐8.03), and 3.14 (1.49‐6.60), respectively. In addition, the expression model of miR‐15a and miR‐16 was special, both of them were lower expressed in PC tissues than that in normal controls, and their expression levels were increased again when PC progressed to malignant metastatic stages.
Finally, we performed subgroup analyses to clarify the diagnostic values of miRNAs based on the data of serum/plasma and urine samples, etc. We found that high‐expression of miR‐375 was significantly associated with a worse OS (HR = 2.93, 95%CI 1.96‐4.40) in serum/plasma subgroup, and its high‐expression was also shown in tissue subgroup (HR = 7.41, 95%CI 6.49‐8.33) and urine subgroup (HR = 1799.29, 95%CI 1796.45‐1802.13). In addition, we processed subgroup analyses of the expression levels of other five miRNAs in serum or urine samples. Among them, the members of miR‐200 family: miR‐141 and miR‐200c were up‐regulated, while the expression levels of miR‐221 and miR‐221 were decreased. The expression of miR‐100 was increased in the urine samples, which was contrary to its expression in patients' tissues. Although the detection of miRNAs in tissues was widely accepted by researchers and doctors to diagnose and predict PC progression, the detection in serum or urine samples was more convenient and uninjurious, which could dynamically monitor the therapeutic effects and patients' prognosis at any time point of the lifetime of PC patients.
The meta‐analysis has some merits. First, we strictly followed the literature inclusion criteria and the quality of enrolled literatures was satisfactory. Second, we conducted subgroup analyses to effectively minimize the influence of heterogeneity among the enrolled studies, and to further explore the scope of application for miRNAs as a prognostic biomarker of malignant tumors. All of these have increased the statistical power of the meta‐analysis. But there are also many shortcomings in the meta‐analysis. First, only a few articles are eligible for a kind of miRNA leading to the relative shortage in subgroup analyses. Secondly, after data integration and subgroup analyses, some miRNAs data still lack statistical significance, such as miR‐15a (P = 0.45), miR‐16 (P = 0.69), miR‐21 (P = 0.49), miR‐25 (P = 0.83), miR‐191 (P = 0.49), and miR‐200c (P = 0.63), etc. Besides, no study is carried out in Africa, which blocks the integrated investigation of the association between miRNAs expression and PC diagnosis and prognosis. Finally, because of the lack of unified cut‐off value of miRNAs expression in different researches, which would reduce the potency of miRNAs as predictive biomarkers. Therefore, the application value of miRNAs as prognostic factors for PC is still controversial, requiring more researches to verify.
5. CONCLUSIONS
The potential use of miRNAs as diagnosis and prognosis factors for PC in the clinic was based on a growing body of investigations in the last decades. Currently, ongoing researches were still controversial that delayed the transformation from bench to bedside. Nevertheless, the potential value of miRNAs used in clinical practice had been generally accepted, which represented not only promising biomarkers for PC but also candidated therapeutic targets. Besides, detecting the expression levels of miRNAs in serum or plasma or urine samples was more exciting than detecting miRNAs in tissues, because of low cost, rapid test, and noninvasion, etc. However, in this meta‐analysis, we found that the expression profiles of miRNAs in the blood samples were different from that of the tissues, and the deviation in the urine samples was more obvious.
Due to the lack of relevant data, further studies in larger sample sizes are needed to conduct more precise stratification between miRNAs expression levels and different progression stages of PC. We will also continue to evaluate and report the clinical value of miRNAs detection when larger studies further verify the validity of miRNAs. The guidelines on study design and sample collection still need to be further improved, in particular, the detecting platforms should be clearly defined. Taken together, the meta‐analysis underline that the use of miRNAs as biomarkers for diagnosis and prediction of PC is promising, though not yet a reality in clinical practice.
Song C‐J, Chen H, Chen L‐Z, Ru G‐M, Guo J‐J, Ding Q‐N. The potential of microRNAs as human prostate cancer biomarkers: A meta‐analysis of related studies. J Cell Biochem. 2018;119: 2763–2786. https://doi.org/10.1002/jcb.26445
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016; 66:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Nadiminty N, Tummala R, Lou W, et al. MicroRNA let‐7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 2012; 287:1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loberg RD, Logothetis CJ, Keller ET, Pienta KJ. Pathogenesis and treatment of prostate cancer bone metastases: targeting the lethal phenotype. J Clin Oncol. 2005; 23:8232–8241. [DOI] [PubMed] [Google Scholar]
- 4. Dall'Era MA, Cooperberg MR, Chan JM, et al. Active surveillance for early‐stage prostate cancer: review of the current literature. Cancer. 2008; 112:1650–1659. [DOI] [PubMed] [Google Scholar]
- 5. Heijnsdijk EA, der Kinderen A, Wever EM, Draisma G, Roobol MJ, de Koning HJ. Overdetection, overtreatment and costs in prostate‐specific antigen screening for prostate cancer. Br J Cancer. 2009; 101:1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grubb RL, 3rd , Black A, Izmirlian G, et al. Serum prostate‐specific antigen hemodilution among obese men undergoing screening in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Cancer Epidemiol Biomarkers Prev. 2009; 18:748–751. [DOI] [PubMed] [Google Scholar]
- 7. Andriole GL, Crawford ED, Grubb RL, 3rd , et al. Mortality results from a randomized prostate‐cancer screening trial. N Engl J Med. 2009; 360:1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crawford ED, Andriole GL, Marberger M, Rittmaster RS. Reduction in the risk of prostate cancer: future directions after the Prostate Cancer Prevention Trial. Urology. 2010; 75:502–509. [DOI] [PubMed] [Google Scholar]
- 9. Chun FK, Epstein JI, Ficarra V, et al. Optimizing performance and interpretation of prostate biopsy: a critical analysis of the literature. Eur Urol. 2010; 58:851–864. [DOI] [PubMed] [Google Scholar]
- 10. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2010; 4:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Nat Acad Sci USA. 2004; 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta‐analysis detected by a simple, graphical test. BMJ. 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005; 58:882–893. [DOI] [PubMed] [Google Scholar]
- 14. DerSimonian R, Laird N. Meta‐analysis in clinical trials. Control Clin Trials. 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 15. Katz B, Reis ST, Viana NI, et al. Comprehensive study of gene and microRNA expression related to epithelial‐mesenchymal transition in prostate cancer. PLoS ONE. 2014; 9:e113700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang B, Liu Z, Ning H, et al. MicroRNA‐21 in peripheral blood mononuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. Cancer Biomark. 2016; 17:223–230. [DOI] [PubMed] [Google Scholar]
- 17. Huang Wei, Kang Xin‐Li, Cen Son, Wang Yang, Chen Xiang. High‐level expression of microRNA‐21 in peripheral blood mononuclear cells is a diagnostic and prognostic marker in prostate cancer. Genet Test Mol Biomarkers. 2015; 19:469–475. [DOI] [PubMed] [Google Scholar]
- 18. Hao Y, Zhao Y, Zhao X, et al. Improvement of prostate cancer detection by integrating the PSA test with miRNA expression profiling. Cancer Invest. 2011; 29:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan Y, Wu Y, Liu Y, Ni J, Nong S. Association of microRNA‐21 expression with clinicopathological characteristics and the risk of progression in advanced prostate cancer patients receiving androgen deprivation therapy. Prostate. 2016; 76:986–993. [DOI] [PubMed] [Google Scholar]
- 20. Cheng HH, Mitchell PS, Kroh EM, et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer‐associated hypoxia. PLoS ONE. 2013; 8:e69239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng F, Wu J, Gao Z, Yu S, Cui Y. Screening the key microRNAs and transcription factors in prostate cancer based on microRNA functional synergistic relationships. Medicine (Baltimore). 2017; 96:e5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leite KR, Tomiyama A, Reis ST, et al. MicroRNA expression profiles in the progression of prostate cancer from high‐grade prostate intraepithelial neoplasia to metastasis. Urol Oncol. 2013; 31:796–801. [DOI] [PubMed] [Google Scholar]
- 23. Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013; 4:350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pang C, Liu M, Fang W, et al. MiR‐139‐5p is increased in the peripheral blood of patients with prostate cancer. Cell Physiol Biochem. 2016; 39:1111–1117. [DOI] [PubMed] [Google Scholar]
- 25. Xu B, Huang Y, Niu X, et al. Hsa‐miR‐146a‐5p modulates androgen‐independent prostate cancer cells apoptosis by targeting ROCK1. Prostate. 2015; 75:1896–1903. [DOI] [PubMed] [Google Scholar]
- 26. Leite KR, Sousa‐Canavez JM, et al. Change in expression of miR‐let7c, miR‐100, and miR‐218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2011; 29:265–269. [DOI] [PubMed] [Google Scholar]
- 27. Leite KR, Tomiyama A, Reis ST, et al. MicroRNA‐100 expression is independently related to biochemical recurrence of prostate cancer. J Urol. 2011; 185:1118–1122. [DOI] [PubMed] [Google Scholar]
- 28. Amankwah EK, Anegbe E, Park H, Pow‐Sang J, Hakam A, Park JY. MiR‐21, miR‐221 and miR‐222 expression and prostate cancer recurrence among obese and non‐obese cases. Asian J Androl. 2013; 15:226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin HM, Mahon KL, Spielman C, et al. Phase 2 study of circulating microRNA biomarkers in castration‐resistant prostate cancer. Br J Cancer. 2017; 116:1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang X, Yuan T, Liang M, et al. Exosomal miR‐1290 and miR‐375 as prognostic markers in castration‐resistant prostate cancer. Eur Urol. 2015; 67:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wach S, Al‐Janabi O, Weigelt K, et al. The combined serum levels of miR‐375 and urokinase plasminogen activator receptor are suggested as diagnostic and prognostic biomarkers in prostate cancer. Int J Cancer. 2015; 137:1406–1416. [DOI] [PubMed] [Google Scholar]
- 32. Hudson RS, Yi M, Esposito D, et al. MicroRNA‐1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012; 40:3689–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang YL, Zhou PJ, Wei L, et al. MicroRNA‐7 inhibits the stemness of prostate cancer stem‐like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015; 6:24017–24031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fendler A, Jung M, Stephan C, et al. MiRNAs can predict prostate cancer biochemical relapse and are involved in tumor progression. Int J Oncol. 2011; 39:1183–1192. [DOI] [PubMed] [Google Scholar]
- 35. Melbø‐Jørgensen C, Ness N, Andersen S, et al. Stromal expression of MiR‐21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS ONE. 2014; 9:e113039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ribas J, Ni X, Haffner M, et al. MiR‐21: an androgen receptor‐regulated microRNA that promotes hormone‐dependent and hormone‐independent prostate cancer growth. Cancer Res. 2009; 69:7165–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Folini M, Gandellini P, Longoni N, et al. MiR‐21: an oncomir on strike in prostate cancer. Mol Cancer. 2010; 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008; 68:1599–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jajoo S, Mukherjea D, Kaur T, et al. Essential role of NADPH oxidase‐dependent reactive oxygen species generation in regulating microRNA‐21 expression and function in prostate cancer. Antioxid Redox Signal. 2013; 19:1863–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li T, Li RS, Li YH, et al. MiR‐21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012; 187:1466–1472. [DOI] [PubMed] [Google Scholar]
- 41. Cai S, Chen R, Li X, et al. Downregulation of microRNA‐23a suppresses prostate cancer metastasis by targeting the PAK6‐LIMK1 signaling pathway. Oncotarget. 2015; 6:3904–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He HC, Zhu JG, Chen XB, et al. MicroRNA‐23b downregulates peroxiredoxin III in human prostate cancer. FEBS Lett. 2012; 586:2451–2458. [DOI] [PubMed] [Google Scholar]
- 43. Majid S, Dar AA, Saini S, et al. MiR‐23b represses proto‐oncogene Src kinase and functions as methylation‐silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 2012; 72:6435–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goto Y, Kojima S, Nishikawa R, et al. The microRNA‐23b/27b/24‐1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget. 2014; 5:7748–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo K, Zheng S, Xu Y, Xu A, Chen B, Wen Y. Loss of miR‐26a‐5p promotes proliferation, migration, and invasion in prostate cancer through negatively regulating SERBP1. Tumour Biol. 2016; 37:12843–12854. [DOI] [PubMed] [Google Scholar]
- 46. Ling XH, Han ZD, Xia D, et al. MicroRNA‐30c serves as an independent biochemical recurrence predictor and potential tumor suppressor for prostate cancer. Mol Biol Rep. 2014; 41:2779–2788. [DOI] [PubMed] [Google Scholar]
- 47. Ling XH, Chen ZY, Luo HW, et al. BCL9, a coactivator for Wnt/β‐catenin transcription, is targeted by miR‐30c and is associated with prostate cancer progression. Oncol Lett. 2016; 11:2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kobayashi N, Uemura H, Nagahama K, et al. Identification of miR‐30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012; 3:1455–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jalava SE, Urbanucci A, Latonen L, et al. Androgen‐regulated miR‐32 targets BTG2 and is overexpressed in castration‐resistant prostate cancer. Oncogene. 2012; 31:4460–4471. [DOI] [PubMed] [Google Scholar]
- 50. Li Q, Lu S, Li X, et al. Biological function and mechanism of miR‐33a in prostate cancer survival and metastasis: via downregulating Engrailed‐2. Clin Transl Oncol. 2017; 19:562–570. [DOI] [PubMed] [Google Scholar]
- 51. Majid S, Dar AA, Saini S, et al. MiRNA‐34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin Cancer Res. 2013; 19:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hagman Z, Larne O, Edsjö A, et al. MiR‐34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010; 127:2768–2776. [DOI] [PubMed] [Google Scholar]
- 53. Hudson RS, Yi M, Esposito D, et al. MicroRNA‐106b‐25 cluster expression is associated with early disease recurrence and targets caspase‐7 and focal adhesion in human prostate cancer. Oncogene. 2013; 32:4139–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi XB, Xue L, Ma AH, et al. Tumor suppressive miR‐124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013; 32:4130–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun X, Liu Z, Yang Z, et al. Association of microRNA‐126 expression with clinicopathological features and the risk of biochemical recurrence in prostate cancer patients undergoing radical prostatectomy. Diagn Pathol. 2013; 8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun X, Yang Z, Zhang Y, et al. Prognostic implications of tissue and serum levels of microRNA‐128 in human prostate cancer. Int J Clin Exp Pathol. 2015; 8:8394–8401. [PMC free article] [PubMed] [Google Scholar]
- 57. Xu S, Yi XM, Zhou WQ, Cheng W, Ge JP, Zhang ZY. Downregulation of miR‐129 in peripheral blood mononuclear cells is a diagnostic and prognostic biomarker in prostate cancer. Int J Clin Exp Pathol. 2015; 8:14335–14344. [PMC free article] [PubMed] [Google Scholar]
- 58. Song Xu, Xiao‐ming Yi, Zheng‐yu Zhang, Jing‐ping Ge, Wen‐quan Zhou. MiR‐129 predicts prognosis and inhibits cell growth in human prostate carcinoma. Mol Med Rep. 2016; 14:5025–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li X, Wan X, Chen H, et al. Identification of miR‐133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res. 2014; 20:2312–2325. [DOI] [PubMed] [Google Scholar]
- 60. Gonzales JC, Fink LM, Goodman OB, Jr , Symanowski JT, Vogelzang NJ, Ward DC. Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate‐specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin Genitourin Cancer. 2011; 9:39–45. [DOI] [PubMed] [Google Scholar]
- 61. Li Z, Ma YY, Wang J, et al. Exosomal microRNA‐141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2015; 9:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Avgeris M, Stravodimos K, Fragoulis EG, Scorilas A. The loss of the tumour‐suppressor miR‐145 results in the shorter disease‐free survival of prostate cancer patients. Br J Cancer. 2013; 108:2573–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dezhong L, Xiaoyi Z, Xianlian L, et al. MiR‐150 is a factor of survival in prostate cancer patients. J Buon. 2015; 20:173–179. [PubMed] [Google Scholar]
- 64. Theodore SC, Davis M, Zhao F, et al. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR‐152 and DNA methyltranferase 1. Oncotarget. 2014; 5:3512–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lichner Z, Fendler A, Saleh C, et al. MicroRNA signature helps distinguish early from late biochemical failure in prostate cancer. Clin Chem. 2013; 59:1595–1603. [DOI] [PubMed] [Google Scholar]
- 66. Liu R, Li J, Teng Z, Zhang Z, Xu Y. Overexpressed microRNA‐182 promotes proliferation and invasion in prostate cancer PC‐3 cells by down‐regulating N‐myc downstream regulated gene 1 (NDRG1). PLoS ONE. 2013; 8:e68982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsuchiyama K, Ito H, Taga M, et al. Expression of microRNAs associated with Gleason grading system in prostate cancer: miR‐182‐5p is a useful marker for high grade prostate cancer. Prostate. 2013; 73:827–834. [DOI] [PubMed] [Google Scholar]
- 68. Zhang H, Qi S, Zhang T, et al. MiR‐188‐5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget. 2015; 6:6092–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hailer A, Grunewald TG, Orth M, et al. Loss of tumor suppressor mir‐203 mediates overexpression of LIM and SH3 Protein 1 (LASP1) in high‐risk prostate cancer thereby increasing cell proliferation and migration. Oncotarget. 2014; 5:4144–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Verdoodt B, Neid M, Vogt M, et al. MicroRNA‐205, a novel regulator of the anti‐apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013; 43:307–314. [DOI] [PubMed] [Google Scholar]
- 71. Kalogirou C, Spahn M, Krebs M, et al. MiR‐205 is progressively down‐regulated in lymph node metastasis but fails as a prognostic biomarker in high‐risk prostate cancer. Int J Mol Sci. 2013; 14:21414–21434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Andersen S, Richardsen E, Moi L, et al. Fibroblast miR‐210 overexpression is independently associated with clinical failure in Prostate Cancer—a multicenter (in situ hybridization) study. Sci Rep. 2016; 6:36573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gordanpour A, Stanimirovic A, Nam RK, Moreno CS, Sherman C, Sugar L. Seth A. miR‐221 Is down‐regulated in TMPRSS2:ERG fusion‐positive prostate cancer. Anticancer Res. 2011; 31:403–410. [PMC free article] [PubMed] [Google Scholar]
- 74. Kneitz B, Krebs M, Kalogirou C, et al. Survival in patients with high‐risk prostate cancer is predicted by miR‐221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 2014; 74:2591–2603. [DOI] [PubMed] [Google Scholar]
- 75. Wei Y, Yang J, Yi L, et al. MiR‐223‐3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. 2014; 4:7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fu H, He HC, Han ZD, et al. MicroRNA‐224 and its target CAMKK2 synergistically influence tumor progression and patient prognosis in prostate cancer. Tumour Biol. 2015; 36:1983–1991. [DOI] [PubMed] [Google Scholar]
- 77. Mavridis K, Stravodimos K, Scorilas A. Downregulation and prognostic performance of microRNA 224 expression in prostate cancer. Clin Chem. 2013; 59:261–269. [DOI] [PubMed] [Google Scholar]
- 78. Lin ZY, Chen G, Zhang YQ, et al. MicroRNA‐30d promotes angiogenesis and tumor growth via MYPT1/c‐JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Cancer. 2017; 6:48. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79. Wei JJ, Wu X, Peng Y, et al. Regulation of HMGA1 expression by microRNA‐296 affects prostate cancer growth and invasion. Clin Cancer Res. 2011; 17:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Damodaran C, Das TP, Papu John AM, et al. MiR‐301a expression: a prognostic marker for prostate cancer. Urol Oncol. 2016; 34:336. e13‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nam RK, Benatar T, Wallis CJ, et al. MiR‐301a regulates E‐cadherin expression and is predictive of prostate cancer recurrence. Prostate. 2016; 76:869–884. [DOI] [PubMed] [Google Scholar]
- 82. Xiong SW, Lin TX, Xu KW, et al. MicroRNA‐335 acts as a candidate tumor suppressor in prostate cancer. Pathol Oncol Res. 2013; 19:529–537. [DOI] [PubMed] [Google Scholar]
- 83. Wang Y, Lieberman R, Pan J, et al. MiR‐375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol Cancer. 2016; 15:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bucay N, Sekhon K, Yang T, et al. MicroRNA‐383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene. 2017; 36:2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mortensen MM, Høyer S, Orntoft TF, Sørensen KD, Dyrskjøt L, Borre M. High miR‐449b expression in prostate cancer is associated with biochemical recurrence after radical prostatectomy. BMC Cancer. 2014; 14:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Colden M, Dar AA, Saini S, et al. MicroRNA‐466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017; 8:e2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tian XM, Luo YZ, He P, Li J, Ma ZW, An Y. Inhibition of invasion and migration of prostate cancer cells by miRNA‐509‐5p via targeting MDM2. Genet Mol Res. 2017; 16. [DOI] [PubMed] [Google Scholar]
- 88. Rane JK, Scaravilli M, Ylipää A, et al. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol. 2015; 67:7–10. [DOI] [PubMed] [Google Scholar]
- 89. Chiyomaru T, Yamamura S, Fukuhara S, et al. Genistein up‐regulates tumor suppressor microRNA‐574‐3p in prostate cancer. PLoS ONE. 2013; 8:e58929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zuo ZH, Yu YP, Ding Y, et al. Oncogenic activity of miR‐650 in prostate cancer is mediated by suppression of CSR1 expression. Am J Pathol. 2015; 185:1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jiao L, Deng Z, Xu C, et al. MiR‐663 induces castration‐resistant prostate cancer transformation and predicts clinical recurrence. J Cell Physiol. 2014; 229:834–844. [DOI] [PubMed] [Google Scholar]
- 92. Saini S, Majid S, Shahryari V, et al. MiRNA‐708 control of CD44(+) prostate cancer‐initiating cells. Cancer Res. 2012; 72:3618–3630. [DOI] [PubMed] [Google Scholar]
- 93. Das DK, Osborne JR, Lin HY, Park JY, Ogunwobi OO. MiR‐1207‐3p is a novel prognostic biomarker of prostate cancer. Transl Oncol. 2016; 9:236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bucay N, Sekhon K, Majid S, et al. Novel tumor suppressor microRNA at frequently deleted chromosomal region 8p21 regulates epidermal growth factor receptor in prostate cancer. Oncotarget. 2016; 7:70388–70403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y, Shao N, Mao X, et al. MiR‐4638‐5p inhibits castration resistance of prostate cancer through repressing Kidins220 expression and PI3K/AKT pathway activity. Oncotarget. 2016; 7:47444–47464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Salido‐Guadarrama AI, Morales‐Montor JG, Rangel‐Escareño C, et al. Urinary microRNA‐based signature improves accuracy of detection of clinically relevant prostate cancer within the prostate‐specific antigen grey zone. Mol Med Rep. 2016; 13:4549–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Song C, Chen H, Wang T, Zhang W, Ru G, Lang J. Expression profile analysis of microRNAs in prostate cancer by next‐generation sequencing. Prostate. 2015; 75:500–516. [DOI] [PubMed] [Google Scholar]
- 98. Kachakova D, Mitkova A, Popov E, et al. Combinations of serum prostate‐specific antigen and plasma expression levels of let‐7c, miR‐30c, miR‐141, and miR‐375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015; 34:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lin D, Cui F, Bu Q, Yan C. The expression and clinical significance of GTP‐binding RAS‐like 3 (ARHI) and microRNA 221 and 222 in prostate cancer. J Int Med Res. 2011; 39:1870–1875. [DOI] [PubMed] [Google Scholar]
- 100. Yaman Agaoglu F, Kovancilar M, Dizdar Y, et al. Investigation of miR‐21, miR‐141, and miR‐221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011; 32:583–588. [DOI] [PubMed] [Google Scholar]
- 101. Casanova‐Salas I, Rubio‐Briones J, Calatrava A, et al. Identification of miR‐187 and miR‐182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol. 2014; 192:252–259. [DOI] [PubMed] [Google Scholar]
- 102. Osipov ID, Zaporozhchenko IA, Bondar AA, et al. Cell‐Free miRNA‐141 and miRNA‐205 as prostate cancer biomarkers. Adv Exp Med Biol. 2016; 924:9–12. [DOI] [PubMed] [Google Scholar]
- 103. Torres‐Ferreira J, Ramalho‐Carvalho J, Gomez A, et al. MiR‐193b promoter methylation accurately detects prostate cancer in urine sediments and miR‐34b/c or miR‐129‐2 promoter methylation define subsets of clinically aggressive tumors. Mol Cancer. 2017; 16:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Stuopelytė K, Daniūnaitė K, Jankevičius F, Jarmalaitė S. Detection of miRNAs in urine of prostate cancer patients. Medicina (Kaunas). 2016; 52:116–124. [DOI] [PubMed] [Google Scholar]
- 105. Stuopelyte K, Daniunaite K, Bakavicius A, Lazutka JR, Jankevicius F, Jarmalaite S. The utility of urine‐circulating miRNAs for detection of prostate cancer. Br J Cancer. 2016; 115:707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Egidi MG, Cochetti G, Guelfi G, et al. Stability assessment of candidate reference genes in urine sediment of prostate cancer patients for miRNA applications. Dis Markers. 2015; 2015:973597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schubert M, Spahn M, Kneitz S, et al. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let‐7 as prognostic marker in high‐risk prostate cancer. PLoS ONE. 2013; 8:e65064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Roberts MJ, Chow CW, Schirra HJ, et al. Diagnostic performance of expression of PCA3, hepsin and miR biomarkers inejaculate in combination with serum PSA for the detection of prostate cancer. Prostate. 2015; 75:539–549. [DOI] [PubMed] [Google Scholar]
- 109. Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011; 77:1265. e9–16. [DOI] [PubMed] [Google Scholar]
- 110. Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008; 68:6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Haj‐Ahmad TA, Abdalla MA, Haj‐Ahmad Y. Potential urinary miRNA biomarker candidates for the accurate detection of prostate cancer among benign prostatic hyperplasia patients. J Cancer. 2014; 5:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sun T, Yang M, Chen S, et al. The altered expression of MiR‐221/‐222 and MiR‐23b/‐27b is associated with the development of human castration resistant prostate cancer. Prostate. 2012; 72:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Budd WT, Seashols‐Williams SJ, Clark GC, et al. Dual action of miR‐125b As a tumor suppressor and oncomiR‐22 promotes prostate cancer tumorigenesis. PLoS ONE. 2015; 10:e0142373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. El Bezawy R, Cominetti D, Fenderico N, et al. MiR‐875‐5p counteracts epithelial‐to‐mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR‐ZEB1 axis. Cancer Lett. 2017; 395:53–62. [DOI] [PubMed] [Google Scholar]
- 115. Nam RK, Amemiya Y, Benatar T, et al. Identification and validation of a five MicroRNA signature predictive of prostate cancer recurrence and metastasis: a cohort study. J Cancer. 2015; 6:1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sun Y, Jia X, Hou L, Liu X. Screening of differently expressed miRNA and mRNA in prostate cancer by integrated analysis of transcription data. Urology. 2016; 94:313. e1–6. [DOI] [PubMed] [Google Scholar]


