Abstract
Introduction:
An important portion of the Trypanosoma cruzi genome is composed of mobile genetic elements, which are interspersed with genes on all chromosomes. The L1Tc non-LTR retrotransposon and its truncated version NARTc are the most highly represented and best studied of these elements. L1Tc is actively transcribed in all three forms of the Trypanosoma parasite and encodes the proteins that enable it to autonomously mobilize. This mini review discusses the enzymatic properties of L1Tc that enable its mobilization and possibly the mobilization of other non-autonomous retrotransposons in Trypanosoma. We also briefly review the Hepatitis Delta Virus-like autocatalytic and 2A self-cleaving viral-like sequences contained in L1Tc that regulate post-transcriptional properties such as relative protein abundance and mRNA stability. Special emphasis is placed on the Pr77 dual system, which is based on the RNA pol II-dependent internal promoter of L1Tc and NARTc and the HDV-like ribozyme activity encoded by the first 77 nucleotides of the element’s DNA and RNA. The high degree of conservation of the Pr77 sequence, referred to as the “Pr77-hallmark”, among different trypanosomatid retroelements suggests that these mobile elements are responsible for the distribution of regulatory sequences within the genome they inhabit.
Conclusion:
We also discuss how the involvement of L1Tc and NARTc in the gene regulatory processes of these parasites could justify their domestication and long-term coexistence in these ancient organisms.
Keywords: Trypanosomatid, Non-LTR retrotransposon, Promoter, AP endonuclease, RNAse H, Nucleic acids chaperone, 2A-self cleaving sequence, HDV-like ribozyme
1. introduction
1.1. Mobile Genetic Elements: Important Components of Trypanosomatid Genomes
Different species of trypanosomatids are responsible for a wide range of human diseases [1-3]. All trypanosomatid genomes sequenced have been found to contain a large number of retrotransposons that compose up to 5% of the nuclear genome [4, 5]. Retrotransposons are mobile genetic elements capable of jumping from site to site in the genome via reverse transcription of their own transcribed mRNA. The most abundant retrotransposons in trypanosomes are non-LTR retrotransposons, which are mobilized by a mechanism known as Target Primed Reverse Transcription (TPRT) [6, 7]. Among non-LTR retrotransposons are Long Interspersed Nucleotide Elements (LINEs), which encode the proteins involved in their mobilization, and Short Interspersed Nucleotide Elements (SINEs), which do not encode proteins and must be mobilized in trans by LINE machinery. The best characterized LINE in trypanosomatids is the L1Tc element of T. cruzi [8, 9] (Fig. 1), which together with its Trypanosoma brucei homologue ingi gives name to the ingi/L1Tc clade [10]. Study of the genomic distribution and organization of T. cruzi L1Tc has shown that this element is distributed along the parasite’s genome in a high copy number; it can be found in single copies or clustered in tandem repeats [11]. Analysis of the chromosomal distribution of L1Tc in various T. cruzi strains suggests that this type of clustering might be a common feature of the genomes of these parasites [11].
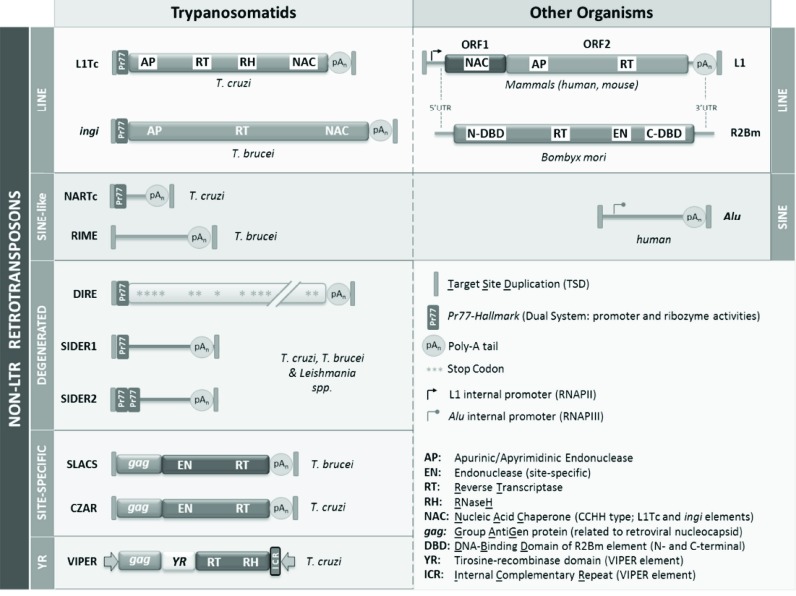
Fig. (1).
Schematic representation of the structure of the well- studied non-LTR retrotransposons from Trypanosomatids and the homologous LINE and SINE in human and mouse. The conserved domains are indicated in each element, in black letters into a white box when the enzymatic activity has been experimentally reported and white letter when the domains have been in silico identified.
Elements homologous to L1Tc and ingi, as well as truncated versions of these retrotransposons, have been identified in the genomes of trypanosomatids such as T. cruzi, T. brucei, Trypanosoma vivax and Trypanosoma congolense (Tvingi, L1Tco, Tcoingi and NARTc, TbRIME, TvRIME, and TcoRIME) (Fig. 1) [12, 13]. Altered versions of LINEs and SINEs that have accumulated mutations disabling their coding capacity also form a part of the trypanosomatid genome. These elements are known as degenerate ingi/L1Tc-related elements (DIREs) and short interspersed degenerate retrotransposons (SIDERs) and are unable to mobilize by themselves (Fig. 1) [14]. SIDERs and DIREs are particularly abundant in Leishmania species, which contains over 2000 SIDERs compared to the 20 found in T. brucei [15]. Remarkably, all of these retroelements contain the conserved Pr77 sequence, which was initially described as part of L1Tc; it is consequently referred to as the “Pr77-hallmark” or “Pr77 signature” [16].
The T. cruzi genome also contains site-specific non-LTR retrotransposons [17], such as CZAR (cruzi-associated retrotransposon) [18] and TcTREZO (T. cruzi tandem repetitive element ZO) [19, 20] (Fig. 1). The insertion of CZAR as well as that of the homologous SLACS sequence in T. brucei [21] occurs between nucleotides 11 and 12 of the miniexon in the spliced-leader (SL) gene locus [18]. TcTREZO elements consist of sequences derived from different regions of the genome and are mostly inserted into MASP pseudogenes [19]. The T. cruzi genome includes several hundred of copies of a tyrosine recombinase (YR) retrotransposon called VIPER (vestigial interposed retroelement) that is frequently located in regions containing other retroelements and retroelement-related genes, such as ingi/L1Tc-like elements and RHS genes (Fig. 1) [22, 23]. It carries an open reading frame (ORF) that harbours three domains encoding a GAG-like, a tyrosine recombinase and a reverse transcriptase-RNase H protein, similar to those present in LTR-retrotransposons [22]. Recently, similarity between the C-terminal portion of the third VIPER protein and those found in syntenic locations in other trypanosomatids such as Leishmania braziliensis, Leishmania panamensis, Leptomonas pyrrhocoris and Crithidia fasciculata have been reported (Fig. 1), suggesting a gene domestication event [23].
2. L1TC encodes enzymatic machinery that confers autonomous character.
2.1. AP Endonuclease
The presence of an AP endonuclease homologue in a non-LTR retrotransposon was first reported for the L1Tc retrotransposon of T. cruzi (Fig. 2) [8, 24, 25]. 212 amino acids at the amino terminal end of L1Tc were found to be 20.3% homologous to the human protein Ap1 [8]. Moreover, L1Tc contains several highly conserved regions with functional domains in positions similar to those described for AP endonuclease [8]. Remarkably, these domains are conserved among L1s, L1-like elements and other non-LTR retrotransposons, suggesting that they share a retrotransposition mechanism [26]. Subsequent studies have provided experimental evidence that the 40-kDa NL1Tc protein encoded by a region of the L1Tc element exhibits AP endonuclease activity [27-29]. A potential biological role of the NL1Tc protein is suggested by its ability to complement the exonuclease III enzyme repair activity of bacteria lacking the coding gene for this enzyme: expression of NL1Tc in these bacteria provides resistance to the DNA damage caused by both alkylating (MMS-induced) and oxidative (H2O2- and t-BuO2H-induced) agents [27].
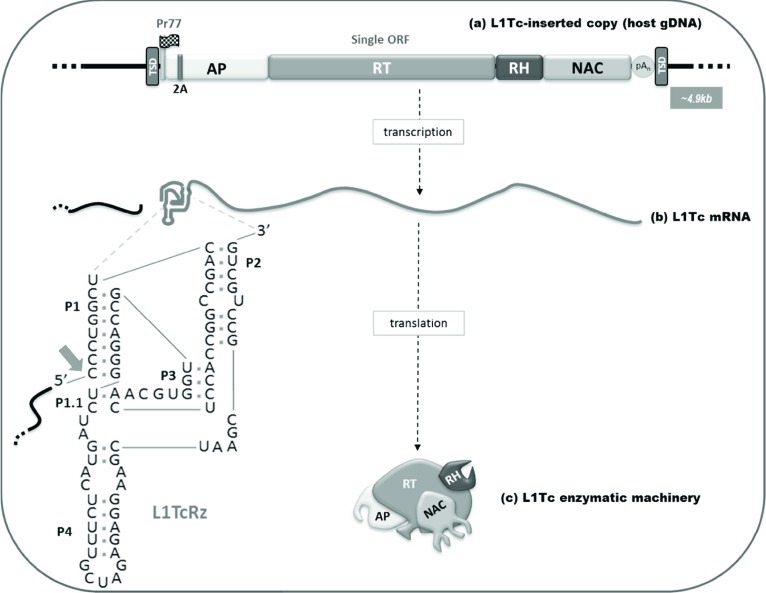
Fig. (2).
Schematic representation of the L1Tc retrotransposon. (a) A single ORF of the L1Tc element is represented by a rectangular box. The functional domains of the element are indicated in colours: AP endonuclease (AP), reverse transcriptase (RT), RNase H (RH) and Nucleic Acids Chaperone (NAC). Pr77 internal promoter is indicated with a flag and the 2A self-cleaving sequence with a purple bar. Poly-A track is located at the 3’ region of the element and is indicated by a grey circle (pAn). Target Site Duplication sequences (TSD) with direct orientation are flanking the element. (b) L1Tc mRNAs bear the HDV-like structure at their 5’ end (L1TcRz). Predicted L1TcRz folding is detailed at nucleotide level and cleavage point is indicated by a red arrow. (c) L1Tc protein encoded proteins constitute the element enzymatic machinery.
Based on the mechanisms proposed for integration of non-site-specific non-LTR retrotransposons [6], it was proposed that the endonuclease activity encoded by the L1Tc element plays a key role in the first stage of transposition (Fig. 3). In this model, NL1Tc generates free 3′-OH sites in chromosomal DNA where the integration of transposable elements would occur. Likewise, the NH2 terminal of the human L1 ORF2, which is highly homologous with the T. cruzi NL1Tc gene and the nuclease family of proteins, exhibits nuclease activity, although it has no preference for AP sites [30]. NL1Tc was also shown to be endowed with 3′-phosphodiesterase and 3′-phosphatase activities, which may allow the 3′-blocking ends to function as targets for the insertion of the L1Tc element [28]. The presence of NL1Tc 3′ repair activity could indicate a possible role for the L1Tc element in repair. In this context, the NL1Tc endonuclease protein reduces the DNA breaks originating from treatment with the anthracycline antibiotic daunorubicin [29]. The repair activity associated with these elements could act as a signal for new transposition and integration events, thereby influencing the rate of LINE retrotransposition. Indeed, it has been reported that human L1 elements can integrate into DNA lesions, resulting in a retrotransposon-mediated DNA repair mechanism in mammalian cells [31].
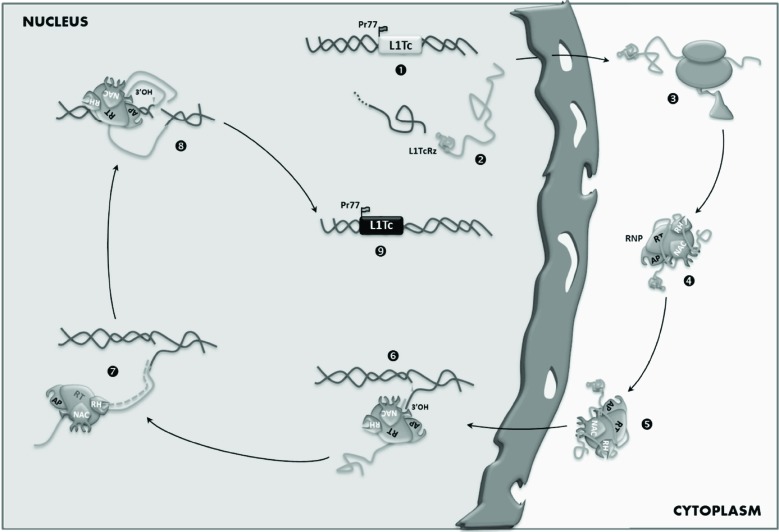
Fig. (3).
Hypothetical model for L1Tc retrotransposition. Pr77-derived L1Tc transcripts (1) are exported towards the cytoplasm (2) where are translated forming a ribonucleoprotein together the element mRNA (RNP) (3). The RNP is transported into the nucleus (4). The AP endonuclease is responsible of the DNA minus-strand cleaving at the target insertion site (5). The cDNA is synthesized by the RTL1Tc employing the L1Tc mRNA as template (6). The RNase H hydrolyses the RNA molecule from the RNA-cDNA duplex (6). The upper DNA strand is probably cleaved by the AP endonuclease (7) and the second-strand cDNA synthesis mediated by the DNA polymerase activity of the RT (7). Thus, a new copy is inserted into a new position of the genome (8).
The identification of a (GAxxAxGaxxxxxtxTATG↑ Axxxxxxxxxxx) conserved pattern in the sequenced T. cruzi genome (CL-Brener strain) preceding most L1Tc and NARTc elements does not appear to be compatible with insertion at apurinic/apyrimidinic sites via APE-mediated repair activity [32]. Interestingly, alignment of the AP protein family consensus sequence and non-LTR elements with sequences present in the SWISS-PROT database revealed high similarities with selected domains of DNase I proteins from different organisms [26]. Thus, 8 out of the 12 amino acids that form the DNA interaction domain of DNase I are conserved in all non-LTR retroelements. The His252 residue proposed by Weston et al. [33] to be involved in DNase I acid-base catalysis, as well as Asp168 and Asn170, which contact the scissile phosphate group, is conserved in all non-LTR retrotransposons [26]. Further evidence of homology has been obtained via crystallographic analyses, which show that the fold of exo III is similar to that of DNase I, despite less than 20% overall sequence similarity between the proteins [26]. Thus, because non-LTR retrotransposons share DNase I conserved domains, it is tempting to think that these elements might recognize into the T. cruzi genome sequence-dependent structural variations similar to those recognized by DNase I which would also act as insertion sites of the elements [26]. Supporting this hypothesis is the fact that all the L1Tc copies analysed in different position in the CL-Brener strain (those found as single copy and those in a tandem repeated form) were flanked by the direct repetition sequence (TGCAGACAT) known as Target Site Duplication (TSD) which is a distinctive feature of the LINE sequences of higher eukaryotes. Interestingly, the same sequence of nine nucleotides was present in a genomic clone from the Maracay strain where the same repetitive sequences were conserved with a similar genomic organization than that found in the CL-Brener strain although the L1Tc element was not present [11]. Thus, it was suggested that this sequence of 9 nts and the surrounding sequences may function as the site of insertion of the element [11]. However, subsequent analyses showed that approximately half of the T. cruzi L1Tc and NARTc elements from the T.cruzi genome (CL-Brener strain) was not flanked by a TSD and that these TSD-less elements were also preceded by the above referred conserved sequence (GAxxAxGaxxxxxtxTATG↑Axxxxxx xxxxx) [32]. This indicated that most, if not all, L1Tc and NARTc elements as the ingi/RIME and human L1/Alu pairs were not randomly distributed in the parasite genomes as previously proposed, but instead showed a relative site specificity probably dictated by the retroelement-encoded endonuclease [32].
2.2. Reverse Transcriptase
The most characteristic feature shared by all the retrotransposons is a long ORF containing an RT-related sequence. The L1Tc element maintains all seven motifs conserved in the RT and in RT-related proteins (Fig. 2) [8, 34], which fit better into the non-LTR retrotransposon branch [8]. Thus, 36 of the 42 conserved amino acids as well as the “Y/FXDD box” catalytic site required for the activity of RTs are found in identical positions in L1Tc [8].
The 65 kDa recombinant protein RTL1Tc exhibited RT activity on different template/primer sets, including heterologous RNAs used as substrates. This activity may be related to its capacity to reverse transcribe and transpose T. cruzi SINE-like sequences (Fig. 3) [35]. The RTL1Tc protein also has a tendency to switch the RNA template during cDNA synthesis, a phenomena known as a “template switching”, which is a hallmark of retroviral reverse transcriptases. Thus, the RTL1Tc is able to synthesize chimeric long cDNAs as a consequence of a continuous reverse transcription and template switching [35].
The RTL1Tc recombinant protein also exhibits DNA-dependent polymerase activity. This DNA polymerase activity allows the synthesis of both the complementary chain to intermediate RNA and the synthesis of a second DNA chain necessary to complete the element’s integration (Fig. 3), as described for RTs from retroviruses and LTR retrotransposons [36, 37]. It has been reported that RT binding to L1 element RNA occurs at or near the poly(A) tail [38]. The mechanism underlying how L1Tc recognizes retrotransposon RNA remains unknown. Independent of sequence conservation between the 3´ ends of LINEs and SINEs, it was recently hypothesized that RT recognition occurs via the 3´-end RNA stem-loop structure, which is known to be recognized by the LINE RT [39, 40].
2.3. RNase H
A sequence sharing significant homology with the RNAse H domain found in retroviruses and Escherichia coli (26.9 and 31.6%, respectively) has been identified downstream of the RT motif domain in the L1Tc ORF (Fig. 2). The three amino acids essential for catalysis by E. coli RNAse H and its neighbouring regions are conserved in L1Tc [41]. Although RNase H activity has not been reported for any non-LTR retrotransposon, this activity could be responsible for removing the RNA template from the RNA/cDNA hybrid during the transposition process in order to facilitate the synthesis of second strand DNA (Fig. 3). The enzymatic activity of the corresponding recombinant protein RHL1Tc was demonstrated using in vitro cleaving assays with various RNA/DNA substrates homologous and heterologous to L1Tc. The RHL1Tc enzyme generated RNA cleavage products similar to those generated by the HIV and E. coli enzymes, although it was found to be 40- and 15-fold less active than homologous proteins [41]. The activity of RHL1Tc is much more pH- and temperature-permissive than its bacterial and viral homologues, which may be related to the life cycle of the parasite and the diverse environmental conditions within hosts.
2.4. Nucleic Acids Chaperone
Analysis of the 3’ end of L1Tc revealed cysteine motifs with the CX2CX12HX3-5H structure that are similar to C2H2 zinc fingers found in transcription factors of high eukaryotic genomes (Fig. 2) [8]. The same structure was also found in trypanosomatid retrotransposons such as VIPER and CZAR. This sequence conservation may indicate a functional relationship with TFIIIA transcription factor motif. Analysis of the L1Tc 3’ end region revealed positively charged regions flanking the two C2H2 motifs as well as the presence of two RRR stretches located immediately upstream and downstream of the C2H2 motifs. Upstream of the first C2H2 motif, an RRRKEK stretch, which functions as a nuclear localization signal (NLS) and a DNA binding region, was also found [42]. The corresponding C2L1Tc protein exhibited binding properties to nucleic acids with different affinities in a sequence independent-manner. However, the RNA-binding capacity of C2L1Tc depends on the structure of the individual RNA. C2L1Tc also bound DNA with a 16-fold higher affinity for ssDNA than for dsDNA. The first zinc finger plays an important role in the binding of the protein to dsDNA, while the second zinc finger is primarily responsible for binding to ssDNA [43]. C2L1Tc exhibited Nucleic Acid Chaperone (NAC) activity on different DNA templates. Thus, C2L1Tc has the ability to accelerate the annealing of two complementary oligonucleotides, prevents melting of perfect DNA duplexes, and facilitates strand exchanges between DNAs to generate the most thermodynamically stable DNA duplex in competitive displacement assays [42]. The regions of C2L1Tc implicated in nucleic-acid binding are the same as those implicated in its NAC activity. The RRR and RRRKEK sequences as well as the upstream C2H2 zinc finger are the main motifs responsible for the protein’s strong affinity for RNA [43]. NAC activity has been described for the non-LTR retrotransposons from the human (bearing CCHC domain) and mouse (lacking zinc finger domain) LINE-1 associated to the named ORF1p protein which is located at the amino terminal end of these elements in contrast to the carboxy terminal location of C2L1Tc (see scheme of Fig. 1) [44].
The NAC activity of C2L1Tc and its stronger affinity for RNA may be implicated in the transposition mechanism of the L1Tc on different levels. Thus, C2L1Tc could mediate the production of a ribonucleoprotein particle composed of L1Tc mRNA and the proteins encoded by the element as well as its transport into the nucleus (Fig. 3). C2L1Tc NAC activity could promote strand exchange and stabilize priming between the L1Tc mRNA poly(A) tail and the T-rich region located at the cleaved target site. This pairing would allow the 3’ end of the cleaved target site to act as a primer for reverse transcription and first-strand cDNA synthesis. Further studies are needed to determine whether NAC activity could also facilitate the strand exchange required by TPRT to synthesize second strand DNA and whether cooperation with RNase H encoded by the element helps to unblock the 5’ end of the generated cDNA, thereby mediating more efficient transfer of the second strand [42].
2.5. 2A-like Self-cleaving Sequence
A 2A-like self-cleaving sequence similar to that described in viral organisms was found at the 5’ end of L1Tc, upstream and in-frame with the proteins encoded by the element (Fig. 2) [45]. To analyse the functional characteristics of this sequence, named L1Tc2A, artificial polyproteins comprising two reporter proteins flanking the L1Tc2A19 sequences containing the consensus domain (-DxExNPGP-) were generated. In vitro transcription and translation experiments showed that the L1Tc2A had self-cleavage activity and that the sequence upstream of L1Tc2A influenced cleavage activity, increasing its efficacy by up to 95.79% [45]. L1Tc2A-mediated cleavage has been shown to play a role in determining the ratio of cleavage products. In a native context, the presence of NL1Tc downstream of the L1Tc2A sequence produced a strong imbalance of the translation products, allowing for a lower level of NL1Tc [45]. Experimental evidence has shown that this imbalance is due to changes in the translation machinery rather than post-translational events or differences in the stability of the translated proteins [45]. The 2A consensus domain is also present at the N terminus of the endonuclease encoded by LINE-like elements in trypanosomatids such as T. congolense, T. brucei gambiense and T. vivax, suggesting an important role in the translation and function of retrotransposons [46].
2.6. The Dual DNA/RNA System of L1Tc’s First 77 Nucleotides
The absence of reports of promoters for coding sequences in the T. cruzi genome as well as the lack of known consensus promoter sequences raised the question of how L1Tc transcription occurs. The fact that its first 77 nt are identical to those of NARTc focused our attention on this sequence. Pr77 showed to behave as a RNA polymerase II-dependent promoter element, generating abundant and translatable transcripts (Fig. 2) [47]. The strong activity of this L1Tc Pr77 sequence is likely responsible for the high abundance of L1Tc mRNA. Pr77-derived transcripts are unspliced and are initiated at nucleotide +1 of L1Tc [47, 48]. Further analyses revealed the existence in Pr77 of a small core sequence described [49] which consists of 4 nucleotides (CGTG). This sequence corresponds to that described for Downstream Promoter Elements known as DPEs [48] and lies in Pr77 at positions +25 to +28 of the nucleotide +1 in L1Tc mRNA. The complete DPE is conserved in terms of composition and location in 99% of closely identical Pr77 sequences of L1Tc elements present in the genomes of different T. cruzi strains (available at http://tritrypdb.org/). The Pr77-DPE is involved in the transcription of Pr77 [48] as well as the first nucleotides of Pr77 and other nucleotides along the Pr77 sequence. These nucleotides are probably involved in binding of transcription factors that mediate Pr77 transcriptional function as nuclear proteins by binding specifically to the Pr77 sequence.
The DPE motif is conserved in terms of sequence composition and distance from the TSS within the Pr77 consensus sequence, independent of their coding or non-coding nature and degree of degeneration [48].
Remarkably, folding and sequence analyses of the first 77 nucleotides of L1Tc mRNA revealed specific characteristics compatible with those of Hepatitis Delta Virus (HDV)-like ribozymes, which have been shown experimentally to be active [50]. Thus, the three helices of HDV-like ribozymes, known as P1, P2 and P4, as well as two pseudoknots were detected during L1Tc RNA folding. In vitro transcription assays and an analysis of co-transcriptional cleavage activity showed that the L1Tc Pr77 RNA sequence possesses HDV-like autocatalytic activity within the sequence predicted to fold into an HDV-like ribozyme conformation. Consequently, this sequence is referred to as L1TcRz [50]. The hydroxyl nature of the 5’ end of the co-transcriptional 3´-products of L1TcRz is consistent with the fast biochemical cleavage reaction of HDV-like ribozymes [50].
L1TcRz self-cleaving activity may be affected by the sequences surrounding these 77 nts, which can induce RNA structural changes that sequester the L1TcRz into a non-catalytic conformation [50]. The precise structure of the L1TcRz may supply a protective cap function to lend stability to Pr77-derived transcripts that lack an SL at their 5´-ends. The existence of an RNAse P-recognition motif, together with the absence of a capped spliced leader structure at the L1Tc RNA 5´ end, suggests a cap-independent translation mechanism for L1Tc that may be IRES-like. Consistently, the non-coding NARTc element lacks an IRES-related RNAse P motif [50].
The cleavage point is located at the 5´ end of the +1 nt of L1Tc (see scheme in Fig. 2), which is consistent with the previously reported Pr77 driven L1Tc mRNAs transcription starting site. Consequently, the in vivo cleavage activity of the L1Tc ribozyme, may be responsible of the L1Tc transcripts starting at the L1Tc nucleotide +1. L1TcRz activity may ensure the production of a precise 5´ end for Pr77-driven RNAs or those released from host co-transcripts and polycistronic transcripts. The L1TcRz will prevent the mobilization of upstream sequences and insure the individuality of the L1Tc/NARTc copies transcribed from associated tandems. Because L1TcRz cleaves upstream of its catalytic domain (Fig. 2), the ribozyme and internal promoter functions persist within the L1Tc mRNAs insuring that the promoter and ribozyme functions travel with L1Tc during retrotransposition [51].
Prediction of HDV-ribozyme secondary structure revealed that the first of two Pr77 signatures found in the SIDER2 subgroup of L. infantum, L. donovani, L. major, L. mexicana and L. braziliensis appear to conform to the HDV-like ribozyme folding of L1TcRz [52]. Pr77 from T. brucei SIDER2 showed hybrid folding of ribozyme L1TcRz and the R2 retrotransposon. The Pr77-hallmark of T. congolense L1Tco and NARTco and of T. vivax SIDER1 also exhibited folding similar to that of L1TcRz. In vitro co-transcriptional activity data indicated that the SIDER2A ribozymes of L. infantum, L. donovani, L. major and L. mexicana but not that of L. braziliensis were functional albeit with moderate activity. The putative ribozymes in Trypanosoma species revealed that TvSIDER2A, L1Tco and NARTco were highly active. However, none of the assayed SIDER ribozymes from T. congolense and T. brucei showed catalytic activity. For L1TcRz, the regions located up- and downstream of the Pr77 sequence may influence Rz activity. Interestingly, the HDV-like ribozymes in Trypanosoma spp were more effective under co-transcriptional conditions, reaching cleaving rates of 100%. In contrast, Leishmania spp ribozymes showed greater cleavage efficiency at 2 h post-transcription than co-transcriptionally [52]. This is consistent with an externally controlled post-transcriptional regulatory function. The ribozyme activity of the largely immobile SIDER elements of Leishmania spp, which are typically located in 3’ UTRs, suggests that these sequences may play a regulatory role [53]. In fact, some of these sequence repeats are involved in the downregulation of their host mRNA via endonucleolytic cleavage prior to deanylation [52]. The existence of a ribozyme in Leishmania suggests that SIDERs may be involved in this cleavage. The mobile nature of Pr77-bearing retrotransposons may have promoted the spread of HDV-like ribozymes throughout trypanosomatids, thereby permitting exaptation events that turned these mobile elements into regulatory sequences.
3. Functions of retrotransposons in host gene regulation
The coexistence of a ribozyme and internal promoter systems within the L1Tc and NARTc elements may play a role in the genetic regulation of the host [51]. Trypanosomatid genomes are organized in large directional polycistronic clusters that are transcribed by RNA polymerase II and are separated by Strand Switch Regions (SSR). Transcription is mainly initiated at SSRs located between diverging clusters [54]. Sequence alignment of different chromosomes from T. cruzi, T. brucei and L. major parasites indicate that all the identified non-LTR or retrotransposon-like sequences appear to be located in areas of chromosome inversion or strand-switch or at chromosome ends [55]. Analysis of retrotransposon distribution in the T. brucei genome clearly demonstrated a strong bias in transposable element (TE) density within strand-switch regions, with ~60 and ~4 times more TEs than in Directional Gene Clusters (DGC) and subtelomeric regions, respectively. A high retrotransposon density was also observed in L. major strand-switch regions [56]. The number of L1Tc elements in sense orientation with respect to the policystronic cluster in which the element is inserted is one order of magnitude higher that the number of elements inserted in an antisense orientation with respect to the transcription orientation of the polycistronic clusters where the elements are located, as deduced from an analysis of L1Tc elements found in T. cruzi genomes (http://tritrypdb.org) [48]. Together, these data led us to suggest that Pr77 could be responsible for or participating in the transcription of adjacent genes and polycistrons [47, 48, 51, 56] and that dispersed copies of L1Tc could prevent the decay of the transcription level of distal regions, thereby ensuring the correct expression of the distal genes of each cluster. In addition, L1TcRz ensures the proper release of any L1Tc or NARTc elements that are co-transcribed as part of a polycistronic RNA. The fast reaction of L1TcRz suggests that RNA cleavage occurs early during element expression, thereby permitting early co-transcriptional cleavage in vivo.
CONCLUSION
Thus, because many functions are encoded by L1Tcs, these elements and their derivatives are likely to mediate several functions in host genomes. Consequently they may constitute an example of domestication and expansion of a whole TE family that has evolved to fulfil an essential role for the organism as it has been suggested for the Leishmania SIDERs [56]. Homologous LINEs have been found and well-studied in higher eukaryotes and insects with common and unique functions and features to L1Tc (Table 1). These elements are mobile and are useful under specific conditions. However, it is likely that the hosts have also evolved to control the expansive mobilization of these elements.
Table 1.
Common and unique characteristics of the best known non-LTR retrotransposons in which the enzymatic activities have been experimentally reported. ORFs (number of Open Reading Frames), AP (AP-endonuclease), RT (Reverse Transcriptase), RH (RNase H), NAC (Nucleic Acid Chaperone), ORF1p protein with NAC activity (the human ORF1 bears CCHC domain and the mouse ORF1p protein lacks zinc fingers), DBD (DNA-Binding Domain at N- and C-terminal), EN (Endonuclease from R2 element, similar to type II restriction enzymes).
| TE | Type |
Size
(kb) |
Transcription
(Mediated by) |
ORFs | Domains | Unique Features |
|---|---|---|---|---|---|---|
|
L1Tc (T. cruzi) |
LINE (autonomous) |
4.9 | Pr77 internal promoter sequence (RNAPII-dependent) | 1 | AP, RT, RH, NAC | • Pr77-dual system: promoter (DPE motif) and HDV-like ribozyme. • 2A-self cleaving sequence. • RNaseH domain. • No antisense promoter activity. |
|
human L1 (Homo sapiens) |
LINE1 (autonomous) |
6 | L1 internal promoter located at 5’UTR of the element (RNAPII-dependent). |
2 | ORF1p, AP, RT | • AP domain shows 3’→5’ exonuclease proofreading activity. • Antisense promoter activity that regulates L1 mobilization. |
|
mouse L1 (Mus musculus) |
LINE1 (autonomous) |
7 | Repeating promoter motif located at 5’ UTR of the element (RNAPII-dependent) and followed by 5’UTR. | 2 | ORF1p, AP, RT | • Translation via dicistronic mRNA that contains an IRES. • Antisense promoter activity that regulates L1 mobilization. |
|
R2Bm (Bombyx mori) |
Site-specific (R2) |
4.2 | The R2 RNA is initially co-transcribed with host ribosomal 28S RNA by RNAPI. | 1 | N-DBD, RT, EN, C-DBD | • HDV-like ribozyme. • 5’ and 3’ regions of the R2 RNA are binding sites for R2 polyprotein. |
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
F. Macias and R. Afonso-Lehmann contributed equally to this work. This work is supported by grant SAF2016-80998-R from the Programa Estatal I+D+i (MINECO), the Network of Tropical Diseases Research RICET (RD16/0027/0005) and FEDER.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.World Health Organization http://www.who.int/mediacentre/factsheets/fs340/en/
- 2.2016 http://www.who.int/mediacentre/factsheets/fs259/en/
- 3.World Health Organization 2016 http://www.who.int/mediacentre/factsheets/fs375/en/
- 4.El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D.C., Haas B.J., Tran A.N., Wortman J.R., Alsmark U.C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J.M., Cerqueira G.C., Creasy T., Delcher A.L., Djikeng A., Embley T.M., Hauser C., Ivens A.C., Kummerfeld S.K., Pereira-Leal J.B., Nilsson D., Peterson J., Salzberg S.L., Shallom J., Silva J.C., Sundaram J., Westenberger S., White O., Melville S.E., Donelson J.E., Andersson B., Stuart K.D., Hall N. Comparative genomics of trypanosomatid parasitic protozoa. 2005 doi: 10.1126/science.1112181. mag.org/content/309/5733/404 [DOI] [PubMed]
- 5.Bringaud F., Rogers M., Ghedin E. Identification and analysis of ingi-related retroposons in the trypanosomatid genomes. Methods Mol. Biol. 2015;1201:109–122. doi: 10.1007/978-1-4939-1438-8_6. https:// link.springer.com/protocol/10.1007/978-1-4939-1438-8_6 [DOI] [PubMed] [Google Scholar]
- 6.Eickbush T.H., Jamburuthugoda V.K. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134(1-2):221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 8.Martin F., Maranon C., Olivares M., Alonso C., Lopez M.C. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: Homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 1995;247(1):49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 9.Thomas M.C., Macias F., Alonso C., Lopez M.C. The biology and evolution of transposable elements in parasites. Trends Parasitol. 2010;26(7):350–362. doi: 10.1016/j.pt.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Bringaud F., Ghedin E., Blandin G., Bartholomeu D.C., Caler E., Levin M.J., Baltz T., El-Sayed N.M. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol. Biochem. Parasitol. 2006;145(2):158–170. doi: 10.1016/j.molbiopara.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Olivares M., del Carmen Thomas M., Lopez-Barajas A., Requena J.M., Garcia-Perez J.L., Angel S., Alonso C., Lopez M.C. Genomic clustering of the Trypanosoma cruzi nonlong terminal L1Tc retrotransposon with defined interspersed repeated DNA elements. Electrophoresis. 2000;21(14):2973–2982. doi: 10.1002/1522-2683(20000801)21:14<2973::AID-ELPS2973>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Bringaud F., Garcia-Perez J.L., Heras S.R., Ghedin E., El-Sayed N.M., Andersson B., Baltz T., Lopez M.C. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002;124(1-2):73–78. doi: 10.1016/s0166-6851(02)00167-6. [DOI] [PubMed] [Google Scholar]
- 13.Bringaud F., Berriman M., Hertz-Fowler C. Trypanosomatid genomes contain several subfamilies of ingi-related retroposons. Eukaryot. Cell. 2009;8(10):1532–1542. doi: 10.1128/EC.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith M., Bringaud F., Papadopoulou B. 2009 doi: 10.1186/1471-2164-10-240. https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-10-240 [DOI] [PMC free article] [PubMed]
- 15.Bringaud F., Berriman M., Hertz-Fowler C. TSIDER1, a short and non-autonomous Salivarian trypanosome-specific retroposon related to the ingi6 subclade. Mol. Biochem. Parasitol. 2011;179(1):30–36. doi: 10.1016/j.molbiopara.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bringaud F., Muller M., Cerqueira G.C., Smith M., Rochette A., El-Sayed N.M., Papadopoulou B., Ghedin E. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3(9):1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksoy S. Site-specific retrotransposons of the trypanosomatid protozoa. Parasitol. Today. 1991;7(10):281–285. doi: 10.1016/0169-4758(91)90097-8. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva M.S., Williams S.P., Beard C.B., Richards F.F., Aksoy S. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell. Biol. 1991;11(12):6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souza R.T., Santos M.R., Lima F.M., El-Sayed N.M., Myler P.J., Ruiz J.C., da Silveira J.F. New Trypanosoma cruzi repeated element that shows site specificity for insertion. Eukaryot. Cell. 2007;6(7):1228–1238. doi: 10.1128/EC.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza R.T., Lima F.M., Barros R.M., Cortez D.R., Santos M.F., Cordero E.M., Ruiz J.C., Goldenberg S., Teixeira M.M., da Silveira J.F. Genome size, karyotype polymorphism and chromosomal evolution in Trypanosoma cruzi. PLoS One. 2011;6(8):e23042. doi: 10.1371/journal.pone.0023042. http://journals.plos.org/plosone/article?id= 10.1371/journal.pone.0023042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aksoy S., Williams S., Chang S., Richards F.F. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 1990;18(4):785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzi H.A., Robledo G., Levin M.J. The VIPER elements of trypanosomes constitute a novel group of tyrosine recombinase-enconding retrotransposons. Mol. Biochem. Parasitol. 2006;145(2):184–194. doi: 10.1016/j.molbiopara.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig A., Krieger M.A. Genomic and phylogenetic evidence of VIPER retrotransposon domestication in trypanosomatids. Mem. Inst. Oswaldo Cruz. 2016;111(12):765–769. doi: 10.1590/0074-02760160224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran J.V., Gilbert N. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 25.Boeke J.D., Stoye J.P. Retroviruses. United States of America: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 26.Martin F., Olivares M., Lopez M.C., Alonso C. Do non-long terminal repeat retrotransposons have nuclease activity? Trends Biochem. Sci. 1996;21(8):283–285. [PubMed] [Google Scholar]
- 27.Olivares M., Alonso C., Lopez M.C. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J. Biol. Chem. 1997;272(40):25224–25228. doi: 10.1074/jbc.272.40.25224. [DOI] [PubMed] [Google Scholar]
- 28.Olivares M., Thomas M.C., Alonso C., Lopez M.C. The L1Tc, long interspersed nucleotide element from Trypanosoma cruzi, encodes a protein with 3′-phosphatase and 3′-phosphodiesterase enzymatic activities. J. Biol. Chem. 1999;274(34):23883–23886. doi: 10.1074/jbc.274.34.23883. [DOI] [PubMed] [Google Scholar]
- 29.Olivares M., Lopez M.C., Garcia-Perez J.L., Briones P., Pulgar M., Thomas M.C. The endonuclease NL1Tc encoded by the LINE L1Tc from Trypanosoma cruzi protects parasites from daunorubicin DNA damage. Biochim. Biophys. Acta. 2003;1626(1-3):25–32. doi: 10.1016/s0167-4781(03)00022-8. [DOI] [PubMed] [Google Scholar]
- 30.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 31.Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 32.Bringaud F., Bartholomeu D.C., Blandin G., Delcher A., Baltz T., El-Sayed N.M., Ghedin E. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site specificity for insertion. Mol. Biol. Evol. 2006;23(2):411–420. doi: 10.1093/molbev/msj046. [DOI] [PubMed] [Google Scholar]
- 33.Weston S.A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J. Mol. Biol. 1992;226(4):1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 34.Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 doi: 10.1038/305827a0. https://www.nature.com/articles/ [DOI] [PubMed]
- 35.Garcia-Perez J.L., Gonzalez C.I., Thomas M.C., Olivares M., Lopez M.C. Characterization of reverse transcriptase activity of the L1Tc retroelement from Trypanosoma cruzi. Cell. Mol. Life Sci. 2003;60(12):2692–2701. doi: 10.1007/s00018-003-3342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilhelm M., Wilhelm F.X. Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci. 2001;58(9):1246–1262. doi: 10.1007/PL00000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulpa D.A., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13(7):655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 39.Kajikawa M., Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111(3):433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 40.Grechishnikova D., Poptsova M. 2016 doi: 10.1186/s12864-016-3344-4. https://bmcgenomics.biomedcentral.com/articles/ [DOI] [PMC free article] [PubMed]
- 41.Olivares M., Garcia-Perez J.L., Thomas M.C., Heras S.R., Lopez M.C. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J. Biol. Chem. 2002;277(31):28025–28030. doi: 10.1074/jbc.M202896200. [DOI] [PubMed] [Google Scholar]
- 42.Heras S.R., Lopez M.C., Garcia-Perez J.L., Martin S.L., Thomas M.C. The L1Tc C-terminal domain from Trypanosoma cruzi non-long terminal repeat retrotransposon codes for a protein that bears two C2H2 zinc finger motifs and is endowed with nucleic acid chaperone activity. Mol. Cell. Biol. 2005;25(21):9209–9220. doi: 10.1128/MCB.25.21.9209-9220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heras S.R., Thomas M.C., Macias F., Patarroyo M.E., Alonso C., Lopez M.C. Nucleic-acid-binding properties of the C2-L1Tc nucleic acid chaperone encoded by L1Tc retrotransposon. Biochem. J. 2009;424(3):479–490. doi: 10.1042/BJ20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin S.L. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7(6):706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heras S.R., Thomas M.C., Garcia-Canadas M., de Felipe P., Garcia-Perez J.L., Ryan M.D., Lopez M.C. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell. Mol. Life Sci. 2006;63(12):1449–1460. doi: 10.1007/s00018-006-6038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roulston C., Luke G.A., de Felipe P., Ruan L., Cope J., Nicholson J., Sukhodub A., Tilsner J., Ryan M.D. ‘2A-Like’ signal sequences mediating translational recoding: A novel form of dual protein targeting. Traffic. 2016;17(8):923–939. doi: 10.1111/tra.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heras S.R., Lopez M.C., Olivares M., Thomas M.C. The L1Tc non-LTR retrotransposon of Trypanosoma cruzi contains an inter nal RNA-pol II-dependent promoter that strongly activates gene transcription and generates unspliced transcripts. Nucleic Acids Res. 2007;35(7):2199–2214. doi: 10.1093/nar/gkl1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macias F., Lopez M.C., Thomas M.C. 2016 https:// bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-2427-6
- 49.Arkhipova I.R., Ilyin Y.V. Properties of promoter regions of mdg1 Drosophila retrotransposon indicate that it belongs to a specific class of promoters. EMBO J. 1991;10(5):1169–1177. doi: 10.1002/j.1460-2075.1991.tb08057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez-Luque F.J., Lopez M.C., Macias F., Alonso C., Thomas M.C. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 2011;39(18):8065–8077. doi: 10.1093/nar/gkr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Luque F., Lopez M.C., Macias F., Alonso C., Thomas M.C. Pr77 and L1TcRz: A dual system within the 5′-end of L1Tc retrotransposon, internal promoter and HDV-like ribozyme. Mob. Genet. Elements. 2012;2(1):1–7. doi: 10.4161/mge.19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez-Luque F.J., Lopez M.C., Carreira P.E., Alonso C., Thomas M.C. 2014 https://bmcgenomics.biomedcentral.com/ articles/10.1186/1471-2164-15-340
- 53.Requena J.M., Rastrojo A., Garde E., Lopez M.C., Thomas M.C., Aguado B. 2016 http://journal-dl.com/downloadpdf/591087f33fbb6e13743
- 54.Martinez-Calvillo S., Nguyen D., Stuart K., Myler P.J. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell. 2004;3(2):506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Wanless D., Von Arx A., Murphy L., Lennard N., Salzberg S., Adams M.D., White O., Hall N., Stuart K., Fraser C.M., El-Sayed N.M. Gene synteny and evolution of genome architecture in trypanosomatids. Mol. Biochem. Parasitol. 2004;134(2):183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 56.Bringaud F., Ghedin E., El-Sayed N.M., Papadopoulou B. Role of transposable elements in trypanosomatids. Microbes Infect. 2008;10(6):575–581. doi: 10.1016/j.micinf.2008.02.009. [DOI] [PubMed] [Google Scholar]