Abstract
The Trypanosomatid family includes flagellated parasites that cause fatal human diseases. Remarkably, protein-coding genes in these organisms are positioned in long tandem arrays that are transcribed polycistronically. However, the knowledge about regulation of transcription initiation and termination in trypanosomatids is scarce. The importance of epigenetic regulation in these processes has become evident in the last years, as distinctive histone modifications and histone variants have been found in transcription initiation and termination regions. Moreover, multiple chromatin-related proteins have been identified and characterized in trypanosomatids, including histone-modifying enzymes, effector complexes, chromatin-remodelling enzymes and histone chaperones. Notably, base J, a modified thymine residue present in the nuclear DNA of trypanosomatids, has been implicated in transcriptional regulation. Here we review the current knowledge on epigenetic control of transcription by all three RNA polymerases in this group of early-diverged eukaryotes.
Keywords: Epigenetics, Transcription, Trypanosomatids, Post-translational modification, Histone variant, Nucleosome
1. INTRODUCTION
The Trypanosomatid family is an ancient group of eukaryotes that includes the genera Leishmania and Trypanosoma. During their life cycles, these parasitic protozoa show several stages that infect mammalian and insect hosts [1]. Around 20 different species of Leishmania cause leishmaniasis, which exhibits three main forms: visceral, cutaneous and mucocutaneous [2]. The parasite is transmitted by the bite of infected sandflies in tropical and subtropical areas of the world. Trypanosoma brucei, transmitted by the tsetse fly, is the etiological agent of human sleeping sickness, which affects more than 500,000 people per year in sub-Saharan Africa [3]. Trypanosoma cruzi, spread mostly by Triatominae insects, produces American trypanosomiasis. It is estimated that several million people are currently infected with T. cruzi in the Americas. In addition to their medical importance, trypanosomatids are relevant in the molecular biology field because they present unique mechanisms of gene expression. Recent work has revealed the importance of nucleosome structure, histone modifications and histone variants in the control of transcription in these protozoan parasites. The purpose of this article is to review the recent literature about epigenetic regulation of transcription by all three RNA polymerases (RNA Pol) in trypanosomatids.
2. HISTONES AND HISTONE VARIANTS
Across Leishmania species, the size of the nuclear genome is conserved, as it varies from 31.9 to 32.8 megabases (Mb) in L. braziliensis and L. major, respectively [4, 5]. In most Leishmania species the genome is distributed into 36 relatively small chromosomes. T. brucei possesses a genome of 26 Mb organized into 11 large chromosomes and an unspecified number of small and intermediate-sized chromosomes [6, 7], while T. cruzi (genome of 60.3 Mb) has 41 chromosomes [8, 9]. Remarkably, protein-coding genes in trypanosomatids are organized into large Polycistronic Transcription Units (PTUs) that usually contain from dozens to hundreds of genes. Unlike operons in bacteria, genes from a PTU normally do not code for functionally-related proteins. The genomes of the trypanosomatids show a noteworthy conservation of gene order, despite an estimated divergence of 200 to 500 million years [10].
Trypanosomatids possess numerous copies of the genes encoding histones H1, H2A, H2B, H3 and H4 [11, 12]. Nevertheless, their sequences are divergent from those found in other organisms, especially within the N-terminal tail domains. Moreover, the trypanosomatid histone H1 is smaller than its orthologues in vertebrates, as it lacks the globular region [13]. However, similarly to other eukaryotes, nucleosomes are the fundamental unit of chromatin in these parasites. Interestingly, chromatin in trypanosomatids does not fold into 30-nm fibers, and chromosomes do not condense during mitosis [14].
In addition to the canonical histones, trypanosomatids contain four histone variants: H2A.Z, H2B.V, H3.V and H4.V. In T. brucei and L. major, H2A.Z and H2B.V are essential for cell viability, and they dimerize exclusively with each other [15, 16]. Sequence identity in T. brucei between H2A.Z and H2A, and between H2B.V and H2B is 43% and 38%, respectively [15]. Histones H3.V and H4.V, which seem to be unique to trypanosomatids, are not essential for viability. In T. brucei, sequence identity between histones H3 and H3.V is only 45%, whereas that between H4 and H4.V is 85% [17]. It is worth noting that an orthologue of histone H4.V has not yet been identified in Leishmania [16].
3. HISTONE-MODIFYING ENZYMES AND EFFECTOR COMPLEXES
As in other organisms, histones in trypanosomatids contain a considerable number of post-translational modifications. Some of these chemical alterations are conserved across evolution, such as the acetylation and methylation of some lysines on histone H4, and methylation at lysine 4 on histone H3 (H3K4me) [18, 19]. Other modifications seem to be specific to trypanosomatids, including a methylated alanine that corresponds to the first residue in histones H2A, H2B and H4, and some acetylated lysines at the C-terminus of histone H2B in T. brucei [20]. Numerous post-translational modifications were recently found in T. cruzi histones, including the acetylation of multiple residues at the C-terminal tail of histone H2A [21, 22]. Notably, several modifications were detected in histone variants H3.V, H2A.Z and H2B.V [21, 22]. Although the function of most of the histone post-translational modifications in trypanosomatids has yet to be determined, some of them have been reported to regulate transcription (see below).
In silico analysis of the trypanosomatid genomes showed the presence of multiple proteins implicated in histone modifications, including acetyltransferases, histone deacetylases and methyltransferases [4, 23, 24]. These enzymes play important roles in fundamental cellular processes, such as transcription regulation, RNA processing, DNA replication, DNA repair and signal transduction. Trypanosomatids encode six highly-divergent Histone acetyl transferases (HATs): four related to the MYST family (HAT1 to HAT4) and two that belong to the GNAT family (ELP3a and ELP3b) [25-27]. In T. brucei, HAT1 is needed for telomeric silencing and growth, while HAT2 participates in H4K10 acetylation and growth [25]. HAT3 is involved in the acetylation of H4K4 [28], whereas HAT4, not present in T. brucei, is responsible for acetylation of H4K4 in L. donovani [29]. On the other hand, ELP3b, which controls RNA Pol II transcription elongation in other organisms, is a nucleolar protein that negatively regulates RNA Pol I transcription elongation of rRNA genes in T. brucei [26]. Similarly, ELP3b is specifically localized to the nucleolus in L. major [30]. The function of ELP3a, located mainly at the nuclear periphery, has not been determined yet [26].
Histone deacetylases are normally involved in repression of transcription. Trypanosomatids possess all three known classes of histone deacetylases: DAC1 and DAC2 (class I), DAC3 and DAC4 (class II), and three Sir2-related proteins (class III) [4]. From these, DAC1 and DAC3 appear to be essential in T. brucei [31]. Interestingly, while Sir2rp1 is involved in telomeric gene silencing, DAC1 antagonizes basal telomeric silencing [32, 33].
Trypanosomatids possess multiple genes that encode for histone methyltransferases of at least three families: DOT1-like, SET domain-containing, and protein arginine methyltransferases (PRMT) [4]. While most organisms have a single DOT1 protein, trypanosomatids contain two homologues (DOT1A and DOT1B), which methylate histone H3 on lysine 76 (the homologue of H3K79 in other eukaryotes) [34]. DOT1A, essential for viability, catalyzes mono- and di-methylation, whereas DOT1B mediates tri-methylation of H3K76 [34]. Trypanosomatids contain five different PRMTs, a relatively large number for a single-celled eukaryote [35]. In vitro methylation assays demonstrated that PRMT-6 utilizes bovine histones as a substrate; and knockdown of PRMT-6 in both procyclic and bloodstream forms of T. brucei led to differential defects in cell division [36]. PRMT-7, which is also expressed in both stages of T. brucei, is a component of several higher order complexes in vivo, suggesting that it plays multiple roles in this organism [37]. Interestingly, the knockout of PRMT-7 in L. major led to an increase in parasite infectivity both in vitro and in vivo [38]. In T. brucei, PRMT-5 is involved in novel methylation-regulated functions that may include RNA processing and translation [35].
Effector complexes read the histone code created by histone modifications to mediate subsequent functional outcomes. To bind to modified histones, constituents of the effector complexes have different domains that include bromodomains, chromodomains and SANT domains [39]. Bromodomain-Containing Factors (BDF) are involved in the recognition of acetylated lysine residues. The trypanosomatid genomes contain five genes with predicted bromodomains (BDF1 to BDF5) [4]. In T. brucei, BDF2, BDF3, and BDF5 localized to the nucleus, but only BDF3 colocalized with H4K10ac at transcription start regions (see below) [17]. Thus, BDF3 in T. brucei is involved in initiation of transcription by RNA Pol II. In contrast, BDF3 in T. cruzi does not seem to participate in transcription, because it is a non-nuclear protein that concentrates in the flagellum and acetylates α-tubulin [40]. Interestingly, BDF2 from T. cruzi may be involved in transcription initiation, as it associates with acetylated histones [41].
4. CHROMATIN-REMODELLING ENZYMES AND HISTONE CHAPERONES
ISWI is a member of the SWI2/SNF2-related chromatin-remodelling complexes that is involved in transcriptional repression [42]. An ISWI orthologue is present in trypanosomatids, which possesses a highly conserved SNF2 N-terminal domain, a conserved helicase domain, and a putative myb-like DNA-binding domain that does not correspond to the SANT-domain subclass that is typically found in ISWI proteins [43]. In T. brucei, ISWI forms a complex with three factors: nucleoplasmin-like protein (NLP), regulator of chromosome condensation 1-like protein (RCCP) and phenylalanine/tyrosine-rich protein (FYRP) [44]. The ISWI complex could be involved in the regulation of transcription by both RNA Pol I and RNA Pol II in T. brucei (see below).
Two histone chaperones, anti-silencing factor 1 (ASF1) and chromatin assembly factor 1b (CAF-1b), are involved in chromatin assembly regulation, since they contribute to the deposition and removal of histones H3 and H4 [45]. Similarly to most species, trypanosomatids have two ASF1 isoforms, ASF1A and ASF1B [46]. In L. major cells that overexpress ASF1A, upregulation of proteins related to chromatin remodelling and physiological stress was observed, and parasites were more susceptible to a DNA damaging agent [47]. Thus, ASF1A seems to participate in chromatin assembly/disassembly and in the response to DNA damage in L. major [47]. In T. brucei, it was shown that ASF1A and CAF-1b are important regulators of cell cycle progression [48]. Interestingly, ASF1A is localized to the cytoplasm, but accumulates in the nucleus in S phase. In contrast, ASF1B is always present in the T. brucei nucleus, and its levels increase during cell cycle progression until cell division, which suggests that ASF1B is involved in nucleosome deposition during DNA replication [46].
The FACT (facilitates chromatin transcription) complex is a histone chaperone that is critical for nucleosome reorganization during transcription, as it helps evict one H2A-H2B dimer to allow passage of RNA polymerases [49]. In yeast, FACT is composed of three subunits: Spt16, Pob3 and Nhp6. The T. brucei FACT complex seems to only contain the Spt16 and Pob3 subunits [50]. Knockdown of Spt16 lead to a G2/early M phase cell cycle arrest in both stages of the parasite. Spt16 is also involved in proper chromosome segregation in T. brucei [51].
5. DNA MODIFICATIONS
DNA methylation is an epigenetic mark that controls gene expression in most organisms [52]. Although DNA methylation has been reported in trypanosomatids [53], its participation in transcription has not been proven. Interestingly, in the nuclear DNA of trypanosomatids and closely related organisms, some thymidines are hydroxylated and glucosylated to produce base J (β-D-glucosyl-hydroxymethyl- uracil) [54]. Thus, base J synthesis occurs in a two-step route: hydroxylation of specific T residues to form hydroxymethyldeoxyuridine (HOMedU), followed by the transfer of glucose to HOMedU to form base J [55]. The first step is performed by thymidine hydroxylases JBP1and JBP2, which belong to the TET/JBP subfamily of Fe2+/αKG enzymes [56, 57], and the second reaction is carried out by a glucosyltransferase [58]. JBP1 is essential in Leishmania [59], whereas removal of JBP1 in T. brucei and T. cruzi results in a substantial reduction of base J [60, 61]. It was estimated that 50% of the total base J in T. brucei is located in the telomeric repeats (being also present in other reiterated sequences and some tandemly-repeated genes) [55]. In contrast, in Leishmania the vast majority of base J (over 98%) is found in the telomeric repeats [62]. Recently, base J has been located in SSRs in trypanosomes and Leishmania, where it has been implicated in transcriptional regulation of RNA Pol II (see below) [63-65]. Interestingly, while base J is found in both the mammalian and the insect stages of T. cruzi and Leishmania, it is not present in the vector form of T. brucei [66].
6. EPIGENETIC CONTROL OF RNA POL II TRANSCRIPTION
The majority of the chromosomes in trypanosomatids have at least two PTUs, which are transcribed by RNA Pol II, generating polycistronic transcripts that are processed by trans-splicing and polyadenylation to produce the mature mRNAs [67]. Trans-splicing is a process that adds a capped 39-nucleotide Spliced Leader (SL) RNA to the 5´ termini of all the mRNAs [68].
The regions flanking PTUs are called Strand Switch Regions (SSRs). It has been shown that RNA Pol II transcription initiates at divergent SSRs, whereas transcription terminates at convergent SSRs (Fig. 1) [69-71]. Transcription start regions do not contain a TATA box or any other typical RNA Pol II core promoter element. Notably, they share several characteristics with TATA-less promoters in vertebrates: they present multiple transcription start sites that span over 50-100 bp, they contain G and C tracts that might direct bidirectional transcription, and they seem to direct constitutive transcription [72].
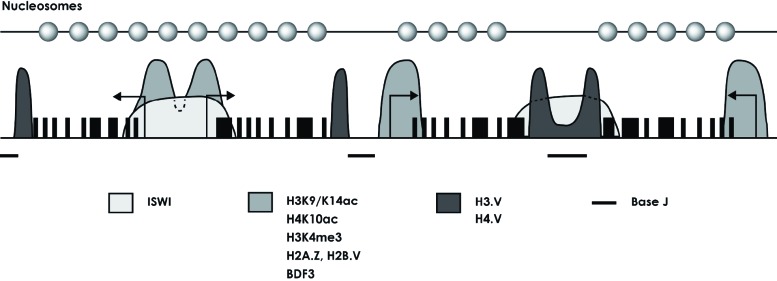
Fig. (1).
Epigenetic regulation of protein-coding genes in trypanosomatids. Schematic representation of a hypothetical chromosome that contains four PTUs, showing the distribution of the epigenetic marks involved in transcription regulation. Transcription of the first and second PTUs (left part of the figure) is divergent, whereas transcription of the third and four PTUs (right part of the figure) is convergent. Nucleosomes located at the vicinity of transcription initiation regions (labelled with arrows) contain histone variants H2A.Z and H2B.V [17]. Histones H3 in such nucleosomes are acetylated at K9/K14 and tri-methylated at K4, while histones H4 are acetylated at K10 [17, 73]. The bromodomain factor BDF3 also binds at transcription initiation regions [17]. Transcription termination regions show enrichment of histone variants H3.V and H4.V [17] and base J [63, 64]. The relative position of nucleosomes along PTUs [78] is indicated at the top of the figure.
Interestingly, a ChIP-chip study showed that all divergent SSRs in the L. major genome contain histone H3 acetylated at K9/K14, which in other organisms is a marker of active transcription [73]. In the complete L. major genome, only 184 peaks of the acetylated histone H3 were observed. Most peaks were found at divergent SSRs, but 54 of them were located at chromosome ends and within PTUs. Transcription factors TRF4 and SNAP50 were also found to be enriched at SSRs in L. major [73]. Another report demonstrated that some modifications related to transcription activation (acetylated H3K9/K14 and H4K5/K8/K12/K16, and tri-methylated H3K4) are associated to SSRs in T. cruzi [74]. Notably, it was reported that SSRs in T. brucei associate with histone H4 acetylated at K10, histone H3 tri-methylated at K4, histone variants H2A.Z and H2B.V, and the bromodomain factor BDF3 (Fig. 1) [17, 75]. Similarly to L. major, most peaks were located upstream of PTUs, but 61 of them were found within PTUs. Interestingly, it was shown that nucleosomes containing histone variants H2A.Z/H2B.V are less stable than canonical nucleosomes, which could facilitate the binding of the transcriptional machinery [17]. Moreover, it was found that histone variants H3.V and H4.V are enriched in regions where transcription ends [17]. Thus, histone modifications and histone variants seem to play important roles in transcription initiation and termination of protein-coding genes in trypanosomatids (Fig. 1) [23].
Notably, the ISWI complex components (ISWI, NLP, RCCP and FYRP) were also found to be enriched in divergent and convergent SSRs in T. brucei [44]. Thus, the ISWI complex might regulate RNA Pol II transcription in trypanosomatids. In other organisms, FACT participates in elongation of RNA Pol II transcription [49]. In T. brucei, FACT subunit Spt16 was found to be evenly distributed in divergent and convergent SSRs and within PTUs [50]. Remarkably, knockdown of Spt16 in this parasite produced a significant loss of histones H3 and H2A in SSRs and within PTUs, and a decrease in the levels of some RNA Pol II transcripts [50]. Interestingly, tandem affinity purifications showed that Spt16 associates with RNA Pol II in L. major [76]. Thus, FACT seems to participate in RNA Pol II transcription in trypanosomatids.
While most J bases are located in telomeric repeats, they are also present in regions where RNA Pol II transcription starts and stops (Fig. 1) [63, 64]. Remarkably, base J in Leishmania is required for proper transcription termination throughout the genome, as loss of base J produces massive readthrough at RNA Pol II termination sites [64, 65]. In contrast, base J in T. brucei does not regulate transcription termination at most convergent SSRs, but it seems to attenuate transcription elongation within specific PTUs and regulate transcription of downstream genes [65]. Interestingly, no defect in RNA Pol II transcription termination was observed upon base J loss in T. cruzi [61]; instead, base J seems to repress RNA Pol II transcription initiation in this parasite [77]. Thus, the role that base J plays in transcription varies across trypanosomatids. A genome-wide analysis of nucleosome positioning in L. major revealed high nucleosome occupancy, but little positioning, along PTUs [78]. Notably, low nucleosomal occupancy was observed at RNA Pol II transcription termination regions, where base J is enriched (Fig. 1) [78].
The SL RNA genes contain the only well-characterized RNA Pol II promoter in trypanosomatids [79]. Numerous transcription factors that help RNA Pol II to synthesize the SL RNA have been studied in T. brucei [80]. However, little is known about epigenetic regulation of these genes. In L. tarentolae, Southern blot analysis with nucleosomal ladders showed that the promoter and transcribed regions of the SL RNA genes are not organized into nucleosomes, but that a consistently positioned nucleosome was found within the non-transcribed intergenic region [81]. Also, a small amount of base J was detected in SL RNA genes in trypanosomes [77, 82]. Remarkably, the loss of base J leads to a decrease in nucleosome abundance, increased acetylation of histones H3 and H4, and increased RNA Pol II occupancy at SL RNA promoter regions in T. cruzi [77].
7. REGULATION OF RNA POL I TRANSCRIPTION
7.1. Ribosomal RNA Genes
In most eukaryotes, the genes encoding the 18S, 5.8S and 28S rRNA molecules are organized as tandem repeats that are separated by intergenic spacers [83]. Trypanosomatids are characterized by the fragmentation of the 28S-like rRNA into multiple independent molecules: 28Sα, β, γ, δ, ε and ζ (Fig. 2) [84]. It was estimated that the L. major genome contains only ∼12 copies of the rRNA gene repeat per haploid genome, organized in head-to-tail tandem arrays [85]. Repetitive elements of 63 bp, contained within the intergenic spacer, separate each gene repeat. A short promoter region was localized immediately downstream of the 63-bp repeats [85]. In T. brucei, class I transcription factor A (CITFA), a trypanosomatid-specific RNA Pol I transcription factor, is essential for rRNA synthesis [86].
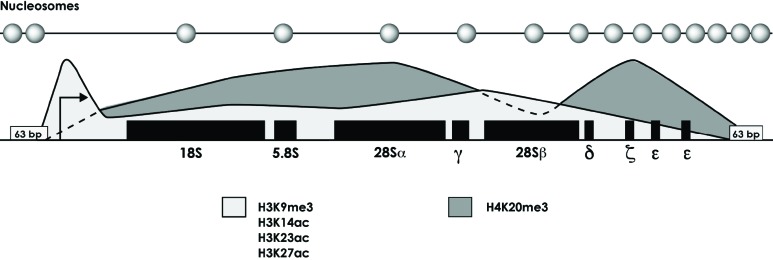
Fig. (2).
Distribution of histone modifications in the rRNA gene repeat of L. major. The promoter region (arrow) is enriched with H3K9me3, H3K14ac, H3K23ac and H3K27ac [87]. The distribution of H4K20me3 is also indicated. 63-bp repeats are present in the intergenic spacers. The relative position of nucleosomes along the rRNA genes is indicated at the top of the figure [87].
The nucleosomal architecture of rRNA genes in L. major was studied by Southern blots of chromatin partially-digested with micrococcal nuclease, using DNA probes spanning the whole rRNA gene repeat [87]. The results showed that the promoter region is practically devoid of nucleosomes, whereas the intergenic spacer presents a tight nucleosomal structure (Fig. 2). The rRNA coding regions contain intermediate levels of nucleosomes. In agreement with these results, ChIP and qPCR assays showed that levels of histone H3 were very low at the promoter, low in most of the transcribed sequence, and high in the 3´ end of the rRNA coding region and the intergenic spacer [87]. The rRNA gene repeat in T. brucei exhibits a similar distribution of histone H3 [88, 89]. Interestingly, the nucleosomal patterns observed in the rRNA genes in L. major strongly resemble those obtained in a yeast mutant that contains only active rRNA genes [90], which suggests that in L. major the ∼12 copies of the rRNA gene repeat might be active in exponentially growing cells [87].
In L. major, ChIP experiments revealed that three histone modifications that are generally associated with activation of transcription (H3K14ac, H3K23ac and H3K27ac) are enriched in the promoter region (Fig. 2) [87]. Interestingly, H3K9me3, which is usually located in transcriptionally silent regions, was also enriched in the promoter region, indicating that this modification could be involved in transcriptional activation of the rRNA genes in this parasite. Notably, H4K20me3, generally related to transcriptional silencing, was enriched all along the rRNA coding region, but was absent from the promoter and the intergenic spacer; which suggests that it does not participate in transcriptional repression of the rRNA genes in L. major. Instead, it might be involved in DNA replication or any other process [87].
Interestingly, in T. brucei the subunits of the ISWI complex are enriched in the intergenic spacer of the rRNA gene repeat, but depleted from the rRNA genes, which suggests that ISWI might regulate transcription of rRNA genes [44]. Also, ChIP data indicate that the FACT subunit Spt16 is present in the rRNA genes, but absent from the promoter region. The FACT complex is apparently involved in the regulation of processivity of RNA Pol I transcription of the rRNA genes [50]. Moreover, the high mobility group protein TDP1 was found to be highly enriched downstream of the rRNA promoter in T. brucei. Interestingly, TDP1 knockdown produced a decrease in rRNA precursor transcripts, which shows that TDP1 plays a key role in RNA Pol I transcription of the rRNA genes in this parasite [91].
7.2. VSG ES Expression
In T. brucei, but not in other trypanosomatids, RNA Pol I not only produces rRNA, but also synthesizes the mRNAs of the variant surface glycoproteins (VSG) and the EP/GPEET procyclins, which are two of the most abundant proteins in the parasite [92]. VSGs participate in antigenic variation, a survival strategy that allows the bloodstream form of the parasite to escape the host immune system by periodically switching its surface coat proteins [93, 94]. T. brucei possesses around 15 telomeric VSG Expression Sites (ESs), but only one of them is active at a given time. The VSG gene is localized at the end of a ~50 kb polycistronic unit that contains other genes known as the Expression-Site-Associated-Genes (ESAGs) (Fig. 3) [95, 96]. To achieve monoallelic expression, RNA Pol I transcription of the VSG genes must be tightly regulated. Interestingly, while active VSG ESs are depleted of nucleosomes, inactive VSG ESs are packed into nucleosomes, which shows that chromatin architecture plays an important role in VSG silencing [88, 89].
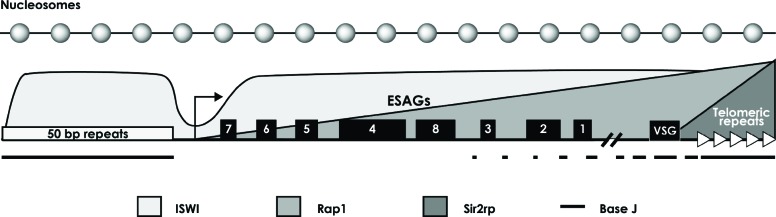
Fig. (3).
Epigenetic VSG ES silencing. Representation of a VSG ES transcription unit, showing the location of the VSG gene (~50 kb downstream of the transcription start site) and the ESAGs. The distribution of ISWI, RAP1, Sir2rp and base J in a silent VSG ES is indicated [32, 97, 101, 102]. Inactive VSG ESs are packed into nucleosomes (top of the figure), unlike active VSG ESs that are depleted of nucleosomes [88, 89].
Several chromatin-related proteins have been shown to regulate VSG ES expression [23, 24, 97, 98]. For instance, histone deacetylase DAC3 is needed for silencing at VSG ES promoters in both bloodstream and insect-stage cells [33]. Also, methyltransferase DOT1B is required to sustain rigorous VSG silencing and to guarantee a fast transcriptional VSG switching [99]. It was also demonstrated that histone chaperones ASF1A and CAF-1b participate in VSG ES silencing [48]. The FACT complex also plays a key role in VSG expression, as the knockdown of its Spt16 subunit results in derepression of VSG ES promoters in both bloodstream and insect forms of T. brucei. Accordingly, Spt16 was found to be enriched only at promoter regions of silent VSG ESs [50]. Knockdown of each of the four members of the chromatin-remodelling ISWI complex (ISWI, NLP, RCCP and FYRP) results in derepression of silent VSG ESs [44]. Notably, ISWI binds active and inactive VSG ESs, but it does not associate with their promoter regions (Fig. 3) [100]. Moreover, the telomeric protein RAP1 associates with telomere DNA, and is essential for silencing VSG ESs (Fig. 3) [101]. Knockdown of RAP1 produced a stronger derepression of genes located within 10 kb from telomeres than genes located further upstream, which indicates that RAP1-dependent silencing antagonizes transcription elongation from ES promoters [101]. In contrast, the Sir2rp1 protein, involved in telomeric gene silencing, does not participate in antigenic variation [32]. Notably, it was found that base J is present in silent VSG ESs, but not in actively transcribed VSG ESs [102]. Base J is principally enriched in the 3´ region of the silent VSG gene transcription unit, in the telomeric repeats and the 50 bp repeats located upstream of the promoter (Fig. 3) [102]. Thus, epigenetic mechanisms play key roles in the regulation of VSG expression.
Interestingly, it has been shown that the CITFA complex predominantly binds the active VSG ES promoter relative to that of a silent ES, and that it is essential for RNA Pol I transcription of the active VSG ES [103]. The only other known protein required for efficient transcription of the active VSG ES is TDP1, whose depletion resulted in ~70% reduction in VSG transcripts, and a concomitant increase in histones on the active VSG ES [91].
8. RNA POL III TRANSCRIPTION
RNA Pol III produces small essential RNAs, such as tRNAs, 5S rRNA and U6 snRNA [104, 105]. In trypanosomatids, RNA Pol III is atypical, as it transcribes all snRNAs genes (not only U6) [106]. Interestingly, snRNA genes in these parasites have a divergently oriented tRNA gene (or a tRNA-like) in their 5´-flanking region, and internal sequences from the neighboring tRNA genes are required for expression of the snRNAs [107, 108]. In L. major, there are 83 tRNA genes distributed on 19 different chromosomes. Most tRNA genes are organized into clusters of 2 to 10 genes, on either top or bottom strand, which may contain other RNA Pol III-transcribed genes [4, 109]. The eleven 5S rRNA genes found in the L. major genome are distributed in six chromosomes, and are always associated to tRNA genes [30]. Interestingly, clusters of RNA Pol III-transcribed genes are usually present at convergent SSRs, and they have been associated to RNA Pol II transcription termination [70].
In trypanosomatids, the knowledge of chromatin structure in genes transcribed by RNA Pol III is scarce. In L. major, nucleosomal ladders analyzed by Southern blots with two RNA Pol III-transcribed genes that contain internal control elements (tRNA-Lys and 5S rRNA) showed a marked smearing in the micrococcal nuclease digestion profile that indicates an open chromatin structure [87]. By contrast, protein-coding genes (transcribed by RNA Pol II) showed a strong and regularly-spaced nucleosomal structure, which might reflect a relatively low transcriptional rate of RNA Pol II. In agreement with these results, a recent genome-wide study showed that clusters of RNA Pol III-transcribed genes in L. major are depleted of nucleosomes [78]. Another genome-wide analysis revealed that peaks of acetylated histone H3 are frequently located in the vicinity of tRNA clusters [73]. Thus, these results demonstrated that, similarly to other organisms, RNA Pol III transcription in trypanosomatids is subjected to epigenetic regulation.
CONCLUSION
It was originally believed that chromatin was not involved in transcriptional control in trypanosomatids. However, numerous publications in the last years have highlighted the importance of epigenetic regulation of gene expression in these organisms. A better knowledge of the proteins and the mechanisms involved in the epigenetic control of transcription initiation and termination in this early-branched group will help us understand the evolution of epigenetic regulation in eukaryotes. It may also help in discovering key targets to control the infections caused by these organisms. Although trypanosomatids seem to share the general aspects of epigenetic regulation, it is becoming evident that they also present some species-specific features. For instance, while base J regulates termination of transcription at the end of each PTU in Leishmania, termination at the end of most PTUs occurs in a J-independent manner in T. brucei [65], and base J does not seem to be involved at all in transcription termination in T. cruzi [61]. It is also worth noting that BDF3 in T. brucei is a nuclear protein that participates in transcription initiation [17], whereas the T. cruzi orthologue concentrates in the flagellum and the flagellar pocket region [40]. These and other differences will require further analysis.
ACKNOWLEDGEMENTS
This work was supported by grant 251831 from CONACyT, and grant IN214715 from PAPIIT (UNAM) to S. Martínez-Calvillo. We thank Ana María Cevallos for critical reading of the manuscript.
LIST OF ABBREVIATIONS
- ASF1
Anti-Silencing Factor 1
- BDF
Bromodomain-Containg Factor
- CAF-1b
Chromatin Assembly Factor 1b
- CITFA
Class I Transcription Factor A
- ES
Expression Site
- ESAG
Expression-Site-Associated-Gene
- FACT
Facilitates Chromatin Transcription
- FYRP
Phenylalanine/Tyrosine-Rich Protein
- HAT
Histone Acetyl Transferase
- NLP
Nucleoplasmin-Like Protein
- PRMT
Protein Arginine Methyltransferase
- PTU
Polycistronic Transcription Unit
- RCCP
Regulator of Chromosome Condensation Protein
- RNA Pol
RNA Polymerase
- SL RNA
Spliced Leader RNA
- SSR
Strand-Switch Region
- VSG
Variant Surface Glycoprotein
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Gull K. The biology of kinetoplastid parasites: Insights and challenges from genomics and post-genomics. Int. J. Parasitol. 2001;31(5-6):443–452. doi: 10.1016/s0020-7519(01)00154-0. [DOI] [PubMed] [Google Scholar]
- 2.Desjeux P. Leishmaniasis: Current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 2004;27(5):305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy P.G. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol. 2013;12(2):186–194. doi: 10.1016/S1474-4422(12)70296-X. [DOI] [PubMed] [Google Scholar]
- 4.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S.M., Bianchettin G., Borzym K., Bothe G., Bruschi C.V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R.M., Cronin A., Cruz A.K., Davies R.M., De G.J., Dobson D.E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A.C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J.C., Muller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'Neil S., Pentony M., Pohl T.M., Price C., Purnelle B., Quail M.A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J.C., Rutter S., Saunders D., Schafer M., Schein J., Schwartz D.C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D.F., Blackwell J.M., Stuart K.D., Barrell B., Myler P.J. 2005. mag.org/content/309/5733/436/F2
- 5.Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M.A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S.L., Faulconbridge A., Jeffares D., Depledge D.P., Oyola S.O., Hilley J.D., Brito L.O., Tosi L.R., Barrell B., Cruz A.K., Mottram J.C., Smith D.F., Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melville S.E., Leech V., Navarro M., Cross G.A. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 2000;111(2):261–273. doi: 10.1016/s0166-6851(00)00316-9. [DOI] [PubMed] [Google Scholar]
- 7.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bohme U., Hannick L., Aslett M.A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U.C., Arrowsmith C., Atkin R.J., Barron A.J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T.J., Churcher C., Clark L.N., Corton C.H., Cronin A., Davies R.M., Doggett J., Djikeng A., Feldblyum T., Field M.C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B.R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A.X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P.J., Moule S., Martin D.M., Morgan G.W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C.S., Peterson J., Quail M.A., Rabbinowitsch E., Rajandream M.A., Reitter C., Salzberg S.L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A.J., Tallon L., Turner C.M., Tait A., Tivey A.R., Van A.S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M.D., Embley T.M., Gull K., Ullu E., Barry J.D., Fairlamb A.H., Opperdoes F., Barrell B.G., Donelson J.E., Hall N., Fraser C.M., Melville S.E., El-Sayed N.M. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. science.sciencemag.org/ content/309/5733/416 [DOI] [PubMed] [Google Scholar]
- 8.Weatherly D.B., Boehlke C., Tarleton R.L. 2009 doi: 10.1186/1471-2164-10-255. central.com/articles/10.1186/1471-2164-10-255 [DOI] [PMC free article] [PubMed]
- 9.El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., Westenberger S.J., Caler E., Cerqueira G.C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D.A., Carrington M., Crabtree J., Darban H., da Silveira J.F., de J.P., Edwards K., Englund P.T., Fazelina G., Feldblyum T., Ferella M., Frasch A.C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M.J., Lorenzi H., Louie T., Machado C.R., McCulloch R., McKenna A., Mizuno Y., Mottram J.C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J.L., Rinta J., Robertson L., Salzberg S.L., Sanchez D.O., Seyler A., Sharma R., Shetty J., Simpson A.J., Sisk E., Tammi M.T., Tarleton R., Teixeira S., Van A.S., Vogt C., Ward P.N., Wickstead B., Wortman J., White O., Fraser C.M., Stuart K.D. Andersson, B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. science.sciencemag.org /content/309/5733/409 [DOI] [PubMed] [Google Scholar]
- 10.Stevens J.R., Noyes H.A., Schofield C.J., Gibson W. The molecular evolution of Trypanosomatidae. Adv. Parasitol. 2001;48:1–56. doi: 10.1016/s0065-308x(01)48003-1. www.genomics.liv.ac.uk/tryps/papers/STEVENS75.pdf [DOI] [PubMed] [Google Scholar]
- 11.Alsford S., Horn D. Trypanosomatid histones. Mol. Microbiol. 2004;53(2):365–372. doi: 10.1111/j.1365-2958.2004.04151.x. [DOI] [PubMed] [Google Scholar]
- 12.Maree J.P., Patterton H.G. 2014 http://www.sciencedirect.com/science/article/pii/
- 13.Espinoza I., Toro G.C., Hellman U., Galanti N. Histone H1 and core histones in Leishmania and Crithidia: Comparison with Trypanosoma. Exp. Cell Res. 1996;224(1):1–7. doi: 10.1006/excr.1996.0105. [DOI] [PubMed] [Google Scholar]
- 14.Hecker H., Gander E.S. The compaction pattern of the chromatin of trypanosomes. Biol. Cell. 1985;53(3):199–208. doi: 10.1111/j.1768-322x.1985.tb00368.x. [DOI] [PubMed] [Google Scholar]
- 15.Lowell J.E., Kaiser F., Janzen C.J., Cross G.A. Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J. Cell Sci. 2005;118(24):5721–5730. doi: 10.1242/jcs.02688. [DOI] [PubMed] [Google Scholar]
- 16.Anderson B.A., Wong I.L., Baugh L., Ramasamy G., Myler P.J., Beverley S.M. Kinetoplastid-specific histone variant functions are conserved in Leishmania major. Mol. Biochem. Parasitol. 2013;191(2):53–57. doi: 10.1016/j.molbiopara.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel T.N., Hekstra D.R., Kemp L.E., Figueiredo L.M., Lowell J.E., Fenyo D., Wang X., Dewell S., Cross G.A. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23(9):1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Cunha J.P., Nakayasu E.S., de Almeida I.C., Schenkman S. Post-translational modifications of Trypanosoma cruzi histone H4. Mol. Biochem. Parasitol. 2006;150(2):268–277. doi: 10.1016/j.molbiopara.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Mandava V., Fernandez J.P., Deng H., Janzen C.J., Hake S.B., Cross G.A. Histone modifications in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;156(1):41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janzen C.J., Fernandez J.P., Deng H., Diaz R., Hake S.B., Cross G.A. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 2006;580(9):2306–2310. doi: 10.1016/j.febslet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 21.de Jesus T.C., Nunes V.S., Lopes M.C., Martil D.E., Iwai L.K., Moretti N.S., Machado F.C., de Lima-Stein M.L., Thiemann O.H., Elias M.C., Janzen C., Schenkman S., da Cunha J.P. Chromatin proteomics reveals variable histone modifications during the life cycle of Trypanosoma cruzi. J. Proteome Res. 2016;15(6):2039–2051. doi: 10.1021/acs.jproteome.6b00208. [DOI] [PubMed] [Google Scholar]
- 22.Picchi G.F., Zulkievicz V., Krieger M.A., Zanchin N.T., Goldenberg S., de Godoy L.M. Post-translational modifications of Trypanosoma cruzi canonical and variant histones. J. Proteome Res. 2017;16(3):1167–1179. doi: 10.1021/acs.jproteome.6b00655. [DOI] [PubMed] [Google Scholar]
- 23.Figueiredo L.M., Cross G.A., Janzen C.J. Epigenetic regulation in African trypanosomes: A new kid on the block. Nat. Rev. Microbiol. 2009;7(7):504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 24.Alsford S., duBois K., Horn D., Field M.C. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. 2012 doi: 10.1017/erm.2012.7. https://www.cambridge.org/core/journals/expert-reviews-in-molecu lar-medicine/article/epigenetic-mechanisms-nuclear-architecture-and-the-control-of-gene-expression-in-trypanosomes/84D5CDBB [DOI] [PubMed]
- 25.Kawahara T., Siegel T.N., Ingram A.K., Alsford S., Cross G.A., Horn D. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol. Microbiol. 2008;69(4):1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alsford S., Horn D. Elongator protein 3b negatively regulates ribosomal DNA transcription in African trypanosomes. Mol. Cell. Biol. 2011;31(9):1822–1832. doi: 10.1128/MCB.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso V.L., Serra E.C. 2012 https://www.hindawi.com/journals/bmri/2012/452934/ref/
- 28.Siegel T.N., Kawahara T., Degrasse J.A., Janzen C.J., Horn D., Cross G.A. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol. Microbiol. 2008;67(4):762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar D., Rajanala K., Minocha N., Saha S. Histone H4 lysine 14 acetylation in Leishmania donovani is mediated by the MYST-family protein HAT4. Microbiology. 2012;158(2):328–337. doi: 10.1099/mic.0.050211-0. [DOI] [PubMed] [Google Scholar]
- 30.Moreno-Campos R., Florencio-Martinez L.E., Nepomuceno-Mejia T., Rojas-Sanchez S., Velez-Ramirez D.E., Padilla-Mejia N.E., Figueroa-Angulo E., Manning-Cela R., Martinez-Calvillo S. Molecular characterization of 5S ribosomal RNA genes and transcripts in the protozoan parasite Leishmania major. Parasitology. 2016;143(14):1917–1929. doi: 10.1017/S0031182016001712. [DOI] [PubMed] [Google Scholar]
- 31.Ingram A.K., Horn D. Histone deacetylases in Trypanosoma brucei: Two are essential and another is required for normal cell cycle progression. Mol. Microbiol. 2002;45(1):89–97. doi: 10.1046/j.1365-2958.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- 32.Alsford S., Kawahara T., Isamah C., Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol. Microbiol. 2007;63(3):724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q.P., Kawahara T., Horn D. Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes. Mol. Microbiol. 2010;77(5):1237–1245. doi: 10.1111/j.1365-2958.2010.07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janzen C.J., Hake S.B., Lowell J.E., Cross G.A. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23(4):497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Pasternack D.A., Sayegh J., Clarke S., Read L.K. Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell. 2007;6(9):1665–1681. doi: 10.1128/EC.00133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisk J.C., Zurita-Lopez C., Sayegh J., Tomasello D.L., Clarke S.G., Read L.K. TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell. 2010;9(6):866–877. doi: 10.1128/EC.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisk J.C., Sayegh J., Zurita-Lopez C., Menon S., Presnyak V., Clarke S.G., Read L.K. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 2009;284(17):11590–11600. doi: 10.1074/jbc.M807279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira T.R., Alves-Ferreira E.V., Defina T.P., Walrad P., Papadopoulou B., Cruz A.K. Altered expression of an RBP-associated arginine methyltransferase 7 in Leishmania major affects parasite infection. Mol. Microbiol. 2014;94(5):1085–1102. doi: 10.1111/mmi.12819. [DOI] [PubMed] [Google Scholar]
- 39.de la Cruz X., Lois S., Sanchez-Molina S., Martinez-Balbas M.A. Do protein motifs read the histone code? BioEssays. 2005;27(2):164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 40.Alonso V.L., Villanova G.V., Ritagliati C., Motta M.C., Cribb P., Serra E.C. Trypanosoma cruzi bromodomain factor 3 binds acetylated alpha-tubulin and concentrates in the flagellum during metacyclogenesis. Eukaryot. Cell. 2014;13(6):822–831. doi: 10.1128/EC.00341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villanova G.V., Nardelli S.C., Cribb P., Magdaleno A., Silber A.M., Motta M.C., Schenkman S., Serra E. Trypanosoma cruzi bromodomain factor 2 (BDF2) binds to acetylated histones and is accumulated after UV irradiation. Int. J. Parasitol. 2009;39(6):665–673. doi: 10.1016/j.ijpara.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Toto M., D’Angelo G., Corona D.F. Regulation of ISWI chromatin remodelling activity. Chromosoma. 2014;123(1-2):91–102. doi: 10.1007/s00412-013-0447-4. [DOI] [PubMed] [Google Scholar]
- 43.Hughes K., Wand M., Foulston L., Young R., Harley K., Terry S., Ersfeld K., Rudenko G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26(9):2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanne T.M., Narayanan M.S., Ridewood S., Ling A., Witmer K., Kushwaha M., Wiesler S., Wickstead B., Wood J., Rudenko G. Identification of the ISWI chromatin remodeling complex of the early branching eukaryote Trypanosoma brucei. J. Biol. Chem. 2015;290(45):26954–26967. doi: 10.1074/jbc.M115.679019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mello J.A., Sillje H.H., Roche D.M., Kirschner D.B., Nigg E.A., Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3(4):329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascoalino B., Dindar G., Vieira-da-Rocha J.P., Machado C.R., Janzen C.J., Schenkman S. Characterization of two different Asf1 histone chaperones with distinct cellular localizations and functions in Trypanosoma brucei. Nucleic Acids Res. 2014;42(5):2906–2918. doi: 10.1093/nar/gkt1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scher R., Garcia J.B., Pascoalino B., Schenkman S., Cruz A.K. Characterization of anti-silencing factor 1 in Leishmania major. Mem. Inst. Oswaldo Cruz. 2012;107(3):377–386. doi: 10.1590/s0074-02762012000300013. [DOI] [PubMed] [Google Scholar]
- 48.Alsford S., Horn D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012;40(20):10150–10160. doi: 10.1093/nar/gks813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler D.D., Luger K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011;286(21):18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Denninger V., Rudenko G. FACT plays a major role in histone dynamics affecting VSG expression site control in Trypanosoma brucei. Mol. Microbiol. 2014;94(4):945–962. doi: 10.1111/mmi.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denninger V., Fullbrook A., Bessat M., Ersfeld K., Rudenko G. The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei. Mol. Microbiol. 2010;78(2):459–474. doi: 10.1111/j.1365-2958.2010.07350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blattler A., Farnham P.J. Cross-talk between site-specific transcription factors and DNA methylation states. J. Biol. Chem. 2013;288(48):34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Militello K.T., Wang P., Jayakar S.K., Pietrasik R.L., Dupont C.D., Dodd K., King A.M., Valenti P.R. African trypanosomes contain 5-methylcytosine in nuclear DNA. Eukaryot. Cell. 2008;7(11):2012–2016. doi: 10.1128/EC.00198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gommers-Ampt J.H., Van L.F., de Beer A.L., Vliegenthart J.F., Dizdaroglu M., Kowalak J.A., Crain P.F., Borst P. beta-D-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75(6):1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 55.Borst P., Sabatini R., Base J. Discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 56.Iyer L.M., Tahiliani M., Rao A., Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cliffe L.J., Hirsch G., Wang J., Ekanayake D., Bullard W., Hu M., Wang Y., Sabatini R. JBP1 and JBP2 proteins are Fe2+/2-oxoglutarate-dependent dioxygenases regulating hydroxylation of thymidine residues in trypanosome DNA. J. Biol. Chem. 2012;287(24):19886–19895. doi: 10.1074/jbc.M112.341974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekar A., Merritt C., Baugh L., Stuart K., Myler P.J. Tb927.10.6900 encodes the glucosyltransferase involved in synthesis of base J in Trypanosoma brucei. Mol. Biochem. Parasitol. 2014;196(1):9–11. doi: 10.1016/j.molbiopara.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Genest P.A., ter Riet B., Dumas C., Papadopoulou B., van Luenen H.G., Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33(5):1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cross M., Kieft R., Sabatini R., Dirks-Mulder A., Chaves I., Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46(1):37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 61.Ekanayake D.K., Minning T., Weatherly B., Gunasekera K., Nilsson D., Tarleton R., Ochsenreiter T., Sabatini R. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol. Cell. Biol. 2011;31(8):1690–1700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genest P.A., ter Riet B., Cijsouw T., van Luenen H.G., Borst P. Telomeric localization of the modified DNA base J in the genome of the protozoan parasite Leishmania. Nucleic Acids Res. 2007;35(7):2116–2124. doi: 10.1093/nar/gkm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cliffe L.J., Siegel T.N., Marshall M., Cross G.A., Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38(12):3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Luenen H.G., Farris C., Jan S., Genest P.A., Tripathi P., Velds A., Kerkhoven R.M., Nieuwland M., Haydock A., Ramasamy G., Vainio S., Heidebrecht T., Perrakis A., Pagie L., van Steensel B., Myler P.J., Borst P. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150(5):909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reynolds D., Cliffe L., Forstner K.U., Hon C.C., Siegel T.N., Sabatini R. Regulation of transcription termination by glucosylated hydroxymethyluracil, base J, in Leishmania major and Trypanosoma brucei. Nucleic Acids Res. 2014;42(15):9717–9729. doi: 10.1093/nar/gku714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Leeuwen F., Taylor M.C., Mondragon A., Moreau H., Gibson W., Kieft R., Borst P. beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. USA. 1998;95(5):2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Calvillo S., Vizuet-de-Rueda J.C., Florencio-Martinez L.E., Manning-Cela R.G., Figueroa-Angulo E.E. Gene expression in trypanosomatid parasites. 2010 doi: 10.1155/2010/525241. https://www.hindawi.com/journals/bmri/ 2010/525241/citations/ [DOI] [PMC free article] [PubMed]
- 68.Michaeli S. Trans-splicing in trypanosomes: Machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6(4):459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 69.Martinez-Calvillo S., Yan S., Nguyen D., Fox M., Stuart K., Myler P.J. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11(5):1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 70.Martinez-Calvillo S., Nguyen D., Stuart K., Myler P.J. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell. 2004;3(2):506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolev N.G., Franklin J.B., Carmi S., Shi H., Michaeli S., Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6(9):e1001090. doi: 10.1371/journal.ppat.1001090. journals.plos.org/plospathogens/ article?id=10.1371/journal.ppat.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anish R., Hossain M.B., Jacobson R.H., Takada S. Characterization of transcription from TATA-less promoters: Identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS One. 2009;4(4):e5103. doi: 10.1371/journal.pone.0005103. journals.plos.org/plosone/article?id=10.1371/journal.pone.0005103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas S., Green A., Sturm N.R., Campbell D.A., Myler P.J. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009;10:152. doi: 10.1186/1471-2164-10-152. https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Respuela P., Ferella M., Rada-Iglesias A., Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J. Biol. Chem. 2008;283(23):15884–15892. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright J.R., Siegel T.N., Cross G.A. Histone H3 trimethylated at lysine 4 is enriched at probable transcription start sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010;172(2):141–144. doi: 10.1016/j.molbiopara.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martinez-Calvillo S., Saxena A., Green A., Leland A., Myler P.J. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int. J. Parasitol. 2007;37(5):491–502. doi: 10.1016/j.ijpara.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ekanayake D., Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: Modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot. Cell. 2011;10(11):1465–1472. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lombrana R., Alvarez A., Fernandez-Justel J.M., Almeida R., Poza-Carrion C., Gomes F., Calzada A., Requena J.M., Gomez M. Transcriptionally driven DNA Replication program of the human parasite Leishmania major. Cell Reports. 2016;16(6):1774–1786. doi: 10.1016/j.celrep.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 79.Gilinger G., Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29(7):1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gunzl A., Vanhamme L., Myler P.J. Transcription in trypanosomes: A different means to the end. In: Bioscience H., editor. Trypanosomes: After the Genome; Barry, J.D., McCulloch, R., Mottram, J.C., Acosta-Serrano, A. Wymonham; 2007. pp. 177–208. [Google Scholar]
- 81.Hitchcock R.A., Thomas S., Campbell D.A., Sturm N.R. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 2007;7:44. doi: 10.1186/1471-2180-7-44. europepmc.org/abstract/ med/17517143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Leeuwen F., Kieft R., Cross M., Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109(2):133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 83.Paule M.R., White R.J. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28(6):1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hernandez R., Cevallos A.M. Ribosomal RNA gene transcription in trypanosomes. Parasitol. Res. 2014;113(7):2415–2424. doi: 10.1007/s00436-014-3940-7. [DOI] [PubMed] [Google Scholar]
- 85.Martinez-Calvillo S., Sunkin S.M., Yan S., Fox M., Stuart K., Myler P.J. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol. Biochem. Parasitol. 2001;116(2):147–157. doi: 10.1016/s0166-6851(01)00310-3. [DOI] [PubMed] [Google Scholar]
- 86.Nguyen T.N., Nguyen B.N., Lee J.H., Panigrahi A.K., Gunzl A. Characterization of a novel class I transcription factor A (CITFA) subunit that is indispensable for transcription by the multifunctional RNA polymerase I of Trypanosoma brucei. Eukaryot. Cell. 2012;11(12):1573–1581. doi: 10.1128/EC.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vizuet-de-Rueda J.C., Florencio-Martinez L.E., Padilla-Mejia N.E., Manning-Cela R., Hernandez-Rivas R., Martinez-Calvillo S. Ribosomal RNA genes in the protozoan parasite Leishmania major possess a nucleosomal structure. Protist. 2016;167(2):121–135. doi: 10.1016/j.protis.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 88.Stanne T.M., Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell. 2010;9(1):136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Figueiredo L.M., Cross G.A. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell. 2010;9(1):148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones H.S., Kawauchi J., Braglia P., Alen C.M., Kent N.A., Proudfoot N.J. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14(2):123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narayanan M.S., Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41(5):2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2(3):542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109(1):5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 94.Pays E., Vanhamme L., Perez-Morga D. Antigenic variation in Trypanosoma brucei: Facts, challenges and mysteries. Curr. Opin. Microbiol. 2004;7(4):369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 95.Berriman M., Hall N., Sheader K., Bringaud F., Tiwari B., Isobe T., Bowman S., Corton C., Clark L., Cross G.A., Hoek M., Zanders T., Berberof M., Borst P., Rudenko G. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;122(2):131–140. doi: 10.1016/s0166-6851(02)00092-0. [DOI] [PubMed] [Google Scholar]
- 96.Vanhamme L., Pays E. Control of gene expression in trypanosomes. Microbiol. Rev. 1995;59(2):223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rudenko G. Epigenetics and transcriptional control in African trypanosomes. Essays Biochem. 2010;48(1):201–219. doi: 10.1042/bse0480201. [DOI] [PubMed] [Google Scholar]
- 98.Gunzl A., Kirkham J.K., Nguyen T.N., Badjatia N., Park S.H. Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: Expression site control from both ends? Gene. 2015;556(1):68–73. doi: 10.1016/j.gene.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Figueiredo L.M., Janzen C.J., Cross G.A. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6(7):e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanne T.M., Kushwaha M., Wand M., Taylor J.E., Rudenko G. TbISWI regulates multiple polymerase I (Pol I)-transcribed loci and is present at Pol II transcription boundaries in Trypanosoma brucei. Eukaryot. Cell. 2011;10(7):964–976. doi: 10.1128/EC.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137(1):99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Leeuwen F., Wijsman E.R., Kieft R., van der Marel G.A., van Boom J.H., Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11(23):3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nguyen T.N., Muller L.S., Park S.H., Siegel T.N., Gunzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res. 2014;42(5):3164–3176. doi: 10.1093/nar/gkt1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 105.Moir R.D., Willis I.M. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta. 2013;1829(3-4):361–375. doi: 10.1016/j.bbagrm.2012.11.001. www.sciencedirect.com/science/journal/18749399/1829/3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fantoni A., Dare A.O., Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 1994;14(3):2021–2028. doi: 10.1128/mcb.14.3.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakaar V., Dare A.O., Hong D., Ullu E., Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 1994;14(10):6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rojas-Sanchez S., Figueroa-Angulo E., Moreno-Campos R., Florencio-Martinez L.E., Manning-Cela R.G., Martinez-Calvillo S. Transcription of Leishmania major U2 small nuclear RNA gene is directed by extragenic sequences located within a tRNA-like and a tRNA-Ala gene. Parasit. Vectors. 2016;9(1):401. doi: 10.1186/s13071-016-1682-3. https://www.ncbi.nlm.nih.gov/pubmed/27430335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Padilla-Mejia N.E., Florencio-Martinez L.E., Figueroa-Angulo E.E., Manning-Cela R.G., Hernandez-Rivas R., Myler P.J., Martinez-Calvillo S. Gene organization and sequence analyses of transfer RNA genes in Trypanosomatid parasites. 2009 doi: 10.1186/1471-2164-10-232. central.com/articles/10.1186/1471-2164-10-232 [DOI] [PMC free article] [PubMed]