Abstract
Trypanosomatids are a group of kinetoplastid parasites including some of great public health importance, causing debilitating and life-long lasting diseases that affect more than 24 million people worldwide. Among the trypanosomatids, Trypanosoma cruzi, Trypanosoma brucei and species from the Leishmania genus are the most well studied parasites, due to their high prevalence in human infections. These parasites have an extreme genomic and phenotypic variability, with a massive expansion in the copy number of species-specific multigene families enrolled in host-parasite interactions that mediate cellular invasion and immune evasion processes. As most trypanosomatids are heteroxenous, and therefore their lifecycles involve the transition between different hosts, these parasites have developed several strategies to ensure a rapid adaptation to changing environments. Among these strategies, a rapid shift in the repertoire of expressed genes, genetic variability and genome plasticity are key mechanisms. Trypanosomatid genomes are organized into large directional gene clusters that are transcribed polycistronically, where genes derived from the same polycistron may have very distinct mRNA levels. This particular mode of transcription implies that the control of gene expression operates mainly at post-transcriptional level. In this sense, gene duplications/losses were already associated with changes in mRNA levels in these parasites. Gene duplications also allow the generation of sequence variability, as the newly formed copy can diverge without loss of function of the original copy. Recently, aneuploidies have been shown to occur in several Leishmania species and T. cruzi strains. Although aneuploidies are usually associated with debilitating phenotypes in superior eukaryotes, recent data shows that it could also provide increased fitness in stress conditions and generate drug resistance in unicellular eukaryotes. In this review, we will focus on gene and chromosomal copy number variations and their relevance to the evolution of trypanosomatid parasites.
Keywords: Copy number variations, Kinetoplastid parasites, Trypanosomatid genomes, Evolution, Genome architecture, Parasitism
1. Introduction
Trypanosomatids are widespread parasites, which infect a wide range of vertebrates, invertebrates and plants [1-6]. Within the Trypanosomatidae family, Trypanosoma cruzi, Trypanosoma brucei and different species of the Leishmania genus stand out due to their importance in public health, being respectively the etiological agents of Chagas disease, African trypanosomiasis and Leishmaniasis, infecting around 20 million people worldwide [7].
Trypanosomatidae family is part of the Excavata supergroup, one of the earliest divergent branches of the eukaryotes [8, 9]. These parasites have some peculiar aspects such as: (I) polycistronic transcription of most of their genes [10, 11]; (II) genes organized in densely packed directional clusters [12]; (III) transcription of protein-coding genes by RNA polymerase I [11, 13-15]; (IV) RNA trans-splicing of polycistronic transcripts to generate mature mRNA molecules [16, 17]; (V) regulation of gene expression mainly by post-transcriptional mechanisms (revised by [18-20]); (VI) RNA editing to generate mitochondrial functional mRNAs [21-25]; and (VII) tolerance to aneuploidies [26-29].
As these parasites have a life cycle that alternates between vertebrate and invertebrate hosts, their distinct life stages are adapted to survive in each of these different environments. In the mammalian host, T. cruzi is able to invade any nucleated cells, while Leishmania is restricted to phagocytic cells and T. brucei does not invade cells, surviving and proliferating in the host bloodstream.
The Leishmania genus is further divided into three distinct subgenera: Leishmania, Viannia and Sauroleishmania. The Leishmania subgenus comprises Old and New World species that are associated with visceral or cutaneous forms of leishmaniasis, and develop in the midgut and foregut of the sand fly vector [30]. Viannia subgenus is constituted by New World species that only cause cutaneous forms of the disease and develop in the vector hindgut [30]. Finally, Sauroleishmania is a subgenus composed by species that infect lizards, and are incapable of replication within mammalian macrophages [31-33]. T. brucei is divided in three subspecies: T. b. brucei, T. b. gambiense and T. b. rhodesiense. Approximately 90% of the human infections in 24 countries of central and west Africa are caused by T. b. gambiense, while T. b. rhodesiense is responsible for acute cases in east and southern Africa and T. b. brucei is usually associated with animal infections, although sporadic human infections have been reported [34]. T. cruzi taxa is further divided into six Discrete Typing Units (DTUs), named TcI-TcVI, where TcV and TcVI are hybrids resulting from the fusion of parental strains from the TcII and TcIII DTUs [35-38].
Since 2005, the sequencing of the Trypanosomatidae genomes provided important datasets for comparative analysis, shedding light on our understanding of mechanisms associated to parasitism and genome evolution within this family [12, 39-45]. Even though they diverged around 200 to 500 million years ago, trypanosomatid species contain large syntenic genomic regions mostly composed by housekeeping genes, suggesting that there is a strong selective pressure to keep the gene order in these parasites [12]. These syntenic regions are interspersed by clusters of species-specific multigene families related to the parasitic lifestyle, structural RNAs and retroelements, which vary greatly in size and content among species and even within species [12, 18, 41-43]. While these species-specific clusters are enriched in subtelomeric regions in T. brucei, they are distributed in internal chromosomes and subtelomeric regions in T. cruzi CL Brener and Leishmania [12].
The advent of next-generation sequencing technologies allowed several trypanosomatid genomes to be sequenced in high depth coverage at a low cost, providing insights into copy number variation events in these parasites [26, 27, 29, 46-48]. In fact, for several trypanosomatid genes there is a correlation between copy number and expression rates [11, 49, 50], which is especially interesting in these organisms that mostly regulate their gene expression post-transcriptionally. Chromosomal Copy Number Variation (CCNV) is a new level of copy number variation, in which a whole chromosome is duplicated or lost. This phenomenon has already been described in several Leishmania species and T. cruzi DTUs [26-29, 46], as well as in yeast and mammalian cells [51-56], and could be related to a rapid adaptation to new environments and stress conditions. In this review, we delve into the gene and chromosomal copy number variations in Trypanosomatids and their biologic implications.
2. Gene copy number variation in Trypanosomatids
Gene copy number variations have gained considerable attention as a potential source of diversity and as an important evolutionary factor affecting fitness [26, 27, 57-63]. This is a well-documented mechanism to alter gene expression and increase sequence variability, through the generation of paralog genes [41, 61-66]. These paralog genes may present an accelerated evolutionary rate [62, 67, 68], resulting in several consequences, such as: (i) neofunctionalization of duplicated genes; (ii) subfunctionalization of duplicated genes by segregation of a multifunctional gene in two genes with single functions; (iii) differential expression of duplicated genes in distinct tissues or development stages by changes in their regulatory regions [62, 63, 69-73].
Trypanosomatid lifecycle involve the transition between different hosts and a series of morphological changes in response to the distinct pressures within their hosts. Recent studies have suggested that in response to these transitions, these parasites have developed adaptive mechanisms such as changes in mRNA levels due to gene dosage. These mechanisms exploit unique characteristics related to the genome organization of these parasites. So far no canonical RNA polymerase II promoter has been identified in these organisms and their genes are transcribed as long polycistronic units [11, 74, 75]. Genes derived from the same polycistron may not be functionally related and may have very distinct mRNA levels. The polycistrons are processed as mature monocistronic mRNAs by the addition of a 39 nt spliced leader RNA to the 5’ of the transcript and polyadenylation to its 3′ end [17, 76, 77]. These features imply that Trypanosomatids mainly rely on post-transcriptional mechanisms to control gene expression. These mechanisms include mRNA processing and stability, translation efficiency, and protein stability [20, 78-80].
Genomic plasticity in trypanosomatids in response to changes in the environment conditions can affect whole chromosomes or be restricted to specific genomic regions and includes aneuploidy, gene amplification or deletion [58, 62]. This phenomenon also occurs in fungi and cancer cells where changes in gene copy number can modulate drug susceptibility, virulence and proliferation [56, 81]. In Leishmania, it has been documented that increased gene copy number modulate gene expression in response to environmental changes within its host as well as after exposition to stressors, such as drug selection [26].
In trypanosomatids, the evolution of parasitism appears to have been shaped by a series of gene losses and gains [61, 62, 82, 83]. Among the genes that were likely lost during the origin of trypanosomatids are diverse enzymes involved in polysaccharide degradation like β-glucosidases or glucoamylases that are required for processing of bacterial prey [83]. In addition, this process of reduction affected several pathways of catabolism, macromolecular degradation and transport.
Adaptation to a parasitic lifestyle in trypanosomatids also included gain of novel functions that could allow survival inside a host. This is revealed by the abundance of gene families that are unique or specific to each of these parasites [83]. Many of these gene families encode surface proteins, which are the main interface between the host and the parasite, suggesting that the divergence of trypanosomatid genomes was ruled at least partially by the evolution of multigene families [82, 83].
In Leishmania parasites, these multigenic families include amastins, GP63, cysteine peptidases and surface antigen proteins [82, 83]. Amastins are poorly functionally characterized surface glycoproteins that are preferentially expressed in the intracellular amastigote stage. Phylogenetic analyses have revealed that this family consists of four subfamilies termed α, β, γ and δ amastins and that an expansion of the latter appear to be key for the evolution of parasitism in Leishmania [84]. The composition of the amastin family varies between species with an increased δ-amastin repertoire in L. (Viannia) braziliensis, L. (Leishmania) mexicana and L. (Leishmania) major than in the other species [82]. These differences in amastin content on Leishmania genomes may be a reflect of the distinct environments for which different human-pathogenic Leishmania species have adapted.
Another important cell surface family that participates in parasite invasion and immune evasion is the zinc-metallopeptidase GP63. This multigene family has a wide substrate specificity and is preferentially expressed in Leishmania promastigotes [85, 86]. Among the diverse functions of this family are midgut attachment, protection of parasites against digestive enzymes inside the sand fly host [85, 87] and inactivation of complement mediated lysis by cleavage of C3b and inhibition of antimicrobial agents’ production by the interaction with JAK kinases in the vertebrate host [88-90]. The GP63 family appears to be organized into three distinct subfamilies that have undergone expansion events during the evolution of trypanosomatids with clear differences in the number and structure of these genes between species [82, 91]. For instance, GP63 of the L. (Sauroleishmania) tarentolae are smaller, lack extracellular regions, present a smaller catalytic domain and appeared to have limited protease activity compared with other species [82, 92]. This limited GP63 functionality in this species is in accordance with its lack of intracellular development, different vector and vertebrate host (lizard). In contrast, the GP63 repertoire of the Viannia species is greater than the other two subgenera and is composed of tandem gene arrays [82]. The presence of these genes in a head-to-tail fashion may correspond to an adaptive mechanism to rapidly increase gene dosage.
Peptidases are key parasite components that mediate differentiation and immune evasion during the initial steps of infection through the interaction with proteoglycans at the host cell surface [93]. These peptidases play immunomodulatory roles, suppressing Th1 type response delaying parasite clearance. Although trypanosomatids appeared to have lost various ancestral cathepsin genes, the remaining cathepsin-L family appeared to have been key for these parasites given the series of expansions it underwent during parasite evolution [83].
Among the trypanosomatids, T. cruzi presents the largest expansion of multigene families, many of them encoding surface proteins, which accounts for its larger genomic size (~55MB), compared to the genomes of L. major (~33MB) and T. brucei (~26MB) [12]. As these surface proteins are directly related to host-parasite interactions, the increased variability of T. cruzi multigene families could be a consequence of the broader range of mammalian hosts that this parasite can infect, as well as its unique ability among the Tritryps to actively infect any nucleated host cell [94-97]. This vast number of host cells/species exposes T. cruzi to different evolutionary pressures, which could be a driving force for diversification [98]. In fact, a significantly larger fraction of T. cruzi protein coding genes shows evidence for positive selection to diversification when compared to Leishmania genes [98].
The content of multigene families also varies among T. cruzi DTUs, where the expansion of these families account for the 5.9 Mb increased size in the genome of the CL Brener (TcVI) strain when compared to the Sylvio X10/1 (TcI) strain [42, 43, 99]. Among the most studied T. cruzi multigene families are the trans-sialidase, MASPs and mucins, which respectively have approximately 1500, 1400 and 850 genes in the hybrid CL Brener genome [41, 64]. Trans-sialidases catalyze the transfer of sialic acid from host glycoconjugates to β-galactopyranose acceptors in the mucin proteins, generating a negative coat that confers protection against the alternative complement and antibody opsonization [99-103]. This family is also enrolled in cellular adhesion and invasion of epithelial [104], neural and glial cells [105] and stimulates anti-apoptotic signals in these infected cells [106, 107]. Mucin glycoproteins covers the whole parasite outer membrane with 2x106 copies [101, 108], protecting the parasite from both the insect and mammalian host’s defense mechanisms and acting in cellular adhesion/invasion processes [99, 101]. MASPs are characterized by conserved N- and C- terminal regions, which respectively encode a signal peptide and a GPI anchor addition site, flanking a hypervariable central region [41, 64]. As these flanking regions are cleaved from the mature protein, only the hypervariable region is exposed at the parasites’ surface [41, 64]. Although the biologic function of this family is still unknown, there is evidence that it could act in cellular adhesion/invasion processes [64, 109, 110], or in immune evasion strategies as expressed members of this family vary in a given population [111] and after successive passages in mice [110].
Interestingly, the T. cruzi closely related parasite T. rangeli, which is not able to establish a productive infection in mammals, has a massive reduction in the number of members of these three multigene families [45], supporting the importance of these families to T. cruzi mammalian cell infection.
Differently to what is found in T. cruzi CL Brener [112] and Leishmania major [12, 40], T. brucei multigene families’ clusters are enriched in sub-telomeric regions [12, 39]. These clusters are mainly composed by Variant Surface Glycoproteins (VSGs) and Expression Site Associated Genes (ESAGs), which are enrolled in T. brucei antigenic variation [12, 39]. VSGs are highly antigenic surface proteins, which contain N-terminal hypervariable domains that are exposed extracellularly and conserved C-terminal domains that are buried into the parasite’s coat [113, 114]. T. b. brucei genome has ~800 different VSGs, where 7% of these genes are functional, 9% are atypical genes with folding limitations, 18% are gene fragments and 66% are full-length pseudogenes (with frameshifts or in-frame stop codons) [12, 39]. T. brucei explores this pseudogene sequence arsenal to increase VSG variability by creating functional chimeric genes through gene conversion [115-118]. As this parasite develops exclusively extracellularly during the mammalian host infection stage, it is constantly exposed to the host’s humoral immune response. To evade antibody complement mediated lysis, T. brucei relies on cell specific monoallelic expression of VSGs, a process called antigenic variation. In a population, each T. brucei cell covers its entire surface with ~107 copies of a single VSG, which shield other invariant antigens present in the parasite surface [39, 115, 119]. As the majority of the population express the same variant, a strong immune response focused on a specific VSG is established and greatly reduce the parasitemia. However, a subset of the population express a different variant, which is not promptly recognized by the hosts’ immune system, and therefore expands in the population. This process generates successive waves of parasitemia and clearance as novel antigenic determinants spread in the parasite population [39, 115, 119]. VSGs sequence also vary among T. brucei subspecies. A comparison between the gene sequences of T. b. brucei VSGs with the unassembled reads from T. b. gambiense revealed that these subspecies shared ~43% of similarity in the VSG repertoire, which suggests that there is a strong positive selection for diversification, or at least a relaxation of purifying selection in T. brucei genes encoding surface proteins [48].
The massive expansions seen in multigenic families in trypanosomatids constitute an important evidence of specialization that these parasites have to undertake to survive, as they have to deal with different stressors within their hosts. While T. cruzi and Leishmania use this arsenal of proteins to invade and survive inside the host cell, the largest T. brucei multigene families are involved in antigenic variation to evade humoral response and survive into the host bloodstream. The continuous selective pressure imposed by the immune system, together with the adaptation to different hosts, provided an ideal environment for microevolution, challenging the ability of the host to control the infection and of the parasite to ensure its survival.
3. Chromosomal Copy Number Variation in Trypanosomatids
Aneuploidy, the presence of abnormal number of chromosomes in a cell, is usually lethal or results in severe abnormalities in superior multicellular eukaryotes, greatly reducing organism fitness [120]. Among its most common examples are the trisomy of the chromosome 21 in Down syndrome, aneuploidies in sexual chromosomes (as the triple X, Klinefelter and XYY syndromes) and tumorigenesis [121-123]. However, some unicellular organisms such as Saccharomyces cerevisiae and Candida albicans appear to rely on aneuploidy as one of the mechanisms to allow rapid adaptation to changing environments, suggesting that the variation in chromosome number could also have a positive fitness effect in stress conditions and drug resistance [53-55]. Extensive aneuploidies have been described in eukaryotes such as mice hepatocyte cells [52], yeast, carcinomatous lung cells [51, 53-55], trypanosomatid parasites of the Leishmania genus [26, 27, 59, 91, 124-126] and in several Trypanosoma cruzi strains [28, 29].
As cytogenetic analyses are hampered in trypanosomatids by the lack of chromosome condensation during mitosis, there are currently two complementary and alternative methodologies to estimate CCNV in these parasites: Fluorescence in situ hybridization (FISH) [26, 124], and Whole Genome Sequencing (WGS) followed by Read Depth Coverage (RDC) and allele frequency [26, 27, 29] analyses. While FISH allows the simultaneous identification of aneuploidies in each cell in a given population, it is laborious and usually restricted to few chromosomes [124, 127]. On the other hand, WGS followed by RDC analyses provides a population level snapshot of all the chromosomes, as well as the simultaneous comparison of CCNV, genotype and allele frequency. However, it lacks the single-cell resolution provided by FISH [27, 127].
Both techniques were employed in parasites from the Leishmania genus, showing that variations in the ploidy appear to be widespread among and within species [27], strains [26, 27] and even within the same parasite population [59, 124, 125]. Based on the read depth coverage and allele frequency, the chromosomal copy number variation among L. major (strains Friedlin and LV39), L. mexicana (strains U1103, M379), L. infantum (JPCM5), L. donovani (strains BPK206/0 and LV9) and L. braziliensis (M2904) was estimated, showing extensive variation in chromosomal gain/loss within Leishmania species [27]. In fact, with the exception of the two L. major strains (Friedlin and LV39), all Leishmania isolates presented a different pattern of aneuploidies, suggesting that chromosomal duplication/loss is a common phenomenon in this parasite.
Interestingly, chromosome 31 was consistently polysomic in all the evaluated Leishmania strains [26, 27, 46, 91]. Gene ontology revealed that this chromosome is enriched with genes enrolled in iron metabolism, electron carrier activity and calcium-dependent cysteine-type endopeptidase activity [91]. Iron-sulfur proteins are crucial for the parasite survival, as it mediates oxi-reduction reactions during mitochondrial electron transport, been also involved in the synthesis of amino acids, biotin and lipoic [128]. The expansion of these genes could be one of the driving forces for chromosome 31 amplification in Leishmania [91]. RNA-seq analysis in L. mexicana showed that chromosome 30, which is also polysomic in this species and corresponds to chromosome 31 in other Leishmania species, is enriched for amastigote upregulated genes. A total of 79 (19%) genes had at least a >2-fold increase in mRNA levels in amastigotes compared to promastigotes, including amastins, amino acid- and other transporters as well as hypothetical proteins [129]. Several of these genes are known to be involved in survival strategies in the mammalian host, such as amino acid transporters, tryparedoxin, ABC transporters and aquaglyceroporin, suggesting that the duplication of this chromosome could favor parasite adaptation to the vertebrate infection [129]. Leishmania chromosome 31 is also syntenic to a duplicated region in T. brucei genome, which comprises the right end of both chromosomes 4 and 8 [62]. Although there is a widespread gene loss in these duplicated regions of T. brucei chromosomes 4 and 8, approximately 47% of the duplicated genes are retained in both chromosomes as paralogous loci [62]. Among these retained paralogs, there is an over representation of secreted and surface-protein encoding genes, suggesting a bias towards important factors at the host-parasite interface [62].
To date, the strain L. braziliensis M2904 is the only trypanosomatid predominately triploid [27], while all the other evaluated strains are usually diploid with some or several duplications or deletions of chromosomes [27, 46, 91]. This suggests that, even though aneuploidies are widespread in Leishmania, the overall ploidy of the population has a tendency to diploidy. Dowins and co-workers showed that the aneuploidy pattern also diverges in 17 L. donovani strains that were recently isolated from patients in Nepal and India [26]. Interestingly, all the 17 Leishmania isolates had a different karyotype, while they presented only 0.011% of variation in their nucleotide sequences. This implies that CCNV events occur frequently during parasite evolution [26, 59]. Besides, the identification of aneuploidies in natural populations reinforces the notion that changes in chromosomal numbers are not caused by long-term maintenance of the parasites in culture [59].
Some of the CCNV of Leishmania and T. cruzi strains estimated based on RDC showed an intermediate value of ploidy, for instance between 2 and 3, which may be a result of a mixed cell population with disomic and trisomic chromosomes [27, 29, 59, 91]. In fact, FISH analysis of seven chromosomes in the L. major strains Friedlin, LEM62 and LEM265 showed that the CCNV pattern of each of these chromosomes were observed in at least two states, among monosomic, disomic or trisomic within a given population [124]. The evaluation of aneuploidy based on FISH analysis of chromosomes in clones and sub-clones from L. major Friedlin, LEM62 and LEM265, revealed that they present a similar chromosomal ploidy pattern as their parental strains, for some, but not all chromosomes [124]. Moreover, the aneuploidy pattern varied among single cells in the same population, a phenomenon that was named mosaic aneuploidy [124, 125].
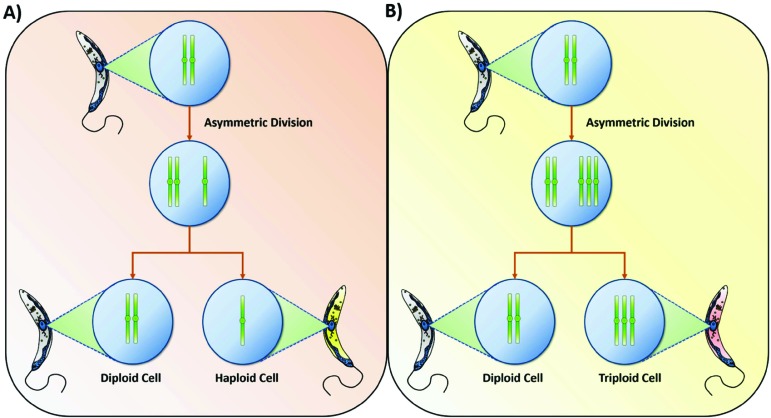
After its observation in different clones and strains from L. major [124], the occurrence of mosaic aneuploidy was also described in several Leishmania species, such as L. infantum, L. donovani, L. amazonensis and L. tropica, showing that this phenomenon is widespread in the parasite genus [126]. To explain the mechanism behind the generation of mosaic aneuploidy, a model based on mis-segregation or stochastic replication of chromosomes was proposed [124-126]. In this model, there is an asymmetric allotment of chromosomes during mitosis, resulting from replication defects that generate aberrant copy numbers of a given chromosome [59, 124]. Chromosomal segregation errors have yielded asymmetric even chromosome numbers (3+1 or 4+0) in SMC3 (central component of a complex that holds together sister chromatids during mitosis) knockdown Trypanosoma brucei [130]. However the total chromosome number in Leishmania in asymmetric sets was usually odd (1+2, 2+3 or 3+4) [124, 125], supporting replication defect events as the major driving force generating Leishmania aneuploidies (Fig. 1). The proportion of different somies observed in interphase and mitotic cells in Leishmania suggests that duplication or loss of chromosomes due to a failure in the replicative process is a usual phenomenon in Leishmania [124, 125]. In fact, it is estimated that only 10% of the cells in a Leishmania population would have the same genotype [124, 125].
Fig. (1).
Mosaic aneuploidy model proposed by Sterkers 2011 and Sterkers 2017 [124, 125]. During mitosis, there is an asymmetric replication of chromosomes followed by unequal division of the duplicated chromosomes to the daughters’ cells. This process generates mosaic aneuploidies inside a Leishmania population. (A) Asymmetric division resulting in one diploid and one haploid cell, which happens when there is a failure in the duplication of a given chromosome. (B) Asymmetric division resulting in one diploid and one triploid cell, which happens when an extra copy of a chromosome is produced during mitosis.
Among the Tritryps, CCNV was also identified in the Leishmania close related parasite T. cruzi, based on tilling arrays [28] and WGS followed by RDC and allele frequency analyses [29]. Due to the presence of large gaps, as well as the extensive content of repetitive sequences in the 41 T. cruzi CL Brener putative chromosomes, the estimation of chromosomal gain/loss in T. cruzi based on RDC was performed by normalizing the sequence coverage using single-copy genes as markers for unique genomic sequences, as proposed in the SCoPE methodology [29]. Similarly to Leishmania, the aneuploidy pattern in T. cruzi also varies among and within DTUs, as seen by variations in TcI, TcII and TcIII strains [29]. Similarly to Leishmania species, the chromosome 31 was the only one consistently duplicated in all the T. cruzi strains [29]. Gene ontology analysis revealed however that this T. cruzi chromosome harbor different set of genes than Leishmania chromosome 31. In T. cruzi, this chromosome is enriched in genes related to glycoprotein biosynthesis and glycosylation processes, especially UDP-GlcNAc-dependent glycosyl-transferase, an enzyme related to the initial steps of mucin glycosylation [29, 101]. As mentioned before, mucins are heavily glycosylated proteins that are present in ~ 2x106 copies covering the whole surface of the parasite. These proteins protect the parasite from both the vector and mammal immune systems, as well as act to ensure the anchorage point and invasion of specific cells and tissues by trypomastigotes [99, 101]. The amplification of chromosome 31 could be related to the need to glycosylate this large set of mucins, and also highlight the importance of glycosylation for T. cruzi biology. Although aneuploidies are frequent events in T. cruzi strains and Leishmania species, is it still unknown whether the mechanism behind the generation of chromosomal duplication/loss is similar in both parasites.
Interestingly, T. brucei appears to be strictly diploid [47]. Whole genome sequencing and RDC estimations of 85 T. b. gambiense group 1 field isolates from East and West Africa revealed no evidence of aneuploidies [47]. Similar results were obtained for the other two T. brucei subspecies, T. b. brucei and T. b. rhodesiense (Almeida et al., unpublished data). This absence of aneuploidies in T. brucei is not a consequence of sexual reproduction, as T. b gambiense group 1 appears to be strictly clonal [47].
One of the restrictions to aneuploidy in T. brucei could be the chromosomal size. T. brucei genome is divided into 11 megabase sized chromosomes, which vary from 1 to 6 Mbp, while T. cruzi and Leishmania genomes are distributed into ~ 34-47 chromosomes, with a size varying from 0.3-3 Mbp [39, 41, 112, 131-133]. Changes in the copy number of specific chromosomes in T. cruzi and Leishmania would therefore alter the dosage of a restricted set of genes, avoiding detrimental consequences of large-scale dosage alterations [59]. This suggests that aneuploidies would be better supported in organisms that have its genome divided into a large number of small chromosomes; however, the evaluation of a CCNV in a broader set of species is necessary to confirm this hypothesis. Also, T. brucei appears to have a similar number of origins of replication (~170) per haploid genome as L. major, L. mexicana and L. donovani (~150-180) [134]. They also share a similar DNA replication speed rate that is 2.4-2.6kb/min for Leishmania and 1.9kb/min for T. brucei, which is in range to the highest replication fork ever reported: 1.9kb/min in transformed JEDD lymphoblastoid cells [134, 135]. Therefore, as there are no significant differences in the chromosomal replication speed and number of origins of replications in Leishmania and T. brucei, mosaic aneuploidy in Leishmania is probably caused by a leaking in the regulation of DNA replication, rather than by unique replication parameters [134].
As DNA replication and transcription share profound functional overlaps in trypanosomatids [136], and as they both frequently start at strand switch regions [136-138], a clash between the transcription and replication machinery could also be relevant to generate the aneuploidies in Leishmania and T. cruzi. In fact, although Leishmania appears to have more than one DNA replication starting site [134], there could be a preferential origin per chromosome, as suggested by genome-wide mapping analysis [138]. If this is the case, a clash between the transcription and replication machinery at the preferential starting site could result in a failure of duplication of a given chromosome. For this reason, an extra copy of a chromosome could be important to assure that at least one copy would be duplicated, avoiding detrimental haploidies.
Genetic exchange events have been documented during the evolution of the parasites Leishmania and T. cruzi [139-146]. As these parasites present an aneuploid number of chromosomes, one of the mechanisms that could be enrolled in their genetic exchange events is a parasexual cycle, similar to what is observed in Candida albicans [53]. According to this model, the fusion of parental aneuploid cells is followed by karyogamy, resulting in a polyploid progeny that undergo reductional mitotic divisions and genome erosion, generating different degrees of chromosomal duplications/losses [125, 147]. This assumption is further supported by the subtetraploidy found in T. cruzi experimental hybrids [148, 149], as well as by their ~70% higher DNA content when compared to the parental strains [150].
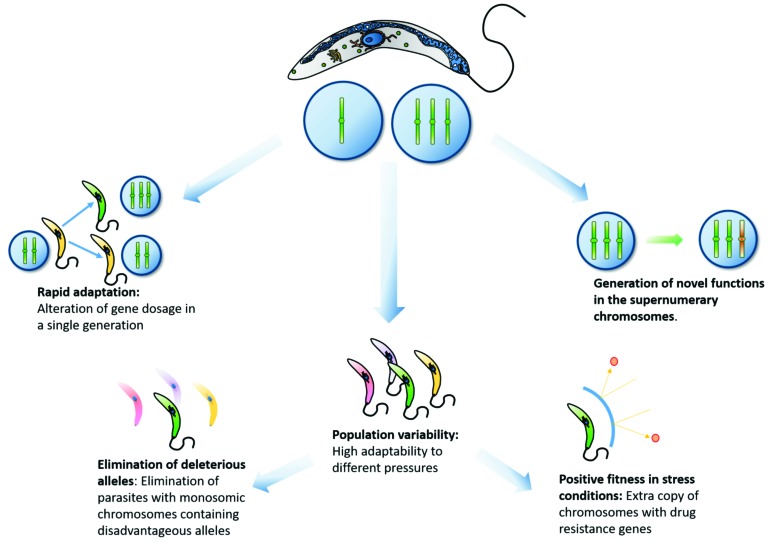
Although usually detrimental in the majority of superior eukaryotes [121-123], CCNV observed in Leishmania species and T. cruzi DTUs could provide several evolutionary advantages to these parasites (Fig. 2). A correlation between antibiotic resistance and CCNV based on transcriptional profiling by microarrays, southern blotting and comparative genomic hybridization was suggested in L. major and L. infantum [49, 50, 58]. Interestingly, these chromosomes reverted back to disomy in the absence of drug pressure, corroborating the extensive genomic plasticity of these parasites [49, 50]. On the other hand, RDC analysis found no clear link between aneuploidy and drug resistance in clinical isolates from L. donovani [26]. Mis-segregation and stochastic replication of chromosomes may also alter gene dosages in a single generation, allowing the parasite to quickly adapt to new environments as well as to the transition between the mammalian and invertebrate hosts [59]. Similarly, a mosaic aneuploidy state could generate a pool of phenotypes in the same population providing an increased adaptability, as distinct combinations of CCNV could be advantageous in different environments. If polysomy is stable for long evolutionary periods, it could also provide a selective advantage to the generation of novel functions in the duplicated chromosomes, as the ancestral gene function would be maintained in the original copy of the chromosome. Homologous recombination could then shuffle genes in aneuploid chromosomes, generating a variety of different phenotypes. As seen in yeast, deleterious mutations found in monosomic chromosomes could be rapidly eliminated [151, 152], while beneficial mutations would be retained or even expanded to triploidy and tetraploidy states [124, 125]. Thus, the presence of mosaic aneuploid provides the parasite the means to rapidly adapt to different environments and stress conditions, as well as combine the advantages of monosomies and polysomies in the same population.
Fig. (2).
Major implications of the mosaic aneuploidy phenomenon. This figure summarizes the expected evolutionary advantages of CCNV and aneuploidies in trypanosomatids.
The presence of aneuploidies in Leishmania and T. cruzi, but not in T. brucei, suggests that the latter has lost the ability to maintain an aberrant copy number of chromosomes. The evaluation of CCNV in other pathogenic and non-pathogenic protozoan species, such as Crithidia, Leptomonas, Endotrypanum, T. rangeli and T. cruzi marinkellei could provide a better view of the distribution and frequency of aneuploidies, as well as its correlation with chromosomal size, further elucidating the mechanisms behind its evolution in trypanosomatids. The simultaneous evaluation of the ploidy of all chromosomes in several cells in a cloned population by Single-Cell Genome Sequencing (SCGS) could provide an accurate estimation of the ratio of CCNV events. Besides, the evaluation of CCNV by SCGS in populations exposed to different stressors could identify aneuploidy trigger agents. Finally, the combination of CCNV and RNA-seq analysis, with DNA and RNA extracted from the same population, could provide insights in the implication of aneuploidies in gene expression.
ConcluSION
Over the last decade, advances in the determination of the biological relevance of gene and chromosomal copy number variations in trypanosomatids have been achieved, contributing to a better understanding of the driving forces behind the evolution of parasitism in this group of organisms. These parasites have developed a massive expansion of species-specific multigene families related to their adaptation to invade and survive in an intracellular host environment or to endure the hosts’ humoral response in the bloodstream. The absence of orthologs for several of these large gene families in the highly syntenic genomes of the parasites T. brucei, T. cruzi and the Leishmania genus denotes the involvement of CNV in species-specific adaptations that happened during trypanosomatid evolution. Besides, the substantial reduction of these families in the non-pathogenic T. rangeli further reinforces the importance of the expansion of multigene families for the establishment of a productive infection in the mammalian host. CCNV is well tolerated and widespread in Leishmania and T. cruzi, allowing variations in gene dosage in a single replication cycle and providing rapid adaptation to different environmental stressors. Different patterns of aneuploidies are found among strains from the same species/DTUs and even within a same population, reinforcing that CCNV generation events occur frequently in these parasites. Interestingly, there is no evidence of aneuploidies in T. brucei, which could be a consequence of its large chromosomes sizes when compared with the fragmented chromosome architecture of T. cruzi and Leishmania. However, if T. brucei lost the ability to sustain aneuploidies, or if Leishmania and T. cruzi independently developed this feature is still unknown. The assessment of aneuploidies in other parasites such as T. rangeli, T. cruzi marinkellei, Crithidia, Leptomonas and Endotrypanum could shed light into the occurrence of CCNV during trypanosomatids’ evolution and its implications for the biology of these parasites.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
This work was funded by Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Instituto Nacional de Ciência e Tecnologia de Vacinas (INCTV)-Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Pró-Reitoria de Pesquisa (PRPq)-Universidade Federal de Minas Gerias (UFMG). CITBM is co-funded by Fondo Nacional de Desarrollo Científico Tecnológico y de Innovación Tecnológica, Perú, under funding agreement No. 195-2016-FONDECYT. DCB is CNPq research fellow. HOV and JLRC received scholarships from CAPES.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Camargo E.P., Kastelein P., Roitman I. Trypanosomatid parasites of plants (Phytomonas). Parasitol. Today. 1990;6(1):22–25. doi: 10.1016/0169-4758(90)90388-k. [DOI] [PubMed] [Google Scholar]
- 2.Podlipaev S.A. Insect trypanosomatids: The need to know more. Mem. Inst. Oswaldo Cruz. 2000;95(4):517–522. doi: 10.1590/s0074-02762000000400013. [DOI] [PubMed] [Google Scholar]
- 3.Tieszen K.L., Molyneux D.H. Transmission and ecology of trypanosomatid flagellates of water striders (Hemiptera: Gerridae). J. Protozool. 1989;36(5):519–523. doi: 10.1111/j.1550-7408.1989.tb01088.x. [DOI] [PubMed] [Google Scholar]
- 4.Votýpka J., Maslov D.A., Yurchenko V., Jirků M., Kment P., Lun Z.R., Lukeš J. Probing into the diversity of trypanosomatid flagellates parasitizing insect hosts in South-West China reveals both endemism and global dispersal. Mol. Phylogenet. Evol. 2010;54(1):243–253. doi: 10.1016/j.ympev.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Votýpka J., Oborník M., Volf P., Svobodová M., Lukes J. Trypanosoma avium of raptors (Falconiformes): Phylogeny and identification of vectors. Parasitology. 2002;125(3):253–263. doi: 10.1017/s0031182002002093. [DOI] [PubMed] [Google Scholar]
- 6.Lopes A.H., Souto-Padrón T., Dias F.A., Gomes M.T., Rodrigues G.C., Zimmermann L.T., Alves Silva T.L., Vermelho A.B. Trypanosomatids: Odd organisms, devastating diseases. Open Parasitol. J. 2010;4(1):30–59. [Google Scholar]
- 7.WHO Research priorities for Chagas disease, human African trypanosomiasis and leishmaniasis. World Health Organ. Tech. Rep. Ser. 2012;975(v-xii):1–100. http://apps.who.int/ iris/bitstream/10665/77472/1/WHO_TRS_975_eng.pdf [PubMed] [Google Scholar]
- 8.Haag J. O’hUigin, C.; Overath, P. The molecular phylogeny of trypanosomes: evidence for an early divergence of the Salivaria. Mol. Biochem. Parasitol. 1998;91(1):37–49. doi: 10.1016/s0166-6851(97)00185-0. [DOI] [PubMed] [Google Scholar]
- 9.Adl S.M., Simpson A.G., Lane C.E., Lukes J., Bass D., Bowser S.S., Brown M.W., Burki F., Dunthorn M., Hampl V., Heiss A., Hoppenrath M., Lara E., Le Gall L., Lynn D.H., McManus H., Mitchell E.A., Mozley-Stanridge S.E., Parfrey L.W., Pawlowski J., Rueckert S., Shadwick L., Schoch C.L., Smirnov A., Spiegel F.W. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Calvillo S., Vizuet-De-Rueda J.C., Florencio-Martínez L.E., Manning-Cela R.G., Figueroa-Angulo E.E. Gene expression in trypanosomatid parasites. J. Biomed. Biotechnol. 2010;59(5):1–15. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton C.E. Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 2016;32:46–51. doi: 10.1016/j.mib.2016.04.018. https://linkinghub.elsevier. com/retrieve/pii/S1369-5274(16)30053-4 [DOI] [PubMed] [Google Scholar]
- 12.El-Sayed N.M. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. science.sciencemag.org/content/309/5733/404.short [ÃÜ]. [DOI] [PubMed] [Google Scholar]
- 13.Günzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2(3):542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M.G., Van der Ploeg L.H. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 1997;51(3):463–489. doi: 10.1146/annurev.micro.51.1.463. views.org/doi/abs/10.1146/annurev.micro.51.1.463 [DOI] [PubMed] [Google Scholar]
- 15.Zomerdijk J.C., Kieft R., Borst P. Efficient production of functional mRNA mediated by RNA polymerase I in Trypanosoma brucei. Nature. 1991;353(6346):772–775. doi: 10.1038/353772a0. http://europepmc.org/abstract/med/1658658 [DOI] [PubMed] [Google Scholar]
- 16.LeBowitz J.H., Smith H.Q., Rusche L., Beverley S.M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7(6):996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 17.Liang X. Haritan, A.; Uliel, S.; Michaeli, S. trans and cis splicing in trypanosomatids: Mechanism, factors, and regulation. Eukaryot. Cell. 2003;2(5):830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teixeira S.M., de Paiva R.M., Kangussu-Marcolino M.M., DaRocha W.D. Trypanosomatid comparative genomics: Contributions to the study of parasite biology and different parasitic diseases. Genet. Mol. Biol. 2012;35(1):1–17. doi: 10.1590/s1415-47572012005000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clayton C. The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog. 2013;9(11):9–12. doi: 10.1371/journal.ppat.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton C., Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156(2):93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Feagin J.E. RNA editing in kinetoplastid mitochondria. J. Biol. Chem. 1990;265(32):19373–19376. [PubMed] [Google Scholar]
- 22.Stuart K. RNA editing in kinetoplastid protozoa. Curr. Opin. Genet. Dev. 1991;1(3):412–416. doi: 10.1016/s0959-437x(05)80308-9. [DOI] [PubMed] [Google Scholar]
- 23.Landweber L.F. The evolution of RNA editing in kinetoplastid protozoa. Biosystems. 1992;28(1-3):41–45. doi: 10.1016/0303-2647(92)90006-k. [DOI] [PubMed] [Google Scholar]
- 24.Read L.K., Lukes J., Hashimi H. Trypanosome RNA editing: The complexity of getting U in and taking U out. Wiley Interdiscip. Rev. RNA. 2016;7(1):33–51. doi: 10.1002/wrna.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aphasizheva I., Aphasizhev R. U-Insertion/Deletion mRNA-editing holoenzyme: Definition in sight. Trends Parasitol. 2016;32(2):144–156. doi: 10.1016/j.pt.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downing T., Imamura H., Decuypere S., Clark T.G., Coombs G.H., Cotton J.A., Hilley J.D., de Doncker S., Maes I., Mottram J.C., Quail M.A., Rijal S., Sanders M., Schönian G., Stark O., Sundar S., Vanaerschot M., Hertz-Fowler C., Dujardin J.C., Berriman M. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21(12):2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., Otto T.D., Sanders M., Seeger K., Dujardin J.C., Berriman M., Smith D.F., Hertz-Fowler C., Mottram J.C. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21(12):2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minning T.A., Weatherly D.B., Flibotte S., Tarleton R.L. Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics. 2011;12(1):139. doi: 10.1186/1471-2164-12-139. https://bmcgenomics.biomedcentral.com/articles/ 10.1186/1471-2164-12-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reis-Cunha J.L., Rodrigues-Luiz G.F., Valdivia H.O., Baptista R.P., Mendes T.A., de Morais G.L., Guedes R., Macedo A.M., Bern C., Gilman R.H., Lopez C.T., Andersson B., Vasconcelos A.T., Bartholomeu D.C. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics. 2015;16(1):499. doi: 10.1186/s12864-015-1680-4. https://bmcgenomics.biomedcentral.com/articles/ 10.1186/s12864-015-1680-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lainson R., Shaw J.J. 1987. Evolution, classification and geographical distribution. [Google Scholar]
- 31.Motazedian H., Noyes H., Maingon R. Leishmania and Sauroleishmania: The use of random amplified polymorphic DNA for the identification of parasites from vertebrates and invertebrates. Exp. Parasitol. 1996;83(1):150–154. doi: 10.1006/expr.1996.0059. [DOI] [PubMed] [Google Scholar]
- 32.Croan D.G., Morrison D.A., Ellis J.T. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 1997;89(2):149–159. doi: 10.1016/s0166-6851(97)00111-4. [DOI] [PubMed] [Google Scholar]
- 33.Taylor V.M., Muñoz D.L., Cedeño D.L., Vélez I.D., Jones M.A., Robledo S.M. Leishmania tarentolae: Utility as an in vitro model for screening of antileishmanial agents. Exp. Parasitol. 2010;126(4):471–475. doi: 10.1016/j.exppara.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Deborggraeve S., Koffi M., Jamonneau V., Bonsu F.A., Queyson R., Simarro P.P., Herdewijn P., Büscher P. Molecular analysis of archived blood slides reveals an atypical human Trypanosoma infection. Diagn. Microbiol. Infect. Dis. 2008;61(4):428–433. doi: 10.1016/j.diagmicrobio.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Westenberger S.J., Barnabé C., Campbell D.A., Sturm N.R. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171(2):527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Freitas J.M., Augusto-Pinto L., Pimenta J.R., Bastos-Rodrigues L., Gonçalves V.F., Teixeira S.M., Chiari E., Junqueira A.C., Fernandes O., Macedo A.M., Machado C.R., Pena S.D. Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2(3):226–235. doi: 10.1371/journal.ppat.0020024. http://journals.plos.org/plospathogens/article?id= 10.1371/journal.ppat.0020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zingales B., Andrade S.G., Briones M.R., Campbell D.A., Chiari E., Fernandes O., Guhl F., Lages-Silva E., Macedo A.M., Machado C.R., Miles M.A., Romanha A.J., Sturm N.R., Tibayrenc M., Schijman A.G. Second Satellite Meeting. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz. 2009;104(7):1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 38.Burgos J.M., Risso M.G., Brenière S.F., Barnabé C., Campetella O., Leguizamón M.S. Differential Distribution of genes encoding the virulence factor trans-sialidase along Trypanosoma cruzi discrete typing units. PLoS One. 2013;8(3):9–11. doi: 10.1371/journal.pone.0058967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berriman M., Ghedin E., Hertz-fowler C. The genome of the African trypanosome, Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. http://journals.plos.org/ plosntds/article?id=10.1371/journal.pntd.0000658 [DOI] [PubMed] [Google Scholar]
- 40.Ivens A.C. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. http://science.sciencemag.org/content/309/5733/436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Sayed N.M. The genome sequence of Trypanosoma cruzi, etiologic agent of chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. http://citeseerx.ist.psu.edu/viewdoc/download? doi=10.1.1.318.9271&rep=rep1&type=pdf [DOI] [PubMed] [Google Scholar]
- 42.Franzén O., Ochaya S., Sherwood E., Lewis M.D., Llewellyn M.S., Miles M.A., Andersson B. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl. Trop. Dis. 2011;5(3):1–9. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franzén O., Talavera-López C., Ochaya S., Butler C.E., Messenger L.A., Lewis M.D., Llewellyn M.S., Marinkelle C.J., Tyler K.M., Miles M.A., Andersson B. Andersson, B. Comparative genomic analysis of human infective Trypanosoma cruzi lineages with the bat-restricted subspecies T. cruzi marinkellei. BMC Genomics. 2012;13:531. doi: 10.1186/1471-2164-13-531. https://bmcgenomics.biomedcentral.com/articles/10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sistrom M., Evans B., Bjornson R., Gibson W., Balmer O., Maser P., Aksoy S., Caccone A. Comparative genomics reveals multiple genetic backgrounds of human pathogenicity in the Trypanosoma brucei complex. Genome Biol. Evol. 2014;6(10):2811–2819. doi: 10.1093/gbe/evu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoco P.H., Wagner G., Talavera-Lopez C., Gerber A., Zaha A., Thompson C.E., Bartholomeu D.C., Lückemeyer D.D., Bahia D., Loreto E., Prestes E.B., Lima F.M., Rodrigues-Luiz G., Vallejo G.A., Filho J.F., Schenkman S., Monteiro K.M., Tyler K.M., de Almeida L.G., Ortiz M.F., Chiurillo M.A., de Moraes M.H., Cunha Ode L., Mendonça-Neto R., Silva R., Teixeira S.M., Murta S.M., Sincero T.C., Mendes T.A., Urmenyi T.P., Silva V.G., DaRocha W.D., Andersson B., Romanha A.J., Steindel M., de Vasconcelos A.T., Grisard E.C. Genome of the avirulent human-infective trypanosome-Trypanosoma rangeli. PLoS Negl. Trop. Dis. 2014;8(9):e3176. doi: 10.1371/journal.pntd.0003176. http://journals.plos.org/plosntds/article?id= 10.1371/journal.pntd.0003176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdivia H.O., Almeida L.V., Roatt B.M., Reis-Cunha J.L., Pereira A.A., Gontijo C., Fujiwara R.T., Reis A.B., Sanders M.J., Cotton J.A., Bartholomeu D.C. Comparative genomics of canine-isolated Leishmania (Leishmania) amazonensis from an endemic focus of visceral leishmaniasis in Governador Valadares, southeastern Brazil. Sci. Rep. 2017;7:40804. doi: 10.1038/srep40804. https://www.nature.com/articles/srep40804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weir W., Capewell P., Foth B., Clucas C., Pountain A., Steketee P., Veitch N., Koffi M., de Meeûs T., Kaboré J., Camara M., Cooper A., Tait A., Jamonneau V., Bucheton B., Berriman M., MacLeod A. Population genomics reveals the origin and asexual evolution of human infective trypanosomes. eLife. 2016;5:e11473. doi: 10.7554/eLife.11473. https://elifesciences.org/articles/11473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson A.P., Sanders M., Berry A., McQuillan J., Aslett M.A., Quail M.A., Chukualim B., Capewell P., Macleod A., Melville S.E., Gibson W., Barry J.D., Berriman M., Hertz-Fowler C. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African Trypanosomiasis. PLoS Negl. Trop. Dis. 2010;4(4):e658. doi: 10.1371/journal.pntd.0000658. http://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leprohon P., Légaré D., Raymond F., Madore É., Hardiman G., Corbeil J., Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ubeda J-M., Légaré D., Raymond F., Ouameur A.A., Boisvert S., Rigault P., Corbeil J., Tremblay M.J., Olivier M., Papadopoulou B., Ouellette M. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9(7):R115. doi: 10.1186/gb-2008-9-7-r115. https://genomebiology.biomedcentral.com/ articles/10.1186/gb-2008-9-7-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doubre H., Césari D., Mairovitz A., Bénac C., Chantot-Bastaraud S., Dagnon K., Antoine M., Danel C., Bernaudin J.F., Fleury-Feith J. Multidrug resistance-associated protein (MRP1) is overexpressed in DNA aneuploid carcinomatous cells in non-small cell lung cancer (NSCLC). Int. J. Cancer. 2005;113(4):568–574. doi: 10.1002/ijc.20617. [DOI] [PubMed] [Google Scholar]
- 52.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. doi: 10.1038/nature09414. https:// www.nature.com/articles/nature09414?message-global=remove [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selmecki A., Forche A., Berman J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell. 2010;9(7):991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abbey D., Hickman M., Gresham D., Berman J. High-resolution SNP/CGH microarrays reveal the accumulation of loss of heterozygosity in commonly used Candida albicans strains. G3 (Bethesda) 2011;1(7):523–530. doi: 10.1534/g3.111.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheltzer J.M., Blank H.H., Pfau S.J., Tange Y., George B.M., Humpton T.J., Brito I.L., Hiraoka Y., Niwa O., Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333(6045):1026–1030. doi: 10.1126/science.1206412. mag.org/content/333/6045/1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfau S.J., Amon A. Chromosomal instability and aneuploidy in cancer: From yeast to man. EMBO Rep. 2012;13(6):515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henrichsen C.N., Chaignat E., Reymond A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 2009;18(R1):R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 58.Laffitte M-C., Leprohon P., Papadopoulou B., Ouellette M., Laffitte M-C., Leprohon P., Papadopoulou B., Ouellette M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000 Res. 2016;5:2350. doi: 10.12688/f1000research.9218.1. https://f1000research.com/articles/5-2350/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mannaert A., Downing T., Imamura H., Dujardin J.C. Adaptive mechanisms in pathogens: Universal aneuploidy in Leishmania. Trends Parasitol. 2012;28(9):370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Navin N., Lucito R., Healy J., Hicks J., Ye K., Reiner A., Gilliam T.C., Trask B., Patterson N., Zetterberg A., Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. http://www.gene-quantification.de/sebat-cnv-2004.pdf [DOI] [PubMed] [Google Scholar]
- 61.Jackson A.P. Tandem gene arrays in Trypanosoma brucei: Comparative phylogenomic analysis of duplicate sequence variation. BMC Evol. Biol. 2007;7:54. doi: 10.1186/1471-2148-7-54. https:// bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson A.P. Evolutionary consequences of a large duplication event in Trypanosoma brucei: Chromosomes 4 and 8 are partial duplicons. BMC Genomics. 2007;8(1):432. doi: 10.1186/1471-2164-8-432. https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-8-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iskow R.C., Gokcumen O., Lee C. Exploring the role of copy number variants in human adaptation. Trends Genet. 2012;28(6):245–257. doi: 10.1016/j.tig.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartholomeu D.C., Cerqueira G.C., Leão A.C., daRocha W.D., Pais F.S., Macedo C., Djikeng A., Teixeira S.M., El-Sayed N.M. Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Res. 2009;37(10):3407–3417. doi: 10.1093/nar/gkp172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Claycomb J.M., Orr-Weaver T.L. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 2005;21(3):149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 66.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., Grassi A.D., Lee C., Tyler-Smith C., Carter N., Scherer S.W., Tavaré S., Deloukas P., Hurles M.E., Dermitzakis E.T. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813 ):848–853. doi: 10.1126/science.1136678. mag.org/content/315/5813/848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H., Yu L., Lai F., Liu L., Wang J. Molecular evidence for asymmetric evolution of sister duplicated blocks after cereal polyploidy. Plant Mol. Biol. 2005;59(1):63–74. doi: 10.1007/s11103-005-4414-1. [DOI] [PubMed] [Google Scholar]
- 68.Jiang H., Liu D., Gu Z., Wang W. Rapid evolution in a pair of recent duplicate segments of rice. J. Exp. Zoolog. B Mol. Dev. Evol. 2007;308(1):50–57. doi: 10.1002/jez.b.21122. [DOI] [PubMed] [Google Scholar]
- 69.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. http://science.sciencemag.org/content/290/5494/1151.full [DOI] [PubMed] [Google Scholar]
- 71.Zhang P. A Segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell. 2000;12(12):2311–2322. doi: 10.1105/tpc.12.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tümpel S., Cambronero F., Wiedemann L.M., Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes). Proc. Natl. Acad. Sci. USA. 2006;103(14):5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roth C., Rastogi S., Arvestad L., Dittmar K., Light S., Ekman D., Liberles D.A. Evolution after gene duplication: Models, mechanisms, sequences, systems, and organisms. J. Exp. Zoolog. B Mol. Dev. Evol. 2007;308(1):58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- 74.Martínez-Calvillo S., Yan S., Nguyen D., Fox M., Stuart K., Myler P.J. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11(5):1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 75.Mottram J.C., Murphy W.J., Agabian N. A transcriptional analysis of the Trypanosoma brucei hsp83 gene cluster. Mol. Biochem. Parasitol. 1989;37(1):115–127. doi: 10.1016/0166-6851(89)90108-4. [DOI] [PubMed] [Google Scholar]
- 76.Parsons M., Nelson R.G., Watkins K.P., Agabian N. Trypanosome mRNAs share a common 5′ spliced leader sequence. Cell. 1984;38(1):309–316. doi: 10.1016/0092-8674(84)90552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haile S., Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007;10(6):569–577. doi: 10.1016/j.mib.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Müller M., Padmanabhan P.K., Rochette A., Mukherjee D., Smith M., Dumas C., Papadopoulou B. Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3′-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res. 2010;38(17):5867–5883. doi: 10.1093/nar/gkq349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McNicoll F., Müller M., Cloutier S., Boilard N., Rochette A., Dubé M., Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 2005;280(42):35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- 80.Bringaud F., Muller M., Cerqueira G.C., Smith M., Rochette A., El-Sayed N.M., Papadopoulou B., Ghedin E. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3(9):1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 82.Valdivia H.O., Scholte L.L., Oliveira G., Gabaldón T., Bartholomeu D.C. The Leishmania metaphylome: A comprehensive survey of Leishmania protein phylogenetic relationships. BMC Genomics. 2015;16(1):887. doi: 10.1186/s12864-015-2091-2. https://bmcgenomics. biomedcentral.com/articles/10.1186/s12864-015-2091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jackson A.P., Otto T.D., Aslett M., Armstrong S.D., Bringaud F., Schlacht A., Hartley C., Sanders M., Wastling J.M., Dacks J.B., Acosta-Serrano A., Field M.C., Ginger M.L., Berriman M. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr. Biol. 2016;26(2):161–172. doi: 10.1016/j.cub.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jackson A.P. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol. Biol. Evol. 2010;27(1):33–45. doi: 10.1093/molbev/msp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao C., Donelson J.E., Wilson M.E. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 2003;132(1):1–16. doi: 10.1016/s0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- 86.Hsiao C.H., Yao C., Storlie P., Donelson J.E., Wilson M.E. The major surface protease (MSP or GP63) in the intracellular amastigote stage of Leishmania chagasi. Mol. Biochem. Parasitol. 2008;157(2):148–159. doi: 10.1016/j.molbiopara.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jecna L., Dostalova A., Wilson R., Seblova V., Chang K-P., Bates P.A., Volf P. The role of surface glycoconjugates in Leishmania midgut attachment examined by competitive binding assays and experimental development in sand flies. Parasitol. 2013;140(8):1026–1032. doi: 10.1017/S0031182013000358. [DOI] [PubMed] [Google Scholar]
- 88.Brittingham A., Morrison C.J., McMaster W.R., McGwire B.S., Chang K.P., Mosser D.M. Role of the Leishmania surface protease gp63 in complement fixation, cell adhesion, and resistance to complement-mediated lysis. J. Immunol. 1995;155(6):3102–3111. [PubMed] [Google Scholar]
- 89.Corradin S., Ransijn A., Corradin G., Roggero M.A., Schmitz A.A., Schneider P., Mauël J., Vergères G. MARCKS-related protein (MRP) is a substrate for the Leishmania major surface protease leishmanolysin (gp63). J. Biol. Chem. 1999;274(36):25411–25418. doi: 10.1074/jbc.274.36.25411. [DOI] [PubMed] [Google Scholar]
- 90.Gomez M.A., Contreras I., Halle M., Tremblay M.L., McMaster R.W., Olivier M. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci. Signal. 2009;2(90):ra58. doi: 10.1126/scisignal.2000213. http://stke.sciencemag.org/content/2/90/ra58 [DOI] [PubMed] [Google Scholar]
- 91.Valdivia H.O., Reis-Cunha J.L., Rodrigues-Luiz G.F., Baptista R.P., Baldeviano G.C., Gerbasi R.V., Dobson D.E., Pratlong F., Bastien P., Lescano A.G., Beverley S.M., Bartholomeu D.C. D.C. Comparative genomic analysis of Leishmania (Viannia) peruviana and Leishmania (Viannia) braziliensis. BMC Genomics. 2015;16(1):715. doi: 10.1186/s12864-015-1928-z. https://bmcgenomics.biomedcentral.com/articles/10.1186/s128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Real F., Vidal R.O., Carazzolle M.F., Mondego J.M., Costa G.G., Herai R.H., Wurtele M., De Carvalho L.M., Ferreira R.C., Mortara R.A., Barbiéri C.L., Mieczkowski P., da Silveira J.F., Briones M.R., Pereira G.A., Bahia D. The genome sequence of Leishmania (Leishmania) amazonensis: Functional annotation and extended analysis of gene models. DNA Res. 2013;20(6):567–581. doi: 10.1093/dnares/dst031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Judice W.A., Manfredi M.A., Souza G.P., Sansevero T.M., Almeida P.C., Shida C.S., Gesteira T.F., Juliano L., Westrop G.D., Sanderson S.J., Coombs G.H., Tersariol I.L. Heparin modulates the endopeptidase activity of Leishmania mexicana cysteine protease cathepsin L-like rCPB2.8. PLoS One. 2013;8(11):e80153. doi: 10.1371/journal.pone.0080153. http://journals.plos.org/plosone/article?id= 10.1371/journal.pone.0080153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burleigh B.A., Andrews N.W. The mechanisms of Trypanosoma Cruzi invasion of mammalian cells. Annu. Rev. Microbiol. 1995;49(1):175–200. doi: 10.1146/annurev.mi.49.100195.001135. [DOI] [PubMed] [Google Scholar]
- 95.Epting C.L., Coates B.M., Engman D.M. Molecular mechanisms of host cell invasion by Trypanosoma cruzi. Exp. Parasitol. 2010;126(3):283–291. doi: 10.1016/j.exppara.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andrade L.O., Andrews N.W. The Trypanosoma cruzi-host-cell interplay: Location, invasion, retention. Nat. Rev. Microbiol. 2005;3(10):819–823. doi: 10.1038/nrmicro1249. [DOI] [PubMed] [Google Scholar]
- 97.Fernandes M.C., Andrews N.W. Host cell invasion by Trypanosoma cruzi: A unique strategy that promotes persistence. FEMS Microbiol. Rev. 2012;36(3):734–747. doi: 10.1111/j.1574-6976.2012.00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flores-Lopez C.A., Machado C.A. Differences in inferred genome-wide signals of positive selection during the evolution of Trypanosoma cruzi and Leishmania spp. lineages: A result of disparities in host and tissue infection ranges? Infect. Genet. Evol. 2015;33:37–46. doi: 10.1016/j.meegid.2015.04.008. https://pdfs.semanticscholar.org/ 75c8/f56e5384280d11259190bb5f5deea141c2c1.pdf [DOI] [PubMed] [Google Scholar]
- 99.De Pablos L.M., Osuna A. Multigene families in Trypanosoma cruzi and their role in infectivity. Infect. Immun. 2012;80(7):2258–2264. doi: 10.1128/IAI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schenkman S., Eichinger D., Pereira M.E., Nussenzweig V. Structural and functional properties of Trans-Sialidase. Annu. Rev. Microbiol. 1994;48:499–523. doi: 10.1146/annurev.mi.48.100194.002435. http://www. annualreviews.org/doi/abs/10.1146/annurev.mi.48.100194.002435 [DOI] [PubMed] [Google Scholar]
- 101.Buscaglia C.A., Campo V.A., Frasch A.C., Di Noia J.M. Trypanosoma cruzi surface mucins: Host-dependent coat diversity. Nat. Rev. Microbiol. 2006;4(3):229–236. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 102.Tomlinson S., Pontes de Carvalho L.C., Vandekerckhove F., Nussenzweig V. Role of sialic acid in the resistance of Trypanosoma cruzi trypomastigotes to complement. J. Immunol. 1994;153(7):3141–3147. [PubMed] [Google Scholar]
- 103.Freire-de-Lima L., Oliveira I.A., Neves J.L., Penha L.L., Alisson-Silva F., Dias W.B., Todeschini A.R. Sialic acid: A sweet swing between mammalian host and Trypanosoma cruzi. Front. Immunol. 2012;3:1–12. doi: 10.3389/fimmu.2012.00356. https:// www.frontiersin.org/articles/10.3389/fimmu.2012.00356/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Butler C.E., de Carvalho T.M., Grisard E.C., Field R.A., Tyler K.M. Trans-sialidase stimulates eat me response from epithelial cells. Traffic. 2013;14(7):853–869. doi: 10.1111/tra.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Melo-Jorge M. PereiraPerrin, M. The Chagas’ disease parasite Trypanosoma cruzi exploits nerve growth factor receptor TrkA to infect mammalian hosts. Cell Host Microbe. 2007;1(4):251–261. doi: 10.1016/j.chom.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 106.Chuenkova M.V. M. Trypanosoma cruzi targets Akt in host cells as an intracellular antiapoptotic strategy. Sci. Signal. 2009;2(97):ra74. doi: 10.1126/scisignal.2000374. mag.org/content/2/97/ra74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chuenkova M.V. PereiraPerrin, M. Trypanosoma cruzi-derived neurotrophic factor: Role in neural repair and neuroprotection. J. Neuroparasitology. 2011;1:55–60. doi: 10.4303/jnp/N100507. https://pdfs. semanticscholar.org/4687/1a10097851cd633ed5157492bcb027f8e6d0.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Serrano A.A., Schenkman S., Yoshida N., Mehlert A., Richardson J.M., Ferguson M.A. The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J. Biol. Chem. 1995;270(45):27244–27253. doi: 10.1074/jbc.270.45.27244. [DOI] [PubMed] [Google Scholar]
- 109.De Pablos L.M., González G.G., Parada J.S., Hidalgo V.S., Lozano I.M., Samblás M.M., Bustos T.C., Osuna A. Differential expression and characterization of a member of the mucin-associated surface protein family secreted by Trypanosoma cruzi. Infect. Immun. 2011;79(10):3993–4001. doi: 10.1128/IAI.05329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.dos Santos S.L., Freitas L.M., Lobo F.P., Rodrigues-Luiz G.F., Mendes T.A. de O., Oliveira A.C., Andrade L.O., Chiari É., Gazzinelli R.T., Teixeira S.M., Fujiwara R.T., Bartholomeu D.C. The MASP family of Trypanosoma cruzi: Changes in gene expression and antigenic profile during the acute phase of experimental infection. PLoS Negl. Trop. Dis. 2012;6(8):e1779. doi: 10.1371/journal.pntd.0001779. http://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seco-Hidalgo V., De Pablos L.M., Osuna A. Transcriptional and phenotypical heterogeneity of Trypanosoma cruzi cell populations. Open Biol. 2015;5(12):150190. doi: 10.1098/rsob.150190. https:// benthamopen.com/tobioj/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weatherly D.B., Boehlke C., Tarleton R.L. Chromosome level assembly of the hybrid Trypanosoma cruzi genome. BMC Genomics. 2009;10:255. doi: 10.1186/1471-2164-10-255. https://bmcgenomics.biomedcentral. com/articles/10.1186/1471-2164-10-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van der Ploeg L.H., Valerio D., de Lange T., Bernards A., Borst P., Grosveld F.G. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 1982;10(19):5905–5923. doi: 10.1093/nar/10.19.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Turner C.M. Antigenic variation in Trypanosoma brucei infections: An holistic view. J. Cell Sci. 1999;112(Pt 19):3187–3192. doi: 10.1242/jcs.112.19.3187. [DOI] [PubMed] [Google Scholar]
- 115.Barry J.D., McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. https://www.sciencedirect.com/ science/article/pii/S0065308X01490373 [DOI] [PubMed] [Google Scholar]
- 116.McCulloch R., Horn D. What has DNA sequencing revealed about the VSG expression sites of African trypanosomes? Trends Parasitol. 2009;25(8):359–363. doi: 10.1016/j.pt.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 117.Roth C., Bringaud F., Layden R.E., Baltz T., Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc. Natl. Acad. Sci. USA. 1989;86(23):9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marcello L., Barry J.D. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17(9):1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pays E., Vanhamme L., Pérez-Morga D. Antigenic variation in Trypanosoma brucei: Facts, challenges and mysteries. Curr. Opin. Microbiol. 2004;7(4):369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 120.Torres E.M., Williams B.R., Amon A. Aneuploidy: Cells losing their balance. Genetics. 2008;179(2):737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hassold T., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 122.Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61(1):437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 123.Lv L., Zhang T., Yi Q., Huang Y., Wang Z., Hou H., Zhang H., Zheng W., Hao Q., Guo Z., Cooke H.J., Shi Q. Tetraploid cells from cytokinesis failure induce aneuploidy and spontaneous transformation of mouse ovarian surface epithelial cells. Cell Cycle. 2012;11(15):2864–2875. doi: 10.4161/cc.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sterkers Y., Lachaud L., Crobu L., Bastien P., Pagès M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell. Microbiol. 2011;13(2):274–283. doi: 10.1111/j.1462-5822.2010.01534.x. [DOI] [PubMed] [Google Scholar]
- 125.Sterkers Y., Crobu L., Lachaud L., Pagès M., Bastien P. Parasexuality and mosaic aneuploidy in Leishmania: Alternative genetics. Trends Parasitol. 2014;30(9):429–435. doi: 10.1016/j.pt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 126.Lachaud L., Bourgeois N., Kuk N., Morelle C., Crobu L., Merlin G., Bastien P., Pagès M., Sterkers Y. Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect. 2014;16(1):61–66. doi: 10.1016/j.micinf.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 127.Dujardin J.C., Mannaert A., Durrant C., Cotton J.A. Mosaic aneuploidy in Leishmania: The perspective of whole genome sequencing. Trends Parasitol. 2014;30(12):554–555. doi: 10.1016/j.pt.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 128.Waller J.C., Alvarez S., Naponelli V., Lara-Nuñez A., Blaby I.K., Da Silva V., Ziemak M.J., Vickers T.J., Beverley S.M., Edison A.S., et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc. Natl. Acad. Sci. USA. 2010;107(23):10412–10417. doi: 10.1073/pnas.0911586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fiebig M., Kelly S., Gluenz E. Comparative life cycle transcriptomics revises Leishmania mexicana genome annotation and links a chromosome duplication with parasitism of vertebrates. PLoS Pathog. 2015;11(10):1–28. doi: 10.1371/journal.ppat.1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bessat M., Ersfeld K. Functional characterization of cohesin SMC3 and separase and their roles in the segregation of large and minichromosomes in Trypanosoma brucei. Mol. Microbiol. 2009;71(6):1371–1385. doi: 10.1111/j.1365-2958.2009.06611.x. [DOI] [PubMed] [Google Scholar]
- 131.Wincker P., Ravel C., Blaineau C., Pages M., Jauffret Y., Dedet J.P., Bastien P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24(9):1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Britto C., Ravel C., Bastien P., Blaineau C., Pagès M., Dedet J.P., Wincker P. Conserved linkage groups associated with large-scale chromosomal rearrangements between old world and new world Leishmania genomes. Gene. 1998;222(1):107–117. doi: 10.1016/s0378-1119(98)00472-7. [DOI] [PubMed] [Google Scholar]
- 133.Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M.A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S.L., Faulconbridge A., Jeffares D., Depledge D.P., Oyola S.O., Hilley J.D., Brito L.O., Tosi L.R., Barrell B., Cruz A.K., Mottram J.C., Smith D.F. Berriman, M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stanojcic S., Sollelis L., Kuk N., Crobu L., Balard Y., Schwob E., Bastien P., Pagès M., Sterkers Y. Single-molecule analysis of DNA replication reveals novel features in the divergent eukaryotes Leishmania and Trypanosoma brucei versus mammalian cells. Sci. Rep. 2016;6:23142. doi: 10.1038/srep23142. https://www.nature.com/articles/srep23142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Técher H., Koundrioukoff S., Azar D., Wilhelm T., Carignon S., Brison O., Debatisse M., Le Tallec B. Replication dynamics: Biases and robustness of DNA fiber analysis. J. Mol. Biol. 2013;425(23):4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- 136.Tiengwe C., Marcello L., Farr H., Dickens N., Kelly S., Swiderski M., Vaughan D., Gull K., Barry J.D., Bell S.D., McCulloch R. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Reports. 2012;2(1):185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lombrana R., Alvarez A., Fernandez-Justel J.M., Almeida R., Poza-Carrion C., Gomes F., Calzada A., Requena J.M., Gomez M. Transcriptionally driven DNA replication program of the human parasite Leishmania major. Cell Reports. 2016;16(6):1774–1786. doi: 10.1016/j.celrep.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 138.Marques C.A., Dickens N.J., Paape D., Campbell S.J., McCulloch R. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol. 2015;16:230. doi: 10.1186/s13059-015-0788-9. https://genomebiology.biomedcentral.com/ articles/10.1186/s13059-015-0788-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Machado C.A., Ayala F.J. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA. 2001;98(13):7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ravel C., Cortes S., Pratlong F., Morio F., Dedet J.P., Campino L. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int. J. Parasitol. 2006;36(13):1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 141.Akopyants N.S., Kimblin N., Secundino N., Patrick R., Lawyer P., Dobson D.E., Beverley S.M., David L. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. 2009;324(5924):265, 268. doi: 10.1126/science.1169464. http://beverleylab.wustl.edu/PDFs/186.%20%20Akopyants%20et%20al%20Leish%20Cross%20Science%202009%20MS+SI.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lewis M.D., Llewellyn M.S., Yeo M., Acosta N., Gaunt M.W., Miles M.A. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl. Trop. Dis. 2011;5(10):e1363. doi: 10.1371/journal.pntd.0001363. http://journals.plos.org/plosntds/article? id=10.1371/journal.pntd.0001363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Messenger L.A., Llewellyn M.S., Bhattacharyya T., Franzén O., Lewis M.D., Ramírez J.D., Carrasco H.J., Andersson B., Miles M.A. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl. Trop. Dis. 2012;6(4):e1584. doi: 10.1371/journal.pntd.0001584. http://journals.plos.org/plosntds/article? id=10.1371/journal.pntd.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Inbar E., Akopyants N.S., Charmoy M., Romano A., Lawyer P., Elnaiem D.E., Kauffmann F., Barhoumi M., Grigg M., Owens K., Fay M., Dobson D.E., Shaik J., Beverley S.M., Sacks D. The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 2013;9(7):e1003672. doi: 10.1371/journal.pgen.1003672. http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baptista R.D., D’Ávila D.A., Segatto M., do Valle Í.F., Franco G.R., Valadares H.M., Gontijo E.D., Galvão L.M., Pena S.D., Chiari E., Machado C.R., Macedo A.M. Macedo, A.M. Evidence of substantial recombination among Trypanosoma cruzi II strains from Minas gerais. Infect. Genet. Evol. 2014;22:183–191. doi: 10.1016/j.meegid.2013.11.021. http://www.sciencedirect.com/science/article/pii/ [DOI] [PubMed] [Google Scholar]
- 146.Messenger L.A., Miles M.A. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015;151(1):150–155. doi: 10.1016/j.actatropica.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sturm N.R., Campbell D.A. Alternative lifestyles: The population structure of Trypanosoma cruzi. Acta Trop. 2010;115(1-2):35–43. doi: 10.1016/j.actatropica.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 148.Gaunt M.W., Yeo M., Frame I.A., Stothard J.R., Carrasco H.J., Taylor M.C., Mena S.S., Veazey P., Miles G.A., Acosta N., de Arias A.R., Miles M.A. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421(6926):936–939. doi: 10.1038/nature01438. https://www.nature.com/articles/nature01438 [DOI] [PubMed] [Google Scholar]
- 149.Ramírez J.D., Llewellyn M.S. Reproductive clonality in protozoan pathogens-truth or artefact? Mol. Ecol. 2014;23(17):4195–4202. doi: 10.1111/mec.12872. [DOI] [PubMed] [Google Scholar]
- 150.Lewis M.D., Llewellyn M.S., Gaunt M.W., Yeo M., Carrasco H.J., Miles M.A. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int. J. Parasitol. 2009;39(12):1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zeyl C., Vanderford T., Carter M. An evolutionary advantage of haploidy in large yeast populations. Science. 2003;299(5606):555–558. doi: 10.1126/science.1078417. http://science.sciencemag.org/content/ 299/5606/555/tab-article-info [DOI] [PubMed] [Google Scholar]