Abstract
Introduction:
Understanding how the nuclear genome of kinetoplastid parasites is replicated received experimental stimulus from sequencing of the Leishmania major, Trypanosoma brucei and Trypanosoma cruzi genomes around 10 years ago. Gene annotations suggested key players in DNA replication initiation could not be found in these organisms, despite considerable conservation amongst characterised eukaryotes. Initial studies that indicated trypanosomatids might possess an archaeal-like Origin Recognition Complex (ORC), composed of only a single factor termed ORC1/CDC6, have been supplanted by the more recent identification of an ORC in T. brucei. However, the constituent subunits of T. brucei ORC are highly diverged relative to other eukaryotic ORCs and the activity of the complex appears subject to novel, positive regulation. The availability of whole genome sequences has also allowed the deployment of genome-wide strategies to map DNA replication dynamics, to date in T. brucei and Leishmania. ORC1/CDC6 binding and function in T. brucei displays pronounced overlap with the unconventional organisation of gene expression in the genome. Moreover, mapping of sites of replication initiation suggests pronounced differences in replication dynamics in Leishmania relative to T. brucei.
Conclusion:
Here we discuss what implications these emerging data may have for parasite and eukaryotic biology of DNA replication.
Keywords: DNA replication, Origin recognition complex (ORC), Origins of replication, Trypanosoma brucei, Leishmania, Kinetoplastid parasites
1. INTRODUCTION
Every cell cycle, the genome must be completely and accurately duplicated before being transmitted to daughter cells. Incomplete or inaccurate replication risks genome instability, a hallmark of cancer [1], and it is vital that the entire genome is normally strictly replicated just once during the S phase of the cell cycle. Hence, DNA replication is a complex, tightly regulated process [2-5] that has been extensively studied in model eukaryotes, in bacteria and, most recently, in archaea [6].
DNA replication is initiated at origins of replication, which are found as a single sequence-defined site in all bacterial genomes. In archaea, origins are also defined by conserved sequences, but their organisation differs from bacteria in that some archaeal genomes possess a few origins. The large genomes of eukaryotes differ yet further, in that each linear chromosome contains many (sometimes hundreds to thousands) of potential origins of replication, which are not all activated in every cell cycle and are rarely defined by sequence elements but, instead, are associated with poorly defined features like chromatin structure and status [7]. Despite these differences, origins of replication across the three domains of life are recognised by initiator factors belonging to the AAA+ superfamily of NTPases [8]: the single DnaA [9] and Orc1/Cdc6 [10] factors in bacteria and archaea, respectively, and the six-subunit (Orc1-Orc6) origin recognition complex (ORC) in eukaryotes [11]. Binding of these factors demarcates the origins of replication and leads to the recruitment and activation of the replicative helicase. Subsequently, the remaining constituents of the replication machinery are in turn recruited, thus starting, and driving, DNA synthesis [3].
Much of the diversity of the eukaryotic domain resides in microbes, many of which display dramatic deviation in core biology from model, primarily opisthokont, organisms. Such diversity extends to nuclear biology, including variations in mitosis [12], gene expression [13], and genome organisation and stability [14]. Though most of our understanding of the machinery and execution of DNA replication in eukaryotes has focused on the opisthokonts, a number of recent studies have begun to explore nuclear replication in kinetoplastids, suggesting that here too there may be surprising variation [15, 16].
2. The kinetoplastid origin recognition complex
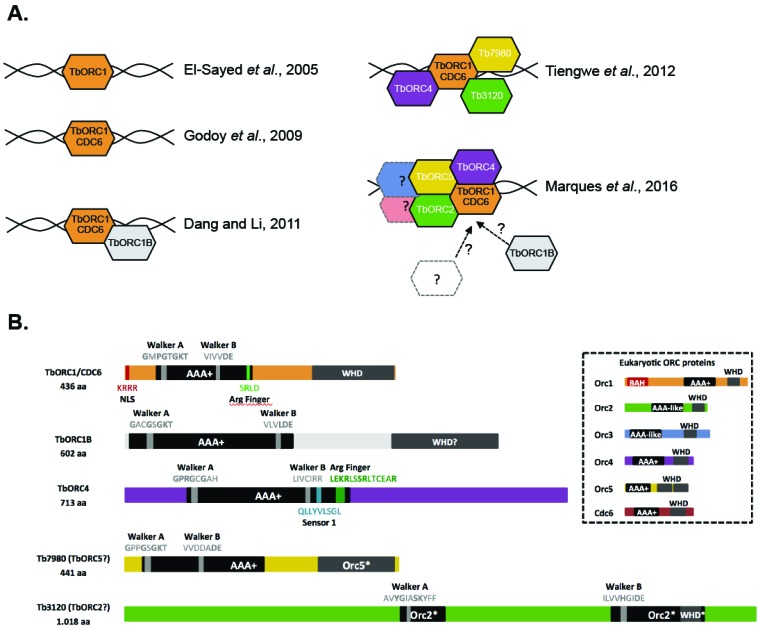
Sequencing of the TriTryp genomes (Trypanosoma brucei, Leishmania major and Trypanosoma cruzi) [17-19] in 2005 provided a spur for the field of kinetoplastid biology [20], including nuclear DNA replication. Prior to genome sequencing, attempts to examine replication dynamics could not consider the whole genome (see below) and little work had examined the replication machinery. Surprisingly, sequence similarity searches were unable to identify various core DNA replication proteins, in particular those involved in the initiation steps of the process: while orthologues of most factors involved in replication fork assembly and DNA synthesis were readily identified, only one ORC-related factor (a putative orthologue of Orc1) could be found [19] (Fig. 1A and 1B). Like in other eukaryotes, this putative Orc1 subunit also shared homology with Cdc6 (an essential factor that interacts with ORC to allow loading of the replicative helicase onto origins of replication) [11]. The lack of identifiable orthologues of the other ORC subunits, Cdc6, and a further helicase loader (Cdt1), suggested that initiation of DNA replication in these parasites might be mechanistically more similar to archaeal organisms than model eukaryotes [19]. In line with this hypothesis, and supported by experimental evidence, Orc1 in T. cruzi and T. brucei was re-named ORC1/CDC6 [21] (Fig. 1A), reflecting the combined ORC and Cdc6 functions that reside in the single protein that binds each origin in archaea, while other studies analysed Orc1 in various Leishmania species [22, 23]. A 2011 study [24], however, questioned whether the kinetoplastid initiator is just a single protein. While focusing on the functional analysis of components of the replicative helicase in T. brucei - the heterohexameric minichromosome maintenance complex (MCM2-7), GINS complex and CDC45 - the authors identified a second, highly divergent orthologue of Orc1 [24] (Fig. 1A and 1B). Labelled as TbORC1B, this putative initiator was shown to interact with TbORC1/CDC6 and TbMCM3, but its role in DNA replication was not explored. Nonetheless, further evidence that T. brucei might not have a single protein initiator factor, but an ORC-like complex, was provided by a study the following year, in which three more TbORC1/CDC6-interacting factors were identified [25] (Fig. 1A and 1B): a highly divergent Orc4-like subunit (TbORC4), and two apparently kinetoplastid-specific factors, Tb7980 and Tb3120, with very limited homology with Orc proteins.
Fig. (1).
The Origin Recognition Complex (ORC) of T. brucei. A) Evolution of our understanding of the origin-binding machinery of T. brucei in the last ten years, from a single factor like in archaea, to a highly divergent ORC-like complex resembling the system in model eukaryotes. B) Schematic representation of T. brucei ORC-like complex subunits and identified domains.
More recent experiments, thus far limited to T. brucei, provide strong evidence of an ORC-like complex. RNA interference (RNAi) shows that loss of TbORC1/CDC6, TbORC1B, TbORC4 or Tb3120 impedes DNA replication [21, 25-27] and leads to similar growth and cell cycle defects [27]. Though loss of Tb7980 also results in the same proliferation defects [25], clear evidence of a role in DNA replication remains to be assessed. TbORC1/CDC6 depletion additionally leads to the expression of silent Variant Surface Proteins (VSGs) in bloodstream form cells [26] and in procyclic cells [28], suggesting that, similar to other eukaryotic Orc1 subunits, and despite lacking a bromo adjacent homology (BAH) domain (Fig. 1B), TbORC1/CDC6 has a role in gene silencing. However, whether these activities are executed within the putative ORC is unknown. TbORC1/CDC6-binding sites have also been mapped genome-wide [28]. Within the core of the chromosomes TbORC1/CDC6 binds mainly at the boundaries of the multi-gene transcription units, where it overlaps with mapped origins of replication (see below for full explanation), supporting a role in replication initiation. In addition, ~60% of TbORC1/CDC6 binding sites locate to the chromosomes’ subtelomeric regions [28]. Whether this dense binding in the subtelomeres is functionally related to binding directly to the telomeres [26], and whether this is related to TbORC1/CDC6’s silencing function(s), is not clear. Finally, gel filtration analysis suggests TbORC1/CDC6 and TbORC4 are present, most likely together, in a high molecular complex (~1011 to 530 kDa) that also seems to include the helicase subunit TbMCM3 [27]. As TbMCM3 has been shown to interact with TbORC1/CDC6 and TbORC1B [24], and its orthologue in yeast mediates the recruitment of the MCM2-7 helicase to the DNA-bound ORC [29], the accumulation of data indicate the presence of an ORC in T. brucei, albeit one that defied simple sequence-based identification. As each putative ORC component is syntenically conserved in T. cruzi and Leishmania, there is a strong likelihood that the diverged composition of T. brucei ORC is a feature of all kinetoplastids. What is less clear is why kinetoplastid ORC might be diverged, and whether this extends to changes in function or regulation.
Most of our knowledge of the processes and molecular machineries involved in the eukaryotic cell cycle has been inferred from studies in a limited group of model organisms, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Xenopus laevis, mouse and human, and extrapolated across the eukaryotic tree. However, because these organisms belong to a single supergroup of the eukaryotic domain, termed the Opisthokonta, it is less clear how well details are conserved in the five other eukaryotic supergroups, and thus, the extent of diversity in replication processes and molecular machineries across the eukaryotic domain is poorly explored [30, 31]. From studies in model eukaryotes, Orc1-5 each appear to be structurally similar, with each subunit containing a central or N-terminal AAA+ ATPase domain and at least one C-terminal winged-helix (WH) DNA-binding domain [6, 11] (Fig. 1B, insert box). Within the AAA+ ATPase domain, conserved Walker A and B motifs (fundamental components of the ATP-binding site and essential for ATPase activity) and a signature arginine finger (involved in ATP hydrolysis of the adjacent AAA+ ATPase subunit in the protein complex) are needed to modulate ORC activity [32, 33]. In contrast, Orc6 appears to have evolved independently and does not possess any of these Orc-characteristic domains [11]. Recently, the ever-growing availability of sequenced genomes from a wide range of organisms, spanning all eukaryotic supergroups, has allowed sequence-based analysis of ORC-orthologues across the eukaryotic domain [30]. Reflecting the putative kinetoplastid ORC composition, not all six subunits of ORC were found across the analysed genomes, suggesting there is variability in the composition of ORC between eukaryotes. However, whether this reflects the existence of simpler ORCs, high divergence at the protein sequence level, less functional conservation of some ORC subunits, or the existence unrelated factors performing the role of Orc-subunits in certain organisms is unknown, and requires experimental investigation.
Recently, the structure of D. melanogaster ORC was solved [34]. Prior to this elegant study, only archaeal ORC1/CDC6 protein structures, and a fragment of the human Orc6 subunit [35], were solved and available on the RCSB Protein Data Bank. This meant that protein structure modelling software could only model eukaryotic ORC subunits structure based on the archaeal ORC1/CDC6 structures. While this had the potential to model Orc4 and Orc5 sequences, as these subunits possess a conserved AAA+ ATPase domain [6, 11], modelling of Orc2 and Orc3 was particularly problematic, as these subunits have highly divergent AAA+ folds containing non-canonical Walker A and B motifs that are, however, conserved amongst different organisms’ Orc2 and Orc3 subunits [36, 37]. Because Orc6 is the least conserved of the Orc subunits in both sequence and function [35, 38-40], it was unlikely that other organisms’ Orc6 subunits could be modelled based on the available human structure. The availability of structures of all D. melanogaster ORC subunits [34], and very recently, of the human counterparts [41], has therefore opened the door for the modelling of Orc2-Orc5 related sequences. From this approach T. brucei Tb7980 and Tb3120 emerge with potential structural similarity to D. melanogaster Orc5 and Orc2 subunits, respectively [27]. Nevertheless, whether these act as functional TbORC5 and TbORC2 orthologues is unknown, and requires further study. Very recently, we have further identified another TbORC1/CDC6-interacting factor, which is a hypothetical zinc-finger protein [42, 43] that appears to possess feeble homology with D. melanogaster Orc3 (unpublished). Again, however, functional analysis is needed to verify such potential orthology. Such functional studies may be of value, since little work has explored if individual ORC subunits provide conserved, discrete functions across eukaryotes. To our knowledge, there is currently no hint for the presence of an Orc6 subunit in kinetoplastids. Nonetheless, if the conformation of kinetoplastid ORC follows that of D. melanogaster, where the subunits are arranged in a Orc1-Orc4-Orc5-Orc3-Orc2 ring, with Orc6 interacting with Orc3 [34], then the greater conservation of Orc1 and Orc4 would suggest these components represent a functionally constrained ‘core’, with greater functional flexibility amongst the other subunits, an arrangement consistent with eukaryote-wide ORC sequence comparisons [30].
Structural analysis is needed to test the above prediction of kinetoplastid ORC architecture, but sequence and experimental analyses provide further clues about subunit activity. Reflecting its early identification [19], ORC1/CDC6 is the subunit showing the greatest conservation with other eukaryotic Orc proteins. Analyses of L. major, T. brucei and T. cruzi ORC1/CDC6 protein sequences [21, 22] were able to identify a putative N-terminal AAA+ ATPase domain, with clearly identifiable Walker A, Walker B and arginine finger motifs, as well as a putative WH domain at the C-terminus [21, 22] (Fig. 1B). Functionality of these domains has been confirmed in T. brucei and T. cruzi ORC1/CDC6 proteins [21], while a Nuclear Localisation Signal (NLS) at the N-terminus has been experimentally confirmed in the L. donovani counterpart [23]. Nonetheless, despite their sequence homology with Orc1 subunits, the kinetoplastid ORC1/ CDC6 proteins are atypical, as none possesses an identifiable BAH domain at their N-terminus [21, 22], a feature thought to be universal across Orc1 subunits and shown to be involved in origin recognition and transcriptional silencing [44-49]. Despite being larger than TbORC1/CDC6, TbORC1B (and its L. major and T. cruzi counterparts) also lacks a BAH domain [24]. An N-terminal AAA+ ATPase domain is seen in TbORC1B, with conserved but widely spaced Walker A and B motifs [24] (Fig. 1B), but no arginine finger signature is found [24]. Whether this explains experimental data suggesting TbORC1B is devoid of ATPase activity [24] is unclear. Finally, sequence alignments with various eukaryotic Orc1 and Cdc6 proteins allows a weak prediction of a WH domain at the C-terminus of TbORC1B [43], but whether it binds to DNA has not been examined. Taken together, it remains unclear if TbORC1/CDC6 and TbORC1B are orthologues of Orc1 and Cdc6 and, since functional data suggest TbORC1B is highly unorthodox (see below), it remains premature to assign such orthology. TbORC4 also seems to possess an AAA+ ATPase domain; although this includes an Orc4 conserved arginine finger motif, both Walker A and B motifs appear degenerate [25] (Fig. 1B), suggesting that TbORC4 is, most likely, devoid of ATPase activity. Nevertheless, Orc4 subunits in other eukaryotes do not possess ATPase activity themselves, but their conserved arginine finger appears to be necessary for Orc1 ATP hydrolysis, by supplying the arginine finger to Orc1’s ATP-binding site [34, 50-52]. Analogously, it is thus possible that TbORC4 might stimulate TbORC1/CDC6 ATPase activity. It has not been possible to assess whether TbORC4 possesses a WH domain [25], and its binding to DNA has not been tested, though S. pombe Orc4 clearly provides such a function through AT-hook motifs [53]. Sequence analysis of Tb7980 suggests the presence of an AAA+ ATPase domain with relatively conserved Walker A and Walker B motifs, but no arginine finger or WH signatures [15, 25, 27] (Fig. 1B). Like TbORC4, Tb7980 ATPase and DNA binding activities have not been assessed. Structural modelling of Tb3120 hints at homology with D. melanogaster Orc2 [27], but this is mainly restricted to their C-termini (where the WH domain of D. melanogaster Orc2 is present) [27] (Fig. 1B). Further alignment of Tb3120 with multiple Orc2 protein sequences revealed that Tb3120 contains non-canonical Walker A and B motifs with characteristic signatures of Orc2 proteins. These are, however, separated by a large insertion (also observed in L. major and T. cruzi Tb3120 orthologues), suggesting that Tb3120 does not possess an intact AAA+ ATPase domain and is, therefore, unlikely to possess ATPase activity. In addition, Tb3120 and its orthologues in L. major and T. cruzi are considerably larger than model eukaryotes’ Orc2 subunits, and appear to have evolved a N-terminal extension with no detectable features or predicted function [27]. Further experimental analysis of Tb3120 will be essential to test whether this protein is the kinetoplastid equivalent of Orc2, and to better understand the functional implications of the sequence features highlighted above.
TbORC1/CDC6 [21], TbORC4, Tb3120 and Tb7980 all localise to the T. brucei nucleus throughout the cell cycle [27]. In striking contrast, TbORC1B localisation and expression is cell cycle dependent, with TbORC1B being detected only in the nucleus of cells in late G1 to late S or G2 phase [27]. Together with the rapid impairment of DNA replication and cell cycle progression upon depletion by RNAi [27], the available data suggest that TbORC1B might act as a positive regulator of DNA replication in T. brucei, rather than being a static member of the putative ORC-like complex. However, TbORC1B expression and localisation dynamics do not resemble any regulatory DNA replication factor described in other eukaryotes, including Cdc6. Indeed, TbORC1B’s lack of ATPase activity renders it an unlikely candidate to provide Cdc6 function, which acts by ATPase-dependent remodelling of ORC [51]. Thus, TbORC1B may represent a truly kinetoplastid-specific adaptation and it remains possible that T. brucei ORC-MCM interactions are archaeal-like in lacking Cdc6/Cdt1 mediation [54], consistent with TbORC1/CDC6 having been shown to be able to complement S. cerevisiae cdc6 temperature-sensitive mutants [21].
Whether or not the emerging data on the putative ORC in kinetoplastids can provide potential targets for drug development against these parasites remains to be seen [15, 16, 55], but unexpected levels of divergence in a core component of genome maintenance is emerging. It is intriguing to note that characterisation of the kinetoplastid kinetochore complex has also required protein interaction-based recovery of novel diverged components [56, 57], with an emerging picture of greatest variation in the subunits that interact with the genome [58]. Perhaps these parallel sets of studies hint that the unusual organisation of the kinetoplastid genome has necessitated widespread innovation in the protein factories that act in chromosome biology.
3. DNA replication dynamics in TRYPANOSOMA BRUCEI and LEISHMANIA
In order to be completely and accurately duplicated within the S phase of the cell cycle, the large linear eukaryotic genomes are replicated from many origins of replication distributed throughout their multiple chromosomes. Increased numbers of origins necessitated the evolution of DNA replication controls to ensure that the genome is duplicated exactly once during the cell cycle, and that under-replication and/or re-replication of parts of the genome is prevented. To this end, the process of eukaryotic DNA replication is divided into two non-overlapping phases: origin licensing and origin firing.
Origin licensing takes place from late mitosis to the end of G1 phase and consists of the loading of the replicative helicase (MCM2-7 double hexamers), in an inactive state, to every potential origin of replication in the genome, which are already demarcated by the binding of ORC [2-5]. During S phase, only a subset of origins is fired, not simultaneously but according to a DNA replication-timing programme [59], by the recruitment of additional replication complexes to the potential origins, establishing bidirectional replication forks at sites distributed across the genome [2-5]. By licensing origins before entry in S phase, already replicated origins cannot be re-licensed during S or G2 phases of the cell cycle, thus avoiding re-replication and the risk of genomic instability [2-5]. However, this implies that before entering S phase, the number of licensed origins must be sufficient to allow the complete replication of the genome, including in the event of replication stress from internal and external sources [1]. Therefore, the licensing of more origins than are needed to fire during S phase provides dormant origins that can be activated as a failsafe to prevent under-replication of regions of the genome and, thus, ensure genomic integrity [60]. If not activated, the excess licensed origins are passively replicated by replication forks established at neighbouring origins. Origin usage in eukaryotes is, therefore, very flexible [4].
With the exception of S. cerevisiae [61], eukaryotic origins of replication are not defined by specific DNA sequences. Instead, ORC-binding to origins and their later activation during S phase seems to depend on the combination of an assortment of less defined markers [2, 4, 7, 62, 63], such as: sequence features, including AT-rich regions [53, 64, 65] and CpG islands [66-71]; DNA topology, like G-quadruplexes [71-76] and negatively supercoiled DNA [77]; chromatin structure and status, such as nucleosome-free chromatin [78-80] and DNase-sensitive sites [76]; histone modifications [76, 81-84]; transcription [85, 86], with regions containing transcriptionally active genes being replicated earlier [87, 88], and some origins found at promoters (including RNA Polymerase II-binding sites) [65, 68, 78]; and the positioning of origin DNA in the nuclear space [89-92], with early fired origins localising to the nuclear interior, and late activated ones locating to the nuclear periphery. Due to their elusive characteristics, identification and mapping of origins of replication in any eukaryotic genome has not been straightforward. However, the development of high throughput sequencing methodologies [93] has increased the availability of sequenced genomes and allowed the development of techniques to map and correlate processes genome-wide, including, amongst others, the chromatin landscape [94-97], gene expression [98-100], and DNA replication dynamics [63, 101] - including origin localisation and usage. Recent studies in kinetoplastids provide an example of how these developments rapidly improved our understanding of DNA replication in these parasites. Prior to genome sequencing, origin mapping relied upon evaluating the ability of genome sequences to enhance episome stability [102] or maintain stability of fragmented chromosomes [103], or on analysis of small T. brucei minichromosomes [104]. The availability of near wholly compiled genomes has, to date, provided a genome-wide view of replication in T. brucei and two Leishmania species.
The T. brucei [17] and L. major [18] genomes (~50-66 Mbp, diploid) are organised in an unconventional architecture: unlike most other eukaryotes, the genomes of these closely related parasites are structured into ~200 multi-gene clusters, each of which is transcribed from its own RNA polymerase (Pol) II transcription start site [105-107]. Genome-wide multigenic transcription means most of the genome is traversed by RNA Pol II and gene expression controls are mainly post-transcriptional [108, 109]. Moreover, the core genomes of these parasites are extensively syntenic [105], despite being organised in strikingly different numbers of chromosomes: 11 stably diploid megabase chromosomes in T. brucei [17], and 36 chromosomes of variable ploidy in L. major [18, 110, 111]. In T. brucei aneuploidy appears much less pervasive than in Leishmania, being limited to the subtelomeres of the megabase chromosomes and to mini- and intermediate chromosomes, all of which act as stores for VSG genes [112].
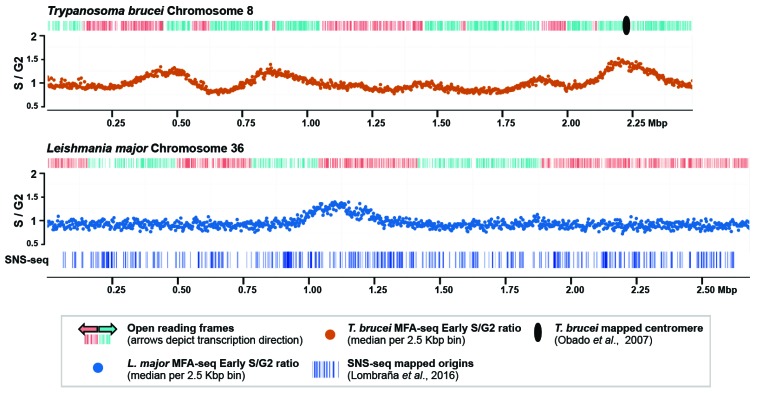
Genome-wide mapping of origins of replication was first performed in T. brucei [28] using deep sequencing marker frequency analysis (MFA-seq), also known as Sort-seq in yeast [113, 114]. Briefly, starting from an unsynchronised population, cells in S and G2 phase are isolated based on their DNA content by fluorescence activated cell sorting (FACS), and their DNA is then purified and sequenced. The resulting reads are mapped to the reference genome, allowing the ratio between S (replicating) and G2 (non-replicating) phases to be calculated. Representing the S/G2 read depth ratios across the chromosomes (Fig. 2) reveals ‘peaks’ that emanate from active origins, while “valleys” denote zones of replication termination [114], thus allowing assessment of origin location and strength genome-wide. In yeast, Sort-seq shows remarkably precise correlation with predicted sequence-conserved origins [113] and with recent replication mapping by Okazaki fragment sequencing (OK-seq) [115]. MFA-seq in T. brucei insect stage form (procyclic forms) cells in early-mid S phase [28] revealed that these parasites comply with the general principles of the eukaryotic DNA replication dynamics model: multiple origins were mapped per chromosome; the number of origins per chromosome correlates with chromosome size; all origins overlap with a subset (~4.4%) of TbORC1/CDC6-binding sites; differences in MFA-peak height suggest origins are activated at different times during S phase, with the earliest activated origins co-localising with mapped centromeres (chromosomes 1 to 8 [116]); and neither active origins nor TbORC1/CDC6-binding sites appear to be defined by specific sequences. Though MFA-seq cannot localise origins with great accuracy, all peaks centred on the so-called strand-switch regions (SSRs) that separate multi-gene transcription units and contain transcription start (divergent SSRs) or end (convergent SSRs) locations, or both (head-to-tail SSRs) [28]. In fact, evidence for DNA replication and transcription functionally intersecting could be seen after RNAi depletion of TbORC1/CDC6, which results in increased transcript levels at the SSRs, suggesting TbORC1/CDC6 might contribute to the outlining of transcription boundaries [28]. However, not all of the putative ORC-like components have been localised in the genome, the manner of ORC recruitment to DNA remains unclear, and MFA-seq could not map origin activity in the chromosome subtelomeres or in mini- and intermediate chromosomes.
Fig. (2).
Origins of replication in T. brucei and Leishmania. Origins of replication in T. brucei chromosome 8 (orange) and L. major chromosome 36 (blue) mapped in early S phase cells by MFA-seq. The top track represents the gene coding sequences, and the graph below illustrates the read depth ratio between early S phase and G2 populations (y-axis), with each dot representing the median S/G2 ratio (y-axis) in 2.5 kbp windows across the chromosome (x-axis); adapted from Marques et al., 2015. In the L. major panel, the vertical blue lines represent origins of replication mapped by SNS-seq; adapted from Lombraña et al., 2016.
Despite localisation of both origins and TbORC1/CDC6-binding sites to SSRs, no common sequence elements have to date been identified, suggesting T. brucei origin demarcation is dissimilar to yeast. Moreover, the number of detectable origins (42 MFA-seq peaks in the ~26 Mbp haploid genome) suggests an inter origin distance (IOD) of ~600 kbp, which is considerably greater than the predicted IOD of ~42 kbp in S. cerevisiae (based on ~280 Sort-seq peaks in a haploid genome of ~12 Mbp) [114]. More recently, MFA-seq was used to map origins in early and late S phase T. brucei cells, in two different strains (TREU927 and Lister 427), and in the two culturable life cycle stages of the parasite (procyclic and bloodstream forms) [43, 117]. In all these settings, essentially the same MFA-seq peaks are observed in the core genome, suggesting that origin location and usage is relatively rigid, though what distinguishes an origin-active SSR from an origin-inactive SSR is unknown. However, one locus shows pronounced changes in replication usage dependent on transcription activity: the single telomeric bloodstream VSG expression site (BES) that is transcriptionally active in bloodstream form cells is replicated early, whereas the remaining ~15 silent BES replicate in late S phase; moreover, all VSG BES are late-replicating in procyclic cell forms, when transcription is suppressed at all the sites [117]. These data strengthen the link between T. brucei DNA replication and transcription, and suggest a potential exploitation of DNA replication to drive antigenic variation, a topic recently reviewed elsewhere [118]. The wide spacing of T. brucei origins and pronounced rigidity in replication dynamics across the chromosomes’ core may result from the parasite’s odd genome organisation in well-defined multi-gene transcription units, meaning there is little flexibility in sites of replication initiation, as origins sited within the transcription units could lead to catastrophic clashes between RNA Pol II and the replisome. Nevertheless, single molecule analysis of DNA replication in chromosome 1 suggests at least some SSRs might be activated after hydroxyurea-induced replicative stress, and some replication might initiate from undefined subtelomeric sites [119]. Whether there is a genome-wide flexibility in origin usage under replicative stress or other conditions requires further investigation.
Given that T. brucei and Leishmania share an unconventional genome architecture and possess high levels of gene synteny, it might be predicted that DNA replication dynamics are comparable in the two parasites. However, MFA-seq in Leishmania insect stage cells (promastigotes) confounds this expectation (Fig. 2) [120]. MFA-seq has so far been performed in two species of Leishmania, L. major (old world) and L. mexicana (new world), whose genomes are, respectively, distributed into 36 and 34 chromosomes [121, 122] of variable ploidy [110]. Strikingly, in both Leishmania species, and in both early and late S cells, only one origin could be identified by MFA-seq per chromosome [120]. If confirmed, origin singularity in Leishmania is unprecedented in eukaryotes studied to date, as this was thought to be exclusive to the smaller genomes of bacteria and some archaea. L. mexicana chromosomes 8 and 20 are each syntenic not to one, but to two L. major chromosomes (29 plus 8, and 36 plus 20, respectively) [110]. Despite this genome reorganisation, only one origin could be mapped to each L. mexicana chromosome, suggesting that a single origin per chromosome is retained in the face of pronounced chromosome rearrangements [120]. Additionally, all detected MFA-seq peaks have a similar height and width, suggesting that all origins are of similar strength/usage and that there is no replication timing programme in Leishmania [120]. Nonetheless, like in T. brucei, all detected origins overlap exclusively with SSRs and, remarkably, ~40% of the identified origins were conserved in location in the two genomes [120]. Though such conservation might indicate a commonality in kinetoplastid origins, no origin-defining sequence, motif or pattern could be identified [120]. Nonetheless, detailed analysis of SSRs with and without origin activity revealed that in Leishmania, but not in T. brucei, the distance between the two most proximal genes to the SSR was significantly larger in SSRs with origin activity than in non-origin active SSRs [120]. This difference even extended to the origin-active SSRs of L. major chromosomes 29 and 36 and their syntenic, but non-origin active SSRs in L. mexicana chromosomes 8 and 20, respectively [120]. These data support the idea that in each Leishmania chromosome a specific SSR is associated with pronounced origin activity, though what causes this effect (e.g. the accumulation of specific factors) has not been investigated. One possibility is that, as in T. brucei, the Leishmania origin-active SSRs overlap with centromeres, but to date centromeres in Leishmania have not been successfully mapped. Many of the Leishmania SSRs, which provide origins of equal strength, are conserved with SSRs that in T. brucei map as origins of variable strength [28, 117], a change in function that also deserves further examination.
The MFA-seq prediction of a single origin in each Leishmania chromosome appears inadequate to explain complete genome replication. Estimations of a 3-4 hour S phase [123] and a ~2.5 kbp.min-1 DNA replication rate [124] suggest that while a single origin might be enough to drive the complete replication of the parasite’s smaller chromosomes (0.28-0.84 Mbp, ~66% of the genome), it is insufficient to fully replicate the larger ones (up to ~3.3 Mbp) [120]. How, then, Leishmania parasites completely replicate their genome is unclear. It is possible that less efficient or less frequently used origins might have escaped MFA-seq detection (detection threshold of ~25% of the activity of the mapped origins) [120]. However, because the detected origins in Leishmania and T. brucei localise exclusively to SSRs, one would predict that other origins would also localise to SSRs. Chromosome 32, of ~1.6 Mbp, for instance, has only four SSRs and, therefore, only four potential origins, but only one displays an MFA-seq peak. Origin activity at the other SSRs, if based on stochastic firing in the population, would therefore exceed the MFA-seq detection threshold. These considerations suggest that other events, perhaps not used in T. brucei, may promote complete Leishmania genome replication (see below). Two more recent studies have investigated DNA replication in Leishmania using different approaches to MFA-seq and the conflicting results may provide clues to how genome replication occurs in these parasites [124, 125].
One study, using the highly sensitive mapping methodology of short nascent leading strand purification coupled with sequencing (SNS-seq) [72] predicts ~5100 DNA replication initiation sites across the L. major genome (Fig. 2), with no preferential localisation at SSRs [125]. This prediction is >100 fold more than the origins mapped by MFA-seq and suggests one potential origin every other gene (~8300 total genes), meaning origins are found throughout the polycistronic transcription units. The huge number of SNS-seq predicted origins may suggest the technique’s sensitivity identifies origins used by only a few cells in the population, perhaps indicating flexibility in origin usage across the cellular population, which contrasts with the rigidity in origin usage and timing in T. brucei [28, 117]. SNS-seq has not to date been applied to T. brucei, but if similar numbers of origins of replication were detected, it would represent 14-fold greater origins than TbORC1/CDC6-binding sites (~360) [28]. DNA fibre stretching analysis, in two independent studies [124, 125], provides evidence that MFA-seq and SNS-seq respectively underestimate and overestimate DNA replication initiation events in Leishmania. Both studies could detect molecules with >1 site of replication initiation, which can be extrapolated to IODs of ~60-300 kbp (with considerably greater variation than fibre analysis predicts in T. brucei) [119]. Such IODs are 10-30 fold greater than the potential IOD of ~6.5 kbp that SNS-seq predicts [125]. Moreover, further extrapolation from the fibre analysis predicts ~150-400 origins, equalling or exceeding the number of mapped SSRs. It is currently impossible to correlate the Leishmania fibre data with MFA-seq, SNS-seq or genomic landmarks as the labelling is unable to localise replication sites within or between chromosomes, or indeed exclude that episomes have been examined. In contrast, fibre analysis linked to fluorescence in situ hybridisation (FISH) in chromosome 1 of T. brucei provides good correspondence with MFA-seq [119].
Can the apparent discord in current Leishmania replication data be explained? The above considerations suggest replication dynamics differ dramatically between T. brucei and Leishmania, despite the common use of SSRs as origins (as revealed by MFA-seq; Fig. 2). Though there are gaps in our understanding in both parasites (e.g. ORC1/CDC6-binding sites have not been mapped in Leishmania; SNS-seq has not been applied to T. brucei), it is attractive to speculate that differing replication strategies are found in the two parasites and that these might explain their differing genome organisations (larger, stable chromosome in T. brucei and smaller, frequently aneuploid chromosomes in Leishmania). At least two hypotheses for differing replication dynamics might be considered.
First, it is possible that MFA-seq hugely underestimates the number of origins in the Leishmania genome and the single peaks observed in each chromosome arise from dense clustering of multiple initiation sites, as revealed by SNS-seq [125, 126]. This model suggests Leishmania adopts a replication strategy similar to metazoans [72, 74, 127], with domains of early and late firing clusters of origins. This may be plausible, as SNS-seq has been widely used in metazoans but shows limited overlap with other established origin-mapping methods, such as ORC chromatin immunoprecipitation (ChIP-seq) or OK-seq mapping [128], and has an apparent bias towards CpG islands and G-quadruplex motifs [63]. However, if this model is correct, it is not clear why only a single, early-replicating origin cluster (MFA-seq peak) is seen per chromosome in Leishmania, whereas multiple putative clusters (MFA-seq peaks) are seen in each T. brucei chromosome. In addition, the model does not readily explain why the origins cluster so precisely around an SSR in each chromosome. In T. brucei, we know that SSRs are bound by ORC1/CDC6 and therefore the MFA-seq peaks, like similar analyses in yeast [113, 115], detect ORC-defined origins and not origin clusters [28]. In considering any such genome-wide mapping approaches it is important to take account of the very different genome sizes being examined in single-celled eukaryotes relative to metazoans [59]. Finally, if all SNS-seq peaks represent replication initiation at a conventional ORC-defined origin, then it is legitimate to ask how Leishmania (and perhaps T. brucei) copes with the likely pronounced impediment to efficient RNA Pol II progression through the multigene transcription units. Given all these questions, it is possible that DNA replication initiation in Leishmania does not rely on ORC, but on transcription-associated events, as postulated by Lombraña et al. (2016), who showed that 87-90% of SNS-seq peaks localise to gene trans-splicing regions, suggesting they correlate with genomic regions where transcription is predicted to decelerate or stall. In contrast, <4% of SNS-seq peaks map to convergent and head-to-tail SSRs, which correspond with ~81-85% of all transcription termination sites, and may be where ORC1/CDC6 localises (by analogy with T. brucei) [125].
A second hypothesis is that Leishmania uses a bi-modal DNA replication strategy. In this model, the MFA-seq peaks represent constitutive activation of a single ORC-bound origin, localised at an SSR, in each chromosome. These initiation events are therefore comparable with the MFA-seq mapping in T. brucei, explaining origin location conservation [129]. The second arm of the bi-modal strategy, necessary for complete duplication of the genome, is stochastic, with DNA replication being randomly initiated at ORC-independent (thus “origin”-independent) sites throughout the chromosomes. What these putative origin-independent replication initiation events are is unclear, but two possibilities can be envisaged, both of which might explain the SNS-seq data. The first possibility was suggested by Lombraña et al. [125], with the SNS-seq peaks representing sites where the MCM helicase accumulates and recruits the replisome without ORC [130]. An alternative, and more radical explanation, is that stochastic, ORC-independent initiation of DNA replication relies on DNA recombination. This suggestion is also compatible with the SNS-seq data, since, similar to mouse and human cells, ~74% of the SNS-seq peaks appeared to be significantly associated with G-quadruplex motifs [125], structures that can lead to DNA Pol slow down/stalling and, consequently, DNA damage [75]. In this model, ORC does not recruit the rest of the replisome, but instead recombination factors perform this role, such as during break-induced replication [131, 132]. Recombination-directed initiation of DNA replication, while unprecedented on such a scale, has the attraction of explaining the pervasive genome plasticity of Leishmania, including the role of recombination in the formation of genome-wide episomes [133-135]. Furthermore, recombination is needed to support continued replication in at least some polyploid archaea in which origins have been deleted [136]. Testing all these models will require genome-wide mapping of origins of replication in Leishmania using techniques other than MFA-seq and SNS-seq, allied to ChIP-seq mapping of key replication and recombination factors. In addition, it will be illuminating to ask if putative ORC-independent events are found in some circumstances in the T. brucei genome, and in other kinetoplastids.
CONCLUSION
Our understanding of DNA replication in kinetoplastids remains limited, but is growing. Most of our knowledge on this vital cellular process has been unravelled in the last ten years. In this period, we have moved from an archaeal-like model of the DNA replication initiator factor to a eukaryotic ORC-like complex, though a highly divergent one in which some subunits still remain to be identified or may be absent, and some appear to display pronounced sequence divergence. Due to its divergence from model eukaryotic ORCs, further analysis of kinetoplastid ORC structure and the functionality of the most diverged subunits, such as Tb3120, may illuminate ORC evolution and activity, as well as their potential as targets for drug development.
Presently, the emerging data on DNA replication in Leishmania is confusing, but we envisage that the current discord will be reconciled in time. Nonetheless, it seems clear that the dynamics of origin usage, replication timing and potentially replication execution in Leishmania are drastically different from T. brucei and other characterised microbial eukaryotes. Comparison of these two related kinetoplastids, and analysis of further organisms in this eukaryotic tree grouping, may reveal fundamental features of the evolution of eukaryotic chromosome replication, while uncovering the processes that shape the biology of Leishmania.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
Our recent work that contributed to this article was supported by the Wellcome Trust [089172, 093589], the BBSRC [BB/K006495/1] and Fundação para a Ciência e Tecnologia (FCT, Portugal) [SFRH/BD/68784/2010]. The Wellcome Centre for Molecular Parasitology is supported by core funding from the Wellcome Trust [104111]. We thank all friends and colleagues with whom we have enjoyed discussing the science explored in this article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Gaillard H., Garcia-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15(5):276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 2.Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. Where, when, and how? Ann. Rev. Biochem. 2010;79(89):130. doi: 10.1146/annurev.biochem.052308.103205. http://www.annualreviews.org/doi/abs/10.1146/annurev.biochem.052308.103205 [DOI] [PubMed] [Google Scholar]
- 3.O’Donnell M., Langston L., Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013;5(7):a010108. doi: 10.1101/cshperspect.a010108. http://cshperspectives.cshlp.org/content/5/7/a010108.full.pdf+html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fragkos M., Ganier O., Coulombe P., Mechali M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015;16(6):360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui K., On K.F., Diffley J.F. Regulating DNA replication in eukarya. Cold Spring Harb. Perspect. Biol. 2013;5(9):a012930. doi: 10.1101/cshperspect.a012930. http://cshperspectives.cshlp.org/content/5/9/a0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Ann. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-052610-094414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard A.C., Mechali M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013;5(10):a010116. doi: 10.1101/cshperspect.a010116. http://cshperspectives.cshlp.org/content/5/10/a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami H., Katayama T., Dna A. ORC, and Cdc6: Similarity beyond the domains of life and diversity. Biochem. Cell Biol. 2010;88(1):49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- 9.Skarstad K., Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 2013;5(4):a012922. doi: 10.1101/cshperspect.a012922. http://cshperspectives.cshlp.org/content/5/4/a012922 [abstract]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelman L.M., Kelman Z. Archaeal DNA replication. Ann. Rev. Genet. 2014;88:71–97. doi: 10.1146/annurev-genet-120213-092148. http://www.annualreviews.org/doi/abs/10.1146/annurev-genet-120213-092148?journal [DOI] [PubMed] [Google Scholar]
- 11.Duncker B.P., Chesnokov I.N., McConkey B.J. The origin recognition complex protein family. Genome Biol. 2009;10(3):214. doi: 10.1186/gb-2009-10-3-214. https://genomebiology.biomedcentral.com/articles/ 10.1186/gb-2009-10-3-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sazer S., Lynch M., Needleman D. Deciphering the evolutionary history of open and closed mitosis. Curr. Biol. 2014;24(22):R1099–R1103. doi: 10.1016/j.cub.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolev N.G., Ullu E., Tschudi C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell. Microbiol. 2014;16(4):482–489. doi: 10.1111/cmi.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao M.C., Chao J.L., Cheng C.Y. Programmed genome rearrangements in Tetrahymena. Microbiol. Spectr. 2014;2(16) doi: 10.1128/microbiolspec.MDNA3-0012-2014. http://www.asmscience.org/content/journal/microbiolspec/10 [DOI] [PubMed] [Google Scholar]
- 15.Tiengwe C., Marques C.A., McCulloch R. Nuclear DNA replication initiation in kinetoplastid parasites: New insights into an ancient process. Trends Parasitol. 2014;30(1):27–36. doi: 10.1016/j.pt.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Calderano S.G., de Melo Godoy P.D., da Cunha J.P., Elias M.C. Trypanosome prereplication machinery: A potential new target for an old problem. Enzyme Res. 2011;2011:518258.. doi: 10.4061/2011/518258. https://www.hindawi.com/journals/er/2011/518258/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bohme U., Hannick L., Aslett M.A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U.C., Arrowsmith C., Atkin R.J., Barron A.J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T.J., Churcher C., Clark L.N., Corton C.H., Cronin A., Davies R.M., Doggett J., Djikeng A., Feldblyum T., Field M.C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B.R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A.X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P.J., Moule S., Martin D.M., Morgan G.W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C.S., Peterson J., Quail M.A., Rabbinowitsch E., Rajandream M.A., Reitter C., Salzberg S.L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A.J., Tallon L., Turner C.M., Tait A., Tivey A.R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M.D., Embley T.M., Gull K., Ullu E., Barry J.D., Fairlamb A.H., Opperdoes F., Barrell B.G., Donelson J.E., Hall N., Fraser C.M., Melville S.E., El-Sayed N.M. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. http://science. sciencemag.org/content/309/5733/416 [DOI] [PubMed] [Google Scholar]
- 18.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S.M., Bianchettin G., Borzym K., Bothe G., Bruschi C.V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R.M., Cronin A., Cruz A.K., Davies R.M., De Gaudenzi J., Dobson D.E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A.C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J.C., Muller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O’Neil S., Pentony M., Pohl T.M., Price C., Purnelle B., Quail M.A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J.C., Rutter S., Saunders D., Schafer M., Schein J., Schwartz D.C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D.F., Blackwell J.M., Stuart K.D., Barrell B., Myler P.J. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. http://science.sciencemag.org/content/309/5733/436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., Westenberger S.J., Caler E., Cerqueira G.C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D.A., Carrington M., Crabtree J., Darban H., da Silveira J.F., de Jong P., Edwards K., Englund P.T., Fazelina G., Feldblyum T., Ferella M., Frasch A.C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M.J., Lorenzi H., Louie T., Machado C.R., McCulloch R., McKenna A., Mizuno Y., Mottram J.C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J.L., Rinta J., Robertson L., Salzberg S.L., Sanchez D.O., Seyler A., Sharma R., Shetty J., Simpson A.J., Sisk E., Tammi M.T., Tarleton R., Teixeira S., Van Aken S., Vogt C., Ward P.N., Wickstead B., Wortman J., White O., Fraser C.M., Stuart K.D., Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. http://science. sciencemag.org/content/309/5733/409 [DOI] [PubMed] [Google Scholar]
- 20.Matthews K.R. 25 years of African trypanosome research: From description to molecular dissection and new drug discovery. Mol. Biochem. Parasitol. 2015;200(1-2):30–40. doi: 10.1016/j.molbiopara.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godoy P.D., Nogueira-Junior L.A., Paes L.S., Cornejo A., Martins R.M., Silber A.M., Schenkman S., Elias M.C. Trypanosome prereplication machinery contains a single functional orc1/cdc6 protein, which is typical of archaea. Eukaryot. Cell. 2009;8(10):1592–1603. doi: 10.1128/EC.00161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar D., Mukherji A., Saha S. Expression and subcellular localization of ORC1 in Leishmania major. Biochem. Biophys. Res. Commun. 2008;375(1):74–79. doi: 10.1016/j.bbrc.2008.07.125. [DOI] [PubMed] [Google Scholar]
- 23.Kumar D., Kumar D., Saha S. A highly basic sequence at the N-terminal region is essential for targeting the DNA replication protein ORC1 to the nucleus in Leishmania donovani. Microbiol. 2012;158(Pt 7):1775–1782. doi: 10.1099/mic.0.055640-0. [DOI] [PubMed] [Google Scholar]
- 24.Dang H.Q., Li Z. The Cdc45.Mcm2-7.GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 2011;286(37):32424–32435. doi: 10.1074/jbc.M111.240143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiengwe C., Marcello L., Farr H., Gadelha C., Burchmore R., Barry J.D., Bell S.D., McCulloch R. Identification of ORC1/CDC6-interacting factors in Trypanosoma brucei reveals critical features of origin recognition complex architecture. PLoS One. 2012;7(3):e32674. doi: 10.1371/journal.pone.0032674. http://journals.plos.org/ plosone/article?id=10.1371/journal.pone.0032674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benmerzouga I., Concepcion-Acevedo J., Kim H.S., Vandoros A.V., Cross G.A., Klingbeil M.M., Li B. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 2013;87(1):196–210. doi: 10.1111/mmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques C.A., Tiengwe C., Lemgruber L., Damasceno J.D., Scott A., Paape D., Marcello L., McCulloch R. Diverged composition and regulation of the Trypanosoma brucei origin recognition complex that mediates DNA replication initiation. Nucleic Acids Res. 2016;44(10):4763–4784. doi: 10.1093/nar/gkw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiengwe C., Marcello L., Farr H., Dickens N., Kelly S., Swiderski M., Vaughan D., Gull K., Barry J.D., Bell S.D., McCulloch R. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Reports. 2012;2(1):185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frigola J., Remus D., Mehanna A., Diffley J.F. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495(7441):339–343. doi: 10.1038/nature11920. https:// www.nature.com/articles/nature11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aves S.J., Liu Y., Richards T.A. Evolutionary diversification of eukaryotic DNA replication machinery. Subcell. Biochem. 2012;62:19–35. doi: 10.1007/978-94-007-4572-8_2. https://link.springer.com/chapter/10 [DOI] [PubMed] [Google Scholar]
- 31.Harashima H., Dissmeyer N., Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013;23(7):345–356. doi: 10.1016/j.tcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Matte A., Delbaere L.T. John Wiley & Sons, Ltd. 2010. ATP-binding motifs. [Google Scholar]
- 33.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146(1-2):11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Bleichert F., Botchan M.R., Berger J.M. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519(7543):321–326. doi: 10.1038/nature14239. https://www.nature.com/articles/nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S., Balasov M., Wang H., Wu L., Chesnokov I.N., Liu Y. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc. Natl. Acad. Sci. USA. 2011;108(18):7373–7378. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speck C., Chen Z., Li H., Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12(11):965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarey M.G., Erzberger J.P., Grob P., Leschziner A.E., Berger J.M., Nogales E., Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 2006;13(8):684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 38.Chesnokov I.N., Chesnokova O.N., Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. USA. 2003;100(16):9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasanth S.G., Prasanth K.V., Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297 (5583):1026–1031. doi: 10.1126/science.1072802. mag.org/content/297/5583/1026/tab-article-info [DOI] [PubMed] [Google Scholar]
- 40.Takara T.J., Bell S.P. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. EMBO J. 2011;30(24):4885–4896. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tocilj A., On K.F., Yuan Z., Sun J., Elkayam E., Li H., Stillman B., Joshua-Tor L. Structure of the active form of human origin recognition complex and its ATPase motor module. eLife. 2017;6:e20818. doi: 10.7554/eLife.20818. https://elifesciences.org/articles/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiengwe C. Analysis of the architecture and function of the nuclear DNA replication apparatus in Trypanosoma brucei. 2010. [Google Scholar]
- 43.Marques C.A. Initiation of nuclear DNA replication in Trypanosoma brucei and Leishmania. 2015. [Google Scholar]
- 44.Hou Z., Bernstein D.A., Fox C.A., Keck J.L. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl. Acad. Sci. USA. 2005;102(24):8489–8494. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozaydin B., Rine J. Expanded roles of the origin recognition complex in the architecture and function of silenced chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30(3):626–639. doi: 10.1128/MCB.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell S.P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262(5141):1844–1849. doi: 10.1126/science.8266072. http:// science.sciencemag.org/content/262/5141/1844 [DOI] [PubMed] [Google Scholar]
- 47.Bell S.P., Mitchell J., Leber J., Kobayashi R., Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83(4):563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 48.Kuo A.J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J.K., Patel D.J., Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484(7392):115–119. doi: 10.1038/nature10956. https://www.nature.com/articles/nature10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noguchi K., Vassilev A., Ghosh S., Yates J.L., DePamphilis M.L. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25(22):5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davey M.J., Jeruzalmi D., Kuriyan J., O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 2002;3(11):826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 51.Sun J., Kawakami H., Zech J., Speck C., Stillman B., Li H. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20(3):534–544. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bleichert F., Balasov M., Chesnokov I., Nogales E., Botchan M.R., Berger J.M. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife. 2013;2:e00882. doi: 10.7554/eLife.00882. https:// elifesciences.org/articles/00882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang R.Y., Kelly T.J. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA. 1999;96(6):2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson R.Y., Abeyrathne P.D., Bell S.D. Mechanism of archaeal mcm helicase recruitment to DNA replication origins. Mol. Cell. 2016;62(1):287–296. doi: 10.1016/j.molcel.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uzcanga G., Lara E., Gutierrez F., Beaty D., Beske T., Teran R., Navarro J.C., Pasero P., Benitez W., Poveda A. Nuclear DNA replication and repair in parasites of the genus Leishmania: Exploiting differences to develop innovative therapeutic approaches. Crit. Rev. Microbiol. 2017;43(2):156–177. doi: 10.1080/1040841X.2016.1188758. [DOI] [PubMed] [Google Scholar]
- 56.Akiyoshi B., Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156(6):1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nerusheva O.O., Akiyoshi B. Divergent polo box domains underpin the unique kinetoplastid kinetochore. Open Biol. 2016;6(3):150206. doi: 10.1098/rsob.150206. https://benthamopen.com/tobioj/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Archivio S., Wickstead B. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J. Cell Biol. 2016 doi: 10.1083/jcb.201608043. http://jcb.rupress.org/content/early/2016/12/28/jcb.201608043 [DOI] [PMC free article] [PubMed]
- 59.Rhind N., Gilbert D.M. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013;5(8):a010132. doi: 10.1101/cshperspect.a010132. http://cshperspectives.cshlp.org/content/5/8/a010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McIntosh D., Blow J.J. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb. Perspect. Biol. 2012;4(10):a012955. doi: 10.1101/cshperspect.a012955. http:// www.lifesci.dundee.ac.uk/groups/julian_blow/publications/ewExternalFiles/CSH_perspect.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bell S.P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357(6374):128–134. doi: 10.1038/357128a0. https:// www.nature.com/articles/357128a0 [DOI] [PubMed] [Google Scholar]
- 62.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 2010;11(10):728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 63.Hyrien O. Peaks cloaked in the mist: The landscape of mammalian replication origins. J. Cell Biol. 2015;208(2):147–160. doi: 10.1083/jcb.201407004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Segurado M., de Luis A., Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4(11):1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacAlpine D.M., Rodriguez H.K., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18(24):3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delgado S., Gomez M., Bird A., Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17(8):2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cadoret J.C., Meisch F., Hassan-Zadeh V., Luyten I., Guillet C., Duret L., Quesneville H., Prioleau M.N. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl. Acad. Sci. USA. 2008;105(41):15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sequeira-Mendes J., Diaz-Uriarte R., Apedaile A., Huntley D., Brockdorff N., Gomez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5(4):e1000446.. doi: 10.1371/journal.pgen.1000446. http://journals.plos.org/plosgenetics/article ?id=10.1371/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., Peiffer I., Rivals E., Puy A., Laurent-Chabalier S., Desprat R., Mechali M. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21(9):1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Costas C., de la Paz Sanchez M., Stroud H., Yu Y., Oliveros J.C., Feng S., Benguria A., Lopez-Vidriero I., Zhang X., Solano R., Jacobsen S.E., Gutierrez C. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat. Struct. Mol. Biol. 2011;18(3):395–400. doi: 10.1038/nsmb.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cayrou C., Coulombe P., Puy A., Rialle S., Kaplan N., Segal E., Mechali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11(4):658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19(8):837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 73.Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintome C., Riou J.F., Prioleau M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33(7):732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cayrou C., Ballester B., Peiffer I., Fenouil R., Coulombe P., Andrau J.C., van Helden J., Mechali M. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015;25(12):1873–1885. doi: 10.1101/gr.192799.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valton A.L., Prioleau M.N. G-Quadruplexes in DNA replication: A problem or a necessity? Trends Genet. 2016;32(11):697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 76.Comoglio F., Schlumpf T., Schmid V., Rohs R., Beisel C., Paro R. High-resolution profiling of Drosophila replication start sites reveals a DNA shape and chromatin signature of metazoan origins. Cell Reports. 2015;11(5):821–834. doi: 10.1016/j.celrep.2015.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Remus D., Beall E.L., Botchan M.R. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23(4):897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.MacAlpine H.K., Gordan R., Powell S.K., Hartemink A.J., MacAlpine D.M. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20(2):201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soriano I., Morafraile E.C., Vazquez E., Antequera F., Segurado M. Different nucleosomal architectures at early and late replicating origins in Saccharomyces cerevisiae. BMC Genom. 2014;15:791. doi: 10.1186/1471-2164-15-791. https://bmcgenomics.biomedcentral.com/articles/10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lubelsky Y., Sasaki T., Kuipers M.A., Lucas I., Le Beau M.M., Carignon S., Debatisse M., Prinz J.A., Dennis J.H., Gilbert D.M. Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res. 2011;39(8):3141–3155. doi: 10.1093/nar/gkq1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Unnikrishnan A., Gafken P.R., Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 2010;17(4):430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crampton A., Chang F., Pappas D.L., Jr, Frisch R.L., Weinreich M. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol. Cell. 2008;30(2):156–166. doi: 10.1016/j.molcel.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 83.Iizuka M., Matsui T., Takisawa H., Smith M.M. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 2006;26(3):1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miotto B., Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010;37(1):57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sequeira-Mendes J., Gomez M. On the opportunistic nature of transcription and replication initiation in the metazoan genome. BioEssays. 2012;34(2):119–125. doi: 10.1002/bies.201100126. [DOI] [PubMed] [Google Scholar]
- 86.Helmrich A., Ballarino M., Nudler E., Tora L. Transcription-replication encounters, consequences and genomic instability. . Nat. Struct. Mol. Biol. 2013;20(4):412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 87.Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224(4650):686–692. doi: 10.1126/science.6719109. http://science.sciencemag.org/content/224/4650/686 [DOI] [PubMed] [Google Scholar]
- 88.Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23(1):1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heun P., Laroche T., Raghuraman M.K., Gasser S.M. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 2001;152(2):385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hansen R.S., Thomas S., Sandstrom R., Canfield T.K., Thurman R.E., Weaver M., Dorschner M.O., Gartler S.M., Stamatoyannopoulos J.A. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. USA. 2010;107(1):139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. https://www.nature.com/articles/nature06947 [DOI] [PubMed] [Google Scholar]
- 92.Wilson R.H., Coverley D. Relationship between DNA replication and the nuclear matrix. Genes Cells. 2013;18(1):17–31. doi: 10.1111/gtc.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zentner G.E., Henikoff S. High-resolution digital profiling of the epigenome. Nat. Rev. Genet. 2014;15(12):814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 95.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 96.Laird P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 97.Park P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15(3):205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 99.Konig J., Zarnack K., Luscombe N.M., Ule J. Protein-RNA interactions: New genomic technologies and perspectives. Nat. Rev. Genet. 2012;13(2):77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 100.Ozsolak F., Milos P.M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gilbert D.M. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat. Rev. Genet. 2010;11(10):673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patnaik P.K. Studies with artificial extrachromosomal elements in trypanosomatids: Could specificity in the initiation of DNA replication be linked to that in transcription? Parasitol. Today. 1997;13(12):468–471. doi: 10.1016/s0169-4758(97)01112-5. [DOI] [PubMed] [Google Scholar]
- 103.Tamar S., Papadopoulou B. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 2001;276(15):11662–11673. doi: 10.1074/jbc.M009006200. [DOI] [PubMed] [Google Scholar]
- 104.Weiden M., Osheim Y.N., Beyer A.L., Van der Ploeg L.H. Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 1991;11(8):3823–3834. doi: 10.1128/mcb.11.8.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D.C., Haas B.J., Tran A.N., Wortman J.R., Alsmark U.C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J.M., Cerqueira G.C., Creasy T., Delcher A.L., Djikeng A., Embley T.M., Hauser C., Ivens A.C., Kummerfeld S.K., Pereira-Leal J.B., Nilsson D., Peterson J., Salzberg S.L., Shallom J., Silva J.C., Sundaram J., Westenberger S., White O., Melville S.E., Donelson J.E., Andersson B., Stuart K.D., Hall N. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. mag.org/content/309/5733/404 [DOI] [PubMed] [Google Scholar]
- 106.Jackson A.P. Genome evolution in trypanosomatid parasites. Parasitology. 2014;142(S1):S40–S56. doi: 10.1017/S0031182014000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ersfeld K. Nuclear architecture, genome and chromatin organisation in Trypanosoma brucei. Res. Microbiol. 2011;162(6):626–636. doi: 10.1016/j.resmic.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 108.Michaeli S. Trans-splicing in trypanosomes: Machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6(4):459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 109.De Gaudenzi J.G., Noe G., Campo V.A., Frasch A.C., Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031. http://essays.biochemistry.org/content/51/31.article-info [DOI] [PubMed] [Google Scholar]
- 110.Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., Otto T.D., Sanders M., Seeger K., Dujardin J.C., Berriman M., Smith D.F., Hertz-Fowler C., Mottram J.C. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21(12):2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mannaert A., Downing T., Imamura H., Dujardin J.C. Adaptive mechanisms in pathogens: Universal aneuploidy in Leishmania. Trends Parasitol. 2012;28(9):370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 112.Glover L., Hutchinson S., Alsford S., McCulloch R., Field M.C., Horn D. Antigenic variation in African trypanosomes: The importance of chromosomal and nuclear context in VSG expression control. Cell. Microbiol. 2013;15(12):1984–1993. doi: 10.1111/cmi.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Muller C.A., Nieduszynski C.A. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 2012;22(10):1953–1962. doi: 10.1101/gr.139477.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muller C.A., Hawkins M., Retkute R., Malla S., Wilson R., Blythe M.J., Nakato R., Komata M., Shirahige K., de Moura A.P., Nieduszynski C.A. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2014;42(1):e3. doi: 10.1093/nar/gkt878. https://academic.oup.com/nar/article/42/1/e3/2437422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McGuffee S.R., Smith D.J., Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol. Cell. 2013;50(1):123–135. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Obado S.O., Bot C., Nilsson D., Andersson B., Kelly J.M. Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol. 2007;8(3):R37. doi: 10.1186/gb-2007-8-3-r37. https://genomebiology.biomedcentral.com/ articles/10.1186/gb-2007-8-3-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Devlin R., Marques C.A., Paape D., Prorocic M., Zurita-Leal A.C., Campbell S.J., Lapsley C., Dickens N., McCulloch R. McCulloch, R. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. eLife. 2016;5:e12765. doi: 10.7554/eLife.12765. https://elifesciences.org/articles/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Devlin R., Marques C.A., McCulloch R. Does DNA replication direct locus-specific recombination during host immune evasion by antigenic variation in the African trypanosome? Curr. Genet. 2017;63(3):441–449. doi: 10.1007/s00294-016-0662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Calderano S.G., Drosopoulos W.C., Quaresma M.M., Marques C.A., Kosiyatrakul S., McCulloch R., Schildkraut C.L., Elias M.C. Single molecule analysis of Trypanosoma brucei DNA replication dynamics. Nucleic Acids Res. 2015;43(5):2655–2665. doi: 10.1093/nar/gku1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marques C.A., Dickens N.J., Paape D., Campbell S.J., McCulloch R. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol. 2015;16:230. doi: 10.1186/s13059-015-0788-9. https://genomebiology.biomedcentral.com/ articles/10.1186/s13059-015-0788-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wincker P., Ravel C., Blaineau C., Pages M., Jauffret Y., Dedet J.P., Bastien P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24(9):1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Britto C., Ravel C., Bastien P., Blaineau C., Pages M., Dedet J.P., Wincker P. Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New world Leishmania genomes. Gene. 1998;222(1):107–117. doi: 10.1016/s0378-1119(98)00472-7. [DOI] [PubMed] [Google Scholar]
- 123.Wheeler R.J., Gluenz E., Gull K. The cell cycle of Leishmania: Morphogenetic events and their implications for parasite biology. Mol. Microbiol. 2011;79(3):647–662. doi: 10.1111/j.1365-2958.2010.07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stanojcic S., Sollelis L., Kuk N., Crobu L., Balard Y., Schwob E., Bastien P., Pages M., Sterkers Y. Single-molecule analysis of DNA replication reveals novel features in the divergent eukaryotes Leishmania and Trypanosoma brucei versus mammalian cells. Sci. Rep. 2016;6:23142. doi: 10.1038/srep23142. https://www.nature.com/articles/srep23142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lombrana R., Alvarez A., Fernandez-Justel J.M., Almeida R., Poza-Carrion C., Gomes F., Calzada A., Requena J.M., Gomez M. Transcriptionally driven DNA replication program of the human parasite Leishmania major. Cell Reports. 2016;16(6):1774–1786. doi: 10.1016/j.celrep.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 126.Rocha-Granados M.C., Klingbeil M.M. Leishmania DNA replication timing: A stochastic event? Trends Parasitol. 2016;32(10):755–757. doi: 10.1016/j.pt.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 127.Picard F., Cadoret J.C., Audit B., Arneodo A., Alberti A., Battail C., Duret L., Prioleau M.N. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10(5):e1004282. doi: 10.1371/journal.pgen.1004282. http://journals.plos.org/plosgenetics/article?id= 10.1371/journal.pgen.1004282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Petryk N., Kahli M., d’Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C.L., Hyrien O. Replication landscape of the human genome. Nat. Comm. 2016;7:10208. doi: 10.1038/ncomms10208. https://www.nature.com/articles/ncomms [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marques C.A., Dickens N.J., Paape D., Campbell S.J., McCulloch R. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol. 2015;16(1):230. doi: 10.1186/s13059-015-0788-9. central.com/articles/10.1186/s13059-015-0788-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hyrien O. How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000Res. 2016;5(F1000 Faculty Rev):2063. doi: 10.12688/f1000research.9008.1. https://f1000research.com/articles/5-2063/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lydeard J.R., Lipkin-Moore Z., Sheu Y.J., Stillman B., Burgers P.M., Haber J.E. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24(11):1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anand R.P., Lovett S.T., Haber J.E. Break-induced DNA replication. Cold Spring Harbor Perspect. Biol. 2013;5(12):a010397. doi: 10.1101/cshperspect.a010397. http://cshperspectives.cshlp.org/content/5/12/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laffitte M.C., Genois M.M., Mukherjee A., Legare D., Masson J.Y., Ouellette M. Formation of linear amplicons with inverted duplications in Leishmania requires the MRE11 nuclease. PLoS Genet. 2014;10(12):e1004805. doi: 10.1371/journal.pgen.1004805. http://journals. plos.org/plosgenetics/article?id=10.1371/journal.pgen.1004805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Genois M.M., Mukherjee A., Ubeda J.M., Buisson R., Paquet E., Roy G., Plourde M., Coulombe Y., Ouellette M., Masson J.Y. Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2012;40(14):6570–6584. doi: 10.1093/nar/gks306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Laffitte M.C., Leprohon P., Hainse M., Legare D., Masson J.Y., Ouellette M. Chromosomal translocations in the parasite Leishmania by a MRE11/RAD50-independent microhomology-mediated end joining mechanism. PLoS Genet. 2016;12(6):e1006117. doi: 10.1371/journal.pgen.1006117. http://journals.plos.org/plosgenetics/article?id= 10.1371/journal.pgen.1006117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hawkins M., Malla S., Blythe M.J., Nieduszynski C.A., Allers T. Accelerated growth in the absence of DNA replication origins. Nature. 2013;503(7477):544–547. doi: 10.1038/nature12650. https:// www.nature.com/articles/nature12650 [DOI] [PMC free article] [PubMed] [Google Scholar]